Abstract
Purpose
Multiple morphological abnormalities of the sperm flagella (MMAF) are important causes of male infertility. Mutations in DNAH1 are the main causative factors proven so far. We aim to determine the mutational landscape of DNAH1 in Chinese patients with MMAF.
Methods
Forty-one Chinese patients with MMAF were enrolled and underwent a 10-gene next-generation sequencing panel screening.
Results
Only the DNAH1 gene was found to have mutations in 12 of these unrelated individuals (29%). Combining published data from two other cohorts of Chinese men with MMAF, we suggest that p.P3909fs*33, p.R868X, p.Q1518X, p.E3284K, and p.R4096L are hotspot mutations. A polymorphism—rs12163565 (G>A)— showed linkage to p.P3909fs*33, suggesting that this involved a founder effect. Four of the 12 patients with DNAH1 mutations were able to use intracytoplasmic sperm injection with their partners and all were successful in obtaining embryos.
Conclusions
Hotspot mutations were identified for Chinese patients with MMAF. MMAF sub-phenotypes might be associated with different combinations of DNAH1 mutations.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-021-02201-5.
Keywords: Morphological abnormalities, Sperm flagella, Teratospermia, Next-generation sequencing panel, Intracytoplasmic sperm injection, Male infertility
Introduction
Sperm tail abnormalities are an important cause of human male infertility. The syndrome of multiple morphological abnormalities of the sperm flagella (MMAF) is characterized by short, absent, bent, coiled, and irregular flagella. To date, nearly 20 genes have been found to be associated with this phenotype [1]. Mutation in the gene encoding the dynein axonemal heavy chain 1 (DNAH1) that encodes for one of the inner axonemal dynein arms (7IDAs) is the first identified cause of MMAF in humans [2]. Multiple ultrastructural defects of the sperm flagella can occur in the same individual, such as lack of IDAs, axonemal disorganization, complete absence of the central doublet microtubule pair, supernumerary dense fibers, and absence of mitochondria [2].
Genetic testing among patients with the MMAF phenotype can have clinical implications. First, any therapies to improve sperm quality will not be recommended for patients with MMAF caused by genetic variants, so they can avoid unnecessary medication. Second, limited evidence suggests that MMAF caused by variants in different genes might be associated with intracytoplasmic sperm injection (ICSI) outcome. For example, good ICSI outcomes can be expected for those patients with MMAF carrying DNAH1 [3], CFAP43, and CFAP44 [4] mutations, while mutations to CFAP65 [5] and CEP135 [6] can impair ICSI outcomes. Third, genetically diagnosed patients need to be informed that any male offspring might also have MMAF and infertility if their spouse also carries a pathogenic variant of the same gene.
Clarification of the proportion of genetic causes in patients with MMAF is important for the design of cost-effective clinical diagnostic next-generation sequencing (NGS) panels. However, screening studies on such cohorts are still limited. Here we used a targeted NGS panel to screen 41 Chinese men with an MMAF phenotype and review previously reported DNAH1 mutations to identify recurrent and novel mutations. This work expands the mutational and phenotypic spectrum of DNAH1 and lays the foundation for the design of MMAF-specific diagnostic screening kits.
Materials and methods
Study subjects
This study was approved by the ethics committees of Renji Hospital, Shanghai Jiao Tong University, School of Medicine and Nanjing Drum Tower Hospital, Nanjing University Medical School. Forty-one patients with primary male infertility were diagnosed with severe asthenozoospermia and MMAF from November 2017 to June 2020. All patients underwent a comprehensive semen analysis according to the World Health Organization laboratory manual [7]. None was from consanguineous families. Some patients underwent light microscopy-based morphological analysis of semen samples. None of them was found to have the typical phenotype of primary ciliary dyskinesia such as chronic respiratory tract infections and abnormally positioned internal organs.
Genetic testing
Genomic DNA was extracted from blood samples collected from all enrolled patients using DNeasy Blood and Tissue Kits (Qiagen, Valencia, CA, USA). We used an NGS panel to perform genetic testing, including ten genes as follows: CFAP44 (OMIM: 617559), CFAP69 (OMIM: 617949), ARMC2 (OMIM: 618424), CFAP43 (OMIM: 617558), AK7 (OMIM: 615364), TTC21A (OMIM: 611430), WDR66 (OMIM: 618146), FSIP2 (OMIM: 615796), DNAH1 (OMIM: 603332), and QRICH2 (OMIM: 618304). The other known MMAF genes are not included because they were discovered after panel design and sample screening, such as DNAH6 [8], DNAH17 [9], CFAP65 [10, 11], CFAP70 [12], DNAH8 [13], CFAP47 [14], and TTC29 [15]. Coding exons and flanking introns (± 10 bp) of these genes were targeted and captured using IDT xGen Lockdown Probes (Integrated DNA Technologies, Coralville, IA, USA). DNA was quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), and the quality of libraries was assessed using 2100 Bioanalyzer High Sensitivity DNA assays (Agilent Technologies, Carlsbad, CA, USA). All DNA libraries were sequenced under 2 × 75-bp paired-end mode on the Illumina NextSeq 500 platform (Illumina Inc., San Diego, CA, USA).
Data analysis
Targeted NGS panel sequences in FASTQ format were filtered and aligned to the human reference sequence (hg19/GRCh37) with the use of BWA v0.7.13 [16]. Variants including single-nucleotide variants (SNVs) and insertion–deletion mutations (InDels) were genotyped from recalibrated BAM files using VarDict [17]. The ANNOVAR software [18] was used to annotate all detected variants. After a preliminary screening to remove silent mutations and polymorphisms (minor allele frequency > 1% in the gnomAD database), the remaining mutations were further evaluated and classified as pathogenic (P), likely pathogenic (LP), variant of unknown significance (VUS), likely benign (LB), or benign (B) following the American College of Medical Genetics (ACMG) guidelines [19]. Copy number variants were identified using the DNAcopy R package [20]. Candidate variants were checked manually using the Integrative Genomics Viewer [21], and then validated by Sanger sequencing to avoid false positives. All compound heterozygous mutations were found in trans by pedigree analyses. DNAH1 mutations were annotated using transcript NM_015512.
Haplotype phasing
The DNAH1 mutations c.11726_11727del (p.P3909fs*33, chr3: 52430998) and an upstream polymorphism rs12163565 (G>A, chr3: 52430526) were amplified at the same amplicon using primers F (5′-CCAGATATGGGTCTCAATGCTC-3′) and R (5′-TGTCTTTGTGGGATGGGATG-3′). To phase the two loci, purified PCR products were cloned into a pMD-18T vector (Takara, Dalian, China) to transform Escherichia coli cells. Transformants were selected randomly and sequenced using universal primers. For the patients who carried c.11726_11727del (p.P3909fs*33), only two recent samples from patients A5389 and A6254 were tested and the remaining DNA specimens were not available.
Results
Genetic mutations
Forty-one Chinese patients with MMAF were enrolled in this study. Of these, 12 (29%) were found to have DNAH1 homozygous or compound heterozygous mutations (Table 1). No candidate pathogenic variations were found in any other tested genes. A total of 17 different mutation types were identified in 12 patients, including 11 loss-of-function (LOF; six stop-gain, two frameshift, and three splicing sites) and six missense mutations. The DNAH1 c.11726_11727del (p.P3909fs*33) and c.4552C>T (p.Q1518X) were identified in five and three unrelated patients, respectively, giving allele frequencies of 7% (6/82) and 4% (3/82).
Table 1.
DNAH1 mutations identified in this study and in two previously reported cohorts
| ID | cDNA change | AA change | Exon/intron | Het/Hom | Clinical significance | SIFT | PolyPhen | AF |
|---|---|---|---|---|---|---|---|---|
| A2191 | c.4552C>T | p.Q1518X | Exon 27 | Het | LP (PVS1+PM2) | - | - | 0.0001669 |
| c.11787+1G>A | NA | Exon 73 | Het | LP (PVS1+PM2) | - | - | 0 | |
| A3010 | c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 |
| c.12089+1G>A | NA | Intron 75 | Het | P (PVS1+PM2+PP4) | - | - | 0 | |
| A3298 | c.4552C>T | p.Q1518X | Exon 27 | Het | LP (PVS1+PM2+PM3) | - | - | 0.0001669 |
| c.12287G>T | p.R4096L | Exon 76 | Het | LP (PM1+PM2+PM3_supporting+PP2+PP3+PP4) | D(0) | D(0.999) | 0 | |
| A3342 | c.7397G>A | p.R2466Q | Exon 48 | Het | VUS (PM1+PM2+PP2) | D(0.05) | B(0.15) | 0 |
| c.12287G>A | p.R4096H | Exon 76 | Het | LP (PM1+PM2+PP2+PP3) | D(0) | D(0.999) | 0 | |
| A5730 | c.6004C>T | p.R2002C | Exon 38 | Het | LP (PM1+PM2+PM3+PP3) | D(0) | D(0.988) | 0 |
| c.10982C>A | p.S3661X | Exon 69 | Het | LP (PVS1+PM2) | - | - | 0 | |
| A6328 | c.2602C>T | p.R868X | Exon 15 | Het | LP (PVS1+PM2) | - | - | 0 |
| c.12748C>T | p.R4250X | Exon 78 | Het | VUS (PM2+PM3) | - | - | 0.0001113 | |
| A3679 | c.5573T>C | p.L1858P | Exon 35 | Het | P (PM1+PM2+PM3+PP2+PP3) | D(0) | D(0.967) | 0 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1) | - | - | 0.001286 | |
| A4218 | c.11726_11727del | p.P3909fs*33 | Exon 73 | Hom | P (PVS1+PM3+PP1) | - | - | 0.001286 |
| A5389 | c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1) | - | - | 0.001286 |
| c.12264_12265del | p.W4089Gfs*51 | Exon 76 | Het | P (PVS1+PM2+PM3) | - | - | 0 | |
| A5601 | c.4552C>T | p.Q1518X | Exon 27 | Het | LP (PVS1+PM2+PM3) | - | - | 0.0001669 |
| c.9685C>T | p.R3229C | Exon 61 | Het | VUS (PM1+PM2+PP3) | D(0) | P(0.858) | 0 | |
| A6137 | c.6526-1G>T | NA | Intron 41 | Het | LP (PVS1+PM2) | - | - | 0 |
| c.9850G>A | p.E3284K | Exon 62 | Het | VUS (PM2+PM3+PP3+PP4) | D(0.01) | P(0.64) | 0.0003371 | |
| A6254 | c.5104C>T | p.R1702X | Exon 32 | Het | P (PVS1+PM2+PM3) | - | - | 0.0003894 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1) | - | - | 0.001286 | |
| P1 | c.4115C>T | p.T1372M | Exon 25 | Het | VUS (PM1+PM2+PP4) | D(0) | D(0.966) | 0 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 | |
| P2 | c.6822C>G | p.D2274E | Exon 43 | Het | VUS (PM1+PP4+BP4) | T(0.55) | B(0.065) | 0.001836 |
| c.9850G>A | p.E3284K | Exon 62 | Het | LP (PM2+PM3+PP3+PP4) | D(0.01) | P(0.64) | 0.0003371 | |
| P3 | c.6212T>G | p.L2071R | Exon 39 | Het | VUS (PM3_supporting+PP4) | D(0) | P(0.839) | 0 |
| c.12200_12202del | p.4067_4068del | Exon 76 | Het | P (PVS1+PM2+PP4) | - | - | 0 | |
| P4 | c.3836A>G | p.K1279R | Exon 22 | Het | B (PP4+BS1+BP4) | T(0.15) | B(0.003) | 0.01207 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 | |
| P5 | c.7377+1G>C | NA | Intron 47 | Hom | P (PVS1+PM2+PM3_supporting+PP4) | - | - | 0 |
| P6 | c.3108G>A | p.W1036X | Exon 19 | Het | P (PVS1+PM2+PM3_supporting+PP4) | - | - | 0 |
| c.5864G>A | p.W1955X | Exon 37 | Het | P (PVS1+PM2+PM3_supporting+PP4) | - | - | 0 | |
| P7 | c.6253_6254del | p.R2085fs*8 | Exon 39 | Het | P (PVS1+PM2+PM3_supporting+PP4) | - | - | 0 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 | |
| P8 | c.2610G>A | p.W870X | Exon 15 | Het | P (PVS1+PM2+PP4) | - | - | 0 |
| c.12287G>T | p.R4096L | Exon 76 | Het | LP (PM1+PM2+PM3_supporting+PP2+PP3+PP4) | D(0) | D(0.999) | 0 | |
| P9 | c.12397C>T | p.R4133C | Exon 76 | Het | LP (PM1+PM2+PM3_supporting+PP4) | D(0) | D(1) | 0.00005564 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 | |
| P10/P11 | c.7066C>T | p.R2356W | Exon 45 | Het | LP (PM1+PM2+PM3+PP4) | D(0) | D(1) | 0.00005585 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 | |
| P12 | c.5766-2A>G | NA | Intron 36 | Het | P (PVS1+PM2+PM3_supporting+PP4) | - | - | 0.0004452 |
| c.10630G>T | p.E3544X | Exon 67 | Het | P (PVS1+PM2+PP4) | - | - | 0 | |
| P1’ | c.7864C>T | p.R2622X | Exon 50 | Het | P (PVS1+PM2+PM3_supporting) | - | - | 0 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 | |
| P2’ | c.7075C>T | p.R2359C | Exon 45 | Het | LP (PM1+PM2+PM3_supporting+PP3) | D(0) | P(0.988) | 0 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 | |
| P3’ | c.10060_10061insATCT | p.E3354fs*28 | Exon 64 | Het | LP (PVS1+PM2) | - | - | 0 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 | |
| P4’ | c.4987C>T | p.R1663C | Exon 31 | Het | LB (PM1+BS1+BP6) | D(0) | B(0.121) | 0.01706 |
| c.7779C>G | p.S2593R | Exon 49 | Het | VUS (PM1+PM2) | D(0.01) | B(0.003) | - | |
| P5’ | c.11726_11727del | p.P3909fs*33 | Exon 73 | Hom | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 |
| P6’ | c.3970G>A | p.A1324T | Exon 23 | Het | VUS (PM1+PM2+PM3_supporting) | D(0.04) | B(0.048) | 0 |
| c.4552C>T | p.Q1518X | Exon 27 | Het | LP (PVS1+PM2+PM3) | - | - | 0.0001669 | |
| P7’ | c.2602C>T | p.R868X | Exon 15 | Het | LP (PVS1+PM2) | - | - | 0 |
| c.11726_11727del | p.P3909fs*33 | Exon 73 | Het | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 | |
| P8’ | c.11726_11727del | p.P3909fs*33 | Exon 73 | Hom | P (PVS1+PM3+PP1+PP4) | - | - | 0.001286 |
P1–P12 [25] and P1′–P8′ [27] were described in previously reported cohorts. The clinical significance of mutations was classified as pathogenic (P), likely pathogenic (LP), variant of unknown significance (VUS), likely benign (LB), or benign (B) based on the evidence outlined in the following brackets. NA not available, Het heterozygous, Hom homozygous; SIFT annotation: D deleterious (0–0.05), T tolerated (> 0.05); PolyPhen annotation: D probably damaging (0.957–1), P possibly damaging (0.453–0.956), B benign (0–0.452); AF allele frequency of East Asians in the gnomAD database
Clinical profiles
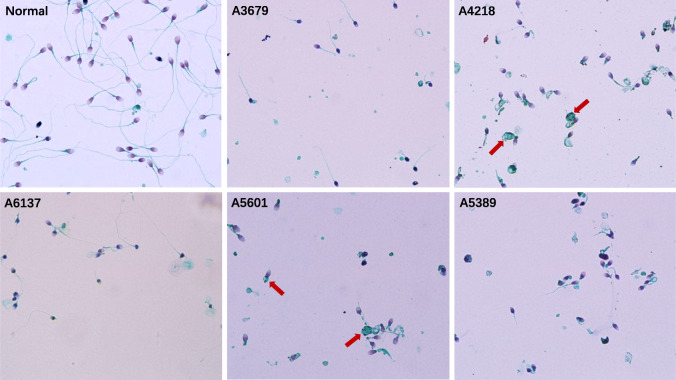
Ten of these twelve patients with DNAH1 mutations were from eastern China, aged between 24 and 41 years. Eight of them had oligospermia (sperm concentration < 5 × 106/mL in semen), of whom patient A3342 had severe oligospermia (< 1 × 106/mL). All patients showed loss of sperm motility and near 100% sperm tail abnormality (Table 2). Five patients underwent sperm morphology analysis, and patients A3679 and A6137 had several elongated sperm flagella visible by light microscopy, but spermatozoa with normal flagella were not found in patients A4218, A5601, and A5389. Instead, A4218 and A5601 had large residual cytoplasmic masses (Fig. 1). Four of the 12 patients underwent assisted reproduction with their partners using ICSI, and all were successful in generating embryos. Two of them had embryos transferred, one delivered successfully and the other experienced two implantation failure.
Table 2.
Semen parameters and ICSI outcomes from 12 patients with MMAF associated with DNAH1 mutations
| Semen parameters | ICSI outcomes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID | Birthplace (China) | Age | Sperm count (×106/mL) | Motility (a + b%) | Sperm tail abnormal rate (%) | Collected oocytes | Embryos | Transferred embryos | Clinical pregnancy | Delivery |
| A2191 | Eastern | 28 | 1.1 | 0 + 0 | 100 | 10 | 5 | 2 | 2 | 2 |
| A3010 | Eastern | 27 | 3.5 | 0 + 1.0 | 100 | 12 | 4 | 4 | 0 | 0 |
| A3298 | Northeast | 41 | 4.5 | 0 + 0 | 100 | 7 | 2 | - | - | - |
| A6137 | Eastern | 33 | 29.1 | 2.8 + 0 | 98 | 13 | 2 | - | - | - |
| A3342 | Central | 27 | <1 | - | - | - | - | - | - | - |
| A5730 | Eastern | 32 | 4.5 | 0 + 0 | 100 | - | - | - | - | - |
| A6328 | Eastern | 28 | 4.5 | 0 + 0 | 89 | - | - | - | - | - |
| A3679 | Eastern | 32 | 1.5 | 2.1 + 0 | 99 | - | - | - | - | - |
| A4218 | Eastern | 25 | 24.7 | 0 + 0 | 95 | - | - | - | - | - |
| A5389 | Eastern | 29 | 33.6 | 0 + 0 | 97 | - | - | - | - | - |
| A5601 | Eastern | 30 | 3.5 | 0 + 0 | 95 | - | - | - | - | - |
| A6254 | Eastern | 24 | 40.5 | 0 + 0 | 99 | - | - | - | - | - |
Semen analysis could not be performed for patient A3342 because of a low sperm concentration. The partner of A3010 experienced two implantation failure. Embryos were frozen and awaiting transfer for A3298 and A6137
ICSI intracytoplasmic sperm injection
Fig. 1.
Spermatozoa with normal and abnormal flagella. Short, absent, bent, coiled, and irregular sperm flagella with excess residual cytoplasm can be seen in high-power fields of view (×1000) for normal semen and five patients with MMAF. Sperms with excess residual cytoplasm are marked with arrows
Haplotype block of p.P3909fs*33
For patients A5389 and A6254, the DNAH1 mutation c.11726_11727del (p.P3909fs*33) was always located on the same allele as rs12163565 (G>A) (Supplementary Figure S1). For patient A4218 with the homozygous mutation c.11726_11727del (p.P3909fs*33), the genotype rs12163565 was also a homozygous locus (G>A) (Supplementary Table S1). This suggests that both loci are in the same haplotype block.
Discussion
Since DNAH1 mutations were first discovered as genetic causes of MMAF [2], the number of identified genes has reached nearly 20 in recent years [1]. However, these genes can only account for about 50% of patients with MMAF [22], which means there are still unknown genetic factors. Homozygous or compound heterozygous mutations of DNAH1 are common causative factors of MMAF, explaining 7.7% [23], 38.9% [2], 41.7% [24], or 57.1% [25] of the etiology in different cohort studies. Here, 41 patients with MMAF were enrolled and screened using a 10-gene NGS panel. Interestingly, only DNAH1 mutations were identified, while mutations of other genes were not found. The DNAH1 mutations explain the etiology of 12/41 (29%) patients with MMAF. The different rates between studies may be due to different inclusion criteria for patients with MMAF, as well as ethnic differences. However, this paper highlights the importance of DNAH1 in the etiology of men with MMAF in China.
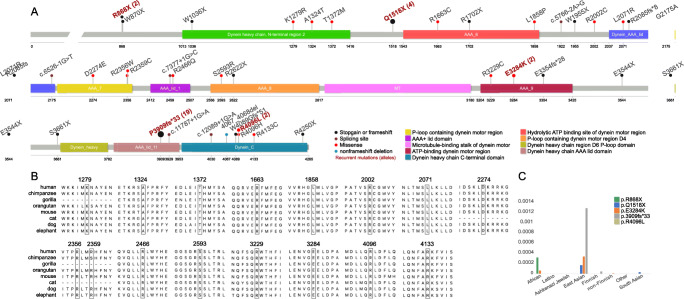
The mutational spectrum of DNAH1 is likely to be ethnically specific. For example, c.11788-1G>A is a hotspot splicing mutation for Tunisian patients with MMAF [2], while p.P3909fs*33 is a recurrent loss-of-function (LOF) mutation for Chinese men [25, 26]. However, large cohort studies are still lacking for clarification of the genetic and phenotypic spectrum of Chinese patients with MMAF. We have summarized the DNAH1 mutations in this study combined with previously reported cohorts of Chinese men with MMAF [25, 27]. The allele frequency of p.P3909fs*33 was 6/82 (7%) in this study, and 5/40 (13%) in Sha, Yang [25] (P10 and P11 were siblings in that study and counted as one sample, so the total allele number = [] × 2 = 40, see Fig. 2a). Wu, Wang [27] reported eight patients with MMAF caused by DNAH1 mutations in their supplementary material, six (75%) of whom carried at least one copy of p.P3909fs*33 (P1′–P8′; Table 1). An additional cohort of nine Chinese patients with MMAF also showed the high prevalence of p.P3909fs [26]. The allele frequency of p.P3909fs*33 is as high as 0.001286 (East Asians in the gnomAD database; https://gnomad.broadinstitute.org). We identified an upstream polymorphism, rs12163565 (G>A) that coexisted with p.P3909fs*33 in our NGS data (Supplementary Table S1) and was subsequently validated by E. coli cloning and sequencing (Supplementary Figure S1). This suggests that p.P3909fs*33 is in the same haplotype block as rs12163565 (G>A) and has been influenced by a founder effect in the East Asian population including Chinese men.
Fig. 2.
Mutational landscape of patients with MMAF with DNAH1 mutations. a Locations of the mutations on the DNAH1 protein domain, mutational types, and hotspot mutations. b All missense mutations exhibit high sequence conservation. c Allele frequencies of five hotspot DNAH1 mutations in various ethnic populations in the gnomAD database
By combining two other previous studies [25, 27], we identified four additional candidate hotspot mutations—p.R868X, p.Q1518X, p.E3284K—and p.R4096L, which were found in two, four, two, and two unrelated individuals, respectively (Fig. 2a). Except for p.R868X and p.R4096L, which do not occur in East Asian populations according to the gnomAD database, the allele frequencies of p.Q1518X and p.E3284K in East Asians are higher than in other populations (Fig. 2c). This implies that these loci might also have been influenced by a founder effect. Although two previously reported mutations c.3836A>G (p.K1279R) and c.4987C>T (p.R1663C) showed a high allele frequency (> 1%) in East Asian populations (Table 1), they were reclassified as B and LB according to the ACMG guidelines [19], so might not be causal MMAF.
Targeted NGS panel may has important clinical implications for patients with MMAF. For all those patients with biallelic pathogenic mutations, they were informed that ICSI is currently the only feasible treatment. Four of the 12 patients with DNAH1 mutations in our study underwent ICSI with their partners and all were successful in obtaining embryos (Table 2). Two couples underwent embryo transfer, one of whom delivered successfully and the other suffered an early miscarriage. Because patients with MMAF caused by DNAH1 mutations may have good ICSI outcomes [3], we encourage the couples to try for another cycle of ICSI. In addition, all these female partners were recommended to be screened for DNAH1 to avoid the possibility of the male offspring having MMAF and none of them was found to carry the pathogenic variant.
Five patients underwent sperm morphology analysis. The degree of flagellar abnormalities appeared to be more severe in patients A4218, A5601, and A5389. In contrast, more than two spermatozoa with elongated tails could be found per microscope field for patients A3679 and A6137 (Fig. 1). There are no published studies to date discussing whether combinations of different DNAH1 mutations result in MMAF sub-phenotypes. Both patients A3679 and A6137 each have one allele with a missense mutation, while both alleles of patients A4218 and A5389 carry LOF mutations. This speculation seems reasonable because LOF affects the integrity of the DNAH1 protein and thus affects the axonemal assembly process, leading to the phenotype of excess residual cytoplasm mainly comprising unassembled axonemal and periaxonemal components [28]. However, this hypothesis does not explain the severe phenotype of patient A5601. To further validate this hypothesis, more samples and further structural studies on spermatozoa are needed to establish the relationship between DNAH1 mutation combinations and MMAF sub-phenotypes.
Conclusion
Forty-one patients with MMAF were enrolled and underwent 10-gene NGS panel screening. Only the DNAH1 gene was found to have mutations in 12 unrelated individuals, giving a diagnostic rate of 29%. In these mutations, p.P3909fs*33 is a widely reported hotspot mutation and is found to be linked to polymorphism rs12163565 (G>A), suggesting that it arose as a founder effect. Moreover, p.R868X, p.Q1518X, p.E3284K, and p.R4096L were also identified as hotspot mutations by combining two previously reported cohorts. Limited evidence suggests that the MMAF sub-phenotypes may be associated with the different DNAH1 mutation combinations, which needs further cohort studies.
Supplementary information
Linkage analysis of theDNAH1p.P3909fs*33 and rs12163565. Mutant loci p.P3909fs*33 (chr3: 52430998) and rs12163565 (G>A, chr3: 52430526) were in the same allele subjected to cloning and sequencing in patients A5389 and A6254. (PDF 656 kb)
TheDNAH1mutation list of 12 diagnosed patients with MMAF. The genomic positions of p.P3909fs*33 and rs12163565 are marked in yellow. (XLSX 85 kb)
Acknowledgements
The authors thank all enrolled patients.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81971376), a grant from the Health Commission of Pudong New Area, Shanghai, People’s Republic of China (PW2020D-7), a grant from Shanghai Municipal Health Commission for advanced and suitable technology promotion projects (2019SY056), and Clinical Research Plan of SHDC (No. SHDC2020CR4035).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wen Yu and Miao An contributed equally to this work and should be considered co-first authors.
Change history
6/17/2021
A Correction to this paper has been published: 10.1007/s10815-021-02256-4
Contributor Information
Hongxiang Wang, Email: dr.whx_renji@163.com.
Zhipeng Xu, Email: xuzhipengyi@163.com.
References
- 1.Toure A, Martinez G, Kherraf ZE, Cazin C, Beurois J, Arnoult C, et al. The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet. 2020. [DOI] [PubMed]
- 2.Ben Khelifa M, Coutton C, Zouari R, Karaouzene T, Rendu J, Bidart M, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wambergue C, Zouari R, Fourati Ben Mustapha S, Martinez G, Devillard F, Hennebicq S, et al. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum Reprod. 2016;31:1164–1172. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]
- 4.Sha YW, Wang X, Su ZY, Mei LB, Ji ZY, Bao H, Li P. Patients with multiple morphological abnormalities of the sperm flagella harbouring CFAP44 or CFAP43 mutations have a good pregnancy outcome following intracytoplasmic sperm injection. Andrologia. 2019;51:e13151. doi: 10.1111/and.13151. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Ma Y, Zou S, Wang S, Qiu J, Xiao Q, Zhou L, Ping P. Comparison and outcomes of nonobstructive azoospermia patients with different etiology undergoing MicroTESE and ICSI treatments. Transl Androl Urol. 2019;8:366–373. doi: 10.21037/tau.2019.04.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sha YW, Xu X, Mei LB, Li P, Su ZY, He XQ, Li L. A homozygous CEP135 mutation is associated with multiple morphological abnormalities of the sperm flagella (MMAF) Gene. 2017;633:48–53. doi: 10.1016/j.gene.2017.08.033. [DOI] [PubMed] [Google Scholar]
- 7.World Health O. WHO laboratory manual for the examination and processing of human semen. World Health Organization: Geneva; 2010. [Google Scholar]
- 8.Tu C, Nie H, Meng L, Yuan S, He W, Luo A, Li H, Li W, du J, Lu G, Lin G, Tan YQ. Identification of DNAH6 mutations in infertile men with multiple morphological abnormalities of the sperm flagella. Sci Rep. 2019;9:15864. doi: 10.1038/s41598-019-52436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sha Y, Wei X, Ding L, Mei L, Huang X, Lin S, Su Z, Kong L, Zhang Y, Ji Z. DNAH17 is associated with asthenozoospermia and multiple morphological abnormalities of sperm flagella. Ann Hum Genet. 2020;84:271–279. doi: 10.1111/ahg.12369. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Wu H, Li F, Tian S, Kherraf ZE, Zhang J, Ni X, Lv M, Liu C, Tan Q, Shen Y, Amiri-Yekta A, Cazin C, Zhang J, Liu W, Zheng Y, Cheng H, Wu Y, Wang J, Gao Y, Chen Y, Zha X, Jin L, Liu M, He X, Ray PF, Cao Y, Zhang F. Biallelic mutations in CFAP65 cause male infertility with multiple morphological abnormalities of the sperm flagella in humans and mice. J Med Genet. 2020;57:89–95. doi: 10.1136/jmedgenet-2019-106344. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Tu C, Nie H, Meng L, Li Y, Yuan S, Zhang Q, du J, Wang J, Gong F, Fan L, Lu GX, Lin G, Tan YQ. Biallelic mutations in CFAP65 lead to severe asthenoteratospermia due to acrosome hypoplasia and flagellum malformations. J Med Genet. 2019;56:750–757. doi: 10.1136/jmedgenet-2019-106031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beurois J, Martinez G, Cazin C, Kherraf ZE, Amiri-Yekta A, Thierry-Mieg N, Bidart M, Petre G, Satre V, Brouillet S, Touré A, Arnoult C, Ray PF, Coutton C. CFAP70 mutations lead to male infertility due to severe astheno-teratozoospermia. A case report. Hum Reprod. 2019;34:2071–2079. doi: 10.1093/humrep/dez166. [DOI] [PubMed] [Google Scholar]
- 13.Liu C, Miyata H, Gao Y, Sha Y, Tang S, Xu Z, Whitfield M, Patrat C, Wu H, Dulioust E, Tian S, Shimada K, Cong J, Noda T, Li H, Morohoshi A, Cazin C, Kherraf ZE, Arnoult C, Jin L, He X, Ray PF, Cao Y, Touré A, Zhang F, Ikawa M. Bi-allelic DNAH8 variants lead to multiple morphological abnormalities of the sperm flagella and primary male infertility. Am J Hum Genet. 2020;107:330–341. doi: 10.1016/j.ajhg.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Tu C, Wang L, Wu H, Houston BJ, Mastrorosa FK, Zhang W, Shen Y, Wang J, Tian S, Meng L, Cong J, Yang S, Jiang Y, Tang S, Zeng Y, Lv M, Lin G, Li J, Saiyin H, He X, Jin L, Touré A, Ray PF, Veltman JA, Shi Q, O’Bryan MK, Cao Y, Tan YQ, Zhang F. Deleterious variants in X-linked CFAP47 induce asthenoteratozoospermia and primary male infertility. Am J Hum Genet. 2021;108:309–323. doi: 10.1016/j.ajhg.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lores P, Dacheux D, Kherraf ZE, Nsota Mbango JF, Coutton C, Stouvenel L, et al. Mutations in TTC29, encoding an evolutionarily conserved axonemal protein, result in asthenozoospermia and male infertility. Am J Hum Genet. 2019;105:1148–1167. doi: 10.1016/j.ajhg.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, Johnson J, Dougherty B, Barrett JC, Dry JR. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 21.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang WL, Tu CF, Tan YQ. Insight on multiple morphological abnormalities of sperm flagella in male infertility: what is new? Asian J Androl. 2020;22:236–245. doi: 10.4103/aja.aja_111_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coutton C, Vargas AS, Amiri-Yekta A, Kherraf ZE, Ben Mustapha SF, Le Tanno P, et al. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat Commun. 2018;9:686. doi: 10.1038/s41467-017-02792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amiri-Yekta A, Coutton C, Kherraf ZE, Karaouzene T, Le Tanno P, Sanati MH, et al. Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum Reprod. 2016;31:2872–2880. doi: 10.1093/humrep/dew262. [DOI] [PubMed] [Google Scholar]
- 25.Sha Y, Yang X, Mei L, Ji Z, Wang X, Ding L, Li P, Yang S. DNAH1 gene mutations and their potential association with dysplasia of the sperm fibrous sheath and infertility in the Han Chinese population. Fertil Steril. 2017;107:1312–1318. doi: 10.1016/j.fertnstert.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Jin H, Han F, Cui Y, Chen J, Yang C, Zhu P, Wang W, Jiao G, Wang W, Hao C, Gao Z. Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin Genet. 2017;91:313–321. doi: 10.1111/cge.12857. [DOI] [PubMed] [Google Scholar]
- 27.Wu H, Wang J, Cheng H, Gao Y, Liu W, Zhang Z, Jiang H, Li W, Zhu F, Lv M, Liu C, Tan Q, Zhang X, Wang C, Ni X, Chen Y, Song B, Zhou P, Wei Z, Zhang F, He X, Cao Y. Patients with severe asthenoteratospermia carrying SPAG6 or RSPH3 mutations have a positive pregnancy outcome following intracytoplasmic sperm injection. J Assist Reprod Genet. 2020;37:829–840. doi: 10.1007/s10815-020-01721-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield M, Thomas L, Bequignon E, Schmitt A, Stouvenel L, Montantin G, Tissier S, Duquesnoy P, Copin B, Chantot S, Dastot F, Faucon C, Barbotin AL, Loyens A, Siffroi JP, Papon JF, Escudier E, Amselem S, Mitchell V, Touré A, Legendre M. Mutations in DNAH17, encoding a sperm-specific axonemal outer dynein arm heavy chain, cause isolated male infertility due to asthenozoospermia. Am J Hum Genet. 2019;105:198–212. doi: 10.1016/j.ajhg.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linkage analysis of theDNAH1p.P3909fs*33 and rs12163565. Mutant loci p.P3909fs*33 (chr3: 52430998) and rs12163565 (G>A, chr3: 52430526) were in the same allele subjected to cloning and sequencing in patients A5389 and A6254. (PDF 656 kb)
TheDNAH1mutation list of 12 diagnosed patients with MMAF. The genomic positions of p.P3909fs*33 and rs12163565 are marked in yellow. (XLSX 85 kb)