Abstract
The intestinal flora is a micro-ecosystem that is closely linked to the overall health of the host. We examined the diversity and abundance of intestinal microorganisms in mice following the administration of andrographolide, a component of the Chinese medical herb Andrographis paniculata. Administration of andrographolide produces multiple beneficial effects including anti-inflammatory, antiviral and antibacterial effects but whether it directly influences the gut microbiota is not known. This study investigated whether the oral administration of andrographolide influences the intestinal microbiota and was compared with amoxicillin treatment as a positive control and water only as a negative control. We examined 21 cecal samples and conducted a high-throughput sequencing analysis based on V3-V4 variable region of the 16S rDNA genes. We found that the diversity and abundance of mouse gut microbiota decreased in direct proportion with the amoxicillin dose whereas andrographolide administration did not affect intestinal microbial community structure. The composition of intestinal microbes following andrographolide treatment was dominated by the Firmicutes while Bacteroidetes dominated the amoxicillin treatment group compared with the negative controls. Specifically, the f__Lachnospiraceae_ Unclassified, Lachnospiraceae_ NK4A136_group and Ruminococcaceae_ UCG-014 were enriched with andrographolide administration while Bacteroides, Klebsiella and Escherichia-Shigella significantly increased in the amoxicillin test groups. Amoxicillin administration altered the microbial community composition and structure by increasing the proportion of pathogenic to beneficial bacteria whereas andrographolide administration led to increases in the proportions and abundance of beneficial bacteria. This study provides a theoretical basis for finding alternatives to antibiotics to decrease bacterial resistance and restore intestinal floral imbalances.
Keywords: high-throughput sequencing, gut microbiota, andrographolide, antibiotics, 16S rRNA
Introduction
The mammalian gut microbiota is a complex micro-ecosystem and is an important contributor to host immunity, metabolism and productivity (1, 2). The activities of intestinal microbes closely parallel host growth and development and are active participates in many physiological processes (3, 4). Importantly, disease states such as obesity-related diabetes type II and other metabolic diseases (5, 6) as well as Alzheimer's, autism and other neurological diseases are tightly linked to the heath of the intestinal microbiota (7–9). Remarkably, the compounds resveratrol and berberine used in Chinese traditional medicine are effective alleviators of disease that work through interactions with the gut microbiota (10). Overall, intestinal microbes are key components of host development and health and play important roles in protecting the body from damage and maintaining health.
Andrographolide (C20H30O5) is the primary bioactive ingredient from the Acanthaceae family member the green chiretta (Andrographis paniculata) and is used in Chinese medicine. Andrographolide is a naturally occurring bicyclic diterpenoid whose other members include the bioactive compounds retinol, phytol and forskolin. Andrographolide has been shown to have anti-inflammatory (11), anti-infective (12), anti-cancer (13), anti-hyperglycemic (14) and anti-angiogenic properties (15). It also can function as an immune stimulator (16) and possesses anti-reproductive and other pharmacological effects (17). However, andrographolide is not water soluble, shows poor oral absorption with low bioavailability and is chemically unstable in body fluids (18, 19). With all these negative attributes, it is unknown whether andrographolide can alter the composition of the gut microbiota. Therefore, the objective of this study was to investigate the effect of this compound on the intestinal microbiota of mice.
Materials and Methods
Animals and Sample Collection
A total of seventy (n = 70) 28 day old healthy mice (initial weight 20 ± 2 g) were purchased from the Experimental Animal Center, Zhengzhou University (Zhengzhou, China). The proportion of males and females in each group was 1:1 to decrease the influence of sex on the microbial community. The mice self-propagated through the experimental animal center and possessed a similar genetic background. All the screened mice were subjected to the same immunization program and were determined to be free of other diseases before the experiment. Mice were raised together for 3 days and then randomly divided into seven groups, each containing ten mice including a control group (CON); high dose amoxicillin group (AMXH, 50 mg/kg), medium-dose amoxicillin group (AMXM, 20 mg/kg), low dose amoxicillin group (AMXL 5 mg/kg); high dose andrographolide group (APH, 20 mg/kg), medium-dose andrographolide group (APM, 20 mg/kg) and low dose andrographolide group (APL, 2 mg/kg). All the mice were raised in plastic cages for 14 days with a recommended standard breeding temperature (33–35°C), humidity (53–57%) and illumination time (12 h/12 h light/dark cycle). Moreover, supplies such as feed and water were supplied ad libitum for all groups throughout the entire experiment. Three mice were randomly selected for euthanasia in each group and the gut was excised from abdominal cavity. The separated guts were transferred to a sterilized Kraft paper and knotted with cotton rope to decrease the cross-pollution in the different intestinal segments. The contents in the intermediate sites of cecum were immediately collected and stored at −80°C until further analysis.
DNA Extraction
DNA from the intestinal samples was extracted using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The quantity of DNA was measured using a NanoDrop 2000 UV spectrophotometer (Thermo Scientific, Waltham, MA, USA). Agarose gel (0.8% w/v) electrophoresis was performed to evaluate the quality and quantity of the extracted DNA.
16S rRNA Amplification and Sequencing
The amplification of the 16S rDNA target region (V3/V4) utilized PCR with primers (5' to 3')_338F: ACT CCT ACG GGA GGC AGC A and 806R: GGA CTA CHV GGG TWT CTA AT. DNA fragments from the gels were recovered using an AxyPrep DNA Gel Extraction Kit (Axygen-Corning, Glendale, AZ, USA). An FLx800 fluorescent microplate reader (BioTek, Winooski, VT, USA) was used for DNA quantification in conjunction with a Quant-iTPico Green dsDNA Assay Kit (Invitrogen, Waltham, MA, USA). The purified PCR products were used for constructing the sequencing library using Illumina TruSeq (Illumina, San Diego, CA, USA) following the manufacturer's specifications. Prior to sequencing, the sequencing libraries were subjected to fluorescence quantification and quality inspection. The qualified libraries were assembled, diluted and mixed in equal proportions. The final libraries were subjected to high-throughput sequencing using a MiSeq sequencing machine (GENEWIZ, Inc.).
Bioinformatics and Statistical Analysis
The paired-end sequences achieved from high-throughput sequencing were merged into tags and the quality of raw reads were evaluated using QIIME software (Qiime1.9.1). The reads that passed the initially quality screening were assigned to the corresponding samples according to the primer and barcode information and interrogative sequences such as ambiguous bases, chimeras and mismatched primers were discarded. The obtained high-quality sequences were clustered in a operational taxonomic unit (OTU) on the basis of 97% similarity. Phylogenetic analysis and classifications were performed for the representative sequences for each OTU. The OTU richness distributions were recorded to calculate the alpha diversity. Beta diversity was performed using QIIME (Version 1.7.0) to identify similarities and differences between different samples. In addition, rarefaction curves were constructed to assess the sequencing depth. Linear discriminant analysis effect size (LEfSe) was generated to investigate the differentially abundant taxon. R (v3.0.3) and GraphPad Prism (version 8.0c) were applied to statistical analysis. The criterion of significance was conducted at p-values < 0.05 and the data was expressed as means ± SD.
Results
Quality Assessment and OTU Classification
We initially performed a quality screening of our high throughput sequencing data to eliminate erroneous and questionable sequences to verify sequence reliability. The data was optimized and produced 1,756,513 high-quality reads with an average of 56,662 reads (range 53,560–106,150) per sample (Table 1) and a sequence length of 443–460 bp (Figure 1).
Table 1.
The sequence information of each sample.
| Sample | Raw_reads | Qualified_Reads | Efective (%) |
|---|---|---|---|
| APL1 | 123,587 | 106,150 | 86.28 |
| APL2 | 95,524 | 87,432 | 85.89 |
| APL3 | 104,216 | 92,830 | 89.07 |
| APM1 | 101,234 | 69,924 | 80.53 |
| APM2 | 100,140 | 91,701 | 87.65 |
| APM3 | 109,961 | 96,342 | 88.05 |
| APH1 | 59,824 | 53,560 | 89.34 |
| APH2 | 101,820 | 93,394 | 82.23 |
| APH3 | 117,461 | 101,512 | 86.42 |
| AMXL1 | 117,503 | 100,193 | 85.26 |
| AMXL2 | 81,257 | 62,843 | 77.33 |
| AMXL3 | 88,733 | 74,240 | 83.66 |
| AMXM1 | 115,031 | 91,710 | 79.72 |
| AMXM2 | 99,838 | 84,482 | 84.61 |
| AMXM3 | 108,419 | 88,871 | 81.96 |
| AMXH1 | 99,561 | 83,134 | 83.50 |
| AMXH2 | 75,849 | 62,213 | 82.02 |
| AMXH3 | 107,177 | 82,495 | 76.97 |
| CON1 | 94,668 | 72,573 | 76.66 |
| CON2 | 106,411 | 93,561 | 87.92 |
| CON3 | 86,873 | 67,353 | 77.53 |
Figure 1.
Effective sequence length distribution.
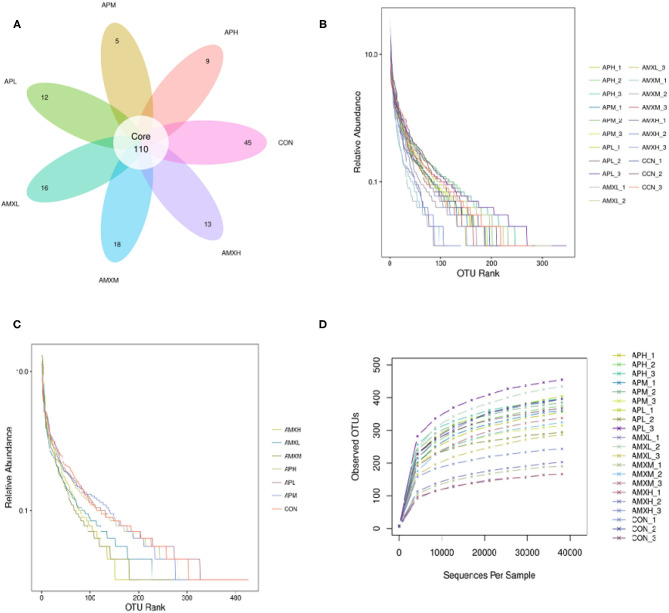
The qualified reads comprised 110 OTUs on the basis of 97% nucleotide-sequence identity (Figure 2A). The rarefaction and rank abundance curves per sample were relatively flat and displayed a tendency to saturate suggesting that the depth and evenness of the sequences meets the requirements for sequencing and analysis (Figures 2B–D).
Figure 2.
Venn diagrams and feasibility analysis (A–D).
Analysis of Microbial Community Diversity
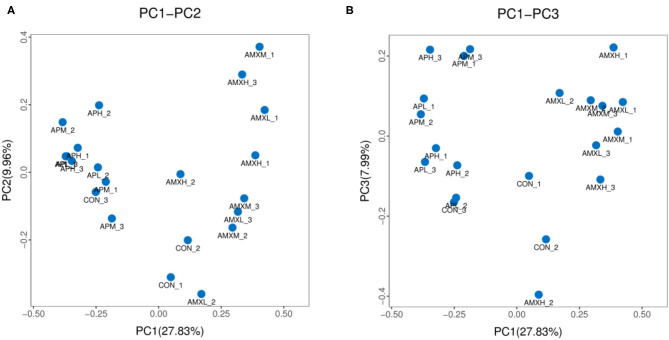
We assessed the alpha diversity of our gut microbiota samples and the Good's coverage estimates varied from 99.8 to 99.9% for all of the samples indicating excellent coverage (Table 2). The Chao1 indices for the experimental groups AMXL, AMXM, and AMXH were 354.61, 323.79, and 250.54 and the corresponding ACE indices were 355.85, 327.99, and 249.51, respectively. Moreover, the averages of Shannon indices for these 3 groups were 4.67, 4.44, and 4.27, respectively. The Chao1, ACE, Shannon, and Simpson indices for these groups treated with AMX displayed a gradual downward trend as the drug concentration was increased indicating that AMX reduced the abundance and diversity of the intestinal microbial community. Remarkably, the three diversity indices (ACE, Chao1, and Shannon) of the control group were higher than those of the AMX treatment groups. In contrast, significant differences in gut microbiota abundance and diversity were only observed between the control and AMXH group. The average for the Chao1 index in the andrographolide treatment groups (APH, APM and APL) ranged from 412.64 to 463.67 while the ACE index ranged from 405.47 to 439.24. Moreover, the average values for the Shannon index in the andrographolide treatment group ranged from 5.50 to 5.70. Interestingly, the Chao1 and ACE index for APH was higher compared with APL and APM and the 4 diversity indices for APM were lower when compared to APL, APM and CON. However, differences between the 4 groups were not significant (P > 0.05). The α-diversity indices revealed no significant difference in the diversity and richness of gut microbiota between the andrographolide and negative control groups. Both the weighted and the unweighted PCoA plots revealed that the samples in each group were clustered separately indicating differences in the gut microbiota for the samples (Figure 3).
Table 2.
Microbial diversity index analysis.
| Sample | Chao1 | ACE | Shannon | Simpson |
|---|---|---|---|---|
| APL | 438.56 ± 80.47 | 437.00 ± 81.78 | 5.57 ± 0.31 | 0.94 ± 0.019 |
| APM | 412.64 ± 20.38 | 405.47 ± 25.64 | 5.63 ± 0.34 | 0.95 ± 0.016 |
| APH | 463.67 ± 22.49 | 439.24 ± 8.56 | 5.50 ± 0.43 | 0.94 ± 0.020 |
| AMXL | 354.61 ± 125.15 | 355.81 ± 121.13 | 4.67 ± 0.92 | 0.89 ± 0.058* |
| AMXM | 323.80 ± 76.82 | 327.99 ± 78.01 | 4.44 ± 0.66* | 0.88 ± 0.041* |
| AMXH | 250.54 ± 38.87* | 249.51 ± 33.95* | 4.27 ± 0.24* | 0.88 ± 0.014* |
| CON | 439.23 ± 14.67 | 444.46 ± 19.06 | 5.75 ± 0.28 | 0.96 ± 0.0070 |
Chao1, ACE, Shannon, and Simpsonwere calculated to assess the alpha diversity of intestinal microbial community. The data are expressed as the mean ± SD.
P < 0.05.
Figure 3.
PCoA analysis of intestinal microbial community in different groups. (A,B) represent PCoA map on the basis of unweighted and weighted uniFrac distance, respectively.
Bacterial Community Composition in Groups
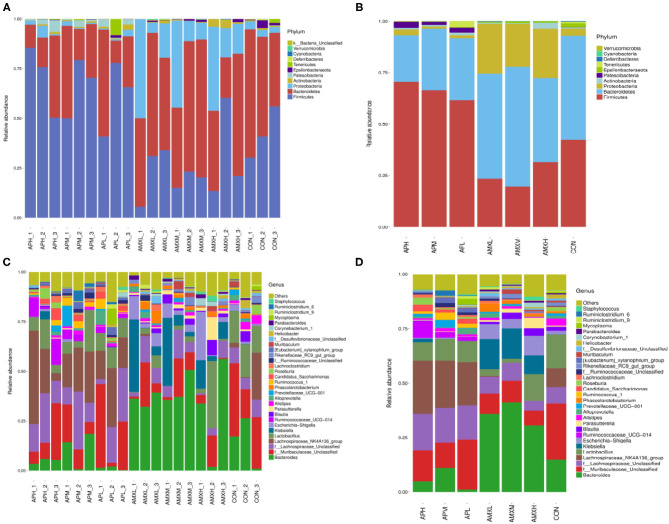
We obtained that were comprised of 11 phyla, 17 classes, 28 orders, 55 families, 157 genera and 168 species. The Firmicutes, Bacteroidetes and Proteobacteria were the 3 dominant phyla for all samples and represented 45.09, 40.46, and 11.12%, of the totals, respectively. These phyla constituted the core of the microbiota and accounted for 96.67% of the taxonomic groups identified. The andrographolide treatment groups were primarily composed of Firmicutes (66.21%) and Bacteroidetes (27.51%) and Proteobacteria (1.83%). The Firmicutes, Bacteroidetes and Proteobacteria content for the AMX treatment groups accounted for 24.88, 50.07, and 22.97% of the total, respectively. The dominant phyla for the negative control group were Bacteroidetes, Firmicutes, and Proteobacteria and were accounted for by 50.55, 40.40, and 3.46% of the totals, respectively (Figure 4).
Figure 4.
The relative abundance of the gut microbiota at the phylum (A,B) and genus (C,D) levels.
The genus Lachnospiraceae_NK4A136 comprised 24.64 and 21.65% of the overall bacterial composition in the APH and APM groups while f_Lachnospiraceae_Unclassified were represented at 16.74 and 15.80%, respectively. The levels of these two genera in the APH and APM groups were significantly higher than the negative control and AMX treatment groups (P < 0.05). In the APL test group, the most numerous genera were f_Muribaculaceae Unclassified and Lachnospiraceae_NK4A136 at 22.79 and 19.77% of the overall bacterial composition, respectively. The levels of the latter genra were significantly higher than the negative control and AMX groups (P < 0.05).
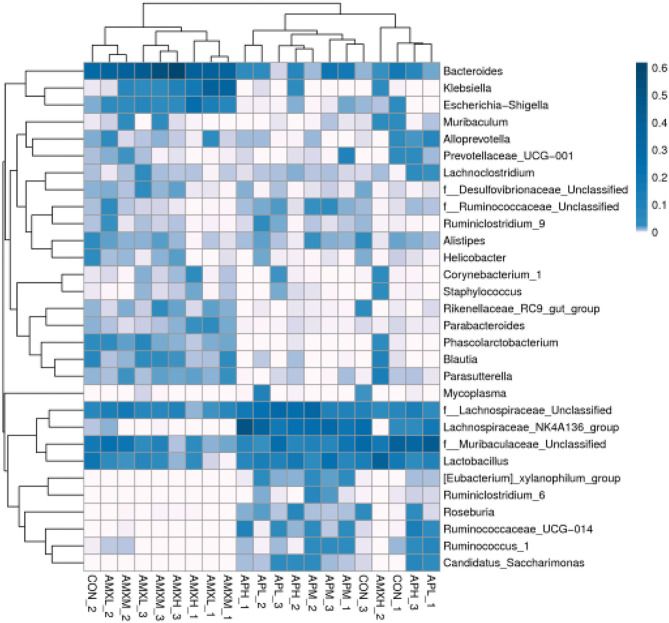
The most abundant genera for the AMX treatment groups were Bacteroides at 35.87, 41.11, and 30.26% for AMXL, AMXM, and AMXH, respectively. Klebsiella was second most abundant for these groups at 13.74, 13.94, and 8.45%, respectively. These levels for Bacteroides and Klebsiella were significantly higher than for the negative control and andrographolide treatment groups (Figure 5).
Figure 5.
Heatmap of the relative richness of bacterial communities at the genus level. Each color-block in the heatmap represents the relative richness of a bacterial genus in a sample.
We performed a metastatic analysis to identify significantly different taxa between the different test groups (Tables 3–11). Our results revealed that at the genus level, the abundance of Candidatus_Saccharimonas increased while the Lactobacillus, Escherichia-Shigella and Alistipes genera decreased significantly in the APL group; The abundance of Ruminococcaceae also increased significantly while Alistipes decreased significantly in the APM and APH groups, respectively. The abundance of Bacteroides in the AMXL and AMXM groups was higher than negative controls while Lactobacillus levels were lower than negative controls. In the latter, Corynebacterium_1 and Klebsiella increased significantly while f_Muribaculaceae_Unclassified and Alipis decreased significantly in the AMXH group. Compared with the AMX test groups, Lachnospiraceae_NK4A136p, Lactobacillus and Candidatus_Saccharimonas abundance in the APL group were significantly higher than for the AMXL group while Bacteroides, Escherichia-Shigella, and Phoscolarctobacterium in the APL group were significantly lower than for the AMXL group. Compared with the AMXM group, the abundance of Lachnospiraceae_NK4A136, Ruminococcus_1, f_Ruminococcaceae_Unclassified and [Eubacterium]_xylanophilum_group increased significantly and was accompanied by a significant decrease in the abundance of Bacteroides Escherichia-Shigella, Blautia and Parasutterella for the APM group. The AMXH and APH groups displayed a significant increase in the abundance of f_Lachnospiraceae_Unclassified, Lachnospiraceae NK4A136_group, Ruminococcaceae_UCG-014, Candidatus_Saccharimonas and Roseburia as well as a significant decrease in the abundance of Phascolarctobacterium and Corynebacterium_1.
Table 3.
Based on Metastats analysis for flora differences between APL and Con groups under the genus.
| Taxon (genus) | Group1_mean | Group1_mean | P-Value |
|---|---|---|---|
| APL | Con | ||
| Lactobacillus | 0.096655015 | 0.154013706 | ≤0.001 |
| Escherichia-Shigella | 0.00046694 | 0.016875919 | 0.030695652 |
| Alistipes | 0.008404823 | 0.031416406 | 0.020913043 |
| Candidatus_Saccharimonas | 0.022779083 | 0.000833917 | 0.047173913 |
Table 11.
Based on Metastats analysis for flora differences between APH and AMXH groups under the genus.
| Taxon (genus) | Group1_mean | Group1_mean | P-Value |
|---|---|---|---|
| APH | AMXH | ||
| f__Lachnospiraceae_Unclassified | 0.167429058 | 0.044311357 | 0.0147 |
| Lachnospiraceae_NK4A136_group | 0.244161782 | 0 | 0.0496 |
| Ruminococcaceae_UCG-014 | 0.078367449 | 0 | 0.03025 |
| Phascolarctobacterium | 0.00053328 | 0.011903551 | 0.03505 |
| Candidatus_Saccharimonas | 0.030343342 | 0 | 0.0448 |
| Roseburia | 0.031571303 | 0.000166727 | 0.0195 |
| Corynebacterium_1 | 0.00036667 | 0.021109588 | 0.0099 |
Table 4.
Based on Metastats analysis for flora differences between APM and Con groups under the genus.
| Taxon (genus) | Group1_mean | Group1_mean | P-Value |
|---|---|---|---|
| APM | Con | ||
| Ruminococcaceae_UCG-014 | 0.028215847 | 0.00100067 | 0.046 |
| Alistipes | 0.013875292 | 0.031416406 | 0.045 |
| Ruminococcus_1 | 0.032751995 | 0.003668828 | 0.0037 |
| f__Ruminococcaceae_Unclassified | 0.020245024 | 0.004902668 | 0.032 |
Table 5.
Based on Metastats analysis for flora differences between APHL and Con groups under the genus.
| Taxon (genus) | Group1_mean | Group1_mean | P-Value |
|---|---|---|---|
| APH | Con | ||
| f__Lachnospiraceae_Unclassified | 0.167429058 | 0.074875512 | 0.0418 |
| Ruminococcaceae_UCG-014 | 0.078367449 | 0.00100067 | 0.049 |
| Alistipes | 0.006536101 | 0.031416406 | 0.02255 |
Table 6.
Based on Metastats analysis for flora differences between AMXL and Con groups under the genus.
| Taxon (genus) | Group1_mean | Group1_mean | P-Value |
|---|---|---|---|
| AMXL | Con | ||
| Bacteroides | 0.359057169 | 0.14836296 | 0.0186 |
| Lactobacillus | 0.031633386 | 0.154013706 | 0.0049 |
| Alistipes | 0.0097429 | 0.031416406 | 0.0282 |
| Phascolarctobacterium | 0.03850549 | 0.007335664 | 0.053 |
Table 7.
Based on Metastats analysis for flora differences between AMXM and Con groups under the genus.
| Taxon (genus) | Group1_mean | Group1_mean | P-Value |
|---|---|---|---|
| AMXM | Con | ||
| Bacteroides | 0.4113589 | 0.14836296 | 0.025473684 |
| Parasutterella | 0.0188766 | 0.004635244 | 0.010263158 |
| Lactobacillus | 0.016674292 | 0.154013706 | ≤0.001 |
Table 8.
Based on Metastats analysis for flora differences between AMXH and Con groups under the genus.
| Taxon (genus) | Group1_mean | Group1_mean | P-Value |
|---|---|---|---|
| AMXH | Con | ||
| f__Muribaculaceae_Unclassified | 0.06852468 | 0.258282341 | 0.048842105 |
| Klebsiella | 0.084603602 | 0.000633583 | 0.043 |
| Alistipes | 0.00333408 | 0.031416406 | 0.010368421 |
| Corynebacterium_1 | 0.021109588 | 0.000867054 | 0.030894737 |
Table 9.
Based on Metastats analysis for flora differences between APL and AMXLon groups under the genus.
| Taxon (genus) | Group1_mean | Group1_mean | P-Value |
|---|---|---|---|
| APL | AMXL | ||
| Bacteroides | 0.012540377 | 0.359057169 | ≤0.001 |
| Lachnospiraceae_NK4A136_group | 0.197847269 | 0.003636974 | 0.013318182 |
| Lactobacillus | 0.096655015 | 0.031633386 | 0.017636364 |
| Escherichia-Shigella | 0.00046694 | 0.068888762 | 0.0405 |
| Phascolarctobacterium | 0 | 0.03850549 | 0.036181818 |
| Candidatus_Saccharimonas | 0.022779083 | 0 | 0.044818182 |
Table 10.
Based on Metastats analysis for flora differences between APM and AMXM on groups under the genus.
| Taxon (genus) | Group1_mean | Group1_mean | P-Value |
|---|---|---|---|
| APM | AMXM | ||
| Bacteroides | 0.111618471 | 0.4113589 | 0.024772727 |
| Lachnospiraceae_NK4A136_group | 0.216624875 | 0.000166763 | 0.004954545 |
| Escherichia-Shigella | 0.005302208 | 0.043795235 | 0.044636364 |
| Blautia | 0.000133387 | 0.028519585 | 0.039681818 |
| Parasutterella | 0.002234237 | 0.0188766 | 0.009909091 |
| Ruminococcus_1 | 0.032751995 | 0.001433477 | 0.014863636 |
| f__Ruminococcaceae_Unclassified | 0.020245024 | 0.002400827 | 0.029727273 |
| [Eubacterium]_xylanophilum_group | 0.021913212 | 0 | 0.019818182 |
Discussion
A large number of bacteria colonize the intestines of mammals and they play an indispensable role in maintaining the overall health of the host (20). The differences between the intestinal floras of different individuals are due to age, diet and health status (21). These intestinal microbes are comprised of both beneficial and harmful members. Adjusting the structure of the intestinal flora and increasing the proportion of probiotics can effectively help the host maintain a healthy state (22, 23). The active ingredients of many traditional Chinese medicines are able to reach the colon and interact with the intestinal flora. By adjusting the composition of the intestinal flora and its metabolites as well as short-chain fatty acids, the functions of the intestinal flora can be improved with beneficial effects on the host. Therefore, studying the composition and structure of intestinal microorganisms is of great significance to disease prevention and treatment (24, 25). The compound andrographolide used in the current study is a potent antibacterial. We found that increasing the levels of andrographolide did not change the alpha diversity of the intestinal flora indicating it does not interfere with the diversity and abundance of the intestinal microbial community in mice. A previous study revealed that baicalin addition to mouse diets increased the diversity index and species abundance of the intestinal flora. The number of beneficial bacteria increased while conditional pathogens such as the enterococci were reduced. Overall, andrographolide induced the emergence of a new and more structurally stable flora (26). The administration of amoxicillin reduced the abundance and diversity of the intestinal flora in mice in a dose-dependent manner in our experiments and this effect of antibiotics in general is well-documented (27, 28). Additionally, the imbalance in the intestinal microbiome caused by antibiotic administration also adversely affects host immune and endocrine systems.
We found that the proportion of Firmicutes in andrographolide-treated mice was significantly higher than that of both the negative control and amoxicillin treatment groups while of the levels of Bacteroidetes and Proteobacteria were less. These results were consistent with previous high-throughput sequencing results in infants treated with antibiotics for bacterial pneumonia (29). We observed that f_Lachnospiraceae_Unclassified, Lachnospiraceae_NK4A136_group and Ruminococcaceae_UCG-014 were abundant in the andrographolide treatment groups while f_Muribaculaceae_Unclassified and Bacteroides levels were the converse. Previous studies have demonstrated that Rumenococcus levels are negatively correlated with inflammatory markers such as C-reactive protein and IL-6. These markers are most likely the result of interactions with pathogen-related molecular patterns, bacterial metabolites, short-chain fatty acids and derivatives of trimethylamine oxide as well as bile acid metabolism. These responses affect intestinal peptide secretion and permeability while regulating the inflammatory state. The presence of Laospirillaceae is closely linked to butyrate metabolism and positively correlated with the presence of inflammatory cells (30, 31). Our results indicate that andrographolide can promote probiotic proliferation while inhibiting pathogenic bacteria. When compared with negative controls, the levels of Bacteroides, Klebsiella and Escherichia-Shigella in the amoxicillin test groups significantly increased while f_Muribaculaceae_Unclassified and Lactobacillus decreased. Klebsiella and Escherichia-Shigella are conditional pathogens. In addition, the presence of Corynebacterium_1 in the high-dose amoxicillin group was also significantly greater than negative controls. This is another example of a reduction of the abundance and diversity of the intestinal microbial community as well as an increase in the levels of pathogenic bacteria due to antibiotic administration.
Conclusions
This study investigated the effects of andrographolide on the intestinal microbial community of mice. The results revealed that the diversity and abundance of gut microbiota treated with AMX undergoes significant alterations that were characterized by elevated levels of harmful bacteria. Conversely, andrographolide administration significantly increased the abundance of beneficial intestinal bacteria including f_Lachnospiraceae_Unclassified, Lachnospiraceae_NK4A136_group and Ruminococcaceae_UCG-014 that are indicators of the stability of the gut microbiota. These results expand the understanding of the potential benefits of andrographolide on the health of the gut microbiome.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at NCBI SRA BioProject, accession no: PRJNA743424.
Ethics Statement
The studies were reviewed and approved by the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People's Republic of China. Written informed consent was obtained from the individual(s), and minor(s)' legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
HW: conceptualization, methodology, formal analysis, data curation, resources, writing-original draft, writing-review and editing, supervision, and project administration. XW: funding acquisition investigation, data curation, and writing-original draft preparation. LH: investigation, validation, data curation, formal analysis, and writing-original draft. XC: investigation, data curation, and resources. JinL: formal analysis, writing-review and editing, supervision, project administration, and funding acquisition. CR: formal analysis, resources, and validation. JicL: funding acquisition and formal analysis. HL: performed manuscript review. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding. This study was supported by Youth teachers Foundation of Xinyang Agriculture and Forestry University (2018LG011), Innovation and Application Special Project of Xinyang (20200016), and Basic Scientific Research Project of Wenzhou in 2019 (No. 2019063).
References
- 1.Pascale A, Marchesi N, Marelli C, Coppola A, Luzi L, Govoni S, et al. Microbiota and metabolic diseases. Endocrine. (2018) 61:357–71 10.1007/s12020-018-1605-5 [DOI] [PubMed] [Google Scholar]
- 2.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. (2013) 500:541–6. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 3.Leshem A, Liwinski T, Elinav E. Immune-microbiota interplay and colonization resistance in infection. Mol Cell. (2020) 78:597–613 10.1016/j.molcel.2020.03.001 [DOI] [PubMed] [Google Scholar]
- 4.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. (2016) 375:2369–79. 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- 5.Sikalidis AK, Maykish A. The gut microbiome and type 2 diabetes mellitus: discussing a complex relationship. Biomedicines. (2020) 8:8. 10.3390/biomedicines8010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitocco D, Di Leo M, Tartaglione L, De Leva F, Petruzziello C, Saviano A, et al. The role of gut microbiota in mediating obesity and diabetes mellitus. Eur Rev Med Pharmacol Sci. (2020) 24:1548–62. 10.26355/eurrev_202002_20213 [DOI] [PubMed] [Google Scholar]
- 7.Pluta R, Ułamek-Kozioł M, Januszewski S, Czuczwar SJ. Gut microbiota and pro/prebiotics in Alzheimer's disease. Aging. (2020) 12:5539–50. 10.18632/aging.102930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roussin L, Prince N, Perez-Pardo P, Kraneveld AD, Rabot S, Naudon L. Role of the gut microbiota in the pathophysiology of autism spectrum disorder: clinical and preclinical evidence. Microorganisms. (2020) 8:1369. 10.3390/microorganisms8091369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cryan JF, O'Riordan KJ, Sandhu K, Peterson V, Dinan TG. The gut microbiome in neurological disorders. Lancet Neurol. (2020) 19:179–94.10. 10.1016/S1474-4422(19)30356-4 [DOI] [PubMed] [Google Scholar]
- 10.Yu L, Xing ZK, Mi SL, Wu X. Regulatory effect of traditional Chinese medicine on intestinal microbiota. Zhongguo Zhong Yao Za Zhi. (2019) 44:34–9. [DOI] [PubMed] [Google Scholar]
- 11.Xu YZ, Tang D, Wang JP. Neuroprotection of andrographolide against microglia-mediated inflammatory injury and oxidative damage in PC12. Neurons Neurochem Res. (2019) 44:2619–30. 10.1007/s11064-019-02883-5 [DOI] [PubMed] [Google Scholar]
- 12.Zhang LL, Bao M, Liu B. Effect of andrographolide and its analogs on bacterial infection: a review. Pharmacology. (2020) 105:3–4. 10.1159/000503410 [DOI] [PubMed] [Google Scholar]
- 13.Ahiwale RJ, Chellampillai B, Pawar AP. Investigation of 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol-sodium (DMPG-Na) lipid with various metal cations in nanocochleate preformulation: application for andrographolide oral delivery in cancer therapy. AAPS Pharmascitech. (2020) 21:279. 10.1208/s12249-020-01801-1 [DOI] [PubMed] [Google Scholar]
- 14.Liang YG. The Effect and Mechanism of Andrographolide Injection on Type 1 Diabetic Rats. ShanXi Medical University (2014). [Google Scholar]
- 15.Guo XL, Zhao MZ, Lin YY. Inhibitory effect of andxogxapholide on angiogenesis induced by the supernatant from cultured tumor cells. J Central South Univ Med Sci. (2018) 3:821–5. 10.11817/j.issn.1672-7347.2018.08.001 [DOI] [PubMed] [Google Scholar]
- 16.Kang RN, He GL, Huang LM. Regulation of andrographolide on pathological lung injury,immune dysfunction and TLR4/NF-KB signaling pathway in klebsiella pneumoniae pneumoniae rats. Chin J Immun. (2020) 36:1453–6. 10.3969/j.issn.1000-484X.2020.12.009 [DOI] [Google Scholar]
- 17.Yang XS, Gao HY, Zhang YX. Research progress in pharmacological action of andrographolide. J Trop Med. (2019) 19:518–22. 10.3969/j.issn.1672-3619.2019.04.032 [DOI] [Google Scholar]
- 18.Zhang SD, Zhao GW, Zhong YQ. Preparation and characterization of andrographolide solid dispersion prepared by Eudragit II. Chin J Hosp Pharm. (2019) 39:2370–4. 10.13286/j.cnki.chinhosppharmacyj.2019.23.03 [DOI] [Google Scholar]
- 19.Guo YS, Dong JL, Yang XF. Rreparation of andrographolide solid dispersions by spray condensation technique. J Chin Med Mater. (2019) 30:2113–7. 10.13863/j.issn1001-4454.2019.09.030 [DOI] [Google Scholar]
- 20.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. (2017) 15:73. 10.1186/s12967-017-1175-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li K, Mehmood K, Zhang H, Jiang X, Shahzad M, Dong X, et al. Characterization of fungus microbial diversity in healthy and diarrheal yaks in Gannan region of tibet autonomous prefecture. Acta Trop. (2018) 182:14–26. 10.1016/j.actatropica.2018.02.017 [DOI] [PubMed] [Google Scholar]
- 22.Aziz Q, Dore J, Emmanuel A, Guarner F, Quigley EM. Gut microbiota and gastrointestinal health: current concepts and future directions. Neurogastroenterol Motil. (2013) 25:4–15. 10.1111/nmo.12046 [DOI] [PubMed] [Google Scholar]
- 23.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. (2012) 489:242–9. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Chen HB, Li SL. Understanding the molecular mechanisms of the interplay between herbal medicines and gut microbiota. Med Res Rev. (2017) 37:1140–85. 10.1002/med.21431 [DOI] [PubMed] [Google Scholar]
- 25.Feng WW, Ao H, Peng C. Gut microbiota, short-chain fatty acids, and herbal medicines. Front Pharmacol. (2018) 9:1354. 10.3389/fphar.2018.01354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song SS. Infiuence of Baicalin on the Mice in the Aspects of the Changes of Intestinal Flora. Guangzhou University of Chinese Medicine (2010). [Google Scholar]
- 27.Du HY, Yanu GY, Zhanugx Tian WY, Wand WJ, Wand P, He GZ. Effect of ampicillin sodium on the structure of intestinal flora in SD rats based on 16S rRNA higlrthroughput amplification. Acta Ecologiae Animalis Domastici. (2019) 41:29–34. 10.3969/j.issn.1673-1182.2020.11.006 [DOI] [Google Scholar]
- 28.Li M, WIu XH, Cao Y. Impact of antibiotics on the murine gut microbiota determined by high throughput sequencing. Chin Microccol. (2019) 31:993–1004. 10.13381/j.cnki.cjm.201909001 [DOI] [Google Scholar]
- 29.Wen XH, He PW, Yu LW, Wen KD, Yue JZ. High-throughput sequencing reveals the change of gut microbiota in infants with pneumonia following antibiotic treatment. Chin J Microecol. (2016) 5:497–500. 10.13381/j.cnki.cjm.201605001 [DOI] [Google Scholar]
- 30.van den Munckhof IC, Kurilshikov A, ter Horst R, Riksen NP, Joosten LA, Zhernakova A, et al. Role of gut microbiota in chronic low-grade inflammation as potential driver for atherosclerotic cardiovascular disease: a systematic review of human studies. Obes Rev. (2018) 19:1719–34. 10.1111/obr.12750 [DOI] [PubMed] [Google Scholar]
- 31.Saresella M, Mendozzi L, Rossi V, Mazzali F, Piancone F, LaRosa F, et al. Immunological and clinical effect of diet modulation of the gut microbiome in multiple sclerosis patients: a pilot study. Front Immunol. (2017) 8:1391. 10.3389/fimmu.2017.01391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at NCBI SRA BioProject, accession no: PRJNA743424.