Abstract
Pituitary neuroendocrine tumors (PitNETs) are the second most common type of intracranial neoplasia. Since their manifestation usually causes hormone hypersecretion, effective management of PitNETs is indisputably necessary. Most of the non-functioning PitNETs pose a real challenge in diagnosis as they grow without giving any signs. Despite the good response of prolactinomas to dopamine agonist therapy, some of these tumors persist or recur; also, about 20% are resistant and 10% behave aggressively. The silent corticotropinomas may not cause symptoms until the tumor mass causes a complication. In somatotropinomas, the possibility of recurrence after transsphenoidal resection is more common in pediatric patients than in adult patients. Therefore, detection of tumors at early stages or identification of recurrence and remission after transsphenoidal surgery would allow wiser management of the disease. Extensive studies have been performed to uncover potential signatures that can be used for preventive diagnosis and/or prognosis of PitNETs as well as for targeted therapy. These molecular signatures at multiple biological levels hold promise for the convergence of preventive approaches and patient-centered disease management and offer potential therapeutic strategies. In this review, we provide an overview of the omics-based biomarker research and highlight the multi-omics signatures that have been proposed as pitNET biomarkers. In addition, understanding the multi-omics data integration of current biomarker discovery strategies was discussed in terms of preventive, predictive, and personalized medicine. The topics discussed in this review will help to develop broader visions for pitNET research, diagnosis, and therapy, particularly in the context of personalized medicine.
Keywords: Pituitary neuroendocrine tumors, Pituitary adenoma, Predictive preventive personalized medicine, Biomarkers, Multi-omics data integration, Omics biomarkers
Introduction
PitNETs were considered a rare condition in the past, but as the number of patients diagnosed is increasing day by day, it seems to be becoming a common disorder [1]. Although these tumors usually behave benignly, some of them have invasive and aggressive features [2]. Moreover, a significant number of patients do not respond to currently used medical therapies including somatostatin analogs (SSAs) or dopamine agonists (DA) [3–5]. Despite these challenging characteristics of PitNETs, previous developments in high-throughput technologies have led to the identification of pitNET-specific biomarkers using genomics, transcriptomics, proteomics, epigenomics, and glycomics. The systems biomedicine perspective can illuminate biomarker discovery by integrating multi-omics data and provide predictive diagnostics, targeted prevention, and personalization of medical services.
Cancer biomarkers play a crucial role in revealing tumor proliferation, progression, invasion and metastasis and could be associated with disease prevention, diagnosis and treatment [6]. The main goal of precision oncology is to decipher molecular mechanisms to efficiently control tumor behavior and its heterogeneity, and to translate these findings into useful means to treat the disease. Translational medicine is a promising bridge between predictive, preventive, and personalized medicine (PPPM or 3PM) science and their real-life application/implementation [7].
In the era of precision medicine, “All for one and one for all” becomes a naïve slogan regarding the heterogeneity of each disease and the variety of lifestyles of the patients. Therefore, person-centered medicine requires logical approaches and validated strategies to reach the desired level of wisdom in health care [7]. This could be possible by the way of the right patient that is treated with the right medication and dose at the right time. To reach this goal an integrative medical approach to improve health care services is a multidisciplinary duty that is composed of cooperation between research, business, and politics [8].
In the last decade, we have witnessed extensive and accelerated advances in omics research related to the analysis of the genome, transcriptome, proteome, metabolome, glycome, metagenome, and epigenome in normal and disease states [9–17]. The growing number of omics-driven studies has provided unparalleled insights into elucidating the molecular mechanisms and key alterations behind each disease. Moreover, the ability to decipher the changes in each phase of omics that affect our understanding of disease pathogenesis and progression has greatly increased. At this point, pattern recognition, an efficient framework to uncover key molecules/panels in cancer, is a crucial concept that should be expressed. Pattern recognition can be described as the use of a set of patterns consisting of different types of biomarkers to increase the accuracy and specificity of tumor prediction, diagnosis, prognosis, and prevention/therapy [6]. The pattern can consist of biomarkers from different omics and can also be combined to form an integrative pattern that leads to the detection of the disease. We now know that, except in certain diseases, each omics stage, either alone or mostly in harmony, has elucidated clinical course, disease progression, and therapeutic efficacy. In addition to the omics-driven repertoire of identified changes, their consequences at the different omics levels could pave the way for logical, systems-based approaches and smart methods to predictive diagnostics, targeted prevention, and personalized treatment modalities to cure the disease [9, 18–21].
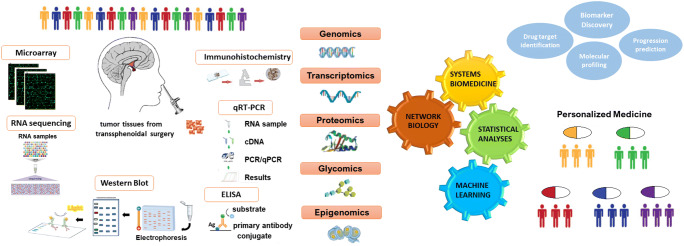
In this study, we present a systems biomedicine-oriented literature review of the current status of pitNET biomarkers with respect to different omics levels (Fig. 1 and Table 1). We reported biomarkers of PitNETs, some of which have been associated with prediction, diagnostic/prognostic evaluation, some as therapeutic targets, and some with the molecular mechanism of PitNETs. In addition, we highlighted how the integration of multi-omics data can uncover novel biomarkers that can be to implement preventive, predictive and personalized medicine strategies used in the future and presented expert recommendations.
Fig. 1.
A conceptual workflow of retrieving disease biomarkers from a variety of “omics” levels
Table 1.
A summary of biomarkers, subtypes, and methodology used in overviewed studies for PitNETs
| Omic level | Omic level description | Sample origin | Altered molecule | Expression pattern | Detection method | Reference study |
|---|---|---|---|---|---|---|
| Genomic signatures | Mutation | Somatotroph PitNETs | GHR (H49L) | - | Sequencing, Western blot, immunoprecipitation | [22] |
| Somatotroph, corticotroph, and NF-PitNET tumor tissues | GNAS, USP8, PABCP1 | - | Exome sequencing | [23] | ||
| Corticotroph PitNET tumor tissue | USP8 | - | Exome sequencing, qRT-PCR | [24] | ||
| Varied pitNET tumor tissues | GNAS, USP8 | - | Exome sequencing, SNP array | [25] | ||
| Varied pitNET tumor tissues | PIK3CA, RAS | - |
Sequencing, qRT-PCR Immunohistochemistry |
[16] | ||
| Varied pitNET tumor tissues | MEN1 | - | RT-PCR, sequencing | [26] | ||
| Varied pitNET tumor tissues | GNAS, USP8, NR3C1, MEN1, KIF5A, GRB10 | - | Whole-exome sequencing | [27] | ||
| Mutation, insertion, deletion | Varied pitNET tumor tissues | ATAD3B, BHLHE22, KDM2B, OR5M1, TTN, VPS13B | - | Whole-exome sequencing | [28] | |
| Epigenomic signatures | DMR | Varied pitNET tumor tissues | POUF1, PIT1 | Hypomethylated | Methylation array | [25] |
| GH-secreting, ACTH-secreting, NF-PitNET tumor tissue | POMC, SSTR5, GH1, GH2 | Hypomethylated | Methylation array | [23] | ||
| NF-PitNET tumor tissue | SFN, STAT5A, DUSP1, PTPRE, FGFR2, ITPKB, CNKSR1 | Hypermethylated | Methylation array, qRT-PCR | [29] | ||
| NF-PitNET tumor tissue | PHYHD1, LTBR, C22orf42, PRR5, ANKDD1A, RAB13, CAMKV, KIFC3, WNT4, STAT6, MYBPHL | Aberrantly methylated | Methylation array | [10] | ||
| NF-PitNET tumor tissue | GALNT9 | Downregulated | Methylation array, qRT-PCR | [30] | ||
| Varied pitNET tumor tissues | TP73, ESR1, MSH6, CADM1, RASSF1, CASP8 | Hypermethylated | qRT-PCR, MS-MLPA | [15] | ||
| Corticotroph tumor cell line, mouse pituitary | NNAT, S100A10 | Hypermethylated | Microarray, COBRA, qRT-PCR | [31] | ||
| Tumor cell line, Varied PitNET tumor tissues | Ikaros | Hypermethylated | Bisulfite sequencing, COBRA, RT-PCR | [32] | ||
| Varied pitNET tumor tissues | RASSF1A, CDH13, CDH1, CDKN2A | Hypermethylated | MSP assay, immunohistochemistry | [33] | ||
| Varied pitNET tumor tissues | KCNAB2 | Hypermethylated | Methylation array | [34] | ||
| Varied pitNET tumor tissues | GSTP1 | Hypermethylated | MSP assay, immunohistochemistry | [35] | ||
| Histone modification | Somatotroph tumor cell line | let-7c-2, mir-23b, mir-29c | Histone citrullinated | RNA sequencing, ChIP | [13] | |
| Varied pitNET tumor tissues | H3K4, H3K9, H3K27 | Histone methylated | MSP assay, immunohistochemistry | [36] | ||
| Varied pitNET tumor tissues | H3K27, H3K9 | Histone methylated/acetylated | ChIP, qRT-PCR, pyrosequencing | [37] | ||
| Transcriptomic signatures | Coding RNA | Lactotroph PitNET tumor tissue | TLE4, ANGPT1,CDH1/2, PCDH9, BAG1, IGFBP3, IGFBP5, ID1/2/4, FOXO1A, GADD45B/G, TGF-β1, TGFBR3, RIS1, NBL1, POUF1, ADAM28, ADAMTS6 | Dysregulated | Microarray, qRT-PCR | [38] |
| Lactotroph PitNET tumor tissue | PTTG1, EGF, EGFR, E2F1, MYC, CCNB1, CCNB2 | Dysregulated | Microarray | [39] | ||
| Lactotroph PitNET tumor tissue | ADAMTS6, CRMP1, PITX1, DCAMKL3, PTTG, ASK, CCNB1, AURKB, CENPE | Dysregulated | Microarray, qRT-PCR | [40] | ||
| Lactotroph PitNET tumor tissue | HMGA2 | Upregulated | FISH, RT-PCR, Western blot | [41] | ||
| NF-PitNET tumor tissues | MTDH | Upregulated | RNA-seq, qRT-PCR | [42] | ||
| NF-PitNET tumor tissues | CEACAM6, CYP4B1, EIF2S2, HID1, IFFO1, MYO18A, PDCD2, SGIP1, SWSAP1 | Dysregulated | Microarray | [9] | ||
| NF-PitNET tumor tissue | IGFBP5, MYO5A, FLT3, NFE2L1 | Upregulated | Microarray, qRT-PCR, immunohistochemistry | [43] | ||
| NF-PitNET tumor tissue | GALNT9 | Downregulated | Methylation array, qRT-PCR | [30] | ||
| NF-PitNET and GH-secreting PitNET tumor tissues | Survivin (BIRC5) | Upregulated | qRT-PCR, immunohistochemistry | [44] | ||
| NF-PitNET tumor tissues | IL-6, STAT3, POUF1, BCL6 | Downregulated | Microarray | [45] | ||
| NF-PitNET tumor tissue | IDH1, PITX2, NOTCH3, DLK1, SFRP1, TLE2 | Dysregulated | Microarray/qRT-PCR | [17] | ||
| Varied pitNET tumor tissues | NR5A1, GATA3 | Dysregulated | RNA-seq | [25] | ||
| Varied pitNET tumor tissues | PHLPP, ENO2, ACTR1A, EHHADH, EHMT2, FOXO1, DLD, CCT2, CSNK1D, CETN2 | Dysregulated | Microarray | [46] | ||
| Varied pitNET tumor tissues | MMP-9 | Upregulated | Western blot, qRT-PCR | [47] | ||
| Varied pitNET tumor tissues | LGALS3, hASH1, ID2, TLE-4 | Dysregulated | Microarray/qRT-PCR | [48] | ||
| Varied pitNET tumor tissues | GAL, LMO4, STAT3, PD-L1, TGFB, and TGFBR3 | Dysregulated | Microarray | [49] | ||
| Varied pitNET tumor tissues | DNMT1, DNMT3A | Upregulated | Immunohistochemistry | [33] | ||
| Gonadotroph and null cell PitNET tumor tissues | GADD45β | Downregulated | Microarray, qRT-PCR | [50] | ||
| Null cell PitNET tumor tissue | SYT, ATP5B, MDH1 | Upregulated | Microarray, qRT-PCR | [51] | ||
| miRNA | Varied pitNET tumor tissues | miR-574, miR-195, miR-497-5p, let-7b, MEG3, miR532 | Dysregulated | RNA-sequencing | [25] | |
| Varied pitNET tumor tissues | miR-212, miR-26a, miR-150, miR-152, miR-191, miR-192, miR-21, miR-141, miR-144, miR-149 | Dysregulated | Microarray/RT-PCR | [52] | ||
| Somatotroph and lactotroph PitNET tumor tissue | miR-15a, miR-16 | Downregulated |
Western blot, qRT-PCR Northern blot |
[53] | ||
| NF-PitNET and somathotroph PitNET | miR-128, miR-155, miR-516a-3p, miR-20a, miR-93 | Downregulated | Western blot, qRT-PCR | [54] | ||
| NF-PitNET tumor tissue | hsa-miR-181a-5p | Upregulated | Microarray, qRT-PCR | [55] | ||
| Invasive and non-invasive PitNETs | miR-424-5p | Downregulated | qRT-PCR, FISH | [56] | ||
| Tumor cell line, varied pitNET tumor tissues | miR-15a, miR-16, miR-132 | Downregulated | qRT-PCR | [57] | ||
| Tumor cell line | miR-106b | Upregulated | qRT-PCR, Western blot | [58] | ||
| Tumor cell line | miR-133 | Upregulated | qRT-PCR, Western blot | [59] | ||
| Exosomal RNA | NF-PitNETs blood | CDK6, RHOU | Upregulated | ddPCR | [60] | |
| Gonadotroph PitNET tumor cell line | hsa-miR-21-5p | Upregulated | RNA-seq, ELISA, qRT-PCR | [61] | ||
| PitNET tumor cell lines | miR-149-5p, miR-99a-3p | Upregulated | Microarray, qRT-PCR, Western blot | [62] | ||
| lncRNA | Somatotroph PitNET tumor tissue | H19, MALAT-1 | Upregulated | qRT-PCR | [63] | |
| Somatotroph, lactotroph, gonadotroph PitNET tumor tissues | RPSAP52 | Upregulated | Microarray, qRT-PCR | [12] | ||
| Lactotroph PitNET tumor tissue | H19 | Downregulated | Microarray, qRT-PCR | [64] | ||
| Invasive PitNET tumor tissue | XIST | Upregulated | Microarray, FISH, RNA immunoprecipitation, Western blot | [56] | ||
| NF-PitNET tumor tissue | HOTAIR, MEG3 | Upregulated | qRT-PCR | [65] | ||
| NF-PitNET tumor tissue | MEG3, ENST00000501583, n334366 | Downregulated | Microarray, qRT-PCR | [66] | ||
| Tumor cell line, varied PitNET tumor tissues | CCAT2 | Upregulated | Knockdown assay, qRT-PCR, ChIP | [67] | ||
| Tumor cell line, varied PitNET tumor tissues | C5orf66-AS1 | Downregulated | Microarray, qRT-PCR | [68] | ||
| circRNA | NF-PitNET tumor tissue | hsa_circ_102597 | Downregulated | Microarray/qRT-PCR | [69] | |
| Varied pitNET tumor tissues | hsa_circ_0001368 | Upregulated | ELISA, circRNA microarray, qRT-PCR | [70] | ||
| NF-PitNET tumor tissue | circOMA1 (hsa_circRNA_0002316) | Dysregulated | qRT-PCR, FISH, Western blot, immunohistochemistry | [14] | ||
| Glycomic signatures | glycoprotein | Varied pitNET tumor tissues | endocan | Upregulated | Immunohistochemistry, RT-PCR | [11] |
| Glycan alterations | NF-PitNET tumor tissues | Glycosylation of Clusterin | Upregulated | Western blot, ChIP assay | [71] | |
| Varied pitNET tumor tissues | Polysialylation of NCAM | Upregulated | Western blot, ELISA | [72] | ||
| Proteomic signatures | Protein | NF-PitNET tumor tissues | NOTCH3, DLK1, SFRP1, TLE2, PITX2, NADP, TPH2, GPX4, GH1, SCGN, GP96, CD59, HSPB1 and other 40 proteins | Dysregulated | MALDI-TOF MS, LC-ESI-Q-IT MS | [17] |
| Varied pitNET tumor tissues | Galectin-3, hASh-1, ID2, TLE4 | Downregulated | Western blot, TMA immunohistochemistry | [48] | ||
| NF-pitNET tumor tissues | IRAK2, GRIP2, ARHGAP5, LILRB4, ZNF432, PRKAR1B, SGPL1, CENB1A, PSMA2, IL-1F6, RHPN2 | Dysregulated | NAC, tandem MS | [73] | ||
| Lactotroph PitNET tumor tissues | NOTCH3, DLK1, HES1, ASCL1, FDZ7, PIT-1, BAG1, TLE4, ANG-1, D-2, RIS1, NBL1, E-cadherin, N-cadherin | Dysregulated | MALDI-TOF MS, LC-ESI-Q-IT MS | [38] | ||
| ACTH-secreting PitNET tumor tissue | LAP3, MDH1/2, AK3, ALDH7A1, EPRS, FASN, PIGR, SND1, ACYL, MPG, LMNA | Dysregulated | NanoLC-MS/MS | [74] | ||
| NF-PitNETs and lactotroph PitNET tumor tissue | hPRL variants | Dysregulated | Western blot, MALDI-TOF MS, LC-ESI-Q-IT MS | [75] | ||
| NF-PitNETs tumor tissue | SRC, AKT1 | Upregulated | Western blot, LC-MS/MS | [76] | ||
| NF-PitNETs tumor tissue | CHGA, CLU | Dysregulated | LC-MS/MS, Western blot | [77] | ||
| NF-PitNETs tumor tissue | YWHAZ (14-3-3 zeta/delta protein) | Upregulated/decreased ubiquitination | LC-MS/MS, Western blot | [78] | ||
| Varied pitNET tumor tissues | HINT-1 | Upregulated | LC-MS/MS, SDS-PAGE, immunohistochemistry | [79] | ||
| Lactotoroph PitNET tumor cell line | Akt, PI3K | Dysregulated | Western blot | [21] | ||
| NF-pitNET tumor tissues | SCG3, TSC2, ALB, AKT1, APOL1, ACACA, SPARCL1, SLC2A4, CHGB, IGFBP5 | Dysregulated | LC-MS/MS | [18] | ||
| NF-pitNET tumor tissues (FSH-positive) | ITGA1, ITGA6, ITGB4, pAKT, FSHR | Upregulated | TMT-labeling, HPLC, LC-MS/MS | [19] |
FISH florescent in situ hybridization, DMR differentially methylated region, TMA tissue microarray, SNP single nucleotide polymorphism, MS-MLPA methylation-specific multiplex ligation-dependent probe amplification, COBRA combined bisulfite restriction analysis, MSP assay methylation-specific PCR, CHIP chromatin immunoprecipitation, NAC nitrotyrosine affinity column, Tandem MS tandem mass spectrometry, ELISA enzyme-linked immunosorbent assay, SDS-PAGE sodium dodecyl sulfate polyacrylamide gel electrophoresis, TMT-labeling tandem mass tag labeling, MALDI-TOF MS matrix-assisted laser desorption ionization time of flight mass spectrometry, LC-MS/MS liquid chromatography with tandem mass spectrometry, LC-ESI-Q-IT MS liquid chromatography with electrospray ionization quadrupole ion trap mass spectrometer
Genomic signatures
With the advent in the era of high-throughput techniques, including next-generation sequencing, genome-level research has peaked and revolutionized molecular science into a deeper horizon where every step of the central dogma is illuminated. At the DNA level, the determination of potential biomarkers leading to the elucidation of disease mechanisms and stratification of patients is possible via the identification of chromosomal aberrations, single nucleotide polymorphisms (SNPs), or copy number variations. In somatotrophic PitNETs, a mutation of GHR was found (histidine to leucine substitution in codon 49) that negatively affects receptor activation, maturation, GH binding, and signal transduction [22]. This mutation has also been associated with the responsiveness to pharmacotherapy, which provided well-matched therapeutic regimens via molecular and morphological studies. In another study, a GNAS mutation was found in somatotrophic PitNETs [23]. The GRB10 mutations in somatotrophic PitNETs have been linked to the pathogenesis of these tumors [27].
Recurrent mutations in the USP8 and PABPC1 genes have been reported for corticotrophic PitNETs [23]. An exome sequencing study of corticotrophic PitNETs identified somatic mutations in the USP8 gene [24]. These mutations have been reported to increase proteolytic cleavage and cause high deubiquitination activity, leading to accumulation of EGFR—a cellular substrate of USP8-, which in turn leads to increased plasma ACTH levels [24]. A novel truncation mutation in corticotrophic PitNETs was found in the NR3C1 gene, which is an unusual player in PitNET pathogenesis [27].
No notable recurrent mutations have been reported in NF-PitNETs [23]. In plurihormonal PitNETs that produced growth and prolactin hormones, two novel loss-of-function mutations were identified. In a whole-exome sequencing study that included all subtypes of PitNETs, patients carrying a MEN1 mutation accounted for 42%. It has been reported that MEN1 mutation carriers may be more aggressive than non-MEN1 patients [25]. Moreover, MEN1 affects cell cycle dysregulations and correlates with more aggressive behavior in PitNETs [26]. KIF5A has been found to be recurrently mutated in PitNETs [27].
PIK3CA is an important oncogene in various cancers and has also been studied in PitNETs for the effects of mutations and insertions [16]. Somatic mutations in PIK3CA have been associated with tumor invasiveness and recurrence.
The presented genome-level biomarkers of PitNETs generally reveal disease mechanism-associated features such as biological behavior and pathogenesis. These efforts could lead to understanding broader visions related to personalization of medical services, as each signature may vary from individual to individual. Through increasing efforts, great progress has been made in genome-level research for PitNETs. However, these reported mutations may not be the sole cause of tumorigenesis, so adopting mutations as the sole diagnostic marker for PitNETs would not lead to influential management of the disease. Integration of genomic data with transcriptomic, proteomic, or other types of omic data is needed to gain broader visions for PitNETs.
Epigenomic signatures
DNA methylation and histone modification are two critical components of the epigenome that result in altered expression of genes, including those involved in cell growth or immune response. DNA methylation is the addition of the methyl group to DNA bases, which normally silences the gene. Histone modification is a covalent posttranslational modification (PTM) to histone proteins that includes methylation, acetylation, ubiquitylation, phosphorylation, and sumoylation of histones. These epigenetic changes can lead to malignant cell transformation and contribute to tumorigenesis through activation of oncogenes or inactivation of tumor suppressors. Epigenetic abnormalities can lead to changes in the phenotype of PitNETs, and therefore, studies on these changes can help in the stratification of PitNETs. Moreover, unlike genetic mutations, epigenetic aberrations have the potential to be reversed and normalized by epigenetic therapy, making such interventions promising and therapeutically useful [80].
There are several biomarker-targeted studies based on methylation profiling being conducted by several initiatives from different countries to provide the most appropriate personalized therapy for PitNET patients (Table 1). Remarkable methylation analysis in PitNETs was performed by Kober et al. who demonstrated DNA hypermethylation in SFN, STAT5A, DUSP1, PTPRE, and FGFR2 genes and the effect of methylation on reducing the expression level of these genes in NF-PitNET patients, suggesting a role of aberrant DNA methylation in the pathogenesis of NF-PitNETs [29]. Their results suggest the influence of aberrant DNA methylation in the dysregulation of cancer-related signaling pathways and the pathogenesis of NF-PitNETs. Another study was designed by Cheng et al. who integrated methylation patterns of genes and mRNA profiles to identify key genes involved in tumor invasiveness. They discovered increased methylation with decreased expression of PHYHD1, LTBR, C22orf42, PRR5, ANKDD1A, RAB13, CAMKV, KIFC3, WNT4, and STAT6, and decreased methylation with increased expression of MYBPHL, suggesting that the invasive behavior of tumors is due to aberrations in the methylation status of these genes in NF-PitNETs [10]. The correlation with CpG hypermethylation and downregulation of the GSTP1 gene was found strikingly significant in a study using methylation-specific polymerase chain reaction (MS-PCR) and immunohistochemical methods in PitNETs [35]. They concluded that GSTP1 inactivation by CpG hypermethylation may be highly involved in the aggressiveness of pituitary tumors. In a study based on methylation patterns of tumor suppressor genes, García-Martínez et al. showed that DNA methylation of tumor suppressor genes has a selective effect on their expression patterns and invasive behavior of PitNETs [15]. Their study revealed different methylation profiles of TP73, MSH6, CADM1, ESR1, RASSF1, and CASP8 genes among PitNET subtypes. Therefore, they suggested that subtypes of PitNETs should be considered distinct entities and molecular clinical trials must be designed considering their differences. This approach might provide personalization of clinical management of each case. Another study also concluded that changes in the methylome profile are responsible for the invasiveness of PitNET subtypes [30]. They also claim that the differential DNA methylome profile of PitNETs, which indicates tumor invasion, correlates with cell adhesion. The other major finding of their study was significant downregulation of the GALNT9 gene independent of DNA methylation in invasive PAs. KCNAB2 is known for its role in ion channel activity signaling pathways and it has been reported that hypermethylation of the KCNAB2 promoter may be associated with the endocrine-inactive status of NF-PitNETs [34].
In an animal study, Zhu et al. suggested that histone methylation and DNA methylation likely have a reinforcing relationship, as both are involved in the control of Ikaros (Ik) gene expression in PitNETs [32]. In another animal study focusing on the strategy of silencing DNA methyltransferase (cytosine 5)-1 (DNMT1), Dudley et al. showed that decreased expression of NNAT and S100A10 genes was associated with increased CpG methylation in the murine PitNET cell line AtT-20 [31]. Supportive results were reported in a study that investigated the role of DNMT1 and DNMT3A in tumor suppressor gene promoter methylation leading to PitNET invasion [33].
A study on the pangenomic classification of PitNETs showed that PitNETs from the POU1F1/PIT1 lineage are associated with common DNA hypomethylation [25]. Salomon et al. designed a study integrating gene expression profiling and epigenetic analysis to gain a deeper understanding of how various omic-level changes interact in PitNETs. This integrated study reported that hypomethylation of promoter regions is associated with increased expression of the GH1, GH2, and SSTR5 genes in GH-secreting PitNETs and the POMC gene in ACTH-secreting PitNETs. Their results support the notion that each subtype is unique and should be evaluated considering the disease-specific etiology in PitNETs [23].
Duong et al. investigated the causal relationship between the reduced expression pattern of the EFEMP1 gene and tumorigenesis of PitNET subtypes from an epigenetic perspective using the ChIP method. ChIP analysis revealed enrichment for the histone modification H3K27Me3 and depletion of the H3K9Ac modification as associated with reduced expression of EFEMP1 in a subtype-independent manner [37]. The tumor suppressor gene RIZ1 directly represses the expression of the c-MYC gene and also has great potential for histone methylation. Methylation of the promoter region of RIZ1 may play an important role in epigenetic silencing of RIZ1 expression, thereby altering histone methylation of H3K4 and H3K27 in pituitary tumors [36]. Another study integrating ChIP array and RNA sequencing methods reported that histone citrullination represses the expression of microRNAs let-7c-2, 23b, and 29c, which directly target the mRNA of oncogenes encoding HMGA, IGF-1, and N-MYC, which play a significant role in the pathogenesis of human prolactinoma/somatoprolactinoma [13].
Most of the reported epigenomic signatures were associated with the molecular mechanisms of PitNETs and provided a deeper understanding of the pathogenesis of the disease. The remaining epigenomic signatures were associated with disease progression and tumor behavior, which could be useful for personalizing the treatment of the disease by providing the ability to distinguish each patient from another.
Transcriptomic signatures
Analysis of protein-coding RNA profiles of different PitNETs has been performed to look for disease-related alterations, whereupon comparative studies between tumor and healthy counterpart tissues have gained great potential as they allow to identify of differences between these two stages. Several studies have reported many potential biomarkers for prognosis, diagnosis, and treatment (Table 1). With the implementation of microarray and RNA sequencing (RNA-seq) technologies, it is now possible to further decipher the molecular alterations that contribute to the development of these tumors, their progression, therapeutic response, and resistance, according to the molecular basis. In this section, significantly altered molecules as biomarkers not only mRNAs but also non-coding RNAs such as long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs) were presented at transcriptome level for different subtypes of PitNETs.
Coding RNAs
Concerning prolactinoma, microarray studies have revealed disease-related mRNA biomarkers [38–40]. TLE4, ANGPT1, DNAJB5, and NOTCH3 were significantly upregulated in microarray mRNA profiling, whereas TGFBR3 was downregulated. qRT-PCR analysis of these five genes confirmed their aberrant expression, with TLE4, ANGPT1, DNAJB5, and NOTCH3 were significantly upregulated 15- to 34-fold, and TGFB3 downregulated 8-fold in prolactinomas compared with normal pituitary controls [38]. In another study, microarray data from rats showed that PTTG1, EGF, EGFR, E2F1, MYC, CCNB1, and CCNB2 were abnormally expressed in prolactinomas. Among these genes, GHRHR and MYC showed convergent expressions between human prolactinomas and rat models [39]. Wierinckx and colleagues reported nine genes confirmed with qRT-PCR to be involved in invasion (ADAMTS6, CRMP1, and DCAMKL3), proliferation (PTTG, ASK, CCNB1, AURKB, and CENPE), or pituitary differentiation (PITX1). These genes, with the exception of PITX1, have also been associated with tumor persistence and resistance [40].
When we consider NF-PitNETs, more studies have been performed using both microarray and RNA-seq methods compared to prolactinomas [9, 17, 30, 42–45]. In one RNA-seq profiling study, tumors were classified into subgroups as slow and fast-growing based on tumor volume doubling time. In the same study, MTDH (metadherin) was found to be an important factor in tumor aggressiveness and was proposed as a promising biomarker for fast-growing tumors. Moreover, MTDH has been suggested as a potential drug target [42]. More recently, a differential co-expression network was reconstructed to evaluate the invasiveness-related signatures of NF-PitNETs and co-expressed and co-regulated mRNA modules were proposed as potential system biomarkers for prognosis and invasiveness of NF-PitNETs [9]. CEACAM6, CYP4B1, EIF2S2, HID1, IFFO1, MYO18A, PDCD2, SGIP1, and SWSAP1 have been suggested as prognostic biomarkers and potential therapeutic targets in future studies [6]. The invasiveness of NF-PitNETs was also investigated in another microarray study. The expression patterns of MYO5A and IGFBP5 confirmed by qRT-PCR were able to distinguish grossly invasive from non-invasive tumors. The upregulation of the protein level of MYO5A was also reported and suggested as a marker of invasiveness [27]. In another study, the expression of GALNT9 was significantly downregulated in invasive NF-PitNETs and may predict tumor invasiveness or recurrent growth features [30]. Survivin (encoded by BIRC5 gene) is another proposed therapeutic biomarker that has been upregulated in NF-PitNETs [44]. The low-level expressions of IL-6, STAT3, POUF1, and BCL6 genes have been linked to the hormone- and immune-related signaling pathways that promote the occurrence of PitNETs [45]. Microarray analysis of NF-PitNETs revealed aberrant expressions of SFRP1, TLE2, PITX2, NOTCH3, and DLK1, which were associated with the developmental Wnt and Notch signaling pathways, suggesting these genes as therapeutic targets in terms of enhancing Notch signaling [17].
In a recent comprehensive study to elucidate the pangenomic classification of PitNETs, silent corticotrophic PitNETs exhibited corticotrophic and gonadotrophic signatures such as GATA3 and NR5A1 [25]. Another integrated bioinformatics analysis of PitNETs revealed ten hub genes, including PHLPP, ENO2, ACTR1A, EHHADH, EHMT2, FOXO1, DLD, CCT2, CSNK1D, and CETN2, which have been proposed as potential biomarkers with diagnostic value in PitNETs [46]. The expression of MMP-9 has been reported to distinguish invasive PitNETs from non-invasive ones, even in the early stages of invasiveness. Moreover, MMP-9 has been proposed as a potential biomarker to predict the invasive behavior of PitNETs and as a therapeutic target to control dural invasion [47]. Differential expression of LGALS3, hASH1, ID2, and TLE-4 genes was found to play an essential role in the development of pituitary carcinomas [48]. In another study, a novel direct data integration strategy was used to comprehensively identify PitNET markers and GAL, LMO4, STAT3, PD-L1, TGFB, and TGFBR3 were suggested as promising target candidates for the development of PitNET immunotherapy [49]. Overexpression of DNA methyltransferases (DNMTs) has also been detected in PitNETs. DNMT1 expression was found significantly increased in macroadenomas and invasive tumors. DNMT3A expression was also detected at elevated levels in invasive tumors and grade IV tumors. There was also a significant association between DNMT1-DNMT3A and high-methylation status after adjusting for clinicopathological features. Therefore, upregulation of DNMT1 and DNMT3A has been associated with aggressive tumor behavior and high-methylation status in PitNETs [33].
With respect to null cell adenomas, a microarray study reported that SYT, ATP5B, and MDH1 were upregulated compared to normal pituitary and suggested as genes with potential oncogenic significance [51]. In another study comparing gonadotrophic and null cell tumors with healthy pituitary, upregulation of GADD45β was reported. It was associated with tumor suppression, decreased colony formation, and blockage of proliferation in mouse gonadotrophic cells. Moreover, GADD45β has been proposed as a novel pituitary tumor suppressor [50].
miRNAs
miRNAs are small non-coding RNAs and are classified as regulatory molecules essential for development and viability [81]. Based on the increasing evidence about miRNAs, their aberrant expression is associated with human diseases, including cancer, and they have been investigated as candidate cancer biomarkers for the diagnosis, prognosis, and treatment of various cancers [82]. Several miRNAs have been proposed as promising biomarkers contributing to diagnosis, prognosis, prediction of progression/recurrence/remission, and postoperative outcomes for PitNETs [83].
Among the different PitNET types, a large number of miRNAs have been reported as biomarkers, including miR-15a, miR-16, miR-133 [52, 57, 59]. In one of the recent studies conducted by Neou and colleagues (2020), thyrotrophic tumors exhibited higher expression of a particular miRNA cluster (including MEG3 and other 84 miRNAs) among all subtypes of PitNETs, whereas another miRNA cluster (miR532-let7 and other 16 miRNAs) was expressed at lower levels in the same tumor subtype [25]. Moreover, the expression of four miRNAs, including miR-574, miR-195, miR-497-5p, and let-7b, was found to negatively correlate with DNA methylation status in thyrotrophic tumors. Differentially expressed miRNAs including miR-212, miR-026a, miR-150, miR-152, miR-191, and miR-192 were overexpressed, while miR-024-1 and miR-098 were downregulated in pitNET samples compared to healthy pituitary tissues. These miRNAs were described as inducers of tumor cell proliferation [52]. Moreover, miR-15a and miR-16-1 were found to be downregulated in PitNETs and associated with tumor growth via an increase in tumor diameter and p43 secretion [53].
miR-516a-3p, miR-93, and miR20a were significantly upregulated and associated with decreased expression of tumor suppression protein Wee1 in NF-PitNETs and gonadotrophic tumors. The significantly upregulated miR-128a, miR-516a-3p, and miR-155 also targeted Wee1 and downregulated its expression in hormonally silent and gonadotrophic PitNETs [54]. Among the six most dysregulated differentially expressed miRNAs, hsa-miR-181b-5p, hsa-miR-181d, hsa-miR-191-3p, and hsa-miR-598 were found upregulated and hsa-miR-3676-5p and hsa-miR-383 were downregulated. The expression of hsa-miR-181a-5p was further verified by qRT-PCR, and the results suggested hsa-miR-181a-5p as an effective biomarker and therapeutic target [55]. Overexpression of miR-424-5p was associated with suppression of proliferation, migration, and invasion, and promoted apoptosis of invasive PitNET cells [56].
In the case of invasive PitNETs, miR-132 and miR-15a/16 were found downregulated in invasive pituitary tumor tissues and reported as tumor suppressors via inhibition of cell proliferation, migration, and invasion. Therefore, these miRNAs have also been described as potential therapeutic targets for invasive PitNETs [57]. Another study focusing on the invasiveness of PitNETs identified upregulation of miR-106b in invasive tumors. Inhibition of miR-106b significantly decreased tumor cell proliferation by cell cycle arrest and tumor invasiveness. It also increased PTEN expression, which affected tumor cell migration and invasion via the PI3K/AKT pathway and MMP-9 activity [58]. In addition to miR-106b, miR-133 was also reported for inhibiting migration and invasiveness in PitNETs. The miR-133 was negatively associated with FOXC1, whose suppressed expression destroyed the increased migration and invasion ability of tumors; thus, miR-133 was proposed as a potential therapeutic target for the treatment of invasive PitNETs [59].
Exosomal RNAs
Exosomes are small extracellular vesicles and contain proteins, lipids, and nucleic acids. In addition to various cell types, tumor cells also secrete exosomes. Therefore, exosomes are promising molecules for the study of disease mechanisms. In addition, exome-based therapeutics are now frequently used as part of personalized medicine approaches. In a recent study, exosomal RNAs from serum samples of NF-PitNET patients were used to investigate the effects of serum exosomes on the invasiveness of NF-PitNETs, and CDK6 and RHOU were detected as invasiveness markers [60]. Moreover, the elevation of exosomal miR-21 isolated from somatotrophic PitNETs and GH3 cells was found to increase osteoblast maturation, matrix protein secretion, and mineralization in vitro. Therefore, miR-21 has a function in the pathogenesis of acromegaly and has been proposed as a new potential therapeutic target [61]. More recently, miR-149-5p and miR-99a have been reported as tumor suppressors for invasive PitNETs and have been proposed as therapeutic agents in the form of exosomes [62].
LncRNAs
LncRNAs are a class of cellular RNAs and are more than 200 base pairs long. They have also evolved in essential basic biological processes. Increasing efforts have indicated that lncRNAs have great potential as diagnostic, prognostic biomarkers, and therapeutic targets in various diseases, including cancer [84].
In somatotrophic PitNETs, the expression of lncRNA H19 was found remarkably upregulated in invasive somatotrophic PitNETs and suggested as a potential target for invasiveness research [63]. RPSAP52 is a novel lncRNA targeting the HMGA2 gene. It was searched in somatotrophic PitNETs and found variable expression in somatotrophic tumors, while it was significantly upregulated in gonadotrophic and lactotrophic tumors. It has been reported that RPSAP52 contributes to tumorigenesis and has been accepted as an essential player for the development of PitNETs via affecting oncogenic proteins such as HMGA1 and HMGA2 [12]. In lactotrophic tumors, significant upregulation of lncRNA H19 was associated with tumor suppression via mTORC1 function in a microarray study both in vivo and in vitro [64].
In another microarray study, the expression of the lncRNA XIST was upregulated and associated with the progression of invasive PitNETs via increased bFGF and decreased mir-424-5p expression. Moreover, XIST has been presented as a novel therapeutic target [56]. The expressions of MEG3 and HOTAIR were investigated to determine their influence on the invasiveness of PitNETs. MEG3 was found to be downregulated, while HOTAIR was upregulated in invasive NF-PitNETs. These lncRNAs were reported as potential diagnostic biomarkers for invasive NF-PitNETs [65].
The co-expression network approach was also performed for NF-PitNETs and revealed potential targets such as MEG3, ENST00000501583, and n334366 for future studies. These lncRNAs were reported to be the most significantly downregulated and n334366 was also associated with the neuroactive ligand-receptor interaction pathway [66]. CCAT2 is a newly identified lncRNA that is upregulated in PitNETs and associated with poor prognosis. CCAT2 has been reported to have a positive correlation with cell proliferation, migration, and invasion. It also has an oncogenic function that promotes tumor development via suppression of PTTG1 degradation [67]. For null cell PitNETs, the expression of C5orf66-AS1 was significantly downregulated in invasive PitNETs compared to non-invasive ones. It also had a negative association with tumor diameter. Due to its ability to inhibit tumor development, C5orf66-AS1 has been proposed as a tumor suppressor [68].
circRNAs
circRNAs have been studied extensively in the last five years. They play many important roles in molecular processes, including cell proliferation, pluripotency, and cell lineage differentiation, and function as miRNA sponges [85]. A limited number of studies reported circRNAs as biomarkers in the case of PitNETs. The circRNA expression profiles were screened by microarray for invasive and non-invasive PitNETs. As a result, the expression of hsa_circ_102597 was found to be downregulated and remarkably associated with tumor size. The expression of hsa_circ_102597 was described as a promising diagnostic and prognostic biomarker for NF PitNETs [69]. Recently, circRNA profiling of somatotrophic PitNETs using a circRNA array was performed and circRNAs were found to be aberrantly expressed in these tumors compared to the normal pituitary gland. The circRNA hsa_circ_0001368 was significantly upregulated in somatotrophic PitNETs and confirmed by qRT-PCR. Knockdown of hsa_circ_0001368 was associated with a significant decrease in proliferation, hormone secretion, and invasion. It also showed a direct correlation with PIT1 and has been proposed as a new potential biomarker and therapeutic target for somatotrophic PitNETs [70]. CircOMA1 (hsa_circRNA_0002316) expression was shown to be associated with significantly decreased miR-145-5p, which plays a role in suppressing cell proliferation and invasion via TPT1 in NF-PitNETs. The interactive interaction of the circOMA1-miR-145-5p-TPT1 axis has been described as a promising treatment target and has the potential to capture the molecular pathogenesis of NF-PitNETs [14].
Considering the transcriptomic signatures reported in this section, most of them were related to uncovering the molecular mechanisms of PitNETs and contributed to new insights into the pathogenesis of the disease. Some transcriptomic signatures were associated with disease progression and tumor behavior (invasiveness, aggressiveness, proliferation), which are crucial parameters for the development of PitNET-specific predictive diagnostics. Since these parameters may be different in each patient due to tumor heterogeneity, a clear understanding of these parameters may be a great asset for the personalization of PitNET treatment regimens in the near future. Some of the transcriptomic signatures have also been reported as therapeutic targets that could be used in treatment modalities. These therapeutic target markers also have the potential to assess treatment efficacy, as their expressions may be affected by treatment.
Proteomic signatures
With the ability to integrate “omics” approaches, it has recently become possible to gain profound insights into the molecular mechanisms underlying disease processes and ultimately to develop successful precision or personalized medicine. Proteomics is an important discipline that aims to study the totality of all proteins in an organism, tissue, cell, or subfraction thereof. The proteome is not fixed; it differs from cell to cell and changes over time. To some extent, the proteome reflects the underlying transcriptome. However, because only 1.5 to 2 percent of the human genome is represented in the transcriptome as protein-coding genes, proteomic studies may allow active proteomes to be distinguished from inactive ones, enabling accurate functional inferences from gene expression data [86]. In terms of translating “omics” approaches to clinical applications, protein-based signatures are promising. Many studies have focused on accessible and non-invasive body fluids such as serum, plasma, urine, and saliva to investigate proteomic signatures as this could be useful for early diagnosis of various tumors in the clinic [87]. Proteomic analysis of PitNETs and healthy tissues has been performed in several studies and has revealed many potential biomarkers for early diagnosis, prognosis, and predictive, preventive, and personalized treatment of patients as clinical therapeutic targets (Table 1).
MS-based comparative proteomic analysis of 11 non-functional PitNETs and eight normal pituitary glands showed that dysregulation of SFRP1, TLE2, PITX2, NOTCH3, and DLK1 proteins indicated activation of Wnt and Notch signaling pathways and could represent potential NF-PitNET biomarkers [17]. Similarly, Evans et al. observed changes in various molecules of the Notch pathway in prolactinomas in their study. They integrated transcriptomic and proteomic strategies and used microarray analysis to identify 726 unique genes that were statistically differentially expressed between prolactinomas and normal glands. They also validated the upregulation of four genes and downregulation of 19 proteins by proteomic analysis. Based on the results of a proteomics strategy, Evans et al. indicated that dysregulations in various molecules (NOTCH3, DLK1, HES1, ASCL1, and FDZ7) of the Notch pathway were observed in prolactinomas, and the expression of the transcription factor Pit-1 and the survival factor BAG1 increased, while the expression of E-cadherin and N-cadherin decreased. In addition, the overexpression of TLE4, ANG-1, D-2, RIS1, and NBL1 genes were also observed in prolactinomas by using microarray or combining with qRT-PCR. Increased expression of multiple MHC class I antigen presenting genes in prolactinomas was also found to be significant [38]. Yu et al. performed a proteomic analysis focusing on differentially expressed molecules associated with NF-PitNET invasion. They examined 29 common DEGs and further validated the differential expression of CHGA and CLU in invasive NF-PitNETs and non-invasive NF-PitNETs, suggesting that they are a potential clinical predictive biomarker and drug-responsive molecule in invasive NF-PitNETs [77]. Another notable proteomics-based study was performed in invasive and non-invasive PA tissues using the 2DE-MS system and discovered the significantly high expression of Hint1 protein in PitNETs, suggesting that Hint1 may be associated with the invasive behavior of PitNETs [88]. Another study focused on proteomic evaluations suggested that LGALS3, hASH1, ID2, and TLE-4 proteins are involved in tumorigenesis and metastasis of pituitary carcinomas [48]. The study focusing on proteomic biomarker discovery by integrating transcriptomic and TMT-based quantitative proteomics approaches in non-functional PitNETs identified 10 highly interacting hub proteins and they further confirmed upregulation of SRC and AKT1 proteins by western blotting as potential biomarkers in non-functional PitNETs [76].
Feng et al. investigated adrenocorticotropic hormone-secreting PitNETs (somatotrophic PitNETs) at both metabolomic and proteomic levels to identify critical networks and signaling pathways potentially involved in the progression of ACTH-producing PitNETs using gas chromatography-mass spectrometry (GC-MS) and nano-liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) methods. Their results showed that glycolysis and fatty acid synthesis were downregulated in this tumor type. They also demonstrated that the Myc signaling pathway plays an important role in the metabolic changes and the formation of ACTH-secreting PitNETs [74]. Based on the results of a study investigating human prolactin (hPRL) protein in PAs by 2DE-MS method, Qian et al. reported that human pituitary glands contain six hPRL variants, and the differential expression patterns of the six hPRL variants differed significantly among subtypes of PitNETs [75].
Protein ubiquitination is an important posttranslational modification associated with several diseases, including PitNETs. Qian et al. performed the first ubiquitination analyses using an anti-ubiquitin antibody (specific for K-ε-GG)-based label-free quantitative proteomics method in PitNETs. Their results suggest that reduced ubiquitination of the 14-3-3 zeta/delta protein in PAs may end with its upregulation in NF-PitNETs, suggesting a contributing role of the 14-3-3 zeta/delta protein in disease progression [78].
Protein nitration is highly involved in various biological processes such as protein degradation, signal transduction, enzyme inactivation, apoptosis, and cell death, thus providing further dysfunctional changes. A remarkable study focuses on pituitary tumor-related nitroproteins intending to identify nitration regions and characterize the function of each nitroprotein using NTAC, MALDI-LTQ MS/MS system. They discovered 9 nitroproteins and 3 proteins that interacted with nitroproteins in NF-PitNET tissues [73]. These results are promising to elucidate the nitroproteome of PitNETs and to achieve a better understanding of the tumorigenesis of PitNETs that might provide to apply 3PM approach by highlighting molecular mechanisms and having the potential to develop preventive diagnostics. More recently, Aydin and colleagues reported protein level expressions of PI3K and Akt were dysregulated according to the treatment of proposed repurposed drugs for the treatment of lactotroph PitNETs and pointed out the importance of these proteins as targets for future treatment strategies [21]. Liu and colleagues performed a large-scale phosphoprotein profiling that reported phosphorylation-related signaling pathway network alterations in NF-PitNETs. The resultant hub molecules are SCG3, TSC2, ALB, AKT1, APOL1, ACACA, SPARCL1, SLC2A4, CHGB, and IGFBP5 that were obtained from proteome and transcriptome and belong to both phosphoproteins and invasive DEGs groups. The findings of the study provided insights into the molecular mechanisms of NF-PitNETs which might lead to the application of 3PM in clinical practice via elucidation of the phosphoprotein biomarkers that can be used as therapeutic targets [18]. Another proteome study unveiled FSH-related proteomic variations and alterations in the molecular networks in FSH-positive NF-PitNETs. They reported three signaling pathways including ECM-receptor interaction, focal adhesion, and PI3K-Akt signaling pathways and corresponding overexpressed proteins such as ITGA1, ITGA6, ITGB4, pAKT, and FSHR. These signaling pathways and proteins were associated with the invasiveness of the tumors. The results of the study showed efficient stratification of post-surgical patients [19].
One of the most important points to mention at the proteome level is the availability of high-throughput methods for detecting posttranslational modifications of proteins. Since proteins are not static molecules, spatiotemporal detection of these modifications in different proteoforms is essential. This topic has been extensively addressed in some studies related to PitNETs [89, 90]. In addition, there is an unmet need to study PitNETs from tissues collected by less invasive methods rather than collecting only PitNET tumor tissues. Tissues that are less invasive to collect, such as CSF, blood plasma, urine, saliva, need to be studied to uncover if there is a correlation between tumor tissue signatures and non-invasive tissue signatures [91].
Glycomic signatures
“Glycome” is the entire repertoire of glycans and glycoconjugates that can be varied in response to specific conditions such as location, biochemical environment, or time in a cell/tissue. Tumor growth, differentiation, transformation, adhesion, metastasis, and immune surveillance are influenced by changes in the glycome profile [92]. Until recently, cancer research has focused mainly on the genome and transcriptome, with relatively little investigation of glycan alterations and glycoprotein biomarkers [93–95]. However, in parallel with the use of state-of-the-art technologies to identify glycan structures, the application of cancer-related glycan studies to determine potential cancer glycan biomarkers has increased over time. Direct analysis of glycan changes without any protein identification represents a new and innovative approach to disease marker discovery. Moreover, changes in proteoglycan profiles are associated with tumor differentiation and biological behavior. In cancer cells, changes in glycome and proteoglycan profiles are functionally important and may provide information for the development of potential diagnostic and therapeutic strategies. In this context, several studies have been reported on glycan changes and glycome profiles in PitNETs (Table 1).
Cornelius et al. studied endocan proteoglycan expression in 18 normal post-mortem pituitaries and 107 patients with PitNETs to investigate the relationship of endocan expression and aggressive behavior of PitNETs using immunohistochemistry and reverse transcription polymerase chain reaction (RT-PCR) techniques. They found that endocan has the potential as a novel and promising biomarker for aggressive behavior in PitNETs [11].
Glycosylation-the addition of a carbohydrate moiety to a protein, lipid, carbohydrate, or other organic molecule-is the most widespread and complex posttranslational modification of proteins and significantly affects protein function. Protein glycosylation plays an important role in cancer cell behavior in regulating cancer development and progression, serves as an important indicator, and provides several specific targets for therapeutic intervention. Chesnokova et al. examined clusterin glycosylation in PitNETs to investigate the mechanism of inhibition of cell proliferation. The study concluded that clusterin can limit the proliferation of benign adenomatous pituitary cells by stimulating the expression of p15 and p16 [71].
Polysialation represents a unique posttranslational modification of the neural cell adhesion molecule (NCAM). Trouillas et al. studied 82 pituitary tumors and 6 healthy tissues to compare the expression levels of NCAM and polysialylated NCAM. The results showed that the expression of NCAM was observed in both healthy and tumor tissues. However, polysialylated NCAM was not expressed in healthy pituitary but was found in 46.3% of typical pituitary tumors and in 85% of tumors selected as highly aggressive. The expression of polysialylated NCAM was significantly correlated with tumor invasion and confirmed the clinical diagnosis of aggressiveness [72].
Given the glycome signatures reviewed in this section, we can conclude that glycoproteins and glycan alterations may have significant potential for identifying disease progression and developing preventive diagnostics. The reported glycome signatures could provide additional information on tumor behavior such as proliferation potential, invasiveness and aggressiveness.
Integrating multi-omics data to converge personalized medicine
Molecules from different levels such as genome (genes), transcriptome (RNAs), proteome (proteins), metabolome (metabolites), lipidome (lipids), glycome (glycoproteins and glycan modifications), and epigenome (histone modifications and aberrant DNA methylations) are interconnected and in wonderful harmony with each other. In the disease stage, some dysregulations occur and this unique orchestra goes into dysfunction. Omics technologies have opened the gateway in which scientists can gain valuable knowledge and a holistic view of the dysfunction stage to elucidate what exactly is going on when a normal cell transition into tumor transformation. Biomarker discovery will help us shed light on this transformation and evaluate potential targets for disease progression, prevention, and therapy.
Theoretically, the basic idea underlying biomarker discovery is quite unproblematic: if we had adequate information about the intrinsic factors causing molecular pathophysiology, it would be likely to uncover disease-promoting pathways and targets to take preventive precautions even before the disease occurs [96, 97]. Since almost all diseases have multifactorial roots, their manifestation is influenced by genetic, epigenetic, environmental, and lifestyle factors. Therefore, knowledge from only one level of omic data can only provide a snapshot. However, disease mechanisms need to be considered from a multi-omics perspective within the systems biomedical approach [91, 96, 98, 99].
In recent decades, high-throughput technologies have contributed undeniable opportunities to unravel the mechanisms underlying not only cancers but also other diseases at multiple molecular levels. The integration and analysis of these multi-level data are essential and play a central role in uncovering disease mechanisms and generating actionable knowledge in the context of precision medicine [98, 100, 101]. Therefore, there is a logical shift in cancer care towards predictive, preventive, and personalized medicine [63, 91]. For the integration of multi-omics data, the most commonly used approaches are network-based methods, but machine learning methods are also becoming popular.
Integrative omics in the study of PitNETs have recently been seen in a couple of studies [20, 102]. It can be seen that research using the integration of multi-omics data is increasing to identify the molecular basis of PitNETs. Recently, Li and colleagues represented the proteoforms of human growth hormone in somatotroph PitNETs via utilization of 2DGE and 2DGE-based Western blot in combination with MS [20]. The findings of the study reported elevated abundance of 30 growth hormone proteoforms, reduced abundance of 5 growth hormone proteoforms, and 11 novel growth hormone proteoforms in somatotroph PitNETs compared to healthy counterparts. In another integrative multi-omics study, Long et al. performed a meta-analysis collecting numerous transcriptome and proteome data and concluded that four major molecular pathways were altered in NF PitNETs, including PI3K/AKT, mTOR, Wnt, and ERK/MAPK [102]. Wei and colleagues performed an integrated analysis of copy number variations, DNA methylation, and mRNA expression in both highly proliferative and low proliferative NF-PitNETs [103]. They concluded that both genetic and epigenetic aberrations altered gene expressions.
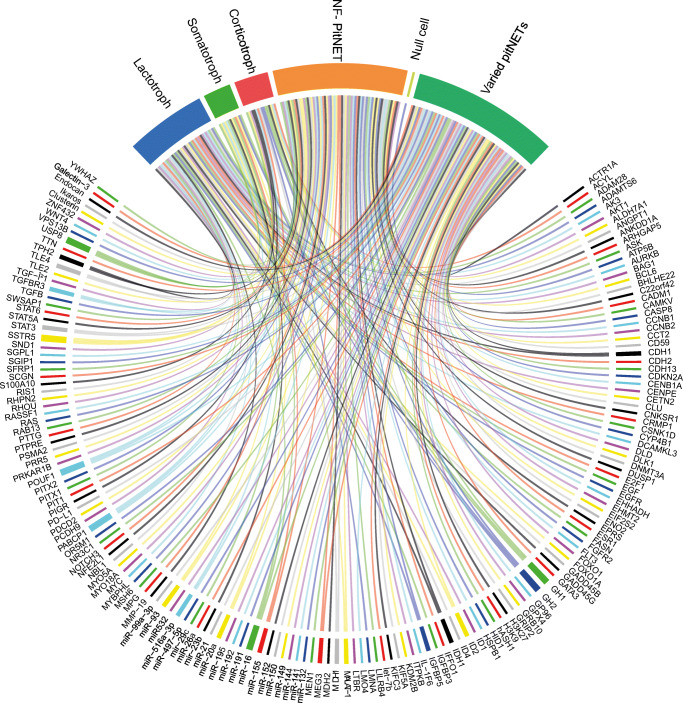
In this study, we gathered and summarized the current biomarkers for PitNETs in the multi-omics level (Fig. 2) and pointed out the importance of the multi-omics approach for the future elucidation of potential diagnostic/prognostic biomarkers, new therapeutic modalities and early detection techniques in the context of predictive diagnostics, targeted prevention, and personalized medicine.
Fig. 2.
The representation of biomarkers with the associated PitNET subtypes. The associations displayed with continuous lines
Moreover, the use of machine learning algorithms is another way to perform multi-omics integration studies. Machine learning-based methods for performing multi-omics data integration have been extensively investigated in various studies [101, 104, 105]. Cancer Integration via Multikernel Learning (CIMLR), a ML-based subtyping method that uses multi-omics data to reveal molecular subtypes of cancer that integrates multi-omics data to stratify cancer by molecular subtypes [106]. It has also been reported that CIMLR is able to predict outcomes and responses to treatments to promote personalized medical therapies. NEighborhood based Multi-Omics clustering (NEMO) is another ML-based novel algorithm for clustering multi-omics data and allows defining cancer subtypes faster and much easier than currently available clustering algorithms [107].
Support vector machines (SVM) are another ML-based unsupervised learning model. Ozer et al. pointed out that SVM and machine learning approaches can create new solutions and pathways in biomedical, bioengineering, and clinical applications to obtain better diagnostic and therapeutic predictive models [108]. MixOmics is an R package used for multivariate analysis of omics data with the goal of integration and biomarker discovery from multi-omics studies [109]. Integrative Network Fusion (INF) is a combination of network-based and ML-based methods and provides a computational workflow for the integration of high-throughput data [110]. It can integrate genomic, transcriptomic, and proteomic data and showed higher predictive performance and revealed more biologically informative biomarkers compared to single-step omics data. INF also analyzed data from histopathological and radiological images.
Despite these advances in computational and technological innovations, PitNETs lack clinically useful biomarkers or promising therapeutic candidates in clinical trial status derived from the integration of multi-omics data. In this step, it is very important and crucial to building collaborations between scientific disciplines such as basic sciences, clinical sciences, and computational sciences to address this unmet need. For the integration of multi-omics data in the context of elucidating disease mechanisms and patient stratification, the technological improvements with the help of ML-based methods, clinical sample selection that perfectly matches the disease with the help of better clinical evaluations, will accelerate the results of the studies so that complex biological problems will be easier to tackle in the near future.
Limitations of omics research
Accumulating evidence demonstrated that numerous studies have been conducted to investigate the PitNET mechanism and decipher molecular signatures. Despite the evidence of too many biomarkers in different studies, the results could be very inconsistent due to the limited number of samples analyzed in each study. Therefore, it is crucial to significantly increase the sample size, which will lead to more robust assessments. Another size issue arises from the signatures. In general, a small signature size makes clinical applicability easier than larger sizes. However, small signature sizes may decrease overall accuracy due to patient/tissue heterogeneity. Therefore, to avoid reduced accuracy and clinical applicability, meta-analysis studies can help to increase robustness and reliability [111].
Despite the contributing sides of “bulk” technologies, they are limited in presenting a specific portrait of the disease because bulk studies are from heterogeneous cell mixtures. Single-cell technologies have emerged to dissect the portrait of individual cells, resulting in gaining valuable insights into the cellular microenvironment of the tumor [112]. Single-cell omics also enable the discovery of rare cells that are the driving force behind disease progression, non-response to drugs, and metastatic/aggressive behavior. Even these advantages, the noise and high dimensionality of single-cell omics create compelling bottlenecks for data analysis [113].
Validation of proposed biomarkers is another pitfall for omics research. In both omics and multi-omics research, biomarkers or biomarker complexes are proposed and should be validated by specific methods to confirm the results. Perturbation by inhibitors, knockdown, or -out by siRNAs (silencing RNAs) can be useful methods for validation. In addition, correlation of the proposed marker(s) with other molecules from other omics layers may be possible to validate the results.
The individual drawbacks of each technology need to be explored. The improvements in the above bottlenecks will make omics-driven research in molecular biology studies and medicine more efficient in the future.
Concluding remarks
As expert recommendations, we would like to make a few points. First, there is no molecular marker for PitNETs from noninvasively collectable tissues such as blood, saliva, hair, or urine (with the exception of biochemical hormone assay from blood). As field researchers, we hope to produce such non-invasive biomarkers that provide a more patient-friendly method of detecting specific disease states for each patient. Second, the integration of multi-omics data holds great potential to study diseases holistically. As knowledge from different omics stages will shed light on disease patterns, assessing disease progression or applying the most appropriate treatment regimens could be possible in a more precise and individualized manner, leading to personalization of health care. This effort can be extended by constructing a specific disease screening panel consisting of multi-omics biomarkers from PitNETs. Since it is not easy to achieve high sensitivity and specificity with only a single biomarker, providing a panel of multi-omics biomarkers will improve the predictive power of diagnostics/prognostics. Such a disease monitoring/screening panel could facilitate accurate 3PM practice in the field. Third, technological advances in high-throughput biology are paving the way for a better understanding of the complex biological architecture of diseases. Since NF-PitNETs are difficult tumors to diagnose, the tumor may evolve into a macroadenoma by the time of diagnosis. Therefore, there is an urgent and unmet need to develop tools for early detection of NF-PitNETs using an integrative multi-omics approach. Fourth, daily lifestyle habits such as eating habits, stress response, smoking status, and heavy metal exposure are undeniable parameters that can influence disease progression. Therefore, a holistic approach that incorporates both daily life parameters and multi-omics patterns will lead to the discovery of precisely obtained treatment modalities. Fifth, imaging data have a great impact on disease diagnosis, therefore, merging imaging data with multi-omics data can provide more effective decisions for the management of PitNETs.
In this review, we have provided an overview of the current status of biomarker discovery in PitNETs. Numerous biomarkers have been identified and some of them give hope, but further confirmation and validation are needed. We also await the discovery of new and promising biomarkers. Disclosing the different levels of omics data is essential to understand the systemic manifestations of PitNETs and to better answer innovative integrative and multivariate questions in this disease. Integrative approaches provide us with the opportunity to assess the flow of information from one omics level to another and help us bridge the gaps from genotype to phenotype. Taking a holistic view of PitNETs can achieve individual response to treatment in multiple ways, increase the predictive and prognostic accuracy of disease phenotypes, and thus ultimately contribute to better treatment and prevention. In summary, a modern, systemic, and holistic understanding of PitNETs that bridges the gaps between diagnostics and therapeutic options will unravel the pathomechanisms under PitNETs and will directly redound to the predictive, preventive, and personalized medicine applied in NF-PitNETs.
Acknowledgements
The scholarships under the YOK 100/2000 Doctoral Fellowship Program and 2211-C Doctoral Fellowship Program under The Scientific and Technological Research Council of Turkey (TUBITAK) provided to Busra Aydin are greatly acknowledged. The authors especially thank Hande Beklen, for her support for the artwork.
Code availability
Not applicable.
Author contribution
BA, AC, and KYA designed the study. BA carried out the literature survey. BA, AC, and KYA interpreted the results. KYA conceived and directed the study. BA and AC drafted the manuscript. KYA revised the manuscript. All authors read and approved the final manuscript.
Data availability
Not applicable.
Declarations
Ethics approval and consent to participate
The authors declare that the information gathered throughout this study belonged to previously published studies; therefore, no ethical statement is required.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro-Oncology, 2020. 22:iv1–96. [DOI] [PMC free article] [PubMed]
- 2.Chatzellis E, Alexandraki KI, Androulakis II, Kaltsas G. Aggressive pituitary tumors. Neuroendocrinology. 2015;101:87–104. doi: 10.1159/000371806. [DOI] [PubMed] [Google Scholar]
- 3.Souteiro P, Karavitaki N. Dopamine agonist resistant prolactinomas: any alternative medical treatment? Pituitary. 2020;23:27–37. doi: 10.1007/s11102-019-00987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gariani K, Meyer P, Philippe J. Implications of somatostatin analogues in the treatment of acromegaly. Eur Endocrinol. 2013;9:132–5. Available from: https://pubmed.ncbi.nlm.nih.gov/29922369 [DOI] [PMC free article] [PubMed]
- 5.Shanik MH. Limitations of current approaches for the treatment of acromegaly. Endocr Pract. 2016;22:210–9. [DOI] [PubMed]
- 6.Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8:51–60. [DOI] [PMC free article] [PubMed]
- 7.Golubnitschaja O, Baban B, Boniolo G, Wang W, Bubnov R, Kapalla M, et al. Medicine in the early twenty-first century: paradigm and anticipation-EPMA position paper 2016. EPMA J. 2016;7:23. doi: 10.1186/s13167-016-0072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golubnitschaja O, Watson ID, Topic E, Sandberg S, Ferrari M, Costigliola V. Position paper of the EPMA and EFLM: a global vision of the consolidated promotion of an integrative medical approach to advance health care. EPMA J. 2013;4:12. doi: 10.1186/1878-5085-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aydin B, Arga KY. Co-expression network analysis elucidated a core module in association with prognosis of non-functioning non-invasive human pituitary adenoma. Front Endocrinol (Lausanne) Frontiers. 2019;10:361. doi: 10.3389/fendo.2019.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng S, Xie W, Miao Y, Guo J, Wang J, Li C, et al. Identification of key genes in invasive clinically non-functioning pituitary adenoma by integrating analysis of DNA methylation and mRNA expression profiles. J Transl Med. 2019;17:407. doi: 10.1186/s12967-019-02148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelius A, Cortet-Rudelli C, Assaker R, Kerdraon O, Gevaert M, Prévot V, et al. Endothelial expression of endocan is strongly associated with tumor progression in pituitary adenoma. Brain Pathol. 2012;22:757–764. doi: 10.1111/j.1750-3639.2012.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Angelo D, Mussnich P, Sepe R, Raia M, del Vecchio L, Cappabianca P, et al. RPSAP52 lncRNA is overexpressed in pituitary tumors and promotes cell proliferation by acting as miRNA sponge for HMGA proteins. J Mol Med. 2019;97:1019–1032. doi: 10.1007/s00109-019-01789-7. [DOI] [PubMed] [Google Scholar]
- 13.DeVore SB, Young CH, Li G, Sundararajan A, Ramaraj T, Mudge J, et al. Histone citrullination represses MicroRNA expression, resulting in increased oncogene mRNAs in somatolactotrope cells. Mol Cell Biol Am Soc Microbiol. 2018;38. [DOI] [PMC free article] [PubMed]
- 14.Du Q, Hu B, Feng Y, Wang Z, Wang X, Zhu D, et al. circOMA1-mediated miR-145-5p suppresses tumor growth of nonfunctioning pituitary adenomas by targeting TPT1. J Clin Endocrinol Metab. 2019;104:2419–2434. doi: 10.1210/jc.2018-01851. [DOI] [PubMed] [Google Scholar]
- 15.García-Martínez A, Sottile J, Sánchez-Tejada L, Fajardo C, Cámara R, Lamas C, et al. DNA methylation of tumor suppressor genes in pituitary neuroendocrine tumors. J Clin Endocrinol Metab. 2019;104:1272–1282. doi: 10.1210/jc.2018-01856. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y, Jiang X, Shen Y, Li M, Ma H, Xing M, et al. Frequent mutations and amplifications of the PIK3CA gene in pituitary tumors. Endocr Relat Cancer. 2009;16:301. doi: 10.1677/ERC-08-0167. [DOI] [PubMed] [Google Scholar]
- 17.Moreno CS, Evans C-O, Zhan X, Okor M, Desiderio DM, Oyesiku NM. Novel molecular signaling and classification of human clinically nonfunctional pituitary adenomas identified by gene expression profiling and proteomic analyses. Cancer Res AACR. 2005;65:10214–10222. doi: 10.1158/0008-5472.CAN-05-0884. [DOI] [PubMed] [Google Scholar]
- 18.Liu D, Li J, Li N, Lu M, Wen S, Zhan X. Integration of quantitative phosphoproteomics and transcriptomics revealed phosphorylation-mediated molecular events as useful tools for a potential patient stratification and personalized treatment of human nonfunctional pituitary adenomas. EPMA J. 2020;11:419–467. doi: 10.1007/s13167-020-00215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Cheng T, Lu M, Mu Y, Li B, Li X, et al. TMT-based quantitative proteomics revealed follicle-stimulating hormone (FSH)-related molecular characterizations for potentially prognostic assessment and personalized treatment of FSH-positive non-functional pituitary adenomas. EPMA J. 2019;10:395–414. doi: 10.1007/s13167-019-00187-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Wang X, Yang C, Wen S, Li J, Li N, et al. Human growth hormone proteoform pattern changes in pituitary adenomas: potential biomarkers for 3P medical approaches. EPMA J. 2021;12:67–89. doi: 10.1007/s13167-021-00232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aydin B, Arslan S, Bayraklı F, Karademir B, Arga KY. miRNA-mediated drug repurposing unveiled potential candidate drugs for prolactinoma treatment. Neuroendocrinology. Switzerland; 2021; Available from: https://www.karger.com/Article/Abstract/515801 [DOI] [PubMed]
- 22.Asa SL, DiGiovanni R, Jiang J, Ward ML, Loesch K, Yamada S, et al. A growth hormone receptor mutation impairs growth hormone autofeedback signaling in pituitary tumors. Cancer Res AACR. 2007;67:7505–7511. doi: 10.1158/0008-5472.CAN-07-0219. [DOI] [PubMed] [Google Scholar]
- 23.Salomon MP, Wang X, Marzese DM, Hsu SC, Nelson N, Zhang X, et al. The epigenomic landscape of pituitary adenomas reveals specific alterations and differentiates among acromegaly, Cushing’s disease and endocrine-inactive subtypes. Clin Cancer Res. 2018;24:4126–4136. doi: 10.1158/1078-0432.CCR-17-2206. [DOI] [PubMed] [Google Scholar]
- 24.Reincke M, Sbiera S, Hayakawa A, Theodoropoulou M, Osswald A, Beuschlein F, et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat Genet. 2015;47:31–38. doi: 10.1038/ng.3166. [DOI] [PubMed] [Google Scholar]
- 25.Neou M, Villa C, Armignacco R, Jouinot A, Raffin-Sanson M-L, Septier A, et al. Pangenomic classification of pituitary neuroendocrine tumors. Cancer Cell. 2020;37:123–134. doi: 10.1016/j.ccell.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Vergès B, Boureille F, Goudet P, Murat A, Beckers A, Sassolas G, et al. Pituitary disease in MEN type 1 (MEN1): data from the France-Belgium MEN1 multicenter study. J Clin Endocrinol Metab. 2002;87:457–465. doi: 10.1210/jcem.87.2.8145. [DOI] [PubMed] [Google Scholar]
- 27.Song Z-J, Reitman ZJ, Ma Z-Y, Chen J-H, Zhang Q-L, Shou X-F, et al. The genome-wide mutational landscape of pituitary adenomas. Cell Res. 2016;26:1255–1259. doi: 10.1038/cr.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bi WL, Horowitz P, Greenwald NF, Abedalthagafi M, Agarwalla PK, Gibson WJ, et al. Landscape of genomic alterations in pituitary adenomas. Clin Cancer Res AACR. 2017;23:1841–1851. doi: 10.1158/1078-0432.CCR-16-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kober P, Boresowicz J, Rusetska N, Maksymowicz M, Goryca K, Kunicki J, et al. DNA methylation profiling in nonfunctioning pituitary adenomas. Mol Cell Endocrinol. 2018;473:194–204. doi: 10.1016/j.mce.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Gu Y, Zhou X, Hu F, Yu Y, Xie T, Huang Y, et al. Differential DNA methylome profiling of nonfunctioning pituitary adenomas suggesting tumour invasion is correlated with cell adhesion. J Neuro-Oncol. 2016;129:23–31. doi: 10.1007/s11060-016-2139-4. [DOI] [PubMed] [Google Scholar]
- 31.Dudley KJ, Revill K, Whitby P, Clayton RN, Farrell WE. Genome-wide analysis in a murine Dnmt1 knockdown model identifies epigenetically silenced genes in primary human pituitary tumors. Mol Cancer Res. 2008;6:1567–1574. doi: 10.1158/1541-7786.MCR-08-0234. [DOI] [PubMed] [Google Scholar]
- 32.Zhu X, Asa SL, Ezzat S. Ikaros is regulated through multiple histone modifications and deoxyribonucleic acid methylation in the pituitary. Mol Endocrinol. 2007;21:1205–1215. doi: 10.1210/me.2007-0053. [DOI] [PubMed] [Google Scholar]
- 33.Ma H-S, Wang EL, Xu W-F, Yamada S, Yoshimoto K, Qian ZR, et al. Overexpression of DNA (cytosine-5)-methyltransferase 1 (DNMT1) and DNA (cytosine-5)-methyltransferase 3A (DNMT3A) is associated with aggressive behavior and hypermethylation of tumor suppressor genes in human pituitary adenomas. Med Sci Monit. International Scientific Literature, Inc.; 2018;24:4841–50. Available from: https://pubmed.ncbi.nlm.nih.gov/30002361 [DOI] [PMC free article] [PubMed]
- 34.Ling C, Pease M, Shi L, Punj V, Shiroishi MS, Commins D, et al. A pilot genome-scale profiling of DNA methylation in sporadic pituitary macroadenomas: association with tumor invasion and histopathological subtype. PLoS One. 2014;9:e96178. doi: 10.1371/journal.pone.0096178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan Y, Qian ZR, Sano T, Asa SL, Yamada S, Kagawa N, et al. Reduction of GSTP1 expression by DNA methylation correlates with clinicopathological features in pituitary adenomas. Mod Pathol. 2008;21:856–865. doi: 10.1038/modpathol.2008.60. [DOI] [PubMed] [Google Scholar]
- 36.Xue Y, Chen R, Du W, Yang F, Wei X. RIZ1 and histone methylation status in pituitary adenomas. Tumor Biol. 2017;39:1010428317711794. doi: 10.1177/1010428317711794. [DOI] [PubMed] [Google Scholar]
- 37.Duong CV, Yacqub-Usman K, Emes RD, Clayton RN, Farrell WE. The EFEMP1 gene: a frequent target for epigenetic silencing in multiple human pituitary adenoma subtypes. Neuroendocrinology. 2013;98:200–211. doi: 10.1159/000355624. [DOI] [PubMed] [Google Scholar]
- 38.Evans C-O, Moreno CS, Zhan X, McCabe MT, Vertino PM, Desiderio DM, et al. Molecular pathogenesis of human prolactinomas identified by gene expression profiling, RT-qPCR, and proteomic analyses. Pituitary. 2008;11:231–245. doi: 10.1007/s11102-007-0082-2. [DOI] [PubMed] [Google Scholar]
- 39.Tong Y, Zheng Y, Zhou J, Oyesiku NM, Koeffler HP, Melmed S. Genomic characterization of human and rat prolactinomas. Endocrinology. 2012;153:3679–3691. doi: 10.1210/en.2012-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wierinckx A, Auger C, Devauchelle P, Reynaud A, Chevallier P, Jan M, et al. A diagnostic marker set for invasion, proliferation, and aggressiveness of prolactin pituitary tumors. Endocr Relat Cancer. 2007;14:887–900. doi: 10.1677/ERC-07-0062. [DOI] [PubMed] [Google Scholar]
- 41.Finelli P, Pierantoni GM, Giardino D, Losa M, Rodeschini O, Fedele M, et al. The high mobility group A2 gene is amplified and overexpressed in human prolactinomas. Cancer Res AACR. 2002;62:2398–2405. [PubMed] [Google Scholar]
- 42.Falch CM, Sundaram AYM, Øystese KA, Normann KR, Lekva T, Silamikelis I, et al. Gene expression profiling of fast-and slow-growing non-functioning gonadotroph pituitary adenomas. Eur J Endocrinol. 2018;178:295–307. doi: 10.1530/EJE-17-0702. [DOI] [PubMed] [Google Scholar]
- 43.Galland F, Lacroix L, Saulnier P, Dessen P, Meduri G, Bernier M, et al. Differential gene expression profiles of invasive and non-invasive non-functioning pituitary adenomas based on microarray analysis. Endocr Relat Cancer. 2010;17:361–371. doi: 10.1677/ERC-10-0018. [DOI] [PubMed] [Google Scholar]
- 44.Németh K, Szücs N, Czirják S, Reiniger L, Szabó B, Barna G, et al. Survivin as a potential therapeutic target of acetylsalicylic acid in pituitary adenomas. Oncotarget Impact J LLC. 2018;9:29180–29192. doi: 10.18632/oncotarget.25650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang W, Xu Z, Fu L, Liu W, Li X. Pathogenesis analysis of pituitary adenoma based on gene expression profiling. Oncol Lett. 2014;8:2423–2430. doi: 10.3892/ol.2014.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng H, Deng Y, Wang L, Cheng Y, Xu Y, Liao J, et al. Identification of potential biomarkers with diagnostic value in pituitary adenomas using prediction analysis for microarrays method. J Mol Neurosci. 2019;69:399–410. doi: 10.1007/s12031-019-01369-x. [DOI] [PubMed] [Google Scholar]
- 47.Gong J, Zhao Y, Abdel-Fattah R, Amos S, Xiao A, Lopes MBS, et al. Matrix metalloproteinase-9, a potential biological marker in invasive pituitary adenomas. Pituitary. 2008;11:37–48. doi: 10.1007/s11102-007-0066-2. [DOI] [PubMed] [Google Scholar]
- 48.Ruebel KH, Leontovich AA, Jin L, Stilling GA, Zhang H, Qian X, et al. Patterns of gene expression in pituitary carcinomas and adenomas analyzed by high-density oligonucleotide arrays, reverse transcriptase-quantitative PCR, and protein expression. Endocrine. 2006;29:435–444. doi: 10.1385/ENDO:29:3:435. [DOI] [PubMed] [Google Scholar]
- 49.Yang Q, Wang Y, Zhang S, Tang J, Li F, Yin J, et al. Biomarker discovery for immunotherapy of pituitary adenomas: enhanced robustness and prediction ability by modern computational tools. Int J Mol Sci. 2019;20:151. doi: 10.3390/ijms20010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michaelis KA, Knox AJ, Xu M, Kiseljak-Vassiliades K, Edwards MG, Geraci M, et al. Identification of growth arrest and DNA-damage-inducible gene β (GADD45β) as a novel tumor suppressor in pituitary gonadotrope tumors. Endocrinology. 2011;152:3603–3613. doi: 10.1210/en.2011-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu J, Song H, Wang X, Shen Y, Chen F, Liu Y, et al. Gene expression profiling in human null cell pituitary adenoma tissue. Pituitary. 2007;10:47–52. doi: 10.1007/s11102-007-0008-z. [DOI] [PubMed] [Google Scholar]
- 52.Bottoni A, Zatelli MC, Ferracin M, Tagliati F, Piccin D, Vignali C, et al. Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J Cell Physiol. 2007;210:370–377. doi: 10.1002/jcp.20832. [DOI] [PubMed] [Google Scholar]
- 53.Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, Degli Uberti EC. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol. 2005;204:280–285. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- 54.Butz H, Liko I, Czirjak S, Igaz P, Khan MM, Zivkovic V, et al. Down-regulation of Wee1 kinase by a specific subset of microRNA in human sporadic pituitary adenomas. J Clin Endocrinol Metab. 2010;95:E181–E191. doi: 10.1210/jc.2010-0581. [DOI] [PubMed] [Google Scholar]
- 55.Wu S, Gu Y, Huang Y, Wong T-C, Ding H, Liu T, et al. Novel biomarkers for non-functioning invasive pituitary adenomas were identified by using analysis of microRNAs expression profile. Biochem Genet. 2017;55:253–267. doi: 10.1007/s10528-017-9794-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhou K, Li S, Du G, Fan Y, Wu P, Sun H, et al. LncRNA XIST depletion prevents cancer progression in invasive pituitary neuroendocrine tumor by inhibiting bFGF via upregulation of microRNA-424-5p. Onco Targets Ther. 2019;12:7095. doi: 10.2147/OTT.S208329. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Renjie W, Haiqian L. MiR-132, miR-15a and miR-16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5. Cancer Lett. 2015;356:568–578. doi: 10.1016/j.canlet.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Zheng Z, Zhang Y, Zhang Z, Yang Y, Song T. Effect of miR-106b on invasiveness of pituitary adenoma via PTEN-PI3K/AKT. Med Sci Monit Int Med J Exp Clin Res. 2017;23:1277. doi: 10.12659/MSM.900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang DS, Zhang HQ, Zhang B, Yuan ZB, Yu ZK, Yang T, et al. miR-133 inhibits pituitary tumor cell migration and invasion via down-regulating FOXC1 expression. Genet Mol Res. 2016;15:1–10. doi: 10.4238/gmr.15017453. [DOI] [PubMed] [Google Scholar]
- 60.Yu S, Wang X, Cao K, Bao X, Yu J. Identification of CDK6 and RHOU in serum exosome as biomarkers for the invasiveness of non-functioning pituitary adenoma. Chin Med Sci J. 2019;34:168–176. doi: 10.24920/003585. [DOI] [PubMed] [Google Scholar]
- 61.Xiong Y, Tang Y, Fan F, Zeng Y, Li C, Zhou G, et al. Exosomal hsa-miR-21-5p derived from growth hormone-secreting pituitary adenoma promotes abnormal bone formation in acromegaly. Transl Res. 2020;215:1–16. doi: 10.1016/j.trsl.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 62.Zhao P, Cheng J, Li B, Nie D, Li C, Gui S, et al. Up-regulation of the expressions of MiR-149-5p and MiR-99a-3p in exosome inhibits the progress of pituitary adenomas. Cell Biol Toxicol. 2021; Available from. 10.1007/s10565-020-09570-0. [DOI] [PubMed]
- 63.Lu T, Yu C, Ni H, Liang W, Yan H, Jin W. Expression of the long non-coding RNA H19 and MALAT-1 in growth hormone-secreting pituitary adenomas and its relationship to tumor behavior. Int J Dev Neurosci. 2018;67:46–50. doi: 10.1016/j.ijdevneu.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 64.Wu ZR, Yan L, Liu YT, Cao L, Guo YH, Zhang Y, et al. Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat Commun. 2018;9:1–14. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, Li C, Liu C, Yu S, Zhang Y. Expression of the long non-coding RNAs MEG3, HOTAIR, and MALAT-1 in non-functioning pituitary adenomas and their relationship to tumor behavior. Pituitary. 2015;18:42–47. doi: 10.1007/s11102-014-0554-0. [DOI] [PubMed] [Google Scholar]
- 66.Xing W, Qi Z, Huang C, Zhang N, Zhang W, Li Y, et al. Genome-wide identification of lncRNAs and mRNAs differentially expressed in non-functioning pituitary adenoma and construction of an lncRNA-mRNA co-expression network. Biol Open. 2019;8:bio037127. doi: 10.1242/bio.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu D, Zhang Y, Cui H. Long noncoding RNA CCAT2 is activated by E2F1 and exerts oncogenic properties by interacting with PTTG1 in pituitary adenomas. Am J Cancer Res. e-Century Publishing Corporation; 2018;8:245–55. Available from: https://pubmed.ncbi.nlm.nih.gov/29511595 [PMC free article] [PubMed]
- 68.Yu G, Li C, Xie W, Wang Z, Gao H, Cao L, et al. Long non-coding RNA C5orf66-AS1 is downregulated in pituitary null cell adenomas and is associated with their invasiveness. Oncol Rep. 2017;38:1140–1148. doi: 10.3892/or.2017.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu Y, Zhang N, Zhang S, Zhou P, Lv L, Richard SA, et al. Differential circular RNA expression profiles of invasive and non-invasive non-functioning pituitary adenomas: a microarray analysis. Medicine (Baltimore) 2019;98:e16148. doi: 10.1097/MD.0000000000016148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du Q, Zhang W, Feng Q, Hao B, Cheng C, Cheng Y, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0001368 is involved in growth hormone-secreting pituitary adenoma development. Brain Res Bull. 2020;161:65–77. doi: 10.1016/j.brainresbull.2020.04.018. [DOI] [PubMed] [Google Scholar]
- 71.Chesnokova V, Zonis S, Zhou C, Ben-Shlomo A, Wawrowsky K, Toledano Y, et al. Lineage-specific restraint of pituitary gonadotroph cell adenoma growth. PLoS One. 2011;6:e17924. doi: 10.1371/journal.pone.0017924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trouillas J, Daniel L, Guigard M-P, Tong S, Gouvernet J, Jouanneau E, et al. Polysialylated neural cell adhesion molecules expressed in human pituitary tumors and related to extrasellar invasion. J Neurosurg. 2003;98:1084–1093. doi: 10.3171/jns.2003.98.5.1084. [DOI] [PubMed] [Google Scholar]
- 73.Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal Biochem. 2006;354:279–289. doi: 10.1016/j.ab.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 74.Feng J, Zhang Q, Zhou Y, Yu S, Hong L, Zhao S, et al. Integration of proteomics and metabolomics revealed metabolite–protein networks in ACTH-secreting pituitary adenoma. Front Endocrinol (Lausanne) 2018;9:678. doi: 10.3389/fendo.2018.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qian S, Yang Y, Li N, Cheng T, Wang X, Liu J, et al. Prolactin variants in human pituitaries and pituitary adenomas identified with two-dimensional gel electrophoresis and mass spectrometry. Front Endocrinol (Lausanne) 2018;9:468. doi: 10.3389/fendo.2018.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng T, Wang Y, Lu M, Zhan X, Zhou T, Li B, et al. Quantitative analysis of proteome in non-functional pituitary adenomas: clinical relevance and potential benefits for the patients. Front Endocrinol (Lausanne) Frontiers. 2019;10:854. doi: 10.3389/fendo.2019.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu S-Y, Hong L-C, Feng J, Wu Y-T, Zhang Y-Z. Integrative proteomics and transcriptomics identify novel invasive-related biomarkers of non-functioning pituitary adenomas. Tumor Biol. 2016;37:8923–8930. doi: 10.1007/s13277-015-4767-2. [DOI] [PubMed] [Google Scholar]
- 78.Qian S, Zhan X, Lu M, Li N, Long Y, Li X, et al. Quantitative analysis of ubiquitinated proteins in human pituitary and pituitary adenoma tissues. Front Endocrinol (Lausanne) 2019;10:328. doi: 10.3389/fendo.2019.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carrillo-Najar C, Rembao-Bojórquez D, Tena-Suck ML, Zavala-Vega S, Gelista-Herrera N, Ramos-Peek MA, et al. Comparative proteomic study shows the expression of Hint-1 in pituitary adenomas. Diagnostics. 2021. [DOI] [PMC free article] [PubMed]
- 80.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015;25:137–147. doi: 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S, et al. miRNA-based biomarkers, therapies, and resistance in cancer. Int J Biol Sci. 2020;16:2628. doi: 10.7150/ijbs.47203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Melmed S. Pathogenesis of pituitary tumors. Nat Rev Endocrinol. 2011;7:257. doi: 10.1038/nrendo.2011.40. [DOI] [PubMed] [Google Scholar]
- 84.Yousefi H, Maheronnaghsh M, Molaei F, Mashouri L, Reza Aref A, Momeny M, et al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2020;39:953–974. doi: 10.1038/s41388-019-1040-y. [DOI] [PubMed] [Google Scholar]
- 85.Yu C-Y, Kuo H-C. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26:29. doi: 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Macmillan Publishers Ltd.; 2001; [DOI] [PubMed]
- 87.Caliskan A, Gulfidan G, Sinha R, Arga KY. Differential interactome proposes subtype-specific biomarkers and potential therapeutics in renal cell carcinomas. J Pers Med. 2021;11:158. doi: 10.3390/jpm11020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carrillo-Najar C, Rembao-Bojórquez D, Tena-Suck ML, Zavala-Vega S, Gelista-Herrera N, Ramos-Peek MA, et al. Comparative proteomic study shows the expression of Hint-1 in pituitary adenomas. Diagnostics. 2021;11:330. doi: 10.3390/diagnostics11020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li J, Zhan X. Mass spectrometry-based proteomics analyses of post-translational modifications and proteoforms in human pituitary adenomas. Biochim Biophys Acta, Proteins Proteomics. 2021;1869:140584. doi: 10.1016/j.bbapap.2020.140584. [DOI] [PubMed] [Google Scholar]
- 90.Zhan X, Qian S. Prolactin proteoform pattern changed in human pituitary adenoma relative to control pituitary tissues. Proteoforms-Concept Appl Med Sci. IntechOpen; 2020.
- 91.Zhan X, Desiderio DM. The use of variations in proteomes to predict, prevent, and personalize treatment for clinically nonfunctional pituitary adenomas. EPMA J. 2010;1:439–459. doi: 10.1007/s13167-010-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kirmiz C, Li B, An HJ, Clowers BH, Chew HK, Lam KS, et al. A serum glycomics approach to breast cancer biomarkers. Mol Cell Proteomics. 2007;6:43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- 93.Özdemir V, Arga KY, Aziz RK, Bayram M, Conley SN, Dandara C, et al. Digging deeper into precision/personalized medicine: cracking the sugar code, the third alphabet of life, and sociomateriality of the cell. Omi A J Integr Biol. 2020;24:62–80. doi: 10.1089/omi.2019.0220. [DOI] [PubMed] [Google Scholar]
- 94.Özdemir V Why are some omics biotechnologies more popular than others? The sociomateriality of glycans offers new clues. Mary Ann Liebert, Inc., publishers 140 Huguenot Street, 3rd Floor New …; 2020. [DOI] [PubMed]
- 95.Kori M, Aydin B, Gulfidan G, Beklen H, Kelesoglu N, Caliskan Iscan A, et al. The repertoire of glycan alterations and glycoproteins in human cancers. Omi A J Integr Biol. Mary Ann Liebert, Inc., publishers 140 Huguenot Street, 3rd Floor New …; 2021 [DOI] [PubMed]
- 96.Aydin B, Arga KY, Karadag AS. Omics-driven biomarkers of psoriasis: recent insights, current challenges, and future prospects. Clin Cosmet Investig Dermatol. 2020;13:611. doi: 10.2147/CCID.S227896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Litman T. Personalized medicine—concepts, technologies, and applications in inflammatory skin diseases. Apmis. 2019;127:386–424. doi: 10.1111/apm.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arga KY. Interview with Prof. K. Yalçın Arga: A pioneer of multi-omics science and health care innovation. Omi A J Integr Biol. 2019;23:460–462. doi: 10.1089/omi.2019.0131. [DOI] [PubMed] [Google Scholar]
- 99.Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9:77–102. doi: 10.1007/s13167-018-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turanli B, Yildirim E, Gulfidan G, Arga KY, Sinha R. Current state of “omics” biomarkers in pancreatic cancer. J Pers Med. 2021;11:127. doi: 10.3390/jpm11020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang S, Chaudhary K, Garmire LX. More is better: recent progress in multi-omics data integration methods. Front Genet. 2017;8:84. doi: 10.3389/fgene.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Long Y, Lu M, Cheng T, Zhan X, Zhan X. Multiomics-based signaling pathway network alterations in human non-functional pituitary adenomas. Front Endocrinol. 2019;10:835. doi: 10.3389/fendo.2019.00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wei Z, Zhou C, Li M, Huang R, Deng H, Shen S, et al. Integrated multi-omics profiling of nonfunctioning pituitary adenomas. Pituitary. 2020; Available from. 10.1007/s11102-020-01109-0. [DOI] [PubMed]
- 104.Grapov D, Fahrmann J, Wanichthanarak K, Khoomrung S. Rise of deep learning for genomic, proteomic, and metabolomic data integration in precision medicine. OMICS. 2018;22:630–6.10.1089/omi.2018.0097 . [DOI] [PMC free article] [PubMed]
- 105.Nicora G, Vitali F, Dagliati A, Geifman N, Bellazzi R. Integrated multi-omics analyses in oncology: a review of machine learning methods and tools. Front Oncol. 2020;10:1030. doi: 10.3389/fonc.2020.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ramazzotti D, Lal A, Wang B, Batzoglou S, Sidow A. Multi-omic tumor data reveal diversity of molecular mechanisms that correlate with survival. Nat Commun. 2018;9:4453. doi: 10.1038/s41467-018-06921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rappoport N, Shamir R. NEMO: cancer subtyping by integration of partial multi-omic data. Bioinformatics. 2019;35:3348–3356. doi: 10.1093/bioinformatics/btz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ozer ME, Sarica PO, Arga KY. New machine learning applications to accelerate personalized medicine in breast cancer: rise of the support vector machines. Omi A J Integr Biol. 2020;24:241–246. doi: 10.1089/omi.2020.0001. [DOI] [PubMed] [Google Scholar]
- 109.Rohart F, Gautier B, Singh A, Lê Cao K-A. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chierici M, Bussola N, Marcolini A, Francescatto M, Zandonà A, Trastulla L, et al. Integrative network fusion: a multi-omics approach in molecular profiling. Front Oncol. 2020;10:1065. doi: 10.3389/fonc.2020.01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Finotello F, Calura E, Risso D, Hautaniemi S, Romualdi C. Editorial: multi-omic data integration in oncology. Front Oncol. Frontiers Media S.A.; 2020;10:1768. Available from: https://pubmed.ncbi.nlm.nih.gov/33042824 [DOI] [PMC free article] [PubMed]
- 112.Griffiths JA, Scialdone A, Marioni JC. Using single-cell genomics to understand developmental processes and cell fate decisions. Mol Syst Biol. 2018;14:e8046. doi: 10.15252/msb.20178046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raimundo F, Papaxanthos L, Vallot C, Vert J-P. Machine learning for single cell genomics data analysis. bioRxiv. Cold Spring Harbor Laboratory; 2021
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.