Abstract
Human immunodeficiency virus type 1 (HIV-1) gene expression is regulated by interactions between both viral and host factors. These interactions are also responsible for changes in the expression of many host cell genes, including cytokines and other immune regulators, which may account for the state of immunological dysregulation that characterizes HIV-1 infection. We have investigated the role of a host cell protein, the transcription factor NFAT1, in HIV-1 pathogenesis. We show that NFAT1 interacts with Tat and that this interaction, which involves the major transactivation domain of NFAT1 and the amino-terminal region of Tat, results in a reciprocal modulatory interplay between the proteins: whereas Tat enhances NFAT1-driven transcription in Jurkat T cells, NFAT1 represses Tat-mediated transactivation of the HIV-1 long terminal repeat (LTR). Moreover, NFAT1 binds to the κB sites on the viral LTR and negatively regulates NF-κB-mediated activation of HIV-1 transcription, by competing with NF-κB1 for its binding sites on the HIV-1 LTR. Tat-mediated enhancement of NFAT1 transactivation may explain the upregulation of interleukin 2 and other cytokines that occurs during HIV-1 infection. We discuss the potentially opposing roles of NFAT1 and another family member, NFAT2, in regulating gene transcription of HIV-1 and endogenous cytokine genes.
Human immunodeficiency virus type 1 (HIV-1) infection produces a state of immunological dysregulation which includes a state of hyperactivation of B and T cells with a general increase in cytokine production (52). These alterations are possibly necessary for the maintenance of virus infection and may be at least partially explained by a direct influence of virus-encoded products on the mechanisms that control immune cell activation.
The transactivator protein Tat of HIV-1 is required for efficient transcription and viral replication (18, 30). Tat binds to the transactivation response (TAR) element, an RNA stem-loop located from positions +1 to +59 of the HIV-1 long terminal repeat (LTR) and exerts its function mainly by increasing the efficiency of transcription elongation (16, 35, 42). Although the exact mechanism by which Tat enhances the rate of transcription elongation is still unknown, it is now clearly established that Tat interacts with different host proteins which are necessary for Tat function. Among those factors, proteins belonging to the general transcription machinery of the host cell have been reported to interact with Tat. These proteins include the core RNA polymerase II (13, 44), whose C-terminal domain is required for Tat-mediated transactivation (50), TAFII55 (12), and TFIIH and CDK7 (5, 14). Tat has also been reported to interact with other nuclear kinase complexes (21, 68), with the transcription factor Sp1, which binds to three tandem sites in the core enhancer element of the HIV-1 LTR (29), and with cyclin T, which has recently been identified as a TAR RNA-binding cofactor for Tat (66). In addition to its role in HIV-1 transcription, Tat has also been shown to upregulate the transcription of several host genes such as those encoding tumor necrosis factor alpha (7), interleukin 2 (IL-2) (51, 63), and IL-6 (2) by interacting with different cellular factors and contributing to some of the altered cytokine production which occurs after HIV-1 infection. Some reports have mapped those Tat-mediated effects to sites that can potentially bind the nuclear factor of activated T cells (NFAT) in genes whose expression is regulated by NFAT proteins (51, 63).
NFAT1 (also called NFATp) (46) is the founding member of the NFAT family of transcription factors, which plays a key role in inducible gene transcription during the immune response (56, 57). Other members of the NFAT family have been described and termed NFAT2 (also called NFATc) (49), NFAT3 (23), and NFAT4 (also called NFATx) (23, 43), with each one having a specific tissue distribution and function. NFAT proteins are activated by stimulation of receptors which induce calcium mobilization and also by calcium ionophores such as ionomycin. Calcium mobilization activates calcineurin, which causes dephosphorylation and subsequent nuclear translocation of NFAT proteins (38), which cooperate in the nucleus with members of the Fos and Jun families of transcription factors (28). This process can be inhibited by the immunosuppressants cyclosporine A (CsA) and FK506, which inhibit calcineurin activity (61). NFAT proteins are involved in the regulation of numerous activation-associated genes that encode cytokines, transcription factors, cell surface receptors, and other signaling proteins (56, 57). The amino-terminal domain of NFAT1 contains its major transactivation domain (40), which is followed by the calcineurin-binding regulatory domain and the DNA-binding domain (DBD), which has similarity at both the sequence and structural levels with Rel proteins (11, 27). Both domains are highly conserved among the different members of the NFAT family (27, 39). NFAT1 is expressed in several immune system cells, including T cells and monocytes (57), as well as in other nonimmune tissues such as the central nervous system (22), all of which are potential targets for HIV-1 infection.
The enhancer element of the HIV-1 LTR contains two tandem NF-κB sites whose function seems to be essential for HIV-1 transcription in both T cells and monocytes (1, 26). NFAT proteins have been shown to bind these sites, and an activating role for one of the NFAT family members, NFATc/NFAT2, in HIV-1 LTR transcription mediated through the κB sites and in HIV-1 replication has recently been described (32, 33). However, other studies have found that in HIV-1 viruses with mutations in the gag gene which render viral replication independent of cyclophilin A, treatment of infected cells with CsA had no inhibitory effect on HIV-1 replication and at some concentrations even produced stimulation of virus replication (6).
In this paper, we have examined the role of the NFAT family member NFAT1 in HIV-1 pathogenesis. We demonstrate that NFAT1 interacts with Tat and that these two proteins modulate each other’s activities. Whereas Tat enhances NFAT1-driven transcription in Jurkat T cells, NFAT1 inhibits Tat-mediated transactivation of the HIV-1 LTR. Moreover, NFAT1 binds the κB sites of the HIV-1 LTR and exerts a negative effect on LTR transcription mediated by NF-κB.
MATERIALS AND METHODS
Plasmids.
The expression plasmids pEFTagNFAT1-C, which bears the gene encoding a hemagglutinin (HA)-tagged murine NFAT1, and pGAL4-NFAT1(1-415), which bears the gene encoding a fusion protein containing the DBD of GAL4 and amino acids 1 to 415 of NFAT1 and pGAL4ΔSP2, have been previously described (39, 40). The expression plasmids pEFTagNFAT1(1–415) and pEFTagNFAT1DBD bear DNAs that encode amino acids 1 to 415 and the DBD (amino acids 398 to 694) of NFAT1, respectively (27, 40). pcTat expresses HIV-1 Tat protein under the control of the cytomegalovirus promoter, and pEFTagTatC22G expresses a mutant Tat with a substitution of Gly for Cys22. pcDNA3-mRelA, which expresses the murine RelA protein under the control of the cytomegalovirus promoter, was a gift from Sankar Ghosh (Yale University). pEGFPTat(1–27) expresses a fusion protein between the green fluorescent protein (GFP) and the first 27 amino acids of HIV-1 Tat in the pEGFP (Clontech) backbone. As reporter plasmids, we used the previously described plasmid pHIV-CAT (48), kindly provided by Gary Nabel (University of Michigan), which contains the HIV-1 LTR controlling the expression of the chloramphenicol acetyltransferase (CAT) gene, and HIV-1 LTR-Luc, which was constructed by subcloning an XhoI-HindIII fragment from pHIV-CAT containing the HIV-1 LTR into the pGL2 luciferase reporter plasmid (Promega). HIV-1 LTR2×3′ κB*-Luc, containing 3′ mutations in both κB sites, was made by PCR-mediated mutagenesis of HIV-1 LTR-Luc with Pfu polymerase (Stratagene). In this plasmid the sequences of both κB sites of the HIV-1 LTR were changed from GGGGACTTTCC to GGGGACTAGTT. The luciferase reporter vector NFAT3×-Luc (20) with three binding sites for NFAT was a gift from David J. McKean (Mayo Clinic). The GAL4 luciferase reporter plasmid GAL4-Luc (10) was kindly provided by Marc Montminy (Joslin Diabetes Center, Boston, Mass.).
Electrophoretic mobility shift assays (EMSAs).
Binding reactions were performed with a solution containing 10 mM HEPES (pH 7.5), 120 mM NaCl, 10% glycerol, 20 μg of poly(dI) · poly(dC) per ml, 0.8 mg of bovine serum albumin per ml, and 0.25 mM dithiothreitol (DTT), in a total volume of 15 μl. Approximately 10,000 cpm of 32P-end-labeled probe (0.1 to 0.4 ng) and 2 ng of purified NFAT1-DBD or p50 NF-κB (kindly provided by Stephen C. Harrison) were used in each binding reaction mixture. In some binding reaction mixtures, we used 1 μg of nuclear extract prepared as described previously (3) from Cl.7W2 (64) cells that had been stimulated for 30 min with 1 μM inomycin instead of purified proteins. Where indicated in the figures, a 100-fold excess of unlabeled probe was included in the binding reaction mixture. Reaction mixtures were incubated at room temperature for 20 min and analyzed on a 4% polyacrylamide gel. The following oligonucleotides were used in the binding reactions: κB-Sp1 wild type, 5′-GATCCGCTGGGGACTTTCCAGGGAGGCGTGGCCTGA; κB-Sp1 5′ mutant, 5′-GATCCGCTAGATCTTTTCCAGGGAGGCGTGGCCTGA; κB-Sp1 3′ mutant, 5′-GATCCGCTGGGGACTAGTTAGGGAGGCGTGGCCTGA; and κB-Sp1 5′+3′ mutant, 5′-GATCCGCTAGATCTTAGTTAGGGAGGCGTGGCCTGA.
Cell culture and transfections.
Jurkat cells and HEK293T cells (15) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 10 mM HEPES, and 2 mM glutamine. Jurkat cells were transfected by electroporation in serum-free medium with pulses of 250 V and 960 μF. Twenty-four hours after transfection, cells were stimulated with 2 μM ionomycin (Calbiochem) and 10 nM phorbol myristate acetate (PMA) (Calbiochem). Eight to fourteen hours after stimulation, cells were harvested and cell extracts were assayed for CAT or luciferase activity as described previously (39). Results from these assays were analyzed by Student’s t test. Cotransfection of a human growth hormone-expressing plasmid (40) was used to determine the efficiency of transfection. HEK293T cells were transfected by a calcium phosphate-DNA precipitation method. Protein concentrations were determined by a colorimetric assay (Bio-Rad Laboratories).
Expression and purification of recombinant proteins.
The NFAT1 DBD [pNFATpXS(1–297) (27)] and different fragments from the amino-terminal domain of NFAT1 were expressed as six-histidine-tagged fusion proteins and purified as described previously (27). pQE-NFAT1(1–415), pQE-NFAT1(67–415), and pQE-NFAT1(140–415), kindly provided by Heidi Okamura, and pQE-NFAT1(1–96), made by subcloning a DNA fragment coding for the first 96 amino acids of NFAT1 in the pQE-31 bacterial expression vector (Qiagen), bear genes that encode different regions of the amino-terminal domain of NFAT1. Glutathione S-transferase (GST)–Tat-1 86R TK, GST–Tat-1 72R, GST–Tat-1 86R C22G, and GST–Tat-1 86R D2–26 were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIAID, NIH), from Andrew Rice and were used to express and purify GST-Tat recombinant proteins by following our own protocol (21, 58).
In vitro binding assays.
In vitro binding reactions were performed with a buffer containing 50 mM Tris-HCl (pH 7.4), 100 mM KCl, 20% glycerol, 0.25% Nonidet P-40, 0.5 mM EDTA, and 5 mM DTT in a total volume of 500 μl. GST-Tat proteins (6 to 8 μg) bound to glutathione-Sepharose beads (Pharmacia) were incubated for 4 h at 4°C with 250 to 500 ng of purified six-His NFAT1 proteins. After the incubation, the beads were washed three times with the same buffer and bound proteins were separated in a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel and analyzed by Western blotting with anti-67.1 and anti-72 (22, 46), which recognize different epitopes in the amino-terminal domain of NFAT1, or with R59 (46), which is directed against the NFAT1 DBD. In every assay a binding reaction mixture with GST protein was included as a negative control.
Immunoprecipitations.
Cellular extracts from HEK293T cells transfected with pcTat and/or pEFTagNFAT1-C were obtained by lysing the cells in a buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 7.4), 0.25% Nonidet P-40, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 20 μM leupeptin, and 10 μM aprotinin. Cell lysates were then precleared with protein A-Sepharose (Pharmacia) and incubated for 4 h at 4°C with the appropriate antibodies. Immunocomplexes were pelleted, and washed and bound proteins were separated on an SDS-polyacrylamide gel and analyzed by Western blotting. Antibodies against the HA epitope tag (12CA5; Boehringer Mannheim) and anti-67.1 were used to immunoprecipitate and detect NFAT1. Antiserum to HIV-Tat (19) and a monoclonal antibody against HIV-1BH10 Tat (amino acids 57 to 71) (8) were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Bryan Cullen and from the Division of AIDS, NIAID, respectively, and were used to immunoprecipitate and detect HIV-1 Tat.
RESULTS
HIV-1 Tat enhances NFAT1-driven transcription.
As HIV-1 Tat had been shown to play a role in the regulation of numerous cytokine genes cooperating with different cellular factors (2, 7, 51, 63), we asked whether it could affect transactivation by NFAT1, a transcription factor involved in regulating the expression of a large number of cytokine genes (57). For that purpose, we studied the effect of HIV-1 Tat on NFAT1-driven transcription in transient-transfection experiments with Jurkat cells. When expressed alone, HIV-1 Tat had little effect on the basal activity of NFAT3×-Luc, which contains three copies of a canonical NFAT1-AP1 site. However, cotransfection of Tat potentiated the activity of NFAT1 to drive reporter expression (Fig. 1A). To exclude a possible effect of Tat on AP1 proteins and to check if this result could be reproduced with only the transactivation domain of NFAT1, a series of experiments were carried out with a fusion protein containing the GAL4 DBD and the first 415 amino acids of NFAT1, which contain its major transactivation domain (40). Tat expression had no effect on the activity of the GAL4 reporter plasmid but significantly (P < 0.02) upregulated GAL4-dependent transactivation mediated by the GAL4-NFAT1(1–415) fusion protein (Fig. 1B), indicating that the potentiation involved the terminal transactivation domain of NFAT1.
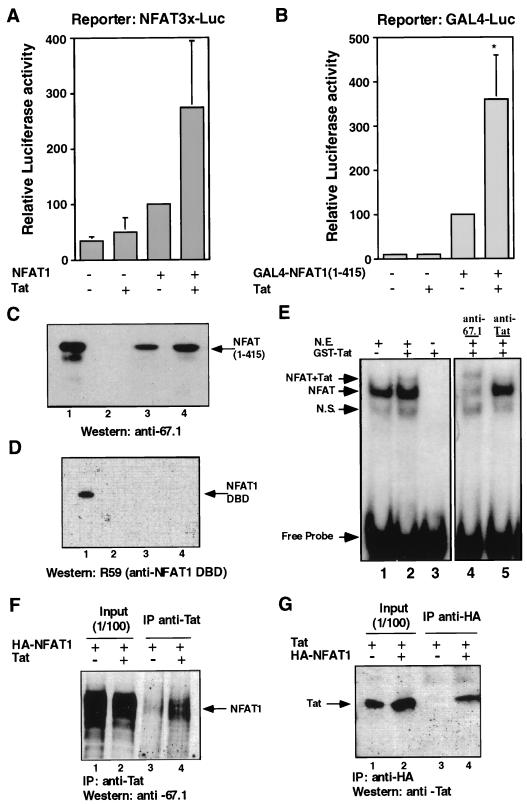
FIG. 1.
HIV-1 Tat interacts with NFAT1 and upregulates NFAT1-mediated transactivation. (A) Jurkat cells were transfected with 2 μg of the reporter plasmid NFAT3×-Luc and expression plasmids for Tat (0.5 μg) and/or NFAT1 (5 μg). Cells were stimulated for 8 h with 10 nM PMA and 2 μM ionomycin. (B) Similar experiments were carried out with the GAL4-Luc reporter plasmid (2 μg) and pGAL4-NFAT1(1–415) (2.5 μg), which expresses a fusion between the GAL4-DBD and the terminal domain of NFAT1. Total amounts of DNA were adjusted by using the appropriate empty vector. Results are shown as percentages of the luciferase activity of the NFAT1- or GAL4-NFAT1(1–415)-transfected cells. Values are the means + standard errors of results from three independent experiments. ∗, P < 0.02. (C and D) GST-Tat one-exon or Tat two-exon recombinant proteins (6 μg) were assayed for their ability to bind recombinant NFAT1(1–415) (C) or the NFAT1 DBD (D). Binding reaction mixtures were run on SDS-polyacrylamide gels and blotted. The antibodies anti-67.1 against the amino-terminal domain of NFAT1 and R59 against the NFAT1 DBD were used to detect bound NFAT1 proteins. Recombinant GST protein was used as a negative control. (E) Nuclear extracts from ionomycin-stimulated Cl.7W2 cells (N.E.) were incubated, in the presence (lane 2) or absence (lane 1) of purified GST-Tat protein, with radiolabeled oligonucleotides containing the adjacent κB and Sp1 sites of the HIV-1 LTR. Antibodies raised against NFAT1 (lane 4) or HIV-1 Tat (lane 5) were used to check the presence of those proteins in the EMSA bands. Lane 3 contains a binding reaction mixture with radiolabeled probe and GST-Tat without nuclear extract. N.S., nonspecific complex. (F) Total cellular extracts of PMA- and ionomycin-stimulated HEK293T cells expressing HA-tagged NFAT1 alone or coexpressing HIV-1 Tat were immunoprecipitated (IP) with anti HIV-1 Tat. Immunoprecipitates were assayed by Western blotting to detect coimmunoprecipitation of NFAT1. (G) Total cellular extracts of PMA- and ionomycin-stimulated HEK293T cells expressing HIV-1 Tat alone or coexpressing HA-tagged NFAT1 were immunoprecipitated with anti-HA. Immunoprecipitates were assayed by Western blotting to detect coimmunoprecipitation of HIV-1 Tat.
Tat interacts with the amino-terminal domain of NFAT1.
To test whether the enhancement of NFAT1 transactivation was mediated via Tat recruitment through a direct protein-protein interaction between the NFAT1 amino-terminal region and Tat, we performed in vitro binding experiments with GST-Tat (one or two exons) and different six-His-tagged fragments of NFAT1. Both forms of Tat protein were able to pull down a fragment of NFAT1 containing its first 415 amino acids (Fig. 1C), although the ability of the Tat two-exon protein to bind NFAT1 appeared somewhat greater than that of the one-exon protein (compare lane 3 to lane 4 in Fig. 1C). When the DBD of NFAT1 was used in similar experiments, no binding to Tat was detected, indicating that the interaction of these two proteins involved specifically the amino-terminal domain of NFAT1 (Fig. 1D). Furthermore, in EMSAs carried out with nuclear extracts from Cl.7W2 cells and an IL-2 promoter ARRE2 site probe, a new slower-migrating complex could be observed when purified GST-Tat was added to the binding reaction mixture (Fig. 1E, compare lanes 1 and 2); this complex was supershifted with antibodies to NFAT1 and HIV-1 Tat (Fig. 1E, lanes 4 and 5). As expected from the lack of interaction of the NFAT1 DBD with Tat (Fig. 1D), no new complex was detected in EMSAs when the binding reaction mixtures contained a combination of the purified NFAT1 DBD and GST-Tat (data not shown).
To confirm that NFAT1 interacted with Tat in cells, HEK293T cells were cotransfected with plasmids expressing HA-tagged NFAT1 and/or HIV-1 Tat. Cells were stimulated with PMA and ionomycin for 6 h to localize NFAT1 to the nucleus, and total cellular extracts were immunoprecipitated with antibodies to Tat or the HA tag. Antibodies against HIV-1 Tat coprecipitated NFAT1 only in cells which had been cotransfected with both plasmids (Fig. 1F); similarly, anti-HA antibodies coprecipitated HIV-1 Tat together with HA-NFAT1 (Fig. 1G). No Tat was immunoprecipitated with the anti-HA antibody in the absence of HA-tagged NFAT1 (Fig. 1G, lane 3), thus ruling out nonspecific binding of HIV-1 Tat to the anti-HA antibody. These experiments demonstrated the existence of an NFAT1–HIV-1 Tat interaction in vivo.
NFAT1 HIV-1 Tat interaction is mediated by the transactivation domain of NFAT1 and the amino-terminal region of Tat.
To localize more precisely the region of the NFAT1 amino-terminal domain involved in the interaction with Tat, we expressed different fragments of this region and tested their capacity to bind Tat. In vitro binding experiments showed that only the most amino-terminal fragment (amino acids 1 to 96) was able to bind Tat (Fig. 2A, lane 12) and that fragments containing C-terminal regions of the NFAT1 amino-terminal domain (amino acids 67 to 415 and 140 to 415) showed no Tat binding activity (Fig. 2A, lanes 6 and 9). Therefore, the region of NFAT1 which interacted with Tat was localized to the first 96 amino acids of NFAT1, which constitute the major, strongly acidic, transactivation domain of NFAT1 (40).
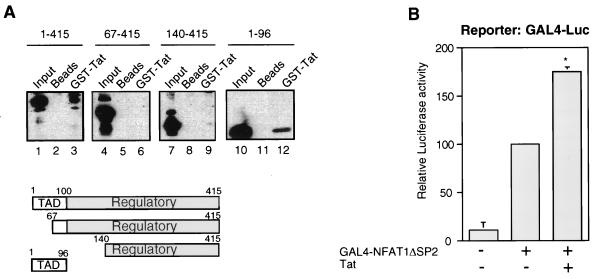
FIG. 2.
The transactivation domain of NFAT1 interacts with HIV-1 Tat. (A) Different recombinant fragments from the amino-terminal domain of NFAT1 (amino acids 1 to 415, lanes 1 to 3; amino acids 67 to 415, lanes 4 to 6; amino acids 140 to 415, lanes 7 to 10; amino acids 1 to 96, lanes 10 to 12) were assayed for binding to the GST–HIV-1 Tat two-exon protein (6 μg) (lanes 3, 6, 9, and 12). Control reaction mixtures with only glutathione-Sepharose beads were included as negative controls (lanes 2, 5, 8, and 11). Anti-72 antibody was used to detect NFAT1 fragments in lanes 1 to 9, and anti-67.1 antibody was used in lanes 10 to 12. TAD, transactivation domain. (B) Jurkat cells were transfected with a GAL4-Luc reporter plasmid (2 μg); pGAL4-NFAT1ΔSP2 (0.25 μg), which expresses a fusion between GAL4-DBD and the amino-terminal domain of NFAT1 with a deletion spanning amino acids 145 to 387; and/or pcTat (0.5 μg). Cells were stimulated for 8 h with 10 nM PMA and 2 μM ionomycin. Total amounts of DNA were adjusted with the appropriate empty vector. Results are shown as percentages of the luciferase activity of the GAL4-NFAT1ΔSP2-transfected cells. Values are the means + standard errors of results from three independent experiments. ∗, P < 0.05.
If the transactivation domain is the region of NFAT1 that makes contact with Tat, the transactivational activity of a fusion protein containing only this domain should also be enhanced by Tat. To test this hypothesis, transient-transfection experiments were performed with Jurkat cells and a GAL4 fusion protein containing an NFAT1 amino-terminal domain with a deletion from amino acids 145 to 387 (GAL4-ΔSP2) and therefore lacking the regulatory domain but maintaining the major transactivation domain. As predicted, coexpression of Tat upregulated GAL4-ΔSP2-mediated transcription from a GAL4 luciferase reporter (Fig. 2B).
We also determined the region of the HIV-1 Tat domain involved in making contact with the transactivation domain of NFAT1. In vitro binding experiments revealed that the NFAT1-Tat interactions involved residues contained in the first 26 amino acids of Tat, since a GST-Tat fusion protein with a deletion of amino acids 2 to 26 did not bind NFAT1 (Fig. 3A, compare lanes 2 and 5). However, a single point mutation in this region, C22 to G, that produces a transcriptionally inactive Tat, was still able to bind the amino-terminal domain of NFAT1 (Fig. 3A, lane 4), suggesting that the interactions of Tat with NFAT1 involved a domain of Tat different than that involved in the interactions of Tat with cyclin T, P-TEFb, or other components of the basal transcriptional machinery (13, 66, 68).
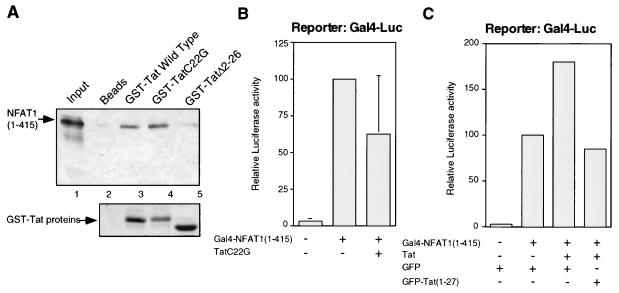
FIG. 3.
HIV-1 Tat enhancement of NFAT1 transactivation requires a transcriptionally active Tat protein and is mediated by the first 26 amino acids of Tat. (A) The binding affinities of two mutated Tat proteins (TatC22G and TatΔ2–26) for the amino-terminal domain of NFAT1 were assayed. Binding reaction mixtures were resolved on an SDS-acrylamide gel and blotted. The bound products were detected with an antibody against an amino-terminal peptide of NFAT1 (67.1). A Ponceau red staining of the blot showing the amounts of Tat proteins used in the binding reaction mixtures is shown below the immunoblot. (B) An inactive Tat protein does not stimulate NFAT1-mediated transactivation. Jurkat cells were transfected with a GAL4-Luc reporter plasmid (2 μg) and pGAL4-NFAT1(1–415) (2.5 μg), with or without a plasmid expressing a mutant (Cys22-to-Gly) Tat protein (0.5 μg). Cells were stimulated for 8 h with 10 nM PMA and 2 μM ionomycin. Total amounts of DNA were adjusted with the appropriate empty vector. Values are means + standard errors of results from three independent experiments. (C) Expression of a GFP-Tat(1–27) fusion protein inhibits Tat-mediated upregulation of NFAT1 transactivation. Jurkat cells were transfected with a GAL4-Luc reporter plasmid (2 μg), pGAL4-NFAT1(1–415) (2.5 μg), pcTat (0.5 μg), and pEGFP or pEGFPTat(1–27) (2 μg). Cells were stimulated for 8 h with 10 nM PMA and 2 μM ionomycin. Values are the means of results from two independent experiments.
We sought to determine whether the functional cooperativity of Tat with NFAT1 required a transcriptionally competent Tat protein by using an inactive Tat protein in which Cys22 had been mutated to Gly. As predicted, no enhancement of NFAT1-driven transcription was observed when this mutant Tat protein was coexpressed with a GAL4-NFAT1(1–415) fusion protein (Fig. 3B). The level of expression of the mutant Tat protein, checked by Western blotting with anti-Tat antibodies, was similar to the levels obtained with the vector expressing wild-type Tat (data not shown). In a second approach, we tested the effect of blocking NFAT1-Tat interaction by overexpressing the first 27 amino acids of Tat as a GFP fusion protein. We predicted that this fragment, which contained the region of Tat involved in the interaction with NFAT1, would displace Tat from its binding site on NFAT1 and therefore would abolish NFAT1-Tat cooperativity. Confirming our hypothesis, cotransfection of a plasmid expressing GFP-Tat(1–27) inhibited Tat enhancement of NFAT1-mediated transactivation (Fig. 3C).
NFAT1 inhibits Tat-mediated activation of HIV-1 LTR transcription.
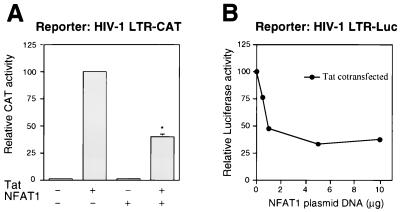
Having shown that the NFAT1-Tat interaction potentiated the transcriptional activity of NFAT1, we asked whether, conversely, NFAT1 would modulate Tat-mediated activation of the HIV-1 LTR (Fig. 4). Transfection assays were performed with Jurkat cells, and as previously reported, Tat expression resulted in a large increase in CAT activity driven by the HIV-1 LTR in transiently transfected Jurkat cells. This increase was significantly reduced (P < 0.01) by addition of NFAT1 (Fig. 4A). This effect showed a direct correlation with the amount of NFAT1 plasmid cotransfected (Fig. 4B). In these experiments, cells were stimulated with PMA and ionomycin to achieve activation and translocation of NFAT1 to the nucleus (38, 39, 61), but the concentration of PMA used in these experiments (10 nM) produced no detectable activation of the HIV-1 LTR, ruling out the possibility that NFAT1 was depressing Tat-mediated activation by downregulating NF-κB or other factors involved in Tat-independent activation of the HIV-1 LTR.
FIG. 4.
Effect of NFAT1 on Tat-mediated activation of the HIV-1 LTR. (A) Jurkat cells were transfected with 1 μg of the reporter plasmid HIV-1 LTR CAT and expression plasmids for Tat (0.25 μg) and/or NFAT1 (10 μg). Cells were stimulated for 12 h with 10 nM PMA and 2 μM ionomycin. Results are shown as percentages of the CAT activity of the Tat-transfected cells. Values are means + standard errors of results from three independent experiments. ∗, P < 0.01. (B) Jurkat cells were cotransfected with 1 μg of HIV-1 LTR-Luc, 0.25 μg of pcTat, and increasing amounts of the NFAT1 expression plasmid. Twenty-four hours after transfection, cells were stimulated for 12 h with 10 nM PMA and 2 μM ionomycin. The results of a representative experiment are shown. In all experiments the total amounts of DNA were maintained at a constant level by cotransfecting balancing amounts of empty pEFTag plasmid.
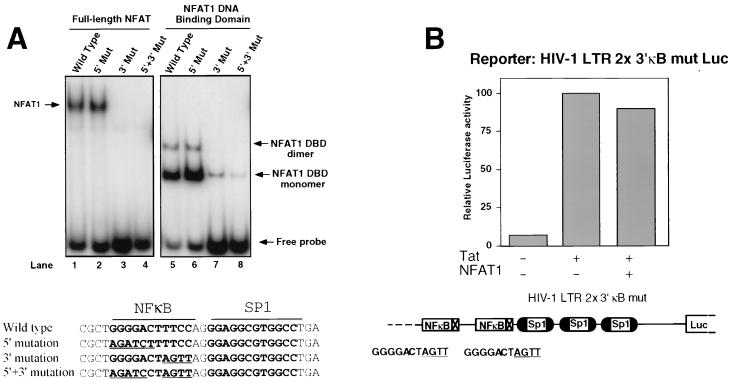
To determine whether NFAT1 binding to DNA was necessary for downregulation of Tat activity, we took advantage of the fact that NFAT1 bound preferentially to the 3′ halves of the κB sites of the HIV-1 LTR (Fig. 5A). Both full-length NFAT1 (lane 1) and a recombinant fragment comprising the DBD of NFAT1 (lane 5) showed binding to a radiolabeled κB-Sp1 oligonucleotide (−66 to −91). As previously shown for the immunoglobulin κ enhancer κB site (45), the recombinant NFAT1-DBD formed two complexes with the probe, which contained one (lower complex) or two (upper complex) molecules of NFAT1-DBD (Fig. 5A, lane 5). Binding of both full-length NFAT1 and NFAT1-DBD was specific, as judged by competition with excess unlabeled wild-type and mutated oligonucleotides (data not shown). NFAT1 bound to the same level (Fig. 5A, lanes 2 and 6) to an oligonucleotide with a mutation in the 5′ half of the κB site, which is known to abolish NF-κB binding (54), but showed greatly reduced binding to an oligonucleotide with a mutation in the 3′ half of the κB site (Fig. 5A, lanes 3 and 7). We then constructed an HIV-1 LTR luciferase reporter plasmid in which both κB sites had been mutated in their 3′ halves to abolish NFAT1 binding (Fig. 4). When this reporter plasmid was used, NFAT1 expression caused no significant reduction of luciferase activity in Tat-transfected cells (Fig. 5B), suggesting that NFAT1-mediated downregulation of Tat activity requires positioning of NFAT1 on the κB sites of the HIV-1 LTR.
FIG. 5.
NFAT1 binding to the κB site of the HIV-1 LTR is necessary for NFAT1-mediated downregulation of Tat transactivation. (A) Nuclear extracts from ionomycin-stimulated Cl.7W2 cells (lanes 1 to 4) or the purified NFAT1 DBD (lanes 5 to 8) were incubated with radiolabeled oligonucleotides containing the adjacent κB and Sp1 sites of the HIV-1 LTR (wild type, lanes 1 and 5) or the indicated 5′ and 3′ mutations (Mut). Arrows indicate the positions of the different complexes. Sequences of the oligonucleotides used in the binding assays are shown below. Sequences in boldface type indicate the actual κB and Sp1 sites on the probe. Mutated bases in the oligonucleotides with 5′ or 3′ mutations are underlined. (B) Jurkat cells were transfected with Tat (0.25 μg) and/or NFAT1 (10 μg) expression plasmids and with luciferase reporter vectors containing a mutated LTR in which both κB binding sites were made unable to bind NFAT1 by mutating their 3′ halves from TTCC to AGTT. The effect of NFAT1 on the transactivation caused by Tat was assayed. Results are shown as percentages of the luciferase activity of the pcTat-transfected cells. Values are the means of results from two independent experiments. In all the experiments, the total amounts of DNA were maintained at a constant level by cotransfecting balancing amounts of empty pEFTag plasmid.
NFAT1 competes with NF-κB for the binding to the κB sites of the HIV-1 LTR.
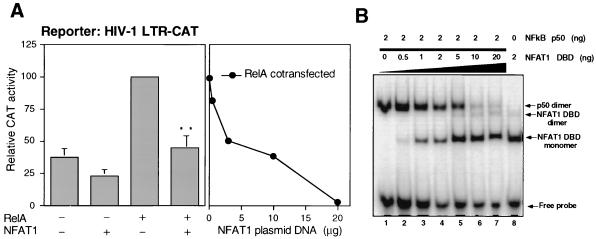
Since NFAT1 bound the κB sites of the HIV-1 LTR but inhibited Tat-mediated transactivation, we also tested the effect of NFAT1 on NF-κB-mediated activation of the HIV-1 LTR. A different NFAT family member, NFAT2, has been shown to synergize with NF-κB for HIV-1 replication and transactivation of the HIV-1 LTR (32, 33). In contrast, we found that NFAT1 inhibited RelA-mediated transactivation of the HIV-1 LTR (Fig. 6A). Transfection of a RelA expression plasmid into Jurkat cells produced an increase in the CAT activity of the HIV-1 LTR-CAT reporter vector, which was significantly reduced in a dose-dependent manner by expression of NFAT1 (Fig. 6A). A possible explanation for this effect is that NFAT1 and NF-κB compete for binding to the κB sites of the HIV-1 LTR (Fig. 6B). A labeled κB-Sp1 probe was incubated with a fixed amount of p50 NF-κB and increasing amounts of NFAT1-DBD. The complex formed by p50 NF-κB disappeared as higher concentrations of NFAT1-DBD were added to the reaction mixture, with the appearance of new bands corresponding to the NFAT1-DBD monomer and dimer (Fig. 6B). No complex containing both NFAT1 and NF-κB was detected at any of the tested concentrations of NFAT1. These binding experiments showed that NF-κB1 and NFAT1 could not bind simultaneously to the same κB sites but that they seemed to compete for them.
FIG. 6.
NFAT1 competes with NF-κB1 for binding to the κB site and downregulates NF-κB-mediated activation of the HIV-1 LTR. (A) RelA-mediated activation. In the left graph, Jurkat cells were cotransfected with 1 μg of the reporter plasmid HIV-1 LTR CAT and expression plasmids for NFAT1 (10 μg) and/or RelA (3 μg). Cells were stimulated for 12 h with 10 nM PMA and 2 μM ionomycin. Results are shown as percentages of the CAT activity of the RelA-transfected cells. Values are means + standard errors of results from six independent experiments. ∗∗, P < 0.01. In the right graph, Jurkat cells were cotransfected with 1 μg of the reporter plasmid HIV-1 LTR CAT, 3 μg of a RelA expression plasmid, and increasing amounts of an NFAT1 expression plasmid. Twenty-four hours after the transfection, cells were stimulated for 12 h with 10 nM PMA and 2 μM ionomycin. A representative experiment is shown. In all the experiments the total amounts of DNA were maintained at a constant level by cotransfecting balancing amounts of empty pEFTag plasmid. (B) A labeled oligonucleotide containing the adjacent κB and Sp1 sites of the HIV-1 LTR was incubated with 2 ng of NF-κB p50 in the presence of increasing amounts of the purified recombinant NFAT1 DBD. Arrows indicate the positions of the different complexes.
DISCUSSION
The viral protein Tat is essential for HIV-1 gene expression and replication and its function is regulated by interactions with several host factors. In addition to being a potent activator of HIV-1 gene transcription, Tat may regulate the expression of other cellular genes whose products influence the course of viral infection, although the exact mechanism underlying most of these effects is still unknown. Specifically, HIV-1 infection is known to produce an altered pattern of cytokine production from both T cells and monocytes (47, 52, 67), and several reports have described a direct involvement of Tat in the regulation of cytokine genes. Expression of IL-6 is induced by an interaction of Tat with the CAAT enhancer binding protein beta (2), and a direct participation of Tat in IL-2 expression has also been reported (51).
In this paper we have addressed the possibility that one of the mechanisms responsible for Tat-mediated regulation of host cellular genes involves the transcription factor NFAT1. We have shown that Tat upregulates NFAT1 transcriptional activity and that the amino-terminal domain of NFAT1 is sufficient for this effect. This result is consistent with previous observations indicating that the increase in IL-2 expression caused by Tat maps to the NFAT sites of the IL-2 promoter (63) and to the CD28 response element (51), which is known to bind either NFAT or Rel proteins (41, 59, 60). Our results show that NFAT1 is able to bind Tat in vivo and that this interaction takes place between the major transactivation domain of NFAT and the amino-terminal region of Tat. Thus, the ability of NFAT1 to recruit Tat to the regulatory regions of cytokine genes may promote the interaction of Tat with TFIID, RNA polymerase II, or other transcription factors or kinases in the cooperative enhancer complex, thus promoting the transcription of these genes. As NFAT1 is a key regulator of gene expression during the immune response (57), the Tat-NFAT1 interaction may be responsible for the upregulation of genes encoding cytokines and other immune modulators during HIV-1 infection (51).
Transient transfection of Jurkat T cells with NFAT1 and Tat-expressing plasmids indicates that NFAT1 inhibits Tat-mediated transactivation of the HIV-1 LTR. Although this effect can be observed by overexpressing the isolated terminal domain of NFAT1 (data not shown), it is augmented by binding of NFAT1 to the κB sites of the HIV-1 LTR. The degree of downregulation produced by NFAT1 coexpression is greater than 50%. Studies performed with different Tat mutants have revealed that a 50% reduction in Tat transcriptional activity suffices for significant impairment of HIV-1 gene transcription and virus replication (65). Many other proteins have been identified as Tat-binding proteins, and some of these, such as Oct2 and p53, have been described as negative regulators which produce degrees of inhibition of transcription of the HIV-1 LTR similar to that produced by NFAT1 (36, 37). The inhibitory effect of NFAT1 on HIV-1 LTR transcription is also consistent with the fact that in mutant HIV-1 viruses which do not require virion-associated cyclophilin A to initiate infection, CsA, which inhibits NFAT1 translocation into the nucleus, can have a stimulatory effect on virus replication (6). The mechanism of NFAT1-mediated Tat inhibition remains to be investigated: it may involve a direct squelching or blocking of Tat-mediated transactivation by NFAT1, or alternatively, the NFAT1-Tat interaction may block the interaction of Tat with other host factors required for Tat to exert its function through the TAR element (18, 66).
Recent reports have indicated that another member of the NFAT family, NFAT2, activates both HIV-1 gene expression and replication (32, 33). In contrast, our results indicate that NFAT1 has a largely downregulatory effect on HIV-1 LTR expression in Jurkat T cells. Differences in the mechanisms of regulation and functions of these two family members may account for their distinct effects on HIV-1 regulation. Indeed, NFAT1 and NFAT2 have been postulated to have opposite effects on T-cell function, based on the immune phenotype of cells lacking either of these two transcription factors. While T cells lacking NFAT1 show a maintained high level of IL-4 after stimulation, which indicates a role for NFAT1 in downregulating IL-4 transcription and thus inhibiting T-helper 2 responses (31), T cells lacking NFAT2 show an impairment in IL-4 production, suggesting a positive role for this family member in IL-4 gene transcription (55). T cells and other immune cells may selectively use different members of the NFAT family to regulate gene expression of HIV-1 and other genes involved in the immune response. Alternatively, the observed differences may result from a hierarchy of transcriptional activity (NF-κB > NFAT2 > NFAT1), and NFAT1 might be capable of upregulating HIV-1 and IL-4 gene expression, especially in cell types (e.g., naive primary T cells) lacking high-level expression of NF-κB and NFAT2.
The fact that NFAT1 and Tat have opposite effects on each other’s activities is not surprising, as a similar regulatory interplay between Tat and Sp1 has been described. A cooperative interaction between NF-κB bound to the κB sites and Sp1 bound to the adjacent Sp1 sites is required for HIV-1 gene expression (53), and Sp1 is also essential for Tat-mediated activation of the HIV-1 LTR (4, 62). However, Tat inhibits the transcription of several Sp1-activated cellular promoters by acting directly or indirectly on Sp1 or Sp1-like proteins bound to their specific binding sites in those promoters (25).
The κB sites of the HIV-1 LTR are essential for viral gene expression and replication in T cells and monocytes (1, 26). We have demonstrated that NFAT1 and NF-κB do not bind cooperatively to the κB sites of the HIV-1 LTR but rather that they compete for binding. Consistent with this observation, NFAT1 inhibits RelA-mediated transactivation of the HIV-1 LTR, presumably by competing with NF-κB for occupancy of the κB sites. A similar interplay between NFAT and NF-κB proteins occurs on the human IL-4 promoter, where the NFAT1-mediated activation of the IL-4 promoter in CD4+ cells is negatively controlled by competitive binding of RelA to the P element on this promoter (9). Thus, the NFAT–NF-κB competition for the κB sites of the HIV-1 LTR may be another example of a more general regulatory mechanism used by cells to modulate the activities of these two families of transcription factors. Other proteins such as HMG-I (Y), which are known to differentially regulate the binding affinities of NFAT and Rel proteins to specific DNA sites, may also be involved in such a mechanism (34). It has been shown that Stat2 modulates HIV gene expression by competing for the coactivator p300 with NF-κB (24); similarly, NFAT1 may also compete with NF-κB for transcriptional coactivators such as p300 (17, 24), whose cellular concentration may be limited.
In summary, we have shown the existence of an interaction between the major transactivation domain of NFAT1 and the amino-terminal region of HIV-1 Tat. NFAT1 interaction with Tat results in potentiation of NFAT1-mediated transactivation; conversely, NFAT1 inhibits both Tat-mediated and RelA-mediated HIV-1 transcription. Other reports have shown that NFAT2 potentiates HIV-1 replication and gene expression (32, 33). Notably, the immune phenotype of T cells deficient in NFAT1 and NFAT2 suggests that these related transcription factors have opposing effects on IL-4 gene transcription as well (55). The contrasting effects of NFAT1 and NFAT2 suggest that NFAT transcription factors exert complex modulatory effects on HIV-1 transcription and the immune response.
ACKNOWLEDGMENTS
We thank S. Ghosh, S. C. Harrison, D. J. McKean, M. Montminy, and H. Okamura for their generous gifts of reagents. The following reagents were obtained through the AIDS Research and Reagent Reference Program, Division of AIDS, NIAID, NIH: GST–Tat-1 86R and GST–Tat-1 72R from A. Rice; antiserum against HIV-1 Tat from B. Cullen; and a monoclonal antibody against HIV-1 Tat from the Division of AIDS, NIAID.
This work was supported by NIH grant CA42471 and a Leukemia Society of America scholar award (to A.R.). F.M. was supported by a postdoctoral fellowship from the Ministry of Education and Culture of Spain.
REFERENCES
- 1.Alcami J, Lain de Lera T, Folgueira L, Pedraza M-A, Jacque J-M, Bachelerie F, Noriega A R, Hay R T, Harrich D, Gaynor R B, Virelizier J-L, Arenzana-Seisdedos F. Absolute dependence on κB responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4+ T lymphocytes. EMBO J. 1995;14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrosino C, Ruocco M R, Chen X, Mallardo M, Baudi F, Trematerra S, Quinto I, Venuta S, Scala G. HIV-1 Tat induces the expression of the interleukin-6 (IL6) gene by binding to the IL6 leader RNA and by interacting with CAAT enhancer-binding protein beta (NF-IL6) transcription factors. J Biol Chem. 1997;272:14883–14892. doi: 10.1074/jbc.272.23.14883. [DOI] [PubMed] [Google Scholar]
- 3.Aramburu J, Azzoni L, Rao A, Perussia B. Activation and expression of the nuclear factor of activated T cells, NFATp and NFATc, in human natural killer cells: regulation upon CD16 ligand binding. J Exp Med. 1995;182:801–810. doi: 10.1084/jem.182.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout B, Jeang K T. Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency type 1 long terminal repeat. J Virol. 1992;66:139–149. doi: 10.1128/jvi.66.1.139-149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domains. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braaten D, Aberham C, Franke E K, Yin L, Phares W, Luban J. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J Virol. 1996;70:5170–5176. doi: 10.1128/jvi.70.8.5170-5176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buonaguro L, Barillari G, Chang H K, Bohan C A, Kao V, Morgan R, Gallo R C, Ensoli B. Effects of the human immunodeficiency virus type 1 Tat protein on the expression of inflammatory cytokines. J Virol. 1992;66:7159–7167. doi: 10.1128/jvi.66.12.7159-7167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campioni D, Corallini A, Zauli G, Possati L, Altavilla G, Barbanti-Brodano G. HIV type 1 extracellular Tat protein stimulates growth and protects cells of BK virus/tat transgenic mice from apoptosis. AIDS Res Hum Retroviruses. 1995;11:1039–1048. doi: 10.1089/aid.1995.11.1039. [DOI] [PubMed] [Google Scholar]
- 9.Casolaro V, Georas S N, Song Z, Zubkoff I D, Abdulkadir S A, Thanos D, Ono S J. Inhibition of NF-AT-dependent transcription by NF-κB: implications for differential gene expression in T helper cell subsets. Proc Natl Acad Sci USA. 1995;92:11623–11627. doi: 10.1073/pnas.92.25.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Shulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/p300 in nuclear receptor signalling. Nature. 1996;383:99–102. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Glover J N M, Hogan P G, Rao A, Harrison S C. Structure of the DNA binding domains from NFAT, Fos and Jun bound to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 12.Chiang C-M, Roeder R G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 13.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. The HIV transactivator Tat binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DuBridge R B, Tang P, Hsia H C, Leong P-M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg M B, Baltimore D, Frankel A D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci USA. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Rodriguez C, Rao A. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators P300/CREB binding protein (CBP) J Exp Med. 1998;187:2031–2036. doi: 10.1084/jem.187.12.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaynor R B. Regulation of HIV-1 gene expression by the transactivator protein Tat. Curr Top Microbiol Immunol. 1995;193:51–77. doi: 10.1007/978-3-642-78929-8_3. [DOI] [PubMed] [Google Scholar]
- 19.Hauber J, Perkins A, Heimer E P, Cullen B R. Trans-activation of human immunodeficiency virus gene expression is mediated by nuclear events. Proc Natl Acad Sci USA. 1987;84:6364–6368. doi: 10.1073/pnas.84.18.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hedin K E, Bell M P, Kalli K R, Huntoon C J, Sharp B M, Mckean D J. Delta-opioid receptors expressed by Jurkat T cells enhance IL-2 secretion by increasing AP-1 complexes and activity of the NF-AT/AP-1-binding promoter element. J Immunol. 1997;159:5431–5440. [PubMed] [Google Scholar]
- 21.Herrmann C H, Rice A P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxy-terminal domain of the large subunit of RNA-polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho A M, Jain J, Rao A, Hogan P G. Expression of the transcription factor NFATp in a neuronal cell line and in the murine nervous system. J Biol Chem. 1994;269:28181–28186. [PubMed] [Google Scholar]
- 23.Hoey T, Sun Y L, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 24.Hottiger M O, Felzien L K, Nabel J N. Modulation of cytokine-induced HIV gene expression by competitive binding of transcription factors to the coactivator p300. EMBO J. 1998;17:3124–3134. doi: 10.1093/emboj/17.11.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howcroft T K, Palmer L A, Brown J, Rellahan B, Kashanchi F, Brady J N, Singer D S. HIV Tat represses transcription through Sp1-like elements in the basal promoter. Immunity. 1995;3:127–138. doi: 10.1016/1074-7613(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 26.Jacque J-M, Fernandez B, Arenzana-Seisdedos F, Thomas D, Baleux F, Virelizier J-L, Bachelerie F. Permanent occupancy of the human immunodeficiency virus type 1 enhancer by NF-κB is needed for persistent viral replication in monocytes. J Virol. 1996;70:2930–2938. doi: 10.1128/jvi.70.5.2930-2938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain J, Burgeon E, Badalian T M, Hogan P G, Rao A. A similar DNA-binding motif in NFAT family proteins and the Rel homology region. J Biol Chem. 1995;270:4138–4145. [PubMed] [Google Scholar]
- 28.Jain J, McCaffrey P G, Miner Z, Kerppola T K, Lambert J N, Verdine G L, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 29.Jeang K T, Chun R, Lin N H, Gatignol A, Glabe C G, Fan H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol. 1993;67:6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 31.Kiani A, Viola J P B, Lichtman A H, Rao A. Down-regulation of IL-4 gene transcription and control of Th2 cell differentiation by a mechanism involving NFAT1. Immunity. 1997;7:849–860. doi: 10.1016/s1074-7613(00)80403-3. [DOI] [PubMed] [Google Scholar]
- 32.Kinoshita S, Chen B K, Kaneshima H, Nolan G P. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell. 1998;95:595–604. doi: 10.1016/s0092-8674(00)81630-x. [DOI] [PubMed] [Google Scholar]
- 33.Kinoshita S, Su L, Amano M, Timmerman L A, Kaneshima H, Nolan G P. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6:235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- 34.Klein-Hessling S, Schneider G, Heinfling A, Chuvpilo S, Serfling E. HMG I(Y) interferes with the DNA binding of NF-AT factors and the induction of the interleukin 4 promoter in T cells. Proc Natl Acad Sci USA. 1996;93:15311–15316. doi: 10.1073/pnas.93.26.15311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laspia M F, Rice A P, Mathews M B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- 36.Li C J, Wang C, Friedman D J, Pardee A B. Reciprocal modulations between p53 and Tat of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:5461–5464. doi: 10.1073/pnas.92.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y-Z, Latchman D S. The octamer-binding proteins Oct-1 and Oct-2 repress the HIV long terminal repeat promoter and its transactivation by Tat. Biochem J. 1997;322:155–158. doi: 10.1042/bj3220155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loh C, Shaw K T, Carew J, Viola J P, Luo C, Perrino B A, Rao A. Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J Biol Chem. 1996;271:10884–10891. doi: 10.1074/jbc.271.18.10884. [DOI] [PubMed] [Google Scholar]
- 39.Luo C, Burgeon E, Carew J A, McCaffrey P G, Badalian T M, Lane W, Hogan P G, Rao A. Recombinant NFAT1 (NFATp) is regulated by calcineurin in T cells and mediates transcription of several cytokine genes. Mol Cell Biol. 1996;16:3955–3966. doi: 10.1128/mcb.16.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo C, Burgeon E, Rao A. Mechanisms of transactivation by nuclear factor of activated T cells. J Exp Med. 1996;184:141–147. doi: 10.1084/jem.184.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maggirwar S B, Harhaj E W, Sun S-C. Regulation of the interleukin-2 CD28-responsive element by NF-ATp and various NF-κB/Rel transcription factors. Mol Cell Biol. 1997;17:2605–2614. doi: 10.1128/mcb.17.5.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marciniak R A, Sharp P A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masuda E S, Naito Y, Tokumitsu H, Campbell D, Saito F, Hannum C, Arai K, Arai N. NFATx, a novel member of the NFAT family that is expressed predominantly in the thymus. Mol Cell Biol. 1995;15:2697–2706. doi: 10.1128/mcb.15.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mavankal G, Ignatius Ou S H, Oliver H, Sigman D, Gaynor R B. Human immunodeficiency virus type 1 and 2 proteins specifically interact with RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:2089–2094. doi: 10.1073/pnas.93.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCaffrey P G, Jain J, Jamieson C, Sen R, Rao A. A T cell nuclear factor resembling NF-AT binds to an NF-κB site and to the conserved lymphokine promoter sequence “Cytokine-1.”. J Biol Chem. 1992;267:1864–1871. [PubMed] [Google Scholar]
- 46.McCaffrey P G, Luo C, Kerppola T K, Jain J, Badalian T M, Ho A M, Burgeon E, Lane W S, Lambert J N, Curran T, Verdine G L, Rao A, Hogan P G. Isolation of the cyclosporine-sensitive T cell transcription factor NFATp. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- 47.Meyaard L, Otto S A, Keet I P, van Lier R A, Miedema F. Changes in cytokine secretion patterns of CD4+ T-cell clones in human immunodeficiency virus infection. Blood. 1994;84:4262–4268. [PubMed] [Google Scholar]
- 48.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 49.Northrop J P, Ho S N, Chen L, Thomas D J, Timmerman L A, Nolan G P, Admon A, Crabtree G R. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- 50.Okamoto H, Sheline C T, Corden J, Jones K A, Peterlin B M. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:11575–11579. doi: 10.1073/pnas.93.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ott M, Emiliani S, Van Lint C, Herbein G, Lovett J, Chirmule N, McCloskey T, Pahwa S, Verdin E. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science. 1997;275:1481–1485. doi: 10.1126/science.275.5305.1481. [DOI] [PubMed] [Google Scholar]
- 52.Pantaleo G, Fauci A S. New concepts in the immunopathogenesis of HIV infection. Annu Rev Immunol. 1995;13:487–512. doi: 10.1146/annurev.iy.13.040195.002415. [DOI] [PubMed] [Google Scholar]
- 53.Perkins N D, Agranoff A B, Pascal E, Nabel G J. An interaction between the DNA-binding domain of RelA (p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. A cooperative interaction between NF-κB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ranger A M, Hodge M R, Gravallese E M, Oukka M, Davidson L, Alt F W, Delabrousse F C, Hoey T, Grusby M, Glimcher L H. Delayed lymphoid repopulation with defects in IL-4-driven responses produced by inactivation of NF-ATC. Immunity. 1998;8:125–134. doi: 10.1016/s1074-7613(00)80465-3. [DOI] [PubMed] [Google Scholar]
- 56.Rao A. NF-ATp: a transcription factor required for the co-ordinate induction of several cytokine genes. Immunol Today. 1994;15:274–281. doi: 10.1016/0167-5699(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 57.Rao A, Luo C, Hogan P G. Transcription factors of the NFAT family—regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 58.Rhim H, Echetebu C O, Herrmann C H, Rice A P. Wild type and mutant HIV-1 and HIV-2 Tat proteins expressed in Escherichia coli as fusions proteins with glutathione S-transferase. J Acquired Immune Defic Syndr. 1994;7:1116–1121. [PubMed] [Google Scholar]
- 59.Rooney J W, Sun Y L, Glimcher L H, Hoey T. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol Cell Biol. 1995;15:6299–6310. doi: 10.1128/mcb.15.11.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shapiro V S, Truitt K E, Imboden J B, Weiss A. CD28 mediates transcriptional upregulation of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF–IL-2B AP-1 sites. Mol Cell Biol. 1997;17:4051–4058. doi: 10.1128/mcb.17.7.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw K T-Y, Ho A M, Raghavan A, Kim J, Jain J, Park J, Sharma S, Rao A, Hogan P G. Immunosuppressive drugs prevent a rapid dephosphorylation of the transcription factor NFAT1 in stimulated immune cells. Proc Natl Acad Sci USA. 1995;92:11205–11209. doi: 10.1073/pnas.92.24.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sune C, Garcia-Blanco M A. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1995;69:6572–6576. doi: 10.1128/jvi.69.10.6572-6576.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vacca A, Farina M, Maroder M, Alesse E, Screpanti I, Frati L, Gulino A. Human immunodeficiency virus type 1 Tat enhances interleukin-2 promoter activity through synergism with phorbol ester and calcium-mediated activation of the NF-AT cis-regulatory motif. Biochem Biophys Res Commun. 1994;205:467–474. doi: 10.1006/bbrc.1994.2689. [DOI] [PubMed] [Google Scholar]
- 64.Valgue-Archer V E, De Villiers J, Sinskey A J, Rao A. Transformation of T lymphocytes by the v-fos oncogene. J Immunol. 1990;145:4355–4364. [PubMed] [Google Scholar]
- 65.Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- 66.Wei P, Garber M E, Fang S M, Fisher W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 67.Yoo J, Chen H, Kraus T, Hirsch D, Polyak S, George I, Sperber K. Altered cytokine production and accessory cell function after HIV-1 infection. J Immunol. 1996;157:1313–1320. [PubMed] [Google Scholar]
- 68.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]