Summary
Objective: To identify and highlight research papers representing noteworthy developments in signals, sensors, and imaging informatics in 2020.
Method: A broad literature search was conducted on PubMed and Scopus databases. We combined Medical Subject Heading (MeSH) terms and keywords to construct particular queries for sensors, signals, and image informatics. We only considered papers that have been published in journals providing at least three articles in the query response. Section editors then independently reviewed the titles and abstracts of preselected papers assessed on a three-point Likert scale. Papers were rated from 1 (do not include) to 3 (should be included) for each topical area (sensors, signals, and imaging informatics) and those with an average score of 2 or above were subsequently read and assessed again by two of the three co-editors. Finally, the top 14 papers with the highest combined scores were considered based on consensus.
Results: The search for papers was executed in January 2021. After removing duplicates and conference proceedings, the query returned a set of 101, 193, and 529 papers for sensors, signals, and imaging informatics, respectively. We filtered out journals that had less than three papers in the query results, reducing the number of papers to 41, 117, and 333, respectively. From these, the co-editors identified 22 candidate papers with more than 2 Likert points on average, from which 14 candidate best papers were nominated after intensive discussion. At least five external reviewers then rated the remaining papers. The four finalist papers were found using the composite rating of all external reviewers. These best papers were approved by consensus of the International Medical Informatics Association (IMIA) Yearbook editorial board.
Conclusions. Sensors, signals, and imaging informatics is a dynamic field of intense research. The four best papers represent advanced approaches for combining, processing, modeling, and analyzing heterogeneous sensor and imaging data. The selected papers demonstrate the combination and fusion of multiple sensors and sensor networks using electrocardiogram (ECG), electroencephalogram (EEG), or photoplethysmogram (PPG) with advanced data processing, deep and machine learning techniques, and present image processing modalities beyond state-of-the-art that significantly support and further improve medical decision making.
Keywords: Sensors, signals, imaging informatics, medical informatics
Introduction
Sensors, signals, and imaging informatics (SSII) continues to be a rapidly growing research field. One could see three independent parts, or at least two, if imaging and signal informatics are considered similar to a biomedical signal as one-dimensional and a medical image as a two- or more-dimensional stream. However, the methods applied here are similar. In contrast, the sensor's part could be seen as more device-oriented. Picard & Wolf define “ sensor informatics ” as new technologies and applications for medical services incorporating wearable sensors, signal processing, machine learning, and data mining techniques [ 1 ]. In our view, the technological development of a sensing device is not part of medical informatics but the integration of such devices in medical information systems and their application in research, clinical trials, and medical care. Unobtrusive health monitoring in private spaces such as the car or the home is based on various sensors and is expeditiously growing in research and applications [ 1 2 3 ]. Witte et al. see “ signal informatics ” as an advanced integrative concept in the framework of medical informatics [ 4 ]. Again, data integration is emphasized. In the medical field, semantical integration is particularly important. In a recent review, Cook defines “ imaging informatics ” via the imaging informaticist as a unique individual who sits at the intersection of clinical radiology, data science, and information technology [ 5 ]. Imaging informatics, however, is the most common term of these three. In 2020, we observed three new reviews in this field [ 5 6 7 ]. Furthermore, the first standardized curriculum for imaging informatics fellowships suggested by the Society for Computer Applications in Radiology (SCAR) in 2004 [ 8 ] has been updated [ 9 ].
In this variety, we faced the daunting task of identifying notable research. Furthermore, thousands of research papers have applied deep learning approaches to well-documented SSII problems – mostly outperforming the classical state of the art. On the one hand, we faced a needle in the haystack problem given the large number of papers, but simultaneously, on the other, we felt as if we were sorting through genetically modified strawberries, where all looked alike. In summary, identifying real novelty among papers in SSII is not easy if considering solely the title or the abstract. As van Ooijen, Nagaraj & Olthof [ 10 ] point out, SSII is more than ‘just’ deep learning. As such, our objective was to catch the spectrum of work—both deep learning and other machine learning techniques—that best represent the developments in SSII in 2020.
2 Paper Selection Process
The process of searching the literature for candidate best papers by the SSII section remained a challenging task, given the broad nature of the SSII category. This year, we overhauled the queries that were applied over the last years [ 11 12 13 ]. First, we developed similar queries to search PubMed and Scopus. We focused on research articles in the English language and excluded all others. We did not include review papers. Then, we built the queries separately for sensors, signals, and images. Each of the six queries was built from two blocks. For sensors, the first block captures all relevant terms for the sensing device, and the second term lists keywords for biomedical signals and vital signs. For signals and images, we used modality names (e.g., computed tomography, magnetic resonance angiography) and processing techniques. In addition, we significantly decreased the number of results for the imaging query by applying the MeSH term “medical informatics”. The queries are listed completely in Appendix 2.
In mid-January 2021, we executed the final query. After removing duplicates and conference proceedings, the query returned a set of 101, 193 and 529 papers for sensors, signals, and imaging informatics, respectively ( Table 1 ). Given these numbers, we sought to further reduce the number of papers needed to be reviewed. During the initial review of abstracts, we noted that several excluded papers were published in journals that were only tangentially related to the topic. We started exploring whether setting a threshold of the minimum number of papers in the results per journal would help. Figure 1 illustrates the effect of setting different thresholds and the number of papers remaining in the imaging informatics set. If we set the threshold to be at least three papers, 333 papers remained. Furthermore, we also discussed focusing on official International Medical Informatics Association (IMIA) journals, impact factors, or the set of journals that are linked to “medical informatics” in MEDLINE or Index Medicus. However, as new open access journals are broadly establishing themselves with broad acceptance by scientists, we kept on the contribution model. Next year, we plan to normalize the threshold by the annual number of papers that were published in the journal.
Table 1. Number of articles recovered from the databases.
| Topic | Pubmed | Scopus | Duplicates + conference | Merged unique | Journal selection |
|---|---|---|---|---|---|
| Sensor Signal Imaging |
97 200 182 |
9 30 397 |
5 37 50 |
101 193 529 |
41 117 333 |
Fig. 1.
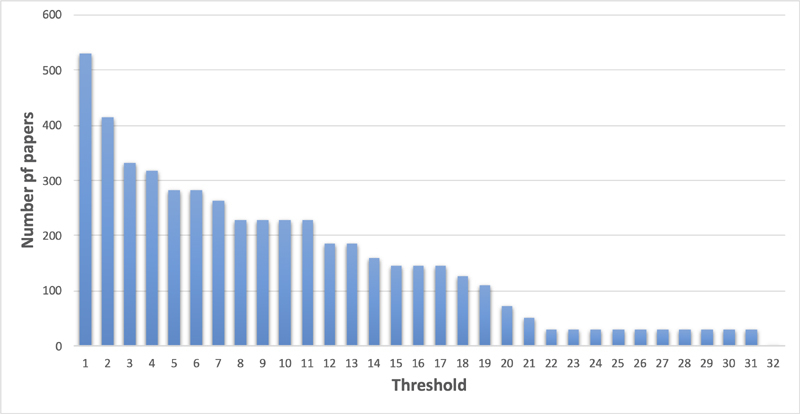
Number of papers remaining according to the number of papers published in the particular journal.
We reviewed the titles and abstracts and independently, ranked them on a three-point Likert scale. Papers were rated from 1 (do not include) to 3 (should be included) for each topical area (sensors, signals, and imaging informatics). Each paper was assessed by two of the section co-editors. In doing so, we identified 22 candidate papers with more than 2 Likert points on average. These papers were read entirely and rescored by all co-editors. After intensive discussion, we nominated 14 candidate best papers. At least five external reviewers then rated the papers. The four finalist papers were were identified based on the composite rating of all external reviewers. These best papers were approved by consensus of the IMIA Yearbook editorial board.
3 Emerging Trends and New Directions
This year's literature yielded a number of interesting developments in the field of SSII. We highlight three emerging trends, particularly in light of the COVID-19 pandemic.
3.1 Extracting Additional Information from Existing Data Sources
One trend has been the use of machine learning to extract additional physiological information from existing measurements. For example, measurements from a photoplethysmogram (PPG), which measures blood volume changes using a pulse oximeter, can potentially provide systolic and diastolic blood pressure measurements. Hsu et al. [ 14 ] showed using a fully connected deep neural network trained on the MIMIC (Multi-parameter Intelligent Monitoring for Intensive Care) II cohort achieves low mean absolute error and root mean squared error when estimating blood pressure from PPG compared to reference measurements using a cuff. Miao et al. [ 15 ] proposed continuous blood pressure measurement using electrocardiogram (ECG) signals using a fusion of a residual network and long short-term memory model. Their methods were trained and evaluated using data from the MIMIC III cohort as well as patients from their institution. Another interesting application is the interpretation of ECG signals to predict hypoglycemic events. Using a commercially available wearable ECG monitor, Porumb et al. [ 16 ] demonstrated one approach using a combined convolutional and recurrent neural network. The outbreak of COVID-19 reinforced the utility and importance of using data from wireless sensors and consumer wearables to track infections. Wearable health monitors such as PPGs have played an important role in providing markers of respiratory health (cough frequency/intensity, respiratory rate/effort). Novel sensing materials and fabrication techniques are yielding ways to unobtrusively monitor symptoms, record lung sound, and measure the respiratory rate. Ding et al. [ 17 ] provide a review of these developments in greater depth, while Wang et al. analyze unobtrusive health monitoring in vehicles [ 2 ] and homes [ 3 ].
3.2 Providing Access to Larger Datasets for Algorithm Development
Advances in SSII have also been driven by the availability of large, diverse, publicly available patient data. MIMIC [ 18 ] remains an important resource for developing and testing algorithms on signals and sensors data collected in an intensive care environment. The UK Biobank is another rich open-access resource that collected multi-modal imaging data on a subset of 100,000 participants [ 19 ]. The pandemic has also spurred the establishment of new resources, focusing on COVID-related diagnosis and treatment. In the United States, the national COVID Cohort Collaborative is building a national data resource to accelerate the development of therapeutics [ 20 ]. Informatics challenges related to indexing, security, standardization, data provenance, and linked clinical outcomes will need to be addressed. The RSNA International COVID-19 Open Radiology Database is another example of annotated and de-identified chest computed tomography (CT) and radiography data on COVID-19-positive patients [ 21 ]. Complementing efforts to collect larger real-world patient datasets and techniques for introducing reasonable simulated data to improve model training are also examined. Augmentation is being pursued to generate additional synthetic examples. Shi et al. [ 22 ] proposed a knowledge-guided adversarial augmentation approach to generate additional training exams for training a model for classifying thyroid ultrasound as benign or malignant. Leveraging standardized terms describing nodules extracted from radiology reports, the authors showed that their knowledge-guided auxiliary classifier generative adversarial network outperforms other deep learning-based comparison methods.
3.3 Ensuring Reliable and Reproducible Results
Despite the large numbers of Artificial Intelligence/Machine Learning (AI/ML) models being published, the translation of these works to clinical practice is slow. One hurdle is the differences between characteristics of the target population relative to the population that the models were trained on. Fu et al. investigated the impact of differences in dose and reconstruction kernel on AI-based detection of simulated pulmonary nodules in chest phantoms scanned using CT [ 23 ]. The authors found that while differences in dose did not affect the volume measurements using the four evaluated AI algorithms, the kernel had a significant effect. Moreover, some models were more adversely impacted than others, but transparency regarding model training (e.g., population characteristics used for training) is generally lacking. With the rapid pace of research into COVID-19, a well-documented concern has been that most published models are poorly reported and at high risk of bias [ 24 ]. Similarly, in signals and sensors research, most research studies are based on data from a single institution, which raises concerns about bias, and many studies do not report performance on an external test cohort [ 25 ]. We anticipate that as the field moves towards clinical translation, the requirements for standardized reporting of AI/ML studies and characterization of model generalization in different target populations will become increasingly important.
4 Summary and Conclusion
In summary, SSII remains a rapidly growing field that increasingly combines a broad range of available imaging and sensor technologies with a significantly rising number of innovative machine learning and AI-based approaches. The main emerging trends can be traced back to the extraction of new or additional information from SSII applications by providing access to larger data sources and repositories, which are urgently needed for the development and validation of new image and signal processing tools. Translating methods into clinical application remains a challenge as new algorithms and tools need to demonstrate their clinical validity by proving scientific validity, analytical validity, and clinical performance, in particular when approaching regulatory approval as software as medical device. This year, we also optimized the process of searching the literature for candidate best papers, which – in our opinion – led to a further increase in the quality of selected papers. This is, of course, an ongoing process and we strive to improve it year on year. Last but not least, 2020 was mainly shaped by the light of the COVID-19 pandemic. Numerous innovations in SSII in the fight against the pandemic have already contributed to improved patient management in this globally unique and challenging situation.
Table 2. Best paper selection of articles for the IMIA Yearbook of Medical Informatics 2021 in the section ‘Sensors, Signals, and Imaging Informatics’. The articles are listed in alphabetical order of the first author's surname.
| Section Sensors, Signals, and Imaging Informatics |
|---|
| ▪ Gemein LAW, Schirrmeister RT, Chrabąszcz P, Wilson D, Boedecker J, Schulze-Bonhage A, Hutter F, Ball T. Machine-learning-based diagnostics of EEG pathology. Neuroimage 2020 Oct 15;220:117021. ▪ Karimi D, Dou H, Warfield SK, Gholipour A. Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis. Med Image Anal 2020 Oct;65:101759. ▪ Langner T, Strand R, Ahlström H, Kullberg J. Large-scale biometry with interpretable neural network regression on UK Biobank body MRI. Sci Rep 2020 Oct 20;10(1):17752. ▪ Saito H, Aoki T, Aoyama K, Kato Y, Tsuboi A, Yamada A, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc 2020 Jul;92(1):144-151.e1. |
Acknowledgments
The section editors would like to thank Adrien Ugon for supporting the external review process and the external reviewers for their input on the candidate best papers.
Appendix 1: Content Summaries of Selected Best Papers for the 2021 IMIA Yearbook, Section Sensors, Signals, and Imaging Informatics (CB).
Gemein LAW, Schirrmeister RT, Chrabąszcz P, Wilson D, Boedecker J, Schulze-Bonhage A, Hutter F, Ball T
Machine-learning-based diagnostics of EEG pathology
Neuroimage 2020 Oct 15;220:117021
The analysis of clinical electroencephalograms (EEGs) is a time-consuming and demanding process and requires years of training. The development of algorithms for automatic EEG diagnosis, such as machine learning (ML) methods, could be a tremendous benefit to clinicians in analyzing EEGs. In this work, end-to-end decoding using deep neural networks was compared with feature-based decoding using a large set of features. Approximately 3,000 recordings from the Temple University Hospital EEG Corpus (TUEG) study were used, representing the largest publicly available collection of EEG recordings to date. For feature-based pathology decoding, Random Forest (RF), Support Vector Machine (SVM), Riemannian geometry (RG), and Auto-Skill Classifier (ASC) were used, while three types of convolutional neural networks (CNN) were applied for end-to-end pathology decoding: the 4-layer ConvNet architecture Braindecode Deep4 ConvNet (BD-Deep4), Braincode (BD) and TCN. The main result of this study was that the EEG pathology decoding accuracy is in a narrow range of 81-86%, also compared to a wide range of analysis strategies, network archetypes, network architects, feature-based classifiers and ensembles, and datasets. Based on the feature visualizations, features extracted in the theta and delta regions of temporal electrode positions were considered informative. Feature correlation analysis showed strong correlations of features extracted at different electrode positions. Besides the fact that there is no statistical evidence that the deep neural networks studied perform better than the feature-based approach, this work presents that a somewhat elaborate feature-based approach can be used to achieve similar decoding results as deep end-to-end methods. The authors recommend decoding specific labels to avoid the consequences of label noise in decoding EEG pathology. This work provides a remarkable and objective comparison between deep learning and feature-based methods based on numerous experiments, including cross-validation, bootstrapping, and input signal perturbation strategies.
Karimi D, Dou H, Warfield SK, Gholipour A
Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis
Med Image Anal 2020 Oct;65:101759
Label noise is unavoidable in many medical image datasets. It can be caused by limited attention or expertise of the human annotator, the subjective nature of labeling, or errors in computerized labeling systems. This is especially concerning for medical applications where datasets are typically small, labeling requires domain expertise and suffers from high inter- and intra-observer variability, and erroneous predictions may influence decisions directly impacting human health. The authors reviewed the state-of-the-art label noise handling in deep learning and investigated how these methods were applied to medical image analysis. Their key recommendations to account for label noise are: label cleaning and pre-processing, adaptions on network architectures, the use of label-noise-robust loss functions, re-weighting data, label consistency checks, and the choice of training procedures. They underpin their findings with experiments on three medical datasets where label noise was introduced by the systematic error of a human annotator, the inter-observer variability, or the noise generated from an algorithm. Their results suggest a careful curation of data for training deep learning algorithms for medical image analysis. Furthermore, the authors recommend integrating label noise analyses in development processes for robust deep learning models.
Langner T, Strand R, Ahlström H, Kullberg J
Large-scale biometry with interpretable neural network regression on UK Biobank body MRI
Sci Rep 2020 Oct 20;10(1):17752
This work presents a novel neural network approach for image-based regression to infer 64 biological metrics (beyond age) from neck-to-knee body MRIs with relevance for cardiovascular and metabolic diseases. Image data were collected from the UK Biobank study, linked to extensive metadata comprising non-imaging properties such as measurements of body composition by dual-energy X-ray absorptiometry (DXA) imaging, patient-related parameters, i.e., age, sex, height and weight, and additional biomarkers for cardiac health including pulse rate, accumulated fat in the liver and grip strength. The authors adapted and optimized a previously presented regression pipeline for age estimation using a ResNet50 architecture, not requiring any manual intervention or direct access to reference segmentations. Based on 31,172 magnetic resonance imaging (MRI) scans, the neural network was trained and cross-validated on simplified, two-dimensional representations of the MR images and evaluated by generated predictions and saliency maps for all examined properties. The work is noteworthy for its extensive validation of both the whole framework and predictions, demonstrating a robust performance and outperforming linear regression baseline in all applied cases. Saliency analysis showed that the developed neural network accurately targets specific body regions, organs, and limbs of interest. The network can emulate different modalities, including DXA or atlas-based MRI segmentation, and on average, correctly targets specific structures on either side of the body. The authors impressively demonstrated how convolutional neural network regression could effectively be applied in MRI and offer a first valuable, fully automated approach to measure a wide range of important biological metrics from single neck-to-knee body MRIs.
Saito H, Aoki T, Aoyama K, Kato Y, Tsuboi A, Yamada A, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T
Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network
Gastrointest Endosc 2020 Jul;92(1):144-151.e1
Wireless capsule endoscopy (WCE) is an established examination method for the diagnosis of small-bowel diseases. Automated detection and classification of protruding lesions of various types from WCE images is still challenging because it takes 1 to 2 hours on average for a correct diagnosis by a physician. In this work, a deep neural network architecture, termed single shot multibox detector (SSD) based on a deep convolutional neural network (CNN) structure with 16 or more layers, was trained on 30,584 WCE images from 292 patients collected from multiple centers and tested on an independent set of 17,507 images from 93 patients, including 7507 images of protruding lesions from 73 patients. All regions showing protruding lesions were manually annotated by six independent expert endoscopists, representing the ground truth for training the network. The CNN performance was evaluated by a ROC analysis, revealing an AUC of 0.911, a sensitivity of 90.7%, and a specificity of 79.8% at the optimal cut-off value of 0.317 for the probability score. In a subanalysis of the categories of protruding lesions, the sensitivities appeared between 77.0% and 95.8% for the detection of polyps, nodules, epithelial tumors, submucosal tumors, and venous structures, respectively. In individual patient analyses, the detection rate of protruding lesions was 98.6%. The rates of concordance of the labeling by the CNN and three expert endoscopists were between 42% and 83% for the different morphological structures. A false positive/negative error analysis was reported, indicating some limitations of the current approach in terms of an imbalanced number of cases, color diversity, and variation of structures in the images. The work is notable for its excellent clinical applicability using a new computer-aided system with good diagnostic performance to detect protruding lesions in small-bowel capsule endoscopy.
Appendix 2: Queries Used for Candidate Paper Retrieval.
The queries used to retrieve literature from PubMed and Scopus differ slightly, as the databases do not use the same data fields and query syntax. A separate query was performed for sensors, signals, and imaging informatics.
2.1 Sensors
2.1.1 PubMed Query
((“2020/01/01”[DP] : “2020/12/31”[DP])AND Journal Article [pt] AND English[lang] AND hasabstract[text] NOT Bibliography[pt] NOT Comment[pt] NOT Editorial[pt] NOT Letter[pt] NOT News[pt] NOT Review[pt] NOT Case Reports[pt] NOT Published Erratum[pt] NOT Historical Article[pt] NOT legislation[pt]NOT “clinical trial”[pt]NOT “evaluation studies”[pt]NOT “technical report”[pt]NOT “Scientific Integrity Review”[pt]NOT “Systematic Review”[pt]NOT “Retracted Publication”[pt]))AND( ( “sensor”[TI] OR “sensors”[TI] OR “sensing”[TI] ) AND ( “vital sign”[TI] OR “vital signs”[TI] OR “biological signal”[TI] OR “biological signals”[TI] OR “biological parameter”[TI] OR “biological parameters”[TI] OR “physiological parameter”[TI] OR “physiological parameters”[TI] OR “physiological signal”[TI] OR “physiological signals”[TI] OR “blood pressure”[TI] OR “temperature”[TI] OR “heart rate”[TI] OR “heartbeat”[TI] OR “heartbeats”[TI] OR “pulse rate”[TI] OR “respiration rate”[TI] OR “respiratory rate”[TI] OR “breathing rate”[TI] OR “ECG”[TI] OR “electrocardiography”[TI] OR “electrocardiogram”[TI] OR “menstrual cycle”[TI] OR “oxygen”[TI] OR “oximetry”[TI] OR “glucose”[TI] OR “end-tidal”[TI] OR “emg”[TI] OR “electromyography”[TI] OR “electromyogram”[TI] OR “ppg”[TI] OR “photoplethysmography”[TI] OR “photoplethysmogram”[TI] OR “pcg”[TI] OR “phonocardiography”[TI] OR “phonocardiogram”[TI] OR “bcg”[TI] OR “ballistocardiography”[TI] OR “ballistocardiogram”[TI] OR “scg”[TI] OR “seismocardiography”[TI] OR “seismocardiogram”[TI] OR “eog”[TI] OR “electrooculography”[TI] OR “electrooculogram”[TI] OR “eda”[TI] OR “electrodermal activity”[TI] OR “GSR”[TI] OR “Galvanic skin response” [TI] OR “eeg”[TI] OR “electroencephalogram”[TI] OR “bci”[TI] OR “brain computer interface”[TI] ) NOT ( “review”[TI] OR “survey”[TI] ))
2.1.2 Scopus Query
TITLE((“sensor” OR “sensors” OR “sensing”) AND (“vital sign” OR “vital signs” OR “biological signal” OR “biological signals” OR “biological parameter” OR “biological parameters” OR “physiological parameter” OR “physiological parameters” OR “physiological signal” OR “physiological signals” OR “blood pressure” OR “temperature” OR “heart rate” OR “heartbeat” OR “heartbeats” OR “pulse rate” OR “respiration rate” OR “respiratory rate” OR “breathing rate” OR “ECG” OR “electrocardiography” OR “electrocardiogram” OR “menstrual cycle” OR “oxygen” OR “oximetry” OR “glucose” OR “end-tidal” OR “emg” OR “electromyography” OR “electromyogram” OR “ppg” OR “photoplethysmography” OR “photoplethysmogram” OR “pcg” OR “phonocardiography” OR “phonocardiogram” OR “bcg” OR “ballistocardiography” OR “ballistocardiogram” OR “scg” OR “seismocardiography” OR “seismocardiogram” OR “eog” OR “electrooculography” OR “electrooculogram” OR “eda” OR “electrodermal activity” OR “GSR” OR “Galvanic skin response” OR “eeg” OR “electroencephalogram” OR “bci” OR “brain computer interface” ))AND PUBDATETXT( “January 2020” OR “February 2020” OR “March 2020” OR “April 2020” OR “May 2020” OR “June 2020” OR “July 2020” OR “August 2020” OR “September 2020” OR “October 2020” OR “November 2020” OR “December 2020”) AND LANGUAGE(english) AND SUBJAREA(MEDI) AND SRCTYPE(j) AND DOCTYPE(ar) AND DOCTYPE(re)
2.2 Signals
2.2.1 PubMed Query
((“2020/01/01”[DP] : “2020/12/31”[DP]) AND Journal Article [pt] AND English[lang] AND hasabstract[text] NOT Bibliography[pt] NOT Comment[pt] NOT Editorial[pt] NOT Letter[pt] NOT News[pt] NOT Review[pt] NOT Case Reports[pt] NOT Published Erratum[pt] NOT Historical Article[pt] NOT legislation[pt] NOT “clinical trial”[pt] NOT “evaluation studies”[pt] NOT “technical report”[pt] NOT “Scientific Integrity Review”[pt] NOT “Systematic Review”[pt] NOT “Retracted Publication”[pt] ) AND (( “biosignal”[TI] OR “biomedical signal”[TI] OR “physiological signal”[TI] OR “ecg”[TI] OR “electrocardiography”[TI] OR “electrocardiogram”[TI] OR “emg”[TI] OR “electromyography”[TI] OR “electromyogram”[TI] OR “ppg”[TI] OR “photoplethysmography”[TI] OR “photoplethysmogram”[TI] OR “pcg”[TI] OR “phonocardiography”[TI] OR “phonocardiogram”[TI] OR “bcg”[TI] OR “ballistocardiography”[TI] OR “ballistocardiogram”[TI] OR “scg”[TI] OR “seismocardiography”[TI] OR “seismocardiogram”[TI] OR “eog”[TI] OR “electrooculography”[TI] OR “electrooculogram”[TI] OR “eda”[TI] OR “electrodermal activity”[TI] OR “Respiration”[TI] OR “Blood Pressure”[TI] OR “eeg”[TI] OR “electroencephalogram”[TI] OR “bci”[TI] OR “brain computer interface”[TI] ) AND ( “processing”[TI] OR “analytics”[TI] OR “analysis”[TI] OR “analyse”[TI] OR “analyze”[TI] OR “analysing”[TI] OR “analyzing”[TI] OR “enhancement”[TI] OR “enhancements”[TI] OR “segmentation”[TI] OR “feature extraction”[TI] OR “feature selection”[TI] OR “classification”[TI] OR “clustering”[TI] OR “measurement”[TI] OR “quantification”[TI] OR “registration”[TI] OR “recognition”[TI] OR “reconstruction”[TI] OR “interpretation”[TI] OR “retrieval”[TI] “augmentation”[TI] OR “data mining”[TI] OR “computer-assisted”[TI] OR “computer-aided”[TI] OR “artificial intelligence”[TI] OR “machine learning”[TI] OR “deep learning”[TI] OR “neural network”[TI] OR “computer vision”[TI] OR “autoencoder”[TI] OR “auto-encoder”[TI] OR “Botzmann”[TI] OR “U-net”[TI] OR “support vector machine”[TI] OR “SVM”[TI] OR “random forest”[TI] ))
2.2.2 Scopus Query
TITLE(( “signal” OR “biosignal” OR “biomedical signal” OR “physiological signal” OR “ecg” OR “electrocardiography” OR “electrocardiogram” OR “emg” OR “electromyography” OR “electromyogram” OR “ppg” OR “photoplethysmography” OR “photoplethysmogram” OR “pcg” OR “phonocardiography” OR “phonocardiogram” OR “bcg” OR “ballistocardiography” OR “ballistocardiogram” OR “scg” OR “seismocardiography” OR “seismocardiogram” OR “eog” OR “electrooculography” OR “electrooculogram” OR “eda” OR “electrodermal activity” OR “Respiration” OR “Blood Pressure” OR “eeg” OR “electroencephalogram” OR “bci” OR “brain computer interface” ) AND ( “processing” OR “analytics” OR “analysis” OR “analyse” OR “analyze” OR “analysing” OR “analyzing” OR “enhancement” OR “enhancements” OR “segmentation” OR “feature extraction” OR “feature selection” OR “classification” OR “clustering” OR “measurement” OR “quantification” OR “registration” OR “recognition” OR “reconstruction” OR “interpretation” OR “retrieval” “augmentation” OR “data mining” OR “computer-assisted” OR “computer-aided” OR “artificial intelligence” OR “machine learning” OR “deep learning” OR “neural network” OR “computer vision” OR “autoencoder” OR “auto-encoder” OR “Botzmann” OR “U-net” OR “support vector machine” OR “SVM” OR “random forest” ) ) AND PUBDATETXT( “January 2020” OR “February 2020” OR “March 2020” OR “April 2020” OR “May 2020” OR “June 2020” OR “July 2020” OR “August 2020” OR “September 2020” OR “October 2020” OR “November 2020” OR “December 2020” ) AND LANGUAGE(english) AND SUBJAREA(MEDI) AND SRCTYPE(j) AND DOCTYPE(ar) AND NOT DOCTYPE(re)
2.3 Imaging Informatics
2.3.1 PubMed Query
((“2020/01/01”[DP] : “2020/12/31”[DP]) AND Journal Article [pt] AND English[lang] AND hasabstract[text] NOT Bibliography[pt] NOT Comment[pt] NOT Editorial[pt] NOT Letter[pt] NOT News[pt] NOT Review[pt] NOT Case Reports[pt] NOT Published Erratum[pt] NOT Historical Article[pt] NOT legislation[pt] NOT “clinical trial”[pt] NOT “evaluation studies”[pt] NOT “technical report”[pt] NOT “Scientific Integrity Review”[pt] NOT “Systematic Review”[pt] NOT “Retracted Publication”[pt]) AND(( “image”[TI] OR “imaging”[TI] OR “video”[TI] OR “X-ray”[TI] OR “X ray”[TI] OR “radiography”[TI] OR “orthopantomography”[TI] OR “fluoroscopy”[TI] OR “angiography”[TI] OR “tomography”[TI] OR “CT”[TI] OR “magnetic resonance”[TI] OR “MRI”[TI] OR “echocardiography”[TI] OR “sonography”[TI] OR “ultrasound”[TI] OR “endoscopy”[TI] OR “arthroscopy”[TI] OR “bronchoscopy”[TI] OR “colonoscopy”[TI] OR “cystoscopy”[TI] OR “laparoscopy”[TI] OR “nephroscopy”[TI] OR “laryngoscopy” [TI] OR “funduscopy”[TI] OR “thermography”[TI] OR “photography”[TI] OR “arthroscopy”[TI] OR “microscopy”[TI] ) AND ( “processing”[TI] OR “analytics”[TI] OR “analysis”[TI] OR “analyse”[TI] OR “analyze”[TI] OR “analysing”[TI] OR “analyzing”[TI] OR “enhancement”[TI] OR “enhancements”[TI] OR “segmentation”[TI] OR “feature extraction”[TI] OR “feature selection”[TI] OR “classification”[TI] OR “clustering”[TI] OR “measurement”[TI] OR “quantification”[TI] OR “registration”[TI] OR “recognition”[TI] OR “reconstruction”[TI] OR “interpretation”[TI] OR “retrieval”[TI] “augmentation”[TI] OR “data mining”[TI] OR “computer-assisted”[TI] OR “computer-aided”[TI] OR “artificial intelligence”[TI] OR “machine learning”[TI] OR “deep learning”[TI] OR “neural network”[TI] OR “computer vision”[TI] OR “autoencoder”[TI] OR “auto-encoder”[TI] OR “Botzmann”[TI] OR “U-net”[TI] OR “support vector machine”[TI] OR “SVM”[TI] OR “r ANDom forest”[TI] )) AND (“medical informatics”[MH])
2.3.2 Scopus Query
TITLE( ( “image” OR “imaging” OR “video” OR “X-ray” OR “X ray” OR “radiography” OR “orthopantomography” OR “fluoroscopy” OR “angiography” OR “tomography” OR “CT” OR “magnetic resonance” OR “MRI” OR “echocardiography” OR “sonography” OR “ultrasound” OR “endoscopy” OR “arthroscopy” OR “bronchoscopy” OR “colonoscopy” OR “cystoscopy” OR “laparoscopy” OR “nephroscopy” OR “laryngoscopy” OR “funduscopy” OR “thermography” OR “photography” OR “arthroscopy” OR “microscopy” ) AND ( “processing” OR “analytics” OR “analysis” OR “analyse” OR “analyze” OR “analysing” OR “analyzing” OR “enhancement” OR “enhancements” OR “segmentation” OR “feature extraction” OR “feature selection” OR “classification” OR “clustering” OR “measurement” OR “quantification” OR “registration” OR “recognition” OR “reconstruction” OR “interpretation” OR “retrieval” “augmentation” OR “data mining” OR “computer-assisted” OR “computer-aided” OR “artificial intelligence” OR “machine learning” OR “deep learning” OR “neural network” OR “computer vision” OR “autoencoder” OR “auto-encoder” OR “Botzmann” OR “U-net” OR “support vector machine” OR “SVM” OR “random forest” ) ) AND PUBDATETXT( “January 2020” OR “February 2020” OR “March 2020” OR “April 2020” OR “May 2020” OR “June 2020” OR “July 2020” OR “August 2020” OR “September 2020” OR “October 2020” OR “November 2020” OR “December 2020” ) AND LANGUAGE(english) AND SUBJAREA(MEDI) AND SRCTYPE(j) AND DOCTYPE(ar) AND NOT DOCTYPE(re)
References
- 1.Picard R, Wolf G.Sensor Informatics and Quantified Self IEEE J Biomed Health Inform2015 Sep;19051531. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Warnecke J M, Haghi M, Deserno T M. Unobtrusive health monitoring in private spaces: the smart vehicle. Sensors. 2020;20(09):2442. doi: 10.3390/s20092442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Spicher N, Warnecke J M, Haghi M, Schwartze J, Deserno T M. Unobtrusive health monitoring in private spaces: the smart home. Sensors. 2021;21(03):864. doi: 10.3390/s21030864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witte H, Ungureanu M, Ligges C, Hemmelmann D, Wüstenberg T, Reichenbach J. Signal informatics as an advanced integrative concept in the framework of medical informatics: new trends demonstrated by examples derived from neuroscience. Methods Inf Med. 2009;48(01):18–28. [PubMed] [Google Scholar]
- 5.Cook T S.The Importance of Imaging Informatics and Informaticists in the Implementation of AI Acad Radiol 2020. Jan;2701113–6. [DOI] [PubMed] [Google Scholar]
- 6.Choudhary A, Tong L, Zhu Y, Wang M D.Advancing Medical Imaging Informatics by Deep Learning-Based Domain Adaptation Yearb Med Inform 2020. Aug;2901129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panayides A S, Amini A, Filipovic N D, Sharma A, Tsaftaris S A, Young A.AI in Medical Imaging Informatics: Current Challenges and Future Directions IEEE J Biomed Health Inform 2020. Jul;24071837–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branstetter B F, 4th, Bartholmai B J, Channin D S.Reviews in radiology informatics: establishing a core informatics curriculum J Digit Imaging 2004. Dec;1704244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makeeva V, Vey B, Cook T S, Nagy P, Filice R W, Wang K C.Imaging Informatics Fellowship Curriculum: a Survey to Identify Core Topics and Potential Inter-Program Areas of Collaboration J Digit Imaging 2020. Apr;3302547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Ooijen P MA, Nagaraj Y, Olthof A.Medical imaging informatics, more than ‘just’ deep learning Eur Radiol 2020. Oct;30105507–9. [DOI] [PubMed] [Google Scholar]
- 11.Hsu W, Baumgartner C, Deserno T M. Notable papers and trends from 2019 in sensors, signals, and imaging informatics. Yearb Med Inform. 2020:139–44. doi: 10.1055/s-0040-1702004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu W, Baumgartner C, Deserno T M. Advancing artificial intelligence in sensors, signals, and imaging informatics. Yearb Med Inform. 2019;28(01):122–26. doi: 10.1055/s-0039-1677943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu W, Deserno T M, Kahn C E., Jr. Sensor, signal, and imaging informatics in 2017. Yearb Med Inform. 2018;27(01):110–13. doi: 10.1055/s-0038-1667084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu Y-C, Li Y-H, Chang C-C, Harfiya L N. Generalized Deep Neural Network Model for Cuffless Blood Pressure Estimation with Photoplethysmogram Signal Only. Sensors. 2020;20(19):5668. doi: 10.3390/s20195668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao F, Wen B, Hu Z, Fortino G, Wang X-P, Liu Z-D. Continuous blood pressure measurement from one-channel electrocardiogram signal using deep-learning techniques. Artif Intell Med. 2020;108:101919. doi: 10.1016/j.artmed.2020.101919. [DOI] [PubMed] [Google Scholar]
- 16.Porumb M, Stranges S, Pescapè A, Pecchia L. Precision Medicine and Artificial Intelligence: A Pilot Study on Deep Learning for Hypoglycemic Events Detection based on ECG. Sci Rep. 2020;10(01):170. doi: 10.1038/s41598-019-56927-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding X, Clifton D, Ji N, Lovell N H, Bonato P, Chen W. Wearable Sensing and Telehealth Technology with Potential Applications in the Coronavirus Pandemic. IEEE Rev Biomed Eng. 2021;14:48–70. doi: 10.1109/RBME.2020.2992838. [DOI] [PubMed] [Google Scholar]
- 18.Johnson A E, Pollard T J, Shen L, Li-Wei H L, Feng M, Ghassemi M. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(01):1–9. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Littlejohns T J, Holliday J, Gibson L M, Garratt S, Oesingmann N, Alfaro-Almagro F. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nat Commun. 2020;11(01):2624. doi: 10.1038/s41467-020-15948-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haendel M A, Chute C G, Bennett T D, Eichmann D A, Guinney J, Kibbe W A. The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2020;28(03):427–43. doi: 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai E B, Simpson S, Lungren M P, Hershman M, Roshkovan L, Colak E. The RSNA International COVID-19 Open Radiology Database (RICORD) Radiology. 2021;299(01):E204–E13. doi: 10.1148/radiol.2021203957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi G, Wang J, Qiang Y, Yang X, Zhao J, Hao R. Knowledge-guided synthetic medical image adversarial augmentation for ultrasonography thyroid nodule classification. Comput Methods Programs Biomed. 2020;196:105611. doi: 10.1016/j.cmpb.2020.105611. [DOI] [PubMed] [Google Scholar]
- 23.Fu B, Wang G, Wu M, Li W, Zheng Y, Chu Z. Influence of CT effective dose and convolution kernel on the detection of pulmonary nodules in different artificial intelligence software systems: A phantom study. Eur J Radiol. 2020;126:108928. doi: 10.1016/j.ejrad.2020.108928. [DOI] [PubMed] [Google Scholar]
- 24.Wynants L, Van Calster B, Collins G S, Riley R D, Heinze G, Schuit E. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J-Y, Shen H, Qu Q, Sun W, Kong X-Q.The application of deep learning in electrocardiogram: Where we came from and where we should go?Int J Cardiol 2021 May 14;S0167-5273(21)00829-9 [DOI] [PubMed]
- 26.Gemein LAW, Schirrmeister R T, Chrabąszcz P, Wilson D, Boedecker J, Schulze-Bonhage A.Machine-learning-based diagnostics of EEG pathology Neuroimage 2020. Oct 15;220117021. [DOI] [PubMed] [Google Scholar]
- 27.Karimi D, Dou H, Warfield S K, Gholipour A.Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis Med Image Anal 2020. Oct;65101759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langner T, Strand R, Ahlström H, Kullberg J.Large-scale biometry with interpretable neural network regression on UK Biobank body MRI Sci Rep 2020. Oct 20;100117752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito H, Aoki T, Aoyama K, Kato Y, Tsuboi A, Yamada A.Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network Gastrointest Endosc 2020. Jul;9201144–1510. [DOI] [PubMed] [Google Scholar]


