Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus that causes coronavirus disease 2019. Angiotensin-converting enzyme 2 (ACE2) is the SARS-CoV binding site and is ubiquitously expressed in endothelial cells of several organs, with the highest levels in the cardiovascular system, kidney and lungs. A disintegrin and metalloproteinase 17 (ADAM17) is involved in ectodomain shedding of ACE2. In the present study, reverse-transcription-quantitative PCR, transfection, TUNNEL assay, dual-luciferase activity assay and western blotting were conducted to investigate the effects of microRNA (miR)-28-3p on ADAM17-dependent shedding of the ACE2 ectodomain following treatment with the spike protein (S-protein) of SARS-CoV-2. It was found that miR-28-3p was significantly downregulated in 293T cells treated with 100 ng/ml of S-protein for 24 h at 37°C, which led to upregulation of ADAM17. In addition, the expression of ADAM17 and miR-28-3p were negatively correlated based on Pearson's correlation test in 293T cells treated with S-protein for 24 h. Overexpression of miR-28-3p and inhibition of ADAM17 regulated 293T cell viability, apoptosis and ACE2 ectodomain shedding. It was also demonstrated that ADAM17 was the target gene of miR-28-3p and that miR-28-3p negatively regulated ADAM17 expression. Notably, the inhibition of ADAM17 expression blocked the effects of miR-28-3p inhibitor on proliferation, apoptosis and ACE2 ectodomain shedding in 293T cells treated with S-protein. The findings of the present study suggested that miR-28-3p inhibits ADAM17-dependent ACE2 ectodomain shedding in 293T cells treated with the S-protein of SARS-CoV-2, which suggested the potential therapeutic role of miR-28-3p mimic in the prevention and treatment of patients with SARS-CoV-2.
Keywords: microRNA-28-3p, angiotensin-converting enzyme 2, A disintegrin and metalloproteinase 17, spike protein
Introduction
At present, infections of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are a global life threatening disease (1). Updated from weekly report of WHO on 13 July 2021, the cumulative deaths from SARS-CoV-2 have surpassed 4 million people and the cumulative cases was up to 186 million people globally (2). S-(S) glycoprotein (spike protein), nucleocapsid protein, membrane protein and envelope protein are all structural proteins of SARS-CoV-2 (3). S-protein is prominent in the viral membrane (3). SARS-CoV-2 uses S-protein binding to angiotensin-converting enzyme 2 (ACE2) to invade the host cells (4). ACE2 is a transmembrane protein with its active site in the N-terminal domain (5), which is also the SARS-CoV binding site. Lung, heart, vessels, gut, testis and brain all express ACE2 receptors (6-9). In particular, ACE2 receptors are abundantly expressed in the kidney (10), which can be infected by SARS-CoV-2 (11).
A disintegrin and metalloproteases (ADAMs) are membrane-bound proteins with disintegrin and metalloproteinase domains (12). A disintegrin and metalloprotease domain 17 (ADAM17) is one of members of the ADAM family of transmembrane proteins containing an N-terminal domain (12). ADAMs are involved in ectodomain shedding of enzymes including ACE2 (12). Cellular (membrane-bound) ACE2 can be cleaved to release the ACE2 ectodomain into the circulation by ADAM17 (13). An increase of ectodomain ACE2 may exacerbate the RAS (renin-angiotensin system) imbalance (ACE/ACE2) in patients with specific comorbidities, such as cardiovascular disease, hypertension, diabetes and chronic lung disease (12). ACE2 is also downregulated by viral transcription (14) and endocytosis together with the virus (15). In addition, a decrease in ADAM17 expression inhibits ACE2 shedding, while introduction of ADAM17 cDNA restores SARS-S-induced ACE2 shedding (16). High levels of shed ACE2 can reduce SARS-CoV virus infection (13,17).
MicroRNAs (miRNAs) are non-coding RNAs which can directly bind to the 3′ untranslated regions (3′-UTRs) of target mRNAs to regulate gene expression posttranscriptionally. miRNAs are powerful regulators of numerous cell processes, including cell growth, differentiation, development and apoptosis (18). miRNAs are involved in various viral infectious diseases, such as human immunodeficiency virus-1 (HIV-1) infection (19), human papillomavirus infection (20), SARS-CoV infection (21) and hepatitis B virus infection (20-22). As mature miRNAs, miR-28-3p and miR-28-5p are derived from the 3′ and 5′ ends, respectively, of pre-miR-28 and then go on to target different mRNAs, such as WD repeat and SOCS box-containing 2 and forkhead box O1 (23-27). There is evidence indicating that miRNAs may block RNA virus replication during viral infection (28). Purohit et al (29) demonstrated that downregulation of miRNA-28 expression increased HIV infection by promoting viral replication (29) and it has also been demonstrated that miR-28-3p inhibits human T cell leukemia virus type 1 replication and virus infection (23). Although the expression and biological function of these miRNAs can suppress the replication of some viruses, the effects of miR-28-3p on SARS-Cov-2 infection remain unknown. In the present study, it was hypothesized that miR-28-3p may exert a regulatory role in ADAM17-dependent ACE2 ectodomain shedding during SARS-Cov-2 infection. Since the present study demonstrated the direct interaction between miR-28-3p and ADAM17, upregulation of miR-28-3p may provide a novel therapeutic strategy for treatment of patients with SARS-CoV-2.
Materials and methods
Cell culture and S-protein treatment
293T cells, with high transfection efficiency and stably expressing hACE2, which are termed tool cells and were used in some studies of SARS-Cov-2 virus (30-33), were purchased from the American Type Culture Collection. Cells were cultured in DMEM (Gibco; Thermo Fisher Scientific Inc.) supplemented with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific Inc.) and 100 mg/ml penicillin/streptomycin (Gibco; Thermo Fisher Scientific Inc.). 293T cells were treated with 100 ng/ml of S-protein for 48 h at 37°C in a humidified incubator with 5% CO2 (cat. no. bs-46008P; BIOSS) (34,35). Cells were maintained in an incubator at 37°C with 5% CO2, and were passaged every 3 days.
Transfection
293T cells were seeded in 6-well plates (1×105 cells/well) and maintained in an incubator at 37°C with 5% CO2 for 12 h to obtain a cell confluence of 30-35%. For transfection, the cell culture medium was replaced with 2 ml Opti-MEM medium (Gibco; Thermo Fisher Scientific Inc.) without FBS and antibiotics 12 h before transfection. Then, a total of 10 µl of Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was diluted with 200 µl of Opti-MEM and the miR-28-3p mimic, miR-28-3p inhibitor, mimic negative control (NC) or inhibitor NC, or with small interfering (si)RNA or siRNA NC, and the mixture was incubated for 5 min at room temperature. Subsequently, the cells were transfected with a final concentration of 20 nM miR-28-3p mimic/inhibitor or miR-28-3p mimic/inhibitor NC, or with 100 pmol si-ADAM17 or siRNA NC (scrambled) at 37°C with 5% CO2 for 16 h, after which the medium was replaced with complete DMEM supplemented with 10% FBS for 72 h in a humidified incubator at 37°C with 5% CO2. At 24, 48 and 72 h after transfection, the transfection efficiency was evaluated by using inverted fluorescence microscope (Ti-S; Nikon Corporation) to observe and count the GFP-positive cells. The miR-28-3p mimic, miR-28-3p mimic NC, miR-28-3p inhibitor and miR-28-3p inhibitor NC were obtained from Guangzhou RiboBio Co. Ltd. The miR-28-3p mimic and miR-28-3p mimic NC, miR-28-3p inhibitor and miR-28-3p inhibitor NC sequences were as follows: miR-28-3p mimic, 5′-UAG AUC ACA GUC CUU UGU UAU-3′; miR-28-3p mimic NC, 5′-AUC UAG UCA GUC CUU UGU UUA U-3′; miR-28-3p inhibitor, 5′-AUC UAG UGU CAG GAA ACA AAU A-3′; and miR-28-3p inhibitor NC, 5′-UAG AUC AGU CAG GAA ACA AAU A-3′. The si-ADAM17 or siRNA NC were obtained from Shanghai GenePharma Co. Ltd. si-ADAM17 and siRNA NC sequences were as follows: si-ADAM17, 5′-TGA GGC AGT CTC TCC TAT TCC TGA CCA GC-3′; and siRNA NC (scramble), 5′-TGA CCA CCC TGA CCT ACG GCG TGC AGT GC-3′.
Western blotting (WB)
293T cells were seeded in 6-well plates (1×105 cells/well) and cultured at 37°C with 5% CO2 for 48 h after transfection (34). Then, cell culture supernatants were collected. Total protein of cell supernatants was concentrated by using Amicon® Ultra Centrifugal Filters (MilliporeSigma). As a control, 10 µg of ovalbumin (OVA) was added to each sample of supernatant and the precipitates were probed with anti-OVA and anti-ACE2 antibodies. Total protein was extracted from cell culture using RIPA buffer (Beyotime Institute of Biotechnology). Protein concentrations were determined using Bradford assays (Thermo Fisher Scientific, Inc.). Total protein (40 µg/lane) was separated by 10% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 5% fat-free skimmed milk for 2 h at room temperature and subsequently incubated with anti-ADAM17 (cat. no. ab39163), anti-GAPDH (cat. no. Ab8245), anti-ACE2 (cat. no. ab108252) and anti-OVA (anti-ovalbumin) (cat. no. ab236590) (all 1:1,000; Abcam) at 4°C overnight and then washed with TBST buffer (0.1% Tween-20 in 1X TBS buffer) three times. The secondary antibody goat anti-rabbit IgG H&L horseradish peroxidase (HRP) (1:3,000; cat. no. ab6721; Abcam) was incubated with the membranes for 2 h at room temperature. The membranes were washed thrice, 5 min each time in TBST, then incubated in ECL substrate chemiluminescent detection reagent (Thermo Fisher Scientific, Inc.) for 5 min. The chemiluminescent blots were imaged first with the ChemiDoc MP imager (Bio-Rad, Inc.) and then on film. GAPDH and OVA were used as the loading controls for cell lysate and cell culture supernatants, respectively. The band analysis tools of ImageJ v.1.8.0 software (National Institutes of Health) were used to calculate the gray value of the bands after subtracting the background density.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
293T cells were seeded in 6-well plates (1×105 cells/well) and cultured at 37°C with 5% CO2. Following transfection for 48 h, TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA on ice and the concentration and purity of RNA were detected using Nanodrop 2000 (Thermo Fisher Scientific Inc.). DNA contamination was removed using DNase I (cat. no. AMPD1, Sigma-Aldrich; Merck KGaA) and then the miRNAs were reversed transcribed into cDNA using the PrimeScript RT kit (Takara Bio Inc.). The reverse transcription program was as follows: 65°C for 5 min, on ice immediately followed by the addition of reverse transcriptase, annealing at 50°C for 15 min followed by extension at 95°C for 5 min and 4°C hold. miScript HiSpec buffer was used to prepare the cDNA and real-time PCR was carried out using iTaq™ SYBR® Green Supermix (Beijing Solarbio Science & Technology Co., Ltd.) according to the manufacturer's protocol. The reaction conditions were as follows: 95°C for 3 min followed by 40 cycles of 3-step PCR (98°C for 10 sec, 60°C for 30 sec and 72°C for 60 sec), 72°C for 10 min, then hold at 4°C. U6 was used as the internal control. The expression of miRNAs was determined using the 2−ΔΔCq method (36). Primer sequences were as follows: miR-28-3p forward, 5′-CGC GCA CTA GAT TGT GAG CT-3′ and reverse, 5′-AGT GCA GGG TCC GAG GTA TT-3′; and U6 forward, 5′-ATT GGA ACG ATA CAG AGA AGA TT-3′ and reverse, 5′-GGA ACG CTT CAC GAA TTT G-3′.
MTT assay
293T cells were seeded in 6-well plates (1×104 cells/well). Following transfection for 48 h, 20 µl MTT (5 mg/ml, Sigma-Aldrich; Merck KGaA) was added to each well and the cells were cultured for another 4 h in an incubator at 37°C with 5% CO2. The solution was discarded after incubation for 4 h and then 150 µl DMSO (Sigma-Aldrich; Merck KGaA) was added to each well and incubated in the dark for 10 min with shaking. A microplate reader (Bio-Rad, Inc.) was used to detect the optical density at 490 nm.
TUNEL assay
TUNEL assay was carried out by using DNA Fragmentation Imaging kit (cat. no. 6432344001; Roche Diagnostics; Merck KGaA). 293T cells were seeded in 6-well plates (1×104 cells/well). Following transfection for 48 h, cells were fixed for 30 min in 4% paraformaldehyde in phosphate-buffered saline solution (PBS) at 4°C, then incubated with 0.1% Triton X-100 in PBS for 2 min on ice. After washing the cells with PBS three times, the cells were incubated with 45 µl of the Reaction Solution, which was combined the Enzyme Solution with the Label Solution, for 60 min in the dark at 37°C and then washed with PBS three times at room temperature. Then, 150 µl of the Nuclei Dye mixture was added and cells were incubated for 5 min at room temperature; a coverslip was added and images were captured under a fluorescence microscope (80i; Nikon Corporation; magnification, ×200). At least 100 cells were counted in 10 random fields and the percentage of positive apoptotic cells was calculated.
Dual-Luciferase activity assay
ADAM17 (3′ UTR) was predicted as the binding site of miR-28-3p by TargetScan v.7.2 (www.targetscan.org) online prediction software. 293T cells were seeded in 6-well plates (1×105 cells/well), and then co-transfected with a pmir-GLO dual-luciferase miRNA target expression vector (Promega Corporation), containing a wild-type (WT) or mutant (Mut) ADAM17 untranslated region (3′UTR) and miR-28-3p mimic (5′-UAG AUC ACA GUC CUU UGU UAU-3′) or mimic-NC (5′-AUC UAG UCA GUC CUU UGU UUA U-3′) using Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc.). Following 48 h of incubation, a dual-luciferase reporter assay (Promega Corporation) was conducted to detect luciferase activity according to manufacturer's instructions. Firefly luciferase activity was normalized by comparison with Renilla luciferase activity.
Statistical analysis
SPSS 18.0 software (IBM Corp.) was used for statistical analyses. Data were shown as means ± standard deviation (SD) from 3 independent experiments. The differences between 2 groups were determined using paired Student's t-tests or one-way analysis of variance followed by the post hoc Tukey's test. The correlation test used was Pearson's correlation test. *P<0.05 was considered to indicate a statistically significant difference.
Results
miR-28-3p expression and ADAM17 and cell viability in S-protein-treated 293T cells
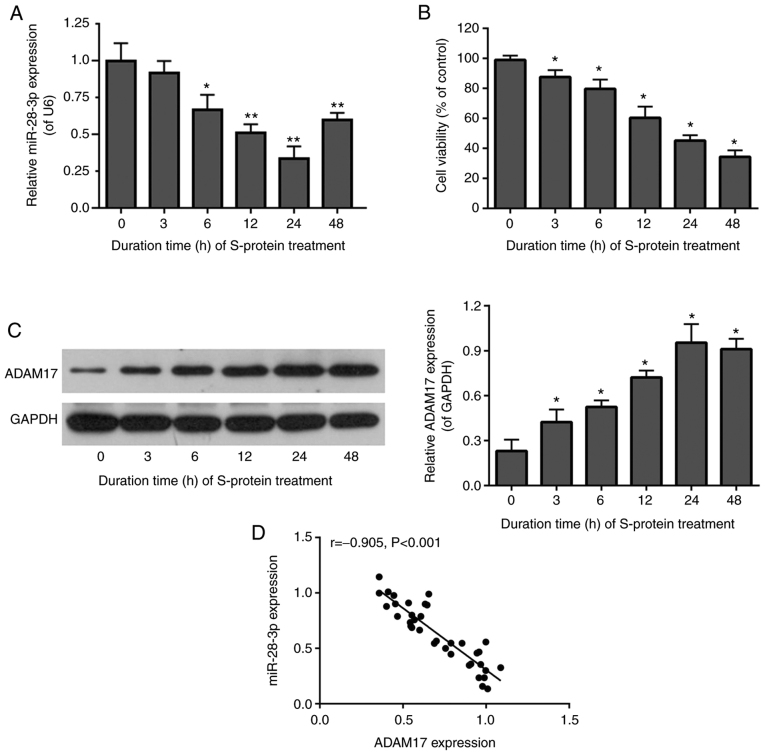
To determine whether miR-28-3p and ADAM17 were related in S-protein-treated 293T cells, miR-28-3p and ADAM17 expression were assessed by RT-qPCR and WB in 293T cells after S-protein treatment at 37°C for 0, 3, 6, 12, 24 and 48 h at 37°C. The level of miR-28-3p was downregulated (Fig. 1A), while ADAM17 protein was upregulated (Fig. 1C) in S-protein-treated 293T cells. Expression of miR-28-3p and ADAM17 was lowest and highest, compared with 0 h group respectively, after treatment for 24 h (Fig. 1A and C). In addition, cell viability of S-protein-treated 293T cells was significantly diminished at 24 h compared with the 0 h group (Fig. 1B). miR-28-3p and ADAM17 were highly and inversely correlated by Pearson's correlation test (Fig. 1D). The above results suggested that S-protein treatment was able to affect cell viability and regulate the expression of both miR-28-3p and ADAM17. In addition, the expression of miR-28-3p and ADAM17 was negatively correlated.
Figure 1.
S-protein inhibits 293T cell viability and expression of miR-28-3p and promotes expression of ADAM17. (A) miR-28-3p expression in in S-protein-treated 293T cells was detected by RT-qPCR. (B) Cell proliferation of 293T cells treated with S-protein was detected by the MTT assay. (C) ADAM17 expression in S-protein-treated 293T cells was detected by WB. (D) Negative correlation of miR-28-3p and ADAM17 by Pearson's correlation test. *P<0.05, **P<0.01, vs. 0 h (untreated) group. WB, western blotting; S-protein, spike protein; ADAM17, A disintegrin and metalloproteinase 17; RT-qPCR, reverse-transcription quantitative polymerase chain reaction.
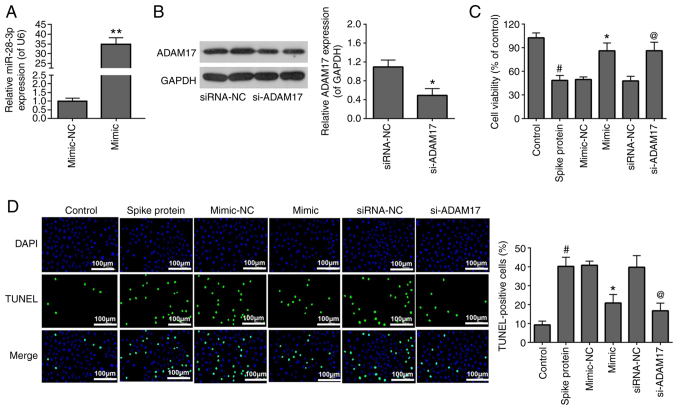
To investigate the effects of miR-28-3p mimic and si-ADAM17 on the viability and apoptosis of S-protein-treated 293T cells, the S-protein-treated 293T cells were transfected with miR-28-3p mimic and si-ADAM17. The transfection efficiency of miR-28-3p mimic (Fig. 2A) and si-ADAM17 (Fig. 2B) were analyzed by RT-qPCR and WB, respectively. For cell viability, 293T treated with S-protein demonstrated lower cell viability when compared with control 293T cells (Fig. 2C). However, both miR-28-3p mimic and si-ADAM17 promoted cell viability in the S-protein-treated 293T cells when compared with mimic-NC or siRNA-NC, respectively (Fig. 2C). For apoptosis, 293T cells treated with S-protein demonstrated a higher apoptosis rate when compared with control 293T cells (Fig. 2D) However, both miR-28-3p mimic and si-ADAM17 could inhibit apoptosis rate in the S-protein-treated 293T cells when compared with mimic-NC or siRNA-NC, respectively. The aforementioned results demonstrated that S-protein treatment affected cell viability and cell apoptosis through both miR-28-3p and ADAM17.
Figure 2.
miR-28-3p mimic and si-ADAM17 reverse the effects of S-protein on proliferation and apoptosis in 293T cells. (A) Expression of miR-28-3p mimic and mimic-NC was measured by RT-qPCR. (B) Expression of si-ADAM17 and siRNA-NC was measured by WB. (C) Cell proliferation in S-protein-treated 293T cells transfected with miR-28-3p mimic, si-ADAM17, mimic-NC or siRNA-NC were measured using the MTT assay. (D) Cell apoptosis in S-protein-treated 293T cells transfected with miR-28-3p mimic, si-ADAM17, mimic-NC or siRNA-NC measured by the TUNEL assay. #P<0.05 vs. control; *P<0.05 vs. mimic-NC, **P<0.01 vs. mimic-NC; @P<0.05 vs. siRNA-NC. WB, western blotting; S-protein, spike protein; ADAM17, A disintegrin and metalloproteinase 17; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; NC, negative control; miR, microRNA; si, small interfering.
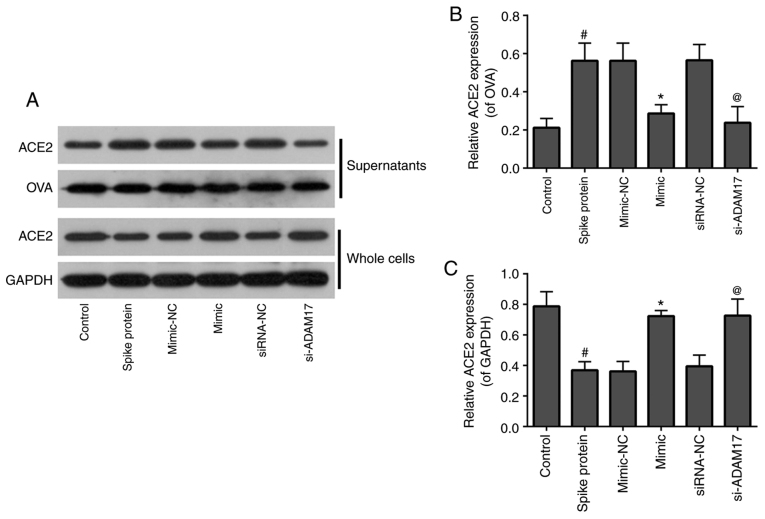
Both miR-28-3p mimic and si-ADAM17 inhibit shedding of the ACE2 ectodomain in 293T cells treated with S-protein
To examine the effects of miR-28-3p mimic and si-ADAM17 on ACE2 shedding in S-protein-treated 293T cells, WB was used to detect the expression of ACE2 in the supernatants and whole cells. As shown in Fig. 3, for supernatants, 293T treated with S-protein demonstrated a higher level of ACE2 when compared with control 293T cells and both miR-28-3p mimic and si-ADAM17 could inhibit ACE2 expression in the S-protein-treated 293T cells when compared with their respective controls (Fig. 3). The expression of ACE2 in the whole cell lysate demonstrated a reverse trend compared with supernatants. 293T cells were treated with S-protein, the expression of ACE2 in cells was lower compared with the control. While 293T cells transfected with miR-28-3p mimic or si-ADAM17, the expression of ACE2 in cells was higher compared with mimic-NC or siRNA-NC, respectively (Fig. 3). These results suggested that S-protein treatment promoted ACE2 shedding. miR-28-3p mimic or si-ADAM17 downregulated ACE2 shedding.
Figure 3.
miR-28-3p mimic and si-ADAM17 inhibit shedding of the ACE2 ectodomain in S-protein-treated 293T cells. (A) Western blotting results of ACE2, OVA, GAPDH expression in supernatants and whole cells of control, spike protein, mimic-NC, mimic, siRNA-NC, si-ADAM17 group. (B) Quantification of ACE2/OVA in supernatants of control, spike protein, mimic-NC, mimic, siRNA-NC and si-ADAM17 groups. (C) Quantification graph of ACE2/GAPDH in whole cells of control, spike protein, mimic-NC, mimic, siRNA-NC and si-ADAM17 groups. #P<0.05 vs. control; *P<0.05 vs. mimic-NC; @P<0.05 vs. siRNA-NC. ACE2, angiotensin-converting enzyme 2; S-protein, spike protein; ADAM17, A disintegrin and metalloproteinase 17; NC, negative control; miR, microRNA; si, small interfering; OVA, ovalbumin.
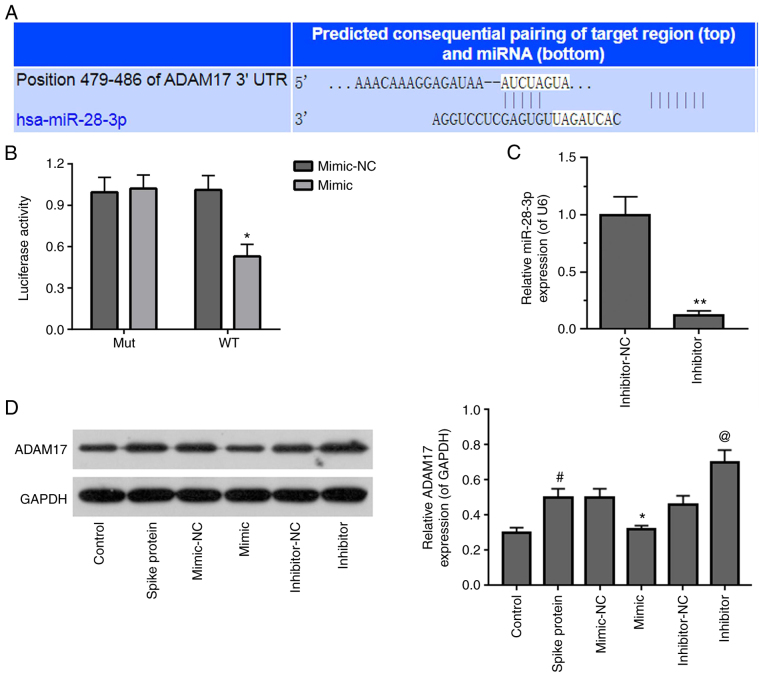
ADAM17 is a potential target of miR-28-3p
TargetScan v.7.2 was used to predict the potential target gene of miR-28-3p. It was found that the ADAM17 3′-UTR contained a potential binding site for miR-28-3p (Fig. 4A). The miR-28-3p mimic significantly suppressed WT, but not Mut 3′-UTR luciferase activity in S-protein-treated 293T cells in dual-luciferase activity assay (Fig. 4B). To verify that ADAM17 is a direct target of miR-28-3p, S-protein-treated 293T cells were transfected with miR-28-3p mimic or miR-28-3p inhibitor. First of all, the inhibitory effect of miR-28-3p inhibitor was verified by RT-qPCR and the inhibitory efficiency was ~50% (Fig. 4C). Subsequently, the ADAM17 expression level was assessed in S-protein-treated-293T cells transfected with miR-28-3p mimic or miR-28-3p inhibitor, respectively. 293T cells treated with S-protein demonstrated higher ADAM17 level when compared with control 293T cells (Fig. 4D). miR-28-3p mimic inhibited ADAM17 expression, while miR-28-3p inhibitor upregulated ADAM17 expression in the S-protein-treated 293T cells when compared with mimic-NC or inhibitor-NC, respectively. These results demonstrated that ADAM17 is a direct target of miR-28-3p during S-protein treatment.
Figure 4.
Identification of ADAM17 as a target of miR-28-3p. (A) Pairing of the 3′-UTR sequence of ADAM17 and miR-28-3p sequence using TargetScan v.7.2. (B) Luciferase assay in S-protein-treated 293T cells with WT-ADAM17 or Mut-ADAM17 and miR-28-3p mimic or mimic-NC. (C) Expression of miR-28-3p inhibitor and inhibitor-NC in 293T cells was measured by RT-qPCR. (D) Expression of ADAM17 in S-protein-treated 293T cells transfected with miR-28-3p mimic, miR-28-3p inhibitor, mimic-NC or inhibitor-NC was detected by WB. #P<0.05 vs control; *P<0.05 vs. mimic-NC, **P<0.01 vs. mimic-NC; @P<0.05 vs. inhibitor-NC. WB, western blotting; S-protein, spike protein; ADAM17, A disintegrin and metalloproteinase 17; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; NC, negative control; miR, microRNA; si, small interfering; WT, wild-type; Mut, mutant; UTR, untranslated region.
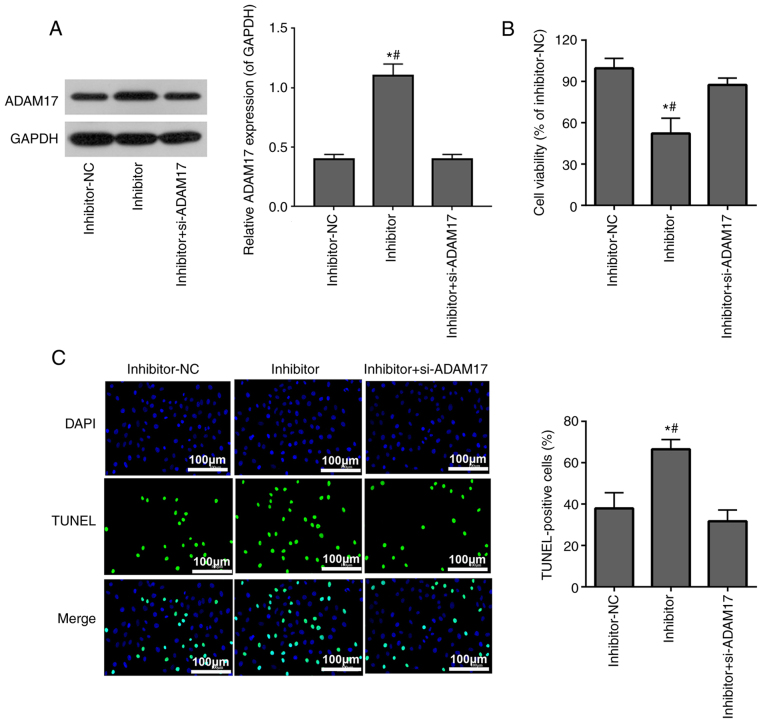
si-ADAM17 inhibits the effects of miR-28-3p inhibitor on cell viability and apoptosis in 293T cells treated with the S-protein
293T cells treated with the S-protein were transfected with miR-28-3p inhibitor alone or co-transfected with si-ADAM17 and miR-28-3p inhibitor. WB confirmed that miR-28-3p inhibitor upregulated ADAM17 protein expression, while co-transfection with si-ADAM17 and miR-28-3p inhibitor successfully reversed the effect of miR-28-3p inhibitor on ADAM17 expression (Fig. 5A). MTT assay demonstrated that miR-28-3p inhibitor inhibited S-protein-treated 293T cell proliferation, while co-transfection with si-ADAM17 and miR-28-3p inhibitor weakened the inhibitory effect of miR-28-3p inhibitor on S-protein-treated 293T cell proliferation (Fig. 5B). TUNEL assay demonstrated that miR-28-3p inhibitor promoted apoptosis of S-protein-treated 293T cells, while co-transfection with si-ADAM17 and miR-28-3p inhibitor weakened the promoted effect of miR-28-3p inhibitor on the apoptosis of S-protein-treated 293T cells (Fig. 5C). The aforementioned results demonstrated that miR-28-3p inhibitor inhibited cell viability and promoted cell apoptosis during S-protein treatment and this inhibition could be abolished by ADAM17 knockdown, which meant that miR-28-3p exerts its function on both cell viability and cell apoptosis through ADAM17.
Figure 5.
si-ADAM17 inhibits the effects of miR-28-3p inhibitor on cell proliferation and apoptosis in S-protein-treated 293T cells. (A) ADAM17 levels were analyzed after transfection of miR-28-3p inhibitor or co-transfection of miR-28-3p inhibitor and si-ADAM17 by WB. (B) S-protein-treated 293T cell proliferation was measured using the MTT assay. (C) S-protein-treated 293T cell apoptosis was measured using the TUNEL assay. *P<0.05 vs. inhibitor-NC; #P<0.05 vs. inhibitor + si-ADAM17. WB, western blotting; S-protein, spike protein; ADAM17, A disintegrin and metalloproteinase 17; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; NC, negative control; si, small interfering.
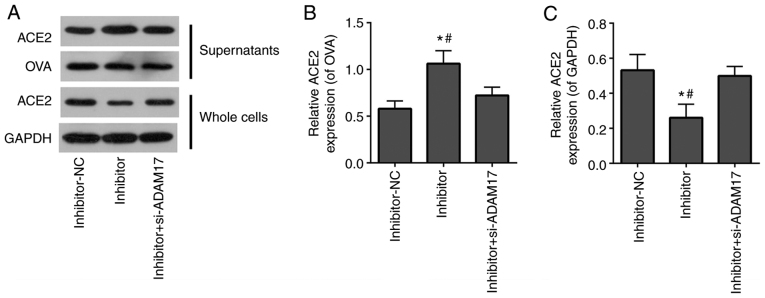
si-ADAM17 reverses the effects of miR-28-3p inhibitor on the shedding of the ACE2 ectodomain in 293T cells treated with the S-protein
To further verify that miR-28-3p regulated the ACE2 ectodomain shedding of S-protein-treated-293T cells by targeting ADAM17, miR-28-3p inhibitor alone or miR-28-3p inhibitor combined with si-ADAM17 was transfected into S-protein-treated 293T cells. WB demonstrated that the miR-28-3p inhibitor elevated ACE2 level in supernatants of S-protein-treated 293T cells and reduced ACE2 level of whole cell lysates (Fig. 6). Co-transfection with miR-28-3p inhibitor and si-ADAM17 reversed the effect of transfected miR-28-3p inhibitor alone on the ACE2 level both in supernatants and in whole cell lysates (Fig. 6). This suggested that miR-28-3p inhibitor enhanced ACE2 shedding of S-protein-treated 293T cells, while si-ADAM17 efficiently blocked miR-28-3p inhibitor-induced ACE2 shedding of S-protein-treated 293T cells. The aforementioned results demonstrated that miR-28-3p inhibitor promoted ACE2 ectodomain shedding during S-protein treatment, which could be abolished by ADAM17 knockdown. miR-28-3p exerts its function on ACE2 ectodomain shedding through ADAM17.
Figure 6.
si-ADAM17 inhibits the effects of miR-28-3p inhibitor on the shedding of the ACE2 ectodomain in 293T cells treated with S-protein. (A) Western blotting results of ACE2, OVA, GAPDH expression in supernatants and whole cells of inhibitor-NC, inhibitor and inhibitor + si-ADAM17 groups. (B) Quantification of ACE2/OVA in supernatants of inhibitor-NC, inhibitor and inhibitor + si-ADAM17 groups. (C) Quantification of ACE2/GAPDH in whole cells inhibitor-NC, inhibitor and inhibitor + si-ADAM17 groups. #P<0.05 vs. inhibitor+si-ADAM17; *P<0.05 vs. mimic-NC. ACE2, angiotensin-converting enzyme 2; S-protein, spike protein; ADAM17, A disintegrin and metalloproteinase 17; NC, negative control; si, small interfering; OVA, ovalbumin.
Discussion
ACE2, a homologue of ACE, serves a pivotal role in balancing responses initiated from ACE, hydrolyses Angiotensin (Ang I) to generate Ang-(1-9) and Ang II to generate Ang-(1-7), which maintain the normal functioning of the cardiovascular system, kidney and lung (37). In addition, ACE2 is a receptor for the S-protein of SARS-CoV-2 (38). The binding affinity of SARS-CoV-2 with ACE2 appears stronger compared with that of SARS-CoV and has been reported to be 10- to 20-fold higher (9,39), which aids the quick spreading of the SARS-CoV-2 virus (40). Meanwhile, similar to SARS-CoV, SARS-CoV-2 invades cells via the binding of S-protein to the ACE2, followed by subsequent downregulation of surface ACE2 expression (40). Previous studies have demonstrated that ADAM17 is involved in regulating of ACE2 ectodomain shedding during SARS-CoV-2 invading cells (3,41,42). ADAM17 cleaves ACE2, which results in shedding of the ACE2 ectodomain into cell culture supernatants and S-protein stimulates ADAM17-dependent ACE2 cleavage (43). ADAM17 is a member of the ADAM family of proteinases, which also includes ADAM10, ADAM9 and ADAM12 (44). Lambert et al (13) found that shedding of the ACE2 ectodomain was significantly inhibited using a mixed ADAM10/ADAM17 inhibitor (GW280264X), but was unaffected by a selective ADAM10 inhibitor (GI254023X) alone. Upregulation of ADAM-17 activity facilitated SARS-CoV viral entry, while knockdown of ADAM17 by siRNA severely attenuated SARS-CoV cellular entry (9). These results suggested ADAM17 may serve a major role in the cleavage and shedding of the ACE2 ectodomain (9,13). However, it is unknown which molecular mechanisms initiate ADAM17 to serve such an important role in ACE2 ectodomain shedding during SARS-CoV-2 virus infection.
The findings of the present study demonstrated that ADAM17 expression was upregulated in S-protein-treated 293T cells compared with the non-treatment control group. In addition, it was also demonstrated that ACE2 ectodomain shedding was increased following treatment with S-protein, while knockdown of ADAM17 expression by siRNA led to a reduction in ACE2 ectodomain shedding, which was consistent with previous research (9,34). The findings of the present study confirmed the previous conclusions that inhibition of ADAM17 may reduce shedding of the ACE2 ectodomain and have a protective effect against SARS-CoV infection. Targeting ADAM17 expression may be a potential treatment strategy for SARS-CoV-2.
It has been known that the S-protein stimulates ADAM17-dependent ACE2 cleavage, which drives virus entry into cells (43). However, there is little known about how the S-protein regulates ADAM17-dependent ACE2 cleavage (12). There has been a recent explosion of knowledge in the field of miRNAs regulating viral infection. For example, miR-27b is widely known as an antiviral miRNA which reduces HIV-1 replication (45) and miR-122 can inhibit hepatitis C viral replication (46). In one study, miR-1246 regulated the expression of ACE2 in patients with SARS-CoV-2 infection (47).
The present study first observed that miR-28-3p expression was downregulated in S-protein treated 293T cells, miR-28-3p expression was negatively correlated with ADAM17 expression (Fig. 1D) and miR-28-3p mimic reduced ACE2 ectodomain shedding (Fig. 3A). Further bioinformatics analysis demonstrated that ADAM17 was a potential target of miR-28-3p (Fig. 4A). It was hypothesized that miR-28-3p participated in ADAM17-dependent ACE2 cleavage by S-protein stimulation. Further experimental findings of the present study suggested that miR-28-3p did participate in ADAM17-dependent ACE2 cleavage by S-protein stimulation.
In conclusion, the present study demonstrated that the S-protein of SARS-Cov-2 promoted ADAM17-dependent ACE2 ectodomain shedding through downregulation of miR-28-3p, which may provide a novel therapeutic strategy for treatment of SARS-CoV-2 infections. However, it remains unclear whether miR-28-3p inhibits ACE2 ectodomain shedding in 293T cells in the absence of S-protein and is miR-28-3p the only one miRNA that is downregulated by S-protein during SARS-Cov-2 invading cells. Future studies should perform infectivity experiments to further verify the findings of the present study and to elucidate the mechanisms of how the S-protein downregulates miR-28-3p.
Acknowledgments
Not applicable.
Abbreviations
- SARS-CoV-2
syndrome coronavirus 2
- ACE2
angiotensin-converting enzyme 2
- S-protein
spike protein
- ADAM17
A disintegrin and metalloproteinase 17
- RT-qPCR
reverse-transcription quantitative polymerase chain reaction
- WB
western blotting
- 3′-UTR
3′ untranslated regions
- WT
wild type
- Mut
mutant
Funding Statement
No funding was received.
Availability of data and materials
The datasets use/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YX was responsible for the concept and study design and provided critical input and reviewed the manuscript for important intellectual content. YL performed the experiments, collected, analyzed and interpreted the data and wrote the manuscript. YX and XL confirmed the authenticity of all the raw data. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Horton R. Offline: 2019-nCoV outbreak-early lessons. Lancet. 2020;395:322. doi: 10.1016/S0140-6736(20)30212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) Coronavirus disease (COVID-2019) situation report. WHO; Geneva: 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Accessed July 19, 2021. [Google Scholar]
- 3.Wan MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM. SARS-CoV-2: Structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao L, Sakagami H, Miwa N. ACE2: The key molecule for understanding the pathophysiology of severe and critical conditions of COVID-19: Demon or angel? Viruses. 2020;12:491. doi: 10.3390/v12050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuba K, Imai Y, Penninger JM. Multiple functions of angiotensin-converting enzyme 2 and its relevance in cardiovascular diseases. Circ J. 2013;77:301–308. doi: 10.1253/circj.CJ-12-1544. [DOI] [PubMed] [Google Scholar]
- 7.Patel VB, Zhong JC, Grant MB, Oudit GY. Role of the ACE2/angiotensin 1-7 axis of the renin-angiotensin system in heart failure. Circ Res. 2016;118:1313–1326. doi: 10.1161/CIRCRESAHA.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner AJ, Hiscox JA, Hooper NM. ACE2: From vasopeptidase to SARS virus receptor. Trends Pharmacol Sci. 2004;25:291–294. doi: 10.1016/j.tips.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, Raizada MK, Grant MB, Oudit GY. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: Celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danilczyk U, Penninger JM. Angiotensin-converting enzyme II in the heart and the kidney. Circ Res. 2006;98:463–471. doi: 10.1161/01.RES.0000205761.22353.5f. [DOI] [PubMed] [Google Scholar]
- 11.Su H, Yang M, Wan C, Yi LX, Tang F, Zhu HY, Yi F, Yang HC, Fogo AB, Nie X, Zhang C. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zipeto D, Palmeira JDF, Argañaraz GA, Argañaraz ER. ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID-19. Front Immunol. 2020;11:576745. doi: 10.3389/fimmu.2020.576745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert DW, Yarski M, Warner FJ, hornhill P, Parkin ET, Smith AI, Hooper NM, Turner AJ. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J Biol Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Guo F, Liu K, Wang H, Rao S, Yang P, Jiang C. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 2008;136:8–15. doi: 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel VB, Clarke N, Wang Z, Fan D, Parajuli N, Basu R, Putko B, Kassiri Z, Turner AJ, Oudit GY. Angiotensin ii induced proteolytic cleavage of myocardial ACE2 is mediated by TACE/ADAM-17: A positive feedback mechanism in the RAS. J Mol Cell Cardiol. 2014;66:167–176. doi: 10.1016/j.yjmcc.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 17.Palau V, Riera M, Soler MJ. ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol Dial Transplant. 2020;35:1071–1072. doi: 10.1093/ndt/gfaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 19.Biswas S, Haleyurgirisetty M, Lee S, Hewlett I, Devadas K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine. 2019;43:307–316. doi: 10.1016/j.ebiom.2019.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S, Eom K, Kim J, Bang H, Wang HY, Ahn S, Kim G, Jang H, Kim S, Lee D, et al. MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC cancer. 2017;17:658. doi: 10.1186/s12885-017-3642-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan MA, Sany MRU, Islam MS, Islam ABMMK. Epigenetic regulator mirna pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front Genet. 2020;11:765. doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G, Dong F, Xu Z, Sharma S, Hu X, Chen D, Zhang L, Zhang J, Dong Q. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC cancer. 2017;17:805. doi: 10.1186/s12885-017-3816-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai XT, Nicot C. miR-28-3p is a cellular restriction factor that inhibits human T cell leukemia virus, type 1 (HTLV-1) replication and virus infection. J Biol Chem. 2015;290:5381–5390. doi: 10.1074/jbc.M114.626325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L, Zhang YF, Hu F. miR-28-5p inhibits the migration of breast cancer by regulating WSB2. Int J Mol Med. 2020;46:1562–1570. doi: 10.3892/ijmm.2020.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu G, Wang Z, Mijiti M, Du G, Li Y, Dangmurenjiafu G. MiR-28-5p promotes human glioblastoma cell growth through inactivation of FOXO1. Int J Clin Exp Pathol. 2019;12:2972–2980. [PMC free article] [PubMed] [Google Scholar]
- 26.Fan HN, Liao XH, Zhang J, Zheng HM. Macrophages promote cell proliferation in colorectal cancer via IL-1β-mediated downregulation of miR-28-3p. J Biol Regul Homeost Agents. 2020;34:1657–1668. doi: 10.23812/20-210-A. [DOI] [PubMed] [Google Scholar]
- 27.Zhou X, Wen W, Shan X, Qian J, Li H, Jiang T, Wang W, Cheng W, Wang F, Qi L, et al. MiR-28-3p as a potential plasma marker in diagnosis of pulmonary embolism. Thromb Res. 2016;138:91–95. doi: 10.1016/j.thromres.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Tiwari A, Mukherjee B, Dixit M. MicroRNA key to angiogenesis regulation: MiRNA biology and therapy. Curr Cancer Drug Targets. 2018;18:266–277. doi: 10.2174/1568009617666170630142725. [DOI] [PubMed] [Google Scholar]
- 29.Purohit V, Rapaka RS, Rutter J, Shurtleff D. Do opioids activate latent HIV-1 by down-regulating anti-HIV microRNAs? J Neuroimmune Pharmacol. 2012;7:519–523. doi: 10.1007/s11481-012-9356-1. [DOI] [PubMed] [Google Scholar]
- 30.Tandon R, Sharp JS, Zhang F, Pomin VH, Ashpole NM, Mitra D, Jin W, Liu H, Sharma P, Linhardt RJ. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J Virol. 2021;95:e01987. doi: 10.1128/JVI.01987-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Hu S, Wang J, Xue Z, Wang C, Wang N. Dexamethasone inhibits SARS-CoV-2 spike pseudotyped virus viropexis by binding to ACE2. Virology. 2021;554:83–88. doi: 10.1016/j.virol.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu S, Wang J, Zhang Y, Bai H, Wang C, Wang N, He L. Three salvianolic acids inhibit 2019-nCoV spike pseudovirus viropexis by binding to both its RBD and receptor ACE2. J Med Virol. 2021;93:3143–3151. doi: 10.1002/jmv.26874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Ding Y, Wang Y, Liang P, Zhang L, Liu R. Oroxylin A is a severe acute respiratory syndrome coronavirus 2-spiked pseudotyped virus blocker obtained from Radix Scutellariae using angiotensin-converting enzyme II/cell membrane chromatography. Phytother Res. 2021;35:3194–3204. doi: 10.1002/ptr.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, Yamamoto N, Sasazuki T, Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc Natl Acad Sci USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang ZY, Huang Y, Ganesh L, Leung K, Kong WP, Schwartz O, Subbarao K, Nabel GJ. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y. Compensation of ACE2 function for possible clinical management of 2019-nCoV-induced acute lung injury. Virol Sin. 2020;35:256–258. doi: 10.1007/s12250-020-00205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore MJ, Dorfman T, Li W, Wong SK, Li Y, Kuhn JH, Coderre J, Vasilieva N, Han Z, Greenough TC, et al. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, et al. Structural and functional basis of SARS-CoV-2 entry by using Human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taneera J, El-Huneidi W, Hamad M, Mohammed AK, Elaraby E, Hachim MY. Expression profile of SARS-CoV-2 host receptors in human pancreatic islets revealed upregulation of ACE2 in diabetic donors. Biology (Basel) 2020;9:215. doi: 10.3390/biology9080215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Loyola MB, Dos Reis TTA, de Oliveira GXLM, da Fonseca Palmeira J, Argañaraz GA, Argañaraz ER. Alpha-1-antitrypsin: A possible host protective factor against Covid-19. Rev Med Virol. 2021;31:e2157. doi: 10.1002/rmv.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zunke F, Rose-John S. The shedding protease ADAM17: Physiology and pathophysiology. Biochim Biophys Acta Mol Cell Res. 2017;1864:2059–2070. doi: 10.1016/j.bbamcr.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Das G, Mukherjee N, Ghosh S. Neurological insights of COVID-19 pandemic. ACS Chem Neurosci. 2020;11:1206–1209. doi: 10.1021/acschemneuro.0c00201. [DOI] [PubMed] [Google Scholar]
- 46.Trobaugh DW, Klimstra WB. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Rostami MR, Leopold PL, Mezey JG, O'Beirne SL, Strulovici-Barel Y, Crystal RG. Expression of the SARS-CoV-2 ACE2 receptor in the human airway epithelium. Am J Respir Crit Care Med. 2020;202:219–229. doi: 10.1164/rccm.202003-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets use/or analyzed during the current study are available from the corresponding author on reasonable request.