Summary
Objective : The current observational research literature shows extensive publication bias and contradiction. The Observational Health Data Sciences and Informatics (OHDSI) initiative seeks to improve research reproducibility through open science.
Methods : OHDSI has created an international federated data source of electronic health records and administrative claims that covers nearly 10% of the world’s population. Using a common data model with a practical schema and extensive vocabulary mappings, data from around the world follow the identical format. OHDSI’s research methods emphasize reproducibility, with a large-scale approach to addressing confounding using propensity score adjustment with extensive diagnostics; negative and positive control hypotheses to test for residual systematic error; a variety of data sources to assess consistency and generalizability; a completely open approach including protocol, software, models, parameters, and raw results so that studies can be externally verified; and the study of many hypotheses in parallel so that the operating characteristics of the methods can be assessed.
Results : OHDSI has already produced findings in areas like hypertension treatment that are being incorporated into practice, and it has produced rigorous studies of COVID-19 that have aided government agencies in their treatment decisions, that have characterized the disease extensively, that have estimated the comparative effects of treatments, and that the predict likelihood of advancing to serious complications.
Conclusions : OHDSI practices open science and incorporates a series of methods to address reproducibility. It has produced important results in several areas, including hypertension therapy and COVID-19 research.
Keywords: Observational research, reproducibility
1 Introduction
The Observational Health Data Sciences and Informatics (OHDSI) initiative [ 1 ] is a multi-stakeholder, interdisciplinary, international collaborative whose mission is to improve health by empowering a community to collaboratively generate the evidence that promotes better health decisions and better care. With a coordinating center at Columbia University, OHDSI has over 300 registered, voluntary collaborators from 30 countries and six continents and over 3,000 participants on its open fora. Participants include experts in informatics, statistics, epidemiology, and clinical sciences with roles in academia, industry, and government. Its federated database holds records on about 600 million unique patients in over 100 databases. At the current rate, OHDSI will soon be at 10% of the world population.
The need for such an initiative comes from several sources. Only a tiny fraction of all possible questions that a clinician could ask about the benefits and risks of drugs and other interventions have been answered in the literature or other knowledge bases. The need for large populations comes from the low rate of important outcomes. Given a specific indication, treatment, population subgroup, and side effect, a database with millions of persons may have only a handful of events.
Furthermore, the literature is replete with contradictions. For example, two groups studied the association of oral bisphosphonates with esophageal cancer using the same observational database and published in two different top journals a month apart; they came to different conclusions on whether or not there was an effect [ 2 , 3 ]. OHDSI took a deep dive on the observational study literature, parsing almost 30,000 observational research results [ 4 ]. The exploration found that 85% of exposure-outcome pairs were positive at standard levels of statistical significance, pointing to severe publication bias that was not explainable even if researchers were perfect at predicting which hypotheses would be positive; the drop at p=0.05 was too steep (see Figure 1 , and see the original publication for methods details [ 4 ]). OHDSI also looked at the over-optimism of p-values and confidence intervals [ 5 ]. Replicating four published studies and using 50 negative control hypotheses, this examination found that “95% confidence intervals” generated by using the studies’ methods covered only 30%, 47%, 60%, and 88% of true values. The consequence of both over-calling positive studies and hiding negative studies is conflicting, unreliable evidence. The publication process effectively becomes a data-dredging machine, resulting in the literature representing a biased evidence base with no comment on most hypotheses and the wrong answer on many others. Evidence becomes subject to comedian Woody Allen’s famous quip, “Boy, the food at this place is really terrible”; “Yeah, I know; and such small portions.” Such limitations lead editors to force authors to sign each paper with the caveat that, “since it is only observational research, the evidence cannot be used for causal assessment.”
Fig. 1.
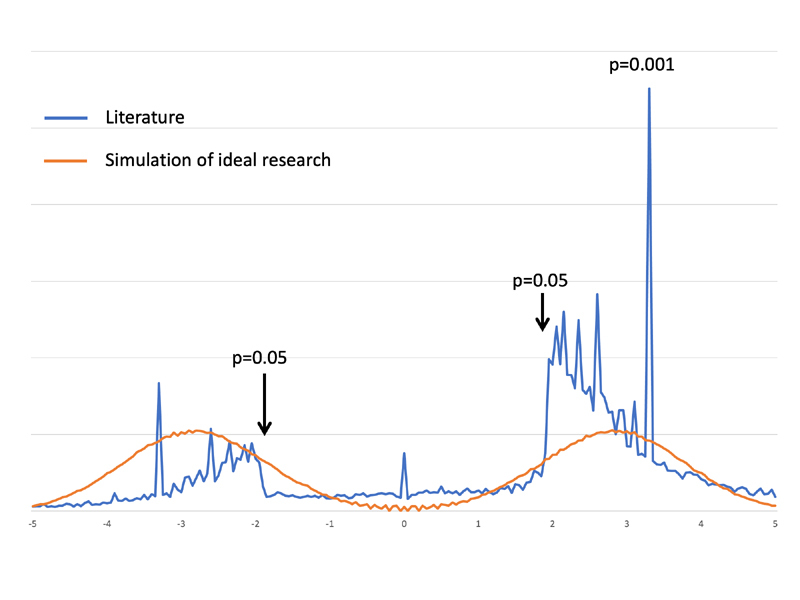
Publication bias in the observational research literature. Based on extracting the statistical significance of 29,982 observational research results from the literature [ 4 ], we plotted (blue line) the relative number of results (y-axis) with a given two-tailed z-score (x-axis) for each study, with a negative z-score indicating that the outcome occurred less often in the primary intervention. The p-values for several z-scores are marked. Note the sharp drop in results for z-scores corresponding to p-values over 0.05. Part of the explanation for few non-significant studies may be that researchers are good at guessing which hypotheses will come out significant. We therefore also plotted (red line) the z-score distribution that one would get if only true hypotheses were studied. Even perfect knowledge cannot duplicate the sharp drop at p=0.05. Publication bias is the only reasonable explanation.
OHDSI seeks to improve the current state with an open-science effort in observational research. All aspects of every study other than access to patient-level data are open to the public. Software is open source, workflows are standardized and transparent, and analytic parameters are derived systematically and are published. Every aspect of a study is made available.
2 Observational Research Infrastructure
OHDSI achieves its large data source by using a federated (distributed) data model, in which each participating organization converts its own data to the OHDSI Observational Medical Outcomes Partnership (OMOP) Common Data Model [ 6 ]. Research questions are translated into analytic code and are distributed over GitHub and run locally, and the aggregate summary results’not patient-level data’are collated centrally (see Figure 2 ). Researchers collaborate to interpret and publish the findings. The OMOP Common Data Model (CDM) is a deep information model, laid out to optimize analysis of extremely large databases, organized with a relatively flat structure such that a novice researcher can comprehend the model quickly, and retaining a flexible table to accommodate new data that are not yet explicitly modeled. The OMOP CDM is maintained by its own workgroup whose deliberations are open to the public. OHDSI maintains a comprehensive vocabulary that includes over 150 source vocabularies from around the world with over nine million concepts that are mapped to a small set of standard vocabularies with which data are stored in the databases. Some examples include the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) for conditions (diagnoses), Logical Observation Identifiers Names & Codes (LOINC) for laboratory tests, and RxNorm for prescription drugs.
Fig. 2.
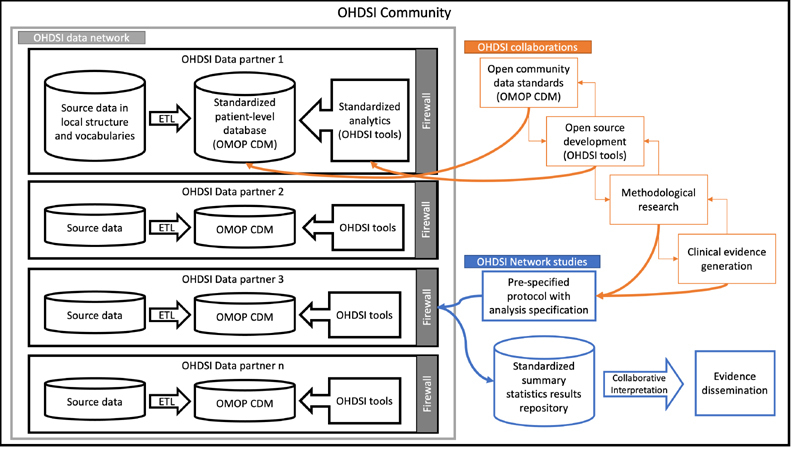
OHDSI research flow. Patient-level data remain private to the OHDSI data partners, but all other OHDSI work is shared publicly. (ETL is extract-transfer-load.)
Agreeing on a database schema and a vocabulary are rarely sufficient to get different sites to actually encode data the same way. There are usually differences in how the data are interpreted and coded. For example, nested results like bacterial culture antibiotic sensitivity can be stored in several ways, and there is more than one way to indicate that a diagnosis is a cause of death. Therefore, OHDSI launched a collaborative effort to develop conventions for how to encode data in the model. The community now has hundreds of specific conventions that are effectively recipes for specific data and contexts. OHDSI also supplies extensive tools to facilitate data conversion. WhiteRabbit profiles the source data to determine where source data fit in the OMOP model. RabbitInAHat maps source structure to OMOP tables and fields, and Usagi maps source codes to OMOP vocabulary. ATHENA provides the vocabularies themselves, including handling licensing issues. ACHILLES profiles the OMOP data to review the progress of the conversion, and the Data Quality Dashboard provides explicit data quality assessments. All of this is supported by the OHDSI online fora for CDM implementers and developers. Some health care data begin as natural language. The OHDSI community has applied natural language processing to translate text into OMOP tables and fields [ 7 – 10 ], but much work remains.
Once the data are in the OMOP CDM, extensive tools facilitate analysis [ 11 ]. ATLAS [ 12 ] provides a graphical user interface to build, visualize, and analyze cohorts. This usually begins with phenotyping, where the user selects the appropriate concepts from the vocabulary to create a concept set that represents the phenotype, which could, for example, represent the treatment or outcome being studied. The user then applies logic and timing to the concept sets to create a cohort, which defines the list of patients that match the criteria over some time span. The cohorts may then be examined patient by patient, summarized and visualized, or used in statistical analyses. Tools to diagnose potential errors in the cohort are provided.
To handle the very large sample sizes (over 100,000,000) with very large numbers of covariates (over 50,000), OHDSI has developed a library for large-scale statistical analysis called CYCLOPS [ 13 ]. It exploits the sparse nature of clinical data and allows inference on data sets with hundreds of millions of patients and hundreds of thousands of columns (variables). It uses cyclic coordinate descent for logistic regression, Poisson regression, and survival analysis. Like all OHDSI tools, it is open source.
Using these tools, OHDSI generates evidence in three forms. (1) Clinical characterization tallies cases to provide proportions and rates with few statistical assumptions. It answers the question, “how often does something happen?”, and it is used in studies such as natural history and quality improvement. An example clinical application of characterization could be answering the question could be “what proportion of diabetes patients start treatment on metformin, as recommended by clinical guidelines?” (2) Population-level estimation identifies associations between exposures and outcomes and assesses causal effects using statistical methods to address bias. This includes comparative effectiveness and safety surveillance. For example, a clinical question for population-level estimation would be, “does metformin cause lactic acidosis more than glyburide?” (3) Patient-level prediction applies machine learning algorithms to classify a target population into those who will or will not experience some future outcome, based on the patient’s baseline characteristics. Patient-level prediction can be used to address clinical questions like “amongst patients initiating metformin, which are at highest risk of developing lactic acidosis?”, and “if my patient takes metformin, what is her probability of a lactic acidosis event?”
3 Generating Evidence
A concrete example of a characterization study is OHDSI’s first network study, on treatment pathways in three common chronic diseases, type 2 diabetes mellitus, hypertension, and depression [ 14 ]. Motivated by a recognition that we do not actually know how often patients take different sequences of medications, OHDSI tallied medications for those diseases in 12 databases, which included about 240 million patient records from five countries. The results showed that metformin was the predominant first-line diabetes drug with about a 75% prevalence in all databases except the one from Japan. Subsequent discussions with a Japanese diabetes physiologist indicated that Japanese doctors prescribe less metformin because they generally believe their patients are less subject to insulin resistance. Hypertension treatment revealed a moderate amount of agreement, with the primary drugs being hydrochlorothiazide and lisinopril, and depression showed the least agreement, with marked differences in drugs even among databases in the same country. The study illustrated the power of a common data model because most of the analyses were completed within three weeks of the conception of the study; this included design, engaging volunteer sites, running the analyses, and transmitting the results centrally.
Asserting causality with population-level estimation is much more difficult due to confounding and other biases. OHDSI takes a multi-pronged approach, recently enumerated as ten principles [ 15 , 16 ], under an initiative called LEGEND. Confounding in comparative research is addressed using propensity score adjustment [ 17 ] like many studies, but it differs in using a systematic approach that is not dependent upon the authors’ knowledge and biases. So-called large-scale propensity score (LSPS) adjustment [ 18 , 19 ] adjusts for all available covariates in the claims or electronic health record database, generally in the tens of thousands. This contrasts with other techniques that attempt to select only the confounders, either through authors’ knowledge or empirical selection. OHDSI employs L1 regularized regression (LASSO) [ 20 ] to handle the challenge that the number of variables usually exceeds the number of subjects available. The technique provides diagnostics to ensure that the adjustment is effective. The treatment and control groups are checked for equipoise (i.e., the degree to which two groups have patients with similar baseline characteristics) using a preference score plot [ 21 ]; if the two groups have poor equipoise (i.e., just a minority of patients in each group share similar baseline characteristics), then the results may not be as generalizable because the analysis will focus on the small subset who overlap. The tens of thousands of covariates are plotted on a balance graph, showing the standardized difference of the mean before and after propensity score adjustment. The generally recognized threshold for sufficient balance is a standardized mean difference less than 0.1 [ 22 , 23 ]. OHDSI found in a large study of half a million hypotheses that 75% of the time, every single one of the tens of thousands of variables achieved balance after matching on the large-scale propensity score [ 16 ]. Furthermore, OHDSI has found that on adjusting for tens of thousands of covariates, important unobserved factors may also become balanced. For example, in studying hypertension therapy, adjusting for many other covariates also adjusted for an important potential confounder, baseline blood pressure, when it was held out of the analysis of the one database that captured it [ 16 , 24 ].
OHDSI checks for residual systematic error that LSPS may have failed to address using negative and synthetic positive control hypotheses [ 5 , 25 ]. The use of negative controls to verify a result is becoming more common, but usually only one to three hypotheses are included. OHDSI uses a large sample (>50) negative controls chosen using semi-automated methods [ 26 ] and a set of synthetic positive controls to create a distribution of estimates with known true values (e.g., hazard ratio of exactly 1 for negative controls). For a 95% confidence interval, 95% of true values should lie inside the calculated confidence intervals. If there is residual systematic error, fewer of the true values will lie inside the confidence intervals. OHDSI diagnoses the problem and also provides an adjustment: the confidence intervals are recalibrated through widening and shifting to achieve actual 95% coverage. This ensures a proper false positive rate corresponding to the selected alpha level (e.g., 5%), with the limitation that it results in fewer true positive results being declared positive.
Credibility and generalizability are both enhanced by using a heterogeneous network that differs in geographic location, practice type, data collection mechanism. In OHDSI’s federated network, analyses are done locally, with no patient-level data pooling across sites, and the results from each site are compared looking for consistency. If the results are not consistent, then that could point to missed biases or important differences among the subpopulations. If the results are consistent, then a combined, meta-analytic summary is estimated.
OHDSI’s openness is an important factor in causal credibility. The study design is released publicly before any analysis is done to ensure that authors cannot steer the results. The code is made freely available on GitHub and all software parameters are published so that other researchers can verify what analysis was done and replicate the study. The results of the study are immediately made available on the Internet at data.OHDSI.org so that other researchers can verify that extreme results were not inappropriately selected for publication.
Using these techniques, OHDSI answers important questions. For example, when the US Food and Drug Administration published a query about whether levetiracetam causes angioedema, OHDSI quickly launched a network study that showed that in fact, the drug is not associated with angioedema, but the comparator, phenytoin, may be responsible for a low rate of the complication [ 27 ].
More recently, OHDSI has moved to carrying out studies at scale with many simultaneous hypotheses. This allows researchers to check the operating characteristics of the analytic pipeline. For example, most drugs do not cause most outcomes. If an analysis produces a large proportion of statistically significant results, then the analytic methods need to be checked. In addition, consideration of a large number of hypotheses allows the medical evidence gap to be filled more efficiently. OHDSI’s first large-scale analysis looked at the side effects of depression medications [ 4 ]. In contrast to the literature’s 85% statistically significant study rate noted above, OHDSI had a statistical significance rate of 11%, with 5% likely due to false positive hypotheses based on the 95% confidence intervals and 6% being true positives. In that study, every one of almost 20 thousand hypotheses underwent full diagnostics to verify whether the result should be trusted. In some cases, hypotheses failed to pass the diagnostics, such as comparing medication treatment for depression to electroconvulsive therapy, implying the treatment groups were too different for propensity scoring to achieve balance.
This highlights a strength of OHDSI: it does not purport to be able to carry out every study, but it is careful to diagnose when its results are likely to be credible or not. Running many hypotheses at once is not an example of data dredging, as long as all the results are revealed to the reader. The literature, with its proven publication bias [ 4 ], hiding most of the studies that are actually performed, is indeed data dredging. Comparing OHDSI’s large-scale systematic approach for confounding adjustment to a more traditional one-study-at-a-time approach, one can ask if it would be better to optimize each study, hand-selecting covariates to adjust for separately and manually for each hypothesis. First, we note that human designers rarely get a list of 10 to 20 variables exactly right (e.g., given the differences in what variables are selected by different authors for seemingly identical studies), and second, it is not possible to assess the operating characteristics of a single study. The single study must be taken on faith that the designers have done a good job, whereas with multiple studies we can review the overall rate of statistical significance, the overall rate of passing diagnostics, etc.
OHDSI next applied these LEGEND methods to a study of hypertension treatment [ 28 ]. The 2017 US hypertension treatment guideline [ 29 ] identifies 58 first- and second-line antihypertensive medications based on the results of 40 randomized trials, observational evidence, and expert opinion. Only about 11% of those recommendations were based on randomized trials and most were based on expert opinion (e.g., assuming class effects). OHDSI sought to fill in the evidence gap with state-of-the-art observational research. With 58 ingredients from 15 drug classes, implying 1,653 possible combination therapies, and 58 outcomes of interest in both effectiveness and safety, OHDSI’s network had data to carry out 587,020 comparisons of the 164,908,500 possibilities (i.e., most possible combinations are not actually feasible). Each of those comparisons is a fully executed study with all diagnostics, including equipoise, balance, Kaplan-Meier curves, etc. Compared to the original 40 randomized trials, OHDSI provided 10,278 comparisons between drug regimens.
The study [ 28 ] first of all largely verified the guideline, with most of the first-line medication classes being indistinguishable on effectiveness and safety, and with beta-blocker classes, which are second-line classes, being inferior to the first-line classes. Also expected was that non-dihydropyridine drugs proved to be inferior. Unexpected was the superiority of thiazide and thiazide-like diuretics showing better effectiveness and safety than angiotensin-converting enzyme inhibitors. This is an important finding, as patients start on angiotensin-converting enzyme inhibitors 48% of the time. The switch to a diuretic could save 1.3 cardiovascular events per 1000 patients. Within-class comparisons were also revealing. For example, while the guideline favors the diuretic chlorthalidone over hydrochlorothiazide, LEGEND found [ 24 , 30 ] that patients started on chlorthalidone suffered significantly worse side effects and no detectable improved effectiveness compared to hydrochlorothiazide, and the result was incorporated into The Medical Letter [ 31 ].
Looking at the operating characteristics, OHDSI compared its results to pre-existing randomized trials [ 16 ]. We found that OHDSI and trials results overlapped in 28 out of 30 hypotheses, noting that a 5% disagreement rate is expected based on the definition of a 95% confidence interval. This included the new diuretic versus angiotensin-converting enzyme inhibitor finding, except that the randomized trial confidence interval was wider than the OHDSI result and overlapped a hazard ratio of one (i.e., was not significant). All the LEGEND results are publicly available on the OHDSI results Web site.
The OHDSI predictive modeling community has developed a suite of open-source tools that runs against the OMOP common data model and implements a vast array of machine learning methods. A 2018 article laid out the vision for OHDSI-scale global patient-level predictive modeling [ 32 ], and many clinical applications are underway [e.g., 33-36].
4 COVID-19
With the emergence of COVID-19, OHDSI swung into action to bring observational research to bear on COVID-19 treatment, starting with an 88-hour Study-a-Thon that was held virtually in place of the previously planned annual European symposium. COVID-19 was particularly challenging to study for several reasons. There were initially no coding standards for the disease and its laboratory tests, and once organizations developed a work-around for it, it was difficult to move to the proper codes even after they were disseminated. Many observational databases go through a conversion and quality assurance process that takes months; COVID-19 was therefore delayed in showing up in most databases. Health care providers were often overwhelmed with patients, and this led to reduced and inaccurate documentation. The timeline for severe cases of the disease moved rapidly so that the time of events needed to be known by minutes instead of days (e.g., did the drug come before intubation). Treatment recommendations for COVID-19 changed rapidly, especially in the early months of the disease, implying the cohort changed over time. OHDSI therefore partnered closely with the data providers to understand and address or account for the resultant difficulties especially at the data conversion and analytic stages.
OHDSI first characterized the disease, comparing it to previous annual influenza as well as H1N1 influenza [ 37 ]. The study showed that while COVID-19 affects older and sicker patients most severely, compared to influenza, it also affects younger, healthier patients. OHDSI found that angiotensin converting enzyme inhibitor drugs and angiotensin receptor blocking drugs, which were suspected of worsening COVID-19, did not in fact pose extra risk, so patients should not stop taking them [ 38 ]. A study of a large cohort of patients without COVID-19 taking hydroxychloroquine and azithromycin [ 39 ] showed that the combination increased risk of sudden death even in the short term, whereas hydroxychloroquine increased risk only with longer exposure. To estimate COVID-19 severity risk, OHDSI trained a predictive model on influenza and validated it on COVID-19 patients from five databases in three countries [ 36 ].
5 Discussion
Thus far, OHDSI has accomplished several things. We have a created a global federated database of electronic health records mapped to common data model that includes nearly 10% of the world’s population. We have assembled a community of hundreds of researchers around the world that have developed open-source tools to enable the data network. That community has embraced open science and has developed an extensive suite of analytic tools that enable the generation of clinical evidence from the data. The first major OHDSI clinical studies have appeared in major medical journals and represent by far the largest observational studies ever conducted. The COVID-19 crisis galvanized the community, and OHDSI has been at the forefront of generating useful evidence at this time. Large-scale adoption of the OMOP common data model has enabled all of these accomplishments.
OHDSI’s federated approach has a number of limitations. Consumers of our studies would need to collaborate with potentially very many data partners to truly reproduce a study. The data network includes some commercial databases that could prove prohibitively expensive to access. Researchers conducting OHDSI studies generally do not have direct access to patient-level data, except perhaps at a local site. This precludes certain model and data diagnostics or at the very least, requires close cooperation of the data owners to conduct some types of analyses. Our approach to causal inference, in particular, has a number of limitations inherent to the observational setting. Our use of positive and negative controls aims to quantify and account for sources of bias but some bias or departure from nominal uncertainty bounds can remain.
While we have a large sample, it is concentrated in developed nations, with a strong emphasis in the US, Europe, and parts of Asia. In a sense, the first 600 million patients were the easy 600 million, i.e., those for whom data were more readily available. We strive to add more representative populations, both in geography and in diversity within covered nations.
We believe the OHDSI collaboration has the potential to truly transform the practice of health care. However, many obstacles remain. The scientific community remains deeply skeptical about observational studies. The COVID crisis has exacerbated this problem because of the glut of hastily conducted observational studies that has entered the literature. The epidemiological community remains largely focused on one-off, handcrafted studies that rely on clinical knowledge to generate “the right answer.” As noted above, this approach does not lend itself to objective evaluation but nonetheless remains deeply embedded in analytical training programs all around the world. Much work remains to improve the paradigm that OHDSI is developing, but the advantages are stark: reproducible results that enable systematic evaluation.
Measuring the real clinical impact of OHDSI findings is difficult. Based on OHDSI’s mission statement, quoted in the Introduction, we seek to go beyond proofs of concept and publication to actually promoting better health decisions and better care. OHDSI is still in the early phase of generating evidence, but we can cite OHDSI’s hypertension evidence being incorporated into the Medical Letter [ 31 ], the European Medicines Agency explicitly citing the OHDSI study on hydroxychloroquine risk [ 40 ] in its deliberation about the drug being used for COVID-19 prophylaxis, and the European Medicines Agency highlighting OHDSI’s study of the (lack of) risk of angiotensin converting enzyme inhibitors and angiotensin receptor blockers in COVID-19, pointing out that OHDSI’s reproducible methods help to address recent doubts about the COVID-19 literature that have arisen from a lack of transparency and uncertainty of research standards in the research community [ 41 ]. True open science demands that we demonstrate that the downstream decisions and care are truly “better”; that is, we should use OHDSI methods and data to measure the downstream effect of our results (an excellent suggestion by a reviewer of this paper). It is difficult to tease apart the actual drivers of policy changes (e.g., COVID-19 decisions were based on multiple sources of evidence), but at least we can in the future determine if practice aligns with our recommendations.
Going forward, OHDSI plans to continually enrich the data network both with data for more patients and also with richer data on each patient that may include sensor data, image data, and genomic data. We will continue to grow the OHDSI researcher network with a long-term view to generating truly impactful clinical evidence to improve the practice of health care. A key focus for OHDSI going forward is to galvanize the methodological research community; the global data resource that OHDSI has assembled will grow and become richer in future years, but dramatic progress is needed in new analytic methods to harness the data.
6 Conclusion
OHDSI demonstrates that it is feasible to create an enormous international network on a voluntary basis with a federated data source that is nearing 10% of the world’s population. Sites have been able to participate despite wide differences in languages, national health care structures, and privacy regulations. OHDSI’s work is completely open other than not providing access to patient-level data, with all methods, tools, models, and results available freely, including a textbook called “The Book of OHDSI” that covers all aspects of its research [ 42 ]. Using novel methods for addressing bias, openness, large-scale analysis, and extensive diagnostics, OHDSI seeks to improve the credibility of observational research, and it has already produced findings that are being incorporated into practice.
Acknowledgments
This work was supported in part by National Institutes of Health grant R01 LM006910 and National Science Foundation grant IIS 1251151.
References
- 1.Hripcsak G, Duke J, Shah N, Reich C, Huser V, Schuemie M. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574–8. [PMC free article] [PubMed] [Google Scholar]
- 2.Cardwell C R, Abnet C C, Cantwell M M, Murray L J. Exposure to oral bisphosphonates and risk of esophageal cancer. JAMA. 2010;304:657–63. doi: 10.1001/jama.2010.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green J, Czanner G, Reeves G, Watson J, Wise L, Beral V. Oral bisphosphonates and risk of cancer of oesophagus, stomach, and colorectum: case-control analysis within a UK primary care cohort. BMJ. 2010;341:c4444. doi: 10.1136/bmj.c4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuemie M J, Ryan P B, Hripcsak G, Madigan D, Suchard M A.Improving reproducibility by using high-throughput observational studies with empirical calibration Philos Trans A Math Phys Eng Sci 2018376(2128):2.0170356E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuemie M J, Suchard M A, Hripcsak G, Ryan P B, Madigan D. Empirical confidence interval calibration for population-level effect estimation studies in observational healthcare data. Proc Natl Acad Sci U S A. 2018;115(11):2571–77. doi: 10.1073/pnas.1708282114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overhage J, Ryan P, Reich C, Hartzema A, Stang P. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19:54–60. doi: 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Anh H, Manion F, Rouhizadeh M, Zhang Y.COVID-19 SignSym–A fast adaptation of general clinical NLP tools to identify and normalize COVID-19 signs and symptoms to OMOP common data modelArXiv 2020;arXiv:2007.10286v3 [DOI] [PMC free article] [PubMed]
- 8.Liu S, Wang Y, Wen A, Wang L, Hong N, Shen F, Bedrick S, Hersh W, Liu H.CREATE: Cohort retrieval enhanced by analysis of text from electronic health records using OMOP common data modelArXiv 2019;arXiv:1901.07601
- 9.Meystre S M, Heider P M, Kim Y, Aruch D B, Britten C D. Automatic trial eligibility surveillance based on unstructured clinical data. Int J Med Inform. 2019;129:13–19. doi: 10.1016/j.ijmedinf.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma H, Mao C, Zhang Y, Vatani H, Yao L, Zhong Y, Rasmussen L, Jiang G, Pathak J, Luo Y.Developing a portable natural language processing based phenotyping system BMC Med Inform Decis Mak 201919(Suppl 3):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.OHDSI. Observational Health Data Sciences and Informatics GitHub Library. Available from: https://github.com/OHDSI/ [Accessed 2021 Feb 7]
- 12.ATLAS – A unified interface for the OHDSI tools. https://www.ohdsi.org/atlas-a-unified-interface-for-the-ohdsi-tools/ [Accessed 2020 Nov 18]
- 13.Suchard M, Simpson S, Zorych I, Ryan P, Madigan D. Massive parallelization of serial inference algorithms for a complex generalized linear model. Transactions on Modeling and Computer Simulation. 2013;23:10. doi: 10.1145/2414416.2414791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hripcsak G, Ryan P B, Duke J D, Shah N H, Park R W, Huser V, Suchard M A, Schuemie M J, DeFalco F J, Perotte A, Banda J M, Reich C G, Schilling L M, Matheny M E, Meeker D, Pratt N, Madigan D. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci U S A. 2016;113:7329–36. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuemie M J, Ryan P B, Pratt N, Chen R, You S C, Krumholz H M. Principles of large-scale evidence generation and evaluation across a network of databases (LEGEND) J Am Med Inform Assoc. 2020;27:1331–7. doi: 10.1093/jamia/ocaa103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuemie M J, Ryan P B, Pratt N, Chen R, You S C, Krumholz H M. Large-scale evidence generation and evaluation across a network of databases (LEGEND): Assessing validity using hypertension as a case study. J Am Med Inform Assoc. 2020;27:1268–77. doi: 10.1093/jamia/ocaa124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbaum P R, Rubin D B. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- 18.Tian Y, Schuemie M J, Suchard M A. Evaluating large-scale propensity score performance through real-world and synthetic data experiments. Int J Epidemiol. 2018;47:2005–14. doi: 10.1093/ije/dyy120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinstein R B, Ryan P, Berlin J A, Berlin J A, Matcho A, Schuemie M. Channeling in the use of nonprescription paracetamol and ibuprofen in an electronic medical records database: evidence and implications. Drug Saf. 2017;40(12):1279–92. doi: 10.1007/s40264-017-0581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B. 1996;58(01):267–88. [Google Scholar]
- 21.Walker A M, Patrick A R, Lauer M S, Hornbrook M C, Marin M G, Platt R. A tool for assessing the feasibility of comparative effectiveness research. Comparative Effectiveness Research. 2013;3:11–20. [Google Scholar]
- 22.Austin P C. Balance diagnostics for comparing the distribution of baseline covariates between treatment groups in propensity-score matched samples. Stat Med. 2009;28:3083–107. doi: 10.1002/sim.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham D J, Reichman M E, Wernecke M, Zhang R, RossSouthworth M, Levenson M. Cardiovascular, bleeding, and mortality risks in elderly Medicare patients treated with dabigatran or warfarin for nonvalvular atrial fibrillation. Circulation. 2015;131:157–64. doi: 10.1161/CIRCULATIONAHA.114.012061. [DOI] [PubMed] [Google Scholar]
- 24.Hripcsak G, Suchard M A, Shea S, Chen R, You S C, Pratt N. Real-world evidence on the effectiveness and safety of chlorthalidone and hydrochlorothiazide. JAMA Intern Med. 2020;180(04):542–51. doi: 10.1001/jamainternmed.2019.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuemie M J, Cepeda M S, Suchard M A, Yang J, Tian Y, Schuler A.How confident are we about observational findings in healthcare: a benchmark studyHarv Data Sci Rev 2020;2(01): 10.1162/99608f92.147cc28e [DOI] [PMC free article] [PubMed]
- 26.Voss E A, Boyce R D, Ryan P B, van der Lei J, Rijnbeek P R, Schuemie M J. Accuracy of an automated knowledge base for identifying drug adverse reactions. J Biomed Inform. 2017;66:72–81. doi: 10.1016/j.jbi.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duke J D, Ryan P B, Suchard M A, Hripcsak G, Jin P, Reich C. Risk of angioedema associated with levetiracetam compared with phenytoin: Findings of the observational health data sciences and informatics research network. Epilepsia. 2017;58:e101–e106. doi: 10.1111/epi.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suchard M A, Schuemie M J, Krumholz H M, You S C, Chen R, Pratt N. Comprehensive comparative effectiveness and safety of first-line antihypertensive drug classes. Lancet. 2019;394:1816–26. doi: 10.1016/S0140-6736(19)32317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whelton P K, Carey R M, Aronow W S, Casey Jr D E, Collins K J, DennisonHimmelfarb C. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension. 2018;71(06):1269–324. doi: 10.1161/HYP.0000000000000066. [DOI] [PubMed] [Google Scholar]
- 30.Hripcsak G, Shea S, Schuemie M J. Chlorthalidone and hydrochlorothiazide for treatment of patients with hypertension-reply. JAMA Intern Med. 2020;180:1133–4. doi: 10.1001/jamainternmed.2020.1736. [DOI] [PubMed] [Google Scholar]
- 31.The Medical Letter, Inc. Drugs for hypertension The Medical Letter on Drugs and Therapeutics 202062(1598):73–80. [PubMed] [Google Scholar]
- 32.Reps J M, Schuemie M J, Suchard M A, Ryan P B, Rijnbeekv P R. Design and implementation of a standardized framework to generate and evaluate patient-level prediction models using observational healthcare data. J Am Med Inform Assoc. 2018;25:969–75. doi: 10.1093/jamia/ocy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Reps J M, Kostka K F, Ryan P B, Zou Y, Voss E A. Development and validation of a prognostic model predicting symptomatic hemorrhagic transformation in acute ischemic stroke at scale in the OHDSI network. PloS One. 2020;15(01):e0226718. doi: 10.1371/journal.pone.0226718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reps J M, Rijnbeek P R, Ryan P B. Identifying the DEAD: development and validation of a patient-level model to predict death status in population-level claims data. Drug Saf. 2019;42(11):1377–86. doi: 10.1007/s40264-019-00827-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reps J M, Williams R D, You S C, Falconer T, Minty E, Callahan A. Feasibility and evaluation of a large-scale external validation approach for patient-level prediction in an international data network: validation of models predicting stroke in female patients newly diagnosed with atrial fibrillation. BMC Med Res Methodol. 2020;20:102. doi: 10.1186/s12874-020-00991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams R D, Markus A F, Yang C, DuarteSalles T, DuVall S L, Falconer T.Seek COVER: Development and validation of a personalized risk calculator for COVID-19 outcomes in an international networkmedRxiv 2020. doi: https://doi.org/10.1101/2020.05.26.20112649 [DOI] [PMC free article] [PubMed]
- 37.Burn E, You S C, Sena A, Kostka K, Abedtash H, Abrahão M TF.Deep phenotyping of 34,128 patients hospitalised with COVID-19 and a comparison with 81,596 influenza patients in America, Europe and Asia: an international network studyNat Commun 2020;11(5009). doi: 10.1038/s41467-020-18849-z [DOI] [PMC free article] [PubMed]
- 38.Morales D R, Conover M M, You S C, Pratt N, Kostka K, Duarte-Salles T. Renin-angiotensin system blockers and susceptibility to COVID-19: an international open science cohort study. Lancet Digit Health. 2021;3(02):e98–e114. doi: 10.1016/S2589-7500(20)30289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lane J CE, Weaver J, Kostka K, Duarte-Salles T, Abrahao T, Alghoul H. Safety of hydroxychloroquine, alone and in combination with azithromycin, in light of rapid wide-spread use for COVID-19: a multinational, network cohort and self-controlled case series study. Lancet Rheumatology. 2020;2(11):e698–e711. doi: 10.1016/S2665-9913(20)30276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.European Medicines Agency. COVID-19: reminder of risk of serious side effects with chloroquine and hydroxychloroquine2020 April 28. Available from: https://www.ema.europa.eu/en/news/covid-19-reminder-risk-serious-side-effects-chloroquine-hydroxychloroquine [cited 2021 Feb 7]
- 41.The European Network of Centres for Pharmacoepidemiology and Pharmacovigilance (ENCePP). Guide on Methodological Standards in Pharmacoepidemiology (Revision 8)EMA/95098/2010. Available from: http://www.encepp.eu/standards_and_guidances/documents/GuideMethodRev8.pdf [Accessed 2021 Feb 7]
- 42.Observational Health Data Sciences and Informatics. The Book of OHDSI; 2020Available from: https://ohdsi.github.io/TheBookOfOhdsi/


