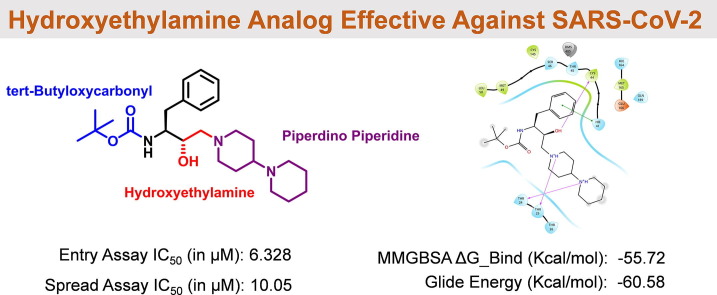
Graphical abstract
Keywords: Hydroxyethylamine compound library, SARS-CoV-2, COVID-19, Virtual screening, MM-GBSA, MD simulation, 3CLpro, Antiviral assay
Abstract
The continued toll of COVID-19 has halted the smooth functioning of civilization on a global scale. With a limited understanding of all the essential components of viral machinery and the lack of structural information of this new virus, initial drug discovery efforts had limited success. The availability of high-resolution crystal structures of functionally essential SARS-CoV-2 proteins, including 3CLpro, supports the development of target-specific therapeutics. 3CLpro, the main protease responsible for the processing of viral polypeptide, plays a vital role in SARS-CoV-2 viral replication and translation and is an important target in other coronaviruses. Additionally, 3CLpro is the target of repurposed drugs, such as lopinavir and ritonavir. In this study, target proteins were retrieved from the protein data bank (PDB IDs: 6 M03, 6LU7, 2GZ7, 6 W63, 6SQS, 6YB7, and 6YVF) representing different open states of the main protease to accommodate macromolecular substrate. A hydroxyethylamine (HEA) library was constructed from harvested chemical structures from all the series being used in our laboratories for screening against malaria and Leishmania parasites. The database consisted of ∼1000 structure entries, of which 70% were new to ChemSpider at the time of screening. This in-house library was subjected to high throughput virtual screening (HTVS), followed by standard precision (SP) and then extra precision (XP) docking (Schrodinger LLC 2021). The ligand strain and complex energy of top hits were calculated by Molecular Mechanics Generalized Born Surface Area (MM/GBSA) method. Promising hit compounds (n = 40) specifically binding to 3CLpro with high energy and average MM/GBSA scores were then subjected to (100-ns) MD simulations. Using this sequential selection followed by an in-silico validation approach, we found a promising HEA-based compound (N,N'-((3S,3′S)-piperazine-1,4-diylbis(3-hydroxy-1-phenylbutane-4,2-diyl))bis(2-(5-methyl-1,3-dioxoisoindolin-2-yl)-3-phenylpropanamide)), which showed high in vitro antiviral activity against SARS-CoV-2. Further to reduce the size of the otherwise larger ligand, a pharmacophore-based predicted library of ∼42 derivatives was constructed, which were added to the previous compound library and rescreened virtually. Out of several hits from the predicted library, two compounds were synthesized, tested against SARS-CoV-2 culture, and found to have markedly improved antiviral activity.
1. Introduction
A novel coronavirus, SARS-CoV-2 was first reported in Wuhan, China in 20191, 2, 3. This pandemic has now spread to almost every country, infecting over 137 million, and killing more than 2.95 million individuals4, 5. The pathogen spreads rapidly from person to person in familial, clinical, and community settings4, 6. Common symptoms of the resulting disease, termed COVID-19, include fever, cough, shortness of breath, diarrhea, and fatigue, with more severe symptoms including atypical pneumonia and stroke3, 7, 8. SARS-CoV-2 is a member of the Coronaviridae family with 7 known human variants (HCoVs) belonging to alpha‐and beta‐coronaviruses: HCoV‐229E, HCoV‐OC43, HCoV‐NL63, HCoV‐HKU1, MERS‐CoV, SARS‐CoV, and SARS‐CoV‐2. The COVID-19 situation continues to evolve rapidly, with the high volume of cases and nosocomial transmission resulting in great strain on global healthcare systems. Importantly, SARS-CoV-2 transmission also occurs in asymptomatic individuals with varied viral loads2, 6. The virus has been detected in blood, oral, and anal specimens, suggesting that it can be shed through various body fluids resulting in transmission through respiratory droplets and fecal-oral transmission9. The SOLIDARITY consortium trial by World Health Organization (WHO), which sourced data from multiple countries concluded no change in mortality rates of COVID-19 treated with either remdesivir, hydroxychloroquine, lopinavir/ritonavir, or interferon regimens10, 11. Further, the vaccines boasting 90–100% efficacy have much lower efficacy in the elderly population (greater than75yrs)12 which has 10–15 times higher mortality than the general population due to COVID-1913. Dexamethasone is the only therapeutic potentially shown to reduce mortality among vulnerable populations14. Trials with Curcumin, Vitamin-D, and heparin have shown mixed results15. Therapeutics like ivermectin are ineffective at FDA-approved dosage and toxic at higher doses16. The use of non-SARS-CoV-2 specific antivirals with unknown efficacies, like Oseltamivir, Nelfinavir, and others, has been widely criticized as false re-purpousing17, 18. Successful and late-stage clinical trial vaccines have proven to be both a great relief and a harbinger of renewed fears of mutant variants19, 20, 21.
As per the viral cycle, upon entry into the host cell, the incoming viral genome is initially translated to produce two large precursor polyproteins 1a (pp1a) and 1ab (pp1ab), that are processed by ORF 1a-encoded viral proteinases, papain-like protease (PLpro), and the 3-chymotrypsin-like cysteine protease (Mpro or 3CLpro) into 16 mature non-structural proteins (NSP1-NSP16)22. Many of the NSPs perform essential functions in viral RNA replication and translation22, 23, 24. The 3CLpro enzyme, a.k.a. non-structural protein 5 (nsp5), self-cleaves the parent polypeptide at its n-terminal (nsp4/5) and c-terminal (nsp5/6) and is liberated to form a homodimer. This homodimer further cleaves the polyprotein using its catalytic dyad that contains the catalytic residues Histidine 41 (His-41) and Cysteine 145 (Cys-145)25. The 3CLpro of SARS-CoV-2 is 96.1% identical to that of SARS-CoV-1, with a 99% similarity suggesting the highest conservancy than any other target within the SARS-CoV-2 genome. However, the inhibitor profile greatly differs due to residue 46 – a serine in SARS-CoV-2 and an alanine in SARS-CoV-1 – which slightly shifts its substrate efficacy, which in turn changes the inhibitor binding affinity26. With the availability of high-resolution X-ray crystal structures, there are numerous successful examples of the implementation of computer-aided drug discovery (CADD) and rational improvement of antiviral activity against 3CLpro27, 28, 29, 30.
Previously, our group implemented a high-throughput CADD approach and screened a pooled library of 15,360 compounds, enabling the discovery of hits that bind to 7 different SARS-CoV-2 targets24. Subsequent screenings of synthetic compounds led to novel hits derived from hydroxyethylamine (HEA) scaffolds against 3CLpro31. We have been able to improve these derivatives/scaffolds based on pharmacophore identification, as well as rational ligand improvement, to generate predicted analogs with improved potency for each series. Using this approach, the designed and synthesized derivatives outperform the initial hits in the antiviral activities. Most virtual screening pipelines are dependent upon docking algorithms that calculate approximations to estimate the binding energy between the drug and target, but our approach employs integrated Molecular Mechanics Generalized Born Surface Area (MM-GBSA) post-processing to shortlist the most viable candidates. MM-GBSA32 accounts for strain on both the ligand and receptor within the complex, considering energies such as Coulomb, covalent-covalent binding, Van-der-Waals, lipophilic, generalized Born electrostatic solvation, and corrections such as hydrogen-bonding, pi-pi packing, and self-contact. Thus, the final hits from our screening gave us a comprehensive binding map for hit ligand improvement. Here, multiple chemotype integrative rational drug design, integrated ligand, and structure-based pharmacophore models were developed for each target hit pool using PharmaGist, with the consensus pharmacophore represented using polar, charged, and hydrophobic (PCH) moieties. The mapping of representative compounds from the synthetic compound libraries of HEA analogs and defined pharmacophores facilitated the integration of new features. The resulting derivative pool was validated through a virtual screening pipeline for 3CLpro. A marked improvement in the biological activity of derivatives against proteases. HEA derivatives have shown a high affinity toward several proteases, including 3CLpro of SARS-CoV-231, 33. SAR analysis demonstrated the importance of robust chemical scaffolds such as HEA in designing and synthesizing inhibitors with potential antiviral efficacy. Molecular dynamics simulation (MDS) studies carried out for 100 ns confirmed the stability of the HEA-enzyme complexes in the active site of 3CLpro34, 35. These studies, centered on the etiologic agent of COVID-19, are a continuation of our commitment to discovering new therapies for infectious diseases by targeting key enzymes33, 36, 37, 38.
2. Result and discussion
2.1. Mutation analysis
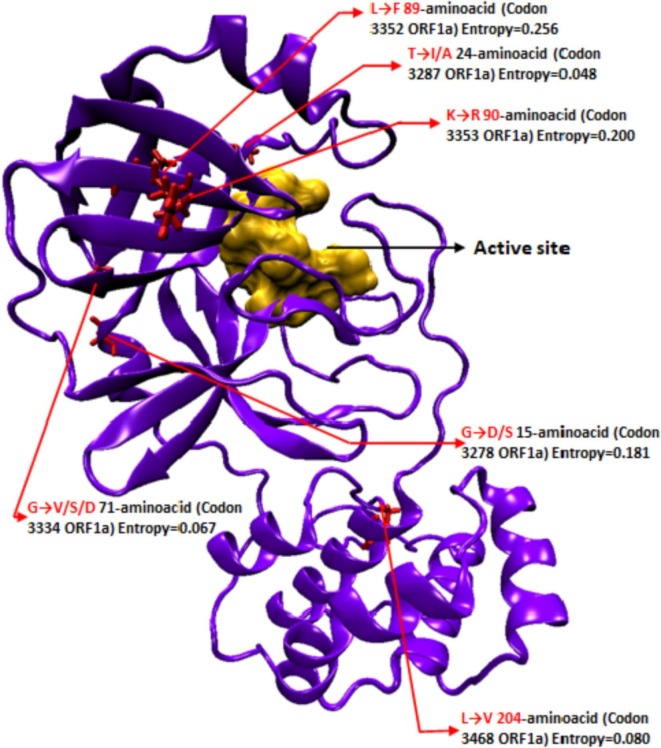
The mutations reported from 35,000 SARS-COV-2 genomes sequenced worldwide (GISAID) analyzed by the Next-strain server (Nextstrain.org) were mapped w.r.t. the active site. Not only did the mutations reported have low entropy, but their locations were also away from the active site. Moreover, most were biochemically synonymous (Fig.1 ). The results show that the 3CLpro is highly conserved, owing to its essential function, and that mutations within the ligand-binding site can have a high cost, reducing the chances of resistance and making it an ideal drug target.
Fig. 1.
Schematic representation of relative locations of mutations in 3CLpro w.r.t the active site; Orange, active site surface model; red, mutant sites; Purple, protein backbone. Active site amino acid residues i.e. 39, 41, 145, 163, 164, and 165. Mutations are labeled with Amino acid substitution, amino acid position in 3CLpro peptide, and codon position in parent ORF1a with current global mutational frequency in form of entropy (GSAID). Mutations with very low frequency (Entropy < 0.05; 3955 genomes sequenced) were not included. Note: None of the mutations are in proximity of the active site/substrate binding region used as the receptor grid zone for the virtual screening of the compound libraries.
2.2. Docking and MD simulations
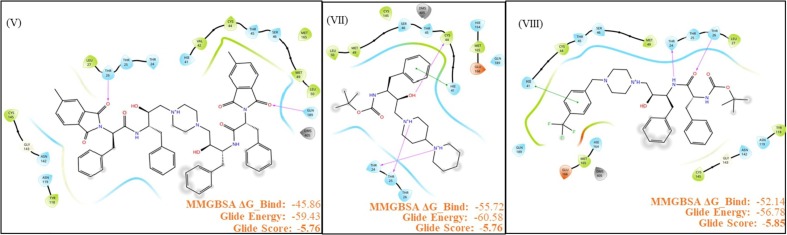
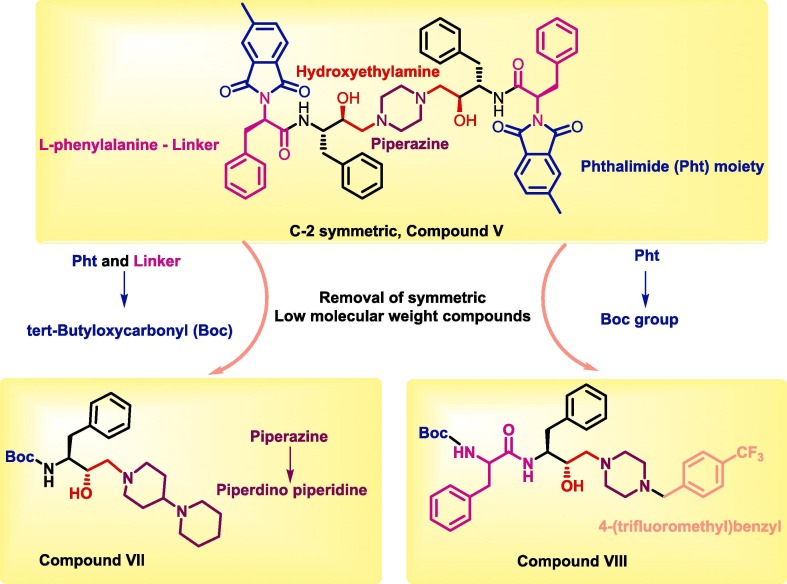
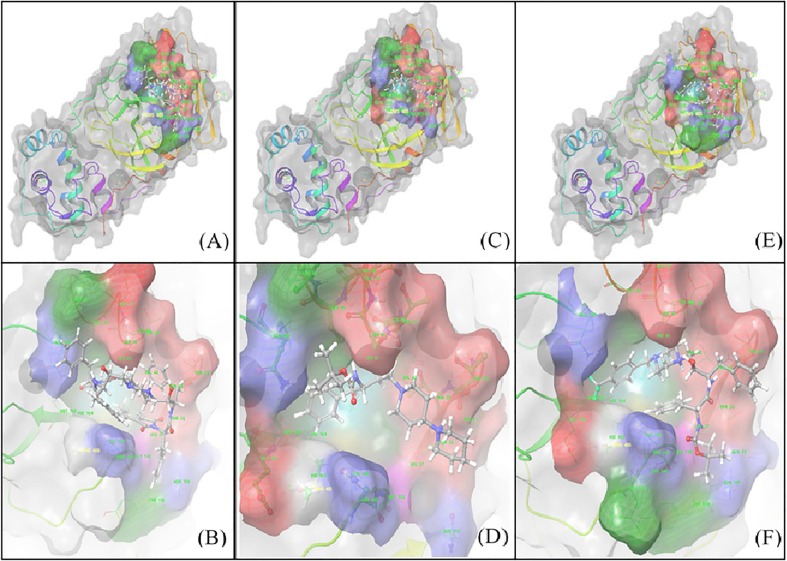
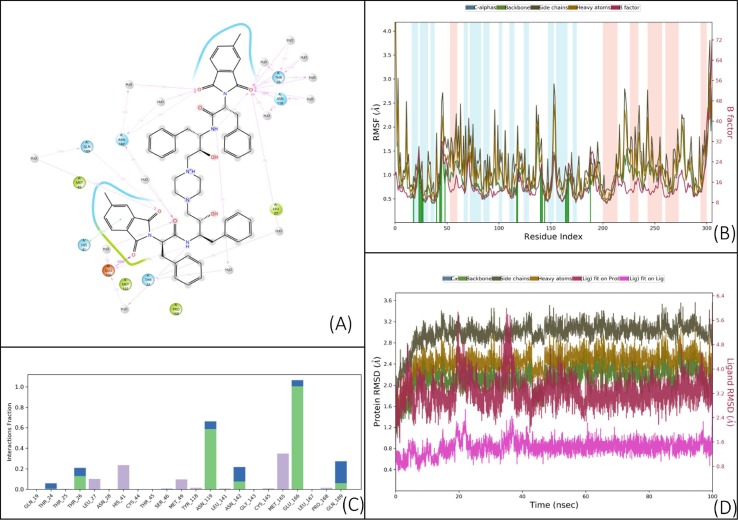
Compound V, which possesses C2 symmetry, exhibited several interactions during molecular docking analysis with enzyme 3CLpro of SARS-CoV-2. The C O group present in the phthalimide (Pht) moiety of the compound showed three strong H-bond interactions with the amino acid residues Gln_189, Thr_26, and Ser_46, while the methyl group present in the Pht moiety showed alkyl interactions with the residues Cys_44, Cys_145, His_41, Leu_27, and Met_49 (Fig.2 ). The other notable interaction was a pi-pi interaction between the aromatic ring of Pht moiety and the residue Met_49. All significant interactions exhibited by compound V were mainly due to the presence of Pht moiety, whereas other pharmacophores present, such as piperazine, HEA, and the amino acid linker (l-phenylalanine), did not display notable interactions. Thus, to explore the potential of other pharmacophores in mediating and perhaps improving enzyme-ligand interactions, compound V was rationally improved via removal of the C-2 symmetry and exchange of key pharmacophores, including replacement of the piperazine moiety of V with a piperidino piperidine and replacement of both the Pht and linker moiety with a Boc group. These modifications resulted in compound VII, as shown in Fig.3 . These rational substitutions helped improve the docking pose of compound VII with enzyme 3CLpro of SARS-CoV-2, as the piperazine in compound V did not show any significant interaction, but piperidino piperidine of compound VII displayed two strong H-bond interactions with residues Thr_24 and Thr_25. The Boc group also showed an important H-bond interaction with residue Ser_46, while the hydroxy group of HEA moiety exhibited strong H-bond interactions with Cys_44. The presence of only one aromatic ring in the molecule also proved beneficial, as it exhibited four pi-pi interactions with the residues Cys_145, His_41, Met_49, and Met_165, in contrast to the single pi-pi interaction of compound V (Fig.2).
Fig. 2.
Schematic representation of 2D interaction maps of MM-GBSA docking outputs of primary hit V and top hits from rationally improved library VII and VIII. Insets are the scores from XP and MM-GBSA energy calculations. The trend in MM-GBSA energy directly correlates with in vitro antiviral activity. Glide energy was much higher for large molecule V.
Fig. 3.
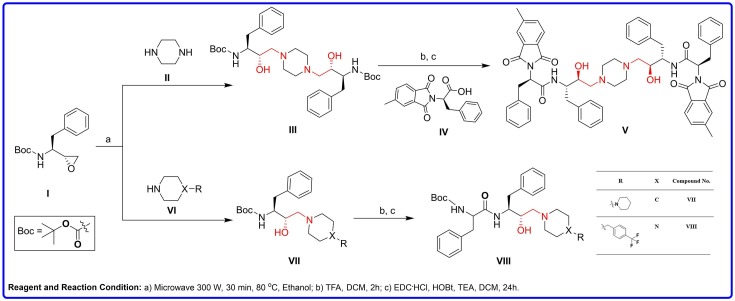
Schematic representation of rational improvement of the parent hit C-2 symmetric compound V to asymmetric and low molecular weight compounds VII and VIII.
Additional rational improvement of compound V was carried out by again removing the C-2 symmetry, replacing the Pht moiety with a Boc group, and introducing a 4-(trifluoromethyl)benzyl to the piperazine ring, while other pharmacophores were kept unchanged viz. the HEA and amino acid linker (Fig.3). In the resulting molecule, compound VIII, the newly introduced 4-(trifluoromethyl)benzyl showed three pi-pi interactions with His_41, Met_49, and Met_165. Also, the trifluoromethyl group present in the moiety exhibited strong H-bond interactions with the residues Gln_189 and Glu_166, as well as a halogen interaction with Glu_166. The presence of amino acid linker (l-phenylalanine) without Pht group also proved beneficial, as the aromatic ring of the linker exhibited a pi-pi interaction (Cys_145), while the C O and N—H group of the moiety displayed strong H-bond interactions with the residues Thr_26 and Thr_24, respectively. Overall, the rational improvement of V to compounds VII and VIII is supported by the observation of improved and enhanced interactions. Rational improvement maintained or even increased the interacting amino acid diversity, and the coverage of molecules involved in an interaction was also significantly improved. Although compound V had a high Glide score, suggesting poor binding, potentially due to its large size, it did evince multiple contacts during MM-GBSA analysis.
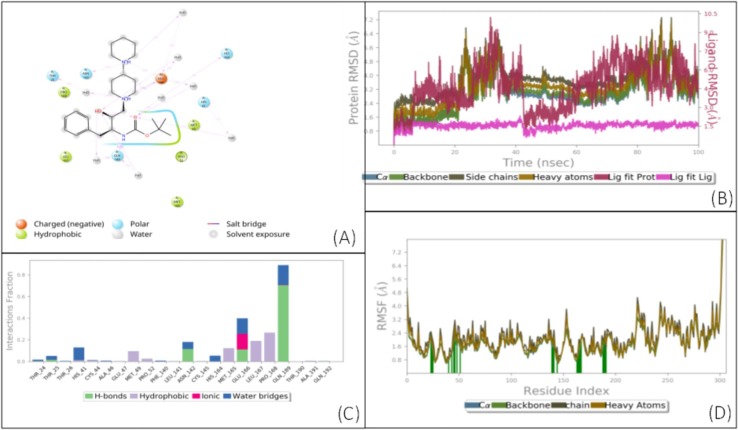
However, a large portion of this molecule was not involved in docking, and MM-GBSA analysis also showed high strain on the docked ligand resulting in ligand as well as receptor strain. This was also seen in the MD simulation, where the complex seemed highly stable throughout the simulation, but consistent fluctuation was observed throughout the simulation (Fig. 4, Fig. 5 ). These data demonstrate that compound V was an ideal initial 3CLpro binding probe whose interactions with the enzyme helped us to understand the active site and potential druggability. While the Glide scores did not improve significantly for compounds VII and VIII, their higher MM-GBSA scores indicate lower ligand strain, and MD simulations show better stability of the complex as the complex stabilized in time (Fig. 6, Fig. 7 ). VIII seems to have more movement within the active site due to its relatively small size and the large active site of 3CLpro, which has a macromolecular substrate. In addition, as these molecules have strong interactions with catalytic Cys145 and with Cys41, which is part of the substrate-binding domain. As such, both the leads could potentially be modified with reactive groups, such as acrylonitrile, to form a reversible covalent inhibitor series, which might be an optimal solution to small molecule wobble in a relatively spacious active site. Therefore, both of these molecules could be starting points for the development of a novel 3CLpro inhibitor series.
Fig. 4.
Schematic representation of 3D interaction maps of MM-GBSA docking outputs of primary hit V (A) & (B) and top hits from rationally improved library VII (C) & (D) and VIII (E) & (F). Both VII and VIII seem to retain binding characteristics of V but are much smaller and have better binding. This represents a successful rational improvement pipeline.
Fig. 5.
Results of a 100 ns (ns) MD simulation of compound V. (A) Schematic 2D representation of bound ligand interactions throughout the simulation. (B) Root mean square fluctuation between the binding site of the target protein and interacting ligand. (C) Critical protein–ligand contacts of amino acid side chain residues with the interaction properties. (D) Root mean square deviations difference between the Main protease (3CLPro) and bound ligand V (<4 Å). The graph was obtained for the RMSF value of ligand (purple line) from the protein backbone (green line). While the ligand was tightly bound to the active site throughout the simulation due to multiple interacting amino acids. The uniform spikes throughout simulation point to the maintenance of strain on both ligand and receptor throughout long simulations. This could be due to strain overpowered by interactions. The large interaction interface does help to map out the binding properties of the target site.
Fig. 6.
Results of a 100 ns (ns) MD simulation of compound VII. (A) Schematic 2D representation of bound ligand interactions throughout the simulation. (B) Root mean square fluctuation between the binding site of the target protein and the interacting ligand. (C) Critical protein–ligand contacts of amino acid side chain residues with the interaction properties. (D) Root mean square deviations difference between the Main protease (3CLPro) and bound ligand VII (<4 Å). The graph was obtained for the RMSF value of ligand (purple line) from the protein backbone (green line).
Fig. 7.
Results of a 100 ns (ns) MD simulation of compound VIII. (A) Schematic 2D representation of bound ligand interactions throughout the simulation. (B) Root mean square fluctuation between the binding site of the target protein and the interacting ligand. (C) Critical protein–ligand contacts of amino acid side chain residues with the interaction properties. (D) Root mean square deviations difference between the Main protease (3CLPro) and bound ligand VIII (<4 Å). The graph was obtained for the RMSF value of ligand (purple line) from the protein backbone (green line). The docked complex quickly stabilized to a very low energy state in 8–9 ns. After that, the ligand was highly stable throughout the simulation except for getting dislodged at 83 ns due to the trifluoride group being pulled by a distant Glu166. The altered state has a rapid decrease in energy and highlights the docking stability as well as the scope of improvement to further strengthen the interactions.
The molecule remained in the active site throughout the simulation. But there was a wobble i.e. movement of the ligand within the active site. Interestingly the change in docking position increases the energy of the receptor too and the initial docking position was re-established due to being energetically favorable. Based on the significantly higher antiviral activity of compound VII, this wobble seems to increase the engagement time of otherwise reversible small inhibitors also it may have a better competitive advantage to the substrate due to interference with more amino acids than its nascent docking due to the wobble within the site.
2.3. In silico ADMET and cytotoxicity evaluations
All the compounds tested in vitro were non-toxic to VERO cell lines used for antiviral testing based on nuclei counts, except for the control drug Ivermectin, which showed a reduction in live VERO cells at 50 µM range. Additionally, in silico absorption, distribution, metabolism, excretion, and toxicity (ADMET) analysis showed no toxic moieties in the ‘Quikprop’ module of the Schrodinger suite, which predicts compound toxicities based on known structural fragment sub-moieties.
2.4. Compound synthesis
The parent compound, V was previously explored as an antiplasmodial agent by our group38 and was resynthesized following slightly modified methods40. Microwave-assisted enantioselective40, 41 ring-opening of (2R,3S)-N-Boc-3-amino-1,2-epoxy-4-phenylbutane (I) with piperazine (II) was carried out in ethanol at 300 W by heating to 80 °C with a 2 min ramp and holding for 30 min to afford di-tert-butyl ((2S,2′S,3S,3′S)-piperazine-1,4-diylbis(3-hydroxy-1-phenylbutane-4,2-diyl))dicarbamate (III) (Scheme 1 ). Deprotection was accomplished with excess of trifluoroacetic acid (TFA) in dichloromethane, and the obtained products were used for coupling reaction with (R)-2-(5-methyl-1,3-dioxoisoindolin-2-yl)-3-phenylpropanoic acid (IV) that afforded (2R,2′R)-N,N'-((2S,2′S,3S,3′S)-piperazine-1,4-diylbis(3-hydroxy-1-phenylbutane-4,2-diyl))bis(2-(5-methyl-1,3-dioxoisoindolin-2-yl)-3-phenylpropanamide) (V) in 59% yield. The chemical structure was characterized with 1H NMR and high-resolution mass spectroscopy (HRMS) spectroscopy (Figure S1 and S2; supporting information) and found in good agreement with previously reported data33.
Scheme 1.
Synthetic pathways for synthesis of potent analogs (V, VII-VIII), showing good inhibitory activity against 3CLpro protein of SARS-CoV-2.
Further, rational structural improvements were performed for compound V to obtain low molecular weight and compact compounds (VII-VIII). During rational improvement and virtual screening of HEA-based library against 3CLpro enzyme of SARS-CoV-2, two compounds (VII-VIII) exhibited good binding affinity. Both VII (Yield = 93%) and VIII (Yield = 67%) were synthesized following the reported procedures33, 39, 42 and their chemical composition was confirmed by 1H, 13C and 2D NMR and HRMS spectroscopic techniques (Figure S3-S10). Further, the purity of compound VIII was determined using HPLC (High Performance Liquid Chromatography) water (A) and acetonitrile (B) as mobile phases with a flow rate of 0.3 mL/min. The mobile phase was started at 60% acetonitrile (0.01% TFA) (Mobile Phase B) to 100% B, with a total run time of 30 min. The purity was found to be 99.8% as illustrated in Figure S11.
2.5. Anti-SARS-CoV-2 activity
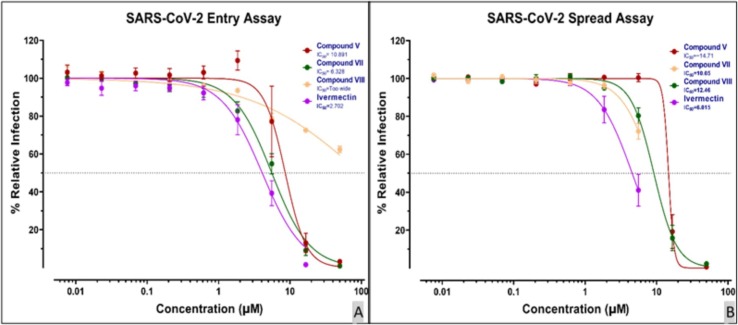
The compounds showed highly potent anti-SARS-CoV-2 activity with <15 µM IC50s (Fig.8 ). The activities of smaller compounds VII and VIII were higher than that of the bulky parent lead compound V. In particular, compound VII exhibited an IC50 value of 10.50 μM in the spread assay. This demonstrates a successful improvement pipeline of targeted drug discovery. As we have observed in our previous studies43, 23, once the 3CLpro is sufficiently expressed even after inhibition of virus, the multiplication cycle continues due to the presence of a small fraction of uninhibited enzyme. Therefore, compound activities are better observed in viral entry assays when initial blocking of freshly expressed 3CLpro occurs and succeeds in blocking the viral cycle more effectively.
Fig. 8.
(A) SARS-CoV-2 antiviral entry assay. Synchronized infections were conducted for viral entry as described in Materials and Methods. Each curve shows a dose–response to the indicated 4 tested compounds (Color-coded; key inset). (B) SARS-CoV-2 antiviral spread assay. Synchronized infections were conducted for a viral spread as described in Materials and Methods. Each curve shows a dose–response to the indicated 4 tested compounds (Color-coded; key inset). The results are presented as the PFU formed in the presence of the drug as a percentage of the PFU formed and each plotted value is the mean with ± standard deviations of an experiment performed in triplicate.
2.6. Anti-3CLpro enzymatic activity
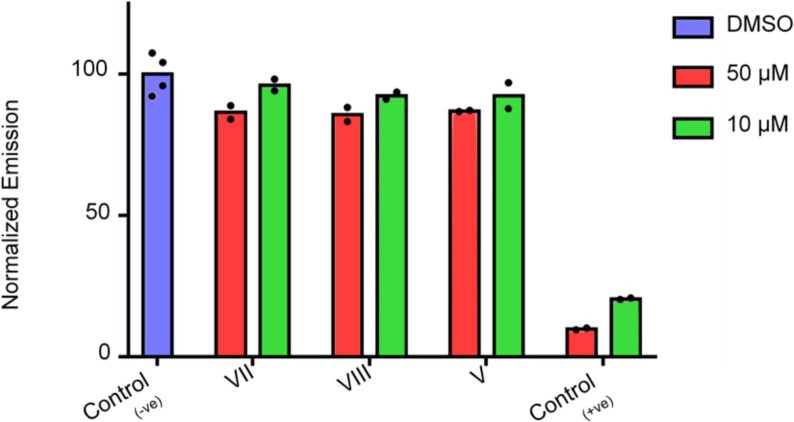
As the compounds tested are competitive inhibitors of the 3CLpro enzyme there was a concentration-dependent activity of the compounds against 3CLpro. There was an average of 5% and 12% inhibition at concentrations of 10 µM and 50 µM, respectively (Fig.9 ). Compared to antiviral activity, the enzyme inhibition activity seemed to be very low. This low activity could be attributed to much higher 3CLpro activity compared to in vivo conditions. In addition, the substrate used was a small peptide compared to the natural large protein substrate. This smaller substrate could be competitively replacing/blocking inhibition by inhibitors due to its small size and abundance. The positive control shows 85% and 90% inhibition at concentrations 10 µM and 50 µM, respectively, but it is incomparable to HEA compounds as ML18829, 44 (16-(R), (R)-N-(4-(tert-butyl)phenyl)-N-(2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl)furan-2-carboxamide, Pubchem CID: 46897844) is an non-covalent inhibitor of the SARS-CoV 3CLpro enzyme. Also, ML188 has moderate in vitro anti-SARS-CoV-2 activity (22 µM) and has a nonspecific non-covalent binding profile blocking PLpro too44. Therefore, while these compounds seem to underperform in the in vitro enzymatic assays, they do show concentration-dependent activity, suggesting specificity and probability of being better competitive inhibitors with large natural substrates. These results highlight both superiority of covalent inhibitors as well as the shortcoming of in vitro enzymatic assays in in mimicking in vivo 3CLpro activities.
Fig. 9.
(A) SARS-CoV-2 3CLpro enzymatic assay. Percent inhibition of 3CLpro activity with a FRET substrate as described in methods. DMSO treated samples are the negative controls and ML188 a known inhibitor44, 51 is a positive control. All compounds were tested at two concentrations of 10 µM and 50 µM in triplicates as described in detail in the methods section. There is a concentration dependant inhibition seen with all the HEA inhibitors though weak as compared to the positive control.
3. Conclusions
While drug compounds are typically produced for use against specific diseases, drug repurposing provides an approach to fast-track new treatments. Despite the persistent search for effective repurposed therapeutics by the medical community to treat COVID-19, clinical use of these now suggests mixed outcomes for their efficacies. This trend has prompted us to rationally design and synthesize target-specific therapeutics and to develop a pipeline to supply potent viral enzyme inhibitors to deal with rapidly spreading variants. As of today, no COVID-19 specific drugs have been approved, but through protein modeling, drug docking, and SAR studies, we have been developing specific derivatives targeting SARS-CoV-2 proteins, specifically 3CLpro. This study stems from a larger study in which we passed 1,000 known chemical structures from existing libraries, as well as hypothetical analogs, through CAAD-based HTVS to identify potent ligands and to define pharmacophores for target-based rational design and synthesis. Using this approach, we now have a continuous pipeline of new analogs to improve the synthesis of potential drug candidates targeting the critical viral protease 3CLpro. This focused experimental paradigm allowed us to employ CADD-based screening to identify HEA-based druggable scaffolds capable of advancing quickly to preclinical evaluation for COVID-19 treatment. When exploring the possible impact of mutations in the active site of the target, our mutational analysis found no changes with current known mutations in the proximity of the active site/substrate binding region used as the receptor grid zone for the virtual screening of the compound libraries. This suggests that rationally modified hits could maintain their potency even in the face of an evolving pathogen. In this study, we identified a lead compound that enabled us to probe the active site of 3CLpro and evaluate pharmacophore-enzyme interactions. Though this parent lead V had more interactions, its ligand strain was greater due to its larger size, and a large portion of this molecule was not involved in docking. The smaller compound VII showed a marked improvement in both energies, interaction coverage of ligand, and size. While compound VIII seemed to be more active than both V and VII, it still showed wobbly interaction due to the large active site of the target and hinge-like movement typical of proteases. Nonetheless, these two highly potent leads, both with nontoxic and drug-like properties, represent starting leads for a targeted inhibitor series with possible reversible covalent inhibitor derivatives. Future studies will need to evaluate these leads in animal models of SARS-CoV-2 to ascertain the therapeutic efficacy.
4. Experimental procedure
4.1. Ligand preparation
Ligands were redrawn from the chemical structure and exported as SDF in the PubChem draw applet45. As a background, a 1652 molecule library of FDA-approved compounds was used to prune low-level binders. The molecules were re-compiled in a chemical library in SDF format and prepared according to the requirements of the Schrodinger32 computer software and used for virtual screening as described below. Ligand processing followed by expansion of protonation and tautomeric states (7.0 + 2.0 pH units) was done by LigPrep along with Epik module35, 46. For each ligand, a total of five stereoisomers with minimum energy conformations were produced, and the lowest energy unique conformers with accurate chirality were used for virtual screening protocol.
4.2. Preparation of target receptor and active site
The 3D X-ray high-resolution crystal structure of SARS Cov2 NendoU protein was obtained from the RCSB PDB47, 48 with PDB IDs. The target protein was retrieved from the protein data bank (PDB, IDs: 5S6Z, 6X1B, 6X4I, & 7KEG) representing different amino acid side chain residue rotations of NendoU to accommodate macromolecular substrate. The target proteins were refined and prepared through the Protein Preparation Wizard, for all crystal structures only protein chains (A in case of multimer model) were preserved. Bond orders were assigned along with the addition of hydrogen atoms. The protein structures were further processed and analyzed to assess conformational stability. Steric clashes, hetero-atoms, and non-essential water molecules were scraped off, as well as hydrogen bonds and proper bond orders were assigned to crystal structure with the help of the Maestro Protein Preparation Wizard Workflow program32, and the default parameters were used. The glide grid was defined covering the active site of 3CLpro enzyme which comprises of a Cysteine–Histidine catalytic dyad (Cys-145 and His-41), in which the thiol side chain group of Cys-145 acts as a nucleophile that is most crucial for the proteolytic hydrolysis reaction. around these the docking receptor grid was generated. The electrostatic grid box was generated around the target receptor via Glide35. The dimensions of the receptor grid were kept as (in angstroms; Å): Inner-box: X:10, Y:10, Z:10, Outer-box: X:30, Y:30, Z:30, and Grid center at Cys293., and OPLS3e forcefield was employed for formal charges. Ligands were prepared with the Ligprep utility at default values and the OPLS_2005 forcefield for energy minimization. Virtual screening studies were performed using Glide. Glide from Maestro suite32 employs a hierarchical function that filters data and selects the most favorable interactions between a protein docking site and a ligand. The software was applied in the first stage using the Standard Precision (SP) mode followed by an Extra Precision (XP) mode which performs an advanced scoring, which in turn, results in an enriched calculation that minimizes false positives. The equation used to calculate the binding energy in the XP mode was: XP Glide Score = Ecoul + Evdw + Ebind + Epenatly where Ecoul and EvdW represent van der Waals and electrostatic terms, respectively. Ebind and Epenalty make reference to contributions that favor binding or penalization of interactions that influence the binding of a ligand. Further, the docked poses were subjected to MM-GBSA pruning and re-scoring with energy calculations.
4.3. MD simulations
The receptor-ligand complex system from top-scoring MM-GBSA output from virtual screening was processed, and side chains of different residues are refined, followed by the strain minimization using the protein preparation interface of Maestro32 before MD simulations. In the course of complex processing, all the missing hydrogen atoms of the system were added and the processed complex was then introduced to the system builder module of Desmond, encompassing ions and solvation tabs. Undesirable water molecules were scraped off from the crystal structure of the complex and the TIP3P water model system was chosen for the solvation of the 3D complex utilizing solvation panel32, 49. Optimized potentials for liquid simulations (OPLS_2005) force-field was utilized for amino acid interaction, encompassing TPI3P explicit solvent archetype for docked complex and orthorhombic water box of dimension (10_10_10 Å). The MD simulations were performed for 100 ns unless otherwise stated and the trajectory (50FPS) was analyzed for intramolecular interactions between protein and ligand.
5. Chemistry
5.1. General
The chemicals and solvents were used as it is without further purification after purchasing. Ethanol (absolute) was purchased from Changshu Hongsheng Fine Chemical Co., Ltd. (Jiangsu, China). Piperazine (CAS No. 110–85-0), 1,4′-bipiperidine (CAS No. 4897–50-1), Boc-l-phenylalanine (CAS No. 13734–34-4) were purchased from Spectrochem Pvt. Ltd. (Mumbai, India). 1-(4-(trifluoromethyl)benzyl)piperazine (CAS No. (107890–32-4) from TCI Chemicals (Hyderabad, India) and 2R,3S)-N-Boc-3-amino-1,2-epoxy-4-phenylbutane (CAS No. 98760–08-8) was procured from GLR Innovation (New Delhi, India). The epoxide ring-opening reactions were carried out in the “Start Synth Microwave Synthesis Labstation” microwave for organic synthesis. The nuclear magnetic resonance (NMR) spectra were recorded in parts per million (ppm) downfield from internal standard tetramethylsilane (TMS) using a JEOL ECX-400P NMR Spectrometer. The chemical composition of all new compounds was confirmed by a high-resolution Biosystems Q-Star Elite time-of-flight electrospray mass spectrometer. The melting point of compound (VIII) was measured in a “BUCHI Labortechnik AG CH-9230”. HPLC purity of the compound (VIII) was analyzed on a Gilson HPLC semi-preparative system using Shodex C-18 column (4.6 mm × 250 mm, 5 µm). The compounds (V, VII, and VIII) were synthesized following optimized methods by our group.33, 50
General experimental procedure. Briefly, a 50 mL round-bottomed was charged with 3.8 mmol of (2R,3S)-N-Boc-3-amino-1,2-epoxy-4-phenylbutane (I), amines (3.8 mmol), 5 mL of ethanol, and the contents were stirred under microwave irradiation for 30 min at 300 W/80 °C. The reaction mixture was then allowed to attain room temperature, and the solvent was concentrated under reduced pressure. Thus, obtained crude product was recrystallized from ethyl acetate and hexane (1:9) and used for the next steps. In the next step, a 100 mL round-bottom flask, the obtained BOC protected intermediate (2.0 mmol) was dissolved in 20 mL of dichloromethane (DCM) and treated with trifluoracetic acid (TFA) (3 mL, 15% of DCM) slowly. The reaction mixture was stirred at room temperature for 4 h and after completion of the reaction, excess of DCM and TFA were removed under reduced pressure. The reaction mixture was adjusted pH, 8–9 using 1 N NaOH, and then extracted with ethylacetate (3x25 mL) and washed with brine solution (3x15 mL). The organic layer was dried over anhydrous sodium sulfate and the excess of ethylacetate was removed under reduced pressure to afford the deprotected intermediates, which was used as such for the next step of the procedure. Next, acid (1.5 mmol) and triethylamine (TEA) (4.5 mmol) was dissolved in 20 mL of DCM and the contents were stirred for 30 min at room temperature, followed by the addition of 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC·HCl) (3.0 mmol). After 30 min. stirring, hydroxybenzotriazole (HOBt) (3.0 mmol) was added followed by the addition of deprotected intermediate (1.0 mmol) at 0 °C. The contents were initially stirred at 0 °C for 30 mins. And then at room temperature for 24 h. After completion of the reaction, excess DCM was removed under reduced pressure and the final coupling product was extracted with ethylacetate (3x25 ml). The organic layer was dried over anhydrous sodium sulfate, the solvent was removed, and the product was purified by column chromatography (70: 30, hexane: ethylacetate).
5.2. Spectroscopic data for compound VIII
tert-Butyl (1-(((2S,3S)-3-hydroxy-1-phenyl-4-(4-(4-(trifluoromethyl)benzyl)piperazin-1-yl)butan-2-yl)amino)-1-oxo-3-phenylpropan-2-yl)carbamate (VIII):
Yield, 67%; melting point (m.p.) 112–114 °C, 1H NMR (400 MHz, CDCl3) δ 7.54 (d, J = 8.1 Hz, 2H), 7.40 (d, J = 8.0 Hz, 2H), 7.30 – 7.16 (m, 9H), 7.13 (d, J = 6.8 Hz, 2H), 6.46 (d, J = 9.1 Hz, 1H), 4.78 (d, J = 6.3 Hz, 1H), 4.25 (d, J = 6.8 Hz, 1H), 3.99 (dd, J = 16.8, 8.1 Hz, 1H), 3.61 (dd, J = 10.8, 3.2 Hz, 1H), 3.49 (s, 2H), 3.07 – 2.94 (m, 1H), 2.94 – 2.80 (m, 3H), 2.43 (dd, J = 72.9, 18.3 Hz, 8H), 2.18 – 2.03 (m, 2H), 1.81 (s, 2H), 1.39 (s, 9H); 13C NMR (101 MHz, CDCl3) δ 171 (s), 142.4 (s), 138.1 (s), 136.6 (s), 129.7 – 129 (m), 128.8 (s), 128.5 (s), 127.1 (s), 125.3, 65.1 (s), 62.4 (s), 53.2 (s), 39 (s), 28.3 (s).
5.3. In vitro antiviral activity assay
In vitro activity assays against SARS-CoV-2 were performed at the United States Army Medical Research Institute for Infectious Diseases (USAMRIID). ATCC Vero E6 cells infected with the SARS-CoV-2 virus were used for performing both entry and spread assays.
5.3.1. Entry assay
Vero E6 cells, in 96 well plates, were pre-treated with compounds starting at a concentration of 50 µM with 3-fold dilutions down to 0.76 nM for approximately 1 h at 37 °C. SARS-CoV-2/MT020880 has then added to the compound-treated cells for 1 h at 37 °C at an MOI of 0.4. After 1 h, the cells were washed with PBS before adding additional compounds back in fresh culture media to the cells for 24 h at 37 °C. Cell media removal, washing of plates, detection of infected cells, and data analysis were performed exactly in the same manner as per reported literature 24.
5.3.2. Spread assay
A similar protocol was utilized for spread assay analysis as described above with little modifications in which virus was used at an MOI of 0.02 and the assay was incubated for 48 h.24
5.4. Validation of 3CLpro enzyme assay with a known 3CLpro inhibitor
From a 10 mM DMSO compound stock, 50 µL aliquots of a known inhibitor ML188 (N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamide; ()) were prepared in assay buffer (Tris-HCl pH 7.3, 1 mM EDTA) at 250 µM, 50 µM, and 5 µM (50 µM, 10 µM, and 1 µM final). Initially, 10 µL of each compound dilution in triplicate was added to a 96-well black wall, clear bottom plate (Corning). No inhibitor control wells in triplicate with 10 µL assay buffer were also prepared. In each well, 20 µL of 1 µM 3CLpro enzyme (0.4 µM final) in the assay buffer was added. Compounds and 3CLpro were incubated at room temperature for 5 min, then 20 µL of 100 µM TVLQ-methyl-amino coumarin (AMC) probe substrate (40 µM final) were added. No enzyme controls were prepared in triplicate with 30 µL assay buffer and 20 µL of 100 µM TVLQ-AMC probe substrate. A multichannel pipette was used for the addition of all components for ease and reproducibility. Reactions were then measured immediately after probe substrate addition for fluorescence emission intensity (excitation λ: 364 nm; emission λ: 440 nm) on a plate reader (i.e. Synergy Neo2 Hybrid). Emission was monitored every 15 min for 1 h and at 3 h post substrate addition.
After all measurements were complete, the average of the no inhibitor control wells at 3 h was calculated. All measurements were normalized to this average, such that the no inhibitor control average is 100 [normalization = (emission/no inhibitor average)*100].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Part of this work was supported by Department of Science & Department of Science & Technology (DST) under BRICS multilateral project (DST/INT/BRICS/ COVID-19/2020). Authors are thankful to the Department of Medicine, Loyola University Medical Center, and Stritch School of Medicine for supporting the Drug Discovery Program and the High Computing Platform. This project was also supported by the Research Funding Committee (RFC), Loyola University Chicago (PK, LU#213627). Opinions, discussions, conclusions, interpretations, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army. The mention of trade names or commercial products does not constitute endorsement or recommendation for use by the Department of the Army or the Department of Defense. SK, CU and NS are thankful to CSIR, New Delhi for providing senior research fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmc.2021.116393.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Maciorowski D, Idrissi SZE, Gupta Y, et al. A Review of the Preclinical and Clinical Efficacy of Remdesivir, Hydroxychloroquine, and Lopinavir-Ritonavir Treatments against COVID-19. SLAS DISCOVERY: Advancing the Science of Drug Discovery. Published online 2020:2472555220958385. [DOI] [PMC free article] [PubMed]
- 2.Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Coronavirus Disease (COVID-19) Dashboard. Accessed January 20, 2021. https://covid19.who.int.
- 5.Mahase E. China coronavirus: WHO declares international emergency as death toll exceeds 200. BMJ. 2020;368 doi: 10.1136/bmj.m408. [DOI] [PubMed] [Google Scholar]
- 6.Bai Y., Yao L., Wei T., et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14):1406. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holshue M.L., DeBolt C., Lindquist S., et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. Published online. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young B.E., Ong S.W.X., Kalimuddin S., et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323(15):1488. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y.i., Li X., Zhu B., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barratt-Due A, Olsen IC, Henriksen KN, et al. Evaluation of Remdesivir and Hydroxychloroquine on Viral Clearance in COVID-19 Patients: Results from the NOR-Solidarity Randomised Trial. Published online 2021.
- 11.Pan H, Peto R, Karim QA, et al. Repurposed antiviral drugs for COVID-19; interim WHO SOLIDARITY trial results. medRxiv. Published online 2020. [DOI] [PMC free article] [PubMed]
- 12.Chagla Z. The BNT162b2 (BioNTech/Pfizer) vaccine had 95% efficacy against COVID-19≥ 7 days after the 2nd dose. Annals of Internal Medicine. Published online 2021. [DOI] [PubMed]
- 13.Raheja H, Chukwuka N, Agarwal C, et al. Should COVID-19 Patients> 75 Years be Ventilated? An Outcome Study. QJM: An International Journal of Medicine. Published online 2021. [DOI] [PMC free article] [PubMed]
- 14.Villar J., Ferrando C., Martínez D., et al. Dexamethasone treatment for the acute respiratory distress syndrome: a multicentre, randomised controlled trial. The Lancet Respiratory Medicine. 2020;8(3):267–276. doi: 10.1016/S2213-2600(19)30417-5. [DOI] [PubMed] [Google Scholar]
- 15.Trieu V, Saund S, Rahate PV, et al. Targeting TGF-β pathway with COVID-19 Drug Candidate ARTIVeda/PulmoHeal Accelerates Recovery from Mild-Moderate COVID-19. medRxiv. Published online 2021.
- 16.Schmith V.D., Zhou J., Lohmer L.R. Published online; Clinical Pharmacology & Therapeutics: 2020. The Approved Dose of Ivermectin Alone is not the Ideal Dose for the Treatment of COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liuzzo G, Patrono C. Re-purposed antiviral drugs without a purpose in COVID-19: a valuable lesson for clinicians. European Heart Journal. Published online 2021. [DOI] [PMC free article] [PubMed]
- 18.Foo CS, Abdelnabi R, Kaptein SJ, et al. Nelfinavir markedly improves lung pathology in SARS-CoV-2-infected Syrian hamsters despite a lack of an antiviral effect. bioRxiv. Published online 2021. [DOI] [PMC free article] [PubMed]
- 19.Garcia-Beltran WF, Lam EC, Denis KS, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. Published online 2021. [DOI] [PMC free article] [PubMed]
- 20.Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 Variants—Clinical, Public Health, and Vaccine Implications. N Engl J Med. Published online. 2021;384(19):1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maciorowski D., Mohama S., Alsawi M.A., et al. Molecular Insights into Severe Acute Respiratory Syndrome Coronavirus 2 Pathobiology: Dissecting the Interplay between Cellular Innate Immunity and Immune Evasion. Critical ReviewsTM in Immunology. 2020;40(6):485–496. doi: 10.1615/CritRevImmunol.2020035855. [DOI] [PubMed] [Google Scholar]
- 22.Snijder E., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta Y., Maciorowski D., Jones K., et al. Bisindolylmaleimide IX: A novel anti-SARS-CoV2 agent targeting viral main protease 3CLpro demonstrated by virtual screening and in vitro assays. Published online. 2020 doi: 10.1016/j.ymeth.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta Y., Maciorowski D., Zak S.E., et al. Bisindolylmaleimide IX: A novel anti-SARS-CoV2 agent targeting viral main protease 3CLpro demonstrated by virtual screening pipeline and in-vitro validation assays. Methods. Published online. 2021 doi: 10.1016/j.ymeth.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimoto F.K. A Biochemical Perspective of the Nonstructural Proteins (NSPs) and the Spike Protein of SARS CoV-2. The protein journal. Published online. 2021;40(3):260–295. doi: 10.1007/s10930-021-09967-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rut W., Groborz K., Zhang L., et al. SARS-CoV-2 M pro inhibitors and activity-based probes for patient-sample imaging. Nat Chem Biol. 2021;17(2):222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- 27.Amin S.A., Banerjee S., Singh S., Qureshi I.A., Gayen S., Jha T. First structure–activity relationship analysis of SARS-CoV-2 virus main protease (Mpro) inhibitors: an endeavor on COVID-19 drug discovery. Molecular diversity. Published online. 2021;25(3):1827–1838. doi: 10.1007/s11030-020-10166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai Y.u., Ye F., Feng Y., et al. Structural basis for the inhibition of the SARS-CoV-2 main protease by the anti-HCV drug narlaprevir. Signal transduction and targeted therapy. 2021;6(1) doi: 10.1038/s41392-021-00468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockbaum G.J., Reyes A.C., Lee J.M., et al. Crystal Structure of SARS-CoV-2 Main Protease in Complex with the Non-Covalent Inhibitor ML188. Viruses. 2021;13(2):174. doi: 10.3390/v13020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manandhar A, Blass BE, Colussi DJ, et al. Targeting SARS-CoV-2 M3CLpro by HCV NS3/4a Inhibitors: In Silico Modeling and In Vitro Screening. Journal of chemical information and modeling. Published online 2021. [DOI] [PubMed]
- 31.Kumar S, Sharma PP, Shankar U, et al. Discovery of new hydroxyethylamine analogs against 3CLpro protein target of SARS-CoV-2: Molecular docking, molecular dynamics simulation, and structure–activity relationship studies. Journal of chemical information and modeling. Published online 2020. [DOI] [PubMed]
- 32.Schrodinger P. Release 2020-2: Maestro. Schrodinger, LLC, New York, NY. Published online 2020.
- 33.Singh S, Rajendran V, He J, et al. Fast-acting small molecules targeting malarial aspartyl proteases, plasmepsins, inhibit malaria infection at multiple life stages. ACS Infect Dis. Published online December 17, 2018. doi:10.1021/acsinfecdis.8b00197. [DOI] [PubMed]
- 34.Release S. NY. Published online; Maestro-Desmond Interoperability Tools, Schrödinger, New York: 2017. 3: Desmond molecular dynamics system, DE Shaw research, New York, NY, 2017. [Google Scholar]
- 35.Small-Molecule S.L. Drug Discovery Suite. 2020;2020–1:2020. [Google Scholar]
- 36.Flynn D.L., Becker D.P., Dilworth V., et al. The herpesvirus protease: mechanistic studies and discovery of inhibitors of the human cytomegalovirus protease. Drug Des Discov. 1997;15(1):3–15. [PubMed] [Google Scholar]
- 37.Kumar P., Achieng A.O., Rajendran V., et al. Synergistic blending of high-valued heterocycles inhibits growth of Plasmodium falciparum in culture and P. berghei infection in mouse model. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-06097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma N., Gupta Y., Bansal M., et al. Multistage antiplasmodial activity of hydroxyethylamine compounds, in vitro and in vivo evaluations. RSC Adv. 2020;10(58):35516–35530. doi: 10.1039/d0ra03997g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S., Upadhyay C., Bansal M., et al. Experimental and Computational Studies of Microwave-Assisted, Facile Ring Opening of Epoxide with Less Reactive Aromatic Amines in Nitromethane. ACS Omega. 2020;5(30):18746–18757. doi: 10.1021/acsomega.0c01760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi L., Chen L., Chen R., Chen L. Synthesis of deuterium-labelled fosamprenavir calcium. Journal of Labelled Compounds and Radiopharmaceuticals: The Official Journal of the International Isotope Society. 2010;53(3):147–151. [Google Scholar]
- 41.Moschona F., Savvopoulou I., Tsitopoulou M., Tataraki D., Rassias G. Epoxide Syntheses and Ring-Opening Reactions in Drug Development. Catalysts. 2020;10(10):1117. [Google Scholar]
- 42.Singh A.K., Rajendran V., Pant A., et al. Design, synthesis and biological evaluation of functionalized phthalimides: a new class of antimalarials and inhibitors of falcipain-2, a major hemoglobinase of malaria parasite. Bioorg Med Chem. 2015;23(8):1817–1827. doi: 10.1016/j.bmc.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 43.Poonam, Gupta Y., Gupta N., et al. Multistage inhibitors of the malaria parasite: Emerging hope for chemoprotection and malaria eradication. Med Res Rev. 2018;38(5):1511–1535. doi: 10.1002/med.2018.38.issue-510.1002/med.21486. [DOI] [PubMed] [Google Scholar]
- 44.Jacobs J., Grum-Tokars V., Zhou Y.a., et al. Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J Med Chem. 2013;56(2):534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.PubChem. PubChem. Accessed July 20, 2020. https://pubchem.ncbi.nlm.nih.gov/.
- 46.Schrodinger L. Small-Molecule Drug Discovery Suite 2019-3. 2017. New York, NY: Schrodinger, LLC; 2019.
- 47.Rose P.W., Prlić A., Altunkaya A., et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic acids research. Published online. 2016:gkw1000. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bank RPD. RCSB PDB: Homepage. Accessed May 2, 2020. https://www.rcsb.org/.
- 49.Fan S, Iorga BI, Beckstein O. Prediction of octanol-water partition coefficients for the SAMPL6-$$$\backslash$log P $$ logP molecules using molecular dynamics simulations with OPLS-AA, AMBER and CHARMM force fields. Journal of Computer-Aided Molecular Design. Published online 2020:1-18. [DOI] [PMC free article] [PubMed]
- 50.Singh AK, Rathore S, Tang Y, et al. Hydroxyethylamine Based Phthalimides as New Class of Plasmepsin Hits: Design, Synthesis and Antimalarial Evaluation. PLoS One. 2015;10(10):e0139347. doi:10.1371/journal.pone.0139347. [DOI] [PMC free article] [PubMed]
- 51.Loffredo Madeline, Lucero Hector, Chen Da-Yuan, et al. The in-vitro effect of famotidine on sars-cov-2 proteases and virus replication. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-84782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.