Abstract
COVID-19 (Corona Virus Disease-2019) is an infectious disease caused by a novel coronavirus, known as the acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This is a highly contagious disease that has already affected more than 220 countries globally, infecting more than 212 million people and resulting in the death of over 4.4 million people. This review aims to highlight the pertinent documentary evidence upon the adverse effects of the SARS-CoV-2 infection on several vital human organs. SARS-CoV-2 primarily targets the lung tissue by causing diffuse alveolar damage and may result in Acute Respiratory Distress Syndrome (ARDS). SARS-CoV-2 infects the cell via cell surface receptor, angiotensin-converting enzyme 2 (ACE2). Besides lungs, SARS-CoV-2 critically damage tissues in other vital human organs such as the heart, kidney, liver, brain, and gastrointestinal tract. The effect on the heart includes muscle dysfunction (acute or protracted heart failure), myocarditis, and cell necrosis. Within hepatic tissue, it alters serum aminotransferase, total bilirubin, and gamma-glutamyl transferase levels. It contributes to acute kidney injury (AKI). Localized infection of the brain can lead to loss or attenuation of olfaction, muscular pain, headaches, encephalopathy, dizziness, dysgeusia, psychomotor disorders, and stroke; while the gastrointestinal symptoms include the disruption of the normal intestinal mucosa, leading to diarrhea and abdominal pain. This review encompassed a topical streak of systemic malfunctions caused by the SARS-CoV-2 infection. As the pandemic is still in progress, more studies will enrich our understanding and analysis of this disease.
Keywords: Acute respiratory syndrome coronavirus 2, Corona Virus Disease-2019, Angiotensin-converting enzyme 2, Vital organs
1. Introduction
Corona Virus Disease-2019 is caused by a novel coronavirus, known as acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Wuhan, Hubei province, China in Dec 2019 [1], [2], [3]. Corona indicates crown-like spikes on the outer surface of the virus; thus, it is known as a coronavirus [4], [5], [6]. The virus is 65–125 nm in diameter, enveloped positive single-stranded RNA virus with a 5′ cap and 3′ poly-A tail (30–32 kb), belongs to the Coronavirus family, resultant from a Beta coronavirus (a novel strain) [4], [7].
The primary modes of human to human transmission are respiratory droplets and aerosols, touching face with contaminated hands, tears, and semen [8]. Mother-to-fetus transmissions are not yet fully confirmed while indirect contact, fecal-oral might be possible [9].
The response of the clinical spectrum of SARS-CoV-2 is quite broad and ranges from being completely asymptomatic to succumbing to sudden death. Most of the SARS-CoV-2 infected patients develop mild, moderate, or no symptoms and recover within a period of 7–14 days. But unfortunately, a subset of patients develop severe symptoms and many of them end up in the intensive care unit [10]. The initial range of the symptoms of COVID-19 patients is cough, low-grade hyperthermia, shortness of breath, loss of smell and taste, nausea, and diarrhea while the critically ill patients indicate severe symptoms and completions such as venous and arterial thrombosis with pulmonary embolism, stroke and myocardial infarction, acute kidney and liver damage, and neurological manifestations [10], [11], [12].
So far, we have evidenced that SARS-CoV-2 primarily affects the lung by causing diffuse alveolar damage with Acute Respiratory Distress Syndrome (ARDS) but the latest studies have shown that the virus also has been associated with damaging impact on other vital organs and tissue such as the heart, brain, large intestine, kidneys and spleen [13], [14], [15].
Published data indicated that people with advanced age and comorbidities are more severely affected [8]. The disease caused by the virus is highly contagious that has affected more than 220 countries globally, infecting more than 212 million people and resulting in a death toll of over 4.4 million people. In the US alone, more than 38.5 million people are infected, around 0.645 million deaths till 23 August 2021, and these counts continue to increase on daily basis [16].
Currently, scientists are keenly working on resolving the challenge of the novel human coronavirus infection. It is of utmost importance to study the underlying mechanism of tissue injuries in COVID-19 patients to know well the reasoning behind the grave outcomes in these patients. Here, we reviewed the route of infection, mode of transmission, and malfunction of the major human organs by SARS-CoV-2.
1.1. Effects on the lungs
Lungs are the most affected organ in COVID-19 patients [17], [18], [19]. In symptomatic patients, pneumonia occurs with obvious signs of viral pneumonia such as decreased oxygen saturation, blood gas deviations, changes visible through chest X‐rays as well as Lymphopenia, and elevation of inflammatory markers (C‐reactive protein and proinflammatory cytokines) [20]. Clinical pathology of autopsy cases of SARS aided in the substantial understanding of the nature of the disease progression or disease outcome. The general pathological alteration in the lungs was of diffuse alveolar injury-causing ARDS. Pathological analysis of pulmonary injuries indicated widespread hemorrhage and necrosis, desquamative pulmonary alveolitis and bronchitis, hemorrhagic infarction, hyaline membrane formation, exudation of protein and monocytes, lymphocytes and plasma cells in alveoli and viral inclusion bodies in alveolar epithelial cells [21].
A retrospective study of 62 confirmed COVID-19 pneumonia patients was done in Wuhan, China. Computerized tomographic examination of COVID-19 pneumonia patients indicated lung parenchyma and the interstitium involved [19]. Injuries presented with a characteristic multifocal distribution in the posterior, middle and lower lung regions. Low lymphocyte count and an elevated high-sensitivity C-reactive protein level (hs-CRP) were noticed [19]. Further pneumonia leads to Acute Respiratory Distress Syndrome (ARDS) in critical illness [22], [23]. The most devastating complication caused by SARS‐CoV‐2 is ARDS with an elevated death rate. In a study of 52 Covid 19 critically ill patients, 35 patients developed ARDS, out of which 32 patients died [24].
Pathological analysis of lung tissue samples from 38 Covid-19 fatalities in Italy showed massive alveolar injury, necrosis of pneumocytes, interstitial and intra-alveolar edema, hyaline membranes, pneumocyte hyperplasia, squamous metaplasia with atypia, and platelet–fibrin thrombi. The immunohistochemical analysis identified macrophage infiltration in the alveolar lumina, and lymphocytes in the interstitium. Electron microscopy indicated virion localization in the pneumocytes [25]. Similarly, the biopsy samples were taken from the lung of a 50-year-old patient who died from severe infection with SARS-C0V-2 [26]. Histological examination indicated bilateral diffuse alveolar injuries with cellular fibromyxoid-organizing exudates. Obvious desquamation of pneumocytes and hyaline membrane formation was noticed in the right lung, indicating acute respiratory distress syndrome while pulmonary edema with hyaline membrane formation was noticed in the left lung, expressive of early-phase ARDS [26]. Interstitial mononuclear inflammatory infiltrates, occupied by lymphocytes, was observed in both lungs [26]. Multi nucleated syncytial cells with atypical enlarged pneumocytes characterized by large nuclei, amphophilic granular cytoplasm, and prominent nucleoli were recognized in the intra-alveolar spaces, indicating viral cytopathic-like alterations [26].
According to the evidence, SARS-CoV2 and other viruses of the corona family enter the human body through the mucosa of the nose, oropharynx and some eventually get deposited in the lungs, SARS-CoV-2 invades the lung cells, damages the cilia (hair-like projection that moves around to keep the airways clear of any debris or mucus), and starts replication. The virus uses angiotensin-converting enzyme 2 (ACE2) as an entry receptor, present on the surface of the alveolar type II cells. ACE2 is a transmural metalloprotein (extent from the outer to the inner surface of the cell membrane), consists of zinc and protein base, and is normally involved in the renin/angiotensin pathway by converting angiotensin-2 to angiotensin 1–7 (a peptide with an antioxidant, anti-inflammatory and vasodilator properties) [27], [28]. ACE2 is the most probable binding site of the SARS-CoV2 particles to the host cell to cause final infection [17], [29]. The peptides are highly expressed by the epithelial cell of the lungs. This may explain the high incidence of pneumonia and bronchitis in COVID-19 patients [29]. A recent study showed that ACE2 is also highly expressed on the mucosa of the oral cavity, particularly in epithelial cells of the tongue [30]. SARS-CoV2 propagates within type II cells, many viral particles are released, and the cells undergo apoptosis and die.
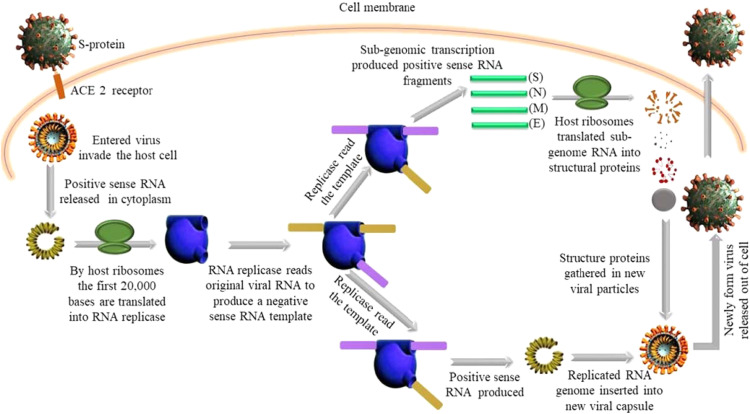
The SARS-CoV-2 must infect living cells to replicate. The genetic material of the virus carries information to make a copy of its self. The exact mechanism of fusion is not known but it is believed that the spike protein (S-protein) or glycoprotein of SARS-CoV-2 attaches to the cellular ACE2 receptor of the host cell via receptor-binding domain (RBD). The S-protein act as a key to enter the host cell. The S-protein can be divided into two domains, S1 and S2. The first one is responsible for (ACE2) recognition while the second one mediates membrane fusion [31]. After attachment of the S1 domain within the spike (S) protein to the cellular ACE2 receptor, induces conformational changes in the S2 domain that facilitates the fusion of the viral envelope with the cell membrane of the host cell by the activation of host cell proteases (TMPRSS2). and make the entry of the virus possible inside the cell through endocytosis. The entered-SARS-CoV-2 will subsequently release its genomic material, single-stranded positive-sense RNA (30,000 bp) in the cytoplasm. Positive sense RNA mimics the mRNA of the host cell and can be directly translated into protein by the host ribosome. The first 20,000 bases are translated into RNA dependant RNA polymerase or RNA replicase (The protein molecules are involved in multiple steps of viral replication). RNA replicase can read viral RNA and make a complementary RNA strand. The new RNA strand is a matching template of original positive sense RNA thus it is known as negative-sense RNA. The negative-sense RNA strand cannot be read by the host ribosome to make protein but can be used as a template by the newly assembled RNA replicase to make new viral positive-sense RNA (Replicating the original RNA). The RNA replicase reads the negative-sense RNA for the second time and produce positive sense RNA fragments (subgenomic) which are translated by the host ribosome into viral structural proteins which include the spike (S), nucleocapsid (N), membrane (M) and envelope (E). The new structural and surface protein assembled in a new viral particle along with a copy of genomic RNA, encapsulated with a membrane within the cytoplasm and released outside of the cell by budding or exocytosis ( Fig. 1, Fig. 2) [32], [33].
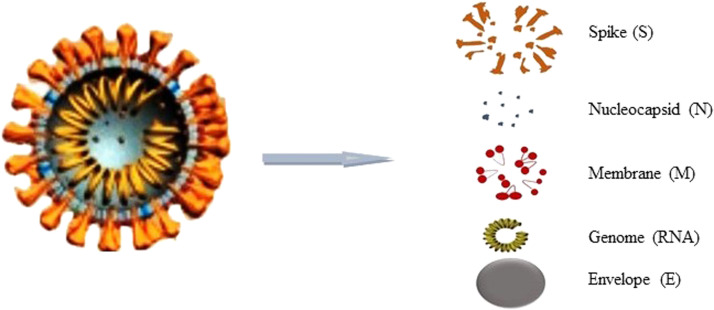
Fig. 1.
Various parts of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The virus consists of structural proteins which include the spike (S), nucleocapsid (N), membrane (M) and envelope (E) and genomic RNA.
Fig. 2.
Hypothetical mechanisms underlying the invasion and replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a host cell. The virus enters the host cell via ACE2 (Angiotensin Converting Enzyme 2) receptor and releases RNA into the cytoplasm. Host ribosomes translate two-thirds of viral RNA (2000 bp) into RNA replicase protein. The RNA replicase acts on viral positive sense-RNA to produce a negative-sense RNA. The RNA replicase reads the negative-sense RNA for the second time and produces positive sense RNA fragments (subgenomic) which are translated by the host ribosome into viral structural proteins. Finally, the new structural and surface protein is assembled in a new viral particle along with a copy of genomic RNA and delivered outside of the cell through exocytosis. Once outside the cell, the process takes place again.
Finally, the lung cells die and shed off adding in the debris and hindering the body ability to keep staff out of the lung and trachea. Inflammation is triggered, the fluid leak into alveolar spaces and as a result no exchange of gases takes place, and the patient is having difficulty breathing along with a dry cough. Other symptoms included fatigue, headache, muscle pain and runny nose. Mild cases will recover within a week while moderate, severe, or critical cases will develop pneumonia which can range from non-threatening to severe. For some severe and critical cases, the symptoms could escalate into ARDS. Due to viral pneumonia and/or ARDS, around 5% of patients usually require critical care and mechanical ventilation. Even the patients who have survived this phase could be left with permanent lung injuries including punching holes in the lungs and giving the honeycomb effect [22, 23].
SARS-CoV-2 has been reported to cause a heightened response in the cell associated with the immune system which encompasses mast cells (MCs) and macrophages that are induced by viruses. The inflammatory response in lung tissue is driven by a complex interaction between networks which include structural cells including fibroblasts, bronchial cells, and both epithelial and endothelian cells. The process of activation involves the induction of MC differentiation as well as the upregulation of pro-inflammatory cytokinins. Leukocyte activation is facilitated by endothelian cells via the secretion of interleukins (IL) IL-1 and IL-6 as well as chemokines. The MC, in turn, releases inflammatory compounds which include prostaglandins, proteases, arachidonic acid compounds and leukotrienes. Stimulation of the Toll Like Receptors (TLR) by SARS-CoV-2 induced production of IL-1 which in turn induced inflammatory interleukins (IL1 and IL6) which lead to life-threatening inflammation. The release of histamine by MCs in combination with IL-1 has been proposed to be the causative process in lung inflammation [34]. IL-1 has been documented to perturb the hematological and metabolic equilibrium in experimental animals. This is characterized by hypotension and attenuation of systemic blood pressure and vascular resistance. IL-1 has also been associated with increased heart rate and the aggregation of leukocytes. The pathogenic infection triggers IL-1 which in turn cause endothelial dysfunction that manifests as increased protein permeability and macrophage intervention. The activation of macrophages by the virus leads to the release of thrombi forming metalloproteinases and proteolytic enzymes which contribute to respiratory dysfunction. This is presented in the lungs as an increase in the products of proteolytic cleavage of fibrinogen, fibrin and D-dimer. The resulting lesions lead to respiratory failure. IL-1 in conjunction with TNF cause thrombosis and pulmonary edema as well as the reduction in blood pressure and bleeding [35]. IL-1 also induced the production of thromboxane B2 which is responsible for systemic inflammation and aggregation of leukocytes which leads to the formation of thrombi and the subsequent failure of organs.
1.2. Effects on the heart
The recent SARS-CoV-2 has been associated with heart abnormalities as compared to the seven-known strain of human coronaviruses, recognized for their impact on the respiratory tract but not on the heart [36]. The complications include muscle dysfunction (acute or protracted heart failure), heart inflammation (myocarditis), cell necrosis and arrhythmias [36]. Besides, some patients, with underlying cardiovascular diseases (CVDs), might have an elevated risk of mortality [36], [37]. The complications were even noticed in the people with mild as well as no symptoms [36], [37].
Recent data from Wuhan, Italy and the United States of America have suggested that CVD is one of the most common comorbidities associated with COVID-19 [38], [39], [40]. Also, the mortality rate of COVID-19 patients with CVD is higher by 10.5% in comparison with diabetes (7.3%), chronic respiratory disease (6.3%) and hypertension (6%) [41].
In a study of 2736 Covid-19 patients admitted to the Mount Sinai Health System in New York between 27 February and 12 April 2020 the plasma troponin I level was measured and 36% of patients were reported with an elevated level of troponin I, compared to normal level (elevated troponin I levels were associated with a higher prevalence of cardiovascular disease) [42]. In another study, 101 patients in Sichuan (admitted in designated COVID-19 treatment centers) between 16 January and 10 March 2020, 15.8% had a plasma troponin I level greater than the normal [43].
Although the specific mechanisms of the myocardial injury induced by SARS-CoV-2 infection are uncertain. It is posited that SARS- CoV-2 can affect the heart in two ways. First, the virus may directly invade the myocardial tissue and cause damage via ACE2 receptors which are widely expressed in the lungs and cardiovascular system [2], [44]. Second, the virus may be eliciting an uncontrolled cytokine storm, resulting in systemic injuries, including the cardiac [37], [45].
1.3. Effects on the liver
Liver impairment has been described as a non-pulmonary manifestation of COVID‐19, the level of liver associated enzymes such as alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL), gamma-glutamyl transferase (GGT) albumin and alkaline phosphatase [ALP] alters to various degrees in COVID-19 patients [46], [47], [48]. However, the exact mechanisms involved in liver damage in COVID-19 patients are not well known. It is hypothesized that liver impairment may be directly caused by the virus since ACE2 receptors are noticed in the hepatocytes and cholangiocytes, thus it leads to the involvement of the liver in SARS- CoV-2 infection [49]. In a study, SARS-CoV-2 RNA has been identified in the stool samples of COVID19 patients with diarrhea [50]. This data indicates the possibility of virus exposure to the liver.
Liver toxicity may indirectly be caused by the use of various medications [47], [48]. In a study, the administration of lopinavir, hydroxychloroquine, redeliver, and tocilizumab increased the odds of liver injury by more than 4-fold [47], [48]. Besides, immune-mediated inflammation including cytokine storm and pneumonia-associated hypoxia may also contribute to a liver impairment or underlying liver ailments such as chronic viral hepatitis, non-alcoholic fatty liver disease in COVID-19 patients. Further, alcohol-related liver disease could also have been responsible for high liver enzyme abnormalities [46], [51].
Patients with abnormal liver tests are at higher risk of progressing to severe disease. In a study of 417 COVID-19 patients, 76.3% had abnormal liver test results and 21.5% had a liver injury during hospitalization. The presence of abnormal liver tests became more noticeable during hospitalization within 2 weeks, with ALT (23.4%), AST (14.8%), TBIL (11.5%) and GGT (24.4%) levels elevated to more than 3× the upper limit of normal, respectively [48]. A study was conducted of 1827 patients with confirmed COVID‐19 in the Yale‐New Haven Health System, United State between March - April 2020, Levels of AST, ALT, ALP, TBI, and albumin were analyzed. Abnormal liver tests were recorded at admission (AST 66.9%, ALT 41.6%, ALP 13.5%, and TBIL 4.3%) and peak hospitalization (AST 83.4%, ALT 61.6%, ALP 22.7%, and TBIL 16.1%) [47]. In another study, the level of GGT and ALP were elevated in thirty (54%) and one (1·8%) of 56 COVID 19 patients during hospitalization [51]. In a small series of other studies, ALT and AST level varies from 16% to 53% [37, 51, 52, 53, 54]. The elevation of liver enzymatic levels in the above cases has indicated that liver morphology is affected in COVID 19 patients.
1.4. Effects on the gastrointestinal tract
Evidence indicates that SARS-CoV-2 affects the digestive system of the infected patient and presents with various symptoms including diarrhea, anorexia, nausea, vomiting and abdominal pain [55]. The gastrointestinal (GI) manifestations of COVID‐19 create big challenges in feeding the patients to obtain their nutrition needs [56]. The lack of contact of the nutrients with the intestinal mucosa could result in atrophy of lymphoid tissue: a decline in the immune system function and intensification in bacterial translocation [57]. In some cases, such as diarrhea, the digestive symptoms, arise before respiratory symptoms, and on rare occasions, maybe the only presenting symptom of COVID-19 [58], [59].
Several studies have shown different GI symptoms induced by SARS-CoV-2 infection [56], [58]. A meta‐analysis consisted of 60 studies (4243 patients) indicated a pooled prevalence of GI symptoms at 17.6%. In this analysis, commonly reported symptoms were anorexia with 26.8%, followed by diarrhea with 12.5%, nausea and vomiting with 10.2%, and abdominal discomfort with 9.2% [60]. In Hubei, China, 204 patients with COVID-19 were analyzed and 50.5% of patients have been reported with GI symptoms including lack of appetite (78.6%), diarrhea (34%), vomiting (3.9%), and abdominal pain (1.9%), the patient, presenting digestive symptoms, have a longer time from onset to admission and a worse prognosis, compared with patients without digestive symptoms [58]. Similarly, in Humanities Research, Italy 292 patients COVID-19 patients had been analyzed from February 22 to March 30, 2020, diarrhea (27.1%) was the most frequent GI symptom [61].
It is believed that the effect of SARS-CoV-2 on the gastrointestinal tract might be related to the abundance of the ACE2 receptor, located in the ileum and colon [46], [62]. ACE2 is highly expressed in the human small intestine particularly in proximal and distal enterocytes [62]. Enterocytes are simple columnar epithelial cells found in the inner surface of the small and large intestines. Thus, enterocytes are directly exposed to foreign pathogens and food particles [62]. SARS‐CoV‐2 virus attachment to the ACE2 receptors of the digestive system is thought to disrupt the normal intestinal flora, resulting in various GI symptoms, particularly diarrhea [58], [63]. The presence of SARS‐CoV‐2 RNA in human feces supports its pathological mechanism in the intestine [58], [63].
1.5. Effects on the kidneys
It has been reported that acute kidney injuries (AKI) are common in patients with COVID-19, associated with increased mortality and most of the patients that survive do not recover kidney function [64]. A retrospective study was conducted of patients admitted to 2 hospitals in Derby, United Kingdom and observed a high incidence of acute kidney injury in patients with COVID-19 that was associated with a 3-fold higher odds of death than COVID-19 without acute kidney injury and a 4-fold higher odds of death than AKI due to other causes [65].
As far as the route of infection is concerned the expression of the ACE2 gene has been reported in human kidneys and bladders [66]. The result indicated that the urinary system is a potential route for 2019-SARCoV infection, along with the respiratory, hepatic and digestion systems. It is also possible that the prescribed medicine (oseltamivir, lopinavir/ritonavir, ribavirin, and chloroquine phosphate or hydroxychloroquine sulfate) for the treatment of COVID 19 may indirectly affect kidneys. As most of the metabolites derived from these medicines are metabolized in the liver and excreted via the kidney. Thus, the liver and kidneys are vulnerable to injuries and toxicity [67].
Primary evidence from Wuhan indicated that initially, the prevalence of AKI in COVID-19 patients was extremely low (3–9%) however, the subsequent analyses indicated an elevation in AKI injuries (15%) [68]. In a study at Tongji Hospital, Wuhan, China of 274 COVID-19 patients (113 dead and 161 fully recovered patients), 29 patients developed AKI after the SARS-CoV 2 infection while four had chronic kidney disease (CKD). High blood urea nitrogen (BUN) concentration (median = 23.52 mg/dL) was noticed in the deceased patients as compared to recovered patients (median = 13.72 mg/dL) Similarly high creatinine concentration was recorded in deceased patients (median = 0.99 mg/dL) compared to recovered patients (median = 0.86 mg/dL). Out of 274 patients, 100 patients (42 deceased, and 58 fully recovered patients) developed proteinuria and urinary occult blood was positive in 84 patients (44 deceased, and 40 recovered patients) [8].
In an observational study of 3235 hospitalized COVID 19 patients in New York City, it has been reported that AKI occurred in 46% of patients and 20% of those patients required dialysis. AKI was associated with elevated mortality and 44% of patients discharged alive had residual acute kidney disease [64]. In another study conducted on 100 COVID 19 patients, admitted to the Ayatollah Alimoradiyan Hospital in Nahavand, Hamadan Province, 35 patients were subsequently found to have elevated plasma creatinine and BUN levels [69]. From these case studies, it has been obvious that the virus is having an impact on the kidney tissues of COVID 19 patients.
1.6. Effects on the brain
Increasing evidence has indicated that the COVID-19 patient exhibits neurologic manifestations. In mild cases, the manifestations include smell dysfunction, muscle pain, headaches, encephalopathy, dizziness and dysgeusia while in severe cases, it is accompanied by seizures, movement disorders, motor and sensory deficits, ataxia, and strokes. Some of these symptoms persist even after treatment and recovery from the infection [70], [71].
The SARS-CoV-2 can reach the brain and utilized angiotensin-converting enzyme-2 (ACE2) as the main entry receptor [33]. The expression of ACE2 protein has been reported in the human brain [72]. ACE2 was highly expressed in some vital parts including the substantia nigra, choroid plexus, middle temporal gyrus, posterior cingulate cortex, olfactory bulb [73] and no expression in endothelial cells, microglia and pericytes have been reported [73]. In addition to ACE2, SARS-CoV-2 may use CD147 (cluster of differentiation 147) also known as basigin (BSG) and neuropilin-1 (NRP1) as entry receptors [74], [75] and the viral cell entry and replication has been facilitated by several proteases such as TMPRSS11A/B, furin (FURIN) and cathepsin B and L [76].
As the exact mechanism of the virus entry from the respiratory tract to the brain is not clear, however, based on other coronaviruses, numerous potential routes of entry for SARS-CoV-2 have been anticipated [77]. The olfactory region (part of the nasal cavity) appears to play a vital role in the neuroinvasion of SARS-CoV-2. Since olfactory nerves are exposed to the outside environment, the associated neurons may act as a carrier for the virus to the brain from the respiratory tract which is also evident from the loss of smell and taste and increased MRI signal in the olfactory cortex of COVID-19 patients. ACE2 and TMPRSS2 have been detected in the nasal mucosa at the RNA and protein levels [78]. SARS-CoV-2 might also reach the brain through blood circulation where the virus could attach to the endothelium via ACE2 receptors and alter the Blood-Brain Barrier (BBB), inducing dispersion of the virus in the central nervous system [73]. Besides SARS-CoV-2-associated cytokines such as interleukin (IL), −1b, IL-17, IL-6 and tumor necrosis factor (TNF) alter the BBB permeability and could enable the entrance of the virus [79], [80].
1.7. Effects on the spleen
The spleen is associated with important physiological functions such as immunological surveillance, removal of aged blood cells, hematopoiesis and the regulation of blood volume [81]. The spleen was reported as a target for the Severe Acute Respiratory Syndrome (SARS) virus in 2006 and it was posited that the collapse of the splenic immune system played a key role in the clinical outcome of patients [82] splenic abscesses were first reported in a patient during the current SARS-CoV-2 outbreak [83] and splenic infarction was also observed in Computed tomography (CT) abdomen [84], [85]. Pathological changes were detected in the spleen where histopathological examination showed decreased cell composition with atrophied cells which presented themselves as a white pulp and a decrease or absence of lymphoid follicles as well as a decrease in the red to white pulp to varying degrees [86]. SARS-CoV-2 was reported to directly neutralize human spleens and lymph nodes through infecting tissue-resident CD169 + macrophages [87].
2. Conclusions
Emerging evidence about the route of entry and the distribution of the SARS-CoV-2019 virus and the infective RNA contains across organs is indicative of the ability of the virus to effectively propagate and replicate in the lungs, brain, heart, spleen, liver and gastrointestinal tract will necessitate looking beyond the paradigm of cellular entry via the ACE2 receptor and investigating alternative mechanisms of cellular entry which may include non-receptor mediated endocytosis. Developing experimental models to test these paradigms will serve as the way forward for future research.
Funding
Research Management Centre, Universiti Malaysia Sabah, Grant number (DKC 2009) entitled “Characterization and Selection of COVID-19 Protein Coding Genes for the Development of DNA Vaccines via a Synthetic Biology Approach” to KFR.
CRediT authorship contribution statement
MDS, MS, and KJR contributed to the conception, writing, and discussion of this review manuscript. ASS, MSK contributed to the discussion of this review manuscript. BAVM contributed to the conception and discussion of this review manuscript. The final version of the manuscript was approved by all authors.
Declaration of conflicting interests
The authors declared no potential conflicts of interest concerning the research, authorship, and/or publication of this article.
References
- 1.Lu H., Stratton C.W., Tang Y. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pixabay, 4,000+ Free Covid & Coronavirus Images - Pixabay, 2021. 〈https://pixabay.com/images/search/covid/〉. (Accessed 17 February 2021).
- 6.Manivannan M., Jogalekar M.P., Kavitha M.S., Venmathi Maran B.A., Gangadaran P. A mini-review on the effects of COVID-19 on younger individuals. Exp. Biol. Med. 2021;246(3):293–297. doi: 10.1177/1535370220975118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roujian L., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., Wang T., Guo W., Chen J., Ding C., Zhang X., Huang J., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C., Ma Q.Y., Zheng Y.H., Yang Y.X. Transmission routes of 2019-novel coronavirus (2019-nCoV) Zhonghua Yu Fang Yi Xue Za Zhi. 2020;54:374–377. doi: 10.3760/cma.j.cn112150-20200216-0016. [DOI] [PubMed] [Google Scholar]
- 10.Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/nejmoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.L. He, M.A. Mäe, L. Muhl, Y. Sun, R. Pietilä, K. Nahar, E.V. Liébanas, M.J. Fagerlund, A. Oldner, J. Liu, G. Genové, L. Zhang, Y. Xie, S. Leptidis, G. Mocci, S. Stritt, A. Osman, A. Anisimov, K.A. Hemanthakumar, M. Räsänen, O. Mirabeau, E. Hansson, J. Björkegren, M. Vanlandewijck, K. Blomgren, T. Mäkinen, X.-R. Peng, T. Arnold, K. Alitalo, L. Eriksson, U. Lendahl, C. Betsholtz, Pericyte-specific vascular expression of SARS-CoV-2 receptor ACE2 – implications for microvascular inflammation and hypercoagulopathy in COVID-19, 2020. https://doi.org/10.1101/2020.05.11.088500.
- 13.Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T., Yarid N., Marshall D.A. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unudurthi S.D., Luthra P., Bose R.J.C., McCarthy J., Kontaridis M.I. Cardiac inflammation in COVID-19: lessons from heart failure. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wichmann D., Sperhake J.P., Lütgehetmann M., Steurer S., Edler C., Heinemann A., Heinrich F., Mushumba H., Kniep I., Schröder A.S., Burdelski C., de Heer G., Nierhaus A., Frings D., Pfefferle S., Becker H., Bredereke-Wiedling H., de Weerth A., Paschen H.R., Sheikhzadeh-Eggers S., Stang A., Schmiedel S., Bokemeyer C., Addo M.M., Aepfelbacher M., Püschel K., Kluge S. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann. Intern. Med. 2020;173:268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Worldometer, COVID Live Update: 212,586,599 Cases and 4,444,470 Deaths from the Coronavirus - Worldometer, 2021. 〈https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?#countries〉. (Accessed 23 August 2021).
- 17.Sadhukhan P., Ugurlu M.T., Hoque M.O. Effect of COVID-19 on lungs: focusing on prospective malignant phenotypes. Cancers. 2020;12:3822. doi: 10.3390/cancers12123822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chau T.-N., Lee K.-C., Yao H., Tsang T.-Y., Chow T.-C., Yeung Y.-C., Choi K.-W., Tso Y.-K., Lau T., Lai S.-T., Lai C.-L. SARS-associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology. 2004;39:302–310. doi: 10.1002/hep.20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou S., Wang Y., Zhu T., Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. Am. J. Roentgenol. 2020;214:1287–1294. doi: 10.2214/AJR.20.22975. [DOI] [PubMed] [Google Scholar]
- 20.Velavan T.P., Meyer C.G. The COVID‐19 epidemic. Trop. Med. Int. Health. 2020;25:278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Y., Wang H., Shen H., Li Z., Geng J., Han H., Cai J., Li X., Kang W., Weng D., Lu Y., Wu D., He L., Yao K. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 2003;200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussain A., Via G., Melniker L., Goffi A., Tavazzi G., Neri L., Villen T., Hoppmann R., Mojoli F., Noble V., Zieleskiewicz L., Blanco P., Ma I.W.Y., Wahab M.A., Alsaawi A., Al Salamah M., Balik M., Barca D., Bendjelid K., Bouhemad B., Bravo-Figueroa P., Breitkreutz R., Calderon J., Connolly J., Copetti R., Corradi F., Dean A.J., Denault A., Govil D., Graci C., Ha Y.-R., Hurtado L., Kameda T., Lanspa M., Laursen C.B., Lee F., Liu R., Meineri M., Montorfano M., Nazerian P., Nelson B.P., Neskovic A.N., Nogue R., Osman A., Pazeli J., Pereira-Junior E., Petrovic T., Pivetta E., Poelaert J., Price S., Prosen G., Rodriguez S., Rola P., Royse C., Chen Y.T., Wells M., Wong A., Xiaoting W., Zhen W., Arabi Y. Multi-organ point-of-care ultrasound for COVID-19 (PoCUS4COVID): international expert consensus. Crit. Care. 2020;24:702. doi: 10.1186/s13054-020-03369-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baksh M., Ravat V., Zaidi A., Patel R.S. A systematic review of cases of acute respiratory distress syndrome in the Coronavirus disease 2019 pandemic. Cureus. 2020;12:8188. doi: 10.7759/cureus.8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carsana L., Sonzogni A., Nasr A., Rossi R.S., Pellegrinelli A., Zerbi P., Rech R., Colombo R., Antinori S., Corbellino M., Galli M., Catena E., Tosoni A., Gianatti A., Nebuloni M. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect. Dis. 2020;20:1135–1140. doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovren F., Pan Y., Quan A., Teoh H., Wang G., Shukla P.C., Levitt K.S., Oudit G.Y., Al-Omran M., Stewart D.J., Slutsky A.S., Peterson M.D., Backx P.H., Penninger J.M., Verma S. Angiotensin converting enzyme-2 confers endothelial protection and attenuates atherosclerosis. Am. J. Physiol. Heart Circ. Physiol. 2008;295:1377–1384. doi: 10.1152/ajpheart.00331.2008. [DOI] [PubMed] [Google Scholar]
- 28.Ohishi M., Yamamoto K., Rakugi H. Angiotensin (1-7) and other angiotensin peptides. Curr. Pharm. Des. 2013;19:3060–3064. doi: 10.2174/1381612811319170013. [DOI] [PubMed] [Google Scholar]
- 29.Ciaglia E., Vecchione C., Puca A.A. COVID-19 infection and circulating ACE2 levels: protective role in women and children. Front. Pediatr. 2020;8:206. doi: 10.3389/fped.2020.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:1–5. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D. Wrapp, N. Wang, K.S. Corbett, J.A. Goldsmith, C.-L. Hsieh, O. Abiona, B.S. Graham, J.S. Mclellan, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, 2019. http://science.sciencemag.org/. (Accessed 4 January 2021). [DOI] [PMC free article] [PubMed]
- 32.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel Coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS Coronavirus. J. Virol. 2020;94 doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conti P., Caraffa A., Tetè G., Gallenga C.E., Ross R., Kritas S.K., Frydas I., Younes A., Di Emidio P., Ronconi G. Mast cells activated by SARS-CoV-2 release histamine which increases IL-1 levels causing cytokine storm and inflammatory reaction in COVID-19. J. Biol. Regul. Homeost. Agents. 2020;34:1629–1632. doi: 10.23812/20-2EDIT. [DOI] [PubMed] [Google Scholar]
- 35.Conti P., Caraffa A., Gallenga C.E., Ross R., Kritas S.K., Frydas I., Younes A., Di Emidio P., Ronconi G., Toniato E. IL-1 induces throboxane-A2 (TxA2) in COVID-19 causing inflammation and micro-thrombi: inhibitory effect of the IL-1 receptor antagonist (IL-1Ra) J. Biol. Regul. Homeost. Agents. 2020;34:1623–1627. doi: 10.23812/20-34-4EDIT-65. [DOI] [PubMed] [Google Scholar]
- 36.Topol E.J. COVID-19 can affect the heart. Science. 2020;370:408–409. doi: 10.1126/science.abe2813. [DOI] [PubMed] [Google Scholar]
- 37.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan W., Liang W., Zhao Y., Liang H., Chen Z., Li Y., Liu X., Chen R., Tang C., Wang T., Ou C., Li L., Chen P., Sang L., Wang W., Li J., Li C., Ou L., Cheng B., Xiong S., Ni Z., Xiang J., Hu Y., Liu L., Shan H., Lei C., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Cheng L., Ye F., Li S., Zheng J., Zhang N., Zhong N., He J. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur. Respir. J. 2020;55 doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Cookingham J., Coppa K., Diefenbach M.A., Dominello A.J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T.G., Hirschwerk D.A., Kim E.J., Kozel Z.M., Marrast L.M., Mogavero J.N., Osorio G.A., Qiu M., Zanos T.P. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA J. Am. Med. Assoc. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., Gong W., Liu X., Liang J., Zhao Q., Huang H., Yang B., Huang C. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020;5:802–810. doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., Ji R., Wang H., Wang Y., Zhou Y. Prevalence of comorbidities and its effects in coronavirus disease 2019 patients: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lala A., Johnson K.W., Januzzi J.L., Russak A.J., Paranjpe I., Richter F., Zhao S., Somani S., Van Vleck T., Vaid A., Chaudhry F., De Freitas J.K., Fayad Z.A., Pinney S.P., Levin M., Charney A., Bagiella E., Narula J., Glicksberg B.S., Nadkarni G., Mancini D.M., Fuster V. Prevalence and impact of myocardial injury in patients hospitalized with COVID-19 infection. J. Am. Coll. Cardiol. 2020;76:533–546. doi: 10.1016/j.jacc.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei J.F., Huang F.Y., Xiong T.Y., Liu Q., Chen H., Wang H., Huang H., Luo Y.C., Zhou X., Liu Z.Y., Peng Y., Xu Y.N., Wang B., Yang Y.Y., Liang Z.A., Lei X.Z., Ge Y., Yang M., Zhang L., Zeng M.Q., Yu H., Liu K., Jia Y.H., Prendergast B.D., Li W.M., Chen M. Acute myocardial injury is common in patients with COVID-19 and impairs their prognosis. Heart. 2020;106:1154–1159. doi: 10.1136/heartjnl-2020-317007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner A.J., Hiscox J.A., Hooper N.M. ACE2: from vasopeptidase to SARS virus receptor. Trends Pharmacol. Sci. 2004;25:291–294. doi: 10.1016/j.tips.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong C.K., Lam C.W.K., Wu A.K.L., Ip W.K., Lee N.L.S., Chan I.H.S., Lit L.C.W., Hui D.S.C., Chan M.H.M., Chung S.S.C., Sung J.J.Y. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agarwal A., Chen A., Ravindran N., To C., Thuluvath P.J. Gastrointestinal and liver manifestations of COVID-19. J. Clin. Exp. Hepatol. 2020;10:263–265. doi: 10.1016/j.jceh.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hundt M.A., Deng Y., Ciarleglio M.M., Nathanson M.H., Lim J.K. Abnormal liver tests in COVID-19: a retrospective observational cohort study of 1,827 patients in a major U.S. hospital network. Hepatology. 2020;72:1169–1176. doi: 10.1002/hep.31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai Q., Huang D., Yu H., Zhu Z., Xia Z., Su Y., Li Z., Zhou G., Gou J., Qu J., Sun Y., Liu Y., He Q., Chen J., Liu L., Xu L. COVID-19: abnormal liver function tests. J. Hepatol. 2020;73:566–574. doi: 10.1016/j.jhep.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chai X., Hu L., Zhang Y., Han W., Lu Z., Ke A., Zhou J., Shi G., Fang N., Fan J., Cai J., Fan J., Lan F. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection. BioRxiv. 2020 doi: 10.1101/2020.02.03.931766. 2020.02.03.931766. [DOI] [Google Scholar]
- 50.Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang C., Shi L., Wang F.-S. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol. Hepatol. 2020;5:428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of Coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/nejmoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ungaro R.C., Sullivan T., Colombel J.F., Patel G. What should gastroenterologists and patients know about COVID-19? Clin. Gastroenterol. Hepatol. 2020;18:1409–1411. doi: 10.1016/j.cgh.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aguila E.J.T., Cua I.H.Y., Fontanilla J.A.C., Yabut V.L.M., Causing M.F.P. Gastrointestinal manifestations of COVID-19: impact on nutrition practices. Nutr. Clin. Pract. 2020;35:800–805. doi: 10.1002/ncp.10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szefel J., Kruszewski W.J., Buczek T. Enteral feeding and its impact on the gut immune system and intestinal mucosal barrier. Przegląd Gastroenterol. 2015;10:71–77. doi: 10.5114/pg.2015.48997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., Li P., Hu B., Wang J., Hu C., Jin Y., Niu X., Ping R., Du Y., Li T., Xu G., Hu Q., Tu L. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am. J. Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee I.-C., Huo T.-I., Huang Y.-H. Gastrointestinal and liver manifestations in patients with COVID-19. J. Chin. Med. Assoc. 2020;83:521–523. doi: 10.1097/JCMA.0000000000000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., Yip C.C.Y., Leung K.H., Fung A.Y.F., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J.X., To K.K.W., Chan K.H., Yuen K.Y., Leung W.K. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aghemo A., Piovani D., Parigi T.L., Brunetta E., Pugliese N., Vespa E., Omodei P.D., Preatoni P., Lleo A., Repici A., Voza A., Cecconi M., Malesci A., Bonovas S., Danese S. COVID-19 digestive system involvement and clinical outcomes in a large academic hospital in Milan, Italy. Clin. Gastroenterol. Hepatol. 2020;18:2366–2368. doi: 10.1016/j.cgh.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang W., Feng Z., Rao S., Xiao C., Xue X., Lin Z., Zhang Q., Qi W. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J.C., Bin Wang S., Xue Y.D. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020;92:680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan L., Chaudhary K., Saha A., Chauhan K., Vaid A., Baweja M., Campbell K., Chun N., Chung M., Deshpande P., Farouk S., Kaufman L., Kim T., Koncicki H., Lapsia V., Leisman S., Lu E., Meliambro K., Menon M., Rein J., Sharma S., Tokita J., Uribarri J., Vassalotti J., Winston J., Mathews K., Zhao S., Paranjpe I., Somani S., Richter F., Do R., Miotto R., Lala A., Kia A., Timsina P., Li L., Danieletto M., Golden E., Glowe P., Zweig M., Singh M., Freeman R., Chen R., Nestler E., Narula J., Just A., Horowitz C., Aberg J., Loos R., Cho J., Fayad Z., Cordon-Cardo C., Schadt E., Levin M., Reich D., Fuster V., Murphy B., He J.C., Charney A., Bottinger E., Glicksberg B., Coca S., Nadkarni G. Acute kidney injury in hospitalized patients with COVID-19. MedRxiv Prepr. Serv. Health Sci. 2020;10:24. doi: 10.1101/2020.05.04.20090944. [DOI] [Google Scholar]
- 65.Kolhe N.V., Fluck R.J., Selby N.M., Taal M.W. Acute kidney injury associated with COVID-19: a retrospective cohort study. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin W., Hu L., Zhang Y., Ooi J., Meng T., Jin P., Ding X., Peng L., Song L., Xiao Z., Ao X., Xiao X., Zhou Q., Xiao P., Fan J., Zhong Y. Single-cell analysis of ACE2 expression in human kidneys and bladders reveals a potential route of 2019-nCoV infection. BioRxiv. 2020 doi: 10.1101/2020.02.08.939892. 2020.02.08.939892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rismanbaf A., Zarei S. Liver and kidney injuries in COVID-19 and their effects on drug therapy; a letter to editor. Arch. Acad. Emerg. Med. 2020;8 doi: 10.22037/aaem.v8i1.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Durvasula R., Wellington T., McNamara E., Watnick S. COVID-19 and kidney failure in the acute care setting: our experience from Seattle. Am. J. Kidney Dis. 2020;76:4–6. doi: 10.1053/j.ajkd.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.H. Mahmoudi, M.Y. Alikhani, N.M. Taheri, A. Behzadi, Assessment of changes in blood urea and creatinine levels in patients with coronavirus disease 2019 (COVID-19), 2020. https://doi.org/10.21203/rs.3.rs-25164/v1.
- 70.Liotta E.M., Batra A., Clark J.R., Shlobin N.A., Hoffman S.C., Orban Z.S., Koralnik I.J. Frequent neurologic manifestations and encephalopathy-associated morbidity in Covid-19 patients. Ann. Clin. Transl. Neurol. 2020;7:2221–2230. doi: 10.1002/acn3.51210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orsucci D., Ienco E.C., Nocita G., Napolitano A., Vista M. Neurological features of COVID-19 and their treatment: a review. Drugs Context. 2020;9:1–12. doi: 10.7573/DIC.2020-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen R., Wang K., Yu J., Howard D., French L., Chen Z., Wen C., Xu Z. The spatial and cell-type distribution of SARS-CoV-2 receptor ACE2 in human and mouse brain. BioRxiv. 2020 doi: 10.1101/2020.04.07.030650. 2020.04.07.030650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cantuti-Castelvetri L., Ojha R., Pedro L., Djannatian M., Franz J., Kuivanen S., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Österlund P., Joensuu M., Meunier F., Butcher S., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and provides a possible pathway into the central nervous system. BioRxiv. 2020 doi: 10.1101/2020.06.07.137802. 2020.06.07.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J., Wang B., Sun X.-X., Wang C.-F., Yang X., Lin P., Deng Y.-Q., Wei D., Yang X.-M., Zhu Y.-M., Zhang K., Zheng Z.-H., Miao J.-L., Guo T., Shi Y., Zhang J., Fu L., Wang Q.-Y., Bian H., Zhu P., Chen Z.-N. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. BioRxiv. 2020 doi: 10.1101/2020.03.14.988345. 2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bergmann C.C., Lane T.E., Stohlman S.A. Coronavirus infection of the central nervous system: host–virus stand-off. Nat. Rev. Microbiol. 2006;4:121–132. doi: 10.1038/nrmicro1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brann D.H., Tsukahara T., Weinreb C., Lipovsek M., Van Den Berge K., Gong B., Chance R., Macaulay I.C., Chou H.J., Fletcher R.B., Das D., Street K., De Bezieux H.R., Choi Y.G., Risso D., Dudoit S., Purdom E., Mill J., Hachem R.A., Matsunami H., Logan D.W., Goldstein B.J., Grubb M.S., Ngai J., Datta S.R. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020;6:5801–5832. doi: 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iadecola C., Anrather J., Kamel H. Effects of COVID-19 on the nervous system. Cell. 2020;183:16–27.e1. doi: 10.1016/j.cell.2020.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Erickson M.A., Banks W.A. Neuroimmune axes of the blood-brain barriers and blood-brain interfaces: bases for physiological regulation, disease states, and pharmacological interventions. Pharmacol. Rev. 2018;70:278–314. doi: 10.1124/pr.117.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mebius R.E., Kraal G. Structure and function of the spleen. Nat. Rev. Immunol. 2005;5:606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- 82.Zhan J., Deng R., Tang J., Zhang B., Tang Y., Wang J.K., Li F., Anderson V.M., McNutt M.A., Gu J. The spleen as a target in severe acute respiratory syndrome. FASEB J. 2006;20:2321–2328. doi: 10.1096/fj.06-6324com. [DOI] [PubMed] [Google Scholar]
- 83.Al-Ozaibi L.S., Alshaikh M.O., Makhdoom M., Alzoabi O.M., Busharar H.A., Keloth T.R. Splenic abscess: an unusual presentation of COVID-19? Dubai Med. J. 2020;3:115–118. doi: 10.1159/000509644. [DOI] [Google Scholar]
- 84.Mariana S.L.P., Lima F.C., Carla, Pimentel F., Carla A., Costa G., Carlos J., Bezerra Holanda J.L. Multisystemic infarctions in COVID-19: focus on the spleen. Eur. J. Case Rep. Intern. Med. 2020;7 doi: 10.12890/2020_001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hassan W., Ramadan H., Omran G. The spleen as an extrapulmonary target of COVID-19. Afro-Egypt. J. Infect. Endem. Dis. 2021;11:96–99. doi: 10.21608/aeji.2021.60611.1133. [DOI] [Google Scholar]
- 86.Feng Z., Diao B., Wang R., Wang G., Wang C., Tan Y., Liu L., Wang C., Liu Y., Liu Y., Yuan Z., Ren L., Wu Y., Chen Y. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. MedRxiv. 2020 doi: 10.1101/2020.03.27.20045427. 2020.03.27.20045427. [DOI] [Google Scholar]
- 87.Ihlow J., Michaelis E., Greuel S., Heynol V., Lehmann A., Radbruch H., Meinhardt J., Miller F., Herbst H., Corman V.M., Westermann J., Bullinger L., Horst D., von Brünneck A.-C., Elezkurtaj S. B cell depletion and signs of sepsis-acquired immunodeficiency in bone marrow and spleen of COVID-19 deceased. Int. J. Infect. Dis. 2021;103:628–635. doi: 10.1016/j.ijid.2020.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]