Abstract
Species identification within the genus Mycobacterium and subsequent antibiotic susceptibility testing still rely on time-consuming, culture-based methods. Despite the recent development of DNA probes, which greatly reduce assay time, there is a need for a single platform assay capable of answering the multitude of diagnostic questions associated with this genus. We describe the use of a DNA probe array based on two sequence databases: one for the species identification of mycobacteria (82 unique 16S rRNA sequences corresponding to 54 phenotypical species) and the other for detecting Mycobacterium tuberculosis rifampin resistance (rpoB alleles). Species identification or rifampin resistance was determined by hybridizing fluorescently labeled, amplified genetic material generated from bacterial colonies to the array. Seventy mycobacterial isolates from 27 different species and 15 rifampin-resistant M. tuberculosis strains were tested. A total of 26 of 27 species were correctly identified as well as all of the rpoB mutants. This parallel testing format opens new perspectives in terms of patient management for bacterial diseases by allowing a number of genetic tests to be simultaneously run.
Tuberculosis (TB), caused by members of the Mycobacterium tuberculosis complex, is one of the most common human infectious diseases, causing three million deaths a year worldwide (20). While the disease is associated with impoverished economic conditions, TB is on the rise in many industrialized nations. The spread in TB is due to immigration, the emergence of drug-resistant strains, and the AIDS epidemic. In advanced stages of AIDS, where TB infections are commonly found, mycobacterial infections due to members of the M. avium-intracellulare complex (MAC) are also on the increase (10). Reduced and compromised immune function as found in newborns, infants, and immunosuppressed individuals allows opportunistic infections caused by mycobacteria other than M. tuberculosis (MOTT). Such species infect many sites within the body but primarily cause pulmonary disease, cervical lymphadenopathy, and localized skin and soft tissue lesions. Mycobacterial species associated with MOTT disease states include M. avium-intracellulare, M. chelonae, M. fortuitum, M. kansasii, M. xenopi, M. marinum, M. scrofulaceum, and M. szulgai.
The increasing number of mycobacterial infections has made it clinically important to quickly identify mycobacteria at the species level. The diagnosis of a pathogenic versus a nonpathogenic species not only has epidemiological implications but is also relevant to the demands of patient management. Individuals with highly contagious infections may be isolated to prevent the spread of the disease. Antibiotic treatments may vary according to the species encountered.
The emergence of drug-resistant M. tuberculosis has created additional concern in the event of TB diagnosis. These strains are resistant to the primary antituberculosis drugs and require a specialized antibiotic treatment. Recently, a number of genetic changes leading to rifampin, isoniazid, pyrazinamide, and ethambutol resistance have been characterized (4). For example, rifampin resistance has been shown to be conferred by missense mutations within a short motif of the beta subunit of DNA-dependent RNA polymerase encoded by the rpoB gene (26) in over 90% of rifampin-resistant isolates (12). The availability of sequence changes resulting in a resistant phenotype allows for the development of probe-based assays for antibiotic resistance.
To date, the design of molecular tests has sped up the diagnostics of this fastidious genus but still suffers some drawbacks. For species identification, a highly polymorphic region of the 16S rRNA gene has been shown to contain species-specific polymorphisms (2, 14). This region is currently used in several commercially available assays, applicable either directly on specimen (Accu-Probe; Gen-Probe, San Diego, Calif.) or after enzymatic amplification of the target for improved sensitivity (AMTD; Gen-Probe; Amplicor MTB, Roche Diagnostics Systems, Somerville, N.J.). However, these kits are mostly designed for the diagnosis of TB and do not take full advantage of the information-rich content of this region. The vast majority of mycobacterial species can be discriminated by this region, as originally reported by Springer et al. (23) and demonstrated directly on clinical specimens by Kirschner et al. (15).
The GeneChip technology, recently developed by Affymetrix (Santa Clara, Calif.), is a promising new method for assessing genetic diversity on a larger scale. This method relies upon the hybridization of the nucleic acid target to large sets of oligonucleotides synthesized at precise locations on a miniaturized glass substrate (7). This technology has been already successfully applied for monitoring gene expression and screening of mutations and polymorphisms in several human and viral genes (3, 6, 17, 18). We have investigated this probe array strategy for bacteriology testing, focusing on mycobacterial diseases. An assay that is able to interrogate the sequence of regions from the 16S rRNA and rpoB loci has been developed. Unique hybridization patterns allow for the identification of Mycobacterium species and the rifampin-resistant alleles.
MATERIALS AND METHODS
Bacterial strains, phenotypic identification, and susceptibility testing.
The strains originated from reference collections (American Type Culture Collection and Deutsche Sammlung von Mikroorganismen) or were clinical isolates obtained through several laboratories. All isolates were grown on either Lowenstein-Jensen or Coletsos medium and were examined for growth rate, gross and microscopic colony morphology, and pigmentation. They were subjected to biochemical tests for niacin, nitrate reduction, catalase (drop method), arylsulfatase, pyrazinamidase, urease, and lipase (Tween 80 hydrolysis) (21) and also to complementary tests for certain isolates. Rifampin resistance testing was performed by the proportion method with medium containing 1 mg of rifampin per ml (13). The isolate was considered resistant if there was more than 1% growth on the antibiotic-containing medium compared with the growth on the drug-free medium.
Bacterial rpoB clones.
Clones were generated from rpoB PCR products that were ligated into pGEM-T Easy plasmids (Promega, Madison, Wis.). The rpoB PCR products were generated from clinical isolates by using the following primer pair; MTX 2281 TB (CCC AGG ACG TGG AGG CGA TCA CAC CGC A) and MTX 2985 TB (ACG TCG CCG CGT CGA TCG CCG) (8). MTX 2281 TB is located at coordinates 2281 to 2308 of GenBank accession no. L27989 sequence. The resulting amplicon was approximately 700 bp in length. The insert sequence was characterized by automated DNA sequencing.
Probe array design and tiling strategy.
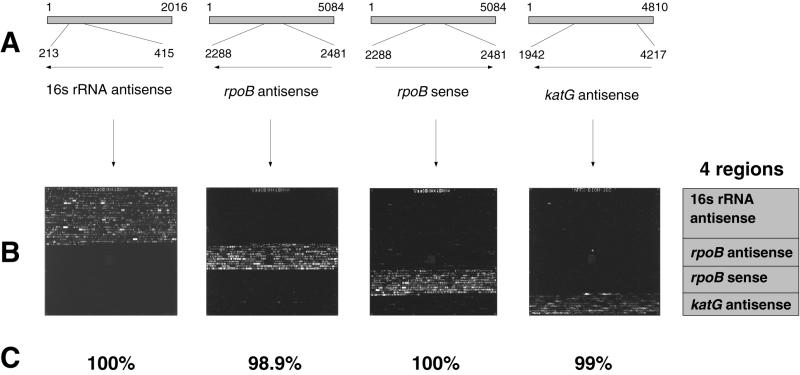
An array tiling strategy similar to that described by Cronin et al. (6), Kozal et al. (17), and Chee et al. (3) was used to identify the sequences differentiating the Mycobacterium species and the rpoB mutations. For every base within a given reference sequence, four probes of equal length are synthesized on the array. The interrogated base is centrally located within the probes, which also have common 3′ and 5′ termini. One probe is an exact complement to the reference sequence, while the three other probes represent the possible single base mismatches to the interrogated base. Base calls are determined by comparing the hybridization intensity of a labeled target to the four probes (see Fig. 2). For the 16S rRNA sequences, probe redundancy was eliminated by synthesizing probes shared by two or more references only once on the array. In addition to the wild-type probes for rpoB, each rifampin resistance mutation is represented by its own set of probes. The array also contains 2.2 kb of the M. tuberculosis katG gene sequence, which encodes catalase peroxidase. The probe array is divided into four distinct zones corresponding to 16S rRNA, rpoB antisense, rpoB sense, and katG antisense sequences, as shown in Fig. 1. The array is divided into specific 50-by-50-μm units or cells over a 1.28- by 1.28-cm area, making a total number of 65,000 different synthesis sites. A database of 82 unique 16S rRNA sequences was utilized to design the array, which enables the discrimination of 54 phenotypically distinct species. Certain species or taxonomic complexes are represented by more than one reference sequence due to the sequence heterogeneity observed in the region of the 16S rRNA tiled on the array (i.e., the MAC). Also, a database of rpoB sequences from 61 rifampin-resistant isolates is represented on the array. Those sequences contain 51 unique rpoB mutations.
FIG. 2.
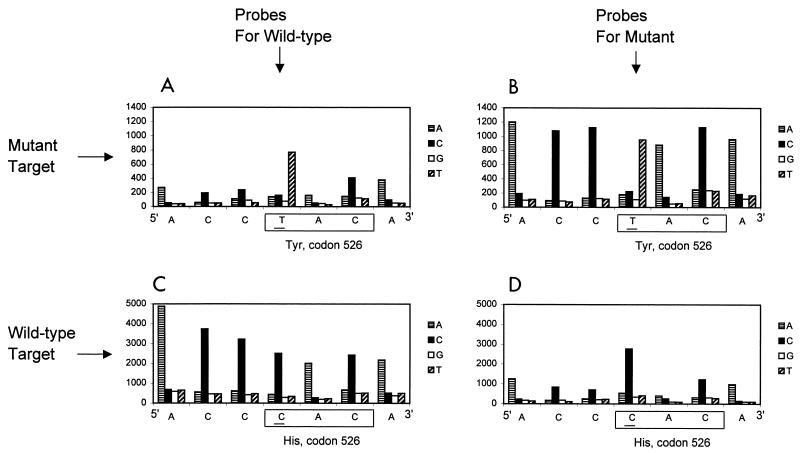
M. tuberculosis rpoB mutation detection with the Mycobacterium probe array. Labeled rpoB targets (sense strand) originating from either a His526Tyr (CAC→TAC) M. tuberculosis rpoB mutant isolate (A and B) or a wild-type M. tuberculosis rpoB sequence (C and D) have been hybridized on the Mycobacterium probe array. Results (signal intensities at each interrogated position for the four different probes) have been obtained by using either the probes specific for the His526Tyr mutant sequence (B and D) or the probes specific for the wild-type sequence (A and C). The signal intensities are shown on the y axis of each panel.
FIG. 1.
The Mycobacterium probe array. The Mycobacterium probe array can be divided into four separate regions, with specific probes for the analysis of either 16S rRNA antisense, rpoB antisense, rpoB sense, or katG antisense targets. (A) Diagram of regions analyzed on the array. First line shows the M. tuberculosis 16S rRNA, rpoB, and katG reference sequences (GenBank accession no. M20940, L27989, and X68081, respectively) with the nucleotide numbers (1 to 2016 for 16S rRNA, 1 to 5084 for rpoB, and 1 to 4810 for katG). Second line shows the region of each reference sequence that is analyzed on the array (213 to 415 for 16S rRNA, 2288 to 2481 for rpoB, and 1942 to 4217 for katG). Arrows indicate whether antisense (←) or sense (→) RNA single-stranded targets are being used. (B) Hybridization experiments with the M. tuberculosis 16S rRNA antisense, rpoB antisense, rpoB sense, and katG antisense targets, described for panel A. The fluorescence images were obtained following hybridization of fluorescein-labeled fragmented RNA, generated by in vitro transcription from PCR amplicons, as described in Materials and Methods (use of hybridization buffer 2). (C) Results (percentages of correct base calling) are given for each experiment and represent performances routinely achieved with MnCl2-imidazole fragmentation protocol and hybridization buffer 2.
Target preparation.
For isolates, one or two freshly grown colonies of bacteria (3- to 5-mm diameter; ca. 108 bacteria) were scraped on the end of a spatula and resuspended into 250 μl of sterile water in a 1.5-ml Eppendorf tube. Total nucleic acids were released from culture material by vortexing the bacterial suspension in the presence of glass beads. A 5-μl aliquot of the lysate was added directly to the PCR. Alternatively, 20 ng of plasmid DNA was added to the PCR. The 16S hypervariable region was PCR amplified with Mycobacterium genus primers (positions 213 to 236 and 394 to 415 on the M. tuberculosis reference sequence M20940 [GenBank]; M. tuberculosis amplicon size is 202 bp). The primers also contained either a bacteriophage T3 or T7 promoter sequence at their 5′ ends (17): T3-M1, 5′-AATTAACCCTCACTAAAGGGAACACGTGGGTGATCTGCCCTGCA, and T7-M2, 5′-GTAATACGACTCACTATAGGGCTGTGGCCGGACACCCTCTCA (promoter sequences in boldface and mycobacterial sequence in lightface). The M. tuberculosis rpoB Rifr locus was amplified with MTX 2281 TB and MTX 2985 TB primers (8), described above, and tailed with T3 or T7 promoter sequences. Amplification of the M. tuberculosis katG region analyzed on the array was performed with T3- or T7-tailed primers 33f (5′-TCACAGCCCGATAACACCAAC) and 2288r (5′-GGCCGATCAACCCGAATCAGC) (positions 1942 to 1962 and 4197 to 4217 on X68081, respectively; M. tuberculosis amplicon size is 2,275 bp).
PCR was carried out in a 100-μl reaction volume containing 50 mM KCl, 10 mM Tris (pH 8.3), 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, 5% (vol/vol) dimethyl sulfoxide, 0.5 μM (each) primer, 200 μM (each) deoxynucleotide triphosphates, and 1.5 U of Taq polymerase (AmpliTaq; Perkin-Elmer, Norwalk, Conn.). PCR was performed in a Perkin-Elmer 2400 thermal cycler with an initial denaturation step at 94°C for 5 min and cycling conditions of 94°C for 45 s, 60°C for 30 s, and 72°C for 30 s (2 min for katG target) for 35 cycles and 72°C for 10 min for the last cycle. PCR products were analyzed by agarose gel electrophoresis.
The promoter-tagged PCR amplicons were used for generating labeled single-stranded RNA targets by in vitro transcription. Each 20-μl reaction mixture contained approximately 50 ng of PCR product; 20 U of T3 or T7 RNA polymerase (Promega); 40 mM Tris acetate (pH 8.1); 100 mM Mg(acetate)2; 10 mM dithiothreitol; 1.25 mM (each) ATP, CTP, and GTP; 0.5 mM UTP; and 0.25 mM fluorescein-UTP. The reaction was carried out at 37°C for 1 h. In vitro-transcribed RNA was fragmented either by adjustment of the concentration of MgCl2 to 30 mM and heating at 94°C for 30 min (17) or by incubation with 30 mM MnCl2 and 30 mM imidazole at 65°C for 30 min. The efficiency of fragmentation was analyzed by denaturing polyacrylamide gel electrophoresis.
Probe array hybridization and analysis.
Hybridizations of the probe arrays were performed with the GeneChip Fluidics Station (Affymetrix). One to five microliters of the fragmented labeled RNA target was diluted in 500 μl of hybridization buffer. Hybridization buffer 1 consisted of 4.5× SSPE (0.675 M NaCl, 45 mM NaH2PO4, 4.5 mM EDTA, pH 7.4) and 0.05% (vol/vol) Triton X-100. Hybridization buffer 2 consisted of 6× SSPE (0.9 M NaCl, 60 mM NaH2PO4, 6 mM EDTA, pH 7.4), 0.05% (vol/vol) Triton X-100, and a proprietary mix of detergents and denaturing agents. The probe array was incubated in the presence of these solutions for 30 min at 50°C and then washed twice in 3× SSPE (0.45 M NaCl, 30 mM NaH2PO4, 3 mM EDTA, pH 7.4)–0.005% (vol/vol) Triton X-100 at 30°C. Fluorescent signal emitted by target bound to the array was detected at a pixel resolution of 6 μm by using the GeneArray scanner (Hewlett-Packard, Palo Alto, Calif.). Probe array cell intensities, nucleotide base call, sequence determination, and reports were generated by functions available on GeneChip software (Affymetrix). A candidate selection index was determined by the percentage of homology between the experimentally derived sequence and all of the reference sequences tiled on the array (Table 1).
TABLE 1.
Mycobacterium species identification
| Isolate reference no. (ATCC-DSM)d | Phenotypic identification | Blind-testing status | Genetic identification (GeneChip results)a
|
|||
|---|---|---|---|---|---|---|
| Highest reference selection | Score | Second reference selection | Score | |||
| 1 | M. tuberculosis | M. tuberculosis (X52917) | 98.2 | M. asiaticum | 94.6 | |
| 2 | M. tuberculosis | + | M. tuberculosis (X52917) | 98.2 | M. asiaticum | 94.0 |
| 3 | M. tuberculosis | M. tuberculosis (X52917) | 98.8 | M. asiaticum | 94.6 | |
| 4 | M. tuberculosis | + | M. tuberculosis (X52917) | 99.4 | M. asiaticum | 94.6 |
| 5 (27294) A | M. tuberculosis | M. tuberculosis (X52917) | 98.2 | M. asiaticum | 95.8 | |
| 6 (clinic) | M. tuberculosis | M. tuberculosis (X52917) | 99.4 | M. asiaticum | 94.6 | |
| 7 (clinic) | M. tuberculosis | M. tuberculosis (X52917) | 100 | M. asiaticum | 94 | |
| 1 | M. bovis | M. tuberculosis (X52917)/M. bovis | 98.8bc | M. asiaticum | 94.6 | |
| 1 | M. avium | M. avium (X52918)/M. paratuberculosis | 100c | M. intracellulare 4 | 94.7 | |
| 2 | M. avium | + | M. avium (X52918)/M. paratuberculosis | 100c | M. intracellulare 4 | 94.1 |
| 3 (43216) D | M. avium | M. avium (X52918)/M. paratuberculosis | 100c | M. intracellulare 4 | 94.1 | |
| 4 (29555) A | M. avium | M. intracellulare 4 (M61683) | 99.4 | M. intracellulare 1 | 98.2 | |
| 1 | M. intracellulare | M. intracellulare 1 (X52927) | 99.4 | Strain sp1 | 98.2 | |
| 2 | M. intracellulare | + | M. intracellulare 1 (X52927) | 99.4 | Strain sp1 | 98.2 |
| 3 (43223) D | M. intracellulare | M. intracellulare 1 (X52927) | 100 | Strain sp1 | 98.8 | |
| 1 (35767) A | M. intracellulare | M. avium (X52918)/M. paratuberculosis | 97c | M. leprae | 94.2 | |
| 1 (25276) A | M. asiaticum | M. asiaticum 2 (X55604) | 98.2 | M. ulcerans 1 | 97 | |
| 2 (25276) A | M. asiaticum | M. asiaticum 2 (X55604) | 100 | M. celatum 2 | 97 | |
| 1 | M. chelonae | M. chelonae 1-fortuitum (X52921) | 98.8 | M. fortuitum-chelonae | 97.6 | |
| 2 (35752) A | M. chelonae | M. chelonae 7 (X82236)/M. abscessus | 99.4c | M. fortuitum 1 | 94.1 | |
| 3 (clinic) | M. chelonae | M. chelonae 1-fortuitum (X52921) | 97 | M. fortuitum-chelonae | 94.7 | |
| 4 (14472) A | M. chelonae | M. chelonae 7 (X82236)/M. abscessus | 97c | M. chelonae 4 | 92.3 | |
| 1 | M. fortuitum | M. fortuitum-chelonae (X52933) | 98.2 | M. chelonae 4 | 97.6 | |
| 2 (49403) A | M. fortuitum | M. fortuitum 1 (X65528)/M. senegalense | 98.2c | M. chelonae 4 | 97 | |
| 1 | M. gordonae | + | M. gordonae (X52923) | 97 | M. shimodei | 91.1 |
| 2 | M. gordonae | M. gordonae (X52923) | 97.7 | Strain MCRO 6 | 92.3 | |
| 3 | M. gordonae | M. gordonae (X52923) | 98.8 | Strain sp. 6 | 92.4 | |
| 4 (14470) A | M. gordonae | M. gordonae (X52923) | 98.8 | M. asiaticum | 94 | |
| 5 (clinic) | M. gordonae | M. gordonae (X52923) | 98.8 | M. shimodei | 94.1 | |
| 6 (clinic) | M. gordonae | + | M. gordonae (X52923) | 97.6 | Strain sp1 | 92.3 |
| 1 (43224) D | M. kansasii | M. kansasii (X15916)/M. gastri | 100c | M. simiae | 98.8 | |
| 2 (12478) A | M. kansasii | M. kansasii (X15916)/M. gastri | 99.4c | M. haemophilum | 96.5 | |
| 1 | M. smegmatis | M. smegmatis (X52922) | 97.7 | Strain MCRO 17 | 95.3 | |
| 1 | M. malmoense | + | M. malmoense (X52930) | 99.4 | M. simiae | 98.8 |
| 2 | M. malmoense | M. malmoense (X52930) | 100 | M. szulgai 2 | 97.6 | |
| 3 | M. malmoense | M. malmoense (X52930) | 100 | M. szulgai 2 | 97.6 | |
| 4 (29571) A | M. malmoense | M. malmoense (X52930) | 100 | M. interjectum | 97 | |
| 5 (29571) A | M. malmoense | M. malmoense (X52930) | 100 | M. szulgai 2 | 98.8 | |
| 1 | M. shimodei | M. shimodei (X82459) | 98.8 | Strain sp.6 | 97.7 | |
| 2 (27962) A | M. shimodei | M. shimodei (X82459) | 98.2 | Strain sp.6 | 97.1 | |
| 3 (clinic) | M. shimodei | M. shimodei (X82459) | 100 | Strain sp.6 | 97.1 | |
| 1 | M. flavescens | M. flavescens (X52932) | 97.7 | M. smegmatis | 96.5 | |
| 1 | M. simiae | M. simiae (X52931) | 100 | M. kansasii/M. gastri | 98.2c | |
| 2 (25275) A | M. simiae | M. simiae (X52931) | 99.4 | M. kansasii/M. gastri | 98.2c | |
| 3 (25275) A | M. simiae | M. simiae (X52931) | 99.4 | M. kansasii/M. gastri | 98.2c | |
| 1 | M. szulgai | M. malmoense (X52930) | 99.4 | M. szulgai | 98.8 | |
| 2 (clinic) | M. szulgai | M. malmoense (X52930) | 99.4 | M. szulgai | 98.8 | |
| 3 (35799) A | M. szulgai | M. malmoense (X52930) | 98.8 | M. szulgai | 98.8 | |
| 1 | M. cookii | M. cookii (X53896) | 96.4 | M. farcinogenes | 93.5 | |
| 1 | M. intermedium | M. intermedium (X67847) | 100 | M. kansasii/M. gastri | 94.1c | |
| 2 | M. intermedium | + | M. intermedium (X67847) | 100 | M. kansasii/M. gastri | 94.1c |
| 1 | M. marinum | M. marinum (X52920)/M. ulcerans | 100c | M. asiaticum | 97 | |
| 2 | M. marinum | + | M. marinum (X52920)/M. ulcerans | 100c | M. asiaticum | 97.6 |
| 1 | M. ulcerans | + | M. ulcerans (X58954)/M. marinum | 99.4c | M. asiaticum | 97.6 |
| 2 | M. ulcerans | + | M. ulcerans (X58954)/M. marinum | 99.4c | M. asiaticum | 98.2 |
| 3 | M. ulcerans | M. ulcerans (X58954)/M. marinum | 99.4c | M. asiaticum | 96.4 | |
| 1 | M. celatum | M. celatum 2 (L08169) | 98.2 | M. asiaticum | 95.8 | |
| 2 | M. celatum | + | M. celatum 2 (L08169) | 98.2 | M. asiaticum | 95.8 |
| 3 (51130) A | M. celatum | M. celatum 1 (L08170) | 99.4 | M. celatum 2 | 95.3 | |
| 4 (51131) A | M. celatum | M. celatum 2 (L08169) | 99.4 | M. celatum 1 | 95.9 | |
| 1 | M. conspicuum | + | M. conspicuum (X88922) | 100 | M. simiae | 97 |
| 1 | M. interjectum | + | M. interjectum (X70961) | 100 | M. haemophilum 1 | 97.1 |
| 1 | M. branderi | + | Mycobacterium sp. strain sp.6 (X82234) | 98.8 | M. shimodei | 91.1 |
| 1 | M. scrofulaceum | + | M. scrofulaceum (X52924) | 99.4 | M. simiae | 98.8 |
| 2 | M. scrofulaceum | + | M. scrofulaceum (X52924) | 99.4 | M. simiae | 98.2 |
| 1 | M. xenopi | + | M. xenopi (X52929) | 97.1 | M. shimoidei | 88.8 |
| 2 (19250) A | M. xenopi | M. xenopi (X52929) | 98.3 | M. shimoidei | 89.9 | |
| 3 (clinic) | M. xenopi | M. xenopi (X52929) | 99.4 | M. paratuberculosis | 89.3 | |
| 1 | M. sphagni | + | M. sphagni (X55590) | 98.2 | M. chitae 2 | 95.3 |
| 1 | M. lentiflavum | + | Mycobacterium sp. strain MCRO 8 (X93034) | 98.2 | Strain MCRO 18 | 97.6 |
Targets have been generated from the various Mycobacterium isolates by in vitro transcription from PCR amplicons and fragmented by the MgCl2 protocol. Hybridization experiments were performed with hybridization buffer 1 as described in Materials and Methods. Sequence giving the highest reference score is referenced both in terms of origin (EMBL accession number) and biological significance (species identification).
M. bovis isolates have the same 16S rRNA sequence as M. tuberculosis isolates.
Same base calling (percent) obtained for two reference sequences (see explanation in Results).
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen.
RESULTS
Optimization of the DNA-probe array hybridization.
Experimental fragmentation and hybridization conditions for the probe array assay were optimized by using wild-type reference targets derived from M. tuberculosis 16S rRNA and from two genes involved in M. tuberculosis drug resistance, i.e., rpoB and katG. Hybridization of these targets to the 20-mer nucleotide probes on the array combined with the MnCl2-imidazole fragmentation procedure and hybridization buffer 2 produced sequence base call accuracies ranging from 98.9 to 100% (Fig. 1). Our assay conditions also produced a high degree of specificity during the hybridization. We observed little cross-hybridization to probes on different regions of the array, despite the use of one set of experimental conditions for several targets hybridizing to thousands of different array sequences (Fig. 1B).
Identification of Mycobacterium species.
We tested 70 mycobacterial isolates from 27 different species characterized by conventional phenotypic methods. For each isolate tested, results are expressed as percent homology to reference sequences tiled on the array as produced by the GeneChip software. Results for the best two scores are presented in Table 1. The highest score was taken as the identification result (range, 96.4 to 100%; mean, 98.97%). Over the approximately 169-nucleotide region analyzed for the 70 isolates, the highest homology scores had less than two discordant base calls on average. Discordant calls occurred all in a conserved region, so there was no effect on identification. Based on our results, all isolates from the following species were unambiguously identified by the probe array (i.e., the highest identification score being the one expected): M. asiaticum, M. chelonae, M. celatum, M. conspicuum, M. cookii, M. fortuitum, M. flavescens, M. gordonae, M. interjectum, M. intermedium, M. malmoense, M. scrofulaceum, M. shimodei, M. simiae, M. smegmatis, M. sphagni, M. tuberculosis, and M. xenopi. Also, 100% of MAC isolates were identified by the probe array as species belonging to the M. avium complex. However, we obtained minor discrepancies between the phenotypic and the genotypic identifications for two of the eight isolates tested. One of the four M. avium isolates was identified as M. intracellulare (score, 99.4%), and one of the four M. intracellulare isolates was identified as M. avium/M. paratuberculosis by the array (score, 97%).
Six species shared the highest reference selection with another species. M. avium and M. paratuberculosis, M. kansasii and M. gastri, M. marinum and M. ulcerans, M. bovis and M. tuberculosis, and M. chelonae and M. abscessus (observed for two of the four M. chelonae isolates) were not individually identified in the study. The identification of both strains was predictable since their respective 16S rRNA sequences are identical in the region tiled on the array. This problem can be solved by expanding the region interrogated by the array for the pair M. avium and M. paratuberculosis (data not shown). This solution, however, will not work for the other species.
Two isolates did not have their respective reference sequences tiled on the array. M. branderi was identified as Mycobacterium sp. strain sp.6 and M. lentiflavum was identified as Mycobacerium sp. strain MCRO 8. The description of M. branderi as a new species includes the Mycobacterium sp. strain sp.6 isolate sequence as a reference (16). M. lentiflavum isolates have previously been described as being phylogenetically related to M. simiae (27) as well as the Mycobacterium sp. strain MCRO 8 isolate sequence (23). The identifications produced by probe array analysis are consistent with the available literature for the two isolates.
Table 1 also includes a blinded study. Twenty PCR amplicons were kindly provided by E. Böttger, and all were accurately identified with the probe array assay. This panel included a mix of pathogenic species (M. tuberculosis, MAC, M. marinum, M. kansasii, and M. xenopi) and nonpathogenic species.
Assuming proper identification for M. branderi and M. lentiflavum, the sequences of 67 of 70 isolates representing 27 species were identified correctly. Only one species, M. szulgai, was not correctly identified. All three M. szulgai isolates were diagnosed as M. malmoense. The reference sequences used to discriminate these two species are identical except at two locations, where a single base difference exists. At one location, an error in the M. szulgai reference sequence created probes that were identical to M. malmoense sequence (data not shown). This error resulted in a base call that incorrectly lowered the M. szulgai score while raising the M. malmoense score. The unique remaining polymorphism differentiating M. malmoense from M. szulgai was efficiently discriminated in all of the samples. The error in the reference sequence has been corrected in a second version of the probe array.
Detection of M. tuberculosis rpoB mutants.
We tested 16 rpoB sequences, generated from 15 rifampin-resistant isolates, and one sensitive M. tuberculosis isolate. To study the influence of the strand polarity on base-calling accuracy, we hybridized rpoB sense and antisense transcripts on separate probe arrays. Mutations included substitutions (single-base and double-base) and deletions (three-base and six-base) (Table 2). All mutations were detected on both strands. Detection of the CAC→TAC point mutation responsible for the His526Tyr substitution is presented in Fig. 2. This mutation accounts for 30% of M. tuberculosis rifampin-resistant strains (12). Redundancy in the probes used in the sequence interrogation provides robustness for the test. The C→T change could be detected by using either the probes specific for the mutation (Fig. 2B) or the probes specific for the wild-type sequence (Fig. 2A). At all positions interrogated by mutant-specific probes, the mutant target hybridized, producing intensities that are roughly the same. However, intensities of the wild-type-specific probes are different for the same target. At the point mutation position, a single probe is perfectly complementary to the target and produces a relatively strong hybridization signal. At the neighboring positions, hybridization to the probes is reduced since there is a one-base mismatch with respect to the target sequence. The opposite experiment, hybridization of the M. tuberculosis rpoB wild-type target to the same probe tiles, is shown in Fig. 2C and D. Hybridization signals to the wild-type C probes are clearly observed.
TABLE 2.
M. tuberculosis rpoB mutant isolates analyzed with the Mycobacterium probe array
| Clinical isolate | Codon | Nucleotide change | Amino acid substitution |
|---|---|---|---|
| G049 | 526 | CAC→GAC | His→Asp |
| G055 | 526 | CAC→CGC | His→Arg |
| G051 | 526 | CAC→TAC | His→Tyr |
| J3919 | 531 | TCG→TTG | Ser→Leu |
| J37 | 513 | CAA→CTA | Gln→Leu |
| 515 | ATG→GTG | Met→Val | |
| J499 | 513 | CAA→GAA | Gln→Glu |
| J535 | 533 | CTG→CCG | Leu→Pro |
| G054 | 531 | TCG→TGG | Ser→Trp |
| J80 | 516–517 | Dela GACCAG | Del Asp-Gln |
| J978 | 526 | CAC→CTC | His→Leu |
| G052 | 511 | CTG→CCG | Leu→Pro |
| G048 | 513 | CAA→CTA | Gln→Leu |
| G050 | 516 | GAC→TAC | Asp→Tyr |
| G047 | 505 | TTC→TTG | Phe→Leu |
| 511 | CTG→CCG | Leu→Pro | |
| 531 | TCG→TGT | Ser→Cys | |
| G053 | 518 | Del AAC | Del Asn |
Del, deletion.
The ability to detect a new point mutation not included in the array design was demonstrated for one isolate (G047). The wild-type probes detected the TTC→TTG mutation responsible for the Phe505Leu substitution (data not shown).
DISCUSSION
The work described in this study represents one of the first applications of high-density DNA probe arrays for bacteriology, focusing on fastidious bacterium diagnostics. The technology allowed us to design an array that contained all of the 16S rRNA polymorphisms over a 200-bp region present in a mycobacterial database. The array also contained 51 M. tuberculosis rifampin resistance-causing rpoB mutations in a 200-bp region and 2.2 kb of the M. tuberculosis wild-type katG gene, this fragment spanning codons where mutations were previously shown to confer resistance to isoniazid (9, 31). An experimental protocol was optimized to perform hybridization on the probe array, scanning, and data analysis in 1 h. The total process from culture, including sample preparation and amplification, takes less than 4 h manually. An additional objective of this study was to evaluate base-calling accuracy by using a single strand of target. Previous studies have combined the information from both strands (3, 6, 17). We found that base-calling accuracies with one target strand ranged routinely from 96.4 to 100%, even though the 16S, rpoB, and katG targets have a high G+C content (>60%) and contain potentially highly stable secondary structures. Moreover, cotesting of independently generated 16S rRNA and rpoB amplicons on the same array provided the same base-calling accuracy as did each target alone (data not shown).
The sequencing of the 16S rRNA gene has been utilized by numerous investigators as a means of discrimination among mycobacterial taxa (27). The hybridization-based probe array assay used here showed a specificity matching the sequence resolution (polymorphisms) of the 16S rRNA marker. In this study, the sequences for 26 of 27 species were correctly identified, with the only discrepancy being an artifact due to an error in the reference sequence. Closely related sequences specific for species of very different clinical importance were clearly differentiated (i.e., M. tuberculosis complex species from M. celatum, M. marinum, M. asiaticum, and M. terrae). Newly described species for which sequences were not tiled on the array (M. lentiflavum and M. branderi) were identified as subspecies variants or closely related species. Recent genetic studies have demonstrated a higher degree of intraspecies sequence diversity in the 16S rRNA locus than was previously believed (8, 23). Given that the assay produces sequence information, a phylogenetic module could be added to the analysis to improve the accuracy of identifying polymorphic variants and strains whose sequences are not exactly represented on the array.
The ability of accurately detecting sequence variation is especially important when the clinical interpretation (i.e., in vitro resistance or sensitive status) depends on the discrimination of a single point mutation as seen in rpoB-mediated rifampin resistance in M. tuberculosis. All tested mutation types were detected by the Mycobacteria probe array, and these include single-base substitutions, double-base substitutions, deletions (three-base and six-base), and insertions (data not shown). The detection of these mutations was also independent of the strand used for hybridization. The ability to place many mutant sequences on the array may have its advantages. A commercially available rifampin resistance test (Inno-LiPA Rif.TB; Innogenetics, Zwijndrecht, Belgium) was reportedly unable to detect a three-base insertion mutation, presumably because the sequence was not present on the strip (5). Interestingly, we have also demonstrated the ability of the probe array to detect new point mutations, neither previously described nor tiled on the array, by simply using the probes defined for the wild-type sequence. Thus, the efficiency of our probe array strategy should not be diminished by the occurrence of a new point mutation or polymorphisms not on the array. This strategy is being advanced by examining katG sequences in isoniazid-resistant M. tuberculosis isolates to identify new mutations (data not shown). In the perspective of testing cultures or direct clinical samples with this molecular approach, we need to position it with growth-based interpretation with more extended studies so that the inference of in vivo resistance status remains clinically pertinent.
Simple, ready-to-use probe assays are commercially available for Mycobacterium identification (Accu-Probe and AMTD, Gen-Probe; Amplicor MTB; Roche Diagnostics Systems; SHARP Signal; Digene Diagnostics Inc., Silver Spring, Md.). However, they are applicable to isolates of the most common disease-causing mycobacterial species (M. tuberculosis and MAC strains). A mycobacterial probe array can expand the existing platform for identification of mycobacterial species and be used to perform complete strain-specific typing of clinical isolates. For example, the probe array could be used, in conjunction with analysis of clinical data, for determining the true incidence of infections due to environmental mycobacteria.
Today, the number of MOTT infections is difficult to assess because there is no system for notification as exists for M. tuberculosis. The current report frequency of these species is likely to be underestimated due to the lack of additional testing in cases of minimal disease or misidentification as M. tuberculosis (11). Despite the likely underestimation of MOTT disease, a growing number of MOTT isolates are submitted to laboratories for identification. This may reflect an increase in the prevalence of opportunistic mycobacterial disease (notably in the AIDS context), or it may also reflect an increase in the number and nature of specimens submitted for culture brought about by a greater awareness of tuberculosis. The increased use of endoscopy for diagnostic purposes and changes in environmental factors affecting the nature, number, and distribution of MOTT species in the environment, including piped water supplies (30), certainly have also contributed to this increase. A single assay that quickly identifies M. tuberculosis, MAC, and other mycobacterial species will clearly aid in documenting their apparent rise in disease states and possibly in new environments.
The potential of the GeneChip probe array strategy for parallel testing of different targets has been demonstrated in this study, as the same hybridization conditions could be used for the three genes tiled on the mycobacterial array (16S rRNA, rpoB, and katG). The platform described here can be expanded to other M. tuberculosis drug resistance determinants since they are being gradually understood. For example, probe array-based mutation analysis of the catalase-peroxidase gene (katG) (31), the promoter region of the inhA and ahpC genes (1, 29), the recently described kasA protein (19) for isoniazid resistance, pyrazinamidase-nicotinamidase gene (pncA) for pyrazinamide resistance (22, 24), and the emb operon for ethambutol resistance (25) could all be done to monitor drug resistance simultaneously. Recently, a probe array interrogating most of these genes has been designed and is currently being used (8). Epidemiological markers could also be added to the array for tracing epidemic or sporadic dissemination of strains. Finally, the potential to perform direct testing on samples providing species identification results (15) and drug resistance genotyping (28) represents the next step in the application of this technology to clinical diagnostics.
ACKNOWLEDGMENTS
We thank E. Böttger for the generous gifts of Mycobacterium samples and for thoughtful discussions; S. Cole for the gift of rpoB clones; M. Mittmann for his assistance with lithographic mask design; D. Wu, E. Wang, and B. Lacroix for software design and development; J. Drenkow for preparation of the rpoB mutant targets; D. Do for technical assistance; A. Lau for graphics preparation; and C. Rogers for his support for this project.
REFERENCES
- 1.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um S K, Wilson T, Collins D, De Lisle G, Jacobs W R. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 2.Böddinghaus B, Rogall T, Flohr T, Blöcker H, Böttger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P A. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 4.Cole S T. Mycobacterium tuberculosis: drug-resistance mechanisms. Trends Microbiol. 1994;2:411–415. doi: 10.1016/0966-842x(94)90621-1. [DOI] [PubMed] [Google Scholar]
- 5.Cooksey R C, Morlock G P, Glickman S, Crawford J T. Evaluation of a Line Probe assay kit for characterization of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis isolates from New York City. J Clin Microbiol. 1997;35:1281–1283. doi: 10.1128/jcm.35.5.1281-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cronin M T, Fucini R V, Kim S M, Masino R S, Wespi R M, Miyada C G. Cystic fibrosis mutation detection by hybridization to light-generated DNA probe arrays. Hum Mutat. 1996;7:244–255. doi: 10.1002/(SICI)1098-1004(1996)7:3<244::AID-HUMU9>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 7.Fodor S P A, Rava R P, Huang X C, Pease A C, Holmes C P, Adams C L. Multiplexed biochemical assays with biological chips. Nature. 1993;364:555–556. doi: 10.1038/364555a0. [DOI] [PubMed] [Google Scholar]
- 8.Gingeras T R, Ghandour G, Wang E, Berno A, Small P M, Drobniewski F, Alland D, Desmond E, Holodniy M, Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 9.Heym B, Alazari P M, Honoré N, Cole S T. Missense mutations in catalase-peroxidase gene, katG, are associated with isoniazid-resistance in Mycobacterium tuberculosis. Mol Microbiol. 1995;15:235–245. doi: 10.1111/j.1365-2958.1995.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 10.Horsburgh C R. Mycobacterium avium complex infection in the acquired immunodeficiency syndrome. N Engl J Med. 1991;324:1332–1338. doi: 10.1056/NEJM199105093241906. [DOI] [PubMed] [Google Scholar]
- 11.Jenkins, P. A. 1991. Mycobacteria in the environment. J. Appl. Bacteriol. Symp. 70(Suppl.):137S–141S. [PubMed]
- 12.Kapur V, Li L L, Iordanescu S, Hamrik M R, Wanger A, Kreiswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase β subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent P T, Kubica G P. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health and Human Services publication no 86-8230. U.S. Washington, D.C: Department of Health and Human Services; 1985. [Google Scholar]
- 14.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F C, Böttger E C. Genotypic identification of mycobacteria by nucleic acids sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschner P, Rosenau J, Springer B, Teschner K, Feldmann K, Böttger E C. Diagnosis of mycobacterial infections by nucleic acid amplification: 18-month prospective study. J Clin Microbiol. 1996;34:304–312. doi: 10.1128/jcm.34.2.304-312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koukila-Kahkola P, Springer B, Böttger E C, Paulin L, Jantzen E, Katila M L. Mycobacterium branderi sp. nov., a new potential human pathogen. Int J Syst Bacteriol. 1995;45:549–553. doi: 10.1099/00207713-45-3-549. [DOI] [PubMed] [Google Scholar]
- 17.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 18.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittman M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 19.Mdluli K, Slayden R A, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane D D, Musser J M, Barry C E., III Inhibition of a Mycobacterium tuberculosis β-ketoacyl ACP synthase by isoniazid. Science. 1998;280:1607–1610. doi: 10.1126/science.280.5369.1607. [DOI] [PubMed] [Google Scholar]
- 20.Raviglione M C, Snider D E, Kochi A. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 21.Roberts G D, Koneman E W, Kim Y K. Mycobacterium. In: Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C: American Society for Microbiology; 1991. pp. 304–339. [Google Scholar]
- 22.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculosis drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 23.Springer B, Stockman L, Teschner K, Roberts G D, Böttger E C. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreevatsan S, Pan X, Zhang Y, Kreiswirth B N, Musser J M. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–640. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sreevatsan S, Stockbauer K E, Pan X, Kreiswirth B N, Moghazeh S L, Jacobs W R, Jr, Telenti A, Musser J M. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob Agents Chemother. 1997;41:1677–1681. doi: 10.1128/aac.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S T, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 27.Tortoli E, Piersimoni C, Kirschner P, Bartoloni A, Burrini C, Lacchini C, Mantella A, Muzzi G, Passerini Tosi C, Penati V, Scarparo C, Tullia Simonetti M, Böttger E C. Characterization of mycobacterial isolates phylogenetically related to, but different from, Mycobacterium simiae. J Clin Microbiol. 1997;35:697–702. doi: 10.1128/jcm.35.3.697-702.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whelen A C, Felmlee T A, Hunt J M, Williams D L, Roberts G D, Stockman L, Persing D H. Direct genotypic detection of Mycobacterium tuberculosis rifampin resistance in clinical specimens by using single-tube heminested PCR. J Clin Microbiol. 1995;33:556–561. doi: 10.1128/jcm.33.3.556-561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson T M, Collins D M. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol Microbiol. 1996;19:1025–1034. doi: 10.1046/j.1365-2958.1996.449980.x. [DOI] [PubMed] [Google Scholar]
- 30.Yates M D, Pozniak A, Uttley A H C, Clarke R, Grange J M. Isolation of environmental mycobacteria from clinical specimens in south-east England: 1973–1993. Int J Tuberc Lung Dis. 1997;1:75–80. [PubMed] [Google Scholar]
- 31.Zhang Y, Heym B, Allen B, Young D, Cole S T. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:501–503. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]