Abstract
Omega-3 polyunsaturated fatty acids (n-3 PUFAs) exert a negative effect on IL-6 production in several liver disorders, including cirrhosis, acute liver failure and fatty liver disease. However, its effect on the production of IL-11, another important IL-6 family cytokine, remains unclear. IL-11 was found to be significantly elevated in acetaminophen (APAP)-induced liver damage. The aim of the present study was to investigate whether and how n-3 PUFAs modulate IL-11 production during APAP-induced liver injury. For that purpose, wild-type (WT) and fat-1 transgenic mice were intraperitoneally injected with APAP to induce liver injury. Serum was collected for ELISA and alanine aminotransferase assay. The hepatocytes of APAP-injected mice were isolated for reverse transcription-quantitative PCR and western blot analyses. For the in vitro study, primary hepatocytes isolated from WT or fat-1 mice were stimulated with APAP. The results revealed that both endogenous and exogenous n-3 PUFAs significantly aggravated APAP-induced liver damage via the downregulation of STAT3 signaling. Notably, n-3 PUFAs inhibited IL-11 expression, but not IL-6 expression in hepatocytes during the APAP challenge. Furthermore, it was demonstrated that limited phosphorylation of ERK1/2 and Fos-like-1 (Fra-1) expression are responsible for the n-3 PUFA-mediated inhibitory effect on IL-11 production in APAP-treated hepatocytes. It was concluded that n-3 PUFAs inhibit IL-11 production and further STAT3 activation in hepatocytes during APAP-induced liver injury. Therefore, ERK1/2-mediated Fra-1 expression is responsible for the effect of n-3 PUFAs on IL-11 expression.
Keywords: acetaminophen, omega-3 polyunsaturated fatty acids, STAT3 signaling, IL-11, ERK1/2-Fos-like-1 signaling
Introduction
IL-11 is a member of the IL-6 cytokine family and has been reported to exert important effects on various liver diseases (1). Lipid-laden hepatocytes secrete IL-11, which acts via autocrine cis-signaling and contributes to hepatocyte lipotoxicity in a reactive oxygen species (ROS)-dependent manner (2). Furthermore, IL-11 receptor agonist enhances the proliferation of hepatocytes and ameliorates oxidative stress during acetaminophen (APAP)-induced liver injury. Among acute liver injury models, IL-11 is markedly upregulated in hepatocytes in response to APAP-induced liver injury (3) and liver ischemia (4). APAP is a commonly used drug for the relief of pain and fever. Although it is considered to be safe at therapeutic concentrations, its overdose can cause acute liver damage (5). STAT3, a member of the STAT family, can be activated by IL-6 family cytokines, and has been found to exert anti-apoptotic and pro-proliferative effects on APAP hepatotoxicity (6). These functions mainly rely on the regulation of genes involved in cell fate, such as the apoptosis regulators Bcl-2 and Bax (7,8).
Omega-3 polyunsaturated fatty acids (n-3 PUFAs), which include eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), are important for human health. Several studies have reported the effect of n-3 PUFAs on various liver diseases (9-11). n-3 PUFAs attenuate systemic inflammation by reducing the circulating level and gene expression of IL-6 (9,12). Dietary n-3 PUFA intake is associated with lower methylation at the IL-6 promoter, leading to a decreased plasma IL-6 concentration (13). Reduced IL-6 production and limited STAT3 phosphorylation have been observed in pancreatic acinar cells treated with n-3 PUFAs during cerulein exposure (14). Notably, previous studies have reported that n-3 PUFAs inhibited the phosphorylation of STAT3 in various disease models, thereby influencing cell differentiation, promoting cell death, inhibiting cell migration and inducing autophagy (15-17). The complex of IL-6 and IL-6 receptor (IL-6R) binds to glycoprotein 130 (gp130), which dimerizes and initiates intracellular STAT3 signaling (18). The presence of n-3 PUFAs may reduce the surface expression of IL-6R and its association with gp130 in lipid rafts, thereby leading to decreased downstream STAT3 activation (16). In addition, n-3 PUFAs may reduce IL-6-induced gp130 dimerization and subsequent STAT3 phosphorylation. Notably, n-3 PUFAs modulate STAT3 signaling by enhancing the expression of Src homology region 2 domain-containing protein tyrosine phosphatase-1, which is a well-known negative regulator of STAT3 signaling (19).
Fat-1 mice, which exhibit increased levels of n-3 PUFAs in their organs and tissues compared with those of their wild-type (WT) counterparts, have been reported to be a reliable animal model to investigate n-3 PUFAs (20). The present study aimed to investigate whether n-3 PUFAs modulate IL-11 expression and downstream STAT3 signaling during APAP hepatotoxicity.
Materials and methods
Mice
WT C57BL/6J mice were obtained from the Laboratory Animal Center of Southern Medical University (Guangzhou, China). Fat-1 transgenic mice, which has been reported to carry the gene that encodes the enzyme that coverts n-6 to n-3 PUFAs endogenously (20), were hybridized with WT C57BL/6J mice, and the fat-1 genotypes of each animal were recognized as we previously described (21). A total of 79 WT C57BL/6J and 39 fat-1 mice (male, 8 weeks old, 20-25 g) were included in this study. Mice included in the present study were housed under a 12 h light/dark cycle condition at a constant temperature (19-23°C) and (55±10%) humidity, fed with commercial diet and had free access to food and water. All animal experiments in this study were approved by the Welfare and Ethical Committee for Experimental Animal Care of Southern Medical University (approval no. L2018234). All mice were euthanized with 5% isoflurane. Mice were sacrificed at 0, 2, 6 and 24 h post-APAP injection. Before being sacrificed, blood (50-100 µl) was obtained from the tail vein. After standing for 4 h, blood was centrifugated at 1,400 × g for 10 min at 4°C, and the supernatant was collected as serum.
APAP-induced liver injury model
APAP (cat. no. sc-203425; Santa Cruz Biotechnology, Inc.) was intraperitoneally injected into 8-week-old male WT or fat-1 mice at 400 mg/kg body weight (overdose) (22,23) or at 600 mg/kg body weight (lethal dose) (24,25) as previously described. To demonstrate the effect of exogenous n-3 PUFAs, WT mice were fed with an n-3 PUFA-enriched diet for 3 weeks before APAP administration. n-3 PUFA-enriched diets were modified to contain 60 g/kg oils from DHA ethyl ester-enriched fish oil (Ocean Nutrition Canada) containing 540 g/kg DHA and 50 g/kg EPA. Commercial mouse food was used for the normal diet (ND) control group.
Reagents
Antibodies (Abs) against phosphorylated (p)STAT3 (rabbit anti-mouse monoclonal; 1:2,000; cat. no. 9145), STAT3 (rabbit anti-mouse monoclonal; 1:1,000; cat. no. 12640), p-p44/42 MAPK (Erk1/2; rabbit anti-mouse monoclonal; 1:1,000; cat. no. 4376), p44/42 MAPK (Erk1/2) (rabbit anti-mouse monoclonal; 1:1,000; cat. no. 4695) and GAPDH (rabbit anti-mouse monoclonal; 1:1,000; cat. no. 5174) were obtained from Cell Signaling Technology, Inc. Anti-IL-11 polyclonal antibody (rabbit anti-mouse; 1:1,000; cat. no. A1902) for immunohistochemistry and immunoblotting was purchased from ABclonal Biotech Co., Ltd. Anti-Fra-1 Ab (mouse anti-mouse monoclonal; 1:100; cat. no. sc-271657) was obtained from Santa Cruz Biotechnology, Inc. The antibody against Bcl-2 (mouse anti-mouse monoclonal; 1:100; cat. no. YM3041) was from ImmunoWay Biotechnology Company. Anti-Bax (rabbit anti-mouse polyclonal; 1:1,000; cat. no. DB123) anti-body was purchased from DB Biotech, spol. s r.o.
Isolation of mouse hepatocytes and hepatic mononuclear cells
Hepatocytes were isolated as previously described (26). Briefly, livers were perfused with calcium-free salt solution and type IV collagenase through the portal vein in situ and then filtered with polyamide mesh. After centrifugation at 50 × g for 1 min at 4°C, the precipitants were collected as hepatocytes for reverse transcription-quantitative PCR (RT-qPCR) and western blotting assays, or for primary hepatocyte culture. The supernatants were harvested for density gradient centrifugation using discontinuous 30/70% (v/v) Percoll gradients to obtain mononuclear cells for RT-qPCR. For primary mouse hepatocyte (PMH) culture, 1×106 hepatocytes were seeded in 6-well dishes coated with mouse tail collagen (cat. no. A1048301; Gibco; Thermo Fisher Scientific, Inc.) in William's E medium (cat. no. A1217601; Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). After incubation for 4 h, the culture was replaced with serum-free RPMI-1640 medium (cat. no. 11875101; Gibco; Thermo Fisher Scientific, Inc.). In some cases, cells were treated with 10 µM Sc144 (cat. no. S7124; Selleck Chemicals), 50 µM U0126 (cat. no. S1102; Selleck Chemicals) or 10 µM C188-9 (cat. no. S8605; Selleck Chemicals) for 2 h before APAP stimulation at 37°C.
Histology
Liver tissues were removed and collected in 1 ml 4% paraformaldehyde for 24 h at room temperature. Sections were cut into 5 µm. Samples were stained with hematoxylin and eosin (H&E) at room temperature for 3 min to observe morphological changes (Nikon Intensilight DS-Ri2; Nikon Corporation). The TUNEL experiment was performed using a commercial kit (cat. no. C1091; Beyotime Institute of Biotechnology) following the manufacturer's protocols. Harris hematoxylin was used at room temperature for 3 min for nuclear staining and sections were mounted with neutral resin. Three fields of view per section were observed (Nikon Intensilight DS-Ri2). For immunohistochemical analysis, specimens were embedded in paraffin and cut into 5-µm thick sections. The sections were deparaffinized in xylene for 15 min at room temperature and rehydrated with a graded series of alcohol, following which a microwave antigen retrieval procedure was performed using 10 mM sodium citrate buffer at 105°C for 10 min. Upon blocking endogenous peroxidase activity, the samples were treated with 5% BSA (cat. no. SRE0096; Sigma-Aldrich; Merck KGaA) at room temperature for 1 h to block non-specific staining. The sections were then incubated with an anti-IL-11 primary antibody (1:50; cat. no. A1902; ABclonal Biotech Co., Ltd.) at 4°C overnight, followed by incubation with the corresponding secondary antibody (HRP-conjugated goat anti-rabbit; 1:200; cat. no. S0001; Affinity Biosciences) at 37°C for 1 h. Peroxidase activity was detected (Nikon Intensilight DS-Ri2) using DAB solution (Beyotime Institute of Biotechnology).
Serum alanine aminotransferase (ALT) assay and cytokine assessment
The serum was collected at the indicated time points after APAP injection. Serum ALT activity was measured with a commercial kit (cat. no. C009-2; Nanjing Jiancheng Bioengineering Institute), based on the manufacturer's instructions. Different cytokine levels in the serum and cell culture supernatants were detected using commercial ELISA kits purchased from eBioscience (Thermo Fisher Scientific, Inc.), including IL-6 (cat. no. BMS603-2), IL-11 (cat. no. EMIL11), IL-13 (cat. no. BMS6015) and IL-22 (cat. no. BMS6022).
Flow cytometry
The Annexin V/PI apoptosis kit was obtained from Hangzhou Multi Sciences (Lianke) Biotech Co., Ltd., cells were stained for 5 min in the dark at room temperature before monitoring. Annexin V+/PI+ was identified as late apoptosis and Annexin V+/PI− was identified as early apoptosis. Cells were stained with JC-1 dye (Nanjing KeyGen Biotech Co., Ltd.) for 30 min at room temperature in the dark to evaluate mitochondrial membrane potential. The cells were analyzed using the FACS LSRFortessa™ flow cytometer (BD Biosciences) with BD FACSDiva™ software (version 8.0.1; BD Biosciences).
Western blotting
Protein samples were obtained from hepatocytes isolated from APAP-treated mice or primary hepatocytes lysed with RIPA buffer (50 mM Tris, 150 mM NaCl and 1% Nonidet P-40, pH 7.4). Protein concentration was measured using a BCA assay kit (cat. no. 23225; Thermo Fisher Scientific, Inc.). Protein samples (20 µg/lane) were separated on 10% SDS-polyacrylamide gels and then transferred onto polyvinylidene fluoride membranes (MilliporeSigma; Merck KGaA). The membranes were blocked with 5% BSA for 1 h at room temperature, and subsequently incubated with the indicated primary antibodies at 4°C overnight. Primary antibodies, including phosphorylated (p)STAT3 (rabbit anti-mouse monoclonal; 1:2,000; cat. no. 9145), STAT3 (rabbit anti-mouse monoclonal; 1:1,000; cat. no. 12640), p-p44/42 MAPK (Erk1/2; rabbit anti-mouse monoclonal; 1:1,000; cat. no. 4376), p44/42 MAPK (Erk1/2) (rabbit anti-mouse monoclonal; 1:1,000; cat. no. 4695) and GAPDH (rabbit anti-mouse monoclonal; 1:1,000; cat. no. 5174) were obtained from Cell Signaling Technology, Inc. Anti-IL-11 polyclonal antibody (rabbit anti-mouse; 1:1,000; cat. no. A1902) was purchased from ABclonal Biotech Co., Ltd. Anti-Fra-1 (mouse anti-mouse monoclonal; 1:100; cat. no. sc-271657) was obtained from Santa Cruz Biotechnology, Inc. The anti-body against Bcl-2 (mouse anti-mouse monoclonal; 1:100; cat. no. YM3041) was from ImmunoWay Biotechnology Company. Anti-Bax (rabbit anti-mouse polyclonal; 1:1,000; cat. no. DB123) antibody was purchased from DB Biotech, spol. s r.o. Subsequently, the membranes were incubated with the corresponding HRP-conjugated secondary antibody (goat anti-rabbit; 1:10,000; cat. no. S0001; Affinity Biosciences) at room temperature for 1 h. The results were visualized with ECL substrate (cat. no. 1705062; Bio-Rad Laboratories, Inc.). Each western blot analysis was performed three times.
RT-qPCR analysis
Isolated hepatocytes were homogenized in 1 ml TRIzol® (Thermo Fisher Scientific, Inc.), and total RNA was extracted based on the manufacturer's instruction. Then, 1,000 ng RNA was synthesized into cDNA using TransScript® Fly First-Stand cDNA Synthesis SuperMix (TransGen Biotech Co., Ltd.) with RNA removal reagent at 50°C for 10 min and 85°C for 5 sec. SYBR Green Real-Time PCR Master Mix (cat. no. A46112; Applied Biosystems; Thermo Fisher Scientific, Inc.) was applied for qPCR on an ABI Prism 7500 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the following thermocycling conditions: Initial denaturation at 94°C for 30 sec, followed by 40 cycles of denaturation at 94°C for 5 sec, and extension at 60°C for 30 sec. The expression levels of the target genes were normalized to GAPDH gene expression and calculated using the 2−∆∆Cq method (27). The primer sequences used in this experiment are shown in Table I.
Table I.
Primers for the target genes.
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| Bcl-2 | GGACTTGAAGTGCCATTGGT | AGCCCCTCTGTGACA GCTTA |
| Bax | GGATGCGTCCACCAAGAAGC | GGAGGAAGTCCAGTGTCCAGCC |
| IL-11 | GTTTACAGCTCTTGATGTCTC | GAGTCTTTAACAACAGCAGG |
| IL-13 | CCTGGCTCTTGCTTGCCTT | GGTCTTGTGTGATGTTGCTCA |
| Il-22 | TGACGACCAGAACATCCAGA | AATCGCCTTGATCTCTCCAC |
| IL-6 | TACCACTTCACAAGTCGGAGGC | CTGCAAGTGCATCATCGTTGTTC |
| Fra-1 | TCATCTGGAGAGGTGGGTCC | CTGCGGTTCTGACTCACTCG |
| GAPDH | CGTCCCGTAGACAAAATGGT | TTGATGGCAACAATCTCCAC |
Fra-1, Fos-like 1.
Statistical analysis
All experiments were independently repeated in triplicate. Statistical analysis was performed using SPSS 12.0 (SPSS, Inc.). The data are presented as the mean ± SD. An unpaired Student's t-test was performed between WT and fat-1 mice or ND control and n-3 PUFA-enriched diet mice at each time point. Comparison of the survival curves was analyzed with the Kaplan-Meier method and log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Endogenous n-3 PUFAs exacerbate APAP-induced liver injury
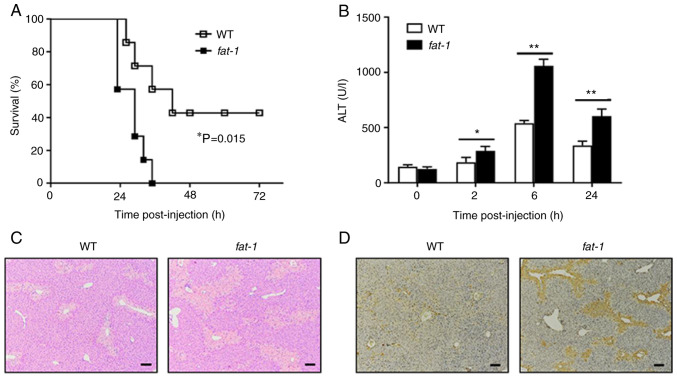
To confirm the role of n-3 PUFAs on APAP-induced liver injury, WT or fat-1 transgenic mice were challenged with a lethal dose of APAP. Animal mortality was markedly increased in fat-1 transgenic mice compared with that of WT mice (Fig. 1A). To further evaluate the effect of n-3 PUFAs on APAP-induced liver injury, WT or fat-1 transgenic mice were intraperitoneally injected with 400 mg/kg APAP, followed by examination of serum ALT activity and liver pathological changes. Serum samples collected from fat-1 transgenic mice showed markedly elevated levels of ALT activity compared with that of serum samples from WT mice following APAP administration (Fig. 1B). Liver H&E staining revealed severe hepatic damage in APAP-treated fat-1 transgenic mice compared with that of the WT controls (Fig. 1C). In addition, TUNEL-positive cells were markedly more abundant in livers from fat-1 transgenic mice than in livers obtained from APAP-treated WT mice (Fig. 1D). Taken together, these results indicated a potential role of endogenous n-3 PUFAs in aggravating the pathogenesis of APAP-induced liver damage.
Figure 1.
N-3 PUFAs exacerbate APAP-induced liver damage. (A) APAP (600 mg/kg) was injected into WT and fat-1 transgenic mice (n=7), and the survival rate of mice was observed (P=0.015). (B-D) WT and fat-1 transgenic mice (n=5) were intraperitoneally injected with APAP (400 mg/kg). (B) Serum ALT activities at different time points were measured. (C) Histological analysis of mouse livers was performed by hematoxylin and eosin staining 24 h post APAP injection. Scale bar, 100 µm. (D) TUNEL staining was performed on paraffin-embedded liver sections 24 h after APAP injection to mark apoptotic cells. Scale bar, 100 µm. *P<0.05, **P<0.01. N-3 PUFAs, omega-3 polyunsaturated fatty acids; APAP, acetaminophen; WT, wild-type; ALP, alanine aminotransferase.
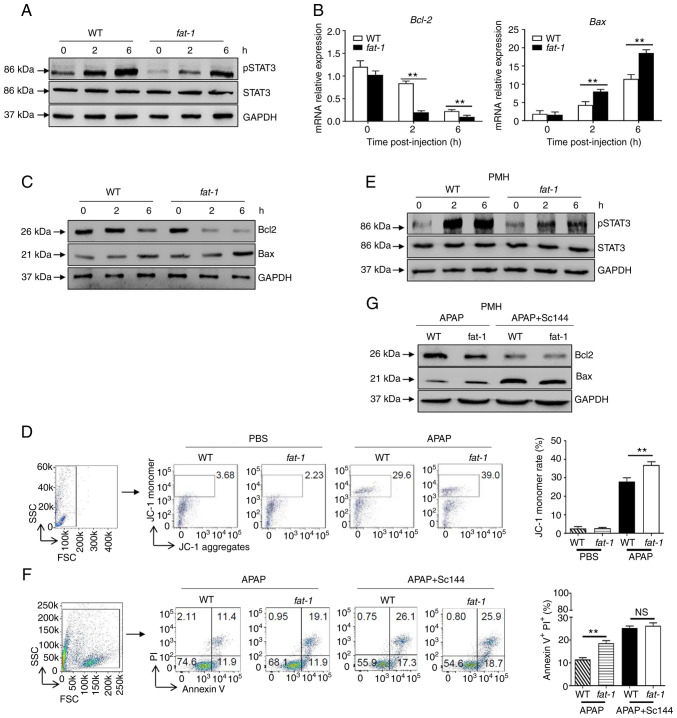
n-3 PUFAs promote APAP-induced hepatocyte apoptosis by inhibiting STAT3 phosphorylation
As hepatic STAT3 signaling is activated by the IL-6 cytokine family and exerts a protective effect against APAP overdose (6), the present study investigated whether the effect of n-3 PUFAs on APAP hepatotoxicity involves STAT3 signaling. The results revealed attenuated protein levels of pSTAT3 in hepatocytes derived from APAP-treated fat-1 transgenic mice compared with those derived from WT controls (Fig. 2A). Similarly, decreased Bcl-2 and enhanced Bax expression were found in hepatocytes obtained from fat-1 mice at both the transcriptional and translational levels compared with those observed in hepatocytes from WT controls (Fig. 2B and C). Considering that the Bcl family plays an essential role in mitochondrial homeostasis (7,8), JC-1 dye was used to detect the mitochondrial membrane potential by flow cytometry. The results showed an increased JC-1 monomer ratio in hepatocytes from fat-1 transgenic mice compared with that found in hepatocytes from WT mice upon APAP injection, which indicated more severe mitochondrial injury in fat-1 transgenic mice (Fig. 2D). To further validate these results, primary hepatocytes from WT or fat-1 mice were isolated and treated with APAP. As expected, attenuated phosphorylation of STAT3 was observed in the hepatocytes obtained from fat-1 mice (Fig. 2E). Furthermore, APAP treatment induced a higher apoptosis ratio in hepatocytes from fat-1 transgenic mice than that observed in hepatocytes from the WT controls. Of note, pretreatment with SC144, a gp130 inhibitor, abrogated the difference in cell apoptosis between hepatocytes from WT and fat-1 mice during APAP exposure (Fig. 2F). Consistently, the protein levels of Bcl-2 and Bax were comparable between APAP-treated WT and fat-1 mice in the presence of SC144 pretreatment (Fig. 2G). Similarly, C188-9, a STAT3-specific inhibitor, also abrogated the effect of n-3 PUFAs on Bcl-2 and Bax expression (Fig. S1). Collectively, these data suggested that STAT3 inactivation was responsible for the effect of n-3 PUFAs on APAP-induced liver injury.
Figure 2.
N-3 PUFAs promote hepatocytes apoptosis through dephosphorylation of STAT3 signaling. (A-D) Hepatocytes were isolated from WT and fat-1 transgenic mice (n=5) at the indicated time after APAP (400 mg/kg) injection. (A) Protein levels of pSTAT3, STAT3 and GAPDH were determined by western blotting analysis. (B) The mRNA levels of Bcl-2 and Bax were measured by reverse transcription-quantitative PCR and expressed as a ratio to GAPDH. (C) The hepatic protein levels of Bcl-2, Bax and GAPDH were evaluated at different time points by western blotting analysis. (D) Mitochondrial membrane potential (ΔΨm) was detected in mouse hepatocytes by flow cytometry 6 h post-APAP injection. (E-G) Primary hepatocytes were isolated from WT and fat-1 mice (n=5) and were stimulated with 20 mM APAP in vitro. In some cases, cells were pretreated with 10 µM Sc144 for 2 h before APAP stimulation. (E) The levels of pSTAT3, STAT3 and GAPDH were detected by western blotting analysis. (F) Cellular apoptosis was measured by Annexin V-PI staining 24 h post-APAP treatment. (G) The protein levels of Bcl-2, Bax and GAPDH expression were determined by western blotting analysis 6 h post-APAP treatment. **P<0.01. NS, not significant; N-3 PUFAs, omega-3 polyunsaturated fatty acids; APAP, acetaminophen; WT, wild-type; p, phosphorylated.
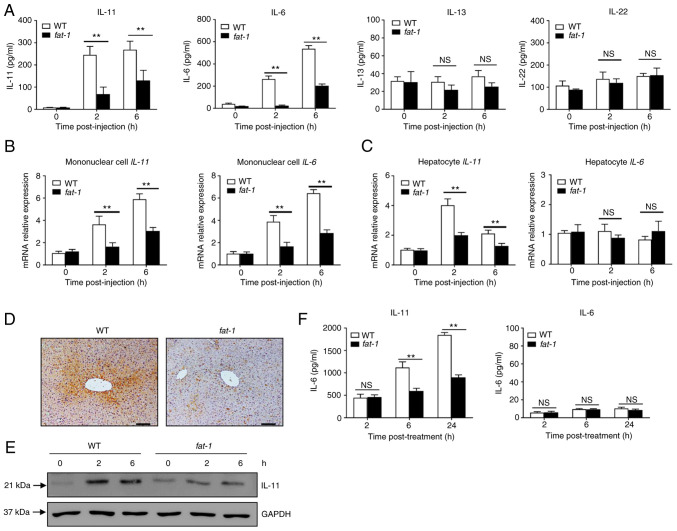
n-3 PUFAs inhibit the production of IL-11 in hepatocytes
Several STAT3-activating cytokines, including IL-6, IL-11, IL-13 and IL-22, are able to directly target hepatocytes (6,7). Therefore, the present study first assessed the levels of cytokines in the serum of APAP-treated mice. Compared with those of WT mice, APAP-challenged fat-1 transgenic mice showed significantly lower levels of IL-6 and IL-11, but not of IL-13 or IL-22 (Fig. 3A). Next, the mRNA expression of cytokines in hepatocytes and mononuclear cells was evaluated. The results showed that the levels of IL-11 and IL-6 were lower in hepatic mononuclear cells from APAP-treated mice compared with those observed in their WT counterparts (Fig. 3B). Notably, hepatocytes from APAP-treated fat-1 transgenic mice produced less IL-11 than those from WT controls (Fig. 3C), while there was no significant difference in the mRNA level of IL-6 (Fig. 3C). Similarly, immunohistochemical staining showed that the hepatic level of IL-11 was lower in fat-1 transgenic mice than in WT mice upon APAP administration (Fig. 3D). In addition, decreased IL-11 expression was found in hepatocytes obtained from fat-1 mice compared with that found in hepatocytes from WT mice (Fig. 3E). Furthermore, ELISA showed decreased IL-11 production in primary hepatocytes from fat-1 transgenic mice compared with that observed in their WT counterparts (Fig. 3F). Taken together, these results suggested that endogenous n-3 PUFAs limited the expression of IL-11 in hepatocytes during APAP-induced liver damage.
Figure 3.
N-3 PUFAs inhibit IL-11 production in hepatocytes. (A-D) WT and fat-1 transgenic mice were injected with 400 mg/kg APAP. (A) The serum levels of IL-11, IL-6, IL-13 and IL-22 were determined at the indicated time points. (B) Liver mononuclear cells and (C) hepatocytes were isolated from WT and fat-1 transgenic mice (n=5) at the indicated time after APAP (400 mg/kg) injection. The levels of IL-11 and IL-6 expression were measured by reverse transcription-quantitative PCR and expressed as a ratio to GAPDH. (D) The protein level of IL-11 in liver tissues was detected by immunohistochemical staining 6 h post-APAP injection. Scale bar, 50 µm. (E) Protein levels of IL-11 and GAPDH were determined by western blotting analysis. (F) Primary hepatocytes were isolated from WT or fat-1 transgenic mice and treated with 20 mM APAP for 24 h in vitro. The supernatants were collected, and the secreted levels of IL-11 and IL-6 were evaluated by ELISA. **P<0.01. NS, not significant; N-3 PUFAs, omega-3 polyunsaturated fatty acids; APAP, acetaminophen; WT, wild-type.
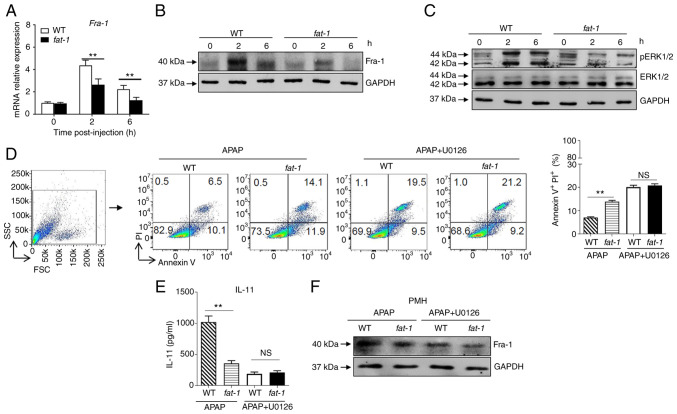
n-3 PUFAs inhibit the production of IL-11 via ERK1/2-dependent decreased Fra-1 levels in hepatocytes
It has been reported that Fra-1, which is recruited to the IL-11 promoter and enhances IL-11 gene transcription, is essential for the production of IL-11 (3). The present results revealed attenuated expression of Fra-1 at the mRNA and protein level in hepatocytes isolated from APAP-treated fat-1 transgenic mice compared with that observed in WT controls (Fig. 4A and B). Considering that Fra-1 expression is mainly dependent on ERK1/2 activity, ERK1/2 activation in hepatocytes from APAP-injected mice was assessed. The results showed that APAP induced a lower level of phosphorylation of ERK1/2 in hepatocytes from APAP-treated fat-1 transgenic mice than that observed in hepatocytes derived from their WT counterparts (Fig. 4C). To identify whether ERK1/2 signaling is involved in the effect of n-3 PUFAs on APAP hepatotoxicity, hepatocytes from WT and fat-1 transgenic mice were treated with U0126, a widely used ERK1/2 inhibitor, to block ERK1/2 activation prior to incubation with APAP. After ERK1/2 inhibition, the level of cell apoptosis was comparable between WT and fat-1 transgenic hepatocytes with U0126 pretreatment (Fig. 4D). Notably, the production of IL-11 (Fig. 4E) and the expression of Fra-1 (Fig. 4F) were similar between hepatocytes from WT mice and those from fat-1 transgenic mice upon APAP stimulation when ERK1/2 signaling was blocked. Taken together, these data suggested that n-3 PUFAs inhibited ERK1/2 phosphorylation following APAP stimulation, which resulted in limited expression of Fra-1 and consequently reduced IL-11 production.
Figure 4.
N-3 PUFAs inhibit IL-11 production by downregulating ERK1/2 pathway. (A-C) Hepatocytes were isolated from WT and fat-1 transgenic mice (n=5) at the indicated time after 400 mg/kg APAP administration. (A) The mRNA expression of Fra-1 was evaluated by reverse transcription-quantitative PCR and expressed as a ratio to GAPDH. (B) The protein level of Fra-1 was determined by western blotting analysis. (C) The protein levels of pERK, ERK and GAPDH were detected by western blotting analysis. (D-F) Primary hepatocytes were isolated from WT and fat-1 mice (n=5) and stimulated with 20 mM APAP for 24 h in vitro. In some cases, the cells were pretreated with 50 µM U0126 for 2 h before APAP stimulation. (D) Cellular apoptosis was measured by Annexin V-PI staining. (E) The culture supernatants were collected for evaluating IL-11 production through ELISA. (F) The protein level of Fra-1 in hepatocytes was confirmed by immunoblotting analysis. **P<0.01. NS, not significant; N-3 PUFAs, omega-3 polyunsaturated fatty acids; APAP, acetaminophen; WT, wild-type; Fra-1, Fos-like-1; p, phosphorylated.
Exogenous DHA exacerbates APAP-induced liver damage
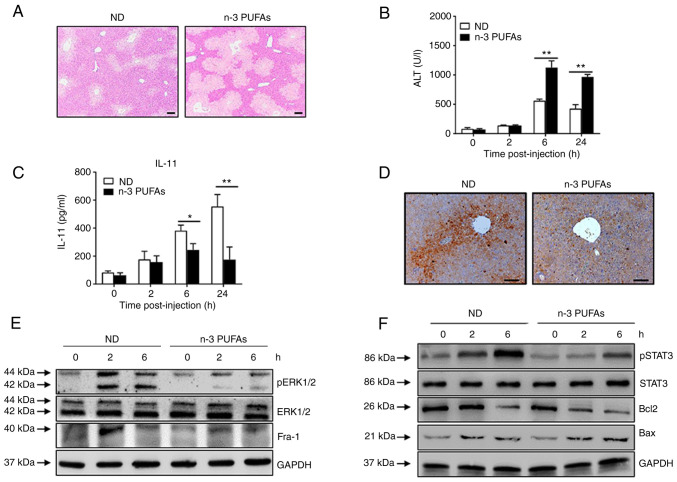
To further demonstrate the effect of n-3 PUFAs on IL-11 production in APAP-induced liver damage, WT mice were fed daily with a n-3 PUFA-enriched diet for 3 weeks prior to APAP administration. As expected, it was found that dietary n-3 PUFAs efficiently enhanced the severity of APAP-induced liver damage, as demonstrated by hepatic pathology (Fig. 5A) and serum ALT levels (Fig. 5B). In addition, a markedly reduced IL-11 level was determined in the serum (Fig. 5C) and liver tissue (Fig. 5D) of mice with a n-3 PUFA-enriched diet compared with that of mice in the ND group. Furthermore, exogenous n-3 PUFAs suppressed ERK1/2 phosphorylation and Fra-1 expression in hepatocytes stimulated with APAP, as evaluated by immunoblotting (Fig. 5E). Notably, exogenous n-3 PUFAs reduced STAT3 phosphorylation and Bcl-2 expression while enhancing Bax expression during the APAP challenge (Fig. 5F). Collectively, these findings indicated that exogenous n-3 PUFAs also exacerbated APAP-induced liver damage and share the same underlying mechanism as endogenous n-3 PUFAs.
Figure 5.
Exogenous DHA aggravate APAP-induced liver damage through dephosphorylated ERK-mediated decreased IL-11 production. (A-D) An overdose of APAP (400 mg/kg) was intraperitoneally injected into WT mice fed with ND or n-3 PUFA-enriched diet (n=5). (A) Histological analysis of mouse livers was performed by hematoxylin & eosin staining. Scale bar, 100 µm. (B) Serum ALT activities at different time points were measured. (C) The serum level of IL-11 was determined at the indicated time points post-APAP injection. (D) The protein level of IL-11 in the liver tissues was detected by immunohistochemical staining 6 h post-APAP injection. Scale bar, 50 µm. (E and F) Hepatocytes were isolated from WT mice fed with ND or n-3 PUFA-enriched diet (n=5) at the indicated time post-400 mg/kg APAP administration. (E) The levels of pERK, ERK, Fra-1 and GAPDH were determined by western blotting analysis. (F) The protein levels of pSTAT3, STAT3, Bcl-2, BAX and GAPDH were detected by western blotting analysis. *P<0.05, **P<0.01. N-3 PUFAs, omega-3 polyunsaturated fatty acids; APAP, acetaminophen; WT, wild-type; ALP, alanine aminotransferase; Fra-1, Fos-like-1; p, phosphorylated; ND, normal diet.
Discussion
n-3 PUFAs have been reported to exert an inhibitory effect on IL-6 production (9,12,13), while their effect on IL-11 expression remains unknown. The present study found that n-3 PUFAs exacerbated APAP-induced liver damage. The data revealed that n-3 PUFAs inhibited IL-11 production in hepatocytes and STAT3 phosphorylation. In addition, it was observed that limited phosphorylation of ERK1/2 and reduced Fra-1 expression were associated with the effect of n-3 PUFAs on IL-11 production.
STAT3 signaling plays an essential role in acute liver injury (6,28). It has been reported that myeloid STAT3 activation inhibits T cell-mediated hepatitis through suppression of type 1 T helper cytokines (28). Furthermore, mice lacking IL-6 or gp130-STAT3 signaling in hepatocytes are more susceptible to supplemented ethionine or lipopolysaccharide-induced liver injury (29,30). The present study found limited STAT3 phosphorylation and Bcl-2 expression levels alongside increased Bax expression in fat-1 mice compared with the findings in their WT counterparts. In addition, n-3 PUFAs did not affect APAP-induced liver injury when STAT3 signaling was inhibited by SC144, indicating that the effect of n-3 PUFAs on APAP hepatotoxicity depends on the regulation of the STAT3 signaling pathway. Notably, SC144 exerts an inhibitory effect on STAT3 phosphorylation through deglycosylation of gp130, the shared β-receptor of IL-6 and IL-11 signaling transduction, which indicates that the effect of n-3 PUFAs on STAT3 phosphorylation may rely on STAT3-inducing cytokines.
IL-11 belongs to the IL-6 family and shares the same gp130 homodimer receptor complex with IL-6. Specificity is gained through an individual IL-6/IL-11 α-receptor (31). Previous studies have demonstrated that IL-6 and IL-11 exert the opposite effect on immune regulation. It has been reported that IL-6 promotes type 2 T helper (Th2) cell response in an asthma model, while IL-11 inhibits Th2 type inflammation (32). IL-6 and IL-11 exhibit similar biological outcomes in the induction of the acute-phase response (33). In a previous study, both IL-11 and IL-6 increased the level of phosphorylated STAT3 in hepatocytes, however, only IL-6, but not IL-11, could induce STAT3 phosphorylation in Kupffer cells, which indicates that hepatocytes may be the target of IL-11 (3). In the present study, decreased levels of IL-6 and IL-11 were observed in the sera of APAP-injected fat-1 mice compared with those found in their WT counterparts, but only IL-11 could be detected, and its expression was found to be attenuated in hepatocytes obtained from fat-1 mice, which indicated that the inhibitory effect of n-3 PUFAs on IL-11 acts via an autocrine loop in hepatocytes during APAP hepatotoxicity.
IL-11 is a pleiotropic cytokine with biological functions in different cell types (2). It is mainly produced by hepatocytes in response to ROS during acute liver injury in mice (3). In a previous study, livers of mice receiving IL-11 exhibited damage, with elevated markers of fibrosis, hepatocyte cell death and inflammation (34). Notably, inhibiting IL-11 signaling by neutralizing antibodies could reduce hepatocyte death, inflammation and hyperglycemia in mouse models of diet-induced steatohepatitis (34). A previous study reported the pathological function of IL-11 in hepatocytes during the early stages of metabolic liver disease (2). MAPK/ERK activation is crucial for the oxidative stress-induced production of IL-11 (3). Treatment with ERK inhibitors can efficiently suppress TNF-α-induced IL-11 induction in murine embryonic fibroblasts (3). Fra-1, a member of the activator protein 1 family, was found to bind to the IL-11 promoter before stimulation, but recruitment of Fra-1 was further enhanced after oxidative stimulation (3,35). Gillies et al (36) found that Fra-1 is expressed in proportion to the amplitude and duration of ERK1/2 activity. A previous study reported that the expression of IL-11 in hepatocytes is regulated by ERK1/2-mediated Fra-1 expression (37). n-3 PUFAs appear to act via receptors or sensors, thereby controlling cellular signaling processes that influence gene expression patterns (38). Several studies have reported that the anticancer properties of n-3 PUFAs involve the altered phosphorylation of ERK1/2 (39,40). It has been reported that n-3 PUFAs induce apoptosis in human breast cancer cells through inhibition of ERK1/2 activation (40). n-3 PUFAs are able to suppress VEGF expression in colon cancer cells by limiting ERK1/2 phosphorylation and hypoxia-inducible factor 1α overexpression (41). In addition, the anti-inflammatory effect of n-3 PUFAs on the endothelium depends on the dephosphorylation of ERK1/2 (42). The present study demonstrated that n-3 PUFAs inhibited ERK1/2 phosphorylation and Fra-1 expression in hepatocytes in response to the APAP treatment. Notably, inhibition of ERK1/2 activation attenuated n-3 PUFA-induced hepatic IL-11 production and subsequent liver injury in APAP-treated mice, suggesting that n-3 PUFAs affect IL-11 production and APAP hepatotoxicity via regulation of the ERK1/2 signaling pathway. However, the specific underlying mechanism by which n-3 PUFAs regulate ERK1/2 signaling needs further investigation. Fluorescence labeled n-3 PUFAs could be used to evaluate whether there is a physical interaction between n-3 PUFAs and ERK1/2 protein in vitro, while IL-11 receptor antagonist or IL-11/IL-11 receptor knockout by CRISPR/CRISPR-associated protein 9 could be used to further identify whether the effect of n-3 PUFAs on APAP-induced liver injury depends on the production of IL-11.
In summary, the present results revealed that n-3 PUFAs inhibit IL-11 production and downstream STAT3 phosphorylation in hepatocytes, thereby aggravating APAP-induced liver injury. The present study demonstrated that the ERK1/2-Fra-1 axis is important for the regulatory function of n-3 PUFAs on IL-11 expression.
Supplementary Data
Acknowledgments
Not applicable.
Funding Statement
This work was supported in part by the National Natural Science Foundation of China (grant nos. 82071781, 81873872 and 81771771), and the Innovation Team of Chronic Kidney Disease with Integrated Traditional Chinese and Western Medicine (grant no. 2019KCXTD014).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YL, ZC and DZ conceived and designed the present study. ZC and DZ provided administrative support. JZ and DZ were responsible for the provision of study materials and patients. YL, JL and YC were responsible for the collection and assembly of data. YL, ZL, JZ, XL, ZC and DZ analyzed and interpreted the data. YL, ZC and DZ confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal protocols in this study were approved by the Welfare and Ethical Committee for Experimental Animal Care of Southern Medical University (approval no. L2018234; Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cook SA, Schafer S. Hiding in plain sight: Interleukin-11 emerges as a master regulator of fibrosis, tissue integrity, and stromal inflammation. Annu Rev Med. 2020;71:263–276. doi: 10.1146/annurev-med-041818-011649. [DOI] [PubMed] [Google Scholar]
- 2.Dong J, Adami E, Chothani SP, Viswanathan S, Ng B, Lim WW, Sing BK, Zhou J, Ko NSJ, Shekeran SG, et al. Autocrine IL11 cis-signaling in hepatocytes is an initiating nexus between lipotoxicity and non-alcoholic steatohepatitis. BioRxiv. 2020 [Google Scholar]
- 3.Nishina T, Komazawa-Sakon S, Yanaka S, Piao X, Zheng DM, Piao JH, Kojima Y, Yamashina S, Sano E, Putoczki T, et al. Interleukin-11 links oxidative stress and compensatory proliferation. Sci Signal. 2012;5:ra5. doi: 10.1126/scisignal.2002056. [DOI] [PubMed] [Google Scholar]
- 4.Zhu M, Lu B, Cao Q, Wu Z, Xu Z, Li W, Yao X, Liu F. IL-11 attenuates liver ischemia/reperfusion injury (IRI) through STAT3 signaling pathway in mice. PLoS One. 2015;10:e0126296. doi: 10.1371/journal.pone.0126296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernal W, Auzinger G, Dhawan A, Wendon J. Acute liver failure. Lancet. 2010;376:190–201. doi: 10.1016/S0140-6736(10)60274-7. [DOI] [PubMed] [Google Scholar]
- 6.Mühl H. STAT3, a key parameter of cytokine-driven tissue protection during sterile inflammation-the case of experimental acetaminophen (paracetamol)-induced liver damage. Front Immunol. 2016;7:163. doi: 10.3389/fimmu.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison DA. The Jak/STAT pathway. Cold Spring Harb Perspect Biol. 2012;4:a011205. doi: 10.1101/cshperspect.a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen M, Kaestel CG, Eriksen KW, Woetmann A, Stokkedal T, Kaltoft K, Geisler C, Röpke C, Odum N. Inhibition of constitutively activated Stat3 correlates with altered Bcl-2/Bax expression and induction of apoptosis in mycosis fungoides tumor cells. Leukemia. 1999;13:735–738. doi: 10.1038/sj.leu.2401415. [DOI] [PubMed] [Google Scholar]
- 9.Schmöcker C, Weylandt KH, Kahlke L, Wang J, Lobeck H, Tiegs G, Berg T, Kang JX. Omega-3 fatty acids alleviate chemically induced acute hepatitis by suppression of cytokines. Hepatology. 2007;45:864–869. doi: 10.1002/hep.21626. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Tang Y, Wang S, Zhou J, Zhou J, Lu X, Bai X, Wang XY, Chen Z, Zuo D. Endogenous n-3 polyunsaturated fatty acids attenuate T cell-mediated hepatitis via autophagy activation. Front Immunol. 2016;7:350. doi: 10.3389/fimmu.2016.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Fernández-Galilea M, Martínez-Fernández L, González-Muniesa P, Pérez-Chávez A, Martínez JA, Moreno-Aliaga MJ. Oxidative stress and non-alcoholic fatty liver disease: Effects of omega-3 fatty acid supplementation. Nutrients. 2019;11:872. doi: 10.3390/nu11040872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley DS, Siegel D, Fedor DM, Adkins Y, Mackey BE. DHA supplementation decreases serum C-reactive protein and other markers of inflammation in hypertriglyceridemic men. J Nutr. 2009;139:495–501. doi: 10.3945/jn.108.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y, Smith CE, Lai CQ, Irvin MR, Parnell LD, Lee YC, Pham LD, Aslibekyan S, Claas SA, Tsai MY, et al. The effects of omega-3 polyunsaturated fatty acids and genetic variants on methylation levels of the interleukin-6 gene promoter. Mol Nutr Food Res. 2016;60:410–419. doi: 10.1002/mnfr.201500436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song EA, Lim JW, Kim H. Docosahexaenoic acid inhibits IL-6 expression via PPARγ-mediated expression of catalase in cerulein-stimulated pancreatic acinar cells. Int J Biochem Cell Biol. 2017;88:60–68. doi: 10.1016/j.biocel.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 15.D'Eliseo D, Di Renzo L, Santoni A, Velotti F. Docosahexaenoic acid (DHA) promotes immunogenic apoptosis in human multiple myeloma cells, induces autophagy and inhibits STAT3 in both tumor and dendritic cells. Genes Cancer. 2017;8:426–437. doi: 10.18632/genesandcancer.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen MJ, Fan YY, Monk JM, Hou TY, Barhoumi R, McMurray DN, Chapkin RS. n-3 PUFAs reduce T-helper 17 cell differentiation by decreasing responsiveness to interleukin-6 in isolated mouse splenic CD4+ T cells. J Nutr. 2014;144:1306–1313. doi: 10.3945/jn.114.194407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasaki S, Horiguchi A, Asano T, Ito K, Asano T, Asakura H. Docosahexaenoic acid inhibits the phosphorylation of STAT3 and the growth and invasion of renal cancer cells. Exp Ther Med. 2017;14:1146–1152. doi: 10.3892/etm.2017.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong A, Yu W, Liu Y, Sanders BG, Kline K. Elimination of ALDH+ breast tumor initiating cells by docosahexanoic acid and/or gamma tocotrienol through SHP-1 inhibition of Stat3 signaling. Mol Carcinog. 2016;55:420–430. doi: 10.1002/mc.22291. [DOI] [PubMed] [Google Scholar]
- 20.Kang JX, Wang J, Wu L, Kang ZB. Transgenic mice: Fat-1 mice convert n-6 to n-3 fatty acids. Nature. 2004;427:504. doi: 10.1038/427504a. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Chen Y, Xie X, Yin A, Yin Y, Liu Y, Dong L, Zhu Z, Zhou J, Zeng Q, et al. Gender difference on the effect of omega-3 polyunsaturated fatty acids on acetaminophen-induced acute liver failure. Oxid Med Cell Longev. 2020;2020:8096847. doi: 10.1155/2020/8096847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson MW, Sparkenbaugh EM, Wang S, Ilich A, Noubouossie DF, Mailer RK, Renné T, Flick MJ, Luyendyk JP, Chen ZL, et al. Plasmin-mediated cleavage of high molecular weight kininogen contributes to acetaminophen-induced acute liver failure. Blood. 2021 Apr 7; doi: 10.1182/blood.2020006198. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha B, Nandi D. Farnesyltransferase inhibitors reduce Ras activation and ameliorate acetaminophen-induced liver injury in mice. Hepatology. 2009;50:1547–1557. doi: 10.1002/hep.23180. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C, Feng J, Du J, Zhuo Z, Yang S, Zhang W, Wang W, Zhang S, Iwakura Y, Meng G, et al. Macrophage-derived IL-1α promotes sterile inflammation in a mouse model of acetaminophen hepatotoxicity. Cell Mol Immunol. 2018;15:973–982. doi: 10.1038/cmi.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres S, Baulies A, Insausti-Urkia N, Alarcón-Vila C, Fucho R, Solsona-Vilarrasa E, Núñez S, Robles D, Ribas V, Wakefield L, et al. Endoplasmic reticulum stress-induced upregulation of STARD1 promotes acetaminophen-induced acute liver failure. Gastroenterology. 2019;157:552–568. doi: 10.1053/j.gastro.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 26.Osawa Y, Uchinami H, Bielawski J, Schwabe RF, Hannun YA, Brenner DA. Roles for C16-ceramide and sphingosine 1-phosphate in regulating hepatocyte apoptosis in response to tumor necrosis factor-alpha. J Biol Chem. 2005;280:27879–27887. doi: 10.1074/jbc.M503002200. [DOI] [PubMed] [Google Scholar]
- 27.Pang Y, Liu Z, Han H, Wang B, Li W, Mao C, Liu S. Peptide SMIM30 promotes HCC development by inducing SRC/YES1 membrane anchoring and MAPK pathway activation. J Hepatol. 2020;73:1155–1169. doi: 10.1016/j.jhep.2020.05.028. [DOI] [PubMed] [Google Scholar]
- 28.Lafdil F, Wang H, Park O, Zhang W, Moritoki Y, Yin S, Fu XY, Gershwin ME, Lian ZX, Gao B. Myeloid STAT3 inhibits T cell-mediated hepatitis by regulating T helper 1 cytokine and interleukin-17 production. Gastroenterology. 2009;137:2125–2135. e1–e2. doi: 10.1053/j.gastro.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroy DC, Beraza N, Tschaharganeh DF, Sander LE, Erschfeld S, Giebeler A, Liedtke C, Wasmuth HE, Trautwein C, Streetz KL. Lack of interleukin-6/glycoprotein 130/signal transducers and activators of transcription-3 signaling in hepatocytes predisposes to liver steatosis and injury in mice. Hepatology. 2010;51:463–473. doi: 10.1002/hep.23322. [DOI] [PubMed] [Google Scholar]
- 30.Streetz KL, Wüstefeld T, Klein C, Kallen KJ, Tronche F, Betz UA, Schütz G, Manns MP, Müller W, Trautwein C. Lack of gp130 expression in hepatocytes promotes liver injury. Gastroenterology. 2003;125:532–543. doi: 10.1016/S0016-5085(03)00901-6. [DOI] [PubMed] [Google Scholar]
- 31.Garbers C, Scheller J. Interleukin-6 and interleukin-11: Same same but different. Biol Chem. 2013;394:1145–1161. doi: 10.1515/hsz-2013-0166. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Homer RJ, Hong L, Cohn L, Lee CG, Jung S, Elias JA. IL-11 selectively inhibits aeroallergen-induced pulmonary eosinophilia and Th2 cytokine production. J Immunol. 2000;165:2222–2231. doi: 10.4049/jimmunol.165.4.2222. [DOI] [PubMed] [Google Scholar]
- 33.Benigni F, Fantuzzi G, Sacco S, Sironi M, Pozzi P, Dinarello CA, Sipe JD, Poli V, Cappelletti M, Paonessa G, et al. Six different cytokines that share GP130 as a receptor subunit, induce serum amyloid A and potentiate the induction of interleukin-6 and the activation of the hypothalamus-pituitary-adrenal axis by interleukin-1. Blood. 1996;87:1851–1854. doi: 10.1182/blood.V87.5.1851.1851. [DOI] [PubMed] [Google Scholar]
- 34.Widjaja AA, Singh BK, Adami E, Viswanathan S, Dong JR, D'Agostino GA, Ng B, Lim WW, Tan J, Paleja BS, et al. Inhibiting interleukin 11 signaling reduces hepatocyte death and liver fibrosis, inflammation, and steatosis in mouse models of nonalcoholic steatohepatitis. Gastroenterology. 2019;157:777–792.e14. doi: 10.1053/j.gastro.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Shin SY, Choi C, Lee HG, Lim Y, Lee YH. Transcriptional regulation of the interleukin-11 gene by oncogenic Ras. Carcinogenesis. 2012;33:2467–2476. doi: 10.1093/carcin/bgs297. [DOI] [PubMed] [Google Scholar]
- 36.Gillies TE, Pargett M, Minguet M, Davies AE, Albeck JG. Linear integration of ERK activity predominates over persistence detection in Fra-1 regulation. Cell Syst. 2017;5:549–563.e5. doi: 10.1016/j.cels.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishina T, Deguchi Y, Miura R, Yamazaki S, Shinkai Y, Kojima Y, Okumura K, Kumagai Y, Nakano H. Critical contribution of nuclear factor erythroid 2-related factor 2 (NRF2) to electrophile-induced interleukin-11 production. J Biol Chem. 2017;292:205–216. doi: 10.1074/jbc.M116.744755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calder PC. Mechanisms of action of (n-3) fatty acids. J Nutr. 2012;142:592S–599S. doi: 10.3945/jn.111.155259. [DOI] [PubMed] [Google Scholar]
- 39.Serini S, Calviello G. Modulation of Ras/ERK and phosphoinositide signaling by long-chain n-3 PUFA in breast cancer and their potential complementary role in combination with targeted drugs. Nutrients. 2017;9:185. doi: 10.3390/nu9030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun H, Hu Y, Gu Z, Owens RT, Chen YQ, Edwards IJ. Omega-3 fatty acids induce apoptosis in human breast cancer cells and mouse mammary tissue through syndecan-1 inhibition of the MEK-Erk pathway. Carcinogenesis. 2011;32:1518–1524. doi: 10.1093/carcin/bgr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calviello G, Di Nicuolo F, Gragnoli S, Piccioni E, Serini S, Maggiano N, Tringali G, Navarra P, Ranelletti FO, Palozza P. n-3 PUFAs reduce VEGF expression in human colon cancer cells modulating the COX-2/PGE2 induced ERK-1 and -2 and HIF-1alpha induction pathway. Carcinogenesis. 2004;25:2303–2310. doi: 10.1093/carcin/bgh265. [DOI] [PubMed] [Google Scholar]
- 42.Liu KL, Yang YC, Yao HT, Chia TW, Lu CY, Li CC, Tsai HJ, Lii CK, Chen HW. Docosahexaenoic acid inhibits inflammation via free fatty acid receptor FFA4, disruption of TAB2 interaction with TAK1/TAB1 and downregulation of ERK-dependent Egr-1 expression in EA.hy926 cells. Mol Nutr Food Res. 2016;60:430–443. doi: 10.1002/mnfr.201500178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.