Abstract
Radiomics is a promising technique for discovering image based biomarkers of therapy response in cancer. Reproducibility of radiomics features is a known issue that is addressed by the image biomarker standardisation initiative (IBSI), but it remains challenging to interpret previously published radiomics signatures. This study investigates the reproducibility of radiomics features calculated with two widely used radiomics software packages (IBEX, MaZda) in comparison to an IBSI compliant software package (PyRadiomics). Intensity histogram, shape and textural features were extracted from 334 diffusion weighted magnetic resonance images of 59 head and neck cancer (HNC) patients from the PREDICT-HN observational radiotherapy study. Based on name and linear correlation, PyRadiomics shares 83 features with IBEX and 49 features with MaZda, a sub-set of well correlated features are considered reproducible (IBEX: 15 features, MaZda: 18 features). We explore the impact of including non-reproducible radiomics features in a HNC radiotherapy response model. It is possible to classify equivalent patient groups using radiomic features from either software, but only when restricting the model to reliable features using a correlation threshold method. This is relevant for clinical biomarker validation trials as it provides a framework to assess the reproducibility of reported radiomic signatures from existing trials.
Subject terms: Cancer imaging, Image processing, Machine learning, Biomarkers
Introduction
Extracting textural features from medical images provides additional information1 to capture changes in tumour heterogeneity that may complement existing shape based metrics2. Radiomics3,4 is the high-throughput extraction of image features from standard-of-care medical images, with the hypothesis that macroscopic image features offer insight into disease process at a molecular level5. Radiomics analysis has been widely adopted in oncology research, showing potential to identify magnetic resonance image (MRI) based biomarkers for clinical outcomes in head and neck cancers (HNC)6. The evolution of radiomics features during treatment, commonly referred to as delta-radiomics, may offer more information than a single time point to identify biomarkers during radiotherapy or chemotherapy and has been explored in HNC with CT imaging7–10, positron emission tomography (PET) imaging11 and more recently in MRI studies12–16.
A review of head and neck cancer studies6 details the investigation of MRI radiomics features for applications such as image segmentation, histopathological classification and prognostic or predictive biomarkers. Previous HNC studies explore a range of MRI sequences, from anatomical imaging such as T1 weighted, T2 weighted and short tau inversion recovery (STIR) to functional imaging such as diffusion weighted (DW-MRI) and dynamic contrast enhanced imaging (DCE-MRI). This study focuses on radiomics features calculated on apparent diffusion coefficient (ADC) maps derived from DW-MRI images. The apparent diffusion coefficient has been correlated to cellularity in many tumour types17 and has been linked to cell proliferation in head and neck squamous cell carcinoma18. During radiotherapy, early changes in ADC have been linked to treatment response outcomes for multiple tumour types, making it a potential candidate for biological image-guided adaptive radiotherapy on MRI guided radiotherapy systems19.
Developing a radiomics model is often considered as a series of discrete tasks each with its own challenges5, with the variability of each task known to impact model performance20–22. In MRI studies, these effects have been investigated with regard to image acquisition23–27, region of interest segmentation23,28–30, image pre-processing28,29,31,32, feature extraction33–35 and feature reduction combined with classifier training13,36,37. To address the known variability issues of features extracted with different software33–35 the image biomarker standardisation initiative (IBSI)38 has proposed a set of feature extraction guidelines. In head and neck cancer, MRI studies have reported feature extraction with software such as MazDa24,39–41, IBEX42 and in-house solutions based on MATLAB37,42–49, none of which adhear to the IBSI guidelines.
Radiomic analysis generates hundreds of features, making feature reduction a crucial step to prevent overfitting when developing a radiomics model. Validation studies50,51 select a small set of features based on previously reported radiomic signatures. The IBSI guidelines should mitigate known feature reproducibility issues33–35 in future studies, but feature uncertainty remains a problem when interpreting previously reported radiomic signatures. This study investigates the correlation between features generated with open-source radiomic software packages (IBEX52 and MaZda53) used in many published studies against an open-source tool (PyRadiomics54) which follows the IBSI guidelines. We then explore the impact of non-reproducible radiomics features on a HNC radiotherapy response model using through therapy ADC radiomics features. Our comparison focuses on DW-MRI of head and neck cancer but provides general confidence on which previously reported radiomics features can be reproduced with software that adheres to the IBSI guidelines.
Results
Variation in radiomic features
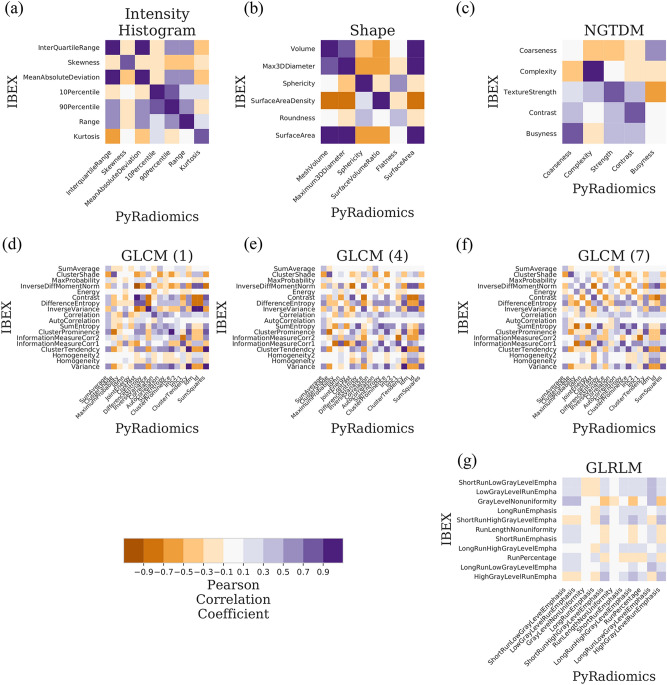
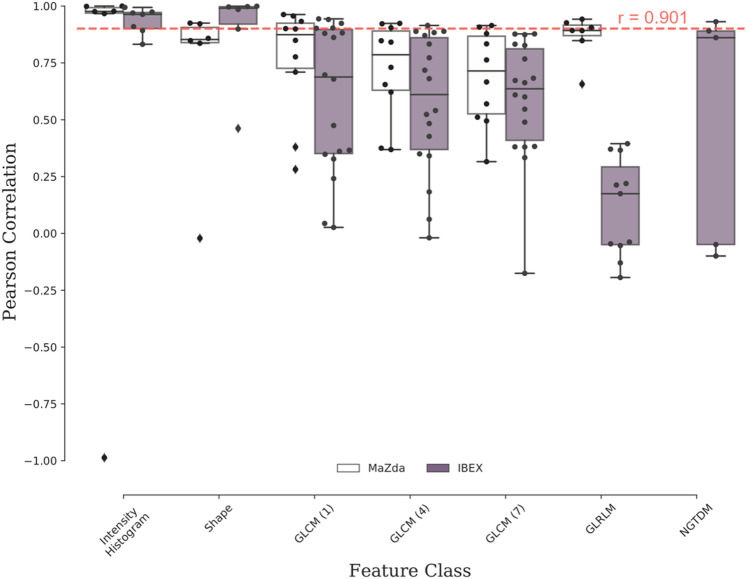
Radiomics features were extracted from 334 apparent diffusion coefficient maps from the prospective PREDICT-HN study59(Fig. 1) with PyRadiomics, IBEX and MaZda. A total of 314 features were extracted per ADC map (PyRadiomics: 125, IBEX: 110, MaZda: 79) including intensity histogram, shape and texture features. Based on name similarity, equation similarity and linear correlation, we identified that PyRadiomics and IBEX have 83 shared features and PyRadiomics and MaZda have 49 shared features. The linear correlation of PyRadiomics and IBEX features (Fig. 2) and PyRadiomics and MaZda features (Supplementary Figure 2) shows high correlation between intensity histogram features with a range of correlation between features in the shape and texture classes. A summary of correlation between features shared with PyRadiomics (Fig. 3) shows that on average IBEX extracts more highly correlated shape features, with MaZda extracting more highly correlated first order, GLCM and GLRM features. Features with a high Pearson’s coefficient () were considered reproducible between software packages. IBEX had 15 reproducible features (intensity histogram: 5, shape: 4, GLCM(1): 4, GLCM(4): 1, NGTDM: 1) and MaZda had 18 reproducible features (intensity histogram: 5, shape: 2, GLCM(1): 3, GLCM(4): 3, GLCM(7): 2, GLRLM: 3). For full detail of the feature names and correlations see Supplementary Tables 2, 3 and 4.
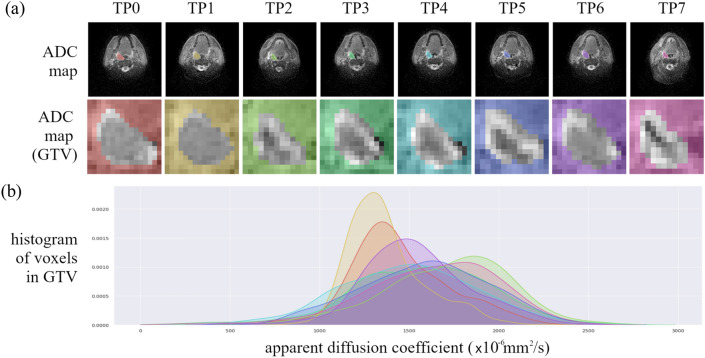
Figure 1.
Apparent diffusion coefficient (ADC) maps of a head and neck cancer patient throughout radiotherapy from the PREDICT-HN prospective clinical trial. (a) ADC maps are displayed with (top row) the gross tumour volume (GTV) highlighted in colour and (middle row) cropped to the GTV to focus on the region of interest for the radiomic analysis. Change in (b) the ADC histogram within the GTV is from baseline (TP0), weekly throughout radiotherapy (TP1–TP6) and post-radiotherapy (TP7) with the histogram colour matched to the GTV contour colour.
Figure 2.
Linear correlation of apparent diffusion coefficient (ADC) radiomics features between IBEX and PyRadiomics software. Correlation matrices are grouped by feature class such as (a) intensity histogram (b) shape (c) NGTDM (d–f) GLCM and (g) GLRLM with colour representing the Pearson correlation coefficient (r). An ideal correlation matrix would have diagonal elements of highly correlated features (r = 1.0, dark purple) between software packages. A list of shared features between software packages is in Supplementary Tables 2–4.
Figure 3.
Summary of linear correlation of apparent diffusion coefficient (ADC) radiomic features between PyRadiomics and (white) MaZda and (purple) IBEX software. The reproducibility threshold (red-dashed line) is defined as greater than a Pearson correlation coefficient of 0.901. This analysis identified a sub-set of reproducible features between IBEX and PyRadiomics from intensity histogram (5/7), shape (4/6), GLCM (neighbourhood 1:4/18, 4:1/18, 7:0/18), GLRLM (0/11) and NGTDM (1/5) categories. The sub-set of reproducible features between MaZda and PyRadiomics is intensity histogram (5/6), shape (2/6), GLCM (neighbourhood 1:3/10, 3:4/10, 7:2/10), GLRLM (3/7).
The PyRadiomics feature set was composed of 18 intensity histogram, 14 shape, 24 GLCM (at three neighbourhoods), 16 GLRLM and 5 NGTDM features. The IBEX feature set had (shared/total) 7/13 intensity histogram, 6/18 shape, 18/21 GLCM (at three neighbourhoods), 11/11 GLRLM and 5/5 NGTDM features. The MaZda feature set had (shared/total) 6/14 intensity histogram, 6/22 shape, 10/12 GLCM (at three neighbourhoods) and 7/7 GLRLM features. Shared features that had dissimilar names included shape features (Flatness, Roundness), (Volume, Area), (VoxelVolume, VoxelSize), (SurfaceArea, Perimeter), (SurfaceVolumeRatio, SurfaceAreaDensity), (FerretDiameter, Maximum3DDiameter), GLCM features (SumSquares, Variance), (InverseDifference, Homogeneity1), (InverseDifferenceMoment, Homogeneity2), (JointEnergy, AngularSecondMoment), (ClusterTendancy, JointEnergy) and one GLRLM feature (RunPercentage, Fraction).
Variation in patient modelling
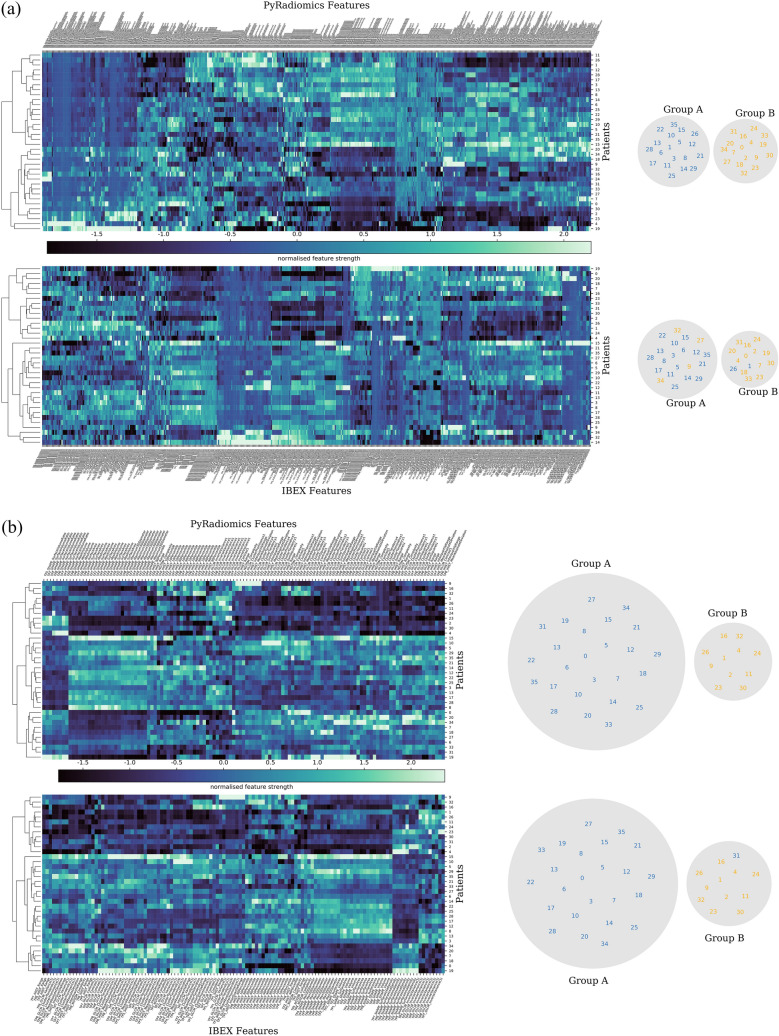
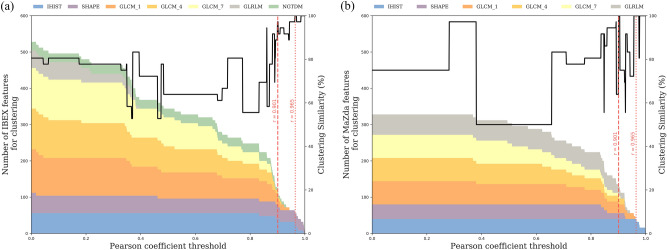
To investigate the impact of feature variability on patient modelling, 36 patients with imaging at all eight time points were clustered into two patient groups using an unsupervised learning method. Radiomic features were calculated throughout radiotherapy on apparent diffusion maps coefficient (ADC) maps, derived from diffusion weighted MRI. Hierarchal clustering based on all shared radiomics features (Fig. 4a, Supplementary Figure 1a) resulted in different patient groups, with six patients classified differently between PyRadiomics and IBEX and fifteen patients classified differently between PyRadiomics and MaZda groups. When clustering with a reduced set of reproducible features we observed nearly identical patient groups, with one patient classified differently between PyRadiomics and IBEX radiomics features (Fig. 4b) and identical PyRadiomics and MaZda groups (Supplementary Figure 2b). The change in clustering similarity over a range of reproducibility thresholds (Fig. 5) highlights that as the reliability threshold increases the number of included radiomics features decreases, with a general trend of increasing clustering similarity for a threshold above 0.90. To cluster identical patient groups in both software a more stringent reproducibility threshold () was required and reduced the IBEX feature set to 7 features (intensity histogram: 3, GLCM(1): 4) and the MaZda feature set to 5 intensity histogram features.
Figure 4.
Comparison of hierarchical clustering of patients with PyRadiomics and IBEX using (a) all shared features and (b) a sub-set of reproducible features (). Unsupervised hierarchical clustering generates a (left) radiomic signature of change in apparent diffusion coefficient (ADC) features after one fraction of radiotherapy in 36 head and neck cancer patients and (right) the resulting patient groups. Clustering with (a) non-reproducible features creates a difference in the patient groups generated from PyRadiomics or IBEX features. Clustering with (b) a sub-set of reproducible features leads to almost identical patient groups generated from PyRadiomics or IBEX features.
Figure 5.
Impact of the reproducibility threshold on the number of (a) IBEX and (b) MaZda radiomics features used for clustering and the resulting clustering similarity. The number and composition of feature types is shown with the coloured area chart and shows a decrease in the number of features as the reproducibility threshold increases. The (black line) clustering similarity is relatively unchanged for a threshold up till 0.85 after which there is a general increase in accuracy for IBEX features. Two reliability thresholds are highlighted where (red dashed line) generates patient groups in IBEX with one patient classified differently and identical patient groups in MaZda and the (red dotted line) generates identical patient groups in both software.
Discussion
Existing studies of variability in radiomics feature extraction33–35 explore a range of image modalities and feature extraction software. A study of mammograms and HNC computed tomography images33 also extracted IBEX and MaZda features, but compared them against one another and with two in-house software packages. One feature extraction study of HNC patients was less comparable as it analysed PET images35 and compared two in-house radiomics software packages. A study of HNC patients who had both CT and MRI imaging34 also extracted PyRadiomics features but compared them to features extracted with Moddicom55 and the radiomics extension to CERR56. Whilst that study34 also extracted features from MRI images of HNC patients, the features were from T2 weighted images, whereas our study is the first, to our knowledge, to investigate feature extraction stability on ADC maps from diffusion weighted MRI.
Our study observed a similar trend of feature extraction reproducibility to existing studies; intensity histogram features have high reproducibility across feature extraction software, with shape and textural features being more software package dependent. The study of mammograms and HNC CT images33 demonstrated high intra-class correlation (ICC) of intensity histogram features and low ICC of GLCM features, though this was partially attributed to the use of default GLCM extraction settings for each software. The study of HNC patients with both CT and MRI images34 showed that CT features had high Spearman correlation of intensity histogram, shape and GLCM features and lower ICC for GLRLM and grey-level size zone matrix (GLSZM) features. The T2 weighted MRI features compared in that study had high Spearman correlation for intensity histogram and shape features between PyRadiomics and CERR and only shape features highly correlated between PyRadiomics and Moddicom; the lack of correlation with intensity histogram features was attributed to Moddicom performing an image intensity correction on the T2 weighted MRI. In our study NGTDM features were extracted with IBEX and had a mixture of well correlated (1/5) and uncorrelated (4/5) features, the previously mentioned PET study35 also extracted NGTDM features but only a percentage of grouped textural features above an ICC threshold was reported. The similarity of our findings with those previously reported may indicate that variability in radiomics features due to the extraction software can be considered independent of the imaging modality.
Two radiomics feature reproducibility studies34,35 have investigated the impact of feature extraction variability on HNC model performance. The joint MRI and CT imaging study34 performed hierarchical clustering of patients for each feature class separately (intensity histogram, GLCM, etc.) and observed consistent clustering of clinical variables (TN category, GTV volume) for radiomics feature classes with a high reproducibility. The PET study35 demonstrated that models of local tumour control built on features from two different radiomics packages can stratify patients into very similar low or high risk recurrence groups, with the model features being highly correlated between the two software packages (ICC > 0.9). These studies and our results demonstrate that it is possible to generate equivalent radiomics models with different feature extraction software, but that care must be taken to ensure that features are highly correlated between the software packages. Additionally, our study extends this observation to patient classification based on radiomics features prior to treatment, at multiple time points during treatment and post-treatment.
A limitation of this and previous studies33–35 is the ambiguity in matching features between different software, firstly identifying shared features and secondly matching the feature extraction settings which often differ between software package. Name similarity does not guarantee equation similarity, for example the shape feature “SurfaceVolumeRatio” is calculated with a voxel based volume in IBEX and a mesh based volume in PyRadiomics. Equation similarity can be difficult to establish due to different notations, such as the formulation of the shape feature “SurfaceArea” between PyRadiomics and IBEX (Supplementary Table 6) which appear quite different but have a high correlation (0.998). We observed that equation similarity does not guarantee strong feature correlation, even with well-matched extraction settings, for a number of IBEX GLCM features such as “AutoCorrelation”, “MaximumProbability” and “JointEnergy” (Supplementary Table 7). Alternatively, minor differences in equations with non-ideally matched extraction settings showed moderate correlation, such as MaZda GLCM features “DifferenceVariance” and “InverseDifferenceMoment” (Supplementary Table 7). The naming issue can be avoided in future for a sub-set of features that have a unique identifier as defined in the IBSI guidelines38. To assist with consistent naming and feature extraction settings an ontology based radiomics workflow has been proposed57.
It is challenging to match the feature extraction settings between PyRadiomics, IBEX and MaZda. We were unable to define a reduced intensity range in MaZda for GLCM and GLRLM features, which may have negatively affected the feature correlation results, though we observed higher correlations in these feature classes from MaZda than with IBEX that had a matched intensity range and bin width. Similarly, we were unable to define identical directions for GLCM and GLRLM feature extraction across the software packages, averaging over different directions may have negatively affected our feature correlation results. A newer version of MaZda (qmazda) was selected for this study as it supports batch feature extraction, from the software documentation it was inferred that there was minimal change to the feature extraction code from earlier MaZda versions. The region of interest was calculated with a Plastimatch derived label map for PyRadiomics and MaZda and was calculated directly from the DICOM structure file (RTSTRUCT) by IBEX, which may have introduced variability in the image region used for feature extraction. We explored radiomics features calculated on the original ADC map to avoid additional variability from image filtration (Wavelet, Laplacian or Gaussian, etc.) between software implementations. Image filtration reproducibility between software using MRI images may be worth investigating as differences in wavelet filtered features from PET images have been previously reported35. Whilst our investigation of feature variability on patient modelling demonstrated general classification differences between software, it is challenging to quantify a clinical impact as the patient groups are not correlated with a clinical outcome. There is some evidence that unsupervised feature clustering is correlated with clinical outcomes in HNC34.
Magnetic resonance imaging offers insight into disease related anatomical and functional changes and is ideal for radiomics analysis as multiple MRI sequences can provide complimentary information58. This study reports the variability of radiomics features extracted from ADC maps, that are a relatively quantitative image, generating features not significantly influenced by scanner manufacturer or magnetic field strength26; radiomics features extracted from T1 and T2 weighted images are less reproducible across scanner and not recommended at this point for multicentre trials24. The increasing uptake of MRI for simulation and therapy in radiotherapy departments offers an unprecedented opportunity to characterise tumour response and personalise patient treatment. This study provides information on how feature extraction software can impact the reproducibility of a radiomics workflow, which we should endeavour to optimise in order to accelerate discovery through reproducible research and data sharing.
Conclusion
This work highlights feature and model reproducibility issues due to different radiomic analysis software. We propose a correlation threshold method to select reproducible features and demonstrate that the identified features from both software generate an equivalent model. This is relevant for the selection of radiomic features in clinical biomarker validation trials as it provides a framework to assess the reproducibility of radiomic signatures from existing studies.
Material and methods
Study cohort
The imaging data used in this study was collected as part of the prospective PREDICT-HN study59 (Fig. 1). The trial imaged 59 patients with head and neck squamous cell carcinoma who were treated with curative intent radiotherapy, patient details are summarised in Table 1. Trial participants were imaged prior to radiotherapy, weekly during radiotherapy (following fraction 5, 10, 15, 20, 25 and 30) and two to three months post-radiotherapy. Imaging data was acquired for all patients prior to radiotherapy (n = 59), with a lower number of images acquired during radiotherapy over the first three weeks (n = 40), at weeks four and five (n = 39), at week six (n = 38) and post-radiotherapy (n = 39).
Table 1.
Patient characteristics.
| Correlation cohort (n = 59) | Cluster cohort (n = 36) | |
|---|---|---|
| Sex | ||
| Male | 50 | 32 |
| Female | 9 | 4 |
| Age (median, range) | 59 (41–81) | 60 (41–81) |
| Primary site | ||
| Oropharynx | 39 | 26 |
| Larynx | 7 | 2 |
| Nasopharynx | 9 | 4 |
| Nasal cavity | 1 | 1 |
| Unknown primary | 3 | 3 |
| T stage | ||
| T0 | 3 | 3 |
| T1 | 8 | 5 |
| T2 | 20 | 12 |
| T3 | 12 | 7 |
| T4 | 16 | 9 |
| N stage | ||
| N0 | 11 | 5 |
| N1 | 9 | 5 |
| N2 | 38 | 26 |
| N3 | 1 | 0 |
| Photon | 42 | 24 |
| Proton | 17 | 12 |
| Radiation dose (cGy, median, range) | 6996 (6600–7000) | |
| Number of fractions | 33 (33–35) | |
Ethics approval and consent to participate
The PREDICT-HN study59 was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board at the University of Texas MD Anderson Cancer Center. The study is registered on clinicaltrials.gov with registration number NCT03491176, date of registration 09/04/2018 (retrospectively registered), date of enrolment of the first participant 30/05/2017. All study participants were 18 years and older, informed consent was obtained from all participants.
Imaging protocol
MRI imaging was performed on a Siemens 1.5 T Aera scanner to acquire both anatomical and functional images. The gross tumour volume (GTV) was contoured on pre-treatment T2 weighted turbo-spin-echo (T2w-TSE) images (voxel size = 0.5 mm, FOV = 256 × 256 mm, axial slices = 12, slice thickness = 2 mm, TE = 80 ms, TR = 4800 ms, FA = 90°, ETL = 15, pixel bandwidth = 300 Hz) by a radiation oncologist. Throughout treatment, the pre-treatment contours were rigidly registered to the through treatment images and manually adjusted to anatomical boundaries, then propagated onto the apparent diffusion coefficient (ADC) maps and visually verified. Diffusion weighted images (DWI) were acquired with the BLADE60,61 sequence (voxel size = 2 mm, FOV = 256 × 256 mm, axial slices = 25, slice thickness = 4 mm, TE = 50 ms, TR = 5400 ms, FA = 90°, b = 0,800 s/mm2, ETL = 15, pixel bandwidth = 1220 Hz). Apparent diffusion coefficient maps were calculated from the DWI images using the default mono-exponential model.
Feature extraction
Features were extracted from all the ADC maps (n = 334) using PyRadiomics (version 2.1.0), IBEX (version 1.0 Beta) and MaZda (qmazda 19.02). All available intensity histogram (IHIST), shape, grey-level co-occurrence matrix (GLCM62), grey-level run length matrix (GLRLM63) and neighbourhood grey-tone difference matrix (NGTDM64) features were calculated on the original ADC map only; the reproducibility of image filtration prior to feature extraction was considered outside the scope of this study. The region of interest for feature extraction was calculated directly from the DICOM radiotherapy contours (RTSTRUCT) with IBEX and an intermediate step to convert the DICOM contours to a binary label map with Plastimatch (version 1.7.3)65 was required for PyRadiomics and MaZda. The feature extraction settings (Supplementary Table 1) were set as the default IBEX settings and matched as closely as possible between the three software packages, based on available documentation. ADC maps were discretised (256 bins, bin width = 16) prior to calculation of IHIST and NGTDM features and with a reduced data range (100 bins, bin width = 21) for GLCM and GLRLM features. GLCM features were calculated at a series of neighbourhoods (1, 4, 7), asymmetric features were calculated with PyRadiomics and IBEX, symmetric features were calculated with MaZda to compensate for a reduced range of direction angles. Symmetric NGTDM features were calculated with a neighbourhood of three. Texture features (GLCM, GLRLM, NGTDM) were calculated in the axial plane over the three dimensional region of interest,an example of this is the NGTDM matrices being constructed with a 2D neighbourhood rather than a 3D neighbourhood. We calculated the average of all GLCM and GLRLM feature directions as PyRadiomics does not report or allow the specification of features for individual directions.
Variation in radiomic features
To determine the relationship between features generated with the established radiomics software (IBEX/MaZda) and the IBSI compliant software (PyRadiomics) we performed a linear regression analysis on radiomics features extracted from ADC maps of all patients at all time points. A linear fit was calculated between PyRadiomics and IBEX or MaZda for every feature in a given feature class (IHIST, SHAPE, GLCM, GLRLM, NGTDM). Features with an invalid value (i.e. infinity due to division by zero) were excluded during the linear regression. A list of shared radiomics features was collated by identifying features extracted by both PyRadiomics and the alternative software package, and was based primarily on name and equation similarity. A small number of features that showed a high linear correlation but had dissimilar names, for example ‘SurfaceArea’ and ‘Perimeter’, were also included. The list of shared features, feature correlations and the number of images used per feature correlation can be found in Supplementary Tables 2, 3 and 4. Full details of the equation comparison are in Supplementary Tables 5–9.
Variation in patient modelling
To demonstrate the potential impact of incorporating non-reproducible features in a radiomics model we used unsupervised learning to identify two groups of patients, based on radiomic features at pre-treatment, throughout radiotherapy and post-radiotherapy. Separate radiomics models were generated based on PyRadiomics and IBEX/MaZda features, first using all features and then with the sub-set of reproducible features. Reproducible features were selected as those with a high Pearson’s correlation coefficient () as calculated for the analysis of variation in radiomics features, the correlation threshold was determined as per the sensitivity analysis described below. Features that contained an invalid number (i.e. infinity) for any patient in the modelling cohort were excluded. Patients (n = 36) with image data for all time points were grouped with SciPy66,67 using Ward's minimum variance clustering method68 on scaled radiomics features (z-score standardisation) with an automatic minimum clustering threshold to generate no more than two clusters.
To test the sensitivity of clustering to the selected correlation threshold, we performed the clustering over a range of thresholds ( to , with an increment of 0.001) and measured the clustering similarity as the percentage of patients clustered by IBEX or MaZda into the same groups as PyRadiomics. We define clustering similarity as,
where and are patient groups from clustering with PyRadiomics features, and are patient groups from clustering with either IBEX or MaZda features, set intersection is denoted with , set union is denoted with and the number of patients in a set is denoted with . Due to the possibility that unsupervised clustering can return similar groups but in a different order, the similarity metric was calculated as,
Supplementary Information
Acknowledgements
This project is supported by funding from the Peter MacCallum Cancer Foundation, RSNA Fellow Grant, Mike Hogg Fund, and RANZCR research grants.
Author contributions
J.K. wrote the main manuscript text and prepared the figures and tables. J.K. and S.P. performed the radiomics feature extraction. J.K., S.P. and N.H. performed the radiomics analysis and data interpretation. S.P., C.C., J.W. and C.F. recruited patients and supervised data collection on the PREDICT-HN study. All authors reviewed the manuscript.
Competing interests
TK has a Research Collaboration Agreement with Varian Medical Systems. NH has a Clinical Research Collaborations Program Grant from Varian Medical Systems. SPN was funded by the Australian Postgraduate Award, the Royal Australian and New Zealand College of Radiologists (RANZCR) Research Grant and the Radiological Society of North America (RSNA) Fellow Grant. CDF received funding support from the National Institutes of Health (NIH)/National Institute for Dental and Craniofacial Research (NIDCR) (1R01DE025248-01/R56DE025248-01). CDF was previously funded via the National Science Foundation (NSF), Division of Mathematical Sciences, Joint NIH/NSF Initiative on Quantitative Approaches to Biomedical Big Data (QuBBD) Grant (NSF DMS-1557679) and is currently supported by the NIH National Cancer Institute (NCI)/Big Data to Knowledge (BD2K) Program (1R01CA214825-01), the NIH/NCI Head and Neck Specialized Programs of Research Excellence (SPORE) Developmental Research Program Career Development Award (P50CA097007-10); the NCI Paul Calabresi Clinical Oncology Program Award (K12 CA088084-06); a General Electric Healthcare/MD Anderson Center for Advanced Biomedical Imaging In-Kind Award; an Elekta AB/MD Anderson Department of Radiation Oncology Seed Grant; the Center for Radiation Oncology Research (CROR) at MD Anderson Cancer Center Seed Grant; and the MD Anderson Institutional Research Grant (IRG) Program. CDF has received speaker travel funding from Elekta AB. Supported in part by the NIH/NCI Cancer Center Support (Core) Grant CA016672 to The University of Texas MD Anderson Cancer Center (P30 CA016672). JCK, CC, JW, HB, BE, RG and LC have no potential competing interests to declare.
Footnotes
The original online version of this Article was revised: The original version of this Article contained an error in the order of the Figures. Figures 1, 2, 3, 4 and 5 were published as Figures 5, 1, 2, 3, and 4 respectively.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/17/2021
A Correction to this paper has been published: 10.1038/s41598-021-98445-3
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-96600-4.
References
- 1.Castellano G, et al. Texture analysis of medical images. Clin. Radiol. 2004;59(12):1061–1069. doi: 10.1016/j.crad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Jaffe CC. Measures of response: RECIST, WHO, and new alternatives. J. Clin. Oncol. 2006;24(20):3245–3251. doi: 10.1200/JCO.2006.06.5599. [DOI] [PubMed] [Google Scholar]
- 3.Kumar V, et al. Radiomics: The process and the challenges. Magn. Reson. Imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambin P, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jethanandani A, et al. Exploring applications of radiomics in magnetic resonance imaging of head and neck cancer: A systematic review. Front. Oncol. 2018;8:21. doi: 10.3389/fonc.2018.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunliffe A, et al. Lung texture in serial thoracic computed tomography scans: Correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development. Int. J. Radiat. Oncol. Biol. Phys. 2015;91(5):1048–1056. doi: 10.1016/j.ijrobp.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fave X, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci. Rep. 2017;7:11. doi: 10.1038/s41598-017-00665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goh V, et al. Assessment of response to tyrosine kinase inhibitors in metastatic renal cell cancer: CT texture as a predictive biomarker. Radiology. 2011;261(1):165–171. doi: 10.1148/radiol.11110264. [DOI] [PubMed] [Google Scholar]
- 10.Rao SX, et al. CT texture analysis in colorectal liver metastases: A better way than size and volume measurements to assess response to chemotherapy? United Eur. Gastroenterol. J. 2016;4(2):257–263. doi: 10.1177/2050640615601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho S, et al. Early variation of FDG-PET radiomics features in NSCLC is related to overall survival—The “delta radiomics” concept. Radiother. Oncol. 2016;118:S20–S21. [Google Scholar]
- 12.Boldrini L, et al. Delta radiomics for rectal cancer response prediction with hybrid 035T magnetic resonance-guided radiotherapy (MRgRT): A hypothesis-generating study for an innovative personalized medicine approach. Radiol. Med. 2019;124(2):145–153. doi: 10.1007/s11547-018-0951-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Y, et al. An investigation of machine learning methods in delta-radiomics feature analysis. PLoS ONE. 2019;14(12):e0226348. doi: 10.1371/journal.pone.0226348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crombe A, et al. T-2-based MRI Delta-radiomics improve response prediction in soft-tissue sarcomas treated by neoadjuvant chemotherapy. J. Magn. Reson. Imaging. 2019;50(2):497–510. doi: 10.1002/jmri.26589. [DOI] [PubMed] [Google Scholar]
- 15.Jeon SH, et al. Delta-radiomics signature predicts treatment outcomes after preoperative chemoradiotherapy and surgery in rectal cancer. Radiat. Oncol. 2019;14:10. doi: 10.1186/s13014-019-1246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang ZJ, et al. A predictive model for distinguishing radiation necrosis from tumour progression after gamma knife radiosurgery based on radiomic features from MR images. Eur. Radiol. 2018;28(6):2255–2263. doi: 10.1007/s00330-017-5154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surov A, Meyer HJ, Wienke A. Correlation between apparent diffusion coefficient (ADC) and cellularity is different in several tumors: A meta-analysis. Oncotarget. 2017;8(35):59492–59499. doi: 10.18632/oncotarget.17752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surov A, et al. Histogram analysis parameters of apparent diffusion coefficient reflect tumor cellularity and proliferation activity in head and neck squamous cell carcinoma. Oncotarget. 2018;9(34):23599–23607. doi: 10.18632/oncotarget.25284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Houdt PJ, Yang Y, van der Heide UA. Quantitative magnetic resonance imaging for biological image-guided adaptive radiotherapy. Front. Oncol. 2021;10(3190):615643. doi: 10.3389/fonc.2020.615643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leger S, et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci. Rep. 2017;7:11. doi: 10.1038/s41598-017-13448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parmar C, et al. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 2015;5:11. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao BS, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci. Rep. 2016;6:7. doi: 10.1038/srep23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiset S, et al. Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother. Oncol. 2019;135:107–114. doi: 10.1016/j.radonc.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Fruehwald-Pallamar J, et al. Texture-based analysis of 100 MR examinations of head and neck tumors—Is it possible to discriminate between benign and malignant masses in a multicenter trial? Rofo-Fortschritte Auf Dem Gebiet Der Rontgenstrahlen Und Der Bildgebenden Verfahren. 2016;188(2):195–202. doi: 10.1055/s-0041-106066. [DOI] [PubMed] [Google Scholar]
- 25.Mayerhoefer ME, et al. Effects of magnetic resonance image interpolation on the results of texture-based pattern classification a phantom study. Investig. Radiol. 2009;44(7):405–411. doi: 10.1097/RLI.0b013e3181a50a66. [DOI] [PubMed] [Google Scholar]
- 26.Peerlings J, et al. Stability of radiomics features in apparent diffusion coefficient maps from a multi-centre test-retest trial. Sci. Rep. 2019;9:10. doi: 10.1038/s41598-019-41344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang F, et al. Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: A simulation study utilizing ground truth. Phys. Med. Eur. J. Med. Phys. 2018;50:26–36. doi: 10.1016/j.ejmp.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Bologna M, et al. Assessment of stability and discrimination capacity of radiomic features on apparent diffusion coefficient images. J. Digit. Imaging. 2018;31(6):879–894. doi: 10.1007/s10278-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duron L, et al. Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS ONE. 2019;14(3):14. doi: 10.1371/journal.pone.0213459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saha A, Harowicz MR, Mazurowski MA. Breast cancer MRI radiomics: An overview of algorithmic features and impact of inter-reader variability in annotating tumors. Med. Phys. 2018;45(7):3076–3085. doi: 10.1002/mp.12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn. Reson. Imaging. 2004;22(1):81–91. doi: 10.1016/j.mri.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Moradmand H, Aghamiri SMR, Ghaderi R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J. Appl. Clin. Med. Phys. 2020;21(1):179–190. doi: 10.1002/acm2.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foy JJ, et al. Variation in algorithm implementation across radiomics software. J. Med. Imaging. 2018;5(4):10. doi: 10.1117/1.JMI.5.4.044505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang ZG, et al. Comparison of radiomics tools for image analyses and clinical prediction in nasopharyngeal carcinoma. Br. J. Radiol. 2019;92(1102):9. doi: 10.1259/bjr.20190271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogowicz M, et al. Post-radiochemotherapy PET radiomics in head and neck cancer—The influence of radiomics implementation on the reproducibility of local control tumor models. Radiother. Oncol. 2017;125(3):385–391. doi: 10.1016/j.radonc.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 36.Qian ZH, et al. Differentiation of glioblastoma from solitary brain metastases using radiomic machine-learning classifiers. Cancer Lett. 2019;451:128–135. doi: 10.1016/j.canlet.2019.02.054. [DOI] [PubMed] [Google Scholar]
- 37.Zhang B, et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017;403:21–27. doi: 10.1016/j.canlet.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Zwanenburg, A. et al. Image biomarker standardisation initiative. arXiv preprint arXiv:1612.07003 (2016).
- 39.Brown AM, et al. Multi-institutional validation of a novel textural analysis tool for preoperative stratification of suspected thyroid tumors on diffusion-weighted MRI. Magn. Reson. Med. 2016;75(4):1708–1716. doi: 10.1002/mrm.25743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fruehwald-Pallamar J, et al. Texture-based and diffusion-weighted discrimination of parotid gland lesions on MR images at 3.0 T. Nmr Biomed. 2013;26(11):1372–1379. doi: 10.1002/nbm.2962. [DOI] [PubMed] [Google Scholar]
- 41.Meyer HJ, et al. MRI texture analysis reflects histopathology parameters in thyroid cancer—A first preliminary study. Transl. Oncol. 2017;10(6):911–916. doi: 10.1016/j.tranon.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou Z, et al. Radiomic analysis in T2W and SPAIR T2W MRI: Predict treatment response to chemoradiotherapy in esophageal squamous cell carcinoma. J. Thorac. Disease. 2018;10(4):2256. doi: 10.21037/jtd.2018.03.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jansen JFA, et al. Texture analysis on parametric maps derived from dynamic contrast-enhanced magnetic resonance imaging in head and neck cancer. World J. Radiol. 2016;8(1):90–97. doi: 10.4329/wjr.v8.i1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, et al. Use of texture analysis based on contrast-enhanced MRI to predict treatment response to chemoradiotherapy in nasopharyngeal carcinoma. J. Magn. Reson. Imaging. 2016;44(2):445–455. doi: 10.1002/jmri.25156. [DOI] [PubMed] [Google Scholar]
- 45.Ouyang FS, et al. Exploration and validation of radiomics signature as an independent prognostic biomarker in stage III–IVb nasopharyngeal carcinoma. Oncotarget. 2017;8(43):74869–74879. doi: 10.18632/oncotarget.20423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren JL, et al. Magnetic resonance imaging based radiomics signature for the preoperative discrimination of stage I–II and III–IV head and neck squamous cell carcinoma. Eur. J. Radiol. 2018;106:1–6. doi: 10.1016/j.ejrad.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Wang GY, et al. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. Eur. J. Radiol. 2018;98:100–106. doi: 10.1016/j.ejrad.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 48.Zhang B, et al. Advanced nasopharyngeal carcinoma: Pre-treatment prediction of progression based on multi-parametric MRI radiomics. Oncotarget. 2017;8(42):72457–72465. doi: 10.18632/oncotarget.19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang B, et al. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin. Cancer Res. 2017;23(15):4259–4269. doi: 10.1158/1078-0432.CCR-16-2910. [DOI] [PubMed] [Google Scholar]
- 50.Leijenaar RTH, et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol. 2015;54(9):1423–1429. doi: 10.3109/0284186X.2015.1061214. [DOI] [PubMed] [Google Scholar]
- 51.Lucia F, et al. External validation of a combined PET and MRI radiomics model for prediction of recurrence in cervical cancer patients treated with chemoradiotherapy. Eur. J. Nucl. Med. Mol. Imaging. 2019;46(4):864–877. doi: 10.1007/s00259-018-4231-9. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, et al. Ibex: An open infrastructure software platform to facilitate collaborative work in radiomics. Med. Phys. 2015;42(3):1341–1353. doi: 10.1118/1.4908210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szczypinski PM, et al. MaZda—A software package for image texture analysis. Comput. Methods Prog. Biomed. 2009;94(1):66–76. doi: 10.1016/j.cmpb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 54.van Griethuysen JJM, et al. Computational radiomics system to decode the radiographic phenotype. Can. Res. 2017;77(21):E104–E107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dinapoli, N. et al. Moddicom: A complete and easily accessible library for prognostic evaluations relying on image features, in 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 771–774 (2015). [DOI] [PubMed]
- 56.Apte AP, et al. Technical note: Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research. Med. Phys. 2018;45(8):3713–3720. doi: 10.1002/mp.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Z, et al. Technical note: Ontology-guided radiomics analysis workflow (O-RAW) Med. Phys. 2019;46(12):5677–5684. doi: 10.1002/mp.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lecler A, et al. Combining multiple magnetic resonance imaging sequences provides independent reproducible radiomics features. Sci. Rep. 2019;9:8. doi: 10.1038/s41598-018-37984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng SP, et al. Predicting treatment response based on dual assessment of magnetic resonance imaging kinetics and circulating tumor cells in patients with head and neck cancer (PREDICT-HN): matching ‘liquid biopsy’ and quantitative tumor modeling. BMC Cancer. 2018;18:8. doi: 10.1186/s12885-018-4808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pipe JG. Motion correction with PROPELLER MRI: Application to head motion and free-breathing cardiac imaging. Magn. Reson. Med. 1999;42(5):963–969. doi: 10.1002/(sici)1522-2594(199911)42:5<963::aid-mrm17>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 61.Wintersperger BJ, et al. Brain magnetic resonance imaging at 3 T using BLADE compared with standard rectilinear data sampling. Investig. Radiol. 2006;41(7):586–592. doi: 10.1097/01.rli.0000223742.35655.24. [DOI] [PubMed] [Google Scholar]
- 62.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973;SMC3(6):610–621. [Google Scholar]
- 63.Galloway MM. Texture analysis using gray level run lengths. Comput. Graphics Image Process. 1975;4(2):172–179. [Google Scholar]
- 64.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man Cybern. 1989;19(5):1264–1274. [Google Scholar]
- 65.Sharp, G. C. et al. Plastimatch-an open source software suite for radiotherapy image processing, in Proceedings of the XVI’th International Conference on the use of Computers in Radiotherapy (ICCR), Amsterdam, Netherlands (2010).
- 66.Virtanen P, et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17(3):261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 68.Ward JH. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58(301):236–244. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.