Abstract
Rationale:
Absence of dystrophin in Duchenne muscular dystrophy (DMD) results in the degeneration of skeletal and cardiac muscles. Owing to advances in respiratory management of DMD patients, cardiomyopathy has become a significant aspect of the disease. While CRISPR/Cas9 genome editing technology holds great potential as a novel therapeutic avenue for DMD, little is known about the potential of DMD correction using CRISPR/Cas9 technology to mitigate cardiac abnormalities in DMD.
Objective:
To define the effects of CRISPR/Cas9 genome editing on structural, functional and transcriptional abnormalities in DMD-associated cardiac disease.
Methods and Results:
We generated induced pluripotent stem cells (iPSCs) from a patient with a deletion of exon 44 of the DMD gene (ΔEx44) and his healthy brother. We targeted exon 45 of the DMD gene by CRISPR/Cas9 genome editing to generate corrected DMD (cDMD) iPSC lines, wherein the DMD open reading frame was restored via reframing (RF) or exon skipping (ES). While DMD cardiomyocytes (CMs) demonstrated morphologic, structural and functional deficits compared to control CMs, CMs from both cDMD lines were similar to control CMs. Bulk RNA-sequencing of DMD CMs showed transcriptional dysregulation consistent with dilated cardiomyopathy, which was mitigated in cDMD CMs. We then corrected dysfunctional DMD CMs by adenoviral delivery of Cas9/gRNA and showed that correction of DMD CMs post-differentiation reduces their arrhythmogenic potential. Single-nucleus RNA-sequencing of hearts of DMD mice showed transcriptional dysregulation in CMs and fibroblasts, which in corrected mice was reduced to similar levels as wildtype mice.
Conclusions:
We show that CRISPR/Cas9-mediated correction of DMD ΔEx44 mitigates structural, functional and transcriptional abnormalities consistent with dilated cardiomyopathy irrespective of how the protein reading frame is restored. We show that these effects extend to postnatal editing in iPSC-CMs and mice. These findings provide key insights into the utility of genome editing as a novel therapeutic for DMD-associated cardiomyopathy.
Keywords: Animal Models of Human Disease, Basic Science Research, Cardiomyopathy, Myocardial Biology, Stem Cells
Graphical Abstract
INTRODUCTION
Duchenne Muscular Dystrophy (DMD) is an X-linked disease caused by mutations in the dystrophin gene, resulting in the degeneration of skeletal and cardiac muscles1. It is the most common form of muscular dystrophy affecting ~1:5,000 boys2. To date, over 3,000 dystrophin mutations have been described, which primarily affect so-called hotspot regions, including the rod domain (exons 43 to 53) and actin-binding domain (exons 6 and 7) of dystrophin3. These mutations mostly involve deletions in the gene, disrupting the open reading frame (ORF) and introducing a premature stop codon, which in turn results in the production of non-functional dystrophin protein4. Dystrophin is a component of the dystrophin-glycoprotein complex (DGC), which links the actin cytoskeleton to the sarcolemma, and is essential for the transmission of extracellular forces to the sarcomeres. In the absence of dystrophin, this link is weakened, and skeletal and cardiac myocytes are exposed to repeated cycles of shearing and tearing of the sarcolemma, resulting in critical damage at the cellular as well as tissue levels1, 5.
Although historically DMD has primarily been regarded as a neuromuscular disease with respiratory failure as the leading cause of death, today cardiomyopathy has become a significant aspect of the disease owing to advances in respiratory medicine6. The onset and progression of DMD-associated cardiomyopathy is highly variable, and often includes successive myocardial atrophy and remodeling, eventually resulting in congestive heart failure, arrhythmia and sudden cardiac death5, 6. Current treatment options focus on medical therapy with the aim to slow the progression of cardiomyopathy6. Novel therapeutic tools are therefore in dire need.
The discovery of new therapeutic avenues for DMD-associated cardiomyopathy has been limited by the lack of mouse models that reflect the clinical phenotype of cardiomyopathy as seen in humans7. Mouse models of DMD are known to develop cardiomyopathy only in a mild form and in much later stages of life, while in humans cardiac dysfunction is readily observed in pre-pubescent boys7, 8. To generate preclinical models that recapitulate the human disease more reliably, induced pluripotent stem cells (iPSCs) from patients carrying a mutation in the DMD gene have been generated to study the pathobiology of DMD-associated cardiomyopathy and to explore novel treatment options for the disease, such as CRISPR/Cas9 genome editing9-11. Our lab and others have previously demonstrated the efficacy of CRISPR/Cas9 genome editing in a number of different DMD models in mice and human cells12-15. These approaches focused on correcting the expression of functional, truncated dystrophin protein through the introduction of small insertions and deletions via non-homologous end joining of double-stranded DNA breaks generated by CRISPR/Cas9, thereby restoring the ORF of the dystrophin transcript.
While our lab and others have shown the potential utility of CRISPR/Cas9 genome editing as a novel therapeutic avenue in DMD-associated cardiomyopathy using iPSCs, the goal of those studies was focused on the assessment of few functional aspects of cardiac biology without providing an in-depth perspective on the correction of the cardiomyopathy phenotype. In addition, because some correction strategies using CRISPR/Cas9 yield single truncated dystrophin proteins via reframing or exon skipping, both of which restore the ORF of the dystrophin transcript16, other strategies have been shown to result in the simultaneous production of multiple forms of truncated dystrophin protein14, 17, 18. Further, it has been shown that some truncated dystrophin proteins may be more functional than others12, 19. It is therefore imperative to determine if these newly engineered proteins perform biologically equally to the full-length dystrophin protein. More importantly, all of these studies have focused on gene correction at the iPSC stage, such that corrected differentiated cardiac myocytes never developed dysfunction due to the absence of dystrophin. It is therefore not known to what extent correction of DMD using CRISPR/Cas9 genome editing technology can mitigate the cardiomyopathy phenotype in dysfunctional DMD cardiomyocytes.
Here, iPSCs from a DMD patient with deletion of exon 44 of the DMD gene (ΔEx44) were generated and differentiated towards the cardiac lineage. Using our previously published gene correction approach, we generated corrected DMD (cDMD) iPSC lines, wherein dystrophin expression was restored by means of reframing (cDMD-RF) or exon skipping (cDMD-ES) via CRISPR/Cas9 genome editing. We evaluate the biological equivalency of these truncated dystrophin proteins to full-length dystrophin protein by assessing differentiated cardiac myocytes from the DMD line, both cDMD lines and a control line derived from the healthy brother of the patient with respect to their structural and functional development. We provide RNA-sequencing data demonstrating that truncated dystrophin proteins prevent transcriptional dysregulation as seen in human dilated cardiomyopathy. We then apply our gene correction approach to differentiated DMD-CMs and demonstrate the mitigation of arrhythmia when dystrophin expression is restored in post-differentiated cardiomyocytes. Finally, we show using single-nucleus (sn)RNA-sequencing of aged ΔEx44 mice that our correction strategy also attenuates cardiac transcriptional abnormalities in vivo.
METHODS
For detailed methodological information, please refer to the Supplemental Material in the Data Supplement. For research materials listed in the Methods, please see the Major Resources Table in the Data Supplement.
Data and materials availability.
All data presented in this study are available in the main text or the supplementary materials. iPSC-CM RNA-seq data have been deposited at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE169190. Mouse snRNA-seq data have been deposited under accession number GSE169551.
Code Availability statement.
The MATLAB code used to perform contractile force measurements of iPSC-CMs is available at https://github.com/DarisaLLC/Cardio. Codes for bulk RNA-seq analysis are available at https://github.com/zwang0715/Atmanli_et_al_RNAseq.
RESULTS
Dystrophin correction restores normal cardiac structural and morphologic development.
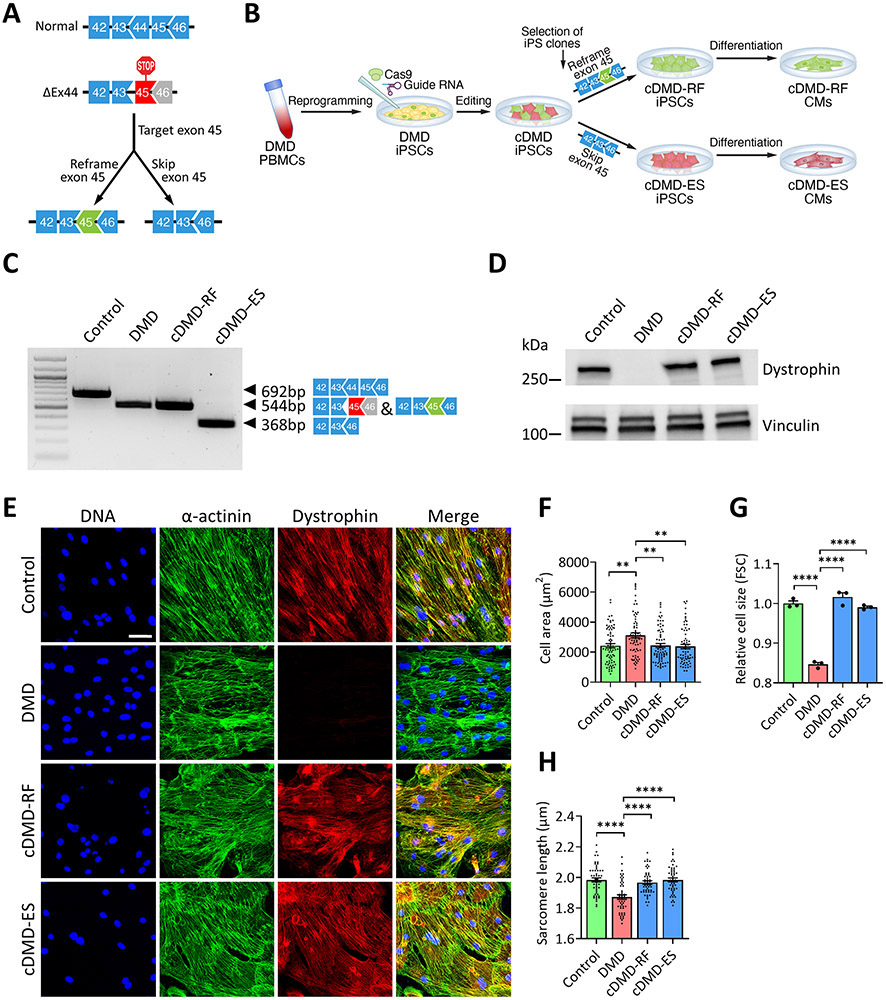
iPSCs from a DMD patient lacking exon 44 of the dystrophin gene and from the patient’s healthy brother (control line) were generated. Deletion of exon 44 disrupts the open reading frame by introducing a premature termination codon. To correct the DMD ΔEx44 genotype, we targeted exon 45 in DMD iPSCs using our previously published CRISPR/Cas9 gene editing approach14. Using this strategy, dystrophin can be corrected by either reframing or skipping of exon 45, thus generating two differently truncated dystrophin proteins (Fig. 1A). To study the biological effects of these truncated dystrophin proteins, we selected, sequenced and expanded individual iPSC clones with the RF or ES genotype (Fig. 1B). We differentiated all four lines towards the cardiac lineage and confirmed successful gene editing by reverse transcription PCR (RT-PCR) using primers targeting exons 42 and 46, respectively, of DMD cDNA (Fig. 1C) and sequencing of RT-PCR products (Online Figure I). We confirmed the presence or absence, respectively, of dystrophin protein by Western blot analysis (Fig. 1D) and immunofluorescence of differentiated iPSC-CMs (Fig. 1E).
Figure 1. Correction of DMD ΔEx44 by CRISPR/Cas9 genome editing rescues structural and morphologic abnormalities in iPSC-derived CMs.
A) Gene editing strategy for DMD ΔEx44 deletion mutation. Deletion of exon 44 (black) generates a premature stop codon in exon 45. The protein reading frame can be restored by disrupting the splice junction of exon 45 (exon skipping) or reframing exon 45 (green).
B) Schematic showing the derivation of DMD patient-derived iPSCs and iPSC-CMs, and generation of corrected iPSC lines.
C) RT-PCR analysis using primers targeting exons 42 and 46 of DMD cDNA from control, DMD and cDMD CMs.
D) Western blot analysis of control, DMD and cDMD CMs for dystrophin. Vinculin served as loading control.
E) Representative immunofluorescence images of control, DMD and cDMD CMs stained for dystrophin and sarcomeric α-F) Quantification of cell area of d35 control, DMD and cDMD CMs using wheat germ agglutinin staining (n = 71 cells for control CMs, n = 70 cells for DMD CMs, n = 71 cells for cDMD-RF CMs, n = 69 cells for cDMD-ES CMs;μm.
F) Quantification of cell area of d35 control, DMD and cDMD CMs using wheat germ agglutinin staining (n = 71 cells for control CMs, n = 70 cells for DMD CMs, n = 71 cells for cDMD-RF CMs, n = 69 cells for cDMD-ES CMs; quantification was performed across three independent batches of differentiation).
G) Quantification of the cell size of DMD and cDMD CMs relative to control CMs using forward scatter (FSC) from flow cytometry (n = 3 samples per group across three independent batches of differentiation; minimum of 50,000 cells per sample recorded).
H) Quantification of sarcomere lengths of d35 control, DMD and cDMD CMs (n = 50 cells per group across three independent batches of differentiation).
Quantified data are shown as mean ± s.e.m. **p<0.01 and ****p<0.0001.
We then asked if the absence of dystrophin protein results in developmental deficits related to the structure and morphology of iPSC-CMs and, conversely, if truncated dystrophin proteins can rescue these defects. We first co-stained iPSC-CMs for wheat germ agglutinin (WGA) and sarcomeric α-actinin, and found DMD CMs to have a larger surface area than control and cDMD CMs (Fig. 1F and Online Figure IIA). Because a two-dimensional analysis does not reveal potential changes in cell volume, we next quantified relative three-dimensional cell sizes by flow cytometry, and interestingly found DMD CMs to be smaller than control and cDMD CMs (Fig. 1G and Online Figure IIB), suggesting that DMD CMs have lower cell volumes, but spread out more on culture surfaces compared to control and cDMD CMs.
We next quantified sarcomere lengths from cells co-stained for WGA and sarcomeric α-actinin, and found DMD CMs to have shorter sarcomeres than control and cDMD CMs (Fig. 1H and Online Figure III), suggesting that in the absence of dystrophin sarcomere biology is altered. Taken together, our observations suggest that DMD results in significant perturbations in the cellular development and maturation of iPSC-CMs akin to dilated cardiomyopathy37, 49-52. In contrast, truncated dystrophin proteins do not affect these processes and cDMD CMs closely resemble normal CMs.
Truncated dystrophin proteins are functionally equivalent to full-length dystrophin protein.
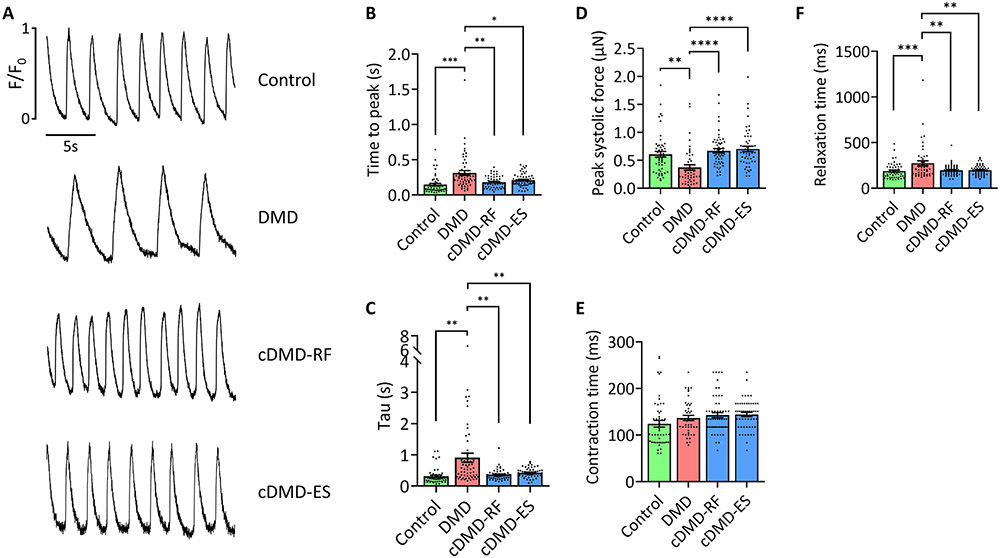
We next asked if and to what extent DMD CMs show functional deficits and if truncated dystrophin proteins restore functionality of iPSC-CMs. We first analyzed spontaneous Ca2+ cycling of single iPSC-CMs on d35 post-differentiation to quantify calcium release and reuptake parameters (Fig. 2A). Consistent with previous reports from our group and others9, 12, DMD CMs showed prolonged calcium transients with increased times to release (Fig. 2B) and to reuptake calcium (Fig. 2C). Although control CMs showed faster contractile rates than DMD and cDMD CMs, differences in beating frequencies showed no correlation with calcium handling parameters (Online Figure IV). Defective calcium signaling in d35 DMD CMs was accompanied by an impaired patterning of the ryanodine receptor 2 (RYR2), with the majority of DMD CMs showing a diffuse distribution pattern of RYR2 as opposed to a correct localization to dyads, as seen in control and cDMD CMs (Online Figure VA and B). Of note, the expression levels of RYR2 were not different between the four lines (Online Figure VC). In summary, DMD CMs show altered calcium signaling and CMs expressing truncated dystrophin proteins exhibit calcium handling characteristics similar to control CMs.
Figure 2. Corrected DMD CMs show normal functional characteristics.
A) Representative calcium traces of single control, DMD and cDMD CMs loaded with the calcium indicator Fluo 4-AM.
B) Quantification of the calcium release phase in single control, DMD and cDMD CMs as shown by time to peak (n = 50 cells for control CMs, n = 56 cells for DMD CMs, n = 52 cells for cDMD-RF CMs, n = 54 cells for cDMD-ES CMs; quantification was performed across three independent batches of differentiation).
C) Quantification of the calcium reuptake phase in single control, DMD and cDMD CMs as shown by tau (n = 50 cells for control CMs, n = 56 cells for DMD CMs, n = 52 cells for cDMD-RF CMs, n = 54 cells for cDMD-ES CMs; quantification was performed across three independent batches of differentiation).
D) Quantification of contractile force of single control, DMD and cDMD CMs (n = 52 cells for control CMs, n = 46 cells for DMD CMs, n = 55 cells for cDMD-RF CMs, n = 52 cells for cDMD-ES CMs; quantification was performed across three independent batches of differentiation).
E) Quantification of contraction time of single control, DMD and cDMD CMs (n = 52 cells for control CMs, n = 46 cells for DMD CMs, n = 55 cells for cDMD-RF CMs, n = 52 cells for cDMD-ES CMs; quantification was performed across three independent batches of differentiation).
F) Quantification of relaxation time of single control, DMD and cDMD CMs (n = 52 cells for control CMs, n = 46 cells for DMD CMs, n = 55 cells for cDMD-RF CMs, n = 52 cells for cDMD-ES CMs; quantification was performed across three independent batches of differentiation).
Functional analyses were performed at d35 post-differentiation. Quantified data are shown as mean ± s.e.m. **p<0.01, ***p<0.001 and ****p<0.0001.
Another hallmark of cardiomyocyte functionality is the generation of contractile force. DMD cardiomyopathy has been shown to result in global cardiac dysfunction, affecting both the systolic and diastolic capacity of the heart53. To investigate the effects of DMD and cDMD on the contractility of iPSC-CMs, we plated CMs on d35 post-differentiation on a soft polydimethylsiloxane (PDMS) matrix at single-cell density and acquired high frame-rate videos of contracting CMs. The PDMS surface allowed iPSC-CMs to undergo auxotonic instead of isometric contraction, as seen on stiff surfaces such as glass or tissue culture polystyrene. Quantification of peak systolic forces of iPSC-CMs revealed that DMD CMs generated two-fold reduced contractile force compared to CMs derived from the control and both cDMD lines (Fig. 2D), suggesting systolic dysfunction. We further quantified contraction and relaxation times and found that while there was no difference in the time to reach peak systole between all lines (Fig. 2E), DMD CMs showed prolonged relaxation times, suggesting diastolic dysfunction, while the control and both cDMD lines relaxed similarly fast (Fig. 2F). This observation is consistent with our finding that DMD CMs have shorter sarcomere lengths (Fig. 1H), further corroborating a deficit in cellular relaxation. Taken together, these results show that the global cardiac dysfunction observed clinically in DMD patients is recapitulated in vitro using CMs derived from iPSCs from a DMD patient. In contrast, control as well as cDMD CMs with truncated dystrophin proteins show no functional deficits. Our results related to the functional, structural and morphologic characteristics of DMD CMs point to a cardiomyopathy phenotype, which is absent in cDMD CMs expressing truncated dystrophin proteins.
Dystrophin correction attenuates a cardiomyopathy transcriptional profile.
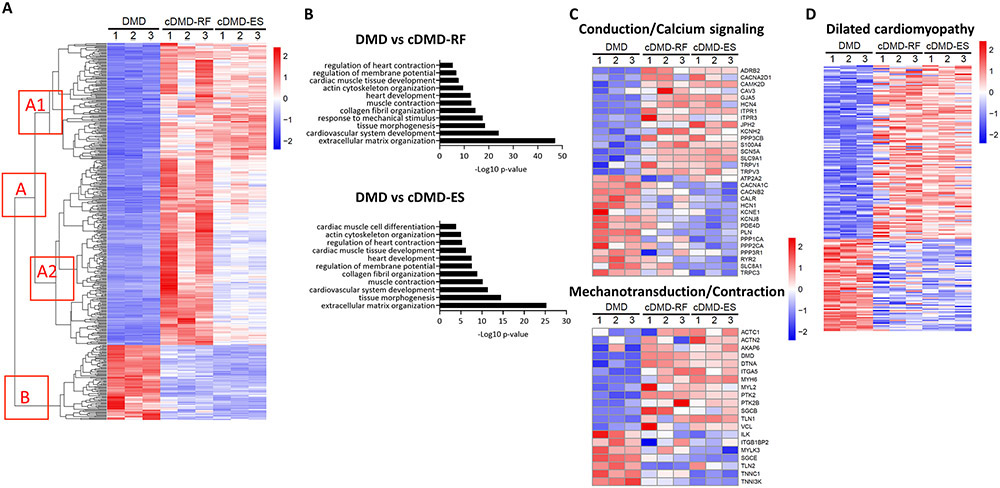
We next sought to investigate the transcriptional state of DMD CMs and how it might differ in CMs expressing truncated dystrophin proteins. Because our control line was not isogenic to the DMD line, we performed RNA-sequencing on CMs from the DMD and both cDMD lines. Principal component analysis revealed that cDMD CMs were transcriptomically different from DMD CMs (Online Figure VIA). Differential gene expression analysis of genes that changed by two-fold or more identified 794 upregulated and 205 downregulated transcripts in cDMD-RF CMs compared to DMD CMs, whereas there were 495 upregulated and 255 downregulated transcripts in cDMD-ES CMs compared to DMD CMs (Fig. 3A and Online Figure VIB). Gene Ontology (GO) analysis of differentially regulated genes between DMD CMs, and cDMD-RF and cDMD-ES CMs, respectively, mapped to extracellular matrix signaling and cardiomyocyte function (Fig. 3B and Online Table III), consistent with previous reports that dystrophin is a key protein within the mechanotransduction process of cardiac contraction54. Indeed, we found 32 transcripts related to cardiac conduction and calcium signaling, and 21 transcripts related to cardiac mechanotransduction and contraction that were differentially regulated between DMD, and cDMD-RF and cDMD-ES CMs (Fig. 3C). We further performed GO analysis of genes whose expression showed a high degree of dissimilarity between cDMD-RF and cDMD-ES CMs despite being differentially regulated compared to DMD CMs (cluster A2 in Figure 3A; Online Figure VII). We found that these genes mapped to the same or similar biological processes as the overall GO analysis, as shown in Fig. 3B, demonstrating that both forms of truncated dystrophin proteins elicit the same biological responses, albeit at different levels.
Figure 3. Dystrophin correction mitigates transcriptional dysregulation in iPSC-CMs.
A) Heatmap showing the expression of differentially regulated genes in DMD, cDMD-RF and cDMD-ES CMs. Hierarchical clustering of differentially expressed genes revealed two major clusters: A, which contains genes upregulated in cDMD CMs; and B, which contains genes downregulated in cDMD CMs. Genes within A cluster can be further divided into A1 cluster, which contains genes that showed similar upregulation in cDMD-RF and cDMD-ES CMs compared with DMD, and A2 cluster, which contains genes that showed a high degree of dissimilarity between cDMD-RF CMs and cDMD-ES CMs despite being differentially regulated compared to DMD CMs.
B) Top GO terms associated with the differentially expressed genes in DMD versus cDMD-RF (top) and DMD versus cDMD-ES (bottom).
C) Heatmap of key genes involved in cardiac conduction and calcium cycling (top), and cardiac mechanotransduction and contraction (bottom).
D) Heatmap of key genes implicated in dilated cardiomyopathy in human patients.
RNA-sequencing was performed on three independent batches of iPSC differentiation.
RNA-sequencing analyses were performed at d50 post-differentiation.
To corroborate our finding that DMD CMs exhibit a cardiomyopathy phenotype and cDMD CMs mitigate it, we examined the expression levels of transcripts that have been implicated in DCM in human patients55, 56. Our transcriptomic analyses of DMD and cDMD CMs identified over 200 known transcripts that have been shown to play a role in DCM and that were differentially regulated between DMD and cDMD CMs (Fig. 3D and Online Table IV). Indeed, two of the hallmark dysregulated transcripts in DCM, NPPA and NPPB, were three-fold downregulated in DMD CMs compared to cDMD CMs (Online Figure VIC), consistent with previous reports that natriuretic peptides are downregulated in in vitro models of DCM as opposed being upregulated as in in vivo models37. Finally, we showed by quantitative PCR (qPCR) that NPPA and NPPB levels in cDMD CMs were similar to those in control CMs (Online Figure VIII), suggesting that truncated dystrophin proteins yield a normal transcriptional profile. Taken together, our transcriptomic data shows that DMD CMs display a transcriptional state consistent with DCM and that truncated dystrophin proteins attenuate the cardiomyopathy profile.
Truncated dystrophin proteins restore membrane tension.
Some of the highest differentially regulated genes between DMD CMs and cDMD CMs were genes involved in membrane biology and actin cytoskeleton organization (Online Figure IXA). Because these cellular processes have been shown to be critical in maintaining proper membrane tension57, 58, which in turn is a key determinant of cardiac myocyte function59, 60, we asked how the absence of dystrophin, and conversely the presence of truncated dystrophin proteins, affect membrane tension. We loaded single CMs with the fluorescent membrane tension probe Flipper-TR and computed the fluorescent lifetime of the probe using fluorescence lifetime imaging microscopy (FLIM). Fluorescent lifetime of Flipper-TR has been shown to be positively correlated with membrane tension29. We found that single DMD CMs have higher membrane tension than single control CMs, while single cDMD CMs had similar membrane tension to single control CMs (Online Figure IXB and C). Analysis of membrane tension in densely grown cellular syncytia of CMs demonstrated similar differences (Online Figure IXD and E). These data show how dystrophin critically regulates membrane biology and cytoskeleton organization in CMs, and how truncated dystrophin proteins are biologically equivalent to full-length dystrophin in this regard.
Postnatal dystrophin correction in DMD CMs reduces their arrhythmogenicity.
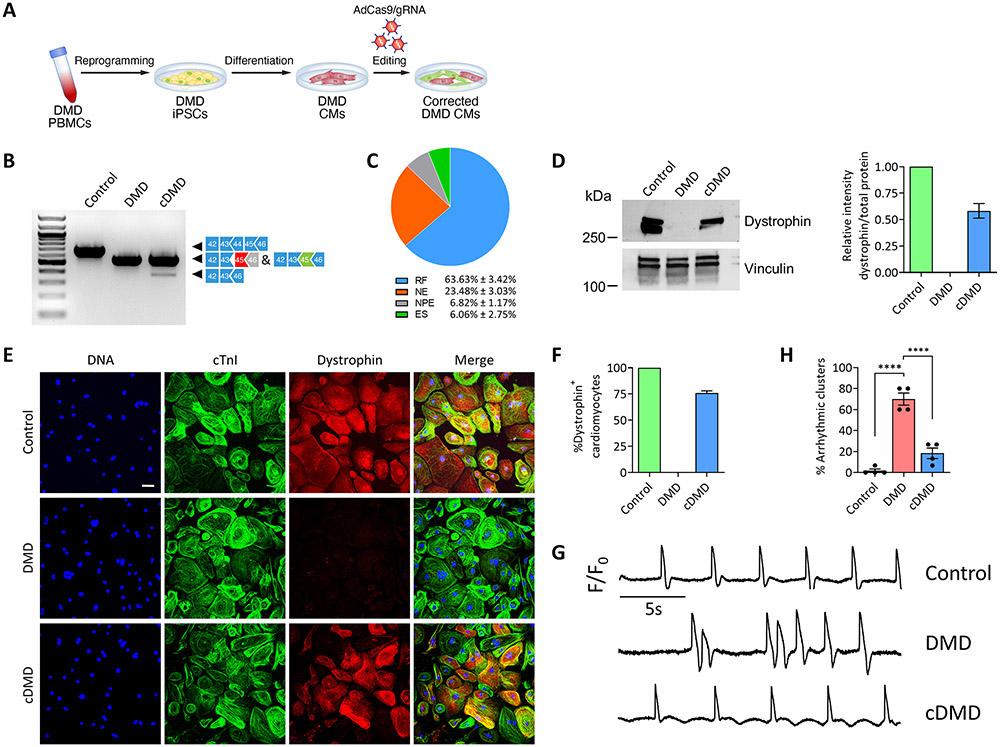
Having established that truncated dystrophin proteins in our ΔEx44 iPSC model prevent a cardiomyopathy profile, we set out to test whether restoring dystrophin expression after a cardiomyopathy phenotype becomes apparent mitigates functional alterations in DMD CMs (Figure 4A). Because our functional data as shown in Fig. 2 showed clear cardiac dysfunction on d35 post-differentiation, we corrected dystrophin expression at that developmental stage in the following experiments. Of note, our iPSC-CM culture system includes the temporal addition of T3, dexamethasone and IGF-1, which have been shown to promote the maturity of iPSC-CMs and to allow them to resemble neonatal cardiac myocytes22, 23. We cloned Cas9 and our gRNA in an adenoviral vector to achieve high transduction efficiency. Indeed, we found that over 90% of iPSC-CMs were successfully transduced as shown by flow analysis seven days post-infection (Online Figure X). We then transduced DMD CMs with AdCas9/gRNA, and transduced control CMs and DMD CMs with AdCas9 as controls. We confirmed successful gene editing by RT-PCR (Figure 4B) and performed TOPO-TA cloning of RT-PCR products. Sequencing of individual bacterial colonies revealed a 70% efficiency of productive gene editing with a 10:1 ratio of reframing versus exon skipping events (Figure 4C), similar to our previous report that reframing is the predominant editing outcome in corrected murine skeletal muscle14. We further validated our gene editing strategy by quantitative Western blot analysis (Figure 4D and Online Figure XIA) and immunofluorescence (Figure 4E) 30 days post-infection with AdCas9/gRNA. Based on immunofluorescence, our approach yielded a correction efficiency of 75% (Figure 4F), in line with our TOPO-TA sequencing results.
Figure 4. Postnatal dystrophin correction reduces arrhythmogenicity of iPSC-CMs.
A) Schematic showing postnatal gene editing in DMD CMs by adenoviral delivery of Cas9 and gRNA.
B) RT-PCR analysis using primers targeting exons 42 and 46 of DMD cDNA from control, DMD and cDMD CMs.
C) Pie chart showing the percentage of gene editing events in iPSC-CMs based on RT-PCR sequence analysis of TOPO-TA generated clones. RF, reframing; NE, no editing; NPE, non-productive editing; ES, exon skipping.
D) (Left) Western blot analysis of control, DMD and cDMD CMs for dystrophin and vinculin. (Right) Quantification of dystrophin expression based on Western blot analysis (n = 3 samples per group across three independent batches of differentiation). Total protein expression served as loading control (Online Fig. XA).
E) Representative immunofluorescence images of control, DMD and cDMD CMs stained for dystrophin and cardiac troponin I. Scale bar 50μm.
F) Quantification of dystrophin-positive CMs in control, DMD and cDMD CMs (n = 1,612 cells for control CMs, n = 1,569 cells for DMD CMs, n = 1,734 cells for cDMD CMs; quantification was performed across three independent batches of differentiation).
G) Representative membrane potential traces of clusters of control, DMD and cDMD CMs loaded with the voltage-sensitive probe FluoVolt.
H) Quantification of the percentage of arrhythmic clusters of control, DMD and cDMD CMs based on FluoVolt imaging (n = 60 clusters per group across four independent batches of differentiation).
iPSC CMs were transduced with adenovirus at d35 post-differentiation and functional analyses were performed at d65 post-differentiation. Quantified data are shown as mean ± s.e.m. ****p<0.0001.
Because a hallmark of DMD cardiomyopathy is the occurrence of deadly arrhythmia8, we asked if the restoration of dystrophin in dysfunctional DMD CMs could mitigate the occurrence of arrhythmia in clusters of CMs as a proxy for a tissue-level effect of dystrophin restoration. We again infected cells on d35 post-differentiation and cultured them for an additional 30 days. We then replated the cells at high-density and loaded the cells with the voltage-sensitive probe FluoVolt to assess the arrhythmogenicity of CMs. While control CMs showed minimal arrhythmogenic potential, 70% of clusters of DMD CMs were arrhythmic, with some DMD CMs spontaneously entering or quitting a tachyarrhythmic episode (Online Figure XII), recapitulating a significant pathophysiologic phenomenon of DMD cardiomyopathy8. Strikingly, restoring dystrophin in a fraction of DMD CMs was sufficient to decrease the presence of arrhythmic clusters to less than 20% (Figure 4G and H), consistent with previous reports that mosaic dystrophin expression in at least 50% of CMs is sufficient to achieve a near-normal cardiac phenotype13. Taken together, our results show that postnatal editing of dystrophin in DMD CMs is efficient and has the potential to resolve the arrhythmogenicity of DMD CMs.
Postnatal dystrophin correction in DMD ΔEx44 mice normalizes cardiac transcriptional regulation.
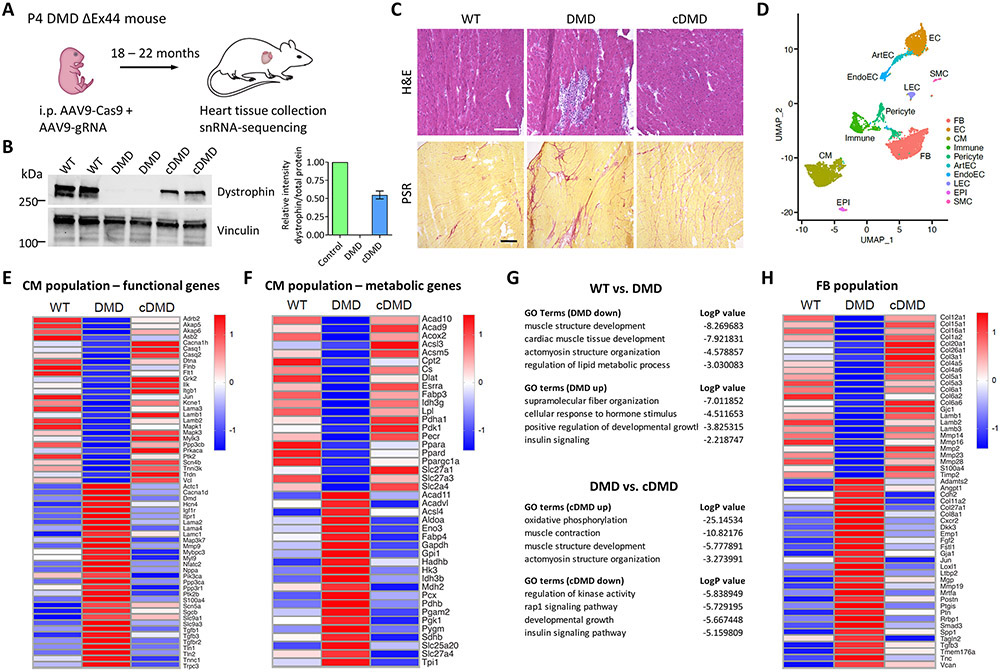
We finally set out to determine if postnatal dystrophin correction in vivo improves cardiac biology of DMD ΔEx44 mice. Because animal models of DMD develop cardiomyopathy only at very late developmental stages and in a mild form7, we asked if DMD mice show cardiac transcriptional dysregulation, and to what extent cardiac dystrophin correction reduces transcriptional abnormalities. We delivered AAV9-Cas9 and AAV9-gRNA to DMD ΔEx44 mice systemically via intraperitoneal injection on P4 and let the mice age for an additional 18 to 22 months (Fig. 5A). We verified successful correction of dystrophin by Western blot analysis (Figure 5B and Online Figure XIB) and immunofluorescence staining (Online Figure XIIIA). H&E and Picrosirius red staining showed that cardiac muscle degeneration and interstitial fibrosis, both histopathologic hallmarks of DMD cardiomyopathy, were present in the hearts of DMD ΔEx44 mice. We could not find evidence of extensive tissue degeneration and interstitial fibrosis in the hearts of wildtype and corrected mice (Fig. 5C and Online Figure XIIIB).
Figure 5. Postnatal dystrophin correction attenuates DMD-associated cardiac transcriptional dysregulation in vivo.
A) Schematic showing postnatal gene editing in DMD ΔEx44 mice using AAV9-Cas9 and AAV9-gRNA on P4. Heart tissues were obtained at 18 to 22 months of age.
B) (Left) Western blot analysis of control, DMD and cDMD CMs for dystrophin and vinculin. (Right) Quantification of dystrophin expression based on Western blot analysis (n = 2 samples per group). Total protein expression served as loading control (Online Fig. XB).
C) Histopathologic H&E (top) and Picro Sirius Red (PSR, bottom) analysis of WT, DMD and cDMD hearts. Scale bars 200μm.
D) UMAP visualization of cardiac cell populations in all snRNA-sequencing samples.
E) Heatmap of key functional genes in the cardiomyocyte (CM) population.
F) Heatmap of key metabolic genes in the CM population.
G) Top GO terms associated with the differentially expressed genes in the comparison WT versus DMD (top) and DMD versus cDMD (bottom).
H) Heatmap of key genes in the fibroblast (FB) population.
Because DMD cardiomyopathy is accompanied by cardiomyocyte dysfunction as well as the occurrence of widespread myocardial fibrosis8, we performed snRNA-sequencing of hearts of DMD ΔEx44 mice. This approach allowed us to separately investigate transcriptomic regulation within cardiomyocytes as well as fibroblasts. To allow cross-sample comparisons, we integrated datasets from all samples for cell type clustering. Cell type identity was assigned based on the top differentially expressed genes and expression of known cellular markers (Online Figure XIVA). We identified all major cardiac cell types, including cardiomyocytes, fibroblasts and endothelial cells, in each sample, as shown in Uniform Manifold Approximation and Projection (UMAP) plots (Figure 5D and Online Figure XV). Similar to our iPSC-CM RNA-sequencing data, dystrophin was expressed in all samples, including DMD hearts, with the highest expression levels in the CM populations and significantly lower expression levels in smooth muscle cells and other vascular cell populations (Online Figure XVI). Population analysis revealed no major differences in the abundance of cell types (Online Figure XIVB). Within the CM population, we identified over 100 genes related to cardiomyocyte function and metabolism that were differentially regulated between DMD mice, and wildtype and corrected mice (Fig. 5E and F). Similar to our bulk RNA-sequencing data from iPSC-CMs (Fig. 3 and Online Figure VI), Nppa, a strong marker of cardiomyopathy, was highly dysregulated in DMD mice, but expression levels restored to similar levels as wildtype mice after dystrophin correction. Further, the metabolic gene panel demonstrated mitochondrial dysfunction in DMD mice as evidenced by upregulation of genes involved in glycolysis and downregulation of genes involved in oxidative phosphorylation, while dystrophin correction prevented the dysregulation of metabolic transcripts (Fig. 5F). The dysregulation of cardiac functional and metabolic genes was further corroborated by GO analysis of differentially regulated genes between DMD CMs, and wildtype and cDMD CMs, respectively, wherein they mostly mapped to cardiac functional and metabolic processes (Fig. 5G).
We next investigated the transcriptomic changes in the fibroblast (FB) population in the hearts of the mice. We found upregulation of genes regulating a fibrotic response in DMD mice, such as TGFβ-signaling, and dysregulation of genes controlling extracellular matrix remodeling, such as periostin (Postn) and several matrix metalloproteinases (Fig. 5H). Dystrophin correction in the hearts of DMD mice mitigated the transcriptional dysregulation in the FB population. Taken together, our data provided evidence that postnatal dystrophin correction in vivo is accompanied by improvements in histopathologic and transcriptomic changes consistent with cardiomyopathy in DMD ΔEx44.
DISCUSSION
The overarching goal of this study was to assess the utility of CRISPR/Cas9 genome editing for the attenuation of cardiac abnormalities in DMD. We first aimed at evaluating the functionality of truncated dystrophin proteins in the setting of correcting dystrophin expression in a DMD ΔEx44 iPSC model. Secondly, we aimed at assessing the potential of DMD gene correction in post-differentiated cardiomyocytes in a human iPSC model as well as murine in vivo model. We showed that truncated dystrophin proteins retaining or lacking exon 45, respectively, that are produced when exon 45 of the dystrophin gene is targeted for the correction of DMD ΔEx44 correct perturbations in the structural, morphological and functional development of DMD CMs to yield a phenotype similar to control CMs. These phenotypic effects were accompanied by reduction of transcriptional dysregulation of key proteins involved in conduction, calcium signaling, mechanotransduction and contraction of CMs as well as proteins implicated in DCM in human patients. We then showed that these effects extended to DMD CMs corrected postnatally in iPSC-CMs as well as to DMD ΔEx44 mice corrected by systemic delivery of Cas9/gRNA, demonstrating that correction of dystrophin using the herein described strategy is a viable option to attenuate cardiac defects in DMD.
CRISPR/Cas9 genome editing has gained significant traction as a potential therapeutic tool for DMD over the past decade12-15, 18, 61-64. Common to CRISPR/Cas9-mediated correction strategies is the restoration of the dystrophin ORF, mostly via reframing or skipping of individual exons. Some correction strategies, however, have been shown to yield simultaneous reframing and exon skipping editing events, such that multiple differently truncated dystrophin proteins are expressed14, 17, 18. Truncated dystrophin proteins have previously been demonstrated to show significant differences with respect to their biochemical stability65, 66 as well as biological function at the cellular and tissue level12, 19. In light of these considerations, a major goal of our study was to test how truncated dystrophin proteins in the context of correcting the DMD exon 44 deletion mutation, which lies within the mutational hotspot in the dystrophin gene, differ from each other and if one outperforms the other. By targeting exon 45 in DMD ΔEx44 iPSCs, we generated iPSC lines and differentiated them to CMs that produce dystrophin protein retaining (reframing) or lacking (exon skipping) exon 45 of the dystrophin gene. In all our assessments, we found cDMD-RF and cDMD-ES CMs to be overall similar to each other and control CMs. Interestingly, in our transcriptional analyses, we found some degree of dissimilarity in the expression profiles of cDMD-RF and cDMD-ES CMs. GO analysis revealed that cDMD-RF CMs showed a stronger upregulation of differentially expressed genes compared to cDMD-ES CMs. It is possible that these differences are the result of inter-clonal variations arisen during the generation of the iPSC lines. It is also possible, however, that the mere presence or absence of a single exon, such as exon 45 in our study, may elicit a different response on the transcriptional activity of cells. Our data could thus point to a broader significance of individual exons in the DMD gene for the transcriptional regulation of cardiomyocytes. We conclude that both truncated dystrophin proteins achieve biological equivalency to full-length dystrophin protein and that both correct the cardiomyopathy phenotype in DMD in vitro to similar levels.
In assaying corrected iPSC-CMs, we found that dystrophin properly localizes to peripheral sarcolemma and to, what we interpret as, T-tubules based on a striated pattern overlaying the staining pattern of sarcomeric α-actinin (Fig. 1E). This is in line with localization patterns of dystrophin as seen in isolated adult murine cardiomyocytes and human ventricular tissues67, 68. However in CMs corrected post-differentiation, dystrophin seemed to localize to T-tubules to a lesser extent and was more concentrated at the peripheral sarcolemma (Fig. 4E). Because we plated iPSC-CMs on glass surfaces for Fig. 1E, but on soft PDMS surfaces for Fig. 4E, it is possible that the biophysical differences between both types of surfaces (elasticity, topography among others) may have contributed to differences in the localization pattern of dystrophin in corrected CMs. Indeed, higher matrix elasticity, such as glass surfaces, has been shown to increase cellular traction stress, which in turn results in remodeling of cell-matrix adhesions69 and given dystrophin’s role in mechanosensation and –transduction in CMs, it is likely that external cues determine its subcellular localization. Future investigation may provide more insight into the biology of dystrophin trafficking, localization and function in corrected CMs.
Our analyses showed that DMD CMs have structural, morphological and functional defects including larger surface areas, lower cell volumes and shorter sarcomere lengths, consistent with data from in vivo and in vitro models of DMD and dilated cardiomyopathy37, 49, 51, 52, 70. Further, our contractility analyses demonstrated systolic and diastolic dysfunction in DMD CMs, in line with clinical findings in DMD patients53 and a previous iPSC model of DMD-associated cardiomyopathy51. Dystrophin functions as an intermediary between membrane and cytoskeleton biology, and it is known that the lack of dystrophin in DMD results in cytoskeletal disruptions, such as microtubule dysregulation and increased cytoskeletal stiffness71, as well as abnormalities in plasma membrane structure and function resulting in calcium overload72, 73. A stiff cytoskeleton may result in increased cellular viscoelastic resistance leading to systolic and diastolic cardiac dysfunction74, possibly by directly impairing the contraction cycle. Similarly, elevated intracellular calcium due to sarcolemmal damage in DMD results in cardiomyocyte hypercontraction, and thus decreased sarcomere lengths as well as low contractility and slow relaxation73.
Our transcriptional data further corroborated existing literature on the cytoskeletal and membrane biological disruptions in DMD. Using RNA-sequencing of iPSC-CMs, we showed that some of the strongest downregulated genes in DMD CMs compared to cDMD CMs were genes involved in membrane biology and actin cytoskeleton organization. These differences at the transcriptional level were accompanied by increased tension at the plasma membrane of DMD CMs, while the membrane tension of cDMD CMs was similar to control CMs. Membrane tension is a mechanical regulator of cell adhesion, motility and contractility75, and it has previously been shown to be increased in cardiomyocytes with defects in membrane biology resulting in decreased sarcomere lengths and contractile defects59, possibly by impairing relaxation kinetics. The significance of cytoskeletal and membrane biology in cardiomyocyte function is further underscored by the fact that mutations in cytoskeletal proteins are known to cause DCM76, and by recent reports that showed that disruptions in the interaction between sarcomeres, the cytoskeleton and the plasma membrane result in decreased sarcomere lengths and contractile defects in DCM50, 70. Given dystrophin’s role as a scaffolding protein for signaling and membrane-associated proteins at the intersection of the intracellular milieu with the extracellular space, it is possible that that altered cardiomyocyte structure, morphology and function in DMD are in part the result of impaired expression and localization of key proteins involved in the regulation of membrane biology and cytoskeletal organization, resulting in non-physiologic cellular stiffness and membrane tension.
Our functional data confirmed previous findings that calcium signaling and contractility are significantly altered in DMD CMs and that correction of dystrophin reverses these functional deficits 9, 12, 13, 51. We found that DMD CMs beat at a slower frequency than control and cDMD CMs, in line with a recent report on an in vitro model of DMD-associated cardiomyopathy51. Our transcriptional data showed multiple instances of dysregulation of genes involved in cardiomyocyte calcium signaling, which may, in conjunction with intracellular calcium overload73, be responsible for dysregulation of the cardiac contraction cycle. Similarly, we found impaired patterning of RYR2 in DMD CMs, consistent with a recent report of a mouse model of α-dystroglycanopathy-associated heart failure77. It is thus possible that increased arrhythmogenicity and reduced systolic force generation in DMD CMs are partly mediated via disrupted localization of RYR2 away from dyads, in addition to complex post-translational modifications78. Interestingly, however, we found an apparent discrepancy between the calcium and contractile kinetics of DMD CMs, whereby an increased time to reach peak intracellular calcium levels did not correspond with increased contraction times (Fig. 2B and E). While technical differences between both experiments may account for these observations, a similar discrepancy has been reported in iPSC models of Noonan syndrome79 and hypertrophic cardiomyopathy80, pointing towards a disconnect between calcium signaling and contractility in damaged CMs. Indeed based on in vivo studies, it has been reported that the responsiveness of contractile proteins to calcium can be blunted in injured myocardium, such that relative contractile properties may be affected for any given calcium transient81, possibly due to changes in expression levels or post-translational modifications. These observations demonstrate that calcium cycling and contractility of CMs can be affected individually and independent from one another. They further underscore the necessity to assess both aspects of cardiac function to gain greater insight into the intricacies of cardiac biology. We therefore believe that our data point to independent disruptions in the calcium handling and contractile machinery of DMD CMs, and may represent an interesting aspect of cardiac biology in DMD.
Finally, we showed that correcting dystrophin expression in post-differentiated cardiomyocytes reduced cardiac abnormalities in vitro as well as in vivo, suggesting that delayed expression of dystrophin has the potential to correct the deficits that have developed until gene correction occurs. Our gene correction approach in vivo as well as in vitro demonstrated efficient mosaic dystrophin expression and in both cases we were able to reach near-normal biological outcomes, consistent with a previous report showing that correcting only ~50% of CMs results in a near-normal cardiac phenotype13. Although for our in vivo work we do not provide cardiac functional data, dystrophin correction by means of gene editing has been shown to restore cardiac function in DMD mice in several reports, despite achieving far inferior correction levels82-84. Similarly microdystrophins, which are truncated dystrophin proteins with approximately one-third the size of full length dystrophin protein and thus significantly shorter than the truncated dystrophin proteins in our study, have been shown to restore cardiac function in DMD mice after gene therapy85, 86. We can therefore speculate based on these reports and our data that our dystrophin correction strategy in DMD mice may similarly achieve improved cardiac function.
We were further able to show that restoring dystrophin expression in DMD mice mitigated cardiac transcriptional dysregulation in DMD. By performing single-nucleus transcriptomics, we were able to separately assess the transcriptional profiles of cardiomyocytes and fibroblasts. While we observed transcriptional dysregulation in functional and metabolic genes in the CM population as well as in fibrosis-related genes in the FB population in DMD mice, we found that these changes were reversed in corrected mice to near-normal levels as seen in wildtype animals. Nonetheless, some differences in the transcriptional activity of wildtype and cDMD mice remained, which could be the result of incomplete dystrophin restoration or the consequence of postnatal gene correction. Although cardiogenesis is not obviously altered in the absence of dystrophin, it has been reported that lack of dystrophin may impair cardiomyocyte maturation51, leaving open whether postnatal dystrophin correction rectifies developmental delays in DMD CMs. Overall, our data demonstrate that our gene correction strategy has the potential to normalize cardiac transcriptional regulation in DMD ΔEx44.
Our study has several limitations. First, although our iPSC-CM culture included biochemical additions to promote the maturity of iPSC-CMs, these have been shown to yield CMs similar to only neonatal cardiomyocytes22, 23. It is therefore possible that not all aspects of the pathophysiology of DMD-associated cardiomyopathy are captured in our model. Further, because DMD-associated cardiomyopathy can be observed as early as in prepubescence8, it will be important to assess whether a similar gene correction efficacy can be detected in more mature CMs. Secondly, because we used a patient-derived DMD iPSC line in this study, we were only able to compare the isogenic DMD CMs and cDMD CMs by RNA-seq in the absence of isogenic control CMs. This approach is similar to previous studies, whereby isogenic gene corrected iPSC lines were generated to study disease biology using patient-derived iPSC-CMs87-90. Further, the use of patient-derived iPSCs presumes that the genetic background is permissive for the disease without major limitations from disease-modifying effectors91. Finally, we employed snRNA-seq because conventional microfluidics-based single-cell RNA-sequencing technologies are not suitable for adult cardiomyocytes. Although for certain tissues a good correlation between nuclear and cytosolic transcripts has been described92, 93, a discrepancy may nonetheless remain. Indeed, our lab has previously shown compensatory upregulation of the dystrophin transcript in skeletal muscle of DMD mice by snRNA-seq, although it is known to exist at low levels in the cytosol39. It is therefore possible that some of the dysregulated transcripts in our snRNA-seq data represent a compensatory response rather than a primary abnormality. Furthermore, our snRNA-seq approach involved mechanical disruption of heart tissues to separate nuclei from cytoplasms. Because fibrotic tissue is stiffer than normal heart tissue, it is possible that during our nuclei isolation approach we introduced sampling bias by not being able to fully isolate nuclei from fibrotic areas of the hearts, such that they may be underrepresented in our data.
In conclusion, in this study we showed that CRISPR/Cas9-mediated correction of DMD ΔEx44 mitigates structural, functional and transcriptional dysregulation consistent with dilated cardiomyopathy irrespective of whether the protein reading frame is restored by reframing or skipping of exon 45. We show that postnatal editing in iPSC-CMs and mice reduces cellular dysfunction and transcriptional dysregulation, respectively. These findings provide key insights into the utility of genome editing as a novel therapeutic for DMD-associated cardiomyopathy.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Duchenne Muscular Dystrophy (DMD) is a genetic neuromuscular disease caused by mutations in the dystrophin gene, resulting in the production of non-functional dystrophin protein in skeletal and cardiac muscle cells.
Cardiomyopathy has become a significant aspect of the disease owing to advances in respiratory medicine and results in congestive heart failure, arrhythmia and sudden cardiac death.
CRISPR/Cas9 genome editing holds great promise as a novel therapeutic avenue in DMD.
What New Information Does This Article Contribute?
CRISPR/Cas9-mediated correction of the DMD gene in induced pluripotent stem cells (iPSCs) generated from a DMD patient with the exon 44 deletion mutation restores morphologic, structural and functional deficits of iPSC-derived cardiomyocytes (iPSC-CMs).
RNA-sequencing of iPSC-CMs from the DMD patient showed transcriptional dysregulation consistent with dilated cardiomyopathy, which was mitigated in iPSC-CMs corrected by CRISPR/Cas9 gene editing.
Single-nucleus RNA-sequencing of hearts of DMD mice showed transcriptional dysregulation in cardiomyocytes and fibroblasts, which in corrected mice was reduced to similar levels as wildtype mice.
DMD is a deadly disease affecting ~1:5,000 boys. Although historically DMD has primarily been regarded as a neuromuscular disease with respiratory failure as the leading cause of death, today cardiomyopathy is recognized to be a significant contributor to morbidity and mortality. Current treatment options only focus on medical therapy with the aim to slow the progression of cardiomyopathy. Our lab and others have previously demonstrated the efficacy of CRISPR/Cas9 genome editing in a number of different DMD models in mice and human cells. However, the goal of these studies was focused on the assessment of few functional aspects of cardiac biology without providing an in-depth perspective on the correction of cardiac abnormalities. In this study, we show that CRISPR/Cas9-mediated correction of DMD in human iPSCs mitigates structural, functional and transcriptional abnormalities consistent with dilated cardiomyopathy. We further show that these effects extend to myocardial DMD gene correction in mice. These findings provide key insights into the utility of genome editing as a novel therapeutic for DMD-associated cardiomyopathy.
ACKNOWLEDGMENTS
We thank Dr. Yi-Li Min for reprogramming of iPSCs and contributing to in vivo experiments; Dr. Francesco Chemello for contributing to in vivo experiments; Dr. Efrain Sanchez-Ortiz for technical assistance in in vivo experiments; Cristina Rodriguez-Caycedo for technical assistance in in vitro experiments; Jose Cabrera for helping with graphics; Alex A. Mireault for helping with tissue processing for histologic analyses; Drs. Jian Xu and Yoon Jung Kim from the Children’s Research Institute at the University of Texas Southwestern Medical Center for performing the Illumina sequencing; John Shelton from the Molecular Histopathology Core for help with histology; Dr. Marcel Metten from the Live Cell Imaging Facility for help with FLIM imaging.
SOURCES OF FUNDING
This work was supported by grants from the NIH (AR-067294, HL-130253, HL-138426, and HD-087351), the Foundation Leducq Transatlantic Networks of Excellence in Cardiovascular Research, and the Robert A. Welch Foundation (grant 1-0025 to E.N.O.).
Footnotes
DISCLOSURES
E.N.O. and R.B.-D. are consultants for Vertex and have filed patents related to CRISPR/Cas9-based correction of DMD.
A.A., R.B.-D., and E.N.O designed the experiments and overall study, and wrote the manuscript. A.A., A.C., M.C., Z.W. and T.N. performed the experiments. M.C. and Z.W. analyzed transcriptomics data. All authors discussed the results and participated in the manuscript preparation and editing.
Publisher's Disclaimer: This article is published in its accepted form. It has not been copyedited and has not appeared in an issue of the journal. Preparation for inclusion in an issue of Circulation Research involves copyediting, typesetting, proofreading, and author review, which may lead to differences between this accepted version of the manuscript and the final, published version.
REFERENCES
- 1.Deconinck N, Dan B. Pathophysiology of duchenne muscular dystrophy: Current hypotheses. Pediatric neurology. 2007;36:1–7 [DOI] [PubMed] [Google Scholar]
- 2.Mendell JR, Lloyd-Puryear M. Report of mda muscle disease symposium on newborn screening for duchenne muscular dystrophy. Muscle & nerve. 2013;48:21–26 [DOI] [PubMed] [Google Scholar]
- 3.Min Y-L, Bassel-Duby R, Olson EN. Crispr correction of duchenne muscular dystrophy. Annu Rev Med. 2019;70:239–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chemello F, Bassel-Duby R, Olson EN. Correction of muscular dystrophies by crispr gene editing. The Journal of Clinical Investigation. 2020;130:2766–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuda T, Fitzgerald KK. Dystrophic cardiomyopathy: Complex pathobiological processes to generate clinical phenotype. Journal of cardiovascular development and disease. 2017;4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyers TA, Townsend D. Cardiac pathophysiology and the future of cardiac therapies in duchenne muscular dystrophy. International Journal of Molecular Sciences. 2019;20:4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yucel N, Chang AC, Day JW, Rosenthal N, Blau HM. Humanizing the mdx mouse model of dmd: The long and the short of it. npj Regenerative Medicine. 2018;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamdar F, Garry DJ. Dystrophin-deficient cardiomyopathy. Journal of the American College of Cardiology. 2016;67:2533–2546 [DOI] [PubMed] [Google Scholar]
- 9.Kamdar F, Das S, Gong W, Klaassen Kamdar A, Meyers TA, Shah P, Ervasti JM, Townsend D, Kamp TJ, Wu JC, Garry MG, Zhang J, Garry DJ. Stem cell-derived cardiomyocytes and beta-adrenergic receptor blockade in duchenne muscular dystrophy cardiomyopathy. Journal of the American College of Cardiology. 2020;75:1159–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin B, Li Y, Han L, Kaplan AD, Ao Y, Kalra S, Bett GC, Rasmusson RL, Denning C, Yang L. Modeling and study of the mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from individuals with duchenne muscular dystrophy. Disease models & mechanisms. 2015;8:457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zatti S, Martewicz S, Serena E, Uno N, Giobbe G, Kazuki Y, Oshimura M, Elvassore N. Complete restoration of multiple dystrophin isoforms in genetically corrected duchenne muscular dystrophy patient-derived cardiomyocytes. Molecular therapy. Methods & clinical development. 2014;1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kyrychenko V, Kyrychenko S, Tiburcy M, Shelton JM, Long C, Schneider JW, Zimmermann W-H, Bassel-Duby R, Olson EN. Functional correction of dystrophin actin binding domain mutations by genome editing. JCI Insight. 2017;2:e95918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long C, Li H, Tiburcy M, Rodriguez-Caycedo C, Kyrychenko V, Zhou H, Zhang Y, Min YL, Shelton JM, Mammen PPA, Liaw NY, Zimmermann WH, Bassel-Duby R, Schneider JW, Olson EN. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Science advances. 2018;4:eaap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min YL, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM, McAnally JR, Amoasii L, Mammen PPA, Bassel-Duby R, Olson EN. Crispr-cas9 corrects duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Science advances. 2019;5:eaav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, Karumbayaram S, Kumagai-Cresse C, Wang D, Zack JA, Kohn DB, Nakano A, Nelson SF, Miceli MC, Spencer MJ, Pyle AD. A single crispr-cas9 deletion strategy that targets the majority of dmd patients restores dystrophin function in hipsc-derived muscle cells. Cell stem cell. 2016;18:533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Long C, Li H, McAnally JR, Baskin KK, Shelton JM, Bassel-Duby R, Olson EN. Crispr-cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Science advances. 2017;3:e1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min YL, Chemello F, Li H, Rodriguez-Caycedo C, Sanchez-Ortiz E, Mireault AA, McAnally JR, Shelton JM, Zhang Y, Bassel-Duby R, Olson EN. Correction of three prominent mutations in mouse and human models of duchenne muscular dystrophy by single-cut genome editing. Molecular therapy : the journal of the American Society of Gene Therapy. 2020;28:2044–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, Harron R, Stathopoulou TR, Massey C, Shelton JM, Bassel-Duby R, Piercy RJ, Olson EN. Gene editing restores dystrophin expression in a canine model of duchenne muscular dystrophy. Science (New York, N.Y.). 2018;362:86–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos JN, Hollinger K, Bengtsson NE, Allen JM, Hauschka SD, Chamberlain JS. Development of novel micro-dystrophins with enhanced functionality. Molecular therapy : the journal of the American Society of Gene Therapy. 2019;27:623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atmanli A, Hu D, Deiman FE, van de Vrugt AM, Cherbonneau F, Black LD 3rd, Domian IJ. Multiplex live single-cell transcriptional analysis demarcates cellular functional heterogeneity. Elife. 2019;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birket MJ, Ribeiro MC, Kosmidis G, Ward D, Leitoguinho AR, van de Pol V, Dambrot C, Devalla HD, Davis RP, Mastroberardino PG, Atsma DE, Passier R, Mummery CL. Contractile defect caused by mutation in mybpc3 revealed under conditions optimized for human psc-cardiomyocyte function. Cell Rep. 2015;13:733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parikh SS, Blackwell DJ, Gomez-Hurtado N, Frisk M, Wang L, Kim K, Dahl CP, Fiane A, Tønnessen T, Kryshtal DO, Louch WE, Knollmann BC. Thyroid and glucocorticoid hormones promote functional t-tubule development in human-induced pluripotent stem cell-derived cardiomyocytes. Circ Res. 2017;121:1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu D, Linders A, Yamak A, Correia C, Kijlstra JD, Garakani A, Xiao L, Milan DJ, van der Meer P, Serra M, Alves PM, Domian IJ. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of hif1α and ldha. Circ Res. 2018;123:1066–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, Haghighi A, Homsy J, Hubner N, Church G, Cook SA, Linke WA, Chen CS, Seidman JG, Seidman CE. Titin mutations in ips cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science (New York, N.Y.). 2015;349:982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sander H, Wallace S, Plouse R, Tiwari S, Gomes AV. Ponceau s waste: Ponceau s staining for total protein normalization. Analytical biochemistry. 2019;575:44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atmanli A, Hu D, Domian IJ. Molecular etching: A novel methodology for the generation of complex micropatterned growth surfaces for human cellular assays. Adv Healthc Mater. 2014;3:1759–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kijlstra JD, Hu D, Mittal N, Kausel E, van der Meer P, Garakani A, Domian IJ. Integrated analysis of contractile kinetics, force generation, and electrical activity in single human stem cell-derived cardiomyocytes. Stem Cell Reports. 2015;5:1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colom A, Derivery E, Soleimanpour S, Tomba C, Molin MD, Sakai N, González-Gaitán M, Matile S, Roux A. A fluorescent membrane tension probe. Nat Chem. 2018;10:1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redford GI, Clegg RM. Polar plot representation for frequency-domain analysis of fluorescence lifetimes. Journal of Fluorescence. 2005;15:805. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Cui M, Shah AM, Ye W, Tan W, Min Y-L, Botten GA, Shelton JM, Liu N, Bassel-Duby R, Olson EN. Mechanistic basis of neonatal heart regeneration revealed by transcriptome and histone modification profiling. Proc Natl Acad Sci U S A. 2019;116:18455–18465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D, Langmead B, Salzberg SL. Hisat: A fast spliced aligner with low memory requirements. Nature methods. 2015;12:357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y, Smyth GK, Shi W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England). 2014;30:923–930 [DOI] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England). 2010;26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature communications. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome biology. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettinato AM, Ladha FA, Mellert DJ, Legere N, Cohn R, Romano R, Thakar K, Chen Y-S, Hinson JT. Development of a cardiac sarcomere functional genomics platform to enable scalable interrogation of human tnnt2 variants. Circulation. 2020;142:2262–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui M, Olson EN. Protocol for single-nucleus transcriptomics of diploid and tetraploid cardiomyocytes in murine hearts. STAR Protocols. 2020;1:100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chemello F, Wang Z, Li H, McAnally JR, Liu N, Bassel-Duby R, Olson EN. Degenerative and regenerative pathways underlying duchenne muscular dystrophy revealed by single-nucleus rna sequencing. Proc Natl Acad Sci U S A. 2020;117:29691–29701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lake BB, Chen S, Hoshi M, Plongthongkum N, Salamon D, Knoten A, Vijayan A, Venkatesh R, Kim EH, Gao D, Gaut J, Zhang K, Jain S. A single-nucleus rna-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nature communications. 2019;10:2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Cui M, Shah AM, Tan W, Liu N, Bassel-Duby R, Olson EN. Cell-type-specific gene regulatory networks underlying murine neonatal heart regeneration at single-cell resolution. Cell Rep. 2020;33:108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu P, Liu J, Zhao J, Wilkins BJ, Lupino K, Wu H, Pei L. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes Dev. 2018;32:1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 2018;36:411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. Dimensionality reduction for visualizing single-cell data using umap. Nature Biotechnology. 2019;37:38–44 [DOI] [PubMed] [Google Scholar]
- 45.Cui M, Wang Z, Chen K, Shah AM, Tan W, Duan L, Sanchez-Ortiz E, Li H, Xu L, Liu N, Bassel-Duby R, Olson EN. Dynamic transcriptional responses to injury of regenerative and non-regenerative cardiomyocytes revealed by single-nucleus rna sequencing. Developmental cell. 2020;53:102–116.e108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burke MA, Chang S, Wakimoto H, Gorham JM, Conner DA, Christodoulou DC, Parfenov MG, DePalma SR, Eminaga S, Konno T, Seidman JG, Seidman CE. Molecular profiling of dilated cardiomyopathy that progresses to heart failure. JCI Insight. 2016;1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu P, Liu J, Zhao J, Wilkins BJ, Lupino K, Wu H, Pei L. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes Dev. 2018;32:1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Morley M, Brandimarto J, Hannenhalli S, Hu Y, Ashley EA, Tang WHW, Moravec CS, Margulies KB, Cappola TP, Li M. Rna-seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105:83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clippinger SR, Cloonan PE, Greenberg L, Ernst M, Stump WT, Greenberg MJ. Disrupted mechanobiology links the molecular and cellular phenotypes in familial dilated cardiomyopathy. Proceedings of the National Academy of Sciences. 2019;116:17831–17840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai Y, Amenov A, Ignatyeva N, Koschinski A, Xu H, Soong PL, Tiburcy M, Linke WA, Zaccolo M, Hasenfuss G, Zimmermann W-H, Ebert A. Troponin destabilization impairs sarcomere-cytoskeleton interactions in ipsc-derived cardiomyocytes from dilated cardiomyopathy patients. Sci Rep. 2020;10:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pioner JM, Guan X, Klaiman JM, Racca AW, Pabon L, Muskheli V, Macadangdang J, Ferrantini C, Hoopmann MR, Moritz RL, Kim DH, Tesi C, Poggesi C, Murry CE, Childers MK, Mack DL, Regnier M. Absence of full-length dystrophin impairs normal maturation and contraction of cardiomyocytes derived from human-induced pluripotent stem cells. Cardiovascular research. 2020;116:368–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong TWY, Ahmed A, Yang G, Maino E, Steiman S, Hyatt E, Chan P, Lindsay K, Wong N, Golebiowski D, Schneider J, Delgado-Olguín P, Ivakine EA, Cohn RD. A novel mouse model of duchenne muscular dystrophy carrying a multi-exonic dmd deletion exhibits progressive muscular dystrophy and early-onset cardiomyopathy. Disease models & mechanisms. 2020;13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mertens L, Ganame J, Claus P, Goemans N, Thijs D, Eyskens B, Van Laere D, Bijnens B, D'Hooge J, Sutherland GR, Buyse G. Early regional myocardial dysfunction in young patients with duchenne muscular dystrophy. Journal of the American Society of Echocardiography. 2008;21:1049–1054 [DOI] [PubMed] [Google Scholar]
- 54.Valera IC, Wacker AL, Hwang HS, Holmes C, Laitano O, Landstrom AP, Parvatiyar MS. Essential roles of the dystrophin-glycoprotein complex in different cardiac pathologies. Advances in Medical Sciences. 2021;66:52–71 [DOI] [PubMed] [Google Scholar]
- 55.Alimadadi A, Munroe PB, Joe B, Cheng X. Meta-analysis of dilated cardiomyopathy using cardiac rna-seq transcriptomic datasets. Genes. 2020;11:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinig M, Adriaens ME, Schafer S, van Deutekom HWM, Lodder EM, Ware JS, Schneider V, Felkin LE, Creemers EE, Meder B, Katus HA, Rühle F, Stoll M, Cambien F, Villard E, Charron P, Varro A, Bishopric NH, George AL, dos Remedios C, Moreno-Moral A, Pesce F, Bauerfeind A, Rüschendorf F, Rintisch C, Petretto E, Barton PJ, Cook SA, Pinto YM, Bezzina CR, Hubner N. Natural genetic variation of the cardiac transcriptome in non-diseased donors and patients with dilated cardiomyopathy. Genome biology. 2017;18:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon C, Caorsi V, Campillo C, Sykes C. Interplay between membrane tension and the actin cytoskeleton determines shape changes. Physical Biology. 2018;15:065004. [DOI] [PubMed] [Google Scholar]
- 58.Le Roux A-L, Quiroga X, Walani N, Arroyo M, Roca-Cusachs P. The plasma membrane as a mechanochemical transducer. Philosophical Transactions of the Royal Society B: Biological Sciences. 2019;374:20180221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hissa B, Oakes PW, Pontes B, Ramírez-San Juan G, Gardel ML. Cholesterol depletion impairs contractile machinery in neonatal rat cardiomyocytes. Sci Rep. 2017;7:43764–43764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sinha B, Köster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler-Browne G, Vedie B, Johannes L, Morone N, Parton RG, Raposo G, Sens P, Lamaze C, Nassoy P. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 2011;144:402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Li H, Min Y-L, Sanchez-Ortiz E, Huang J, Mireault AA, Shelton JM, Kim J, Mammen PPA, Bassel-Duby R, Olson EN. Enhanced crispr-cas9 correction of duchenne muscular dystrophy in mice by a self-complementary aav delivery system. Science advances. 2020;6:eaay6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS. Muscle-specific crispr/cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for duchenne muscular dystrophy. Nature communications. 2017;8:14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA. In vivo genome editing improves muscle function in a mouse model of duchenne muscular dystrophy. Science (New York, N.Y.). 2016;351:403–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science (New York, N.Y.). 2016;351:400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Findlay AR, Wein N, Kaminoh Y, Taylor LE, Dunn DM, Mendell JR, King WM, Pestronk A, Florence JM, Mathews KD, Finkel RS, Swoboda KJ, Howard MT, Day JW, McDonald C, Nicolas A, Le Rumeur E, Weiss RB, Flanigan KM. Clinical phenotypes as predictors of the outcome of skipping around dmd exon 45. Annals of neurology. 2015;77:668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruszczak C, Mirza A, Menhart N. Differential stabilities of alternative exon-skipped rod motifs of dystrophin. Biochimica et biophysica acta. 2009;1794:921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaprielian RR, Stevenson S, Rothery SM, Cullen MJ, Severs NJ. Distinct patterns of dystrophin organization in myocyte sarcolemma and transverse tubules of normal and diseased human myocardium. Circulation. 2000;101:2586–2594 [DOI] [PubMed] [Google Scholar]
- 68.Masubuchi N, Shidoh Y, Kondo S, Takatoh J, Hanaoka K. Subcellular localization of dystrophin isoforms in cardiomyocytes and phenotypic analysis of dystrophin-deficient mice reveal cardiac myopathy is predominantly caused by a deficiency in full-length dystrophin. Exp Anim. 2013;62:211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Janmey PA, Fletcher DA, Reinhart-King CA. Stiffness sensing by cells. Physiological Reviews. 2020;100:695–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manalo A, Schroer AK, Fenix AM, Shancer Z, Coogan J, Brolsma T, Burnette DT, Merryman WD, Bader DM. Loss of cenp-f results in dilated cardiomyopathy with severe disruption of cardiac myocyte architecture. Sci Rep. 2018;8:7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kerr JP, Robison P, Shi G, Bogush AI, Kempema AM, Hexum JK, Becerra N, Harki DA, Martin SS, Raiteri R, Prosser BL, Ward CW. Detyrosinated microtubules modulate mechanotransduction in heart and skeletal muscle. Nature communications. 2015;6:8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Borgne F, Guyot S, Logerot M, Beney L, Gervais P, Demarquoy J. Exploration of lipid metabolism in relation with plasma membrane properties of duchenne muscular dystrophy cells: Influence of l-carnitine. PLoS One. 2012;7:e49346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Law ML, Cohen H, Martin AA, Angulski ABB, Metzger JM. Dysregulation of calcium handling in duchenne muscular dystrophy-associated dilated cardiomyopathy: Mechanisms and experimental therapeutic strategies. J Clin Med. 2020;9:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coleman AK, Joca HC, Shi G, Lederer WJ, Ward CW. Tubulin acetylation increases cytoskeletal stiffness to regulate mechanotransduction in striated muscle. Journal of General Physiology. 2021;153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gauthier NC, Fardin MA, Roca-Cusachs P, Sheetz MP. Temporary increase in plasma membrane tension coordinates the activation of exocytosis and contraction during cell spreading. Proceedings of the National Academy of Sciences. 2011;108:14467–14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hershberger RE, Cowan J, Jordan E, Kinnamon DD. The complex and diverse genetic architecture of dilated cardiomyopathy. Circ Res. 2021;128:1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ujihara Y, Kanagawa M, Mohri S, Takatsu S, Kobayashi K, Toda T, Naruse K, Katanosaka Y. Elimination of fukutin reveals cellular and molecular pathomechanisms in muscular dystrophy-associated heart failure. Nature communications. 2019;10:5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Q, Wang W, Wang G, Rodney GG, Wehrens XH. Crosstalk between ryr2 oxidation and phosphorylation contributes to cardiac dysfunction in mice with duchenne muscular dystrophy. Journal of molecular and cellular cardiology. 2015;89:177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hanses U, Kleinsorge M, Roos L, Yigit G, Li Y, Barbarics B, El-Battrawy I, Lan H, Tiburcy M, Hindmarsh R, Lenz C, Salinas G, Diecke S, Müller C, Adham I, Altmüller J, Nürnberg P, Paul T, Zimmermann W-H, Hasenfuss G, Wollnik B, Cyganek L. Intronic crispr repair in a preclinical model of noonan syndrome–associated cardiomyopathy. Circulation. 2020;142:1059–1076 [DOI] [PubMed] [Google Scholar]
- 80.Cohn R, Thakar K, Lowe A, Ladha FA, Pettinato AM, Romano R, Meredith E, Chen Y-S, Atamanuk K, Huey BD, Hinson JT. A contraction stress model of hypertrophic cardiomyopathy due to sarcomere mutations. Stem Cell Reports. 2019;12:71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao WD, Atar D, Backx PH, Marban E. Relationship between intracellular calcium and contractile force in stunned myocardium. Circ Res. 1995;76:1036–1048 [DOI] [PubMed] [Google Scholar]
- 82.Hakim CH, Wasala NB, Nelson CE, Wasala LP, Yue Y, Louderman JA, Lessa TB, Dai A, Zhang K, Jenkins GJ, Nance ME, Pan X, Kodippili K, Yang NN, Chen S-j, Gersbach CA, Duan D. Aav crispr editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight. 2018;3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Refaey ME, Xu L, Gao Y, Canan BD, Adesanya TMA, Warner SC, Akagi K, Symer DE, Mohler PJ, Ma J, Janssen PML, Han R. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res. 2017;121:923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu L, Lau YS, Gao Y, Li H, Han R. Life-long aav-mediated crispr genome editing in dystrophic heart improves cardiomyopathy without causing serious lesions in mdx mice. Molecular Therapy. 2019;27:1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Howard ZM, Dorn LE, Lowe J, Gertzen MD, Ciccone P, Rastogi N, Odom GL, Accornero F, Chamberlain JS, Rafael-Fortney JA. Micro-dystrophin gene therapy prevents heart failure in an improved duchenne muscular dystrophy cardiomyopathy mouse model. JCI Insight. 2021;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kolwicz SC Jr., Hall JK, Moussavi-Harami F, Chen X, Hauschka SD, Chamberlain JS, Regnier M, Odom GL. Gene therapy rescues cardiac dysfunction in duchenne muscular dystrophy mice by elevating cardiomyocyte deoxy-adenosine triphosphate. JACC. Basic to translational science 2019;4:778–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Estes SI, Ye D, Zhou W, Dotzler SM, Tester DJ, Bos JM, Kim CSJ, Ackerman MJ. Characterization of the cacna1c-r518c missense mutation in the pathobiology of long-qt syndrome using human induced pluripotent stem cell cardiomyocytes shows action potential prolongation and l-type calcium channel perturbation. Circulation: Genomic and Precision Medicine. 2019;12:e002534. [DOI] [PubMed] [Google Scholar]
- 88.Higgins EM, Bos JM, Dotzler SM, Kim CJ, Ackerman MJ. mras variants cause cardiomyocyte hypertrophy in patient-specific induced pluripotent stem cell-derived cardiomyocytes. Circulation: Genomic and Precision Medicine. 2019;12:e002648. [DOI] [PubMed] [Google Scholar]
- 89.Jaffré F, Miller CL, Schänzer A, Evans T, Roberts AE, Hahn A, Kontaridis MI. Inducible pluripotent stem cell–derived cardiomyocytes reveal aberrant extracellular regulated kinase 5 and mitogen-activated protein kinase kinase 1/2 signaling concomitantly promote hypertrophic cardiomyopathy in raf1-associated noonan syndrome. Circulation. 2019;140:207–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feyen DAM, Perea-Gil I, Maas RGC, Harakalova M, Gavidia AA, Arthur Ataam J, Wu TH, Vink A, Pei J, Vadgama N, Suurmeijer AJ, Te Rijdt WP, Vu M, Amatya PL, Prado M, Zhang Y, Dunkenberger L, Sluijter JPG, Sallam K, Asselbergs FW, Mercola M, Karakikes I. The unfolded protein response as a compensatory mechanism and potential therapeutic target in pln r14del cardiomyopathy. Circulation. 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Musunuru K, Sheikh F, Gupta RM, Houser SR, Maher KO, Milan DJ, Terzic A, Wu JC. Induced pluripotent stem cells for cardiovascular disease modeling and precision medicine: A scientific statement from the american heart association. Circulation: Genomic and Precision Medicine. 2018;11:e000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bakken TE, Hodge RD, Miller JA, Yao Z, Nguyen TN, Aevermann B, Barkan E, Bertagnolli D, Casper T, Dee N, Garren E, Goldy J, Graybuck LT, Kroll M, Lasken RS, Lathia K, Parry S, Rimorin C, Scheuermann RH, Schork NJ, Shehata SI, Tieu M, Phillips JW, Bernard A, Smith KA, Zeng H, Lein ES, Tasic B. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One. 2018;13:e0209648–e0209648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khrameeva E, Kurochkin I, Han D, Guijarro P, Kanton S, Santel M, Qian Z, Rong S, Mazin P, Sabirov M, Bulat M, Efimova O, Tkachev A, Guo S, Sherwood CC, Camp JG, Pääbo S, Treutlein B, Khaitovich P. Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. Genome research. 2020;30:776–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.