Abstract
Heterochromatin represents a specialized chromatin environment vital to both the repression and expression of certain eukaryotic genes. One of the best-studied heterochromatin-associated proteins is Drosophila HP1. In this report, we have disrupted all somatic copies of the Tetrahymena HHP1 gene, which encodes an HP1-like protein, Hhp1p, in macronuclei (H. Huang, E. A. Wiley, R. C. Lending, and C. D. Allis, Proc. Natl. Acad. Sci. USA 95:13624–13629, 1998). Unlike the Drosophila HP1 gene, HHP1 is not essential in Tetrahymena spp., and during vegetative growth no clear phenotype is observed in cells lacking Hhp1p (ΔHHP1). However, during a shift to nongrowth conditions, the survival rate of ΔHHP1 cells is reduced compared to that of wild-type cells. Upon starvation, Hhp1p becomes hyperphosphorylated concomitant with a reduction in macronuclear volume and an increase in the size of electron-dense chromatin bodies; neither of these morphological changes occurs in the absence of Hhp1p. Activation of two starvation-induced genes (ngoA and CyP) is significantly reduced in ΔHHP1 cells while, in contrast, the expression of several growth-related or constitutively expressed genes is comparable to that in wild-type cells. These results suggest that Hhp1p functions in the establishment and/or maintenance of a specialized condensed chromatin environment that facilitates the expression of certain genes linked to a starvation-induced response.
Gene expression in eukaryotes relies upon the accessibility of the DNA template to a variety of components in the transcription apparatus, which is controlled in turn by a locus-specific chromatin structure. Chromatin of higher eukaryotes is often discussed in terms of two cytologically distinct domains: euchromatin and heterochromatin. Heterochromatin is classically distinguished from euchromatin by its paucity of genes, its highly condensed chromatin structure throughout the cell cycle, its late replication in S phase, and its enrichment in repetitive DNA sequences (25, 32). However, genes have been identified that reside within heterochromatic regions (21, 27), and the expression of some of these genes is dependent upon their placement in heterochromatin. Furthermore, recent evidence has shown that heterochromatin plays important roles in long-range intra- and interchromosomal interactions that affect nuclear organization (8, 10), chromosome segregation (9, 33, 46), and locus-controlled patterns of transcriptional activity (42).
Heterochromatin and euchromatin represent two different structural environments, and each has profound effects on gene expression (55, 58). A large body of genetic evidence suggests that heterochromatin interferes with the expression of normally euchromatic genes. In yeast cells, for example, mitotically transmissible silencing of silent mating-type loci depends upon nearby sequences, as well as on histone and nonhistone chromosomal components (44). Sequences not related to mating type become transcriptionally repressed when placed at or near this specialized chromatin environment, and similar positional silencing occurs near telomeres (24). Likewise in Drosophila melanogaster, heterochromatic position-effect variegation (PEV) is characterized by mitotically transmissible silencing and position-dependent gene silencing (55, 57, 58). Silencing by pericentric heterochromatin has also been observed in transgenic mice (18). Thus, the underlying mechanism leading to positional silencing appears to exist in yeast, flies, and mammals.
PEV in flies is influenced by modifiers (both enhancers and suppressors), and these gene products have been proven important in dissecting the composition of heterochromatin. One of the PEV modifier genes, Su(var)205, encodes HP1, which is perhaps the best-studied structural protein associated with heterochromatin (11, 12). A loss-of-function mutation in Su(var)205 leads to increased expression of euchromatic genes that suffer from position effect (i.e., White), while overproduction of HP1 results in decreased expression of these genes (12). Opposite results have been obtained for heterochromatin-positioned genes (i.e., Light), suggesting that genes localized in heterochromatin differ in their regulatory requirements (26). Conservation of HP1 throughout evolution (50) and the lethality of HP1 null alleles (13) suggest that HP1 is important for cell viability and development in flies. In support of this, Swi6p, a chromodomain-containing protein from fission yeast, while not essential, is required to maintain transcriptional repression at the silent mating-type loci and centromeres (2, 15, 38). Recent studies of Δswi6 cells have shown a reduced spore viability, along with altered expression of some meiotic genes, suggesting that proteins of this general family play an important yet poorly understood role in gene expression (37).
Like other ciliates, Tetrahymena spp. exhibit nuclear dimorphism, wherein each cell contains both a germ line micronucleus and a somatic macronucleus (22, 23). Even though these nuclei coexist as neighbors in a common cytoplasm, they differ considerably in function and provide a useful model for studying the function of HP1-like proteins. Recently, we reported the identification of an HP1-like protein, Hhp1p, that is absent from transcriptionally silent micronuclei but is modestly enriched in the condensed chromatin regions (referred to as chromatin bodies) that punctuate transcriptionally active macronuclei (30). Here, we report the disruption of all somatic (macronuclear) copies of the endogenous HHP1 genes in Tetrahymena thermophila. Our results demonstrate that, unlike Drosophila HP1, Hhp1p is not essential. Although no phenotypic difference is observed in vegetative cells lacking Hhp1p, a clear phenotypic difference is observed when cells are starved for nutrients. During a shift to nongrowth (starvation) conditions, the survival rate of cells lacking Hhp1p (ΔHHP1) is reduced relative to wild-type cells, morphological changes in chromatin body size fail to occur, and the activation of two starvation-induced genes is markedly reduced. These results suggest a model wherein the loss of Hhp1p leads to aberrant assembly of the condensed chromatin in macronuclei during the transition from growth to nongrowth conditions. We suspect that failure to properly assemble condensed chromatin domains leads to the reduced expression of starvation-induced genes that reside in and utilize condensed chromatin in a positive fashion to bring about their expression.
MATERIALS AND METHODS
Cell culture.
A genetically marked strain of T. thermophila, CU428 (Chx/Chx-[cy-s]VII), was used as the wild-type cell strain in all of the experiments reported here. It was generously provided by Peter Bruns (Cornell University, Ithaca, N.Y.).
Plasmid construction.
Genomic DNA was digested with SpeI and religated, and the HHP1 5′ and 3′ flanking regions were obtained by inverse PCR with the following primers: 5′-GCTCGGCCCGGGCTTTTGTCATATTTGATGC-3′ and 5′-GCTCGGATCGATGCTAATCCGCTGATTAG-3′. A 0.5-kb fragment of the 5′ flanking region and a 0.4-kb fragment of the 3′ flanking region were released by digesting the inverse PCR product with SpeI. The 0.4-kb fragment was repaired with the Klenow fragment of Escherichia coli DNA polymerase I and then digested with ClaI and ligated to a ClaI-HincII restriction fragment of p4T2-1 (containing a 1.5-kb drug resistance marker driven by an HHF1 promoter, followed by the Neo gene and the BTU2 terminator [see reference 19]). The resulting plasmid is referred to as pNeo-3′. The 0.5-kb fragment of the 5′ flanking region was cloned into the T/A vector pCRTMII (Invitrogen Corp.), released by SpeI digestion, and ligated to SpeI-linearized pNeo-3′. The final construct, referred to as pHHP1-Neo, was cut with SacI and XhoI, and a 2.4-kb insert containing the HHP1 5′ and 3′ sequences interrupted by the drug resistance cassette was released and used for transformation.
Gene replacement with the Biolistic gun.
Transformation was performed according to the protocol of Cassidy-Hanley et al. (6) by using the DuPont Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) except that 1.0-μm gold beads (Bio-Rad) were used instead of tungsten particles. CU428 cells were grown in 1% (wt/vol) enriched Proteose Peptone (1× SPP) medium to 2 × 105 cells/ml, harvested, and then starved in 10 mM Tris-HCl (pH 7.5) for 18 h. For each transformation attempt, 107 cells were washed and resuspended in 1 ml of Tris (10 mM) buffer. The gold particles were coated immediately before use with 1.5 μg of DNA/100 μl of particle suspension (60 μg of gold beads/ml of sterile H2O). The conditions for bombardment have been described previously (6). After bombardment, cells were immediately resuspended in 20 ml of SPP medium and plated on four 96-well microtiter plates. Paromomycin (Sigma Chemical Co.) was added 3 h later to a final concentration of 50 μg/ml. Two transformants were obtained after 50 to 60 generations of growth under selection with paromomycin. The final drug resistance of both HHP1 knockout strains was 2.5 mg of paromomycin per ml; wild-type cells typically cannot grow in the presence of more than 0.08 mg of paromomycin per ml. After complete phenotypic assortment had occurred, cells were grown in the absence of paromomycin (1× SPP) for at least 1 month before experiments were initiated with the knockout strain.
Southern blot analyses.
Total genomic DNA was isolated as described by Gaertig and Gorovsky (19) from wild-type CU428 cells and ΔHHP1 cells. Genomic DNA (15 μg) was digested with HindIII. To probe for HHP1, a 0.5-kb fragment from the 5′ flanking region of HHP1 (see Fig. 1A) was labeled with [α-32P]dATP by random priming (39). To probe for the Neo gene, a 1.3-kb fragment containing the Neo coding sequence was used as a probe (see Fig. 1A). Southern blotting was performed according to the standard procedure (39). Hybridization was carried out at 42°C in hybridization buffer containing 50% formamide.
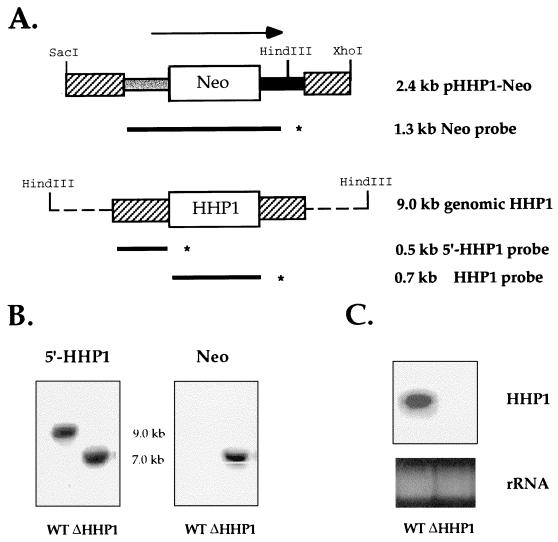
FIG. 1.
HHP1 gene is completely replaced by a disruption construct. (A) Disruption construct and macronuclear genomic map of HHP1 loci. A 2.4-kb SacI-XhoI fragment of pHHP1-Neo construct and a 9.0-kb HindIII fragment of the genomic HHP1 gene are shown. A 0.5-kb fragment of the 5′ flanking region (shown as a striped box on left) and a 0.4-kb fragment of the 3′ flanking region of the HHP1 gene (shown as a striped box on the right) were produced by PCR. These two PCR fragments were then inserted into a gene disruption cassette (20) containing a 1.5-kb drug resistance marker driven by an HHF1 promoter (shown as a gray box), followed by the Neo gene and the BTU2 terminator (shown as a solid box). A 1.3-kb fragment of the Neo drug resistance marker, a 0.5-kb fragment of the 5′ flanking region of HHP1, and a 0.7-kb fragment of the coding region of HHP1, shown as bold lines with asterisks at their ends, were used as Neo, 5′-HHP1, and HHP1 probes, respectively, in the Southern and Northern blot analyses. (B) Total genomic DNA from wild-type and ΔHHP1 cells was digested with HindIII and analyzed by Southern blot analyses with 5′-HHP1 probe or Neo probe. (C) Northern blot analysis of total cell RNA in wild-type (WT) and ΔHHP1 strains with HHP1 probe. The bottom panel shows ethidium bromide-stained rRNA as a loading control.
Northern blot analyses.
Total RNA was isolated by Trizol extraction according to the manufacturer’s protocol (GIBCO-BRL). Cells (6 × 106) were extracted in 1 ml of Trizol, followed by standard protocols as provided by GIBCO-BRL. RNA (10 μg) was electrophoresed on a 2.2 M formaldehyde–1% agarose gel. Gels were blotted onto Magnagraph nylon membranes (MSI, Inc.), and hybridization was carried out under the same conditions as for the Southern analyses (see above). HHT1, HHT3, CyP, and ngoA probes were generously provided by the laboratory of Martin Gorovsky at the University of Rochester.
Western analyses.
Macronuclear acid-soluble proteins were extracted from logarithmically growing or starved cultures of ΔHHP1 or CU428 strains and subjected to electrophoresis and immunoblotting analyses under conditions described previously (30).
Growth analyses.
ΔHHP1 and CU428 cells were used to inoculate 50 ml of 1× SPP medium at starting densities of 0.5 × 104 to 1 × 104 cells/ml. Cultures were grown at 30°C with shaking. Aliquots of the cultures (100 to 200 μl) were counted with a hemocytometer at frequent intervals.
Viability tests during starvation.
ΔHHP1 and CU428 cells were starved at a density of 2 × 105 to 3 × 105 cells/ml in 10 mM Tris-HCl (pH 7.4) at 30°C without shaking. To determine cell densities, cells were fixed with 1% formaldehyde and counted with a hemocytometer every 5 h for up to 40 h after the initial starvation period. The survival rate of approximately 100 hand-picked cells, after defined periods of starvation, was determined as described by Yu and Gorovsky (60). To evaluate the kinetics of regrowth in food, cells starved for 24 h were transferred to 1× SPP medium at a starting density of 104 cells/ml, and the cell density was determined at 5-h intervals.
Indirect immunofluorescence and analysis of DAPI-stained interphase nuclei.
ΔHHP1 and CU428 cells in logarithmic growth (2 × 105 to 3 × 105 cells/ml) were starved for 24 h at 30°C. Equal amounts of ΔHHP1 cells and CU428 cells were mixed and fixed in periodate-lysine-paraformaldehyde (PLP) (see reference 30). Fixed cells were then subjected to indirect immunofluorescence analyses by using α-Hhp1p and rhodamine-conjugated secondary antibodies, followed by staining with 0.2 μg of the DNA-specific dye 4′,6-diamidino-2-phenylindole (DAPI) per ml. Micrographs were obtained for both DAPI and α-Hhp1p antibody staining, and prints were digitized by using a Hewlett-Packard ScanJet II cx/T scanner. The DAPI-stained areas of randomly chosen macronuclei of ΔHHP1 or CU428 were measured by using NIH Image 1.59 software. Then, 100 nuclei from each cell strain were measured from six individual micrographs taken in two independent experiments.
Ultrastructural analyses.
Tetrahymena cells were fixed and processed for ultrastructural evaluation as previously described (51). Images were captured on negatives, which were directly digitized by using a Hewlett-Packard ScanJet II cx/T scanner. The number of chromatin bodies per unit area and the size of individual chromatin bodies were determined with the NIH Image 1.59 software.
Alkaline phosphatase treatment.
Total acid-soluble proteins from growing or starved cells were dried, incubated with or without alkaline phosphatase as described before (30), and resolved on a long (30 cm) acid-urea gel.
RESULTS
Somatic gene replacement of HHP1.
To disrupt the single-copy HHP1 gene in somatic macronuclei, a disruption plasmid was constructed (pHHP1-Neo [see Fig. 1A]) and used to transform starved Tetrahymena cells according to standard methods (6). The HHP1 disruption construct containing a previously described gene disruption cassette (19) flanked by a 0.5-kb 5′- and a 0.4-kb 3′-HHP1 flanking sequence was used to replace the macronuclear gene encoding Hhp1p (see Fig. 1A and Materials and Methods). Two transformants (referred to as ΔHHP1.2 and a ΔHHP1.6) were obtained after 50 to 60 generations of growth under selection with paromomycin. Southern blot analyses were performed on both strains to determine if all of the somatic copies of the HHP1 gene were completely replaced by the disruption construct. Identical results were obtained with both transformants; the results obtained with ΔHHP1.6 are shown in Fig. 1.
When a 0.5-kb fragment from the 5′ flanking region of the HHP1 gene (Fig. 1A) was used as a probe against the total genomic DNA from wild-type cells, a 9.0-kb band corresponding to the intact HHP1 gene was detected (Fig. 1B, left panel). Alternatively, a 7.0-kb band corresponding to the expected size of the replacement fragment alone was detected in both knockout strains (Fig. 1B, left panel). After the same blot was reprobed with a 1.3-kb Neo probe, this 7.0-kb band was still detected in knockout cells, while no signal was observed in wild-type cells, confirming that this 7.0-kb fragment was derived from the disruption construct (Fig. 1B, right panel).
To rule out the possibility that any wild-type HHP1 genes remained in the polyploid macronucleus (i.e., the transcriptionally active nucleus) of knockout cells after phenotypic assortment, Northern blot analyses were performed to evaluate steady-state message levels in both wild-type and knockout strains. With a 0.7-kb fragment from the HHP1 coding sequence used as a probe, the expected 1.0-kb message was detected in wild-type cells but not in knockout strains (Fig. 1C). These results demonstrate that all copies of the endogenous HHP1 gene were disrupted and that HHP1 mRNA is absent in the disruption strains.
HHP1 disruption strains (ΔHHP1) lack Hhp1p.
To further verify that Hhp1p is not synthesized in our HHP1 disruption strains (ΔHHP1), Western blot and immunofluorescence analyses were performed. Total acid-soluble protein from macronuclei of wild-type or ΔHHP1 cells was resolved on sodium dodecyl sulfate (SDS) gels and stained with Coomassie blue (Fig. 2A, left panel) or transferred to nitrocellulose and probed with α-Hhp1p antiserum. Although equivalent amounts of protein were extracted from both cell strains (Fig. 2A, left panel), Hhp1p was detected only in wild-type macronuclear extracts and not in protein extracted from the ΔHHP1 cells (Fig. 2A, right panel).
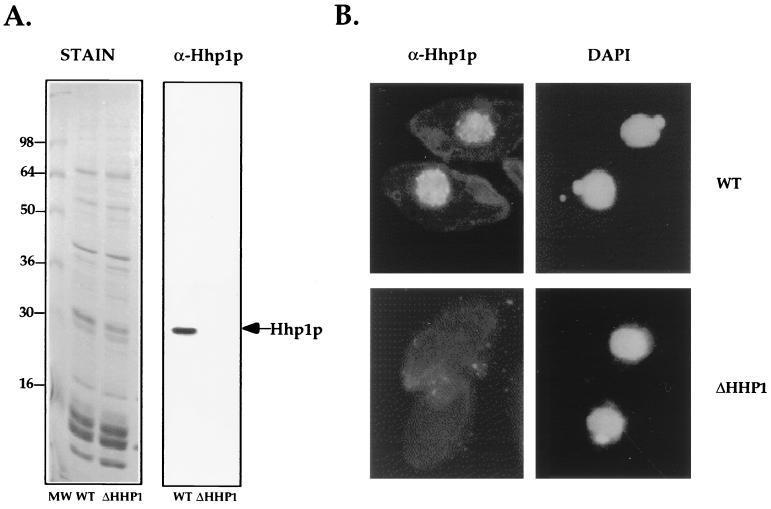
FIG. 2.
ΔHHP1 cells lack Hhp1p. (A) Western blot analysis. Total acid-soluble macronuclear protein, extracted from vegetatively growing wild-type (WT) or ΔHHP1 cells, was resolved on SDS–12% polyacrylamide gels. Gels were either stained with Coomassie blue (left panel) or blotted onto nitrocellulose and immunoblotted by using α-Hhp1p antibodies (right panel). As expected, Hhp1p was only detected in wild-type cells and not in ΔHHP1 cells (see arrow). (B) Indirect immunofluorescence analyses. Vegetatively growing cells from wild-type or ΔHHP1 strains were fixed and processed for immunofluorescence by using α-Hhp1p and rhodamine-conjugated secondary antibodies. Cells were also stained with the DNA-specific dye DAPI.
Previous studies have shown that Hhp1p is modestly enriched in condensed chromatin bodies compared to the core histone H2A used to indicate the distribution of “general” chromatin (30). Vegetatively growing cells from wild-type or ΔHHP1 cells were fixed and processed for indirect immunofluorescence analyses with α-Hhp1p antiserum. As expected, macronuclei in wild-type cells stained intensely for Hhp1p (Fig. 2B, upper panels), while no staining was detected in either of the ΔHHP1 disruption strains (Fig. 2B, lower panels). These results confirm that Hhp1p is not expressed and strongly suggest that Hhp1p alone accounts for the punctate immunostaining pattern of α-Hhp1p observed in macronuclei during vegetative growth.
ΔHHP1 cells exhibit reduced viability during starvation.
The finding that HHP1 expression is completely eliminated in our disruption strains suggests that, unlike HP1 in Drosophila, Hhp1p is not essential in Tetrahymena. Moreover, the growth rate of ΔHHP1 cells from two independent transformants (Fig. 3A) is indistinguishable from that of wild-type cells.
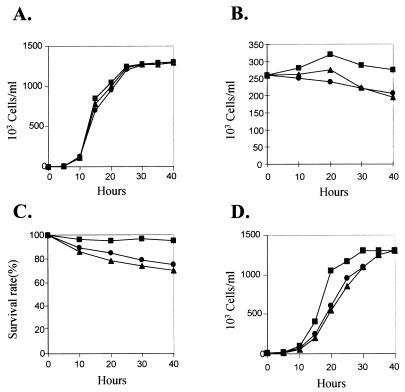
FIG. 3.
ΔHHP1 cells exhibit reduced viability during starvation. (A) Growth curve of wild-type and two ΔHHP1 transformants, ΔHHP1.2 and ΔHHP1.6. Cells were grown at 30°C in 1× SPP medium. (B) Changes in cell density of wild-type and ΔHHP1 cells during starvation. Cells in the logarithmic phase of growth were starved in 10 mM Tris-HCl (pH 7.4) at 30°C, and the cell densities were measured at the indicated times. (C) Survival of wild-type and ΔHHP1 cells during prolonged periods of starvation. Cells were starved at 30°C for the indicated times, at which time ∼100 single cells were isolated and transferred to 1× SPP medium. After 2 days, drops containing numerous cells were counted as positive for survival (60). (D) Recovery of wild-type and ΔHHP1 cells after starvation. Cells were starved at 30°C for 24 h and then were transferred to 1× SPP medium at a starting density of 104 cells/ml. The cells densities were measured at the indicated times. Symbols: ■, wild type; ▴, ΔHHP1.2; ●, ΔHHP1.6.
In Tetrahymena, starvation is a physiological state that is necessary to induce the sexual pathway, termed conjugation, during which a unique program of genetic information, which is not present during vegetative growth, is expressed (41, 52). To possibly uncover a more subtle phenotype of cells lacking Hhp1p, knockout cells were also examined under nongrowth (starvation) conditions (60).
Wild-type and ΔHHP1 cells were depleted of nutrients (i.e., washed into 10 mM Tris [pH 7.5]), and the cell density was examined over a subsequent 40-h period as shown in Fig. 3B. Relative to similarly treated wild-type cells, a significant difference in cell density was observed in both ΔHHP1 transformants over the first 40 h of starvation (Fig. 3B). While more than 95% of the wild-type cells were recovered after 40 h of starvation, only ∼80% of ΔHHP1 cells survived these physiological conditions (Fig. 3B). This result suggests that the absence of Hhp1p directly or indirectly affects cell survival during prolonged periods of starvation. This conclusion is further supported by directly examining the survival rate (Fig. 3C) and the kinetics of regrowth of cells after a 24-h starvation period (Fig. 3D). Our results indicate that the survival rate of ΔHHP1 transformants decreases 25 to 30% after 40 h of starvation, while that of wild-type cells decreases 0 to 5% (Fig. 3C). Similarly, the rate of regrowth of ΔHHP1 cells after 24 h of starvation is reproducibly lower than that of wild-type cells (Fig. 3D), although their growth rate will eventually be as same as that of wild-type cells after 2 to 3 days of recovery in nutrient-rich medium (Fig. 3A). Thus, we conclude that ΔHHP1 cells exhibit reduced viability during a transient period after a shift from growth to nongrowth (starvation) conditions.
ΔHHP1 cells display abnormally large macronuclei upon starvation.
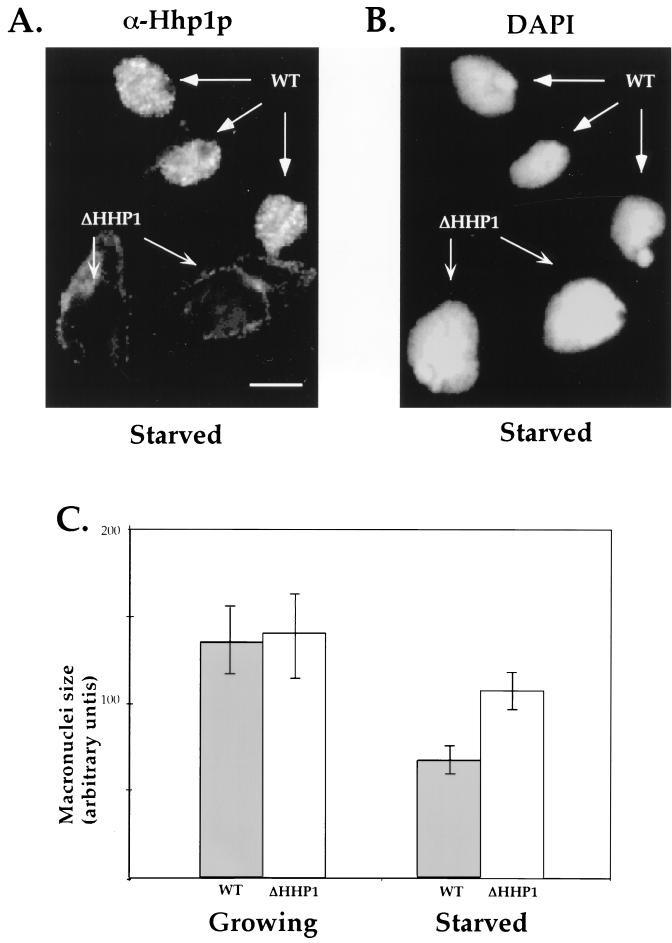
Earlier studies utilizing linker histone knockout strains of Tetrahymena have shown that H1 is involved in chromatin condensation in vivo based on the finding that the DAPI-stained area of macronuclei in H1 disruption strains is increased (48). In contrast, during periods of prolonged starvation, macronuclei in wild-type cells become smaller concomitant with an increase in the size of condensed chromatin bodies and a general decrease in gene expression and other cellular activities (31). To determine whether Hhp1p is involved in any of these morphological changes, starved ΔHHP1 or wild-type cells were intentionally mixed together, fixed and stained with both DAPI (Fig. 4B) and α-Hhp1p antiserum (Fig. 4A), and examined by fluorescence microscopy. As shown in Fig. 4B, the DAPI-stained area of macronuclei in ΔHHP1 cells (indicated by the absence of α-Hhp1p staining [Fig. 4A, left panel]) is significantly larger (1.5-fold) than that of wild-type cells. Interestingly, this difference in macronuclear size is not observed in vegetatively growing cells, a result quantitated in Fig. 4C. Thus, while no significant difference in macronuclear size has been detected between wild-type and ΔHHP1 cells under nutrient-rich (growing) conditions, a 1.5-fold difference is detected under nutrient-poor (starvation) conditions. In keeping with published in vivo data on H1 knockout strains (48), these data suggest that Hhp1p may facilitate macronuclear chromatin condensation during a physiological response to starvation.
FIG. 4.
ΔHHP1 cells display abnormally large macronuclei upon starvation. Wild-type (WT) and HHP1 knockout (ΔHHP1) cells were starved in 10 mM Tris for 24 h, mixed at a 1:1 ratio, and stained in situ with α-Hhp1p antibodies (A) and DAPI (B). As expected, a punctate Hhp1p staining pattern was observed in the macronuclei of wild-type cells, while no staining was detected in ΔHHP1 cells (see arrows in panel A). Note the relatively larger size of macronuclei in the ΔHHP1 cells. Bar, 10 μm. (C) The DAPI-stained cross-sectional areas of 100 to 200 macronuclei from growing or starved wild-type or ΔHHP1 cells were measured; the nuclear size is presented in arbitrary units. The mean and standard error values for three independent experiments are shown. Note that no significant difference between the macronuclei of wild-type and ΔHHP1 cells is detected under growing conditions, while an approximately 1.5-fold difference is detected between cell strains after starvation.
Altered morphology of chromatin bodies observed in wild-type cells upon starvation does not occur in ΔHHP1 cells.
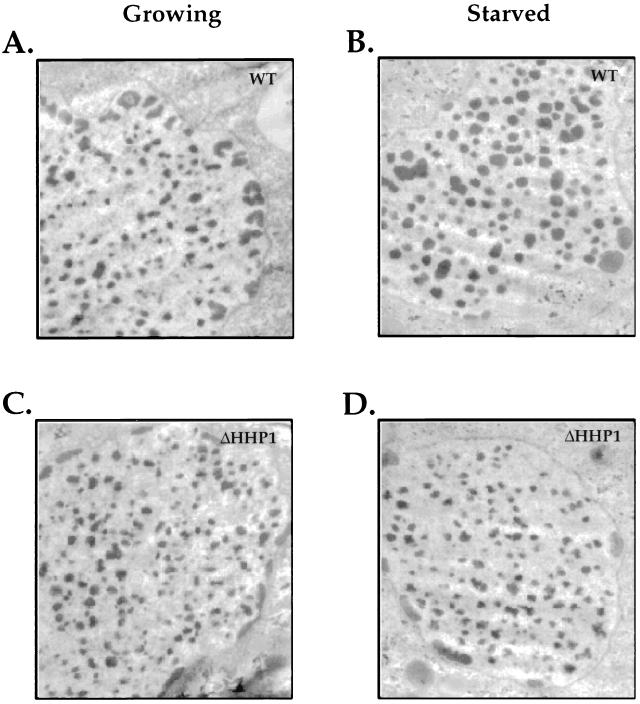
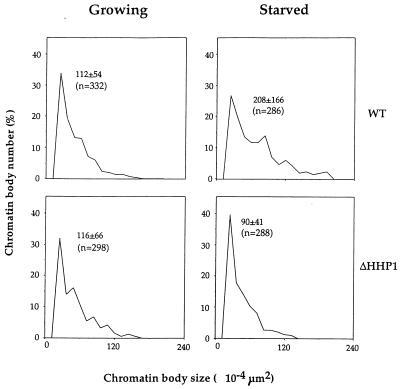
Previous studies have shown that the size of the condensed chromatin bodies in macronuclei is increased in wild-type cells upon prolonged starvation (31), and our recent studies have shown that Hhp1p is modestly enriched in these electron-dense structures (30). To examine more directly the state of the condensed chromatin in macronuclei from cells lacking Hhp1p, ultrastructural analyses were performed on wild-type and ΔHHP1 cells under both nutrient-rich and starvation conditions. Consistent with our observations at the light microscopic level, no obvious differences were observed in the ultrastructure of the macronuclei chromatin. In particular, the size and number of the electron-dense chromatin bodies were the same when the wild-type and ΔHHP1 cells were maintained under growth conditions (compare Fig. 5A and 5C; see also Fig. 6). However, while the number of chromatin bodies per unit area remains roughly the same when cells lacking Hhp1p are subjected to starvation conditions, the average chromatin body size in the ΔHHP1 strain is about half of that observed in the wild-type cells (compare Fig. 5B and D; see also Fig. 6). Our quantitative data, presented in Fig. 6, suggest that the documented increase in chromatin body size observed in wild-type cells upon starvation (31) does not occur in the ΔHHP1 cells.
FIG. 5.
Ultrastructural analyses of chromatin bodies in macronuclei of wild-type and ΔHHP1 cells. Growing (A and C) or starved (B and D) wild-type (A and B) or ΔHHP1 (C and D) cells were fixed and processed for ultrastructural analysis by transmission electron microscopy. Note the difference in size between the chromatin bodies of the wild-type and ΔHHP1 cells, particularly in the starved cells. No consistent differences in the size or morphology of the multiple, peripherally located nucleoli were observed between the strains.
FIG. 6.
Distribution of chromatin body size in growing and starved cells. The size of chromatin bodies from growing and starved wild-type (WT) or ΔHHP1 cells was measured. The mean ± the standard error and the number of measurements (indicated as “n”) are shown in each graph. Note that the size distributions of chromatin bodies from growing wild-type cells (upper left panel) and growing ΔHHP1 cells (lower left panel) are comparable. In contrast, chromatin bodies from starved wild-type cells (upper right) are significantly larger than those from starved ΔHHP1 cells (lower right).
Hhp1p is hyperphosphorylated in starved cells.
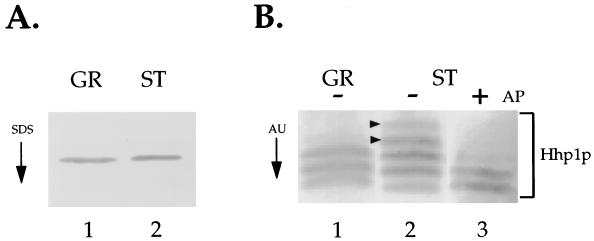
Since, in contrast to wild-type cells, the relative size of the chromatin bodies fails to increase in ΔHHP1 transformants upon starvation, we next asked whether the amount of Hhp1p is altered upon starvation in cells containing Hhp1p. Western blot analysis with α-Hhp1p antiserum failed to detect any significant difference in the amount of Hhp1p extracted from growing and starved cells (Fig. 7A). However, the phosphorylation state of many chromosomal proteins, including Drosophila HP1, is dynamically changing in response to different physiological or developmental conditions (14, 29), and our previous studies have shown that Hhp1p is multiply phosphorylated in vegetatively growing cells (30). To determine whether the phosphorylation state of Hhp1p is modified in response to starvation, acid-soluble macronuclear protein from vegetatively growing or starved wild-type cells was resolved on long acid-urea gels, blotted to a membrane, and probed with α-Hhp1p antiserum (Fig. 7B).
FIG. 7.
Hhp1p is hyperphosphorylated in starved cells. (A) The amount of Hhp1p remains the same upon starvation. Total acid-soluble macronuclear protein, extracted from vegetatively growing (GR) or 24-h-starved (ST) wild-type cells, was resolved on an SDS–12% polyacrylamide gel, blotted onto nitrocellulose, and immunoblotted with α-Hhp1p antibodies. (B) Proteins from growing or starved wild-type cells were resolved on an acid-urea (AU) gel and analyzed by Western blot analysis with α-Hhp1p antibodies. In the sample shown in lane 3, Hhp1p from starved cells was incubated with alkaline phosphatase (+AP) before electrophoresis on a long acid-urea gel. Note the appearance of two slower-migrating bands in the sample from starved cells (indicated by arrowheads in lane 2).
As shown previously (30), Hhp1p extracted from growing cells resolves into several distinct bands corresponding to a single, faster-migrating dephosphorylated isoform and a collection of slower-migrating phosphorylated isoforms (Fig. 7B, lane 1). In starved cells, however, two additional slower-migrating bands are consistently detected (see arrowheads in Fig. 7B, lane 2), and most of these slower-migrating bands “collapse” to the position of the single faster-migrating band when the samples are first treated with alkaline phosphatase (Fig. 7B, lane 3). These data suggest that these additional slow-migrating species represent new, multiply phosphorylated isoforms of Hhp1p that are induced upon starvation. Thus, although the amount of Hhp1p remains the same during periods of prolonged starvation, the steady-state phosphorylation level of Hhp1p increases as part of the response to this physiological state.
Activation of two starvation-induced genes is reduced in ΔHHP1 cells.
Heterochromatin is often associated with gene silencing, leading to the general belief that a more condensed chromatin environment causes decreased accessibility of transcription factors to target genes. However, recent studies with strains lacking linker histone H1 have elegantly shown that the absence of linker histone can impose opposing effects on the regulation of at least certain genes in vivo (49). It was of interest then to determine whether similar results would be obtained with cells lacking Hhp1p. More specifically, our finding that the assembly of condensed chromatin is compromised in ΔHHP1 cells, at least during starvation, led us to ask if transcriptional regulation in macronuclei is affected by a lack of Hhp1p under this physiological condition. Under nongrowth conditions, many growth-related genes are repressed in Tetrahymena, whereas a new set of starvation-related genes is induced (5, 52). To assess the possible role of Hhp1p in starvation-induced gene regulation, three different types of genes were examined in wild-type and ΔHHP1 cells: growth-related genes, starvation-induced genes, and genes that are constitutively expressed in both conditions.
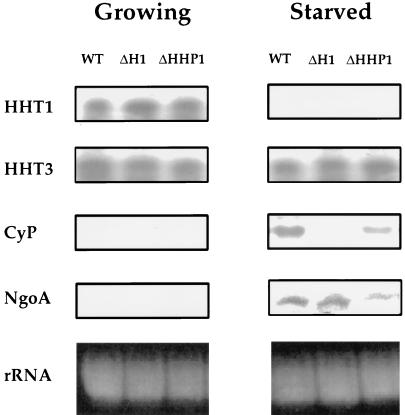
Northern blot analyses were performed on total RNA isolated from growing and starved cell populations of wild-type and ΔHHP1 strains (Fig. 8). For reference, an H1 disruption strain (ΔH1) was also analyzed (49). One growth-associated gene selected for examination was the HHT1 gene, which encodes histone H3 and is highly expressed in growing cells but is repressed in starved cells (3, 52, 60). Relative to wild-type cells, no difference was observed in the steady-state level of H3 mRNA in either H1 or HHP1 disruption strains during growth or starvation. Similarly, no differences were detected in mRNA levels in the different cell lines when either β-tubulin or actin genes were used as probes in similar analyses (data not shown). These data suggest that the regulation of several growth-associated genes is unaffected by the loss of Hhp1p. Unlike HHT1, the HHT3 gene encodes a relatively minor constitutively expressed or “basal” histone H3 variant whose expression is detected in both growing and starved cells (3, 60). When the mRNA level of HHT3 was examined in ΔHHP1 cells, an expression pattern nearly identical to that of wild-type cells was observed (Fig. 8). These data suggest that the regulation of “constitutive” genes is also unaffected by the loss of Hhp1p.
FIG. 8.
Activated expression of two starvation-induced genes is reduced in ΔHHP1 cells. Northern analyses of HHT1, HHT3, ngoA, and CyP expression were performed. Total RNA (10 μg) was isolated from wild-type, ΔH1, and ΔHHP1 cells at 250,000 cells/ml during log-phase growth (Growing) or after a 24-h starvation (Starved). The bottom panels show ethidium bromide-stained rRNA as a loadings control.
ngoA and CyP are the only two known starvation-induced genes in Tetrahymena that are not transcribed to a large extent during vegetative growth (41). While the function of ngoA is unknown (41), CyP encodes a cysteine protease whose level increases dramatically upon starvation (34). As shown in Fig. 8, neither ngoA nor CyP is detected in growing cells from any of the strains examined, suggesting that, during growth, both genes are maintained in a repressed state in the absence of either Hhp1p or H1. However, in starved cells, while ngoA and CyP mRNA is clearly present in wild-type cells, message levels for both genes are considerably less abundant in cells lacking Hhp1p. In agreement with published studies with H1 knockout cells (49), the ngoA message level remains roughly the same in ΔH1 cells compared to the wild-type cells while, in contrast, the expression of CyP is dramatically decreased in ΔH1 cells. Quantitation of three independent sets of Northern blot data reveals that mRNA levels of ngoA and CyP in ΔHHP1 cells are reduced by approximately two- and fourfold, respectively, relative to wild-type cells. Our results are consistent with the possibility that Hhp1p facilitates the expression of at least some starvation-induced genes, perhaps through a phosphorylation-mediated pathway leading to the assembly of a condensed chromatin state.
DISCUSSION
Hhp1p is not essential during vegetative growth, but its absence affects cell viability during starvation.
Widespread conservation of HP1-like proteins throughout evolution (50) and the lethality of HP1 null alleles in Drosophila (13) suggest that members of this family are important for cell viability and development. Although the specific molecular details underlying the function of HP1-like proteins remain unclear, several lines of evidence support the hypothesis that HP1-like proteins function in the assembly and maintenance of heterochromatin, a specialized chromatin environment that is most often associated with gene silencing (16). Besides this more-accepted role in gene silencing, HP1 also appears to play an important role during mitosis, with possible functions in transferring the inner centromere protein INCENP from centromeres to the spindle (1) and in binding to telomeres and preventing telomere fusions (17). Therefore, the absence of HP1 in flies may result from abnormal patterns of chromosome segregation that lead to lethality in Drosophila (35).
In this study, we have successfully disrupted the single-copy gene encoding an HP1-like protein, Hhp1p, in the ciliated protozoan Tetrahymena. Unlike Drosophila HP1, complete elimination of Hhp1p from somatic macronuclei does not appear to have an obvious effect on vegetative cell growth. In contrast to the situation in flies, nuclear division in Tetrahymena macronuclei is amitotic, occurring without chromosome condensation and segregation. Thus, the loss of Hhp1p need not affect the proper segregation of chromosomes during growth, a property imparted exclusively to micronuclei that lack Hhp1p completely (7, 30, 45).
While no obvious phenotypic difference was detected in ΔHHP1 cells under nutrient-rich (growing) conditions, several phenotypic differences were observed during starvation. Our data indicate that ΔHHP1 cells exhibit a reduced viability during starvation (Fig. 3B to D). While these data suggest a potentially important role of Hhp1p in mediating the expression of a subset of starvation-induced genes, reduced viability of ΔHHP1 cells could lead to cell lysis and the generation of cell debris that, in turn, could be used as a nutrient source and so influence our results. We feel that this explanation is unlikely for several reasons. First, the mating pathway or conjugation in Tetrahymena is a process that is extremely sensitive to nutrient and ionic conditions (4) and, in the laboratory, initiation of conjugation is conveniently induced by starvation. Mating-efficiency tests, defined by monitoring the percentage of paired cells after starved cells of opposite mating types are mixed, were performed between ΔHHP1 cells and wild-type partners (mixed after 18 to 24 h of starvation). These tests indicate that the remaining viable ΔHHP1 cells are able to enter the conjugation pathway as efficiently as wild-type control partners (data not shown). Thus, as initiation of conjugation is generally taken as a sensitive indicator of “sufficiently starved” cells, it seems unlikely that the observed reduction in the expression of the starvation-induced genes in ΔHHP1 cells is due to a significant degree of “refeeding” from cellular lysis. Second, several growth-specific genes (i.e., HHT1 and HHT3) remain suppressed in ΔHHP1 cells during starvation (Fig. 8), again suggesting that cell lysis and refeeding are not sufficient to induce the expression of these genes. Thus, based on results from both a biological and a molecular assay to monitor the starvation state of the cell, it seems likely that the starvation-induced phenotypic differences that we have observed in ΔHHP1 cells are due, directly or indirectly, to the absence of Hhp1p and not to a failure to sufficiently starve the cells.
Our studies also show that the removal of Hhp1p from macronuclei causes a significant increase in the size of macronuclei relative to those of the wild-type cells and, interestingly, this effect is only seen in starved cells. In some ways this phenotype is reminiscent of previous analyses of cells lacking H1, where an increase of macronuclear size was noted in growing cells and suggested to correlate with decondensation of the chromatin fiber in the absence of linker histone (48). Similarly, we suspect that the increase in the DAPI-stained area of macronuclei observed in ΔHHP1 strains is directly related to incomplete condensation and compaction of the chromatin fiber seen in starved cells.
Our data are consistent with a putative function of Hhp1p in a pathway of chromatin assembly that leads to the generation of larger chromatin bodies upon prolonged starvation. During this physiological state, cells lacking Hhp1p have significantly smaller chromatin bodies than their wild-type counterparts. However, given that chromatin bodies exist in cells lacking Hhp1p, it seems unlikely that Hhp1p is solely responsible for initiating the assembly of these electron-dense structures. Our results are most consistent with Hhp1p participating in a starvation-induced pathway of condensed chromatin assembly that leads to an increase in chromatin body size, an increase that may be mediated by a phosphorylation-induced conformational change or a novel protein-protein association. Alternative explanations are also possible. For example, potential interactions between HP1 and the nuclear envelope have been reported (40, 59), and these interactions might be disrupted in mutant cells, resulting in an increase in the size of the macronucleus.
Gene regulation mediated by Hhp1p-associated effects.
In Tetrahymena, total cellular RNA and poly(A)-containing RNA decrease upon starvation to approximately 20% of that observed in growing cells (5). The finding that chromatin body size increases upon starvation in an Hhp1p-dependent fashion led us to predict initially that this alteration in chromatin structure might be linked to transcriptional repression of growth-related genes during starvation. Although contrary to our initial expectations, the finding that the expression of a limited number of growth-related genes (histone, tubulin, and actin genes) remains unchanged in cells lacking Hhp1p is consistent with several observations: ΔHHP1 cells grow at a wild-type rate under nutrient-rich conditions, and the morphology and ultrastructure of the macronuclei are indistinguishable from those of wild-type cells in this physiological state.
While the molecular details remain to be defined, our studies point to a more complex picture of Hhp1p function wherein a more condensed chromatin environment potentially produces a positive effect on gene expression: the activation of two starvation-induced genes, ngoA and CyP, is decreased in ΔHHP1 cells compared to wild-type or ΔH1 strains (Fig. 8). It remains a formal possibility that secondary effects, produced in the sick or dying ΔHHP1 cells, affect the regulation of the starvation-induced genes that we have selected for study. Several considerations suggest that this is unlikely. If cell death was causing global changes in gene regulation, it seems likely that we would have observed misregulation with some of the genes that we assayed, and this is not the case. Moreover, our experiments detected misregulation of only starvation-induced genes and not of growth-dependent or constitutively expressed genes, a finding consistent with the starvation-induced phenotype that we observed in ΔHHP1 cells. Even if the effect of the loss of Hhp1p on ngoA and CyP is indirect, the finding that Hhp1p is not essential during vegetative growth but exerts a clear starvation phenotype likely reveals an important physiological function(s) of Hhp1p in vivo. It is also worth noting that another HP1-like protein, Swi6p, is also not essential in yeast cells (2) and, interestingly, the absence of the SWI6 gene affects sporulation and meiosis (37), a process triggered by starvation in yeast.
Unfortunately, the in situ location of ngoA or CyP with respect to chromatin bodies is not known due to technical limitations. Nonetheless, our findings are reminiscent of results with Drosophila HP1, wherein mutations in HP1 lead to a decreased expression of variegating heterochromatic genes (e.g., Light [26]). On the basis of these data it has been suggested that the expression of heterochromatin-located genes requires the existence of an appropriate heterochromatin environment or compartment (55, 58). In vivo studies with Tetrahymena have shown that linker histone H1 is required for the activated expression of CyP in starved cells (see reference 49 and Fig. 8). This surprising result is consistent with the idea that H1, a histone thought to facilitate higher-order chromatin structure, can act as both a positive and a negative gene regulator. Along this line, genome-wide expression analyses have recently begun to dissect the identity of genes whose expression, coordinately regulated under diverse physiological conditions, depends on the functions of key transcriptional regulators, including those that modify chromatin components (28). Using this technology, up- and downregulation have been observed in the absence of certain regulators, suggesting that the transcriptional effects of various knockouts are likely to be gene specific. Inasmuch as chromatin effects are also likely to be gene specific, it remains of interest to know to what extent the loss of Hhp1p affects the genome-wide expression (up or down) of other starvation-induced genes in this organism.
ACKNOWLEDGMENTS
We are grateful to Jody Bowen for her technical assistance in obtaining the ΔHHP1 transformants and for sharing many reagents, including the gene disruption cassette p4T2-1, HHT1, HHT3, ngoA, and CyP probes. All ultrastructural analyses were carried out in the electron microscopy facility of the Biology Department at Syracuse University.
This research was supported by a grant from the National Institutes of Health to C.D.A. (GM40922).
REFERENCES
- 1.Ainsztein A M, Kandels-Lewis S E, Mackay A M, Earnshaw W C. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Biol. 1998;143:1763–1774. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allshire R C, Javerzat J-P, Redhead N J, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 3.Bannon G A, Calzone F J, Bowen J K, Allis C D, Gorovsky M A. Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena. Nucleic Acids Res. 1983;11:3903–3917. doi: 10.1093/nar/11.12.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruns P J, Brussard T B. Pair formation in Tetrahymena pyriformis, an inducible developmental system. J Exp Zool. 1974;188:337–344. doi: 10.1002/jez.1401880309. [DOI] [PubMed] [Google Scholar]
- 5.Calzone R J, Stathopoulos V A, Grass D, Gorovsky M A, Angerer R C. Regulation of protein synthesis in Tetrahymena: RNA sequence sets of growing and starved cells. J Biol Chem. 1983;258:6899–6905. [PubMed] [Google Scholar]
- 6.Cassidy-Hanley D, Bowen J, Lee H J, Cole E, VerPlank A L, Gaertig J, Gorovsky M A, Bruns P J. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyne R S, Yao M C. Evolutionary conservation of sequences directing chromosome breakage and rDNA palindrome formation in tetrahymenine ciliates. Genetics. 1996;144:1479–1487. doi: 10.1093/genetics/144.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csink A K, Henikoff S. Genetic modification of heterochromatin association and nuclear organization in Drosophila. Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- 9.Dernburg A F, Sedat J W, Hawley R S. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 10.Dernburg A F, Broman K W, Fung J C, Marshall W F, Philips J, Agard D A, Sedat J W. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 11.Eissenberg J C, James T C, Foster-Hartnett D M, Hartnett T, Ngan V, Elgin S C R. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eissenberg J C, Morris G D, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eissenberg J C, Hartnett T. A heat shock-activated cDNA rescues the recessive lethality of mutations in the heterochromatin-associated protein HP1 of Drosophila melanogaster. Mol Gen Genet. 1993;240:333–338. doi: 10.1007/BF00280383. [DOI] [PubMed] [Google Scholar]
- 14.Eissenberg J C, Ge Y-W, Hartnett T. Increased phosphorylation of HP1, a heterochromatin-associated protein of Drosophila, is correlated with heterochromatin assembly. J Biol Chem. 1994;269:21315–21321. [PubMed] [Google Scholar]
- 15.Ekwall K, Javerzat J-P, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 16.Elgin S C R. Heterochromatin and gene regulation in Drosophila. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 17.Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 18.Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 19.Gaertig J, Gorovsky M A. Efficient mass transformation of Tetrahymena thermophila by electroporation of conjugants. Proc Natl Acad Sci USA. 1992;89:9196–9200. doi: 10.1073/pnas.89.19.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaertig J, Gu L, Hai B, Gorovsky M A. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 1994;22:5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatti M, Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- 22.Gorovsky M A. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973;20:19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- 23.Gorovsky M A. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- 24.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 25.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 26.Hearn M G, Hedrick A, Grigliatti T A, Wakimoto B T. The effect of modifiers to position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics. 1991;128:785–797. doi: 10.1093/genetics/128.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilliker A Y, Appels R, Schalet A. The genetic analysis of D. melanogaster heterochromatin. Cell. 1980;21:607–619. doi: 10.1016/0092-8674(80)90424-9. [DOI] [PubMed] [Google Scholar]
- 28.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eucaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 29.Huang D W, Fanti L, Pak D T S, Botchan M R, Pimpinelli S, Kellum R. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol. 1998;142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, Wiley E A, Lending R C, Allis C D. An HP1-like protein is missing from transcriptionally silent micronuclei but is enriched in condensed macronuclear chromatin. Proc Natl Acad Sci USA. 1998;95:13624–13629. doi: 10.1073/pnas.95.23.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeter J R, Pavlat W A, Cameron I L. Changes in the nuclear acidic proteins and chromatin structure in starved and refed Tetrahymena. Exp Cell Res. 1975;93:79–88. doi: 10.1016/0014-4827(75)90425-5. [DOI] [PubMed] [Google Scholar]
- 32.John B. The biology of heterochromatin. In: Verma R S, editor. Heterochromatin: molecular and structure aspects. New York, N.Y: Cambridge University Press; 1988. pp. 1–147. [Google Scholar]
- 33.Karpen G H, Le M H, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- 34.Karrer K M, Peiffer S L, DiTomas M E. Two distinct gene subfamilies within the family of cysteine protease genes. Proc Natl Acad Sci USA. 1993;90:3063–3067. doi: 10.1073/pnas.90.7.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellum R, Alberts B M. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995;108:1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- 36.Kimmerly W, Buchman A, Kornberg R, Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988;7:2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leem S-H, Chung C-H, Sunwoo Y, Araki H. Meiotic role of SWI6 in Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:3154–3158. doi: 10.1093/nar/26.13.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorentz A, Osterman K, Fleck O, Schmidt H. Switching gene swi6, involved in the repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene. 1994;143:139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- 39.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Marshall W F, Fung J C, Sedat J W. Deconstructing the nucleus: global architecture from local interactions. Curr Opin Genet Dev. 1997;7:259–263. doi: 10.1016/s0959-437x(97)80136-0. [DOI] [PubMed] [Google Scholar]
- 41.Martindale D W, Bruns P J. Cloning of abundant mRNA species present during conjugation of Tetrahymena thermophila: identification of mRNA species present exclusively during meiosis. Mol Cell Biol. 1983;3:1857–1865. doi: 10.1128/mcb.3.10.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milot E, Strouboulis J, Trimborn T, Wijgerde M, deBoer E, Langeveld A, Tan-Un K, Vergeer W, Yannoustsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 43.Nicol L, Jeppesen P. Human autoimmune sera recognize a conserved 26-kD protein which associated with mammalian heterochromatin that is homologous to heterochromatin protein 1 of Drosophila. Chromosome Res. 1990;2:245–253. doi: 10.1007/BF01553325. [DOI] [PubMed] [Google Scholar]
- 44.Pillus L, Grunstein M. Chromatin structure and epigenetic regulation in yeast. In: Elgin S C R, editor. Chromatin structure and gene expression. Oxford, England: IRL Press; 1995. pp. 123–146. [Google Scholar]
- 45.Prescott D M. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renauld H, Gasser S M. Heterochromatin: a meiotic matchmaker? Trends Cell Biol. 1997;7:201–205. doi: 10.1016/S0962-8924(97)01034-9. [DOI] [PubMed] [Google Scholar]
- 47.Reuter G, Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- 48.Shen X, Yu L, Weir J W, Gorovsky M A. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 49.Shen X, Gorovsky M A. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 50.Singh P B, Millwe J R, Rearce J, Kothary R, Buton R D, Paro R, James T C, Gaunt S J. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991;19:789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smothers J F, Madireddi M T, Warner F D, Allis C D. Programmed DNA degradation and nucleolar biogenesis occur in distinct organelles during macronuclear development in Tetrahymena. J Eukaryot Microbiol. 1997;44:79–88. doi: 10.1111/j.1550-7408.1997.tb05942.x. [DOI] [PubMed] [Google Scholar]
- 52.Stargell L A, Karrer K M, Gorovsky M A. Transcriptional regulation of gene expression in Tetrahymena thermophila. Nucleic Acids Res. 1990;18:6637–6639. doi: 10.1093/nar/18.22.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stone E M, Pillus L. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J Cell Biol. 1996;135:571–583. doi: 10.1083/jcb.135.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sweet M T, Jones K, Allis C D. Phosphorylation of linker histone is associated with transcriptional activation in a normally silent nucleus. J Cell Biol. 1996;135:1219–1228. doi: 10.1083/jcb.135.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wakimoto B T. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 56.Wakimoto B T, Hearn M G. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wallrath L L. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- 58.Weiler K S, Wakimoto B T. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 59.Ye Q, Worman H J. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 60.Yu L, Gorovsky M A. Constitutive expression, not a particular primary sequence, is the important feature of the H3 replacement variant hv2 in Tetrahymena thermophila. Mol Cell Biol. 1997;17:6303–6310. doi: 10.1128/mcb.17.11.6303. [DOI] [PMC free article] [PubMed] [Google Scholar]