Abstract
Purpose:
Improved risk stratification and predictive biomarkers of treatment response are needed for non–muscle-invasive bladder cancer (NMIBC). Here we assessed the clinical utility of targeted RNA and DNA molecular profiling in NMIBC.
Experimental Design:
Gene expression in NMIBC samples was profiled by NanoString nCounter, an RNA quantification platform, from two independent cohorts (n = 28, n = 50); targeted panel sequencing was performed in a subgroup (n = 50). Gene signatures were externally validated using two RNA sequencing datasets of NMIBC tumors (n = 438, n = 73). Established molecular subtype classifiers and novel gene expression signatures were assessed for associations with clinicopathologic characteristics, somatic tumor mutations, and treatment outcomes.
Results:
Molecular subtypes distinguished between low-grade Ta tumors with FGFR3 mutations and overexpression (UROMOL-class 1) and tumors with more aggressive clinicopathologic characteristics (UROMOL-classes 2 and 3), which were significantly enriched with TERT promoter mutations. However, UROMOL subclasses were not associated with recurrence after bacillus Calmette-Guérin (BCG) immunotherapy in two independent cohorts. In contrast, a novel expression signature of an inflamed tumor microenvironment (TME) was associated with improved recurrence-free survival after BCG. Expression of immune checkpoint genes (PD-L1/PD-1/CTLA-4) was associated with an inflamed TME, but not with higher recurrence rates after BCG. FGFR3 mutations and overexpression were both associated with low immune signatures.
Conclusions:
Assessment of the immune TME, rather than molecular subtypes, is a promising predictive biomarker of BCG response. Modulating the TME in an immunologically “cold” tumor warrants further investigation. Integrated transcriptomic and exome sequencing should improve treatment selection in NMIBC.
Translational Relevance.
We performed RNA-based profiling by NanoString nCounter on non–muscle-invasive bladder cancer (NMIBC) clinical specimens and found that a novel expression signature of an inflamed tumor microenvironment (TME), but not molecular subtyping, was associated with improved recurrence-free survival after bacillus Calmette-Guérin (BCG) immunotherapy. We further demonstrated that immune checkpoint gene expression was not associated with higher recurrence rates after BCG. These findings were externally validated in a large RNA sequencing dataset of NMIBC, suggesting our immune signature could be a robust predictive biomarker for BCG response and that an immunologically “cold” TME is a mechanism of resistance to BCG. Our results also raise concerns about treatment strategies combining BCG and immune checkpoint blockade in NMIBC and instead support approaches focused on modulating the TME. Our integrated transcriptomic and panel sequencing found FGFR3 overexpression and mutations to be associated with a “cold” TME, further supporting investigations into FGFR inhibitors for NMIBC.
Introduction
Current risk stratification for non–muscle-invasive bladder cancer (NMIBC) is inadequate and relies on clinicopathologic features, with limited ability to accurately predict which tumors are most likely to recur and/or progress to muscle-invasive disease (1). Hence, patients with NMIBC undergo intensive surveillance with frequent, invasive cystoscopies, rendering bladder cancer among the most expensive cancers to manage (2). Thus, there is a critical need for prognostic and predictive biomarkers in NMIBC. Most urgently needed are molecular biomarkers that are predictive for response to immunotherapies. Intravesical bacillus Calmette-Guérin (BCG) immunotherapy has been a standard of care in NMIBC for over 40 years, but there exists no biomarker to prioritize patients for optimal treatment in this era of recurrent BCG shortages. The anti-PD-1 immune checkpoint inhibitor pembrolizumab was recently FDA approved for BCG-unresponsive NMIBC based on a single-arm phase II trial demonstrating an initial response rate of 40% at 3 months. However, only 20% of patients remained disease free at 12 months (3) and PD-L1 expression by IHC staining was not associated with response, leaving little guidance on who is likely to benefit (3).
A potential approach to risk stratify patients with NMIBC and predict treatment responses is transcriptomic profiling. This strategy has led to the identification and validation of luminal and basal-like molecular subtypes of bladder cancer, similar to the luminal and basal-like breast cancer subtypes (4, 5). For muscle-invasive bladder cancer (MIBC), multiple retrospective studies indicate that molecular subtyping is prognostic and predictive of response to chemotherapy and immunotherapy, and a consensus molecular classification has recently been established (4–6). In contrast, while early unsupervised microarray analyses of gene expression had focused on NMIBC, there have been only a few recent gene expression efforts in NMIBC using more contemporary expression profiling platforms (7–9). The largest of these was UROMOL, a prospective multicenter European study that established three major molecular subtypes of NMIBC (7). However, the 460 NMIBC specimens analyzed represent only 34% of the initial 1,372 fresh-frozen specimens obtained, after applying strict criteria on RNA quality and carcinoma cell percentage. Inclusion of only the highest-quality tumor tissue increases confidence in the resulting biologic insights but can limit the clinical translation of these findings. Most NMIBC specimens available for clinical testing are archival, formalin-fixed, paraffin-embedded (FFPE) tissues, from which RNA can be difficult to extract and is generally of poor quality (10). Thus, a clinically feasible method of multiplex gene expression profiling in NMIBC must overcome the limited amount of available tissue, low cellularity, and poor RNA quality inherent in the majority of these specimens.
RNA counting methods, such as nCounter (NanoString Technologies, Inc), that do not require enzymatic reactions allow for FFPE samples to be used in expression profiling (11), but the feasibility and value of these approaches have not been robustly assessed in NMIBC. Thus, we sought to evaluate gene expression signatures determined by RNA counting to identify associations with clinicopathologic characteristics, assess prognostic and predictive significance, and evaluate the added value of gene expression data over established DNA mutation sequencing methods in NMIBC.
Materials and Methods
Patient samples
Clinical NMIBC specimens were collected from two sites, University of North Carolina at Chapel Hill (UNC, Chapel Hill, NC) and Memorial Sloan Kettering Cancer Center (MSK, New York, NY). All specimens from UNC and MSK were archival FFPE NMIBC tissue samples procured by transurethral resection of bladder tumor (TURBT) from treatment-naïve, newly diagnosed patients and were reviewed by a genitourinary pathologist (S.E. Wobker, H.A. Al-Ahmadie) to confirm grade, stage, and urothelial histology. Specimens in the UNC cohort were collected through the Institutional Review Board–approved UNC Health Registry/Cancer Survivor Cohort Study, which prospectively ascertained persons with newly diagnosed with cancer seen in the UNC Hospital system. Enrolled patients gave informed consent for use of biospecimens, clinical data, and questionnaire data for approved research. Specimens in the MSK cohort were collected through an Institutional Review Board–approved sequencing effort, in which tumor specimens and matched germline DNA were profiled by targeted panel sequencing using a panel of 341 (later updated to 410) cancer-associated genes within a Clinical Laboratory Improvement Amendments–certified laboratory as previously described by Pietzak and colleagues and available on cbioportal.org (http://www.cbioportal.org/study/summary?id=blca_nmibc_2017; ref. 12). We included all tumors with available FFPE for RNA extraction and gene expression analysis from Pietzak and colleagues. Treatment and management were at the discretion of the treating urologic oncologist. Patients treated with BCG immunotherapy received 6 weekly full doses of TICE BCG with only 8% receiving additional maintenance BCG. Patients were then followed every 3 months with cystoscopy and urine cytology for the first year, then every 3–6 months. All HGT1 tumors had restaging TURBTs with confirmation of uninvolved detrusor muscle at initial diagnosis. Recurrence was defined as histologically proven cancer on biopsy or TURBT.
Expression analysis
RNA extraction and gene expression methods have been published previously (13). Briefly, RNA was isolated from a 1-mm FFPE core or two 10-μm unstained FFPE slides using the Qiagen RNeasy FFPE Kit and protocol (catalog no. 73504). RNA was quantitated using a Thermo Fisher Scientific NanoDrop 2000 Spectrophotometer (catalog no. ND-2000) and Agilent 4200 TapeStation. After excluding samples with low concentration or low percentages of RNA molecules >300 nucleotides long, the remaining 90% of samples were processed by the UNC Translational Genomics Laboratory using the NanoString nCounter platform. Samples were run on a custom codeset that included gene sets for the Hedegaard classifier (7), for p53 pathway defects (14), and for 52 immune-related genes (15). Samples were randomized to batch and two Stratagene Universal Human Reference RNA samples were included to assess batch variability. Batch variability was low, with correlations between reference standards exceeding 0.98.
After counting RNA, QC procedures eliminated samples with low tumor gene expression. Four steps were used to identify such samples. (i) Expression values below the limit of detection, defined as the average of a sample's negative probe mean, was set to zero. (ii) Correlations between six housekeeping genes (ACTB, GUSB, KU70/XRCC6, NAGA, PGK1, RPS10) were evaluated across samples. Pairwise correlations were all above 80%, with an overall mean correlation of 87.2%, and therefore all six genes were retained in the dataset. (iii) The relationship between housekeeping gene missingness and endogenous gene missingness was assessed, where missingness was defined as having an expression below the limit of detection. Missingness in endogenous genes was correlated with housekeeping gene missingness. Samples with greater than half of the endogenous genes marked as missing were removed from further analysis. Principal component analysis (PCA) was then used to identify outliers. (iv) Finally, we used the R package remove unwanted variation to normalize the data (16). Dimensions of unwanted variation (k) from 1 to 5 were assessed and k = 1 was selected on the basis of sample PCA plots. Upon normalization, nearest-neighbor averaging imputation was applied to all samples via the impute R package (16). Samples from MSK and UNC were assessed for quality concurrently. When duplicate assays both passed QC, these were averaged to produce a single expression vector for each patient.
Gene expression signatures
Genes for the BASE47 gene signature were as originally described by Damrauer and colleagues (4). Gene expression data were clustered and Prediction Analysis of Microarrays (PAM) was used to select a parsimonious set of genes that represented the three UROMOL subclasses described by Hedegaard and colleagues (7) For UROMOL subclasses, a total of 117 genes were included in the classifier, though the same clusters were recapitulated with 110 genes. This smaller gene set was used to cluster the samples. Classification by the p53 pathway defect expression signature originally described in breast cancer (14) and recently shown to be associated with TP53 status in MIBC (17) was according to published methods (14, 17). Finally, we developed a panel of immune genes based on work previously described by Bindea and colleagues (15). As NMIBC specimens either had high expression of all genes in the panel or low expression of all genes, the immune score for a tumor was defined as the average of the counts of genes comprising the immune signature panel. Tumors were grouped into tertiles by ranking tumors by gene scores and identifying the 33rd and 66th percentile thresholds adapted from Miller and colleagues (18). For recurrence analyses, the top two tertiles were combined into a “high” immune signature score and compared with the bottom tertile, considered a “low” immune score. Proliferation was determined from the median expression of proliferation-related genes in Parker and colleagues (19). Java TreeView was used to visualize the data according to TP53 status and immune score (11).
Comparison of RNA-based classes with DNA mutations
Targeted panel sequencing of MSK samples was performed using the MSK-IMPACT assay (12, 20, 21). Coding and promoter mutations and indels, excluding silent mutations, were considered. Mutation calls were not assessed for samples that failed MSK-IMPACT QC. Specific genes were curated from Pietzak and colleagues based on genes mutated at >10% in NMIBC (12). Missense mutations and amplifications in known oncogenes were deemed significant if the variant was recurrent or if existing literature reported it as a functionally validated activating alteration. Alterations in tumor suppressor and DNA damage repair genes were deemed deleterious if truncating mutations (nonsense, frameshift indels), recurrent missense mutations, or homozygous deletions. Recurrent missense mutations were defined as those reported in either the Catalogue of Somatic Mutations in Cancer or in the cBioPortal for Cancer Genomics more than 10 times. Missense mutations reported by the MSK-IMPACT bioinformatics pipeline, but not meeting our definition for recurrent alterations, were reported as “novel missense mutations”/“missense mutations of unknown significance” and not included in statistical analyses of clinicopathologic and recurrence associations.
Validation cohorts
UROMOL Cohort: Publicly available RNA sequencing (RNA-seq) data from the UROMOL Project (prediction of bladder cancer disease course using risk scores that combine molecular and clinical risk factors) were downloaded from the Supplementary Methods and Materials section from Hedegaard and colleagues (7). and from www.medrxiv.org (https://www.medrxiv.org/content/10.1101/2020.06.19.20054809v1) to determine BCG treatment history and recurrence outcomes. Only 19% of the UROMOL cohort received BCG and maintenance BCG was infrequently given (7).
Northwestern Cohort: Publicly available RNA-seq data from GSE154261 were downloaded from Robertson and colleagues (9). This cohort compromised of 73 primary T1 tumors treated at Northwestern University (Evanston, IL) with all patients receiving induction BCG, 64% receiving some maintenance BCG, and 84% having a restaging TURBT before BCG (9).
Statistical analyses
Clustering of gene expression data were performed in R using the heatmap.plus R package (11). Pearson distance measures and complete linkage were used to determine clusters. Java TreeView was used to visualize the clustered data and generate heatmaps (11). The compareGroups R package was used to generate descriptive tables and to determine the statistical significance of associations among variables (11). Continuous variables were assessed using t tests or ANOVA, where appropriate, and categorical variables were assessed using chi-squared tests or Fisher exact tests. Cox regression modeling was used to determine the association between gene signatures and recurrence after BCG. The Kaplan–Meier method and log-rank test were used for estimations of recurrence-free survival (RFS).
Results
Patient and tumor characteristics
RNA was extracted from tumor samples of 41 UNC patients and 68 MSK patients, of which 28 samples (68%) and 50 samples (74%), respectively, met QC criteria. In general, samples with low (<30%) tumor cellularity more often failed QC (Supplementary Table S1; Supplementary Fig. S1). Patient demographics and clinicopathologic characteristics for profiled samples are described in Table 1. Recurrence outcomes were available only for the MSK cohort. Clinicopathologic characteristics were not associated with recurrence risk in this cohort (Supplementary Table S2), consistent with our prior study (12). One patient was treated with immediate cystectomy, so their tumor was analyzed but they were excluded from recurrence analyses. Median follow-up among the MSK cohort was 39 months [interquartile range (IQR), 13–66.2], with recurrence occurring in 23 patients. Median follow-up among the 38 patients (76%) receiving BCG immunotherapy was 38.2 months (IQR, 10.7–66.6), with recurrence occurring in 18 patients.
Table 1.
Patient and tumor characteristics.
| All | UNC | MSK | MSK BCG treated | |
|---|---|---|---|---|
| n = 78 | n = 28 | n = 50 | n = 38 | |
| Age (years) | 65.0 (12.5) | 63.9 (12.1) | 65.6 (12.7) | 66.8 (11.4) |
| Male sex | 59 (75.6%) | 22 (78.6%) | 37 (74.0%) | 29 (76.3%) |
| Stage and gradea | ||||
| T1 high-grade | 35 (46.1%) | 18 (64.3%) | 17 (34.0%) | 14 (36.8%) |
| Ta high-grade | 25 (32.0%) | 0 (0.0%) | 25 (50.0%) | 23 (60.5%) |
| Ta low-grade | 18 (21.1%) | 10 (35.7%) | 8 (16.0%) | 1 (2.6%) |
| Carcinoma in situ (CIS) | 22 (28.2%) | 8 (28.5%) | 14 (28.0%) | 14 (36.8%) |
| Tumor cellularitya | ||||
| 0–40 | 33 (43.4%) | 13 (46.4%) | 18 (36.0%) | 17 (44.7%) |
| 50–90 | 43 (56.7%) | 11 (39.3%) | 32 (64.0%) | 21 (55.3%) |
| UROMOL subclass | ||||
| 1/ Luminal | 26 (33.3%) | 7 (25.0%) | 19 (38.0%) | 10 (26.3%) |
| 2/ CIS-like | 28 (35.9%) | 8 (27.6%) | 20 (40.0%) | 17 (44.7%) |
| 3/ Early basal-like | 24 (30.85%) | 13 (46.4%) | 11 (22.0%) | 11 (30.0%) |
| Immune scoreb | ||||
| Low | 20 (25.6%) | 6 (21.4%) | 14 (28.0%) | 11 (30.0%) |
| Medium | 20 (25.6%) | 5 (17.9%) | 15 (30.0%) | 8 (21.0%) |
| High | 38 (48.7%) | 17 (60.7%) | 21 (42.0%) | 19 (50.0%) |
| Treatment | ||||
| BCG | 38 (76%) | 38 (100%) | ||
| Mitomycin | 5 (10%) | 0 | ||
| TURBT only | 6 (12%) | 0 | ||
| Cystectomy | 1 (2%) | 0 | ||
| Tumor size ≥3 cm | 26 (52%) | 18 (47.4%) | ||
| Multiple tumors | 19 (38%) | 18 (47.4%) | ||
Note: Data are presented as mean (SD) or n (%).
Abbreviations: CIS, carcinoma in situ; MSK, Memorial Sloan Kettering Cancer Center; rankBCG, bacillus Calmette-Guérin; SD, standard deviation; TURBT, transurethral resection of the bladder tumor; UNC, University of North Carolina.
aMissing values omitted.
bGrouped into tertiles based on score.
Molecular subtyping by RNA counting
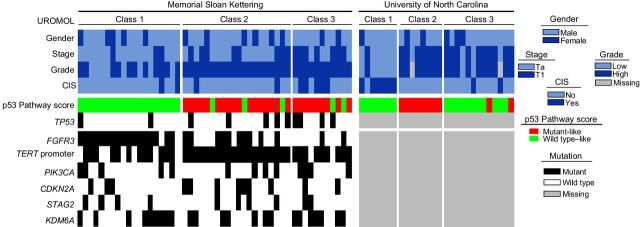
We first sought to determine molecular subtype using the BASE47 classifier developed to separate MIBC into luminal and basal-like subtypes (4). We found limited discriminatory capacity as only four NMIBC tumors were classified as basal like, whereas the remainder were luminal. We therefore focused our subsequent analyses on a gene expression classifier derived from the large UROMOL cohort of NMIBC samples (7). Our combined NMIBC sample cohort was well distributed across the three previously described UROMOL subclasses (Fig. 1) and clinical characteristics differed across the groups (Table 2). UROMOL class 1, previously described as luminal-like/well differentiated, was enriched in low-grade Ta tumors that had greater tumor cellularity due to their predominantly papillary architecture. As expected, these tumors had significantly higher FGFR3 gene expression levels in both the UNC and MSK cohorts (Supplementary Fig. S2; ref. 7). Consistent with UROMOL class 2 tumors being the most aggressive subtype, they had the highest proliferation expression scores and expression of the proto-oncogene forkhead box M1 (FOXM1) in both cohorts (Supplementary Fig. S3). UROMOL class 3, previously described as being an early basal-like subtype, was enriched in high-grade T1 tumors.
Figure 1.
Heatmap of patient and tumor genetic characteristics according to cohort and UROMOL subclass.
Table 2.
Descriptive statistics by UROMOL signature classification.
| 1/Luminal | 2/Luminal CIS-like | 3/Early basal-like | ||
|---|---|---|---|---|
| n = 26 | n = 28 | n = 24 | P | |
| Institution/cohort | 0.08 | |||
| UNC | 7 (26.9%) | 8 (28.6%) | 13 (54.2%) | |
| MSK | 19 (73.1%) | 20 (71.4%) | 11 (45.8%) | |
| Age (years) | 61.6 (14.9) | 65.3 (11.2) | 68.3 (10.3) | 0.159 |
| Male sex | 20 (76.9%) | 20 (71.4%) | 19 (79.2%) | 0.797 |
| Stage | 0.001 | |||
| T1 | 4 (15.4%) | 15 (53.6%) | 16 (66.7%) | |
| Ta | 22 (84.6%) | 13 (46.4%) | 8 (33.3%) | |
| Gradea | <0.001 | |||
| High | 12 (46.2%) | 27 (96.4%) | 21 (87.5%) | |
| Low | 14 (53.8%) | 0 (0.00%) | 2 (8.33%) | |
| Carcinoma in situ | 8 (30.8%) | 2 (7.14%) | 7 (29.2%) | 0.063 |
| Tumor cellularitya | 0.004 | |||
| 0–40 | 6 (23.1%) | 11 (40.7%) | 16 (69.6%) | |
| 50–90 | 20 (76.9%) | 16 (59.3%) | 7 (30.4%) |
Note: Data are presented as mean (SD) or n (%).
Abbreviations: CIS, carcinoma in situ; MSK, Memorial Sloan Kettering Cancer Center; SD, standard deviation; UNC, University of North Carolina.
aMissing values omitted.
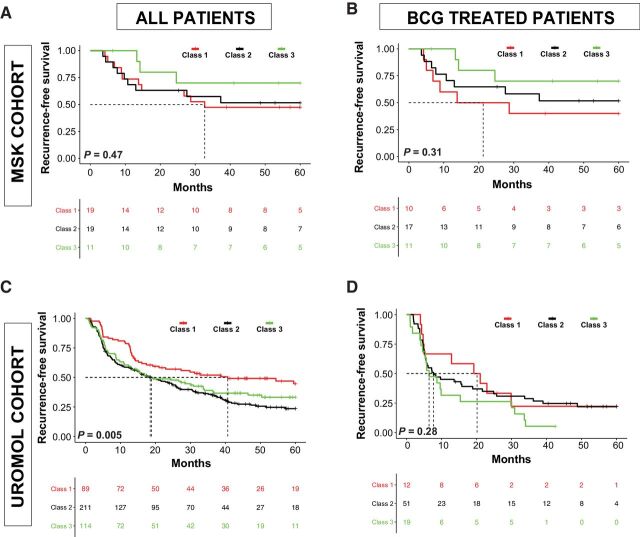
There were very few progression events within the MSK cohort, preventing formal analysis, but all 3 patients with progression to secondary MIBC had UROMOL class 2 tumors, consistent with the original study (7). There was no correlation between UROMOL classes and RFS within the entire MSK cohort or the subset of 38 patients treated with BCG (Fig. 2A and B). To rule out the possibility that this lack of association was the result of small sample size, we analyzed publicly available data from the UROMOL study and found that class 2 was associated with worse RFS in the UROMOL study (n = 438), but subtype was not associated with recurrence rates in the subset of patients treated with BCG (n = 83; Fig. 2C and D). These results suggest that the UROMOL classifier is prognostic of outcome but not predictive of BCG response.
Figure 2.
RFS stratified by UROMOL subclass. A, Entire MSK cohort. B, MSK patients treated with BCG. C, Entire UROMOL cohort. D, UROMOL patients treated with BCG.
Immune gene expression and signatures
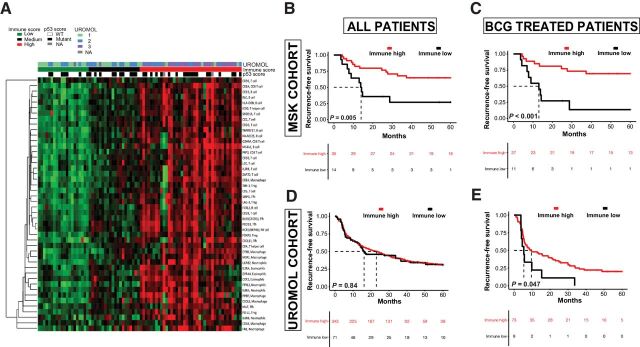
As UROMOL classes were not predictive of recurrence following BCG treatment among either the MSK or UROMOL cohorts, we sought to evaluate other RNA-based signatures as potential predictive biomarkers of clinical benefit from BCG immunotherapy. Previous biomarker studies have identified higher pretreatment T-cell infiltration and an inflamed TME as associated with improved response to systemic immunotherapies in various cancer types, including bladder cancer (15, 22, 23). We evaluated the expression of known immune-related genes to determine the degree of immune cell infiltration and inflammation within the TME (15). NMIBC tumors could be broadly classified as having either low expression or higher expression across all immune genes studied, with no groups emerging with more complex patterns (i.e., innate immune response only; Fig. 3A). Therefore, the median expression of all immune genes was used to calculate an immune score. Tumors were grouped into tertiles by ranking tumors by gene scores and identifying the 33rd and 66th percentile thresholds adapted from Miller and colleagues (18). These scores varied among UROMOL classes, with basal-like UROMOL class 3 tumors having the highest expression of immune-related genes, whereas class 1 tumors had the lowest (Supplementary Fig. S4). Tumors with high immune scores (top two tertiles) were associated with improved RFS in the MSK cohort [HR = 0.33; 95% confidence interval (CI), 0.14–0.78; P = 0.01) as well as among the subset treated with BCG (HR = 0.23; 95% CI, 0.09–0.59; P = 0.002; Fig. 3B and C; Supplementary Table S2). We also sought to externally validate the immune score using the UROMOL cohort (Supplementary Fig. S5) and found that higher immune scores were associated with improved RFS among the 83-patient UROMOL subgroup treated with BCG (HR = 0.5; 95% CI, 0.24–1.00; P = 0.05), but not among the entire 438-patient UROMOL cohort (HR = 0.96; 95% CI, 0.70–1.33; P = 0.84; Fig. 3D and E). No statistically significant difference was seen for high immune score with improved RFS in the Northwestern HGT1 cohort (HR = 0.7; 95% CI, 0.52–3.6; P = 0.4; Supplementary Fig. S6).
Figure 3.
RNA expression–based immune score differs among UROMOL subclasses and is associated with RFS after BCG. A, Average expression counts for markers of various immune cell types for each patient. B–E, RFS by immune score in the entire MSK cohort (B); MSK patients treated with BCG (C); entire UROMOL cohort (D), UROMOL patients treated with BCG (E).
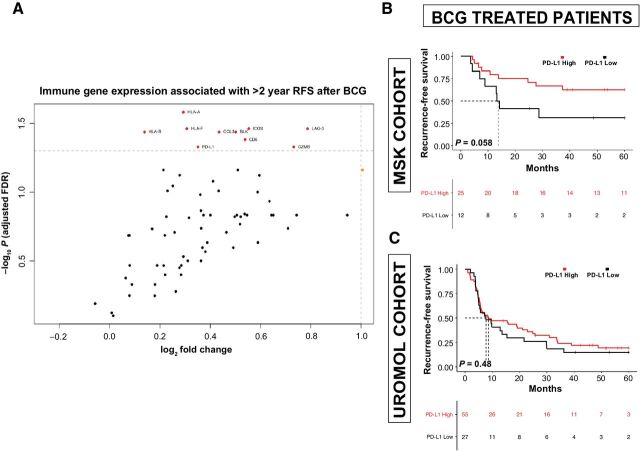
We next sought to determine which individual immune-related genes were associated with favorable outcomes following treatment with BCG. Correlates of remaining free from high-grade recurrences >2 years after BCG treatment included high expression of HLA class I histocompatibility antigens, granzyme B, and inducible T-cell co-stimulator. Surprisingly, we found high expression of the immune checkpoint genes PD-L1 and LAG-3 to also be associated with remaining recurrence free after BCG (Fig. 4A).
Figure 4.
Immune correlates of recurrence following BCG treatment. A, Volcano plot demonstrating correlation of expression of individual immune-related genes with probability of remaining free of a high-grade recurrence beyond 24 months after BCG (only genes statistically significant after Benjamini–Hochberg false discovery correction are in red and labeled). RFS by PD-L1 (CD274) expression among BCG-treated MSK patients (B) and patients in the UROMOL cohort (C).
As this finding contradicts a prevailing hypothesis that immune checkpoint expression leads to BCG treatment failure, and there is growing interest in anti-PD-L1/PD-1 immunotherapy for patients with NMIBC, we further assessed the significance of expression of targetable immune checkpoints (PD-L1, PD-1, and CTLA-4) within our cohort. Expression of these immune inhibitory molecules was highest in UROMOL class 3 tumors and lowest in class 1 tumors (Supplementary Fig. S7). Higher expression of PD-L1 (P = 0.058), PD-1 (P < 0.01), and CTLA-4(P < 0.01) in pretreatment NMIBC specimens was associated with lower recurrence rates after BCG immunotherapy in our cohort, suggesting that these molecules correspond to increased immune infiltration and are not associated with resistance to BCG (Fig. 4B; Supplementary Fig. S7). Examining pretreatment expression levels of these immune checkpoint genes in the 83-patient BCG-treated UROMOL cohort found no association with RFS in BCG-treated patients (Fig. 4C; Supplementary Fig. S7). Furthermore, expression of PD-L1, PD-1, and CTLA-4 in pretreatment specimens from the Northwestern HGT1 cohort were also not associated with RFS after BCG (Supplementary Fig. S7).
Association between gene expression–based signatures and somatic DNA mutations
As the UROMOL study inferred somatic mutations from RNA expression data alone (7), which can lead to false-positive deleterious mutation calls (24), we evaluated correlations between UROMOL subclasses and somatic DNA mutations using tumor-normal target panel data available for the MSK cohort. FGFR3 mutations were enriched among luminal class 1 tumors, while infrequently altered in basal-like class 3 tumors (Fig. 1; Table 3). Alterations in ERBB2, a gene known to be mutually exclusive with FGFR3 (12), were absent in class 1 tumors but seen in 20% of class 2 tumors and 36% of class 3 tumors (P = 0.02; Table 3). Mutations in the chromatin remodeling gene TET2 were enriched in basal-like class 3 tumors (Table 3). Somatic mutations in the TERT promoter were highly enriched in UROMOL class 2 tumors, found to be present in all 20 samples (100%; Fig. 1). UROMOL class 2 tumors also had the highest fraction of the genome altered (Supplementary Fig. S8).
Table 3.
Gene alterations across UROMOL subclasses.
| Gene | 1/Luminal (n = 19) | 2/Luminal CIS-like (n = 20) | 3/Early basal-like (n = 11) | P |
|---|---|---|---|---|
| FGFR3 | 17 (89%) | 10 (50%) | 2 (18%) | <0.001 |
| KRAS | 1 | 2 | 0 | 0.8 |
| HRAS | 1 | 0 | 2 | 0.1 |
| ERBB2 | 0 | 4 (25%) | 4 (36%) | 0.02 |
| ERBB3 | 2 | 1 | 1 | 0.8 |
| PIK3CA | 5 | 3 | 4 | 0.4 |
| TSC1 | 1 | 4 | 1 | 0.4 |
| NF1 | 0 | 4 | 1 | 0.1 |
| TP53 | 2 | 5 | 4 | 0.3 |
| RB1 | 0 | 1 | 1 | 0.7 |
| MDM2 | 1 | 3 | 0 | 0.5 |
| CCND1 | 0 | 2 | 1 | 0.4 |
| CDKN2A | 3 | 7 | 2 | 0.3 |
| CDKN1A | 3 | 3 | 1 | 1 |
| STAG2 | 4 | 4 | 2 | 1 |
| KDM6A | 9 | 6 | 7 | 0.2 |
| ARID1A | 2 | 5 | 2 | 0.6 |
| KMT2A | 2 | 1 | 0 | 0.6 |
| KMT2C | 2 | 3 | 0 | 0.5 |
| KMT2D | 6 | 5 | 1 | 0.4 |
| EP300 | 4 | 1 | 2 | 0.3 |
| CREBBP | 1 | 5 | 1 | 0.2 |
| TERT | 12 (63%) | 20 (100%) | 9 (82%) | 0.007 |
| ERCC2 | 2 (11%) | 2 (10%) | 5 (45%) | 0.044 |
| ATM | 0 | 4 | 2 | 0.1 |
| BRCA1 | 0 | 2 | 0 | 0.3 |
| BRCA2 | 0 | 1 | 1 | 0.7 |
| TET2 | 0 | 1 (5%) | 3 (27%) | 0.03 |
| MGA | 1 | 3 | 0 | 0.5 |
| BCOR | 0 | 0 | 1 | 0.2 |
| BRD4 | 0 | 0 | 0 | 1 |
| DNAJB1 | 0 | 0 | 1 | 0.2 |
| IDH1 | 0 | 0 | 0 | 1 |
| JAK3 | 0 | 0 | 1 | 1 |
| ARID1B | 0 | 1 | 2 | 0.2 |
| BAP1 | 1 | 3 | 2 | 0.6 |
| PBRM1 | 2 | 2 | 0 | 0.6 |
| NFE2L2 | 0 | 1 | 0 | 1 |
| TBX3 | 1 | 0 | 0 | 0.6 |
| NCOR1 | 1 | 0 | 1 | 0.5 |
Note: Bold indicates statistical significance.
Abbreviation: CIS, carcinoma in situ.
Alterations in TP53 and cell-cycle genes are among the most common genetic events in MIBC and high-risk NMIBC, yet biomarker investigations into p53 status to guide clinical management have reported conflicting results (12, 25). We explored a mutant p53-pathway gene signature, developed and validated in breast cancer (14), within our NMIBC cohort. This signature was recently shown to be associated with TP53 status in MIBC (17), but has little overlap with the “p53-like” gene expression signature identified by Choi and colleagues in MIBC (Supplementary Fig. S9; ref. 6). The mutant p53-pathway gene signature was highest in class 2 tumors, followed by class 3 tumors; these two subclasses had the most TP53 mutations (Fig. 1; Supplementary Fig. S9). RNA-based p53 pathway assessment detected defects in the p53 pathway more than TP53 DNA mutations alone and had fair correlation with DNA alterations in an expanded set of commonly altered cell-cycle pathway genes (TP53, MDM2, RB1, CDKN2A, CDKN1A, CCND1; Supplementary Fig. S10). Neither p53 mutant-like gene expression nor TP53 DNA alterations were associated with recurrence in the entire MSK cohort or those treated with BCG. In addition, mutant p53-pathway gene signature tumors had high immune scores, whereas p53-wild-type-like tumors had intermediate immune scores (Supplementary Fig. S10). Higher mutant p53-pathway gene signatures also correlated with increased expression of proliferation and FOXM1 (Supplementary Fig. S11).
Finally, as the pretreatment TME inferred by immune score was associated with BCG response, we sought to determine whether there was a correlation between genomic alterations in individual genes and immune score. We found that FGFR3 mutations by targeted panel sequencing and FGFR3 overexpression by RNA profiling were both associated with lower immune scores (Supplementary Table S3; Supplementary Fig. S2). FGFR3 mutations remained significantly associated with low immune score even after adjusting for multiple comparisons. Conversely, ERBB2 alterations were enriched in immune-score-high tumors (38% vs. 0 vs. 0, P = 0.001; Supplementary Table S3).
Discussion
There are several barriers that must be overcome before molecular biomarkers identified within the context of retrospective clinical studies can be successfully translated to clinical practice. These include the limited quantity and often poor quality of tissue available for many patients with NMIBC. Here, we show that gene expression profiling by RNA counting is feasible using archival FFPE NMIBC clinical samples, despite relatively limited tumor tissue and low tumor cellularity.
To build upon prior work with the NMIBC-specific UROMOL gene expression classifier (7), we combined targeted panel sequencing of tumor-normal pairs along with transcriptomic profiling and observed that TERT promoter mutations were significantly enriched in UROMOL class 2 (100%) and class 3 (82%) tumors. This novel observation demonstrates the potential molecular insights that can be obtained by integrated RNA and DNA molecular profiling and supports the development of urine-based screening and surveillance biomarkers that detect TERT promoter mutations in urinary cell-free DNA to identify aggressive bladder tumors earlier (26). We also found UROMOL molecular subtypes correlated with expected clinicopathologic characteristics but were not associated with BCG response. UROMOL subtypes can be recapitulated within bladder cancer cell lines, suggesting that these molecular subtypes are tumor cell “intrinsic” signatures (7). We hypothesize that molecular subtypes based on tumor cell phenotype, independent of signals from infiltrating stromal and immune cells, provide important biologic insights but are not likely to be robust predictive biomarkers for response to immunotherapy such as BCG.
To develop a better predictive biomarker for BCG response, we assessed immune cell infiltration and inflammation within the TME using a novel gene expression signature. We observed that a high immune score was associated with improved RFS after BCG immunotherapy in the MSK and UROMOL cohorts, demonstrating the importance of the pretreatment TME in determining BCG response, consistent with the work of several other groups (27, 28). While BCG is a live, attenuated bacteria known to provoke an influx of innate and adaptive immune cells within the bladder wall, the recruitment of these immune cells appears unable to overcome an existing immunologically “cold” TME. Notably, no statistical difference in RFS after BCG was seen within the Northwestern cohort for the immune score. This may be due to it being a HGT1 only cohort compared with the MSK and UROMOL cohorts which also included LGTa and HGTa tumors. It is also possible that the addition of maintenance BCG may attenuate the negative effects of a “cold” pretreatment TME, as 64% of the Northwestern cohort received maintenance BCG compared with only a few patients within the MSK and UROMOL cohorts. Maintenance BCG is recommended in multiple guidelines as it reduces the risk of recurrence by 19% at 5 years compared with induction BCG alone (29, 30), but many patients do not receive it due to the persistent global BCG shortage and concerns over treatment-related toxicity (29–31). If prospectively validated, the immune score may provide a rational, biomarker-driven approach to selecting which patients would most benefit from induction and maintenance BCG. This could be invaluable in helping to alleviate the global BCG shortage.
Further investigation into the causes of a cold TME in NMIBC is needed, but our study suggests that combination strategies that modulate the TME to promote antitumor immune cell recruitment hold promise in NMIBC. Our finding that mutation and overexpression of FGFR3 are associated with a low immune score in NMIBC is consistent with data from studies of MIBC, upper tract urothelial carcinoma, and mouse models of bladder cancer (23, 32, 33), providing further support for investigations into the potential immunomodulating benefits of FGFR3-targeted therapies. Interestingly, our data also suggest that FGFR3 and ERBB2 are not only mutually exclusive in NMIBC but result in contrasting differences in the TME. Both FGFR3 and ERBB2 are attractive “targets,” given the prevalence of these genomic alterations in patients with NMIBC. As more targeted therapies with demonstrable activity in metastatic urothelial cancer are evaluated in clinical trials for patients with NMIBC, these differences in the TME will likely become even more relevant.
Our analysis also found that high gene expression of PD-L1, PD-1, and CTLA-4 was not associated with worse BCG response in either the MSK, UROMOL, or Northwestern cohorts. While this may seem contrary to the prevailing theory that PD-1/PD-L1 expression is a mechanism of resistance to BCG (34, 35), others have reported high PD-L1 gene expression to be associated with favorable outcomes with intravesical therapy in T1 NMIBC (36). These observations might be from differences in pre-analytic preparations or posttranslational modifications between gene level and protein level expression of PD-1/PD-L1 (37). However, IHC-based studies assessing the role of PD-1/PD-L1 expression in NMIBC by comparing BCG “responders” with “nonresponders” have had conflicting results, varying considerably depending on the antibody and cutoff used, and studies supporting PD-1/PD-L1 as a mechanism of resistance to BCG generally do not adjust for differences in important clinical factors that are known to affect both BCG response and PD-L1 expression levels, such as concomitant carcinoma in situ (34–36, 38). The first and most highly cited study suggesting PD-L1 expression may be a mechanism of immune evasion to BCG was by Inman and colleagues, where 11 of 16 patients with recurrent tumors had associated post-BCG granulomata with strong PD-L1 expression by IHC staining (35). However, the authors did not assess PD-L1 expression in post-BCG granulomata from patients without recurrence, which is particularly relevant as post-BCG granulomata are associated with a favorable response to BCG (39) and comprised of macrophages that often express PD-L1 even in healthy tissue (40). Pretreatment specimens from the prospective NMIBC cohort within Inman and colleagues showed no difference in recurrence rates after BCG by PD-L1 expression [PD-L1+ 30% (3/10) vs. PD-L1− 38% (13/34), P = 0.2] (35). Taken together with our data, PD-1/PD-L1 expression is unlikely to mediate resistance to BCG immunotherapy in treatment-naïve NMIBC, and high expression of PD-L1, PD-1, or CTLA-4 should not preclude treatment with BCG. Our results raise concerns about ongoing trials combining BCG and immune checkpoint blockade, as they indicate that an immunologically “cold” TME may be a shared mechanism of resistance to both BCG and checkpoint inhibitors (23).
As p53 defects by IHC staining has been one of the most heavily investigated biomarkers in NMIBC with numerous conflicting reports of significance (25), we sought to investigate the additive value of DNA and RNA molecular profiling for p53 pathway alterations in NMIBC. While we found overlap between RNA-based p53 pathway assessment and genomic alterations in cell-cycle pathway genes, this does not appear to be clinically relevant for recurrence after BCG. Whether defects in the p53 pathway are associated with progression to secondary MIBC remains unclear, but warrants further investigation (41).
Limitations of our study include the use of targeted sequencing panels rather than whole-exome/transcriptomic sequencing, and that clinical outcomes were available only for the MSK cohort. These limitations may have caused us to miss subtle but important associations with gene expression signatures and precluded our ability to perform robust multivariable analyses. In addition, large prospective validation that account for differences in tumor cell purity is required and ongoing at our center and elsewhere.
In sum, the current analysis demonstrates the feasibility of performing RNA-based subtyping on clinical NMIBC specimens, even those with relatively low tumor cellularity. Gene expression signatures can provide novel biologically and clinically relevant information on the TME (i.e., immune score) and tumor-intrinsic properties (i.e., mutant p53 pathway signature) that are additive and complementary to analysis of genomic DNA. The integration of transcriptomic tumor profiling with exome sequencing is a promising approach to improve risk stratification and treatment selection for patients with NMIBC.
Authors' Disclosures
G. Iyer reports grants and personal fees from Mirati Therapeutics; grants from Janssen and Seagen, Inc; and personal fees from Basilea Pharmaceuticals and DAVA Oncology outside the submitted work. D.B. Solit reports personal fees from Pfizer, Loxo Oncology at Lilly, Vividion Therapeutics, Scorpion Therapeutics, BridgeBio, and Syros Pharmaceuticals outside the submitted work. M.I. Milowsky reports grants from Merck, Roche/Genentech, Bristol-Myers Squibb, Astellas Pharma, Clovis Oncology, Inovio Pharmaceuticals, Mirati Therapeutics, Constellation Pharmaceuticals, Syndax, Incyte, Amgen, Regeneron, Arvinas, Seagen, Pfizer, and Johnson & Johnson/Janssen outside the submitted work. W.Y. Kim reports stock and other ownership interests with AbbVie, Abbott, Amgen, Arvinas, BeiGene, Bristol-Myers Squibb, Bluebird Bio, FibroGen, Illumina, Johnson & Johnson, Myovant, Natera, Oramed, Syndax, and Zentalis and receives honoraria from H3 Biomedicine and Takeda; reports consulting or advisory role with Foundation Medicine, GeneCentric; reports research funding support for Acerta, Foundation Medicine, GeneCentric, and Merck; and reports travel, accommodations, and expenses grant with H3 Biomedicine and Takeda. M.E. Nielsen reports other support from American College of Physicians, American Urological Association, and Grand Rounds outside the submitted work. H.A. Al-Ahmadie reports personal fees from AstraZeneca, Janssen Biotech, Bristol Myers Squibb, and Paige.ai outside the submitted work. E.J. Pietzak reports grants from Sidney Kimmel Center for Prostate and Urologic Cancers, Marie-Josée and Henry R. Kravis Center for Molecular Oncology, NIH Cancer Center Support Grant P30 CA008748, NCI SPORE in Bladder Cancer P50 CA221745, MSKCC Bladder Cancer SPORE Career Enhancement Award, Bochner-Fleisher Scholars Award, Memorial Sloan Kettering Cancer Center/Weill Medical College of Cornell University Clinical and Translational Science Methodology NIH/NCATS grant number UL1-TR002384, NIH/NCI K12 Paul Calabresi Career Development Award for Clinical Oncology (K12 CA184746), and Wofchuck Family Young Investigator Award during the conduct of the study, as well as personal fees from UpToDate, Janssen Pharmaceuticals, Merck & Co Inc, QED Therapeutics, and Chugai Pharmaceutical outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Table 1, 2, 3
Supplemental Figure 1 shows the quality check failure rate by Tumor Cellularity and by Stage and Grade.
Supplemental Figure 2A shows FGFR3 Expression by UROMOL Subclasses in MSK and UNC cohorts and Supplemental Figure 2B demonstrates the Inverse Relationship between FGFR3 expression and mutation with Immune Signature Score.
Supplemental Figure 3 shows the relationship for UROMOL Subclasses in MSK and UNC cohorts between expression of Proliferation markers and FOXM1.
Supplemental Figure 4 shows the relationship between Immune Signature score by Patient Cohort and UROMOL subtype.
Supplemental Figure 5 shows immune score signature using the publicly available UROMOL.
Supplemental Figure 6 shows Recurrence Free Survival in the Northwestern HGT1 cohort by immune score stratified by (1) High, Medium, Low; (2) High/Medium vs. Low.
Supplemental Figure 7 shows PDL1 (CD274) Expression, PD1 (PDCD1) Expression, and CTLA4 Expression by UROMOL Subclasses in MSK and UNC cohorts and RFS (by high, medium, low tertiles) in MSK, UROMOL, & Northwestern Cohorts.
Supplemental Figure 8 shows the relationship between the Fraction of Genome Altered by UROMOL subgroups within the MSK cohort.
Supplemental Figure 9A demonstrates the overlap between "p53-associated" signature from Troester et al. vs. "p53-like" signature from Choi et al; Supplemental Figure 9B shows the "p53-like" signature from Choi et (MDA) by UROMOL Subclasses in MSK and UNC cohorts; and Supplemental Figure 9C shows the "p53-associated" signature from Troester et al. by UROMOL Subclasses in MSK and UNC cohorts.
Supplemental Figure 10 shows the comparison of DNA based alterations in TP53 and other cell cycle regulation genes compared to "p53-associated" signature from Troester et al. (TP53 score), as well as the association between RFS with TP53 Mutation (DNA) & p53 RNA signature.
Supplemental Figure 11 shows the correlation between P53 Score by immune score, FOXM1 expression, Proliferation marker expression.
Acknowledgments
The authors would like to thank Jessica Moore and Margaret McPartland from the MSKCC Urology Editorial and Grant Services for their editorial assistance with this article.
This work was supported by the UNC Center for Environmental Health and Susceptibility, Center Support grant P30 ES010126 (UNC), and NCI grant R01 CA253450 (M.A. Troester), the MSKCC Sidney Kimmel Center for Prostate and Urologic Cancers, Cycle for Survival, Marie-Josée and Henry R. Kravis Center for Molecular Oncology at MSKCC, NCI Cancer Center Support Grant P30 CA008748 (MSKCC), NCI Specialized Programs of Research Excellence (SPORE) in Bladder Cancer P50 CA221745 (MSKCC), NIH T32ES007018 (K.R. Roell), NCI of the NIH (T32-CA057726, to H.C. Benefield, F30-CA236199, to H.C. Benefield), Gertrude B. Elion Mentored Medical Student Research Award of Triangle Community Foundation (H.C. Benefield), MSKCC Bladder Cancer SPORE Career Enhancement Award (E.J. Pietzak), NIH/NCI K12 Paul Calabresi Career Development Award for Clinical Oncology K12 CA184746 (E.J. Pietzak), MSKCC Department of Surgery Research Award (E.J. Pietzak), Bochner-Fleisher Scholars Award (E.J. Pietzak), Memorial Sloan Kettering Cancer Center/Weill Medical College of Cornell University Clinical and Translational Science Methodology NIH/NCATS grant number UL1-TR002384 (E.J. Pietzak), and the Wofchuck Family Young Investigator Award (E.J. Pietzak).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
J.S. Damrauer: Conceptualization, resources, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. K.R. Roell: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. M.A. Smith: Conceptualization, resources, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. X. Sun: Data curation, investigation, methodology, writing–review and editing. E.L. Kirk: Resources, data curation, formal analysis, investigation, writing–review and editing. K.A. Hoadley: Resources, formal analysis, supervision, investigation, writing–review and editing. H.C. Benefield: Resources, data curation, formal analysis, writing–review and editing. G. Iyer: Data curation, supervision, writing–review and editing. D.B. Solit: Resources, supervision, funding acquisition, writing–review and editing. M.I. Milowsky: Resources, supervision, investigation, methodology, writing–review and editing. W.Y. Kim: Resources, formal analysis, supervision, investigation, methodology, writing–review and editing. M.E. Nielsen: Resources, supervision, writing–review and editing. S.E. Wobker: Resources, data curation, investigation, writing–review and editing. G. Dalbagni: Resources, writing–review and editing. H.A. Al-Ahmadie: Resources, data curation, investigation, writing–review and editing. A.F. Olshan: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. B.H. Bochner: Conceptualization, resources, supervision, funding acquisition, investigation, project administration, writing–review and editing. H. Furberg: Conceptualization, resources, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M.A. Troester: Conceptualization, resources, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. E.J. Pietzak: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Rieken M, Shariat SF, Kluth L, Crivelli JJ, Abufaraj M, Foerster B, et al. Comparison of the EORTC tables and the EAU categories for risk stratification of patients with nonmuscle-invasive bladder cancer. Urol Oncol 2018;36:8.e17–8.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mossanen M, Wang Y, Szymaniak J, Tan WS, Huynh MJ, Preston MA, et al. Evaluating the cost of surveillance for non-muscle-invasive bladder cancer: an analysis based on risk categories. World J Urol 2019;37:2059–65. [DOI] [PubMed] [Google Scholar]
- 3. Keytruda® (pembrolizumab) for the treatment of patients with Bacillus Calmette-Guerin (BCG)-unresponsive, high-risk, non-muscle invasive bladder cancer (NMIBC) with carcinoma in-situ (CIS) with or without papillary tumors who are ineligible for or have elected not to undergo cystectomy. Oncologic Drugs Advisory Committee (ODAC) Meeting. Silver Spring, Maryland; 2019.
- 4. Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A 2014;111:3110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kamoun A, de Reynies A, Allory Y, Sjodahl G, Robertson AG, Seiler R, et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol 2020;77:420–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014;25:152–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hedegaard J, Lamy P, Nordentoft I, Algaba F, Hoyer S, Ulhoi BP, et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell 2016;30:27–42. [DOI] [PubMed] [Google Scholar]
- 8. Hurst CD, Alder O, Platt FM, Droop A, Stead LF, Burns JE, et al. Genomic subtypes of non-invasive bladder cancer with distinct metabolic profile and female gender bias in KDM6A mutation frequency. Cancer Cell 2017;32:701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robertson AG, Groeneveld CS, Jordan B, Lin X, McLaughlin KA, Das A, et al. Identification of differential tumor subtypes of T1 bladder cancer. Eur Urol 2020;78:533–37. [DOI] [PubMed] [Google Scholar]
- 10. Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One 2014;9:e98187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bhattacharya A, Hamilton AM, Furberg H, Pietzak E, Purdue MP, Troester MA, et al. An approach for normalization and quality control for NanoString RNA expression data. Brief Bioinform 2021;22:bbaa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pietzak EJ, Bagrodia A, Cha EK, Drill EN, Iyer G, Isharwal S, et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol 2017;72:952–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Troester MA, Sun X, Allott EH, Geradts J, Cohen SM, Tse CK, et al. Racial differences in PAM50 subtypes in the carolina breast cancer study. J Natl Cancer Inst 2018;110:176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Troester MA, Herschkowitz JI, Oh DS, He X, Hoadley KA, Barbier CS, et al. Gene expression patterns associated with p53 status in breast cancer. BMC Cancer 2006;6:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- 16. Hastie T, Tibshirani R, Narasimhan B, Chu G. Impute: imputation for microarray data. R package version 1.64.0. 2020.
- 17. Sun X, Hoadley KA, Kim WY, Furberg H, Olshan AF, Troester MA. Age at diagnosis, obesity, smoking, and molecular subtypes in muscle-invasive bladder cancer. Cancer Causes Control 2017;28:539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller LD, Chou JA, Black MA, Print C, Chifman J, Alistar A, et al. Immunogenic subtypes of breast cancer delineated by gene classifiers of immune responsiveness. Cancer Immunol Res 2016;4:600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sweis RF, Spranger S, Bao R, Paner GP, Stadler WM, Steinberg G, et al. Molecular drivers of the non-T-cell-inflamed tumor microenvironment in urothelial bladder cancer. Cancer Immunol Res 2016;4:563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yizhak K, Aguet F, Kim J, Hess JM, Kubler K, Grimsby J, et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 2019;364:eaaw0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmitz-Drager BJ, Goebell PJ, Ebert T, Fradet Y. p53 immunohistochemistry as a prognostic marker in bladder cancer. Playground for urology scientists? Eur Urol 2000;38:691–9. [DOI] [PubMed] [Google Scholar]
- 26. Chaudhuri AA, Pellini B, Pejovic N, Chauhan PS, Harris PK, Szymanski JJ, et al. Emerging roles of urine-based tumor DNA analysis in bladder cancer management. JCO Precis Oncol 2020;4:PO.20.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pichler R, Fritz J, Zavadil C, Schafer G, Culig Z, Brunner A. Tumor-infiltrating immune cell subpopulations influence the oncologic outcome after intravesical Bacillus Calmette-Guerin therapy in bladder cancer. Oncotarget 2016;7:39916–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chevalier MF, Trabanelli S, Racle J, Salome B, Cesson V, Gharbi D, et al. ILC2-modulated T cell-to-MDSC balance is associated with bladder cancer recurrence. J Clin Invest 2017;127:2916–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lamm DL, Blumenstein BA, Crissman JD, Montie JE, Gottesman JE, Lowe BA, et al. Maintenance bacillus Calmette-Guerin immunotherapy for recurrent TA, T1 and carcinoma in situ transitional cell carcinoma of the bladder: a randomized Southwest Oncology Group Study. J Urol 2000;163:1124–9. [PubMed] [Google Scholar]
- 30. Chang SS, Boorjian SA, Chou R, Clark PE, Daneshmand S, Konety BR, et al. Diagnosis and treatment of non-muscle invasive bladder cancer: AUA/SUO guideline. J Urol 2016;196:1021–9. [DOI] [PubMed] [Google Scholar]
- 31. Lenis AT, Donin NM, Litwin MS, Saigal CS, Lai J, Hanley JM, et al. Association between number of endoscopic resections and utilization of bacillus calmette-guerin therapy for patients with high-grade, non-muscle-invasive bladder cancer. Clin Genitourin Cancer 2017;15:e25–e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robinson BD, Vlachostergios PJ, Bhinder B, Liu W, Li K, Moss TJ, et al. Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat Commun 2019;10:2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Foth M, Ismail NFB, Kung JSC, Tomlinson D, Knowles MA, Eriksson P, et al. FGFR3 mutation increases bladder tumourigenesis by suppressing acute inflammation. J Pathol 2018;246:331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kates M, Matoso A, Choi W, Baras AS, Daniels MJ, Lombardo K, et al. Adaptive immune resistance to intravesical BCG in non-muscle invasive bladder cancer: implications for prospective BCG-unresponsive trials. Clin Cancer Res 2020;26:882–91. [DOI] [PubMed] [Google Scholar]
- 35. Inman BA, Sebo TJ, Frigola X, Dong H, Bergstralh EJ, Frank I, et al. PD-L1 (B7-H1) expression by urothelial carcinoma of the bladder and BCG-induced granulomata: associations with localized stage progression. Cancer 2007;109:1499–505. [DOI] [PubMed] [Google Scholar]
- 36. Breyer J, Wirtz RM, Otto W, Erben P, Worst TS, Stoehr R, et al. High PDL1 mRNA expression predicts better survival of stage pT1 non-muscle-invasive bladder cancer (NMIBC) patients. Cancer Immunol Immunother 2018;67:403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tretiakova M, Fulton R, Kocherginsky M, Long T, Ussakli C, Antic T, et al. Concordance study of PD-L1 expression in primary and metastatic bladder carcinomas: comparison of four commonly used antibodies and RNA expression. Mod Pathol 2018;31:623–32. [DOI] [PubMed] [Google Scholar]
- 38. Delcourt C, Gemival P, Nouhaud FX, Gobet F, Gillibert A, Ferlicot S, et al. Clinical interest of PD-L1 immuno-histochemistry expression as a predictive factor of Bacillus Calmette Guerin (BCG) efficacy in refractory high-risk non-muscle-invasive bladder cancer (NMIBC). World J Urol 2020;38:1517–24. [DOI] [PubMed] [Google Scholar]
- 39. Jallad S, Goubet S, Symes A, Larner T, Thomas P. Prognostic value of inflammation or granuloma after intravesival BCG in non-muscle-invasive bladder cancer. BJU Int 2014;113:E22–7. [DOI] [PubMed] [Google Scholar]
- 40. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity 2018;48:434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bellmunt J, Kim J, Reardon B, Perera-Bel J, Orsola A, Rodriguez-Vida A, et al. Genomic predictors of good outcome, recurrence, or progression in high-grade T1 non-muscle-invasive bladder cancer. Cancer Res 2020;80:4476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1, 2, 3
Supplemental Figure 1 shows the quality check failure rate by Tumor Cellularity and by Stage and Grade.
Supplemental Figure 2A shows FGFR3 Expression by UROMOL Subclasses in MSK and UNC cohorts and Supplemental Figure 2B demonstrates the Inverse Relationship between FGFR3 expression and mutation with Immune Signature Score.
Supplemental Figure 3 shows the relationship for UROMOL Subclasses in MSK and UNC cohorts between expression of Proliferation markers and FOXM1.
Supplemental Figure 4 shows the relationship between Immune Signature score by Patient Cohort and UROMOL subtype.
Supplemental Figure 5 shows immune score signature using the publicly available UROMOL.
Supplemental Figure 6 shows Recurrence Free Survival in the Northwestern HGT1 cohort by immune score stratified by (1) High, Medium, Low; (2) High/Medium vs. Low.
Supplemental Figure 7 shows PDL1 (CD274) Expression, PD1 (PDCD1) Expression, and CTLA4 Expression by UROMOL Subclasses in MSK and UNC cohorts and RFS (by high, medium, low tertiles) in MSK, UROMOL, & Northwestern Cohorts.
Supplemental Figure 8 shows the relationship between the Fraction of Genome Altered by UROMOL subgroups within the MSK cohort.
Supplemental Figure 9A demonstrates the overlap between "p53-associated" signature from Troester et al. vs. "p53-like" signature from Choi et al; Supplemental Figure 9B shows the "p53-like" signature from Choi et (MDA) by UROMOL Subclasses in MSK and UNC cohorts; and Supplemental Figure 9C shows the "p53-associated" signature from Troester et al. by UROMOL Subclasses in MSK and UNC cohorts.
Supplemental Figure 10 shows the comparison of DNA based alterations in TP53 and other cell cycle regulation genes compared to "p53-associated" signature from Troester et al. (TP53 score), as well as the association between RFS with TP53 Mutation (DNA) & p53 RNA signature.
Supplemental Figure 11 shows the correlation between P53 Score by immune score, FOXM1 expression, Proliferation marker expression.