Highlights
-
•
The syndecan ectodomains are flexible random-coil intrinsically disordered proteins.
-
•
Their extended conformations have the same size as their cell surface partners.
-
•
Their binding-motifs to cells and receptors are not accessible in all conformations.
-
•
Only some of conformations of the ectodomains may be biologically active.
-
•
The syndecan-4 ectodomain forms a disulfide-bonded dimer with extended conformations.
Keywords: Syndecans, Intrinsically disordered proteins, Conformations, Cell-matrix interactions
Abbreviations: CD, circular dichroism; CCS, collision cross section; CSD, charge state distribution; DLS, dynamic light scattering; DTT, dithiothreitol; ED, ectodomain; ESI-MS, electrospray ionization - mass spectrometry; GAG, glycosaminoglycan; IDP, intrinsically disordered protein; ESI-IM-MS, electrospray ionization - ion mobility - mass spectrometry; MoRF, molecular recognition feature; PAGE, polyacrylamide gel electrophoresis; PMG, pre-molten globule; RC, random-coil; SASA, solvent accessible surface area; SAXS, small angle X-ray scattering; SDC, syndecan; SDS, sodium dodecyl sulfate; SEC, size exclusion chromatography; TFE, trifluoroethanol
Abstract
Syndecans are membrane proteoglycans regulating extracellular matrix assembly, cell adhesion and signaling. Their ectodomains can be shed from the cell surface, and act as paracrine and autocrine effectors or as competitors of full-length syndecans. We report the first biophysical characterization of the recombinant ectodomains of the four human syndecans using biophysical techniques, and show that they behave like flexible random-coil intrinsically disordered proteins, and adopt several conformation ensembles in solution. We have characterized their conformational landscapes using native mass spectrometry (MS) and ion-mobility MS, and demonstrated that the syndecan ectodomains explore the majority of their conformational landscape, from minor compact, globular-like, conformations to extended ones. We also report that the ectodomain of syndecan-4, corresponding to a natural isoform, is able to dimerize via a disulfide bond. We have generated a three-dimensional model of the C-terminus of this dimer, which supports the dimerization via a disulfide bond. Furthermore, we have mapped the NXIP adhesion motif of syndecans and their sequences involved in the formation of ternary complexes with integrins and growth factor receptors on the major conformations of their ectodomains, and shown that these sequences are not accessible in all the conformations, suggesting that only some of them are biologically active. Lastly, although the syndecan ectodomains have a far lower number of amino acid residues than their membrane partners, their intrinsic disorder and flexibility allow them to adopt extended conformations, which have roughly the same size as the cell surface receptors (e.g., integrins and growth factor receptors) they bind to.
Introduction
The four mammalian syndecans (SDC-1–4) are membrane proteoglycans, which regulate numerous biological processes including cell signaling [1], [2], cell adhesion, migration, and interactions with their microenvironment [3], angiogenesis [4], lipid metabolism [5], growth factor recruitment and presentation to their receptors [6], extracellular matrix assembly [7], [8], and tissue regeneration [9]. They exert their functions via a large interaction network, which comprises 347 partners [10]. Additionally, numerous studies pointed out the role of the syndecans in several pathological processes like tumorigenesis [2], host-pathogen interactions [11], neurodegenerative diseases [12], [13] and inflammation [14].
The syndecans comprise an ectodomain (ED) ranging from 20 to 40 kDa, a transmembrane domain and a cytosolic domain containing a variable region flanked by two constant regions. They bear glycosaminoglycan (GAG) chains, which are covalently attached to their protein core through conserved serine-glycine motifs. The four syndecans contain heparan sulfate chains at their N-terminus but SDC-1 and −3 also carry chondroitin sulfate chains in the juxtamembrane region [1], [2]. Their shedding by various proteinases leads to the release of their soluble ectodomains in the extracellular milieu, where they can act as paracrine or autocrine effectors and competitive inhibitors of cell surface proteoglycans [15], [16]. The syndecan ectodomains play a role in wound repair, and modulate the behavior of cancer cells and their microenvironment [1]. Shed syndecan-2 inhibits angiogenesis [4], and enhances tumorigenic activities of cancer cells [17] whereas shed syndecan-1 translocates to the nucleus of cells delivering growth factors and inhibiting histone acetylation [18]. This ectodomain is also involved in chemotherapy resistance via the epidermal growth factor receptor in cancer [19], and inhibits early stages of liver fibrogenesis by binding to transforming growth factor β1 and interfering with its action [20]. Shed syndecan-4 promotes immune cell recruitment, extracellular matrix remodeling and mitigates cardiac dysfunction in response to lipopolysaccharide [21]. Shed syndecans have been proposed as biomarkers or as prognostic predictors in several diseases [22].
However, despite the variety and importance of their biological roles, very few structures are available for the syndecans [10], which prevents detailed studies of their molecular mechanisms of action. Indeed, the structures of the transmembrane domain of SDC-2 and SDC-4 and of the cytoplasmic domain of SDC-1 and -4 alone or in complex with other proteins have been determined (reviewed in [10]), but no structural data are available for the ectodomains of syndecans, which have been predicted to lack ordered structure [23]. This prompted us to characterize the ectodomains of the four human syndecans (ED 1-4) by several biophysical methods (circular dichroism, CD, dynamic light scattering, DLS, and size exclusion chromatography - small angle X-ray scattering, SEC-SAXS) to determine their content in disorder and secondary structure, their shape and size (i.e., the maximum dimension Dmax, the radius of gyration Rg and their hydrodynamic radius Rh), and their flexibility. Their conformational landscape, ranging from compact, globular-like conformations, to extended conformations was further explored by native mass spectrometry (ESI-MS) and ion-mobility mass spectrometry (ESI-IM-MS) [24]. The fact that the NXIP adhesion motif and the sequences involved in the formation of ternary complexes with integrins and growth factor receptors are not accessible in all the ectodomain conformations suggests that only some of them are biologically active. The extended conformations of these small disordered proteins (20–40 kDa) have approximately the same size as the large cell surface receptors they bind to (i.e., integrins and growth factor receptors).
Results
The syndecan-4 ectodomain is able to dimerize
The human ectodomains ED-1–4 were expressed as recombinant proteins in E. coli with a 6xHis tag at the N-terminus and a FLAG tag at the C-terminus (Supplementary Table S1). Since the ectodomains were expressed in E. coli they did not bear GAG chains but it has been previously shown that recombinant syndecan EDs lacking GAG chains are biologically active by supporting cell adhesion (ED-1, [25]), mediating heparinase-enhanced migration of myeloma cells [26], and inhibiting osteoclast differentiation [27]. Furthermore, several syndecan partners (e.g. integrins) bind to the protein core of syndecans but not to their GAG chains. The natural isoform 2 of SDC-4, lacking the transmembrane and cytosolic domains and mimicking the proteolytic ectodomain shedding [28], was used to study the ectodomain of SDC-4, and is referred to as ED-4 throughout this study. The ectodomains were purified by two affinity chromatography steps using Ni-NTA agarose and anti-FLAG agarose. An additional step of purification by size exclusion chromatography (SEC) was carried out for SAXS, MALLS, DLS, ESI-MS and ESI-IM-MS experiments as described in the Methods section.
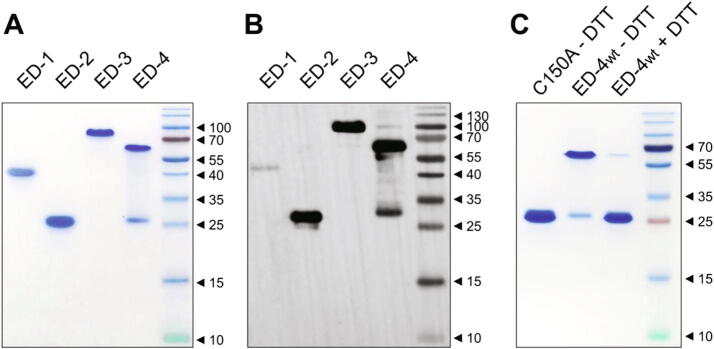
A single band was detected by SDS-PAGE (Fig. 1A) and Western blot (Fig. 1B) for ED-1, ED-2 and ED-3. In contrast, two major bands migrating with an apparent molecular weight of 28 kDa and 63 kDa were observed for ED-4 (Fig. 1A and 1B). The reduction of ED-4 by dithiothreitol (DTT) led to a marked decrease in the 63-kDa band, and an increase in the 28-kDa band suggesting that the dimerization may occur via disulfide bonding. The unique cysteine residue of syndecan-4 was mutated into alanine (C150A), and the mutant migrated as a single band confirming that the role of the disulfide bond in the dimerization process (Fig. 1C). Full-length syndecans homo- and heterodimerize via a conserved transmembrane GxxxG motif [29], but this is the first report that the ectodomain of syndecan-4 is able to dimerize.
Fig. 1.
SDS-PAGE and Western blot analysis of the recombinant ectodomains of human syndecans. 2 µg of proteins purified by affinity chromatography and gel filtration were analyzed in non-reducing conditions by SDS-PAGE and stained by Coomassie blue (A), and by Western blot with an anti-FLAG antibody and chemiluminescence detection (B). The wild-type ectodomain of syndecan-4 (±2 mM DTT), and the C150A mutant were analyzed by SDS-PAGE (C). Molecular masses indicated are in kDa.
The molecular weight of the syndecan EDs were higher than the theoretical values when calculated by SDS-PAGE and size-exclusion chromatography but consistent with them and with the existence of a dimer of ED-4 when determined by ESI-MS and SEC-MALLS (Supplementary Table S2 and Supplementary Fig. S1). The anomalous migration of syndecan EDs on SDS-PAGE was previously attributed to the high proline content of the ectodomains [30] but this could be likely due to the presence of intrinsic disorder [23], predicted in the four EDs by a metapredictor of disorder (metaPrDOS, Supplementary Fig. S2A). ED-3 migrated in SDS-PAGE much slower than to the other ectodomains with an apparent Mw of 89 kDa. This was consistent with the apparent Mw of murine ED-3 (90–120 kDa) [30] but far larger than the theoretical Mw of ED-3. Its slow migration could not be attributed solely to its higher number of amino acid residues (Supplementary Table S2) and/or to its content in intrinsic disorder calculated from the deconvolution of CD spectra (69%, Fig. 2), which was identical to that of ED-1. Its mucin-like sequence of approximately 200 amino acids rich in threonine, serine and proline residues (amino acid residues 115–302), which accounts for 14% of ED-3 amino acid residues and for less than 10% for the other ectodomains, likely contribute to its slower electrophoretic mobility.
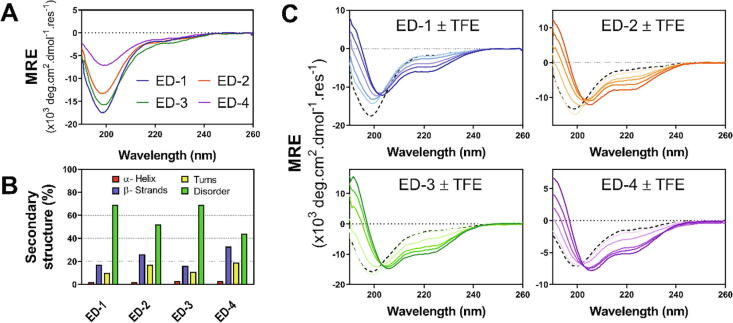
Fig. 2.
Circular dichroism experiments. A) CD spectra of the syndecan ectodomains (MRE: mean residue molar ellipticity). B) Percentage of secondary structures and random coil calculated from the deconvolution of CD spectra using Dichroweb as described in the Methods section. C) CD spectra of the syndecan ectodomains recorded in presence of increasing concentrations of trifluoroethanol (TFE) from 0 % (black dotted line) to 60 % v/v (the darkest color).
The four syndecan ectodomains are enriched in intrinsic disorder
To experimentally estimate their amount of intrinsic disorder, the syndecan ectodomains were then analyzed by circular dichroism (Fig. 2A). The largest ectodomains, ED-1 and ED-3, were the most disordered (69 % of random coil), whereas the smallest ones, ED-2 and ED-4, contained 52 % and 44 % of random coil respectively (Fig. 2B). The content in α-helix of the four ectodomains was very low (<3 %), but they contained a significant amount of β-strands, which was higher in ED-2 and ED-4 (26 % and 33 % respectively) than in ED-1 and ED-3 (16 % and 17 % respectively) (Fig. 2B). β-strands were predicted to be located in the C-terminal half of ED-1 and ED-4 but distributed along ED-2 and ED-3 sequences (Supplementary Fig. S2A). Although they are extensively disordered, the syndecan EDs contain a significant amount of hydrophobic residues (37–52 %) of the total amino acid residues, which are usually depleted in IDPs [31].
To determine if the ectodomains were able to undergo a disorder-to-order transition, the Molecular Recognition Features (MoRFs), which are short disordered sequences able to undergo this transition upon binding to a partner, were predicted. Two major MoRFs were predicted at the N- and C-termini of ED-1 and ED-3 and a short one in their central part (Supplementary Fig. S2A). In contrast, almost the entire sequences of ED-2 and ED-4 were predicted to be a MoRF, except for the N- and C-termini (Supplementary Fig. S2A). CD experiments were performed in presence of increased concentrations of 2,2,2-trifluoroethanol (TFE) to check if the ectodomains were able to undergo a disorder-to-order transition. An increase in α-helix in ED-1 (15 %) and ED-3 (31 %) in presence of 60 % TFE was associated with a decrease in disorder (22 % and 29 % respectively in ED-1 and ED-3) (Fig. 2C, Supplementary Fig. S3), suggesting that they might undergo a disorder-to-order transition. The gain of α-helix in syndecan-3 could be a cooperative process. In contrast, the increase in α-helix in ED-2 (21 %) and ED-4 (12 %) was associated with a decrease in β-strands (9 % and 11 % respectively) and a low or negligible decrease in disorder (6 % and 2 %, respectively) (Fig. 2C).
CD spectra were also collected in presence of 3 M and 5 M guanidinium chloride, which destabilizes folded proteins but stabilizes polyproline II helix [32]. A small increase in signal in the 220–230 nm region was observed for the four ectodomains in presence of guanidinium chloride suggesting that they were not able to form a significant amount of polyproline II helix (Supplementary Fig. S2B). CD spectra were also collected in presence of two chemical chaperones in order to determine if they were able to stabilize or favor any conformation of the ectodomains. The stabilizing osmolyte chaperone trimethylamine N-oxide, which shifts the folding-unfolding equilibrium toward the folded state, induced changes in the CD signal (Supplementary Fig. S2C), which could be related to a decrease in disorder. The hydrophobic chaperone 4-phenylbutyric acid, which prevents aggregation, did not induce significant changes in CD spectra (Supplementary Fig. S2C).
The syndecan ectodomains are highly extended and flexible in solution
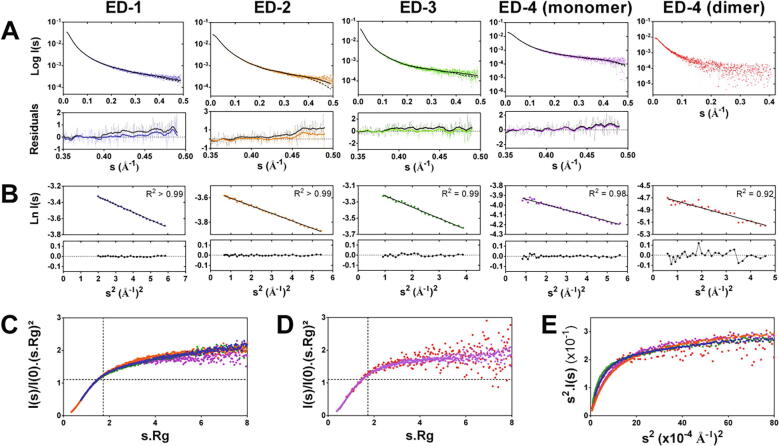
The four syndecan ectodomains were analyzed by SEC-small-angle X-ray scattering (SEC-SAXS) (Fig. 3 and Supplementary Fig. S4). ED-2 and the monomeric form of ED-4 had a similar radius of gyration (42.9 ± 0.33 Å and 41.53 ± 0.67 Å respectively), and a similar maximum intramolecular distance Dmax (185 Å and 180 Å respectively) (Table 1 and Supplementary Fig. S5). Their pair distribution functions were superimposed indicating that they share similar conformations despite their low sequence identity (<20 %) (Supplementary Fig. S5). ED-3 had the largest Rg (65.0 ± 0.62 Å) and Dmax (300 Å) whereas the Rg of ED-1 was 52.7 ± 1.43 Å, and its Dmax was 230 Å (Table 1 and Supplementary Fig. S5). The experimental values of the radius of gyration of the four ectodomains were close to the theoretical values calculated using the Flory’s equation for a random coil protein containing the same number of amino acid residues (Table 1). Furthermore, the Flory exponential scaling factors calculated for the syndecan EDs were 0.58 and 0.59, similar to the value calculated for chemically disordered proteins (0.60) which are more extended than IDPs [33]. The hydrodynamic radius (Rh) of the ectodomains, determined by size-exclusion chromatography-dynamic light scattering (SEC-DLS), ranged from 34.1 Å (ED-2) to 50.1 Å (ED-3), and their Rg/Rh ratio, that provides information about protein shape and compactness, ranged from 1.16 (ED-4 monomer) to 1.31 (ED-4 dimer) (Table 1). These values were far higher than those of globular proteins (0.778) and close to those reported for prolate ellipsoids (1.36–2.24) [34] (Table 1). The Rh, Rg and Dmax of the ED-4 dimer were larger than that of the monomer (Table 1 and Supplementary Fig. S5). The shape of the normalized Kratky plots of the four EDs (Fig. 3C) and of the ED4-dimer (Fig. 3D) was consistent with those of IDPs [35]. The Porod exponents of the EDs were similar (1.7 for ED-1, ED-3 and the monomer of ED-4, 1.8 for ED-2, and 1.9 for the dimer of ED-4) indicating that they all are flexible, the Porod exponent being equal to 2 for fully flexible systems [36]. The EDs flexibility was also demonstrated by the plateau observed in the Kratky-Debye plots (Fig. 3E). The syndecan ectodomains are thus disordered, flexible, and extended proteins in solution.
Fig. 3.
SEC-SAXS analysis of the syndecan ectodomains. A) SEC-SAXS scattering intensities (colored curves) and fitting to random coil (RC, black solid line) and pre-molten globule (PMG, black dotted line) models using EOM. The residual plots for the PMG (black) and RC models (colored) are indicated as well as the smoothed residuals (thick lines with the same color legend). B) Determination of the radius of gyration according to the Guinier approximation at low angle. The residual plots and the coefficient of determination (R2) are indicated. Normalized Kratky plots of C) the four syndecan ectodomains (EDs), and D) the monomeric and dimeric forms of the ectodomain of syndecan-4. E) Scaled Kratky-Debye plots of the syndecan ectodomains (blue: ED-1, orange: ED-2, green: ED-3, purple: ED-4 monomer, red: ED-4 dimer).)
Table 1.
Size, shape and compactness of the syndecan ectodomains. The radii of gyration were calculated experimentally from SEC-SAXS data by Guinier approximation and theoretically using Flory’s equation (see Methods section). Their hydrodynamic radii (Rh) were determined by SEC-DLS. The C150A mutant was used to calculate the Rh of the ED-4 monomer.
| ED-1 | ED-2 | ED-3 | ED-4 (monomer) | ED-4 (dimer) | |
|---|---|---|---|---|---|
| SAXS | |||||
| Rg (Å) (Guinier approximation Mean ± error) | 53.30 ± 0.3 | 42.90 ± 0.33 | 65.00 ± 0.62 | 41.53 ± 0.67 | 59.25 ± 2.27 |
| Dmax (Å) (Pair distribution function ) | 230 | 185 | 300 | 180 | 240 |
| Theoretical Rg calculated using Flory’s equation | |||||
| Folded protein | 18.5 | 15.9 | 20.5 | 16.1 | 19.7 |
| Pre-molten globule (Mean ± error) | 46.3 ± 0.6 | 35.2 ± 0.4 | 55.9 ± 0.8 | 36.3 ± 0.5 | 52.1 ± 0.7 |
| Random coil (Mean ± error) | 54.2 ± 11.8 | 39.6 ± 7.8 | 67.4 ± 15.7 | 40.9 ± 8.2 | 62.2 ± 14.1 |
| Hydrodynamic properties (SEC-DLS) | |||||
| Rh (Å) (Mean ± error) | 44.9 ± 0.8 | 34.1 ± 0.7 | 50.1 ± 1.3 | 35.7 ± 0.8 | 45.4 ± 1.2 |
| Shape factor Rg/Rh | 1.19 | 1.26 | 1.30 | 1.16 | 1.31 |
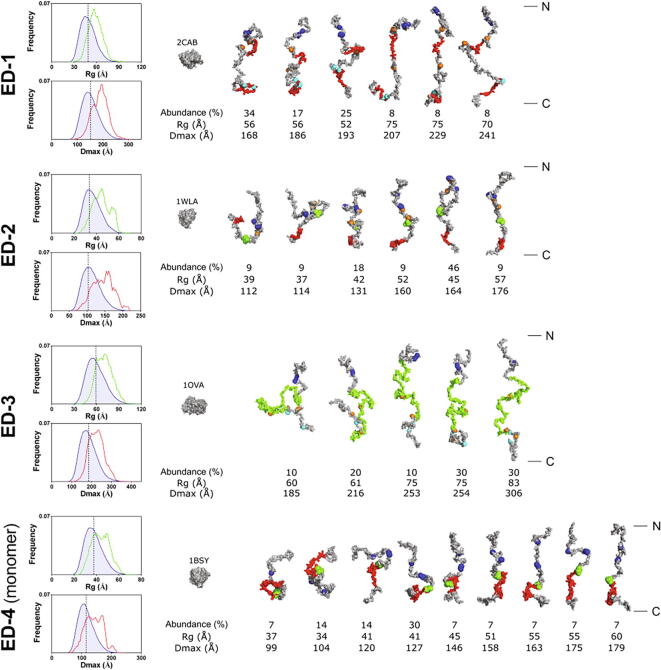
SAXS scattering intensities were then analyzed using the Ensemble Optimization Method (EOM, [37]) in order to confirm that they behave as random coil IDPs, and to explore their conformational landscape. Two different pools of conformers corresponding to premolten globule (PMG) and random coil (RC) proteins were generated. The PMG model failed to correctly fit scattering intensities at high angles (>4 Å) whereas the RC model perfectly fitted the experimental data (Fig. 3A and Table 2), and was thus selected for further analysis of the syndecan EDs. The values of Rflex for the selected ensembles of the four ectodomains (81–86 %) were very close to those of the pools of random coil conformers (84–86 %) (Table 2), in agreement with the high entropy and conformational heterogeneity of IDPs [31]. The distributions of Rg and Dmax of the ensembles fitting experimental SAXS data were shifted to higher values than those of the random coil ensemble. The distributions of Rg and Dmax of the selected ensembles were roughly centered on Rg ~ 56 Å and Dmax ~ 193 Å for ED-1 and on Rg ~ 70 Å and Dmax ~ 225 Å for ED-3. The multimodal distribution of both Rg and Dmax for ED-2 and ED-4 might indicate the existence of several subpopulations of conformations for these two ectodomains (Fig. 4). Selected models of the conformation ensembles of ED1-4 are presented in Fig. 4. Biologically active sequences of syndecans (i.e., NxIP sequences promoting cell adhesion and synstatin sequences involved in the formation of binary and ternary complexes with growth factor receptors and/or integrins [26], [38], [39], [40]) were mapped onto the models of the ectodomains generated from EOM analysis. As shown on Fig. 4, these sequences are not accessible in all the conformations of the ectodomains, some of them being partially masked or buried in some conformations. This suggests that only certain conformations of the syndecan ectodomains are biologically active at the cell surface.
Table 2.
Ensemble Optimization Method (EOM) analysis of SAXS data. Fitting assessment between the experimental and theoretical scattering intensities generated by EOM for the pre-molten globule and the random coil ensembles. The CorMap test is a goodness-of-fit test [41].
| EOM analysis | ED-1 | ED-2 | ED-3 | ED-4 (monomer) |
|---|---|---|---|---|
| Pre-molten globule (PMG) model | ||||
| Reduced χ2 | 0.904 | 0.947 | 0.765 | 0.636 |
| CorMap p-value | 0.0002 | 0.000 | 0.008 | 0.004 |
| Random-coil (RC) model | ||||
| Reduced χ2 | 0.616 | 0.632 | 0.663 | 0.597 |
| CorMap p-value | 0.224 | 0.227 | 0.0645 | 0.0618 |
| Rflex ensemble (%) | 82.71 | 86.17 | 81.18 | 84.02 |
| Rflex pool (%) | 84.31 | 85.60 | 84.59 | 83.69 |
| Rσ | 0.92 | 2.47 | 0.85 | 1.01 |
Fig. 4.
Ensemble Optimization Method analysis of SAXS data. The Rg and Dmax distributions derived from EOM analysis are shown on the left. The blue line shows the distribution of the initial pool of random coil conformers, whereas the green and red lines represent respectively the Rg and Dmax distributions of the selected conformer ensembles fitting the SAXS data. The dotted vertical black line represents the average Rg and Dmax of the random coil initial pool. The models issued from EOM deconvolution carried out with the parameters generated for RC proteins are represented with their relative abundance (%), Rg and Dmax. Backbone and side chains of EOM conformers were reconstructed to visualize biologically relevant sequences or amino acid residues. Serine residues bearing heparan sulfate or chondroitin sulfate chains in the proteoglycan form of syndecans are blue and cyan spheres respectively. The synstatin sequences of ED-1 (residues 93–120 and 210–240) and ED-4 (residues 87–131) are in red. The binding site of CD148 on ED-2 (residues 123–140) is in red and the NXIP motif of ED-2 (residues 95–98) and ED-4 (residues 87–90) is represented as green spheres. The mucin-like sequence of ED-3 is in green (residues 115–302), whereas the residues corresponding to single nucleotide polymorphisms are represented as orange spheres. Globular proteins with SASA values similar to those of the most compact conformations of the EDs are drawn to scale with their PDB code (PDB 2CAB: carbonic anhydrase, 1WLA: myoglobin, 1OVA: ovalbumin, 1BSY: β lactoglobulin). N- and C-termini of the EDs conformers are indicated on the right.
The conformational landscape of syndecan ectodomains
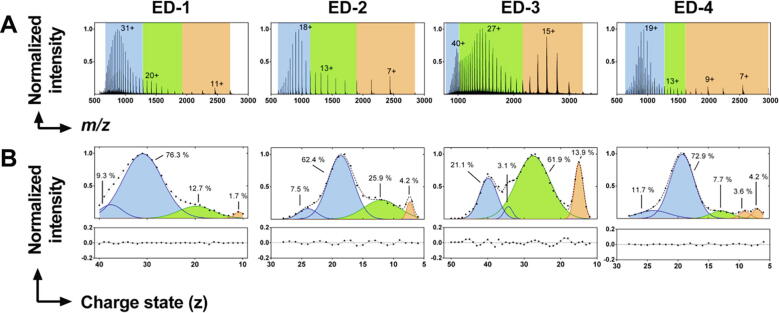
Electrospray ionization-mass spectrometry (ESI-MS) in native conditions provides information on the variable degrees of compactness of IDPs. The charge state distributions (CSDs) determined by ESI-MS are related to the global compactness of proteins upon their transfer from solution to gas phase, the highest charge states corresponding to the most extended conformations [42]. The CSDs of the four ectodomains were trimodal, showing that the ectodomains of syndecans could adopt preferential conformations (Fig. 5A). Gaussian functions were fitted to the CSD envelopes to determine the relative proportions of the three major conformational ensembles. The most extended conformations (i.e. the highest charge states) were the most abundant for ED-1 (86 %), ED-2 (70 %) and ED-4 (85 %) but accounted for only 24 % for ED-3, in which the population with intermediate charge states predominated (Fig. 5B). Most compact conformations were found in minor amount for ED-1 (2 %), ED-2 (4 %), and ED-4 (8 %) and in larger amount for ED-3 (14 %).
Fig. 5.
Analysis of the syndecan ectodomains by native ESI-MS. A) Charge state distribution (CSD) of the syndecan ectodomains (EDs). Blue: extended conformations, green: intermediate conformations, orange: more compact conformations. The charge states on which the populations are centered are indicated. B) Deconvolution of the CSDs by Gaussian fitting, and their respective residual plots. The relative abundance (%) of each population is indicated. The color legend is the same as in A).
The average solvent-accessible surface area (SASA) of the ectodomains and of their most extended and most compact conformations were calculated (Supplementary Table S3). The average SASA values of ED-2 (16 074 Å2) and of the monomer of ED-4 (17 780 Å2) were in the same range, as were those of ED-1 (29 653 Å2) and ED-3 (28 624 Å2) although the ED-3 sequence is 30 % longer than the ED-1 sequence. The average SASA values being related to the extent of protein ionization, this is consistent with the fact that ED-3 might adopt more compact conformations than ED-1 as mentioned above. However, the SASA values of the most extended conformations of the ectodomains were directly proportional to the number of their amino acid residues, ranging from 18 269 Å (ED-2) and 18 844 Å (ED-4 monomer) to 31 104 Å (ED-1) and 42 071 Å (ED-3) confirming that ED-3 is also able to adopt extended conformations.
The average charges of the most compact conformations of the ectodomains were very close (±1 proton) to that reported for globular proteins with a similar number of amino acids residues [43]. The SASA values of the most compact conformation of ED-1 and ED-3 were 17 % and 23 % higher than those of the globular proteins carbonic anhydrase I and ovalbumin (see details in the Methods section) (Supplementary Table S3). In contrast, the SASA values of the most compact conformation of ED-2 and ED-4 were slightly lower than those of the globular proteins myoglobin and β-lactoglobulin (Supplementary Table S3), suggesting that ED-2 and ED-4 are able to adopt compact, globular-like, conformations. In conclusion, ED-1, −2 and −4 primarily adopted fully extended conformations in the gaseous phase as they did in solution, whereas ED-3 forms partially extended conformations with a low but significant contribution of more compact states. In addition, the most compact conformations of ED-2 and ED-4, present in minor amounts, are globular-like. This suggests that the syndecan EDs cover a large part of their conformational landscape, which was further defined by ion-mobility mass spectrometry (ESI-IM-MS) experiments as detailed below. The C150A mutant of ED-4, unable to dimerize, was used for these experiments, and the dimer of ED-4 was studied by selecting odd numbers of charge states for ED-4.
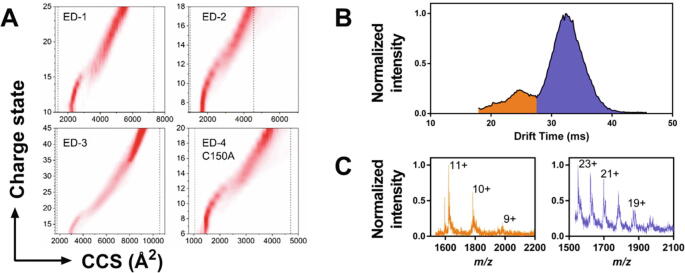
The collision cross-section (CCS) distributions of the ectodomains, calculated by ESI-IM-MS, increased as a function of their charge state due to Coulomb repulsion as expected (Fig. 6A). The boundaries of the accessible CCS space of the ectodomains were roughly estimated using a simple model based on the volume of the amino acid residues in either a sphere or a cylinder as representatives of the most compact and the most extended conformations respectively [44]. ED-1 explored approximately half of its accessible CCS space (54 %) whereas ED-2 (73 %), ED-3 (70 %) and ED-4 (68 %) explored about 70 % of their CCS space (Fig. 6A). These results, in combination with the large range of charge states detected, are consistent with the high flexibility of the four EDs, and confirm that they can adopt extended elongated conformations. The predominance of high charge states with relatively large CCS values suggests that these elongated structures represent a majority of the sampled conformational space. In addition, low charge states with low CCS values confirmed the existence of a more compact conformational ensemble (Fig. 6A).
Fig. 6.
Ion mobility-mass spectrometry (ESI-IM-MS) experiments. A) Distributions of the collision cross sections (CCS) of the syndecan ectodomains as a function of charge state. The dashed lines indicate the limits of the CCS space as estimated in each case from the model of Beveridge et al. [44]. B) Global distribution of ED-4, containing both the monomeric (orange) and dimeric (blue) forms. Their extracted charge state distributions are displayed in (C).
A transition from a compact conformational ensemble to a more extended one occurred in the four ectodomains as the charge state z increased, yielding to a bimodal CCS distribution. Beveridge et al. [45] proposed that such transition may be related to the distinct desolvation mechanisms of compact and elongated conformations in the electrospray ionization process. The transition between the compact and extended states occurs when the radius of an ion of charge state z reaches the maximum size of a solvent droplet bearing a charge z, the so-called Rayleigh limit. It occurred at z = 8–13 for ED-2 and ED-4, at z = 11–15 for ED-1, and at z = 16–18 for ED-3. The charge state where the transition in the CCS distributions arose for each ED corresponded to 80 to 85 % of the corresponding Rayleigh limit estimated according to Mora et al. [46]. These results were consistent with the existence of two different conformational ensembles of the syndecan ectodomains, each following a single but different desolvation mechanism, and one transitional state where both desolvation mechanisms occur [45]. No transition was observed in the CCS distribution of the ED-4 dimer suggesting that compact conformations were not favored for the dimer. Its conformations were thus investigated further.
The ED-4 dimer adopts extended conformations
Two peaks were observed in the global ESI-IM-MS arrival time distribution recorded for ED-4 (Fig. 6B). The mass spectrum extracted for the first peak (drift time 25 ms) displayed a charge state distribution consistent with 9–11 positive charges and the mass of ED-4 monomer. The mass spectrum extracted for the second peak (drift time 32 ms) corresponded to 18–23 positive charges and to the mass of the ED-4 dimer (Fig. 6C). The drift time of the dimer was higher than that of the monomer with half charge. The drift time of a molecular ion being proportional to the ratio of its CCS to its charge z, this indicated that the CCS of the ED-4 dimer was at least twice that of the monomer. This is consistent with the association of two ED-4 monomers retaining a relatively extended conformation, which was supported by the values of the hydrodynamic radius of ED-4 monomer (36 Å) and dimer (45 Å), and of the radius of gyration (41.5 Å and 59.3 Å for ED-4 monomer and dimer respectively). The monomers could bind to each other via their C-terminus containing the unique cysteine residue, with both flexible ectodomains retaining their flexibility and pointing in opposite directions. Molecular dynamics simulations were carried then out on the 40 amino acid residues located at the C-terminus of ED-4 to investigate this possibility. The ensemble-averaged properties of the system, including the plots of heat capacity, ensemble-averaged fractions of secondary structure, fraction of disulfide bonds, and the radius of gyration were described in Supplementary Fig. S6A-D. According to these data, the system was largely unstructured at room temperature, with an almost equal amount of α-helical and β-sheet structure, and remained a dimer even at high temperatures. Because the fraction of disulfide bonds was substantial even at higher temperatures, they probably largely contribute to the stabilization of the dimer. The dimer having been experimentally studied at room temperature, we analyzed its conformational ensemble at 300 K, which also corresponded to the heat-capacity peak and to the switch between β-sheet and α-helical structure midpoint (Supplementary Figs S6A and S6B).
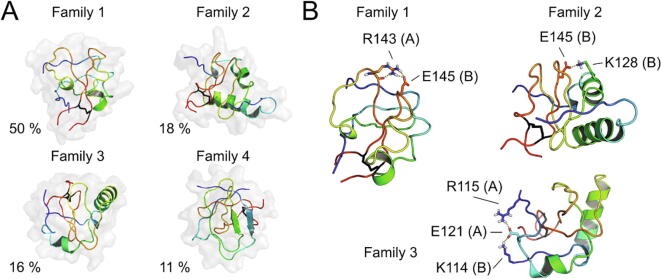
Four families constituted 95 % of the ensemble and their representatives, after conversion to all-atom representation and refinement, are shown in Fig. 7A. The disulfide-bonded C-termini were extended and parallel to each other in all conformations but did not form a regular parallel β-sheet. Family 1 (50 % of the population) was largely disordered, with only two short α-helices in chains A (Ser133-Gly137) and B (Ser130-Gly137) where A and B serve only to distinguish both chains of the dimer (Fig. 7A). There was also a distorted β-hairpin in the Pro145-Ser132 region of chain A (not marked in the Fig.). The helical sections were largely extended in the representative conformation of family 2 (18 % population) from Glu123 to Gly137 in chain A and from Asp124 to Ser126 in chain B. Chain B also contained a distorted β-hairpin from Ile140 to Leu147 in chain B (Fig. 7A). Similarly, sections of the same regions remained α-helical in the representative conformation of family 3 (16 % population), extending from Glu123 to Asn127 in chain A and from Glu123 to Gly137 in chain B, the helix in chain A being remarkably shorter (Fig. 7A). In the representative conformation of family 4 the only regular structure was a short anti-parallel β-sheet composed of the Glu145-Ala148 sequence in chain A with Ser130-Ser133 forming a part of a distorted β-hairpin that extended from Ser130 to Ala148. Another distorted β-hairpin extending from Asn139 to Val146 was observed in chain B (Fig. 7A).
Fig. 7.
Models of the C-terminus of the ED-4 dimer. A) Representative conformations of the dominant conformational families of the 40 last residues of ED-4 in a dimeric form, which constitute 95 % of the ensemble, with their respective abundance. The disulfide bond (present in all conformations) is shown as black sticks. Each chain is colored from blue to red from the N- to the C-terminus. B) The representative conformations of families 1, 2, and 3 with side chains forming salt bridges shown in detailed stick representation. The orientation of the dimer differs from the orientation displayed in A to expose the side chains forming salt bridges.
Consistent with the small value of the ensemble-averaged radius of gyration (Supplementary Fig. S6D), all structures encompassing the C-terminal 40 residues were compact. The maximum particle dimension (Dmax) and radius of gyration (Rg) of the whole syndecan-4 dimer calculated from SAXS data were 240 Å and 59.25 Å respectively, suggesting a highly extended structure. This elongated structure could be formed provided that the N-termini of the dominant conformations of the C-terminal section studied here were not restricted to point in the same direction. As shown in Supplementary Fig. S7A-D, where the representative conformations were drawn in tube representation (the width of the tube being related to the flexibility of a region), and where other conformations of a family were superimposed, the N-termini were very flexible in the dominant families 1–3 so that they could assume any orientation, while the less flexible N-terminal regions of the structures of family 4 pointed in different directions. The sequence of the C-terminal part of ED-4 contains numerous positively and negatively charged residues and polar residues. Therefore, the compact shape of the dimer C-terminus at 300 K was likely stabilized by salt bridges in addition to the Cys150A-Cys150B disulfide bond. Inter- and intra-chain salt bridges, shown in the representative conformations of families 1, 2, and 3, occurred on the protein surface (Fig. 7B). In family 1, the strongest one was the inter-chain bridge between Arg143A and Glu145B, which connected two loop regions exhibiting reduced flexibility compared to the other parts of the structure (Fig. 7B). In the representative structure of family 2, Glu145B forms an intra-chain bridge with Lys128A (Fig. 7B). However, the sequence of chain A containing Arg143 was close to Glu145B and, therefore, the Arg143A-Glu145B salt bridge could also be formed in the conformations of family 2. In the representative conformation of family 3, there was an inter-chain salt bridge between Lys114B and Glu121A, which was also engaged in an intra-chain salt bridge with Arg115A (Fig. 7B).
Discussion
We report here the first characterization of size, shape, secondary structure, and conformational landscape of the ectodomains of the four human syndecans, that play major roles as paracrine or autocrine effectors or as competitors of full-length syndecans and are involved in angiogenesis and in diseases such as cancer and fibrosis. We have shown that they are enriched in intrinsic disorder, and provided experimental support to in silico predictions [23]. The juxtamembrane region of the four syndecan ectodomains are predicted to be highly disordered, which is consistent with this region being highly sensitive to proteolytic cleavage, including the ED shedding sites [10], [16]. Further cleavage sites by metalloproteinases exist along the sequence of ED-1, ED-2, and ED-4 [10]. The NXIP motif, required for cell adhesion, and found in the sequence of human ED-2 and ED-4, is also intrinsically disordered. The ectodomains contain from ~ 15 % (ED-1 and ED-3) to ~ 30 % (ED-2 and ED-4) of β-strands, but few, if any α-helices. However, they are able to form α-helices in presence of trifluoroethanol. This is in agreement with the partial folding into α-helix of the ectodomain of syndecan-4 during molecular dynamic simulations carried out at the glycocalyx level [47].
Full-length syndecans form homo-and heterodimers [29] but our study is the first one to report that the isoform-2 of syndecan-4 (ED-4) dimerizes via a disulfide bridge mediated by a unique cysteine residue located at its C-terminus. Indeed, the mutation of this cysteine residue into alanine (C150A) nearly abolishes the dimerization process. The dimer of ED-4 remains fully disordered and flexible as shown by SAXS and MS experiments. Ion-mobility MS data suggests that ED-4 dimerizes via their C-terminus with the monomers extending in opposite directions, which is consistent with the values of the radius of gyration and of the hydrodynamic radius calculated for the dimer. No dimer of ED-2 has been detected, and minute amount of a dimer of ED-1 and ED-3 has been identified during ESI-MS experiments, which is consistent with the self-association of syndecan 3 mediated by a motif in the transmembrane domain and the ectodomain flanking region [48]. The ectodomains 1, 2 and 3 do not contain any cysteine residue.
SEC-SAXS data show that the syndecan ectodomains are highly flexible and extended in solution, and behave like random coils rather than pre-molten globule IDPs. The four ectodomains display three major ensembles of conformations with various degrees of compactness in the gas phase as shown by ESI-MS and ESI-IM-MS. ED-1 explores a reduced part of its conformational space (54 %) compared to the three other ectodomains (~70%), suggesting that it might adopt a restricted number of conformations. The most compact conformations are the less abundant for the four EDs. The most extended conformations account for 70 % to 85 % of the conformations for ED-2, ED-1 and the monomeric form of ED-4. In contrast, ED-3 exists mostly as partially extended conformations with a low but significant contribution of more compact states, although it contains the same amount of disorder but is longer than ED-1. Moreover, ED-3 is able to form more compact conformations than the other ectodomains as shown by its SASA and the intensity of low and intermediate charge states by ESI-MS. ESI-MS detects different conformers of a protein with variable degree of compactness and the distribution of these molecular species without averaging them in contrast to SAXS. The intrinsically disordered proteins sample a 3 to 5-fold broader conformational space in the gas phase than in solution. The absence of water in ESI-MS increases intramolecular electrostatic interactions, which can trigger changes in protein conformations and generate additional subpopulations including fully collapsed structures, which are not detected in solution [49]. This might explain why the most compact conformations of the ectodomains were only detected by ESI-MS, and why the extended conformations were predominant both in SAXS and ESI-MS experiments for the ectodomains ED-1, ED-2 and ED-4. In contrast, ED-3 adopted extended conformations in solution whereas more partially extended conformations were predominantly detected by ESI-MS together with a significant amount of compact conformations. This might be due to the presence in ED-3 of the mucin-like sequence rich in polar serine and threonine residues, which is predicted to be intrinsically disordered and might participate in electrostatic interactions, and/or in changes induced by the ionization process [50], which might affect ED3 to a higher extent than the other ectodomains.
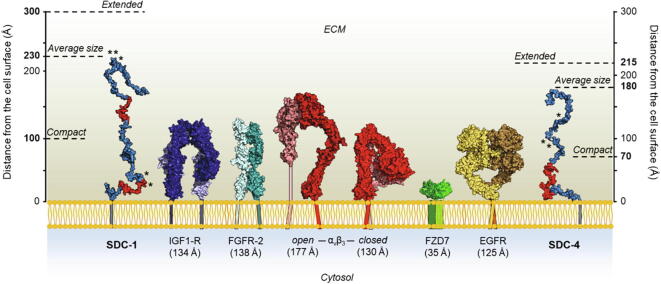
The ectodomains of syndecans have a smaller molecular weight than their cell surface partners such as αvβ3, αvβ5, α3β1 and α4β1 integrins and growth factor receptors including the insulin growth factor receptor (IGFR1), the epidermal growth factor receptor (EGFR), the receptor tyrosine-protein kinase erbB-2 (HER2) and the vascular endothelial growth factor receptor 2 (VEGFR2) [26], [39], [40]. However, the size of their ectodomains, evaluated by the Dmax calculated by SEC-SAXS, varies from ~ 180 Å (ED-4 and ED-2) to 230 Å (ED-1) and 300 Å (ED-3). This is of the same order of magnitude than the size of ligand-bound integrins, such as αIIbβ3 integrin, which extends to 180–200 Å in length upon binding to fibrin [51], and αvβ3 integrin in its open form (177 Å), and of their other cell surface partners (Fig. 8). The intrinsic disorder of the syndecan EDs is thus an important feature allowing them to accommodate to their partner size at the cell surface.
Fig. 8.
Syndecans and their partners at the cell surface. The dimeric form of the ectodomains of IGF1-R (PDB: 5U8R), FGFR-2 (PDB: 1EV2 and 4HWU), αvβ3 integrin in open (PDB: 6AVU) and closed (PDB: 6MS1) conformations, frizzled-7 (FZD7, PDB: 5T44) and EGFR (PDB: 3NJP) in active conformation are drawn to scale. The size of the ED-1 and ED-4 conformers (blue) corresponds to the Dmax determined by SAXS. The synstatin sequences are in red. The sizes of the most extended conformation (extended) and the most compact conformations (compact) of the EDs determined by EOM analysis are indicated. (*): serine residues bearing the GAGs chains. The cytosolic domain of the EDs and of the receptors are not represented.
The conformations of the syndecan ectodomains may be modulated upon binding to their numerous partners [10]. The docking of their partners to the core protein ectodomain might stabilize particular conformations of the ectodomains. This will be investigated by studying complexes formed by syndecan EDs and their ligands. On the other hand, the disorder of syndecan EDs might lead to the formation of fuzzy complexes with their partners. This might be the case for the highly disordered synstatin sequences of human ED-1, namely residues 93–120, which form a ternary complex with IGFR1 and αvβ3 integrin [52], and residues 210–240, which form a ternary complex with VEGFR2 and α4β1 integrin [26]. Mechanical forces could also regulate the conformations of the four ectodomains because conditional disorder may be initiated by such forces [31], and because the syndecans, which link the extracellular matrix to the actin cytoskeleton, act as mechanosensors as reported for syndecan 4 in cardiac fibroblasts [53], and for syndecans 1 and 4 in the endothelial glycocalyx [47], [54]. The glycosaminoglycan chains covalently attached to the syndecan ectodomains can also modulate their conformational space, and this regulation process might be fine-tuned by the number, length or number/type of sulfate groups which vary depending on the cell type, and the physiopathological context. The mutual impact of syndecan-1 protein core and its GAG chains is difficult to analyze [55]. Ongoing experiments with the EDs expressed in eukaryotic cells and therefore bearing their GAG chains will provide new insights on the influence of GAG chains, which are themselves flexible, on the conformations and flexibility of the syndecan EDs. It has been suggested that the fully glycosylated chondroitin sulfate-attachment region of human aggrecan would be less flexible and rather more extended than its non-glycosylated counterpart [56], raising the possibility than GAGs could modulate the flexibility and size of the aggrecan protein core. The chondroitin sulfate chains of biglycan affect the secondary structure of its protein core and may stabilize it during biosynthesis, whereas the GAG chain of decorin has no detectable effect on the structure or conformational stability of its protein core [57]. The role of GAGs on the structure, conformation, and stability of the protein core of proteoglycans is thus difficult to predict, and is likely to be GAG- and protein core-dependent. The ultimate goal will be to determine how ligand binding to the protein core and/or the GAG chains of the syndecan ectodomains influence dimerization of the transmembrane domain, and cell signaling.
Another crucial challenge will be to determine i) if the conformational ensembles of the syndecan ectodomains identified by SAXS and MS are all biologically active whatever the biological context, ii) if only some of them are able to fulfill biological functions, or iii) if the different conformational ensembles are associated with specific functions in a particular context. Indeed, the mapping of biologically active sequences of syndecans (i.e. NxIP sequences of promoting cell adhesion and synstatin sequences involved in the formation of ternary complexes with growth factor receptors and integrins) on the conformations identified by EOM analysis of SAXS data show that these sequences are not accessible in all these conformations. This suggests that only some of them are biologically active. The study of cell-matrix interactions, and cell signaling in presence of the chemical chaperone trimethylamine N-oxide, which induces a decrease in disorder in the syndecan ectodomains, should provide new insights into the biological role of syndecan conformations.
Materials and methods
Expression of the recombinant ectodomains of human syndecans
The constructs for human ectodomains of syndecans (ED-1 to -3), the isoform 2 of syndecan-4 (ED-4), and the ED-4 C150A mutant were synthesized and subcloned in the pET-15b vector by GenScript (https://www.genscript.com). They were then expressed as recombinant proteins containing an N-terminal 6xHis tag and a C-terminal FLAG tag in Escherichia coli BL21 (DE3) cells (Supplementary Table S1) and purified by two affinity chromatography steps followed by size exclusion chromatography for some experiments as detailed below.
Escherichia coli BL21 (DE3) cells were transformed with 80 ng of plasmid, and colonies were grown at 37 °C on ampicillin-containing LB-agar plates. Cells were suspended in LB broth Lennox (Sigma-Aldrich, L3022) with ampicillin 100 µg/ml until the absorbance at 600 nm reached 0.6–0.8. The expression of the EDs was induced by 1 mM isopropyl-β-D-thiogalactopyranoside for 4 h at 37 °C. After centrifugation at 6 000 × g for 10 min, 4 °C, the pellets were resuspended in 10 mM HEPES, 150 mM NaCl pH 7.4 (HEPES buffered saline - HBS) containing 2.5 mM MgCl2, 2.5 mM CaCl2, 100 µg/ml DNAse I (Sigma, D5025), 1 mM CHAPS (Roche), and a protease inhibitors cocktail (Complete EDTA-free, Roche) and lysed for 8 × 30 s with 100 µm-diameter glass beads in a bead beater (BioSpec). The lysates were then centrifuged twice at 10 000 × g for 10 min and the supernatant was loaded on Ni-NTA agarose resin (Qiagen). The purification steps were carried out at 4 °C with a flow rate of 20 ml/h. After washing with HBS containing 10 mM and 20 mM imidazole, the recombinant syndecan EDs were eluted with 100 and 300 mM imidazole in HBS. Eluates were loaded on anti-FLAG M2 agarose resin (Sigma-Aldrich, A2220) and the ectodomains were eluted by the FLAG peptide at 200 µg/ml in HBS. The purified ectodomains were then concentrated with Amicon devices (MWCO 10 kDa). For SEC-SAXS, SEC-MALLS, SEC-DLS, ESI-MS and ESI-IM-MS experiments, the ectodomains were further purified by size exclusion chromatography on a Superdex S200 Increase 10/300 GL column (GE Healthcare) at 10 °C, with a flow rate of 0.5 ml/min. The eluted fractions were concentrated in Amicon devices (MWCO 10 kDa), centrifuged 20 min at 18 000 × g and stored at −20 °C until use.
SDS-PAGE and Western blot
The purified ectodomains were analyzed by SDS-PAGE on a 15 % acrylamide gel and stained overnight with InstantBlue (ISB1L, Sigma-Aldrich). For Western blotting, proteins were transferred on a polyvinylidene fluoride membrane (IPVH20200, Millipore) for 2 h at 250 mA and the membrane was saturated with phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 11.9 mM phosphate, Biosolve) containing 5 % (w/v) fat milk and 0.05 % (v/v) Tween-20 for 90 min at 37 °C. The membrane was incubated overnight at 4 °C with a monoclonal anti-FLAG antibody (F3165, Sigma-Aldrich, 76 ng/ml in blocking buffer), washed 4 times with the blocking solution and incubated with a secondary antibody conjugated to horseradish peroxidase (40 ng/ml, 172–1011, Bio-Rad) for 1 h at room temperature. Finally, the membrane was washed twice in PBS containing 0.05 % (v/v) Tween-20, and twice in PBS alone. The immunoreactive bands were detected by chemiluminescence with the Amersham ECL Prime kit (GE Healthcare), and the signal recorded with a ChemiDoc XRS + system (Biorad) after 5 to 60 s of exposure.
Circular dichroism (CD)
Far-UV CD spectra were recorded in a quartz cuvette at 20 °C with a path length of 0.1 cm in a Chirascan spectrometer (Applied Photophysics, SFR BioSciences UMS3444/US8, Protein Science Facility, Lyon, France), from 260 to 190 nm with a step of 0.5 nm, a response time of 1 s and a bandwidth of 0.5 nm. The purified ectodomains were diluted in 10 mM potassium phosphate pH 7.5 at 150 µg/ml for ED-1 and ED-4 and 100 µg/ml for ED-2 and ED-3. Five spectra were collected and averaged for each sample and the buffer signal was subtracted. CD signal was recorded in millidegrees (mdeg) and converted to mean residue molar ellipticity per residue (deg.cm2.dmol−1.residue-1). Some experiments were carried out in presence of increasing concentrations (10, 20, 30, 45 and 60 % (v/v)) of trifluoroethanol (TFE). Spectra were deconvolved using the Dichroweb server (http://dichroweb.cryst.bbk.ac.uk), and the CDSSTR algorithm with the set 7 containing five chemically denatured proteins [58] as the reference set.
Size exclusion chromatography - small angle X-ray scattering (SEC-SAXS)
Scattering intensities were collected at SOLEIL (SWING beamline, Gif-sur-Yvette, France) with the scattering intensity I as a function of the modulus of the scattering vector s with where θ is the half of the scattering angle and λ the X-ray wavelength (Table 3). The calibration was made with water and the protein samples were centrifuged at 18 000×g for 20 min at 4 °C before injection on the size exclusion chromatography column. The concentrations of the ectodomains were determined by measuring the absorbance at 280 nm and using the theoretical molar extinction coefficient calculated by ProtParam (https://web.expasy.org/protparam/protpar-ref.html). Data reduction and frames subtraction were done manually with the beamline software FOXTROT. Buffer blank frames were taken in the dead volume of the column.
Table 3.
Experimental setup for the collection of SEC-SAXS scattering intensities.
| Data acquisition | ||||
| Instrument | SWING beamline (SOLEIL, France) | |||
| Detector | Dectris EIGER 4 M | |||
| Wavelength (Å) | 1.03324 | |||
| Energy (keV) | 12 | |||
| Sample to detector distance (m) | 2.0 | |||
| s-range (Å−1) | 0.0041 – 0.5597 | |||
| SEC-SAXS experiments | ||||
| ED-1 | ED-2 | ED-3 | ED-4 | |
| SEC column | Superdex S200 Increase 5/150 GL (GE Healthcare) | |||
| Injection volume (µl) | 50 | |||
| Protein concentration (mg.ml−1) | 7.7 | 5.0 | 14.6 | 8.3 |
| Flow rate (ml.min−1) | 0.2 | |||
| Temperature (°C) | 20 | |||
| Exposure | Continuous: 1 frame/s (990 ms exposure and 10 ms dead time) | |||
| Buffer | HBS (10 mM HEPES, 150 mM NaCl pH 7.4) | |||
| smin (Å -1) | 0.0114020 | 0.00821002 | 0.00961379 | 0.00912193 |
| I(0) (cm−1) | 0.0430 | 0.02892 | 0.04588 | 0.02070 |
| ± 1.8 x10-4 | ± 0.94 x10-4 | ± 3.24 x10-4 | ± 1.42 x10-4 | |
| Guinier analysis smax.Rg / number of points | 1.29 / 23 | 0.998 / 34 | 1.28 / 24 | 0.985 / 33 |
Gaussian decompositions were performed with the UltraScan solution modeler (US-SOMO) HPLC-SAXS module [59] after conversion from buffer-subtracted I(s) vs s chromatograms to I(t) vs t (t, time). Scattering curves of the syndecan EDs were then back-converted to I(s) vs s curves. These frames (at least 29) were averaged with PRIMUSqt to create the final scattering data file after curves comparison with the CorMap test implemented in the program [41]. Data points differing from the Guinier fitting at low angles were deleted and the useful data range was determined with SHANUM [41]. The radius of gyration was calculated at low angles according to the Guinier approximation using the equation with s.Rg < 1.0 or 1.3 depending of the number of points. The theoretical radius of gyration (Rg) was calculated with the Flory’s equation where N is the number of amino acid residues, R0 a constant and ν an exponential scaling factor. R0 = 2.54 ± 0.01 and ν = 0.522 ± 0.01 for intrinsically disordered proteins, R0 = 1.927 ± 0.27 and ν = 0.598 ± 0.028 for more extended proteins such as chemically denatured proteins [60], while R0 = √(3/5) × 4.75 and ν = 0.29 for folded proteins [61]. The flexibility of the proteins was assessed with the dimensionless Kratky plot and the Krakty-Debye plot .
Ensemble Optimization Method (EOM)
Intrinsically disordered proteins do not adopt a single conformation in solution and each of the conformations contribute to the collected scattering intensities. This ensemble of conformation is described according to the following equation:
where νk and Ik(s) are the volume fraction and the scattering intensity of the k-th conformers respectively [33]. SEC-SAXS data were processed with the Ensemble Optimization Method (EOM 2.0) [37], which computes a large pool of possible conformations from protein sequence in order to cover its conformational space. Gaussian distributions of Rg and Dmax of the generated conformers as well as their respective theoretical scattering profile were automatically calculated with CRYSOL [41]. A genetic algorithm was run to select an ensemble of conformations which best fits the experimental scattering intensities. The best ensemble of conformations was described as a distribution of Rg and Dmax. The flexibility was evaluated by Rflex (100 % corresponding to the highest flexibility), with the pool representing a reference for flexibility [37]. Rσ corresponds to the ratio of the standard deviations for the distributions of the selected ensemble and of the pool respectively [37]. Two pools of 10,000 conformers corresponding to the pre-molten globule and random-coil models were generated with specific dihedral angle distribution [37]. The number of harmonics was set to 15 and 100 cycles were performed on the generated pool by the genetic algorithm. EOM fitting to the experimental scattering intensities was assessed with the CorMap test implemented in PRIMUSqt [41]. Protein backbone was reconstructed from alpha-carbon traces using SABBAC [62].
Multi angle laser light scattering and dynamic light scattering
SEC-MALLS-DLS experiments were carried out at SOLEIL (SWING beamline, Gif-sur-Yvette, France) with a Wyatt Dawn Heleos light scattering instrument equipped with an Optilab rEX refractometer and a spectrophotometer for molecular mass measurements. The hydrodynamic radii (Rh) were measured with a Wyatt QELS + . For ED-1 and ED-2, the measurements were carried out on line after X-ray exposure and injection on a Superdex S200 Increase 5/150 GL column (GE Healthcare) as described above. 50 µl of ED-3 (6.5 mg/ml) and ED-4 (5.2 mg/ml) were injected on a Superdex S200 Increase 10/300 GL column (GE Healthcare) at a flow rate of 0.5 ml/min and at 20 °C in HBS. Data analysis was carried out with the Astra software and default parameters (refractive index: 1.331, viscosity: 0.992 cP).
Native electrospray ionization mass spectrometry (ESI-MS)
ESI-MS experiments were carried out in pseudo-native conditions, i.e. by minimizing potential sources of denaturation in solution and during the desolvation/ionization process [63]. A Superdex S200 Increase 10/300 GL column (GE Healthcare) was used to exchange proteins buffer from HBS to 50 mM ammonium acetate pH 7.4 and then the ectodomain solutions were concentrated as described above. 200 µl of ED-1 (600 µg/ml), ED-2 (590 µg/ml), ED-3 (420 µg/ml) and ED-4 (180 µg/ml) in 50 mM ammonium acetate pH 7.4 were injected at a flow rate of 10 µl/min in an Impact II mass spectrometer (Bruker) equipped with an Apollo II ionization source (Bruker, ICBMS UMR5246, Villeurbanne, France). The instrument parameters were set as follows: ESI-source temperature 200 °C, capillary voltage 3 kV, nebulization gas pressure 41 Psi allowing the detection of species between 500 and 3,100 Da, and acquisition frequency 2 Hz. The average solvent-accessible surface areas (SASA) were calculated according to Li et al. [64]. Briefly, the average charge zav was calculated according to:
where A and B are the lowest and highest charge states of the distribution respectively. I and Itot are the peak intensity at a given charge state, and the sum of the intensities of the charge state distribution of the conformers respectively. zav is directly proportional to SASA for extended conformers according to: [65]. SASA from globular proteins were extracted from[43], and calculated for the syndecan EDs according to . ESI-MS data were deconvolved as described in [42] using Origin software (OriginLab). I(m/z) vs m/z graphs were transformed into I(z) vs z and intensities were normalized. Gaussian fitting was performed until the fit reached at least R2 = 0.99. The SASA of the most compact conformation of the ectodomains were compared to those of globular proteins with the same, or a very close, number of amino acid residues. ED-1 (260 residues) was compared to carbonic anhydrase I (PDB 2CAB, 260 residues), ED-2 (154 residues) to myoglobin (PDB 1WLA, 153 residues), ED-3 (373 residues) to ovalbumin (PDB 1OVA, 386 residues), and ED-4 (163 residues) to β-lactoglobulin A (PDB 1BSY, 162 residues).
Electrospray ionization – ion mobility – mass spectrometry (ESI-IM-MS)
In ion mobility – mass spectrometry, electrosprayed ions are driven by a weak electric field through a cell filled with a buffer gas. Ions with higher charge state generally fly faster. Moreover, the velocity of the ions is affected by the strength of their interaction with the buffer gas, depending of their collision cross section (CCS). Since helium was used as a buffer gas, this CCS is, in good approximation, a measure of the geometrical area of an ion. Ions with a compact structure are less prone to collision with the gas, and therefore they travel faster through the cell. Ion populations can then be separated as a function of the compactness of their structure in an arrival time distribution. The arrival time distributions were converted into CCS distributions to ease the comparison between different charge states. Due to Coulomb repulsion between charges, CCS generally increases with charge state. IM-MS experiments were performed using a home-built dual-drift tube instrument previously described in details [66]. IM-MS separation was achieved by injecting ion bunches in a 79-cm long drift tube filled with 4 Torr helium at room temperature. The drift voltage across the tube varied from 200 to 600 V. Mass spectra were recorded as a function of the arrival time of the ions using a time-of-flight mass spectrometer (Maxis Impact Bruker, Bremen, Germany). This allowed the extraction of either arrival time distributions (ATDs) for ions at the mass-to-charge ratios (m/z) of interest, or of mass spectra for species within a selected arrival time range. CCS values were derived from drift time measurements at different drift voltages, based on the Mason-Schamp relation [67].
Simulation procedure
The calculations were carried out using the physics-based coarse-grained UNRES model of polypeptide chains [68], [69], [70], in which the polypeptide chain is represented as a sequence of α-carbon (Cα) atoms linked by virtual bonds, with united side chains (SC) attached to the Cα's and united peptide groups (p) positioned halfway between the consecutive Cαs. Only the SC’s and p’s are interaction sites, while the Cαs only define chain geometry. The latest version of the force field, calibrated with 9 training proteins, was used [71]. The force field handles the formation and breaking of disulfide bonds [72], this option was used in the present simulations. The full procedure is detailed in Supplementary Material.
Data deposition
SAXS data were deposited in the Small Angle Scattering Biological Data Bank (SASBDB) [73], SASBDB IDs SASDL69 (ED-1), SASDL79 (ED-2), SASDL89 (ED-3), SASDL99 (ED-4 monomer) and SASDLA9 (ED-4 dimer) and structural ensembles from SAXS data in the Protein Ensemble Database [74], PED: P00185 (ED-1), PED00186 (ED-2), PED00187 (ED-3) and PED00188 (ED-4).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
SEC-SAXS experiments were carried out on the BM29 BioSAXS beamline (ESRF, Grenoble, France, MX-1841 and MX-1920), and SWING beamline (SOLEIL, Université Paris-Saclay, France, project 20170906). We thank Martha Brennich (BM29, ESRF) and Aurélien Thureau (SWING, Soleil) for their skillful assistance and for their expert suggestions on SAXS data processing, and Cy Jeffries (EMBL, Hamburg, Germany) for sharing with us his expertise in SAXS theory. We are also grateful to Alexandra Berlioz-Barbier (ICBMS, UMR 5246, Villeurbanne, France) for her help in native ESI-MS experiments. We thank John Couchman (University of Copenhagen, Denmark) for sharing with us his expertise of syndecans, and for stimulating discussions. We thank Lionel Ballut (UMR 5086, Lyon, France), Philippe Dugourd and Luke MacAleese (Institut Lumière Matière, UMR 5306, Villeurbanne, France) for their critical reading of the manuscript, and Federica Quaglia (University of Padova, Italy) for curating the structure ensembles of syndecan ectodomains and for depositing them in the PED database.
Funding
This work was supported by a grant from the Fondation pour la Recherche Médicale n°DBI20141231336 to SRB, and by an ANR grant n°17-CE29-0013-01 to ALF and FC.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mbplus.2021.100081.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Couchman J.R., Gopal S., Lim H.C., Nørgaard S., Multhaupt H.A.B. Fell-Muir Lecture: Syndecans: from peripheral coreceptors to mainstream regulators of cell behaviour. Int. J. Exp. Pathol. 2015;96:1–10. doi: 10.1111/iep.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afratis N.A., Nikitovic D., Multhaupt H.A.B., Theocharis A.D., Couchman J.R., Karamanos N.K. Syndecans – key regulators of cell signaling and biological functions. FEBS J. 2017;284(1):27–41. doi: 10.1111/febs.13940. [DOI] [PubMed] [Google Scholar]
- 3.Mitsou I., Multhaupt H.A.B., Couchman J.R. Proteoglycans, ion channels and cell-matrix adhesion. Biochem. J. 2017;474:1965–1979. doi: 10.1042/BCJ20160747. [DOI] [PubMed] [Google Scholar]
- 4.De Rossi G., Evans A.R., Kay E., Woodfin A., McKay T.R., Nourshargh S., Whiteford J.R. Shed syndecan-2 inhibits angiogenesis. J. Cell Sci. 2014;127:4788–4799. doi: 10.1242/jcs.153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonova E.I., Galzitskaya O.V. Role of Syndecans in Lipid Metabolism and Human Diseases. Adv. Exp. Med. Biol. 2015;855:241–258. doi: 10.1007/978-3-319-17344-3_10. [DOI] [PubMed] [Google Scholar]
- 6.Kwon M.-J., Jang B., Yi J.Y., Han I.-O., Oh E.S. Syndecans play dual roles as cell adhesion receptors and docking receptors. FEBS Lett. 2012;586:2207–2211. doi: 10.1016/j.febslet.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 7.Klass C.M., Couchman J.R., Woods A. Control of extracellular matrix assembly by syndecan-2 proteoglycan. J. Cell Sci. 2000;113(Pt 3):493–506. doi: 10.1242/jcs.113.3.493. [DOI] [PubMed] [Google Scholar]
- 8.Yang N., Friedl A. Syndecan-1-induced ECM fiber alignment requires integrin αvβ3 and syndecan-1 ectodomain and heparan sulfate chains. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0150132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung H., Multhaupt H.A.B., Oh E.-S., Couchman J.R. Minireview: Syndecans and their crucial roles during tissue regeneration. FEBS Lett. 2016;590:2408–2417. doi: 10.1002/1873-3468.12280. [DOI] [PubMed] [Google Scholar]
- 10.Gondelaud F., Ricard‐Blum S. Structures and interactions of syndecans. FEBS J. 2019;286(15):2994–3007. doi: 10.1111/febs.v286.1510.1111/febs.14828. [DOI] [PubMed] [Google Scholar]
- 11.Bartlett A.H., Park P.W. Proteoglycans in host-pathogen interactions: molecular mechanisms and therapeutic implications. Expert Rev. Mol. Med. 2010;12 doi: 10.1017/S1462399409001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letoha T., Hudák A., Kusz E., Pettkó-Szandtner A., Domonkos I., Jósvay K., Hofmann-Apitius M., Szilák L. Contribution of syndecans to cellular internalization and fibrillation of amyloid-β(1–42) Sci. Rep. 2019;9:1393. doi: 10.1038/s41598-018-37476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudák A., Kusz E., Domonkos I., Jósvay K., Kodamullil A.T., Szilák L., Hofmann-Apitius M., Letoha T. Contribution of syndecans to cellular uptake and fibrillation of α-synuclein and tau. Sci. Rep. 2019;9:16543. doi: 10.1038/s41598-019-53038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gopal S. Syndecans in inflammation at a glance. Front. Immunol. 2020;11:227. doi: 10.3389/fimmu.2020.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asundi V.K., Erdman R., Stahl R.C., Carey D.J. Matrix metalloproteinase-dependent shedding of syndecan-3, a transmembrane heparan sulfate proteoglycan, in Schwann cells. J. Neurosci. Res. 2003;73(5):593–602. doi: 10.1002/(ISSN)1097-454710.1002/jnr.v73:510.1002/jnr.10699. [DOI] [PubMed] [Google Scholar]
- 16.Manon-Jensen T., Itoh Y., Couchman J.R. Proteoglycans in health and disease: the multiple roles of syndecan shedding. FEBS J. 2010;277:3876–3889. doi: 10.1111/j.1742-4658.2010.07798.x. [DOI] [PubMed] [Google Scholar]
- 17.S. Choi, Y. Choi, E. Jun, I.-S. Kim, S.-E. Kim, S.-A. Jung, E.-S. Oh, Shed syndecan-2 enhances tumorigenic activities of colon cancer cells, Oncotarget. 6 (2015) 3874–3886. 10.18632/oncotarget.2885. [DOI] [PMC free article] [PubMed]
- 18.Stewart M.D., Ramani V.C., Sanderson R.D. Shed syndecan-1 translocates to the nucleus of cells delivering growth factors and inhibiting histone acetylation: a novel mechanism of tumor-host cross-talk. J. Biol. Chem. 2015;290:941–949. doi: 10.1074/jbc.M114.608455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Zuo D., Chen Y., Li W., Liu R., He Y., Ren L., Zhou L., Deng T., Wang X., Ying G., Ba Y. Shed Syndecan-1 is involved in chemotherapy resistance via the EGFR pathway in colorectal cancer. Br. J. Cancer. 2014;111(10):1965–1976. doi: 10.1038/bjc.2014.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regős E., Abdelfattah H.H., Reszegi A., Szilák L., Werling K., Szabó G., Kiss A., Schaff Z., Kovalszky I., Baghy K. Syndecan-1 inhibits early stages of liver fibrogenesis by interfering with TGFβ1 action and upregulating MMP14. Matrix Biol. 2018;68–69:474–489. doi: 10.1016/j.matbio.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Strand M.E., Aronsen J.M., Braathen B., Sjaastad I., Kvaløy H., Tønnessen T., Christensen G., Lunde I.G. Shedding of syndecan-4 promotes immune cell recruitment and mitigates cardiac dysfunction after lipopolysaccharide challenge in mice. J. Mol. Cell. Cardiol. 2015;88:133–144. doi: 10.1016/j.yjmcc.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Bertrand J., Bollmann M. Soluble syndecans: biomarkers for diseases and therapeutic options. Br. J. Pharmacol. 2018 doi: 10.1111/bph.14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonova E.I., Galzitskaya O.V. Cell communication using intrinsically disordered proteins: what can syndecans say? J. Biomol. Struct. Dyn. 2015;33(5):1037–1050. doi: 10.1080/07391102.2014.926256. [DOI] [PubMed] [Google Scholar]
- 24.Stuchfield D., Barran P. Unique insights to intrinsically disordered proteins provided by ion mobility mass spectrometry. Curr. Opin. Chem. Biol. 2018;42:177–185. doi: 10.1016/j.cbpa.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 25.McFall A.J., Rapraeger A.C. Characterization of the high affinity cell-binding domain in the cell surface proteoglycan syndecan-4. J. Biol. Chem. 1998;273(43):28270–28276. doi: 10.1074/jbc.273.43.28270. [DOI] [PubMed] [Google Scholar]
- 26.Jung O., Trapp-Stamborski V., Purushothaman A., Jin H., Wang H., Sanderson R.D., Rapraeger A.C. Heparanase-induced shedding of syndecan-1/CD138 in myeloma and endothelial cells activates VEGFR2 and an invasive phenotype: prevention by novel synstatins. Oncogenesis. 2016;5 doi: 10.1038/oncsis.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J.-M., Lee K., Kim M.Y., Shin H.-I., Jeong D. Suppressive effect of syndecan ectodomains and N-desulfated heparins on osteoclastogenesis via direct binding to macrophage-colony stimulating factor. Cell Death Dis. 2018;9:1119. doi: 10.1038/s41419-018-1167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing Y., Xu Q., Lee C. Widespread production of novel soluble protein isoforms by alternative splicing removal of transmembrane anchoring domains. FEBS Lett. 2003;555:572–578. doi: 10.1016/s0014-5793(03)01354-1. [DOI] [PubMed] [Google Scholar]
- 29.Choi Y., Kwon M.-J., Lim Y., Yun J.-H., Lee W., Oh E.-S. Trans-regulation of Syndecan Functions by Hetero-oligomerization. J. Biol. Chem. 2015;290(27):16943–16953. doi: 10.1074/jbc.M114.611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park P.W. Isolation and functional analysis of syndecans. Methods Cell Biol. 2018;143:317–333. doi: 10.1016/bs.mcb.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uversky V.N. Intrinsically disordered proteins and their “mysterious” (meta)physics. Front. Phys. 2019;7 doi: 10.3389/fphy.2019.00010. [DOI] [Google Scholar]
- 32.Chemes L.B., Alonso L.G., Noval M.G., de Prat-Gay G. Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains. Methods Mol. Biol. 2012;895:387–404. doi: 10.1007/978-1-61779-927-3_22. [DOI] [PubMed] [Google Scholar]
- 33.Kikhney A.G., Svergun D.I. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015;589:2570–2577. doi: 10.1016/j.febslet.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 34.Brewer A.K., Striegel A.M. Characterizing the size, shape, and compactness of a polydisperse prolate ellipsoidal particle via quadruple-detector hydrodynamic chromatography. Analyst. 2011;136(3):515–519. doi: 10.1039/C0AN00738B. [DOI] [PubMed] [Google Scholar]
- 35.Burger V.M., Arenas D.J., Stultz C.M. A structure-free method for quantifying conformational flexibility in proteins. Sci. Rep. 2016;6:29040. doi: 10.1038/srep29040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brosey C.A., Tainer J.A. Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr. Opin. Struct. Biol. 2019;58:197–213. doi: 10.1016/j.sbi.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tria G., Mertens H.D.T., Kachala M., Svergun D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2(2):207–217. doi: 10.1107/S205225251500202X10.1107/S205225251500202X/fc5007sup1.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beauvais D.M., Rapraeger A.C. Syndecan-1 couples the insulin-like growth factor-1 receptor to inside-out integrin activation. J. Cell Sci. 2010;123:3796–3807. doi: 10.1242/jcs.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Jin H., Rapraeger A.C. Syndecan-1 and syndecan-4 capture epidermal growth factor receptor family members and the α3β1 integrin via binding sites in their ectodomains: novel synstatins prevent kinase capture and inhibit α6β4-integrin-dependent epithelial cell motility. J. Biol. Chem. 2015;290:26103–26113. doi: 10.1074/jbc.M115.679084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beauvais D.M., Ell B.J., McWhorter A.R., Rapraeger A.C. Syndecan-1 regulates alphavbeta3 and alphavbeta5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. J. Exp. Med. 2009;206:691–705. doi: 10.1084/jem.20081278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franke D., Petoukhov M.V., Konarev P.V., Panjkovich A., Tuukkanen A., Mertens H.D.T., Kikhney A.G., Hajizadeh N.R., Franklin J.M., Jeffries C.M., Svergun D.I. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santambrogio C., Natalello A., Brocca S., Ponzini E., Grandori R. Conformational characterization and classification of intrinsically disordered proteins by native mass spectrometry and charge-state distribution analysis. Proteomics. 2019;19 doi: 10.1002/pmic.201800060. [DOI] [PubMed] [Google Scholar]
- 43.Kaltashov I.A., Mohimen A. Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal. Chem. 2005;77(16):5370–5379. doi: 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beveridge R., Covill S., Pacholarz K.J., Kalapothakis J.M.D., MacPhee C.E., Barran P.E. A mass-spectrometry-based framework to define the extent of disorder in proteins. Anal. Chem. 2014;86(22):10979–10991. doi: 10.1021/ac5027435. [DOI] [PubMed] [Google Scholar]
- 45.Beveridge R., Phillips A.S., Denbigh L., Saleem H.M., MacPhee C.E., Barran P.E. Relating gas phase to solution conformations: lessons from disordered proteins. Proteomics. 2015;15:2872–2883. doi: 10.1002/pmic.201400605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mora J.F.d.l., Van Berkel G.J., Enke C.G., Cole R.B., Martinez-Sanchez M., Fenn J.B. Electrochemical processes in electrospray ionization mass spectrometry. J. Mass Spectrom. 2000;35(8):939–952. doi: 10.1002/1096-9888(200008)35:8<939::AID-JMS36>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 47.Jiang X.Z., Luo K.H., Ventikos Y. Principal mode of Syndecan-4 mechanotransduction for the endothelial glycocalyx is a scissor-like dimer motion. Acta Physiol. (Oxf). 2020;228 doi: 10.1111/apha.13376. [DOI] [PubMed] [Google Scholar]
- 48.Asundi V.K., Carey D.J. Self-association of N-syndecan (syndecan-3) core protein is mediated by a novel structural motif in the transmembrane domain and ectodomain flanking region. J. Biol. Chem. 1995;270(44):26404–26410. doi: 10.1074/jbc.270.44.26404. [DOI] [PubMed] [Google Scholar]
- 49.Borysik A.J., Kovacs D., Guharoy M., Tompa P. Ensemble methods enable a new definition for the solution to gas-phase transfer of intrinsically disordered proteins. J. Am. Chem. Soc. 2015;137(43):13807–13817. doi: 10.1021/jacs.5b06027. [DOI] [PubMed] [Google Scholar]
- 50.Natalello A., Santambrogio C., Grandori R. Are charge-state distributions a reliable tool describing molecular ensembles of intrinsically disordered proteins by native MS? J. Am. Soc. Mass Spectrom. 2017;28(1):21–28. doi: 10.1007/s13361-016-1490-1. [DOI] [PubMed] [Google Scholar]
- 51.Dai A., Ye F., Taylor D.W., Hu G., Ginsberg M.H., Taylor K.A. The structure of a full-length membrane-embedded integrin bound to a physiological ligand. J. Biol. Chem. 2015;290(45):27168–27175. doi: 10.1074/jbc.M115.682377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rapraeger A.C. Synstatin: a selective inhibitor of the syndecan-1-coupled IGF1R-αvβ3 integrin complex in tumorigenesis and angiogenesis. FEBS J. 2013;280:2207–2215. doi: 10.1111/febs.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samarel A.M. Syndecan-4: a component of the mechanosensory apparatus of cardiac fibroblasts. J. Mol. Cell. Cardiol. 2013;56:19–21. doi: 10.1016/j.yjmcc.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voyvodic P.L., Min D., Liu R., Williams E., Chitalia V., Dunn A.K., Baker A.B. Loss of syndecan-1 induces a pro-inflammatory phenotype in endothelial cells with a dysregulated response to atheroprotective flow. J. Biol. Chem. 2014;289(14):9547–9559. doi: 10.1074/jbc.M113.541573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eriksson A.S., Spillmann D. The mutual impact of syndecan-1 and its glycosaminoglycan chains–a multivariable puzzle. J. Histochem. Cytochem. 2012;60(12):936–942. doi: 10.1369/0022155412460242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jowitt T.A., Murdoch A.D., Baldock C., Berry R., Day J.M., Hardingham T.E. Order within disorder: aggrecan chondroitin sulphate-attachment region provides new structural insights into protein sequences classified as disordered. Proteins. 2010;78:3317–3327. doi: 10.1002/prot.22839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnan P., Hocking A.M., Scholtz J.M., Pace C.N., Holik K.K., McQuillan D.J. Distinct secondary structures of the leucine-rich repeat proteoglycans decorin and biglycan. Glycosylation-dependent conformational stability. J. Biol. Chem. 1999;274(16):10945–10950. doi: 10.1074/jbc.274.16.10945. [DOI] [PubMed] [Google Scholar]
- 58.Whitmore L., Wallace B.A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 59.Brookes E., Vachette P., Rocco M., Pérez J. US-SOMO HPLC-SAXS module: dealing with capillary fouling and extraction of pure component patterns from poorly resolved SEC-SAXS data. J. Appl. Crystallogr. 2016;49:1827–1841. doi: 10.1107/S1600576716011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernadó P., Svergun D.I. Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol. BioSyst. 2012;8(1):151–167. doi: 10.1039/C1MB05275F. [DOI] [PubMed] [Google Scholar]
- 61.Receveur-Brechot V., Durand D. How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 2012;13:55–75. doi: 10.2174/138920312799277901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maupetit J., Gautier R., Tufféry P. SABBAC: online Structural Alphabet-based protein BackBone reconstruction from Alpha-Carbon trace. Nucl. Acids Res. 2006;34:W147–151. doi: 10.1093/nar/gkl289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leney A.C., Heck A.J.R. Native mass spectrometry: what is in the name? J. Am. Soc. Mass Spectrom. 2017;28(1):5–13. doi: 10.1007/s13361-016-1545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J., Santambrogio C., Brocca S., Rossetti G., Carloni P., Grandori R. Conformational effects in protein electrospray-ionization mass spectrometry. Mass Spectrom. Rev. 2016;35:111–122. doi: 10.1002/mas.21465. [DOI] [PubMed] [Google Scholar]
- 65.Testa L., Brocca S., Šamalikova M., Santambrogio C., Alberghina L., Grandori R. Electrospray ionization-mass spectrometry conformational analysis of isolated domains of an intrinsically disordered protein. Biotechnol. J. 2011;6(1):96–100. doi: 10.1002/biot.v6.110.1002/biot.201000253. [DOI] [PubMed] [Google Scholar]
- 66.Simon A.-L., Chirot F., Choi C.M., Clavier C., Barbaire M., Maurelli J., Dagany X., MacAleese L., Dugourd P. Tandem ion mobility spectrometry coupled to laser excitation. Rev. Sci. Instrum. 2015;86 doi: 10.1063/1.4930604. [DOI] [PubMed] [Google Scholar]
- 67.Revercomb H.E., Mason E.A. Theory of plasma chromatography/gaseous electrophoresis. Review. Anal. Chem. 1975;47(7):970–983. doi: 10.1021/ac60357a043. [DOI] [Google Scholar]
- 68.Voth G.A. Coarse-Graining of Condensed Phase and Biomolecular Systems. CRC Press. 2008 doi: 10.1201/9781420059564. [DOI] [Google Scholar]
- 69.Czaplewski C., Karczynska A., Sieradzan A.K., Liwo A. UNRES server for physics-based coarse-grained simulations and prediction of protein structure, dynamics and thermodynamics. Nucl. Acids Res. 2018;46:W304–W309. doi: 10.1093/nar/gky328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krupa P., Karczyńska A.S., Mozolewska M.A., Liwo A., Czaplewski C. UNRES-Dock - protein-protein and peptide-protein docking by coarse-grained replica-exchange MD simulations. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa897. [DOI] [PubMed] [Google Scholar]
- 71.Liwo A., Sieradzan A.K., Lipska A.G., Czaplewski C., Joung I., Żmudzińska W., Hałabis A., Ołdziej S. A general method for the derivation of the functional forms of the effective energy terms in coarse-grained energy functions of polymers. III. Determination of scale-consistent backbone-local and correlation potentials in the UNRES force field and force-field calibration and validation. J. Chem. Phys. 2019;150 doi: 10.1063/1.5093015. [DOI] [PubMed] [Google Scholar]
- 72.Chinchio M., Czaplewski C., Liwo A., Ołdziej S., Scheraga H.A. Dynamic formation and breaking of disulfide bonds in molecular dynamics simulations with the UNRES force field. J. Chem. Theory Comput. 2007;3(4):1236–1248. doi: 10.1021/ct7000842. [DOI] [PubMed] [Google Scholar]
- 73.Kikhney A.G., Borges C.R., Molodenskiy D.S., Jeffries C.M., Svergun D.I. SASBDB: towards an automatically curated and validated repository for biological scattering data. Protein Sci. 2020;29:66–75. doi: 10.1002/pro.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lazar T., Martínez-Pérez E., Quaglia F., Hatos A., Chemes L.B., Iserte J.A., Méndez N.A., Garrone N.A., Saldaño T.E., Marchetti J., Rueda A.J.V., Bernadó P., Blackledge M., Cordeiro T.N., Fagerberg E., Forman-Kay J.D., Fornasari M.S., Gibson T.J., Gomes G.-N.-W., Gradinaru C.C., Head-Gordon T., Jensen M.R., Lemke E.A., Longhi S., Marino-Buslje C., Minervini G., Mittag T., Monzon A.M., Pappu R.V., Parisi G., Ricard-Blum S., Ruff K.M., Salladini E., Skepö M., Svergun D., Vallet S.D., Varadi M., Tompa P., Tosatto S.C.E., Piovesan D. PED in 2021: a major update of the protein ensemble database for intrinsically disordered proteins. Nucl. Acids Res. 2021;49(2021):D404–D411. doi: 10.1093/nar/gkaa1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.