Abstract
During the winter season from November 1996 to May 1997, 550 fecal specimens were submitted from 94 outbreaks of gastroenteritis occurring in East Anglia, United Kingdom. These specimens were tested for the presence of small round-structured viruses (SRSVs) by electron microscopy, reverse transcriptase PCR, or both methods. SRSVs were shown to be associated with 64 of 94 (68%) of these outbreaks, of which 16 (25%) outbreaks occurred at a single location (Southend) within the region. Twenty-four specimens from 13 of the 16 SRSV-positive outbreaks occurring in Southend were available for genomic analysis, in which divergence within the RNA polymerase region of the SRSV genome was investigated. A further 27 specimens from 17 other SRSV-associated outbreaks, occurring at different locations within East Anglia but at the same time as those at Southend, were also studied. Fifty of the total of 51 (98%) specimens studied were shown to belong to genogroup II, and within this genogroup, 49 of 50 (98%) specimens were shown to be Grimsby-like viruses, with only one Mexico-like strain. Furthermore, phylogenetic analysis of the Grimsby-like viruses indicated clusterings according to the geographical location of the outbreak. One specimen contained a virus belonging to genogroup I, and this had the greatest sequence identity (83%) with Southampton virus.
Winter vomiting disease (hyperemesis hiemis) was first reported in 1929 (44) and was described as a nonbacterial, epidemic gastroenteritis, but no etiological agent was identified at that time. Norwalk virus was identified in specimens from affected patients more than 40 years after the first report (27), and since then many other similar viruses have been identified as causative agents (see, for example, references 8 and 33). These agents, collectively known as small round-structured viruses (SRSVs) on the basis of their morphology (4), have high attack rates among both children and adults, especially in semiclosed communities such as families (39), hospitals (5), residential homes (24), schools (29), universities (28), and cruise ships (37). Outbreaks resulting from the ingestion of contaminated water (12) and food (21), especially shellfish (6), have also been reported. Seasonal epidemics of gastroenteritis caused by SRSVs occur, as do sporadic cases throughout the year. These viruses are positive-sense single-stranded RNA viruses (22) and have been classified as members of the Caliciviridae family (25). Recently, the family Caliciviridae has been subdivided into three genogroups and SRSVs have been assigned to genogroups I and II, while classical caliciviruses belong to genogroup III (17, 43). The SRSV genome has three open reading frames (ORFs) (25, 31) that encode a polyprotein precursor to nonstructural proteins including the RNA-dependent RNA polymerase (ORF 1), capsid protein (ORF 2) (25, 26), and a protein of unknown function (ORF 3) (17).
Since attempts to culture SRSVs in vitro have been unsuccessful to date and no animal model of infection and disease exists, laboratory diagnosis relies largely upon electron microscopy (EM) and reverse transcriptase (RT) PCR (RT-PCR). Some laboratories have developed enzyme-linked immunosorbent assay-based detection techniques, but these are strain specific (11, 20, 36). Although useful as a screening technique, EM is labor intensive and is relatively insensitive, requiring a high number of virus particles to be present in the fecal specimens for successful detection (106 to 107 particles per gram). More recently, RT-PCR with broadly reacting primers has been introduced. The Ni-E3 primer pair detects approximately 90% of SRSV genotypes circulating in the United Kingdom (13), with increased sensitivity compared with that of EM. SRSV RNA can be detected in up to 30% of EM-negative specimens from outbreaks (data not shown).
Little has so far been published on the molecular epidemiology of SRSV strains circulating within a defined region during a given season. In this study, a representative selection of 24 specimens from 13 of 16 (81%) SRSV-associated outbreaks occurring in a single location (Southend, United Kingdom) during the course of the 1996–1997 season are compared with a further 27 specimens from 17 representative SRSV-associated outbreaks occurring elsewhere in East Anglia at the same time as those in Southend. The polymerase gene, which has been shown to display diversity between different members of the same genogroups and different genogroups (17, 43), was chosen as a marker for genetic variation in this study. Phylogenetic analysis of the RNA-dependent RNA polymerase genes from a diverse population of positive-sense viruses has divided the enzymes into three supergroups, prompting the proposal that comparison of the RNA polymerase genes is the most reliable method for phylogenetic analysis (30).
Point mutations occur at a particularly high frequency in positive-sense RNA viruses, with rates equivalent to an average of one mutation per genome replication. This is a result of the error-prone nature of the RNA polymerase enzyme. Consequently, these viruses can adjust rapidly to selective pressures, with adaptively superior variants succeeding under adverse host or environmental conditions. In the long term, functional constraints of the proteins, together with changes in the host such as acquisition of immunity, control evolution.
Sequence analysis of the RNA polymerase region of the genomes of all 51 specimens has been performed to obtain a molecular epidemiological snapshot of the strains cocirculating during this period and to identify changes in the polymerase gene in viruses isolated from outbreaks occurring in different locations and the divergence of strains during the course of the season.
MATERIALS AND METHODS
Fecal specimen preparation.
Fecal specimens were obtained from affected individuals, and 10% suspensions of feces were prepared in 2 ml of 199 balanced salt solution (Sigma, Dorset, United Kingdom). A total of 5 μl of the 10% suspension was mixed with an equal volume of 3% phosphotungstic acid (pH 6.3), placed on a Formvar carbon-coated copper grid, and examined at ×50,000 magnification with a JEOL CX100 electron microscope. The remaining material was stored at −20°C until it was tested by RT-PCR.
Outbreak selection.
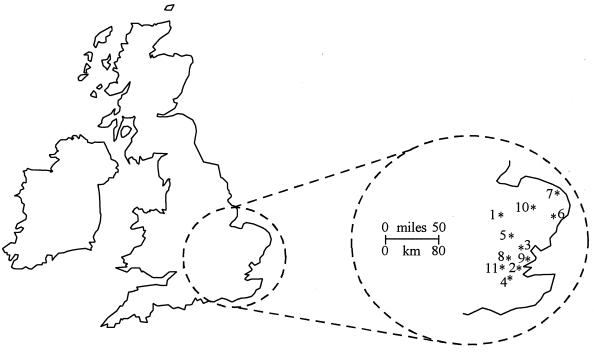
Submitted specimens were screened by EM. An outbreak was deemed positive for SRSV if at least one specimen from an outbreak contained virus particles. If all specimens from an outbreak were negative by EM, representative specimens were subjected to RT-PCR with the broadly reactive primer pair Ni-E3 (13) to detect SRSV RNA. The center providing specimens from the most SRSV-associated outbreaks during the season (Southend) was chosen for closer investigation (Table 1). For geographical comparison, specimens collected from outbreaks occurring during four 7-day periods were chosen to coincide with at least one outbreak occurring at Southend (Table 2; Fig. 1). In total, 51 SRSV-positive fecal specimens were obtained from 30 outbreaks.
TABLE 1.
Specimens obtained from SRSV-associated gastroenteritis outbreaks occurring in Southend, United Kingdom
| Date of onset of outbreak (day/mo/yr)a | Community location of outbreak | No. of specimens in outbreak | No. (%) of specimens EM positive | No. (%) of specimens Ni-E3 RT-PCR positive/total no. of specimens tested (%) | No. of specimens in study |
|---|---|---|---|---|---|
| 26/11/1996 | Hospital | 10 | 1 (10) | 3/10 (30) | 4 |
| 2/12/1996(a) | Hospital | 23 | 1 (4) | 2/16 (13) | 1 |
| 2/12/1996(b) | Hospital | 15 | 1 (7) | 5/15 (33) | 2 |
| 3/12/1996(a) | Hospital | 10 | 1 (10) | 5/9 (56) | 3 |
| 3/12/1996(b) | Hospital | 17 | 0 | 6/16 (38) | 2 |
| 10/12/1996 | Hospital | 8 | 1 (13) | 1/8 (13) | 1 |
| 12/12/1996 | Hospital | 5 | 1 (20) | 2/4 (50) | 1 |
| 20/1/1997 | Hospital | 7 | 0 | 3/7 (43) | 1 |
| 5/2/1997 | Hospital | 7 | 0 | 5/7 (71) | 2 |
| 20/2/1997 | Hospital | 8 | 1 (13) | 4/8 (50) | 4 |
| 21/3/1997 | Hospital | 6 | 0 | 4/6 (67) | 1 |
| 19/4/1997 | Residential home | 10 | NT | 7/10 (70) | 1 |
| 14/5/1997 | Hospital | 2 | NT | 2/2 (100) | 1 |
| Total | 128 | 7 | 49 | 24 |
When more than one outbreak occurred on the same date but at different locations within the same institution, outbreaks are identified as (a) and (b).
NT, specimens were not tested.
TABLE 2.
Specimens obtained from SRSV-associated gastroenteritis outbreaks occurring in East Anglia, United Kingdom, at locations other than Southend
| Location of outbreak | Date of onset of outbreak (day/mo/yr)a | Community location of outbreak | No. of specimens in outbreak | No. (%) of specimens EM positive | No. (%) of specimens Ni-E3 RT-PCR positive | No. of specimens in study |
|---|---|---|---|---|---|---|
| Ipswich | 24/11/1996 | Hospital | 2 | 1 (50) | 1 (50) | 1 |
| Basildon | 24/11/1996 | Hospital | 3 | 1 (33) | 3 (100) | 3 |
| Chelmsford | 27/11/1996 | Hospital | 5 | 0 | 4 (80) | 1 |
| Lowestoft | 12/12/1996 | Residential home | 4 | 1 (25) | 1 (25) | 1 |
| Ipswich | 12/12/1996 | Hospital | 4 | 0 | 4 (100) | 1 |
| Chelmsford | 13/12/1996 | Hospital | 2 | 0 | 1 (50) | 1 |
| Herts & Essex | 18/1/1997 | Hospital | 4 | 2 (50) | 4 (100) | 3 |
| Basildon | 18/1/1997 | Hospital | 2 | 0 | 2 (100) | 1 |
| Chelmsford | 19/1/1997 | Hospital | 3 | 0 | 2 (67) | 1 |
| Addenbrooke’s | 20/1/1997 | Hospital | 10 | 2 | 5 (56)b | 2 |
| Chelmsford | 22/1/1997 | Hospital | 3 | 0 | 3 (100) | 2 |
| Dulwich | 14/2/1997 | Hotel | 22 | 0 | 6 (27) | 2 |
| West Suffolk | 14/2/1997(a) | Hospital | 8 | 1 (13) | 2 (25) | 2 |
| West Suffolk | 14/2/1997(b) | Hospital | 6 | 0 | 5 (83) | 2 |
| Whipps Cross | 15/2/1997 | Residential home | 3 | 0 | 3 (100) | 1 |
| Romford | 15/2/1997 | Hospital | 7 | 0 | 5 (71) | 1 |
| West Suffolk | 17/2/1997 | Hospital | 3 | 0 | 2 (67) | 2 |
| Total | 91 | 8 | 53 | 27 |
When more than one outbreak occurred on the same date but at different locations within the same institution, the outbreaks are identified as (a) and (b).
Only nine specimens in total were tested.
FIG. 1.
Map of the United Kingdom with an inset showing East Anglia. The locations of outbreaks of gastroenteritis included in this study are indicated: 1, Addenbrooke’s; 2, Basildon; 3, Chelmsford; 4, Dulwich; 5, Herts and Essex; 6, Ipswich; 7, Lowestoft; 8, Romford; 9, Southend; 10, West Suffolk; and 11, Whipps Cross.
Oligonucleotide primers.
The primers used for PCR amplification were Ni (5′-GAA TTC CAT CGC CCA CTG GCT-3′) (13) and E3 (5′-ATC TCA TCA TCA CCA TA-3′) (13), which amplify a 113-bp product for the detection of both genogroup I and II SRSV RNA in fecal material; GI (5′-TCN GAA ATG GAT GTT GG 3′) (14) and GII (5′ AGC CNT NGA AAT NAT GGT 3′) (14), which, together with antisense primer E3, amplify 190- and 270-bp products of the RNA polymerase gene of genogroup I and genogroup II viruses, respectively; Ando (5′-GTG AAC AGY ATA AAY CAY TGG-3′), a degenerate primer based on SR48, SR50, and SR52 (1) which, together with antisense primer E3, amplifies a 115-bp product of the RNA polymerase gene; SM51 (5′-TGG GAC TCA ACA CAA AAT A-3′) (38) with SM31 (5′-CGA TTT CAT CAT CAC CAT A-3′) (38), which amplify a 304-bp product of the polymerase gene; and primer pair SM52 (5′-TGT GAT GGA TGT AGG TGA TT-3′) (38) and SM32 (5′ TGA CTT CAG ATA GTG CAC AG-3′) (38), which amplify a 127-bp product of the RNA polymerase gene. For the detection and sequencing of cloned PCR products, the primers for the cloning vector were pTAG-5′ (5′-GCT ATG ACC ATG ATT ACG CCA A-3′) and pTAG-3′ (5′-TGT CCC ACG GCC AGT GAA-3′), which amplified the portion of the transformed plasmid containing the viral insert, together with flanking regions.
RNA extraction and RT-PCR.
RNA was extracted from 100 μl of the 10% fecal suspension by the method of Boom et al. (3), described previously for fecal specimens (15), and eluted in 26 μl of RNase-free sterile water containing 40 U of RNase inhibitor (RNAsin; Promega, Madison, Wis.). One microliter of random primer (20 mU; PdN6; Pharmacia Biotech) was added to the extracted RNA, the mixture was heated at 70°C for 5 min and chilled on ice, and 14 μl of the reaction mixture was added, yielding a total volume of 35 μl of the PCR mixture consisting of 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 5 mM MgCl2, each deoxynucleotide triphosphate (dNTP; Boehringer Mannheim, Mannheim, Germany) at a concentration of 50 μM, and 200 U of Moloney murine leukemia virus (M-MuLV) RT (FPLC-pure, cloned M-MuLV; Life Technologies, Gaithersburg, Md.). Reverse transcription was performed at room temperature for 10 min, followed by incubation at 37°C for 1 h, after which the reaction was terminated by incubation at 95°C for 5 min followed by chilling on ice. Five microliters of cDNA was then added to 45 μl of the PCR mixture, yielding a total of 50 μl of a reaction mixture consisting of 18 mM Tris-HCl (pH 8.4), 45 mM KCl, 2 mM MgCl2, each dNTP at a concentration of 35 μM, 1 U of Taq polymerase (Life Technologies), and 20 pmol of each primer (primers Ni and E3). After denaturation at 94°C for 2 min, 30 temperature cycles each consisting of 94°C for 1 min, 40°C for 1 min, and 72°C for 1 min were performed, followed by a final extension at 72°C for 10 min.
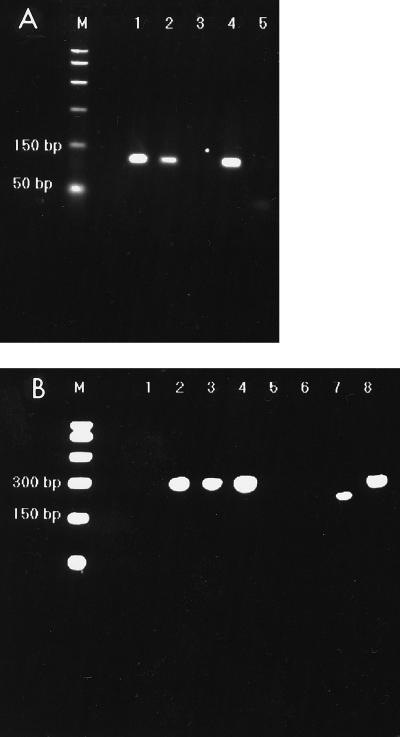
Amplification products were examined by gel electrophoresis of 10 μl of the reaction mixture in agarose gels (3% Nusieve 3:1; Flowgen) containing ethidium bromide (4 μg ml−1 of agarose gel; Fig. 2A).
FIG. 2.
(A) Representative gel of Ni-E3 PCR products (113 bp). Lanes 4 and 5, positive and negative controls, respectively; Lane M, a 1-kb molecular size marker. (B) Representative gel of GI-GII-E3-PCR products. Lanes 7 and 8, GI and GII positive controls, respectively. The GI-E3 PCR product is 190 bp, and the GII-E3 product is 270 bp. Lane 6, negative control; lane M, 1-kb molecular size marker.
Appropriate positive and negative controls to confirm the sensitivity and specificity of the PCR and to detect contamination were included in each assay run.
Genogrouping RT-PCR.
All specimens found to be positive with the primer pair Ni-E3 (13) were genogrouped by RT-PCR with the primers GI-GII-E3 (14). Briefly, 5 μl of cDNA was added to a reaction mixture yielding a total reaction volume of 50 μl containing 18 mM Tris-HCl (pH 8.4), 45 mM KCl, 0.1 mM MgCl2, each dNTP at a concentration of 35 μM, 1 U of Taq polymerase, and 20 pmol of each primer (primers GI, GII, and E3). The GI and GII (sense) primers are genogroup specific and, when used in combination with the antisense E3 primer, allow differentiation between genogroup I and genogroup II strains on the basis of amplicon size. The GI-E3 and GII-E3 amplicons yield product sizes of 190 and 270 bp, respectively (Fig. 2B).
Two EM-positive, Ni-E3 RT-PCR-negative specimens were subjected to RT-PCR with the additional primer pairs Ando-E3, SM51-SM31, and SM52-SM32. RT-PCR with these primer pairs was performed under the same conditions used for the Ni-E3 RT-PCR.
Cloning.
When required, cloning was carried out on RT-PCR products with the LigAtor cloning kit (Ingenius, Madison, Wis.). Transformation of bacterial cells and growth on agar plates were performed according to the manufacturer’s instructions. The subcultured cells were incubated at 37°C for 16 h, followed by incubation at 4°C for 2 h prior to screening of the colonies.
Five white colonies from each plate were subcultured onto Luria-Bertani agar plates containing ampicillin (50 μg ml−1) and were screened for the presence of the pTAg vector by PCR with a reaction mixture consisting of 18 mM Tris-HCl (pH 8.4), 45 mM KCl, 1.5 mM MgCl2, each dNTP at a concentration of 35 μM, 1 U of Taq polymerase, and the 3′ and 5′ primers specific for the pTAg vector (20 pmol of each). PCR consisted of 32 temperature cycles of 94°C for 15 s, 40°C for 45 s, and 72°C for 1 min. Reaction products were detected by gel electrophoresis (2% agarose) and were deemed positive if a band (stained with ethidium bromide) of the correct molecular weight for the particular insert was observed. The subcultured bacteria were incubated at 37°C overnight, and those giving positive PCRs were subcultured onto Dorset egg agar slopes for long-term storage.
DNA sequencing.
The RT-PCR products required for sequencing were purified by centrifugation through Centricon-30 concentrator columns (Amicon, Beverley, Mass.) with double-processed tissue-culture-grade water (Sigma). Approximately 30 ng of the purified PCR products was sequenced in both directions with the same primer pairs used in the positive RT-PCR and the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer Applied Biosystems Foster City, Calif.) according to the manufacturer’s instructions. The extension products that were obtained by sequencing were purified to remove excess dye terminators by ethanol precipitation.
Sequence analysis.
Sequence data were initially analyzed by using SeqEd, version 1.0.3, prior to importing the data into the DNAStar package (DNAStar Ltd., London, United Kingdom) for alignment by using the MegAlign program. Phylogenetic analyses were carried out with the PHYLIP set of programs (10) and application of the maximum-likelihood procedure with a Quartet Puzzling program (40).
RESULTS
During the epidemic season under investigation (November 1996 to May 1997), The Cambridge laboratory received 550 fecal specimens from 94 suspected outbreaks of SRSV-associated gastroenteritis. The majority of these outbreaks occurred in either nursing homes or hospitals (87%), with only 7 (7%) associated with food outlets. SRSVs were confirmed as the etiological agents in 27 of 86 (31%) outbreaks by EM and in 54 of 71 (76%) outbreaks by PCR. Of specimens from 61 outbreaks screened by both methods, EM identified only 19 (31%) outbreaks in which SRSVs were the etiological agents, whereas PCR identified 45 (74%). SRSVs were not detected by either EM or RT-PCR in specimens from 30 of 94 (32%) outbreaks. No other etiological agent associated with gastroenteritis was identified in specimens collected from these outbreaks. Of the 64 confirmed SRSV-associated outbreaks, 16 (25%) occurred at Southend. Twenty-four specimens were taken from 13 of the 16 (81%) SRSV-positive Southend outbreaks; specimens were no longer available from the other three outbreaks. (Table 1). Four 7-day periods during which at least one outbreak occurred at Southend and the greatest number of EM- or RT-PCR-confirmed SRSV-associated outbreaks occurred at other locations were selected. Outbreaks occurring during these 7-day periods were deemed to occur simultaneously. Of the 20 SRSV-positive outbreaks occurring simultaneously at different locations, material was available from 17 (85%). A representative selection of 27 specimens from these 17 outbreaks was included in the study (Table 2; Fig. 1).
RT-PCR screening was performed with the broadly reactive primer pair Ni-E3, which detects approximately 90% of the SRSVs currently circulating in the United Kingdom (13). An attempt to genogroup the cDNA from all 51 specimens was performed by genogroup-specific RT-PCR with the primers GI and GII and the antisense primer E3. Twenty-nine of the 51 (57%) specimens were positive for the 270-bp GII-E3 product (and so the strains were designated as being of genogroup II). Strains from the remaining 22 specimens failed to be amplified with this GI-GII-E3 primer combination.
All PCR products were subjected to direct sequencing in both directions with the primer pairs from which the amplicons were obtained. Direct sequence data were available for viruses from 38 of the 51 specimens, and the viruses from the remaining 13 specimens required cloning of the PCR product prior to sequencing. Clones were sequenced with primers specific for the vector pTAg DNA close to the insertion site.
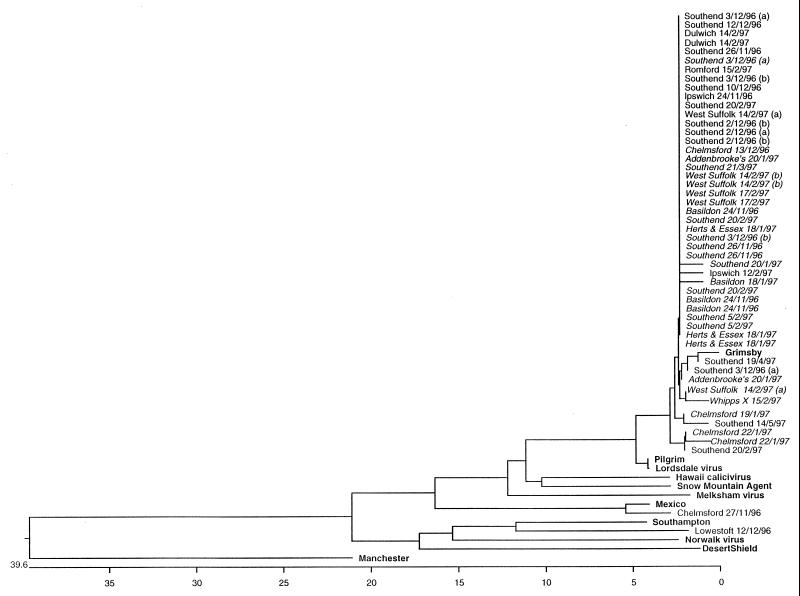
An overall comparison of all sequence data obtained for viruses from the 50 specimens from which cDNA was successfully amplified with the Ni-E3, GI-GII-E3, or Ando-E3 primers was made. The larger GII-E3 sequencing products were trimmed to match the same region of ORF 1 to which the shorter Ni-E3 and Ando-E3 products match and were subjected to distance alignment with the MegAlign software of the DNAstar package. Sequence data available for a wide range of members of the family Caliciviridae in the GenBank database, including the genogroup III Manchester calicvirus (35) and SRSVs belonging to genogroups I and II (Pilgrim [16], Grimsby [16], Lordsdale [7], Hawaii [41], Snow Mountain Agent [8], Melksham [18], Mexico [23], Southampton [31], Desert Shield [32], and Norwalk virus [25, 27]) were included in the alignment. The resulting phylogenetic tree is shown in Fig. 3. Strains from 49 of the 50 (98%) specimens analyzed by this method were shown to belong to genogroup II, and within this genogroup, 48 of 49 (98%) specimens were shown to contain viruses with greater than 96% identity with Grimsby virus.
FIG. 3.
Phylogenetic alignment of all sequence data obtained from outbreak specimens from this study and data available from the GenBank database for typical members of genogroup I (Norwalk, Southampton, and Desert Shield), genogroup II (Grimsby, Pilgrim, Lordsdale, Hawaii, Snow Mountain agent, Mexico, and Melksham), and genogroup III (Manchester, a classical calicivirus). These strains are indicated in boldface type. Outbreak strains are identified by the location and date of onset of the outbreak. Specimens in italic type are those for which GII-E3 sequence data were available and whose data were therefore used in the phylogenetic analysis whose results are presented in Fig. 4.
In addition to these 48 specimens yielding Grimsby-like virus, a further genogroup II virus was identified from an outbreak in Chelmsford (27 November 1996). RNA extracted from this specimen amplified in the initial screening PCR with the Ni-E3 primer pair but failed to amplify with the genogroup-specific primers. MegAlign distance analysis methods of the Ni-E3 amplicon DNA sequence data revealed 96% identity with the genogroup II Mexico virus over the 75-base interprimer region.
During the 1993–1994 season, Mexico virus was the predominant strain isolated from outbreaks occurring in the United Kingdom (20, 34). Since then, Grimsby virus has been predominant, with few outbreaks associated with this earlier epidemic strain.
Of the 51 specimens included in the study, 2 (4%) were EM positive for SRSV but negative for SRSV RNA with both the broadly reactive primer pair Ni-E3 and the genogroup-specific primer set GI-GII-E3 in the RT-PCR. One of these specimens, isolated from an outbreak occurring in Lowestoft (12 December 1996), was successfully amplified with primer pair Ando-E3. The 130-bp product was subjected to DNA sequence analysis following cloning. Sequence analysis indicated that this was a genogroup I strain with greatest identity with Southampton virus (83% over the 77-bp interprimer sequence) (Fig. 3).
Similarly, a second EM-positive and Ni-E3 RT-PCR- and GI-GII-E3-RT-PCR-negative specimen, from an outbreak occurring at Southend (26 November 1996) was successfully amplified with the SM52-SM32 primer pair, yielding a 127-bp product. The region of the polymerase gene amplified in this reaction is different from that obtained with the Ni-E3, GI-GII-E3, and Ando-E3 primers, so comparison of sequence data for this strain with that for the other outbreak strains was not possible, and this strain is therefore not included in Fig. 3. The sequence of the virus obtained from this specimen was, however, compared with the appropriate region of the available GenBank sequences, and this analysis showed the greatest identity with Grimsby virus (96% over the 87-bp interprimer region). This is perhaps not surprising because sequence data for viruses from the three other specimens analyzed from this outbreak were also Grimsby-like. Thus, of the 51 specimens investigated, 49 contained Grimsby-like strains, 1 contained a Mexico-like strain, and 1 contained a genogroup I strain, which is most closely related to Southampton virus.
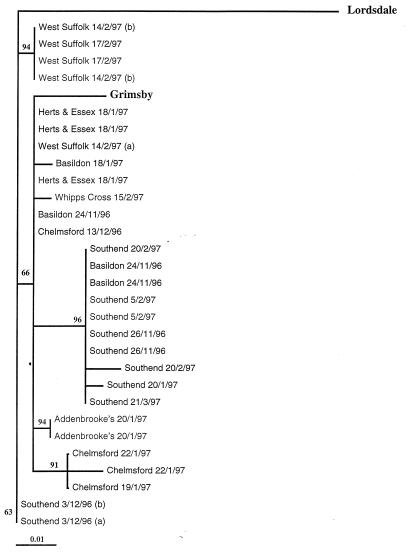
A further analysis of the GII-E3 amplicon sequences was performed in order to detect low-level sequence changes among the Grimsby-like strains. Twenty-nine sequences were analyzed in this way, and the final sequence data were compared with those for Grimsby virus (16) and Lordsdale virus (7), both of which are members of genogroup II. In order to obtain robust phylogenetic analyses, three sequence analysis programs were used: MegAlign (DNAStar) with distance methods, Quartet Puzzling with maximum-likelihood methods, and PHYLIP with phylogeny interference. All three programs showed that the 29 sequences aligned most closely with that of Grimsby virus, as predicted above, and that further clustering occurred within the group (Fig. 4).
FIG. 4.
Alignment of all sequence data from GII-E3 products. The sequence data for the outbreak strains were compared with the sequence data for two typical genogroup II SRSVs, Grimsby and Lordsdale, from the GenBank database. The numbers in boldface type represent bootstraps for the alignment, and outbreak specimens are identified by date and location of outbreak. The calibration bar denotes divergence of 1%.
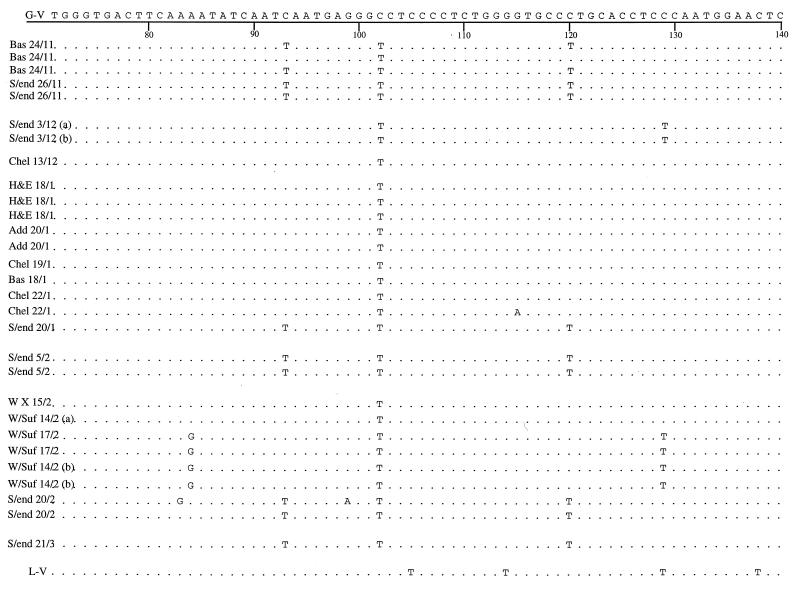
Sequence alignment data for the GII product sequences were further analyzed for single point mutations over the region sequenced. Sequence data were compared with those available for Grimsby virus, which was taken as the reference strain, and the Lordsdale virus sequence was included for comparison. These data were analyzed for geographical and temporal variations (Fig. 5).
FIG. 5.
Alignment of sequences obtained for genogroup II (positive GII-E3 PCR products) specimens isolated from outbreaks occurring at all locations studied in this work during the 1996–1997 season compared with the sequence available for Grimsby virus (G-V) and Lordsdale virus (L-V). The sequence of the Grimsby virus is shown along the top, and consensus with this sequence in the other strains is indicated by a period. Single point mutations are identified by the replacement base. Numbering refers to the base position in the sequence. Specimens are identified by the date and location of outbreaks. Locations of outbreaks are abbreviated as follows: Bas, Basildon; S/end, Southend; Chel, Chelmsford; H&E, Herts and Essex; Add, Addenbrooke’s; W X, Whipps Cross; and W/Suf, West Suffolk.
DISCUSSION
It is evident from the distance alignment (Fig. 3) that a single strain similar to Grimsby virus was associated with the majority of outbreaks of SRSV-associated gastroenteritis during the 1996–1997 season. Among the 49 specimens in which Grimsby-like virus RNA was detected, there was little variation within the region sequenced during the course of the season, indicating that there is a relatively low mutation rate in this region of the RNA. It is unlikely that this dominance of a single strain is due to primer selection since representative specimens were selected from more than 80% of the eligible outbreaks occurring in the region at the time.
Analysis of genogroup clustering (Fig. 4) with geographical location and the date of onset of each outbreak indicated that most outbreaks occurring in one center over the course of the season were frequently caused by the same virus, whereas outbreaks occurring in different locations frequently formed separate clusters. For example, during the season, three outbreaks (six specimens, of which data for five are included in Fig. 4) occurred at West Suffolk Hospital, and two occurred on the same date but at different locations within the hospital. Of these, the two outbreaks which occurred on the same date were caused by different viruses, one of which was also responsible for the third outbreak. Similarly, from the GII-derived sequence data available for seven outbreaks occurring in Southend, five outbreaks were caused by very closely related viruses. The other two outbreaks, which occurred simultaneously but at different locations within the hospital, were caused by an identical virus that was different from that causing the other Southend outbreaks.
Further geographical and temporal analysis of the GII-E3 amplicon sequences (Fig. 5) show a number of point mutations; e.g., G→A (position 15) and A→T (position 50) are consistently seen in all strains. The mutation from C to T at position 102 is present only in the outbreak viruses sequenced in this work, and the Lordsdale sequence retains the base present in Grimsby virus. With the exception of only two viruses (both isolated from one outbreak at Basildon), the replacement of C by T at positions 36, 93, and 120 is detected only in strains from Southend. However, two strains from Southend taken on the same date (3 December 1996) but from different locations within the hospital and the third isolate from the Basildon outbreak (24 November 1996) have retained the cytosine base at positions 36, 93, and 120 of the Grimsby virus. This pattern is clearly reflected in the alignment tree in Fig. 4, as is the occurrence of an outbreak in Southend (20 February 1997) caused by two closely related but distinct viruses.
Sequence data were obtained for viruses from two EM-positive, Ni-E3 RT-PCR-negative specimens with additional primer pairs (SM52-SM32 and Ando-E3). It is likely that the cDNA from these specimens failed to amplify with the Ni-E3 screening primers as a result of a single point mutation, occurring at the point of elongation of the E3 primer for the Grimsby-like virus isolated from an outbreak in Southend, (26 November 1996) (which was subsequently amplified in the SM52-SM32 reaction) and the point of elongation of Ni for the genogroup I virus isolated from an outbreak at Lowestoft (which was subsequently amplified in the Ando-E3 reaction). In the latter isolate, genogroup-specific primers probably failed to amplify due to the lower sensitivity of the reaction. Other work has indicated that although genogroup I viruses cocirculate with genogroup II, they cause relatively few outbreaks of gastroenteritis, being more commonly associated with sporadic cases, whereas genogroup II viruses more commonly cause outbreaks (15, 19). This observation is consistent with the data presented in this work, which indicate that only 1 of 30 (3%) outbreaks studied was associated with a genogroup I virus.
In The Netherlands, 60 outbreaks associated with SRSV were analyzed in a study by Vinje et al. (42). Those workers concluded that there was limited genetic variability of outbreak strains during the 12-month period studied, with a predominance of a single strain, as found here in East Anglia. However, Vinje et al. (42) also reported that there was a significant shift in circulating virus strains at the end of the period studied, one of which would become the dominant strain in the following year. Although this appears to be in contrast to the findings of the current study, it is noted that the period studied in The Netherlands covered two seasons (January to December), whereas here, only one complete season was investigated.
In another study of nine outbreaks in the southwestern region of the United Kingdom occurring between 1992 and 1995, Green et al. (19) investigated capsid sequence diversity using primers broadly reactive with the capsid regions of both genogroup I and genogroup II viruses. In close similarity to the present study, Green et al. (19) reported a predominance of genogroup II SRSV-associated outbreaks, the most prevalent strain being Bristol virus. This is in contrast to the current study, in which Grimsby virus was predominant, most probably reflecting either a change in the predominant virus circulating during different seasons or the fact that different virus strains are circulating in different regions of the United Kingdom.
During the winter season 1996–1997, a total of 30 outbreaks of gastroenteritis associated with SRSVs were investigated for differences in circulating strains over the East Anglian region of the United Kingdom. It was evident that during this period one strain of SRSV was predominant in causing outbreaks in semiclosed communities. This genogroup II strain was most closely related to Grimsby virus. Little variation or divergence occurred over the part of the RNA-dependent RNA polymerase gene sequenced during the course of the season, and any genetic variations were generally consistent within outbreaks occurring at particular locations. Furthermore, single point mutations were not generally expressed at the amino acid level. In the two instances in which the mutations were expressed in the amino acid sequence, the replacement amino acid residue bore a side chain with the same functional group present in the Grimsby virus, thus retaining the same biological and structural functionality of the protein. This suggests that there is no selective pressures on the virus to evolve, such as evasion of the host immune response or changes in the environment. The absence of the occurrence of mutations and evolution in the Grimsby-like SRSVs is consistent with the fact that the virus has adapted to its ecological and physiological niches and with the fact that the functional constraints imposed by the polymerase enzyme ensure that only viruses with silent mutations in the polymerase gene will survive.
SRSVs cause an acute, self-limiting infection with a very short incubation period (15 to 36 h) and a high secondary attack rate. This is the first report of the detailed molecular epidemiology of SRSVs in a small geographical region. The findings of small but significant differences between SRSV strains identified in towns in close geographic proximity over a 7-month period are consistent with local circulation of strains and may reflect the short incubation period and duration of illness, limiting opportunities for wide dissemination.
The prevalence in the population of caliciviruses, both the predominant type and other types cocirculating in the same geographical locations, must be determined in order to monitor the diversity of virus strains and the efficacy of any calicivirus vaccines developed in the future (2, 9). The Grimsby virus and Grimsby-like viruses have been isolated in the United Kingdom for the past 5 years, and from the results of this study, it may be suggested that the Grimsby virus is endemic and is circulating in different communities as relatively stable variants. Phylogenetic analysis of viruses isolated during consecutive seasons and from more diverse geographical locations would be required in order to determine whether Grimsby virus is truly endemic in the United Kingdom.
ACKNOWLEDGMENTS
Gillian Storey at the School of Biological Sciences, University of Durham, Durham, United Kingdom, is acknowledged for her help in running the sequencing gels.
A.J.M. was supported by a trainee grant of the Public Health Laboratory Service.
REFERENCES
- 1.Ando T, Monroe S S, Gentsch J R, Jin Q, Lewis D C, Glass R I. Detection and differentiation of antigenically distinct small round-structured (Norwalk-like) viruses by reverse transcriptase PCR and Southern hybridization. J Clin Microbiol. 1995;33:64–71. doi: 10.1128/jcm.33.1.64-71.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball J M, Estes M K, Hardy M E, Conner M E, Opekun A R, Graham D Y. Recombinant Norwalk virus-like particles as an oral vaccine. Arch Virol Suppl. 1996;12:243–249. doi: 10.1007/978-3-7091-6553-9_26. [DOI] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salismans M M M, Jansen C L, Wertheim-van Dillen P M E, van den Noordaa J. Rapid and simple method for the purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caul E O, Appleton H. The electron microscopic and physical characteristics of small round human faecal viruses: an interim scheme for classification. J Med Virol. 1982;9:257–265. doi: 10.1002/jmv.1890090403. [DOI] [PubMed] [Google Scholar]
- 5.Chadwick P R, McCann R. Transmission of a small round structured virus by vomiting during a hospital outbreak of gastroenteritis. J Hosp Infect. 1994;26:251–259. doi: 10.1016/0195-6701(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 6.Communicable Disease Report Weekly. Outbreaks of gastroenteritis in England and Wales associated with shellfish: 1996 and 1997. Commun Dis Rep CDR Weekly. 1998;8:21. . (Editorial.) [PubMed] [Google Scholar]
- 7.Dingle K E, Lambden P R, Caul E O, Clarke I N. Human enteric calcivirus: the complete genome sequence and expression of virus-like particles from a genetic group II small round-structured virus. J Gen Virol. 1995;76:2349–2355. doi: 10.1099/0022-1317-76-9-2349. [DOI] [PubMed] [Google Scholar]
- 8.Dolin R, Reichman R C, Roessner K D, Tralker T S, Schooley R T, Gary W, Morens D. Detection by immune electron microscopy of the Snow Mountain Agent of acute viral gastroenteritis. J Infect Dis. 1982;146:184–189. doi: 10.1093/infdis/146.2.184. [DOI] [PubMed] [Google Scholar]
- 9.Estes M K, Ball J M, Crawford S E, O’Neal C, Opekun A R, Graham D Y, Conner M E. Virus-like particles for mucosal immunization. Adv Exp Med Biol. 1997;412:387–395. doi: 10.1007/978-1-4899-1828-4_61. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP—Phylogeny interference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Gary G W, Jr, Kaplan J E, Stine S E, Anderson L J. Detection of Norwalk virus antibodies and antigen with a biotin-avidin immunoassay. J Clin Microbiol. 1985;22:274–278. doi: 10.1128/jcm.22.2.274-278.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray J J, Green J, Cunliffe C, Gallimore C, Lee J V, Neal K, Brown D W G. Mixed genogroup SRSV infections among a party of canoeists exposed to contaminated recreational water. J Med Virol. 1997;52:425–429. [PubMed] [Google Scholar]
- 13.Green J, Gallimore C I, Norcott J P, Lewis D, Brown D W G. Broadly reactive reverse transcriptase polymerase chain reaction for the diagnosis of SRSV-associated gastroenteritis. J Med Virol. 1995;47:392–398. doi: 10.1002/jmv.1890470416. [DOI] [PubMed] [Google Scholar]
- 14.Green J, Henshilwood K, Gallimore C I, Brown D W G, Lees D N. A nested reverse transcriptase PCR assay for detection of small round-structured viruses in environmentally contaminated molluscan shellfish. Appl Environ Microbiol. 1998;64:858–863. doi: 10.1128/aem.64.3.858-863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green J, Norcott J P, Lewis D, Arnold C, Brown D W G. Norwalk-like viruses in the UK: demonstration of genomic diversity by PCR sequencing. J Clin Microbiol. 1993;31:3007–3012. doi: 10.1128/jcm.31.11.3007-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green J, Vinje J, Lewis D C, Gallimore C I, Koopmans M, Brown D W G. Proceedings of the 1st International Symposium on Caliciviruses. Addlestone, United Kingdom: European Society for Veterinary Virology; 1996. Genomic diversity among human caliciviruses; pp. 37–49. [Google Scholar]
- 17.Green S M, Dingle K E, Lambden P R, Caul E O, Ashley C R, Clarke I N. Human enteric Caliciviridae: a new prevalent SRSV group defined by RNA-dependent RNA polymerase and capsid diversity. J Gen Virol. 1994;75:1883–1888. doi: 10.1099/0022-1317-75-8-1883. [DOI] [PubMed] [Google Scholar]
- 18.Green S M, Lambden P R, Caul E O, Ashley L R, Clarke I N. Capsid diversity in small round-structured viruses: molecular characterization of an antigenically distinct human enteric calicivirus. Virus Res. 1995;37:271–283. doi: 10.1016/0168-1702(95)00041-n. [DOI] [PubMed] [Google Scholar]
- 19.Green S M, Lambden P R, Caul E O, Clarke I N. Capsid sequence diversity in small round structured viruses from recent UK outbreaks of gastroenteritis. J Med Virol. 1997;52:14–19. [PubMed] [Google Scholar]
- 20.Hale A D, Lewis D, Green J, Jiang X, Brown D W G. Evaluation of an antigen capture ELISA based on recombinant Mexico virus capsid protein. Clin Diagn Virol. 1996;5:27–35. doi: 10.1016/0928-0197(95)00200-6. [DOI] [PubMed] [Google Scholar]
- 21.Hicks N J, Beynon J H, Bingham P, Soltanpoor N, Green J. An outbreak of viral gastroenteritis following a wedding reception. Commun Dis Rep CDR Rev. 1996;6:R136–R139. [PubMed] [Google Scholar]
- 22.Jiang X, Graham D Y, Wang K, Estes M K. Norwalk virus genome cloning and characterization. Science. 1990;250:1580–1583. doi: 10.1126/science.2177224. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X, Matson D O, Velazquez F R, Zhong W, Hu J, Ruiz-Palacios G, Pickering L K. A study of Norwalk-related virus in Mexican children. J Med Virol. 1995;47:309–316. doi: 10.1002/jmv.1890470404. [DOI] [PubMed] [Google Scholar]
- 24.Jiang X, Turf E, Hu J, Barrett E, Dai X M, Monroe S, Humphrey C, Pickering L K, Matson D O. Outbreaks of gastroenteritis in elderly nursing homes and retirement facilities associated with human caliciviruses. J Med Virol. 1996;50:335–341. doi: 10.1002/(SICI)1096-9071(199612)50:4<335::AID-JMV9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Jiang X, Wang M, Estes M K. Sequence and genomic organisation of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X, Wang M, Graham D Y, Estes M K. Expression, self-assembly and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapikian A Z, Wyatt R G, Dolin R, Thornhill R S, Kalica A R, Chanock R M. Visualisation by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol. 1972;10:1075–1081. doi: 10.1128/jvi.10.5.1075-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilgore P E, Belay E D, Hamlin D M, Noel J S, Humphrey C D, Gary Jr H E, Ando T, Monroe S S, Kludt P E, Rosenthal D S, Freeman J, Glass R I. A university outbreak of gastroenteritis due to a small round-structured virus. J Infect Dis. 1996;173:787–793. doi: 10.1093/infdis/173.4.787. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi S, Morishita T, Yamashita T, Sakae K, Nishio O, Miyake T, Ishihara Y, Isomura S. A large outbreak of gastroenteritis associated with a small round structured virus among schoolchildren and teachers in Japan. Epidemiol Infect. 1991;107:81–86. doi: 10.1017/s0950268800048706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 31.Lambden P R, Caul E O, Ashley C R, Clarke I N. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259:516–518. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- 32.Lew J F, Kapikian A Z, Jiang X, Estes M K, Green K Y. Molecular characterization and expression of capsid protein of a Norwalk-like virus recovered from a Desert Shield troop with gastroenteritis. Virology. 1994;200:319–325. doi: 10.1006/viro.1994.1194. [DOI] [PubMed] [Google Scholar]
- 33.Lewis D C. Norwalk agent and other small round-structural viruses in the UK. J Infect. 1991;23:220–222. doi: 10.1016/0163-4453(91)92567-o. [DOI] [PubMed] [Google Scholar]
- 34.Lewis D C, Hale A, Jiang X, Eglin R, Brown D W G. Epidemiology of Mexico virus, a small round-structured virus in Yorkshire, United Kingdom, between January 1992 and March 1995. J Infect Dis. 1997;175:951–954. doi: 10.1086/513998. [DOI] [PubMed] [Google Scholar]
- 35.Liu B, Clarke I N, Caul E O, Lambden P R. Human enteric caliciviruses have a unique genome structure and are distinct from the Norwalk-like viruses. Arch Virol. 1995;140:1345–1356. doi: 10.1007/BF01322662. [DOI] [PubMed] [Google Scholar]
- 36.Madore H P, Treanor J J, Pray K A, Dolin R. Enzyme-linked immunosorbent assays for Snow Mountain and Norwalk agents of viral gastroenteritis. J Clin Microbiol. 1986;24:456–459. doi: 10.1128/jcm.24.3.456-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McEvoy M, Balke W, Brown D, Green J, Cartwright R. An outbreak of viral gastroenteritis on a cruise ship. Commun Dis Rep CDR Rev. 1996;6:R188–R192. [PubMed] [Google Scholar]
- 38.Norcott J P, Green J, Lewis D, Estes M K, Barlow K L, Brown D W G. Genomic diversity of small round structured viruses in the United Kingdom. J Med Virol. 1994;44:280–286. doi: 10.1002/jmv.1890440312. [DOI] [PubMed] [Google Scholar]
- 39.Pickering L K, DuPont H L, Blacklow N R, Cukor G. Diarrhea due to Norwalk virus in families. J Infect Dis. 1982;146:116–117. doi: 10.1093/infdis/146.1.116. [DOI] [PubMed] [Google Scholar]
- 40.Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 41.Thornhill T S, Wyatt R G, Kalica A R, Dolin R, Chanock R M, Kapikian A Z. Detection by immune electron microscopy of 26–27 nm virus-like particles associated with 2 family outbreaks of gastroenteritis. J Infect Dis. 1977;135:20–27. doi: 10.1093/infdis/135.1.20. [DOI] [PubMed] [Google Scholar]
- 42.Vinje J, Altena S A, Koopmans M P G. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J Infect Dis. 1997;176:1374–1378. doi: 10.1086/517325. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Jiang X, Madore H P, Gray J, Desselberger U, Ando T, Seto Y, Oishi I, Lew J F, Green K Y, Estes M K. Sequence diversity of small, round-structured viruses in the Norwalk virus group. J Virol. 1994;68:5982–5990. doi: 10.1128/jvi.68.9.5982-5990.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zahorsky J. Hyperemesis hiemis or the winter vomiting disease. Arch Pediatr. 1929;46:391–395. [Google Scholar]