Abstract
Classical (CKp) and hypervirulent (hvKp) Klebsiella pneumoniae are two different circulating pathotypes. The aim of this study was to assess the prevalence, epidemiology and molecular relatedness of hvKps using a systemic review and meta-analysis. The data extracted from Medline, Embase, and Web of Science and finally 14 studies met the eligible criteria. To combine prevalence proportions of all studies, we performed the metaprop command embedded in the Meta package software. Totally, of 1814 K. pneumoniae isolates, 21.7% (394/1814) were hvKp. The molecular typing showed that all hvKp isolates were grouped into 50 different sequence types (STs) of them ST23, ST11, ST65 and ST86 were common. K1, K2 and K64 were dominant capsule serotypes that strongly related to ST23, ST65 and ST11, respectively. It seems that clonal group 23 (CG23) is associated with liver abscess and CG11 related to various clinical sources.
Keywords: Hypervirulent Klebsiella pneumonia, Virulence factors, Molecular typing, Pathotypesi genotypes
INTRODUCTION
Klebsiella pneumoniae has two different circulating pathotypes in the world including classical (cKp) and hypervirulent (hvKp). These two pathotypes differ in their treatment options, epidemiology, antibiotic resistance pattern, and identification tests (1, 2).
HvKp can cause various infections such as liver abscess, pneumonia, endophthalmitis, and osteomyelitis even in healthy and young people with normal immunity. This bacterium has various plasmid-encoded virulence factors such as aerobactin, salmochelin, resistance to heavy metals, and mucoid capsule. The presence or absence of these plasmid-encoded virulence factors can distinguish hvKp from CKp strains and these features are unique to hvKp (3–6).
The regulator mucoid phenotype gene, rmpA, is a positive regulator for transcription of the capsular polysaccharide gene and increase capsule production and cause hypermucoviscous phenotype (7, 8). On the other hand, some of the K. pneumoniae capsular serotypes are associated with hvKp strains. For example, it was indicated that the incidence of K1 and K2 capsular serotypes is more frequent among hvKp isolates (9). Moreover, it was shown that some sequence types are predominant in hvKp isolates. For example, ST11 and ST23 have been reported as the two most common STs among hvKps and some capsular serotypes have linked to special ST (10, 11). The identification of hvKps is a controversial issue. String test as commonly used to identify hypervirulent strains, has low sensitivity and specificity (4, 12). Recently, molecular marker was proposed for detection of hvKp. For example, in many studies aerobactin encoded by iuc/iut genes, has been considered as molecular marker of hvKp (13, 14).
The aim of the current study was to assess the prevalence of hvKp strains and their molecular epidemiology using a systemic review and meta-analysis based on the preferred reporting global items. This meta-analysis provides more detailed data to overview the status of the hvKp prevalence and common genotypes.
MATERIALS AND METHODS
Search strategy.
We performed a systematic search for: “hypervirulent Klebsiella pneumoniae” and “hypermucoviscous Klebsiella pneumoniae” and “K serotype in Klebsiella pneumoniae” as keywords using different electronic (MeSH) databases including Medline, Embase, and Web of Science from 2000 to January 2020. Unpublished records and grey literature such as conference papers, theses, and patents were excluded.
Eligibility criteria.
Studies were selected according to these criteria: i) having full-text of publications, ii) studies should be conducted on clinical isolates, iii) identification of hvKp strains should be performed based on detection of aerobactin (iuc or iut) genes. We excluded review articles, congress or poster abstracts, and duplicate publications of the same study.
Data extraction.
In this study hvKp was defined based on the presence of iut or iuc genes. Data extraction was done by two independent reviewers and data were extracted as follows: the first author’s name, year of publication, country, total strain number in each study, the number of hypermucoviscous K. pneumoniae strains, string test results, K serotype, MLST sequence type and source of each strain.
Statistical analysis.
The meta-analysis was undertaken using the R software version 3.6.1 statistical computing environment and Meta package version 4.11-0 (15). For combining prevalence’s proportions of all studies, we performed the metaprop command embedded in the Meta package and utilized a generalized linear mixed model (GLMM) to calculate an overall proportion (16).
RESULTS
Study selection.
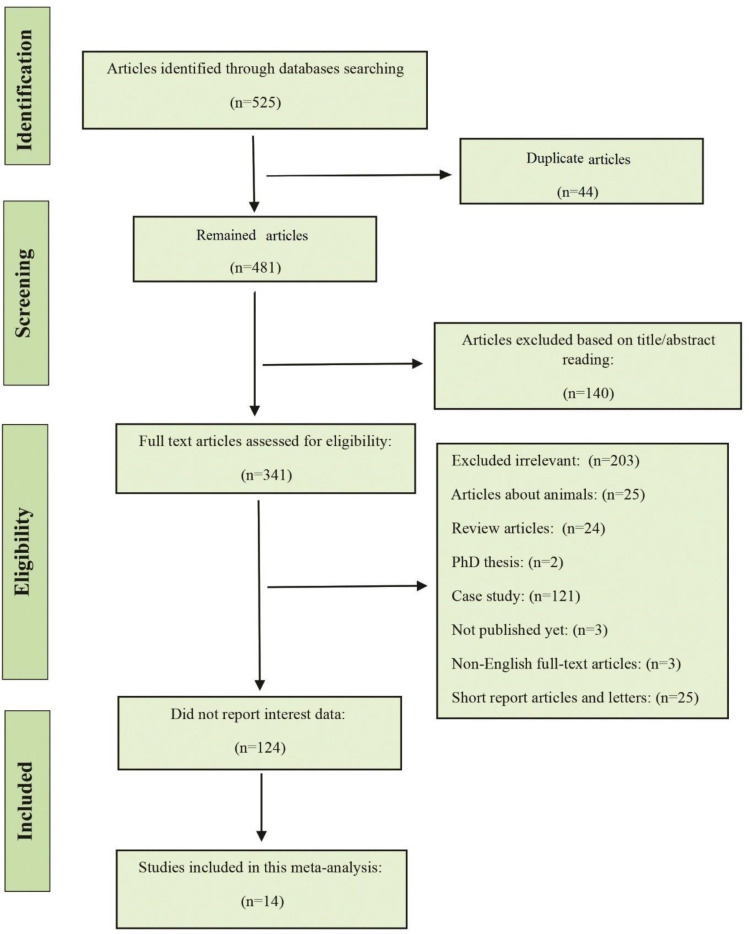
Initially, 525 articles were found trough the database searching. The duplicate studies, reviews, unrelated topics, were removed and 138 studies reminded. In the second screening, we evaluated 138 articles for detailed full-text evaluation. From these articles, 124 did not report data of interest (e.g. number of string test positive hvKp isolates was not presented, or MLST was not conducted). Eventually, the 14 studies met the eligible criteria for statistical analysis and were considered in this study (Fig. 1).
Fig. 1.
The PRISMA flowchart of the study identification and selection criteria.
Study characteristics.
The characteristics of all isolates studies have been presented in Table 1. Out of the 14 studies, 8 of them were performed in China and the rest were done in other countries (Japan, Denmark, Singapore, Taiwan, Korea and France). Totally, 1814 K. pneumoniae isolates were evaluated in 14 studies and 394 strains of K. pneumoniae containing aerobactin were considered as hvKp strains (21.7%). The largest sample size belonged to the Yawei Zhang study (1052 K. pneumoniae isolates) while the study of Benjamin Rossi et al. has the lowest sample size (23 K. pneumoniae isolates). The prevalence of hvKp in the study by Yawei Zhang (5.2%) was less than other studies. While the results of the Russel study showed the most prevalent hvKp (91.4%). Hypermocuviscosity of the hvKp strain defined according to string test in all studies. Out of 394 hvKp isolates, 273 of them (69.3%) were positive for string test. Among 394 hvKp isolates, the most isolates were reported from liver abscess (49.5%) followed by sputum (19.3%), blood (14.5%), CSF (5.6%), urine (3%), stool (2.3%), wound (1%), drainage (0.5%) and other samples (4.3%).
Table 1.
Characteristics of 14 selected studies in this meta-analysis.
| Author | Published year | Country | Total sample size | hvKp (n) | HmvKp (n) | Common K serotype | Common ST type | Source of samples |
|---|---|---|---|---|---|---|---|---|
| Yawei Zhang et al. (26) | 2020 | China | 1052 | 55 | 6 | K64 | ST11 | Blood, sputum, other source |
| Lingling Zhan et al. (27) | 2017 | China | 140 | 13 | 13 | Knt | ST11 | Sputum, urine, blood, drainage |
| Sohei Harada et al. (28) | 2019 | Japan | 102 | 26 | 18 | K1 | ST23 | Blood |
| Carsten Struve et al. (29) | 2015 | Denmark | 80 | 30 | 28 | K1 | ST23 | Liver abscess |
| Russel Lee et al. (23) | 2016 | Singapore | 70 | 64 | 63 | K1 | STnt | Liver abscess |
| Jizi Zhao et al. (30) | 2016 | China | 65 | 16 | 16 | K2 | ST65 | Liver abscess, stool, sputum, blood |
| Yangjy Luo et al. (31) | 2014 | China | 51 | 44 | 32 | K1 | ST23 | Liver abscess |
| Q. Yan et al. (32) | 2016 | China | 49 | 14 | 13 | K1 | ST23 | Sputum |
| Meiping Ye et al. (19) | 2016 | China | 44 | 40 | 27 | K1 | ST23 | Liver abscess |
| Li Fu et al. (33) | 2018 | China | 43 | 16 | 7 | K2 | ST11 and ST107 | Blood, Wound, Sputum, Urine |
| Yee-Huang Ku et al. (34) | 2017 | Taiwan | 33 | 22 | 18 | K2 | ST23 and ST373 | CSF |
| S. W. JUNG et al. (35) | 2013 | Korea | 33 | 16 | 14 | K1 | ST23 | Blood |
| Yajie Zhao et al. (36) | 2019 | China | 29 | 24 | 4 | Knt | ST11 | Blood, Wound, Sputum, Urine, Stool, Drainage |
| Benjamin Rossi et al. (37) | 2019 | France | 23 | 14 | 14 | K1 | ST23 | Liver abscess |
hvKp, hypervirulent K. pneumoniae; HmvKp, hypermucoviscous K. pneumoniae; ST, Sequence Type.
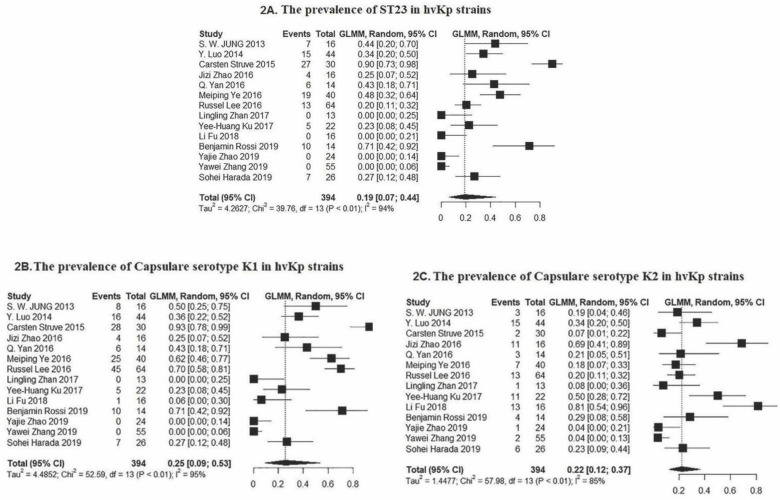
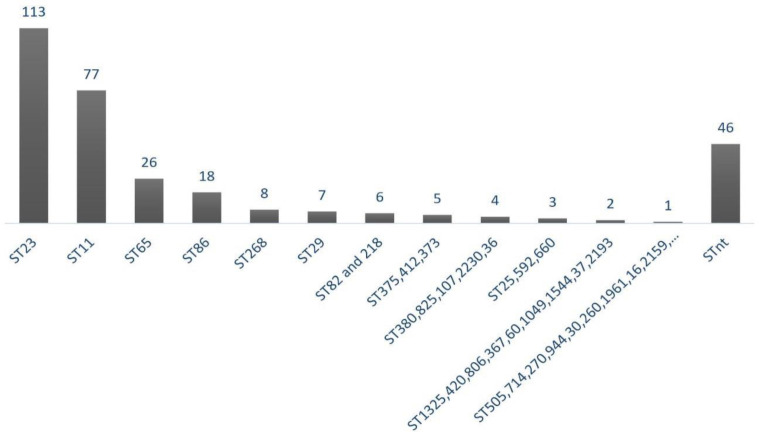
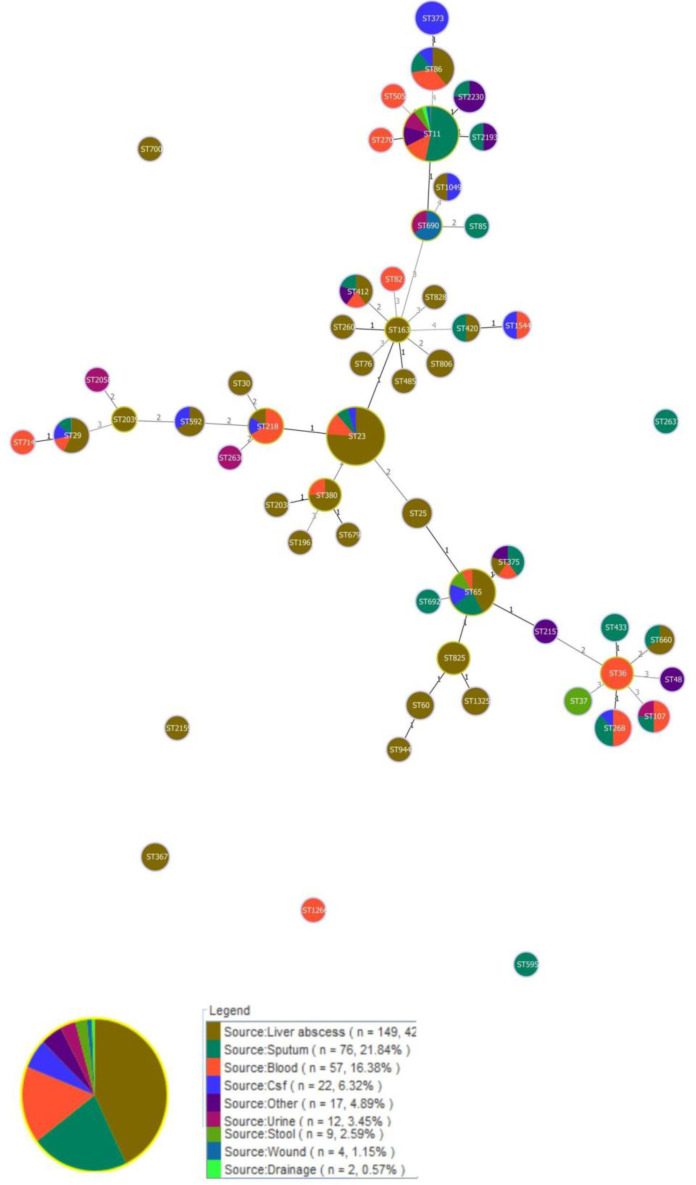
The molecular typing by MLST showed that the 394 hvKp isolates were grouped into 50 different STs and 46 isolates were non-typeable. Among them, 113 isolates belonged to ST23 which has been recognized as dominant sequence types. The Forest plot of meta-analysis for the prevalence of ST23 was shown in Fig. 2. Moreover, the frequency of each ST was depicted in Fig. 3 as follows: ST23 (n=113), ST11 (n=77), ST65 (n=26), ST86 (n=18), ST268 (n=8), ST29 (n=7), ST82 and 218 (n=6), ST373, 375, and 412 (n=5), ST380, 825, 107, 2230, and 36 (n=4), ST25, 592, and 660 (n=3), ST1325, 420, 806, 367, 60, 1049, 1544, 37, and 2193 (n=2), ST505, 714, 270, 944, 30, 260, 1961, 163, 2159, 700, 485, 76, 2038, 2039, 828, 692, 595, 679, 2187, 48, and 1266 (n=1). Out of 113 ST23, 86 of them were isolated from the liver abscess, while ST11 were identified from different sources. Minimum spanning tree (MST) method based upon allelic profiles to investigate the clonal relationship among hvKp isolates was shown in this meta-analysis study. The association between ST types and sample sources were shown in Fig. 4.
Fig. 2.
Forest plot of the meta-analysis on prevalence of (2A) common sequence type-23 (2B) common capsule serotype K1 and (2C) K2 among hvKp strains. P Value <0.01 and confidence interval (CI= 95%).
Fig. 3.
The proportion of STs among 394 hvKp isolates. The collections of STs which have the same frequency were considered in a graph bar. ST, Sequence Type; nt, non-typeable.
Fig. 4.
Minimum spanning tree (MST) analysis of multilocus sequence typing (MLST) data for hvKp isolates. Each circle corresponds to a sequence type (ST) that ST number is given inside each circle. The area of each circle is proportional to the number of isolates. The numbers on the connecting lines between STs correspond to the numbers of allelic differences. The circle was colored according to clinical source. The lines between STs indicate clonal relationships. MST is drawn based on the 3 allelic differences.
The capsular serotypes K1 (39.3%), K2 (23.3%), and K64 (12.7%) were the most prevalent. Other capsular serotypes identified in this meta-analysis included K64 (50/394), K20 (13/394), K57 (11/394), K5 (8/394), K54 (6/394), K62 (4/394), K35 (1/394), K16 (1/394) and also 53 of 394 that were non-typeable (Fig. 2). All strains within ST23 (n=113) belonged to K1 serotype. Also, other STs including ST 367, ST107, ST163, ST2159, ST260, ST700, ST82, and ST1961 were identified as K1 serotype. The proportions of STs among different capsule serotypes have been shown in Table 2. Based on Funnel plot (supplementary data), some evidence of publication bias for the prevalence of hvKps, dominant capsule type and ST was evaluated among the selected studies.
Table 2.
The distribution of STs among different capsule serotypes of 394 hvKp isolates.
| K serotype | Sequence Types |
Total
(n=394) |
|---|---|---|
| K1 | ST23 (n=113), ST367 (n=2), ST107, ST163, ST2159, ST260, ST700, ST82, ST1961 (n=1), and STnt (n=33) | 155 |
| K2 | ST65 (n=25), ST86 (n=18), ST373 (n=5), ST11 (n=4), ST825 (n=4), ST380 (n=4), ST375 (n=4), ST690 (n=3), ST433(n=2), ST690 (n=3), ST25 (n=3), ST1325 (n=2), ST85, ST679, ST2636, ST2637, ST2157, ST2058, ST2038, ST2039, ST107 (n=1), and STnt (n=9), | 92 |
| K64 | ST11 (n=44), ST2191 (n=2), and ST2230 (n=4) | 50 |
| K20 | ST11 (n=2), ST1544 (n=2), ST268 (n=7), ST29, and ST420 (n=1) | 13 |
| K57 | ST218 (n=5), ST412(n=2), ST592 (n=2), ST107, and STnt (n=1) | 11 |
| K5 | ST1049 (n=2), ST60 (n=2), ST76, ST825, ST485, and STnt (n=1) | 8 |
| K54 | ST29 (n=3), ST218, ST412, and ST714 (n=1) | 6 |
| K62 | ST36 (n=4) | 4 |
| K16 | ST660 (n=1) | 1 |
| K35 | ST1266 (n=1) | 1 |
| Knt | ST11 (n=27), ST29 (n=3), ST37 (n=2), ST412 (n=2), ST660 (n=2), ST806 (n=2) ST107, ST268, ST270, ST30, ST375, ST420, ST48, ST505, ST592, ST595, ST65, ST692, ST944 (n=1), and STnt (n=2) | 53 |
DISCUSSION
The hvKp is a pathotype of K. pneumoniae that has recently emerged and has higher virulence, causing high mortality in patients (17, 18). In this meta-analysis study, of all K. pneumoniae, 21.7% were hvKp, of which 69.3% were positive for the string test. These results showed that most hvKp isolates have a hypermucoviscous phenotype and this rate was significant (P<0.01). Conversely, 30.7% of the isolates did not exhibit the hypermucoviscous phenotype. These results indicated that other factors must be considered in identification and screening to avoid missing the number of hvKp. According to source of isolation data, we can conclude that although much of the strains are related to liver abscesses, this bacterium can also cause blood and respiratory infections. Moreover, clonal relatedness of strains is showing the high heterogeneity among hvKp isolates.
Genomic analysis of CC23 showed that virulence factor genes such as pagO, luxR and shiF (related to SAM -dependent methyltransferase) can increase the virulence of ST23 in liver abscesses. The prevalence of these genes in CC23 isolates obtained from liver abscesses, urine and sputum was 100% and 2–11%, respectively (19). On the other hand, half of the ST11 isolates obtained from sputum samples, and the rest were in other samples. For example, in some studies ST11 was detected as the dominant genotype in blood (70%) and urine (57.4%) (20). It seems that there is no significant relationship between the prevalence of ST11 and isolation source. Surprisingly enough, ST23 and ST11 have been identified as two common virulent clones in eastern countries, while few studies have been conducted on K. pneumoniae-induced pyogenic liver abscess (KP-PLA) in other geographical regions (11, 21).
Capsular serotypes K1 and K2 as high prevalent serotypes along with other factors, play an important role in increasing the virulence of hvKp. The reason for the high prevalence of K1 among hvKp isolates is not yet clear, so far. However, serum resistance of hvKp isolates with K1 and K2 capsular serotype may be a logical reasoning for increasing their prevalence (22). On the other hand, intestinal carriage has a key role in the spreading of K1 serotype. A study conducted by Russel Lee in Singapore showed that intestinal carriage of K1 was higher in Chinese than in Malays, Indians and Caucasians. However, there are few data on intestinal carriages from non-Chinese people (11, 23). For example, in a study by Aghamohammad et al. in Iran, K1 was not detected in any K. pneumoniae isolate from fecal carriages (24). High prevalence of K1 serotype in China, and given that 8 out of 14 included studies in this meta-analysis were from China, the heterogeneity of the meta-analysis study cause K1 to be known as a common serotype. However, in this study, other serotypes such as K64, K62 and K35 were also found among hvKp isolates, and K64 was recognized as the third common serotype after K1 and K2. Moreover, 53 isolates were non-typeable serotype. In other studies, beside of K1 and K2 serotypes, the other capsular serotypes such as K5, K16, K20, K54, K57 and KN1 were introduced as common serotypes among the hvKp isolates (23).
Some STs are associated with a particular capsule serotype, e.g. ST23 belonged to clonal group 23 (CG23) and serotype K1, also 44 out of 50 isolates with K64 capsule serotype belonged to ST11 and all four hvKp isolates with K62 capsule type belonged to ST36, but we found more diversity of STs among K2, we identified more than 20 different STs among 92 hvKp isolates with K2 capsule type. ST65 and ST86 were also associated with the K2 capsule serotype. Two systematic reviews by Catalán-Nájera et al. and Bialek-Davenet have shown the association between STs and K serotypes. These authors reported that clonal group 23 (CG23) belonged to serotype K1 in hvKp isolates. In addition, CG86, CG65, CG375 and CG380 were associated with hypervirulent variants of serotype K2 (11, 25).
CONCLUSION
The molecular epidemiology showed that ST23 and K1 recognized as the most common STs and capsule types among the hvKp isolates. However, only about one-third of hvKp isolates, was identified as ST23-K1 genotype. It means that most isolates had genotype diversity. On the other hand, ST23-K1 is a common genotype in China and most of the studies included in this meta-analysis have been reported from China. Therefore, the results of molecular epidemiology can be affected by the geographic regions. Also, K64 identified as the third most common capsular serotype in this meta-analysis. In general, the expansion of high-risk hvKp clone and also the emergence of antibiotic-resistant hvKp isolates increase the mortality of infections associated with this bacterium. Therefore, rapid identification, molecular studies of these strains, and their medically relevant features is necessary.
Finally, it should be mentioned that this study has some limitations. First, we focused on only detection methods based on the presence of iuc or iut genes and studies that used other detection methods were excluded. Second, there was publication bias and heterogeneity among the selected studies, as with other similar systematic reviews. Third, we cannot fully demonstrate the prevalence of hvKp capsule types and sequence types because there is insufficient information on this topic worldwide, as major studies have been conducted in Asian countries, particularly in China.
ACKNOWLEDGEMENTS
The authors would like to thank the personnel in the Bacteriology Department of Pasture Institute of Iran for their help. This research was supported by Pasture Institute of Iran.
REFERENCES
- 1.Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob 2019;18:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sellick JA, Russo TA. Getting hypervirulent Klebsiella pneumoniae on the radar screen. Curr Opin Infect Dis 2018;31:341–346. [DOI] [PubMed] [Google Scholar]
- 3.Lee C-R, Lee JH, Park KS, Jeon JH, Kim YB, Cha C-J, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol 2017;7:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 2015;71:553–560. [DOI] [PubMed] [Google Scholar]
- 5.Tabrizi A, Badmasti F, Shahcheraghi F, Azizi O. Outbreak of hypervirulent Klebsiella pneumoniae harbouring blaVIM-2 among mechanically-ventilated drug-poisoning patients with high mortality rate in Iran. J Glob Antimicrob Resist 2018;15:93–98. [DOI] [PubMed] [Google Scholar]
- 6.Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 2018;56(9):e00776–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Sun S, Zhao ZY, Sun Y. The pathogenicity of rmpA or aerobactin-positive Klebsiella pneumoniae in infected mice. J Int Med Res 2019;47:4344–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Q, Lan P, Huang D, Hua X, Jiang Y, Zhou J, et al. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol 2018;18:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remya P, Shanthi M, Sekar U. Occurrence and characterization of hyperviscous K1 and K2 serotype in Klebsiella pneumoniae. J Lab Physicians 2018;10:283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Bao C, Liu J, Hao X, Cao J, Ye L, et al. Microbiological characterisation of Klebsiella pneumoniae isolates causing bloodstream infections from five tertiary hospitals in Beijing, China. J Glob Antimicrob Resist 2018;12:162–166. [DOI] [PubMed] [Google Scholar]
- 11.Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence 2017;8:1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu F, Lv J, Niu S, Du H, Tang Y-W, Bonomo RA, et al. In vitro activity of ceftazidime-avibactam against carbapenem-resistant and hypervirulent Klebsiella pneumoniae isolates. Antimicrob Agents Chemother 2018;62 (8):e01031–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russo TA, Olson R, MacDonald U, Metzger D, Maltese LM, Drake EJ, et al. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 2014;82:2356–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey DC, Alexander E, Rice MR, Drake EJ, Mydy LS, Aldrich CC, et al. Structural and functional de lineation of aerobactin biosynthesis in hypervirulent Klebsiella pneumoniae. J Biol Chem 2018;293:7841–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balduzzi S, Rücker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evidence-based mental health. Evid Based Ment Health 2019; 22:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stijnen T, Hamza TH, Özdemir P. Random effects meta-analysis of event outcome in the framework of the generalized linear mixed model with applications in sparse data. Stat Med 2010; 29:3046–3067. [DOI] [PubMed] [Google Scholar]
- 17.Bengoechea JA, Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev 2019;43:123–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajabnia R, Asgharpour F, Shahandashti EF, Moulana Z. Nosocomial emerging of (VIM1) carbapenemase-producing isolates of Klebsiella pneumoniae in North of Iran. Iran J Microbiol 2015;7:88–93. [PMC free article] [PubMed] [Google Scholar]
- 19.Ye M, Tu J, Jiang J, Bi Y, You W, Zhang Y, et al. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol 2016;6:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko KS, Lee J-Y, Baek JY, Suh J-Y, Lee MY, Choi JY, et al. Predominance of an ST11 extended-spectrum β-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J Med Microbiol 2010;59:822–828. [DOI] [PubMed] [Google Scholar]
- 21.Dong N, Yang X, Zhang R, Chan EW-C, Chen S. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg Microbes Infect 2018;7:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh K-M, Chiu S-K, Lin C-L, Huang L-Y, Tsai Y-K, Chang J-C, et al. Surface antigens contribute differently to the pathophysiological features in serotype K1 and K2 Klebsiella pneumoniae strains isolated from liver abscesses. Gut Pathog 2016;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee IR, Molton JS, Wyres KL, Gorrie C, Wong J, Hoh CH, et al. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci Rep 2016;6:29316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aghamohammad S, Badmasti F, Solgi H, Aminzadeh Z, Khodabandelo Z, Shahcheraghi F. First report of extended-spectrum betalactamase-producing Klebsiella pneumoniae among fecal carriage in Iran: high diversity of clonal relatedness and virulence factor profiles. Microb Drug Resist 2020;26:261–269. [DOI] [PubMed] [Google Scholar]
- 25.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard A-S, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 2014;20:1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Jin L, Ouyang P, Wang Q, Wang R, Wang J, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother 2020;75:327–336. [DOI] [PubMed] [Google Scholar]
- 27.Zhan L, Wang S, Guo Y, Jin Y, Duan J, Hao Z, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol 2017;7:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harada S, Aoki K, Yamamoto S, Ishii Y, Sekiya N, Kurai H, et al. Clinical and molecular characteristics of Klebsiella pneumoniae isolates causing bloodstream infections in Japan: occurrence of hypervirulent infections in health care. J Clin Microbiol 2019;57(11):e01206–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 2015;6 (4):e00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Chen J, Zhao M, Qiu X, Chen X, Zhang W, et al. Multilocus sequence types and virulence determinants of hypermucoviscosity-positive Klebsiella pneumoniae isolated from community-acquired infection cases in Harbin, North China. Jpn J Infect Dis 2016;69:357–360. [DOI] [PubMed] [Google Scholar]
- 31.Luo Y, Wang Y, Ye L, Yang J. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect 2014;20: O818–824. [DOI] [PubMed] [Google Scholar]
- 32.Yan Q, Zhou M, Zou M, Liu W-e. Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur J Clin Microbiol Infect Dis 2016;35:387–396. [DOI] [PubMed] [Google Scholar]
- 33.Fu L, Huang M, Zhang X, Yang X, Liu Y, Zhang L, et al. Frequency of virulence factors in high biofilm formation blaKPC-2 producing Klebsiella pneumoniae strains from hospitals. Microb Pathog 2018;116:168–172. [DOI] [PubMed] [Google Scholar]
- 34.Ku Y-H, Chuang Y-C, Chen C-C, Lee M-F, Yang Y-C, Tang H-J, et al. Klebsiella pneumoniae isolates from meningitis: epidemiology, virulence and antibiotic resistance. Sci Rep 2017;7:6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung S, Chae H, Park Y, Yu J, Kim S, Lee H, et al. Microbiological and clinical characteristics of bacteraemia caused by the hypermucoviscosity phenotype of Klebsiella pneumoniae in Korea. Epidemiol Infect 2013;141:334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Zhang X, Torres VVL, Liu H, Rocker A, Zhang Y, et al. An outbreak of carbapenem-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a major teaching hospital in Wenzhou, China. Front Public Health 2019;7:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi B, Gasperini ML, Leflon-Guibout V, Gioanni A, de Lastours V, Rossi G, et al. Hypervirulent Klebsiella pneumoniae in cryptogenic liver abscesses, Paris, France. Emerg Infect Dis 2018;24:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]