Abstract
Background
Recent evidence shows that inducing ferroptosis may improve efficacy of tumor therapy. However, ferroptosis-related genes have been little studied in patients with breast cancer especially in the neoadjuvant setting. ACSL4 and GPX4 have been well established as the positive and negative regulator of ferroptosis, respectively. This study aimed to explore the predictive value of ACSL4 and GPX4 for patients with breast cancer administered neoadjuvant chemotherapy.
Methods
This study included patients treated with paclitaxel-cisplatin-based neoadjuvant chemotherapy. Immunohistochemistry staining of ACSL4 and GPX4 was carried out on the core needle biopsy specimens. Logistic regression was performed to explore the predictive biomarkers of pathological complete response (pCR). Survival analyses were examined by log-rank test and Cox proportional hazard regression.
Findings
A total of 199 patients were included for the analyses. Both ACSL4 expression and ACSL4/GPX4 combination status could serve as independent predictive factors for pCR. The interaction for pCR was observed between ACSL4 and clinical tumor stage. Besides, ACSL4 expression, GPX4 expression, and their combination status were independent prognostic factors for disease-free survival. Analyses of the Kaplan-Meier Plotter database suggested that higher ACSL4 expression is related to better overall survival, and higher GPX4 expression is related to better distant metastasis-free survival. Pathway analyses revealed that ACSL4 and GPX4 might function in crucial pathways including apoptosis, autophagy, cell adhesion, lipid metabolism, etc.
Interpretation
This study revealed the critical value of ACSL4 and GPX4 serving as novel predictive and prognostic biomarkers for patients with breast cancer receiving neoadjuvant chemotherapy. It might be a novel strategy to induce ferroptosis to promote chemosensitivity. Future studies are required to elucidate the potential mechanisms.
Funding
This work was supported by Shanghai Natural Science Foundation [grant number 19ZR1431100], Clinical Research Plan of Shanghai Hospital Development Center [grant numbers SHDC2020CR3003A, 16CR3065B, and 12016231], Shanghai “Rising Stars of Medical Talent” Youth Development Program for Youth Medical Talents - Specialist Program [grant number 2018-15], Shanghai “Rising Stars of Medical Talent” Youth Development Program for Outstanding Youth Medical Talents [grant number 2018-16], Shanghai Collaborative Innovation Center for Translational Medicine [grant number TM201908], Multidisciplinary Cross Research Foundation of Shanghai Jiao Tong University [grant numbers YG2017QN49, ZH2018QNA42, and YG2019QNA28], Nurturing Fund of Renji Hospital [grant numbers PYMDT-002, PY2018-IIC-01, PY2018-III-15, and PYIII20-09], Science and Technology Commission of Shanghai Municipality [grant numbers 20DZ2201600 and 15JC1402700], and Shanghai Municipal Key Clinical Specialty.
Keywords: Breast cancer, Neoadjuvant chemotherapy, Pathological complete response, Ferroptosis, ACSL4, GPX4
Research in context.
Evidence before this study
In recent years, ferroptosis has gained a lot of interest as a new form of programmed cell death, characterized by iron-dependent accumulation of lipid reactive oxygen species to lethal levels. Reportedly, ferroptosis can be induced by chemotherapeutic drugs, targeted therapeutic agents, and radiotherapy. ACSL4 and GPX4 are the key genes that promote and inhibit the occurrence of ferroptosis, respectively. However, few studies focused on their predictive values of chemosensitivity for patients with breast cancer especially in the neoadjuvant setting.
Added value of this study
This study reveals the critical value of ACSL4 and GPX4 as novel predictive and prognostic biomarkers for patients with breast cancer receiving neoadjuvant chemotherapy. This might help not only screen the candidate responders but also determine the optimal strategy.
Implications of all the available evidence
It is of highly practical value for a high ACSL4/low GPX4 profile to predict pathological complete response for neoadjuvant chemotherapy in breast cancer.
Alt-text: Unlabelled box
1. Introduction
Neoadjuvant chemotherapy has become one of the most important strategies for patients with locally advanced breast cancer, since it not only reduces tumor burden and thereby provides surgical and even breast-conserving opportunities, but also facilitates assessment of individual response at early time [1]. A great number of large clinical trials have revealed that patients who achieve pathological complete response (pCR) after neoadjuvant chemotherapy have better prognosis than those who do not [2]. However, patients are still trapped in a low proportion of pCR currently [3]. To improve pCR rate is a vital goal of neoadjuvant chemotherapy [4,5]. Therefore, identifying patients with superior response to neoadjuvant chemotherapy in early time naturally becomes the main focus in the research on breast cancer at this stage.
In recent years, ferroptosis has gained a lot of interest as a new form of programmed cell death, characterized by iron-dependent accumulation of lipid reactive oxygen species (ROS) to lethal levels [6]. Till date, emerging genes have been identified as related to the crucial process of ferroptosis. For instance, glutathione peroxidase 4 (GPX4) inhibits the occurrence of ferroptosis by reducing ROS level through converting potentially toxic lipid hydroperoxides (L-OOH) to non-toxic lipid alcohols (L-OH) [7,8]. Inactivation of GPX4 through erastin or RSL3 overwhelmingly enhances lipid peroxidation and ultimately results in ferroptosis [7,9]. Interestingly, Hangauer et al. demonstrated that drug-tolerant persister cells are vulnerable to GPX4 inhibition in breast cancer BT474 cell line, which might indicate a promising therapeutic strategy of targeting GPX4 to reverse acquired drug resistance [10]. On the other hand, acyl-CoA synthetase long chain family member 4 (ACSL4) catalyzes the acetylation of long-chain polyunsaturated fatty acids to produce lipid peroxides that arise esterification via interaction with membrane phospholipids and then leads to ferroptosis [11,12]. Highly expressed ACSL4 remarkably increases cellular sensitivity to ferroptosis [13,14]. Nevertheless, the clinical significance of such ferroptosis-related genes has never been studied in breast cancer especially in the neoadjuvant setting.
Reportedly, ferroptosis can be induced by chemotherapeutic drugs (cisplatin [15,16] and paclitaxel [17,18]), targeted therapeutic drugs (lapatinib [19] and siramesine [20]), and radiotherapy [6]. As a programmed cell death mode, ferroptosis may potentially reflect patients’ sensitivity to chemotherapy as well as other treatments. Since ACSL4 and GPX4 have been well established as the positive and negative regulator of ferroptosis, respectively [21], we hypothesized that their expression and disequilibrium status might help predict neoadjuvant chemosensitivity for patients with breast cancer, which was elucidated in this retrospective study of our prospective clinical trials.
2. Methods
2.1. Study population
In this study, we included women aged 18 to 70 years with histologically confirmed locally advanced invasive breast cancer (T2-4 or N1-3) from two separately registered clinical trials, SHPD001 (NCT02199418) and SHPD002 (NCT02221999). The study protocols were published previously [22]. Briefly, paclitaxel at 80 mg/m² on day 1, 8, 15, and 22 combined with cisplatin at 25 mg/m² on day 1, 8, and 15 was given intravenously every 28 days for 4 cycles for all the patients. For human epidermal growth factor receptor 2 (HER2)-positive patients, trastuzumab was recommended at a loading dose of 4 mg/kg, followed by a maintenance dose of 2 mg/kg on day 1 weekly for 16 weeks. In SHPD002, patients with hormone receptor (HorR)-positive breast cancer were randomized to chemotherapy combined with endocrine therapy according to their menstrual status (letrozole for postmenopausal women and ovarian function suppression for premenopausal women) or chemotherapy alone. Premenopausal patients with triple negative breast cancer (TNBC) were randomized to chemotherapy with or without ovarian function suppression in SHPD002. Planned surgery was given sequentially after neoadjuvant chemotherapy at Department of Breast Surgery, Renji Hospital, School of Medicine, Shanghai Jiao Tong University.
Between March 2014 and February 2019, 199 patients with qualified biopsy specimens before neoadjuvant chemotherapy and treated with surgery were available for the analysis.
2.2. Data collection
Baseline information was collected prospectively including patients’ age, menopausal status, body mass index (BMI), family history, tumor size, clinical tumor (T) stage, clinical nodal (N) stage, estrogen receptor (ER), progesterone receptor (PR), HER2, and Ki-67. The high expression levels of ACSL4 and GPX4 were determined by the cutoff values of 0.107 and 0.159, respectively, calculated by Maxstat analysis. Family history was defined as any cancer diagnosed in the first-degree relatives [23]. The follow-up data was also prospectively collected.
All the biopsy tissues were confirmed as invasive breast cancer at Department of Pathology, Renji Hospital. ER and PR positivity were defined as ⩾ 1% of tumor cells with positive nuclear staining. HER2 positivity was defined as immunohistochemistry (IHC) 3+ or amplification confirmed by florescent in situ hybridization [24]. The cutoff value of Ki-67 was 30%. Molecular subtypes were defined as follows according to the St. Gallen Consensus [25]: luminal A-like (ER and/or PR positive, HER2 negative, and Ki-67 < 15%), luminal B-like (ER and/or PR positive; HER2 negative and Ki-67 ⩾ 15% or HER2 positive), HER2-enriched (ER and PR negative, and HER2 positive), and basal-like (ER, PR, and HER2 negative).
2.3. Outcomes
In this study, the definition of pCR was only a few scattered tumor cells remained or residual tumor less than 0.5 cm in diameter in the breast [26]. Disease-free survival (DFS) was calculated as the time from surgery to the first occurrence of locoregional, ipsilateral, contralateral, distant recurrence, and death from any cause.
2.4. IHC
IHC was performed on formalin-fixed paraffin-embedded tissue sections. ACSL4 and GPX4 were detected using the rabbit anti-ACSL4 antibody (1:100; RRID: AB_2714020; Cat#ab155282, Abcam, UK) and anti-GPX4 antibody (1:100; RRID: AB_10973901; Cat#ab125066, Abcam, UK), respectively [27], [28], [29], [30]. Slides were incubated with primary antibodies overnight at 4°C followed by wash and incubation with goat anti-rabbit secondary antibodies (1:1; Cat#ab214880, Abcam, UK) at room temperature for 45 min. Negative controls were treated identically without the primary antibodies.
Biopsy tissue sections were photographed using Leica Aperio CS2 digital pathology scanner (Leica Microsystems Imaging Solutions Ltd, Germany), and analyzed with 4 representative regions of tumor cells randomly captured at ×200 magnification using Leica ImageScope v12.3.2 (Leica Microsystems Imaging Solutions Ltd, Germany). The IHC staining intensity of ACSL4 and GPX4 was analyzed by Image-Pro Plus v6.0 (Media Cybernetics Inc, USA). Protein expression was presented by mean optical density, which was calculated as the integrated optical density (IOD) of positively stained area divided by tumor area in each image [31].
2.5. Cell culture
The human breast cancer cell lines MCF7 (RRID: CVCL_0031; Cat#HTB-22), MDA-MB-231 (RRID: CVCL_0062; Cat#HTB-26), and Hs578T (RRID: CVCL_0332; Cat#HTB-126) were obtained from the American Type Culture Collection (ATCC), authenticated by Short tandem repeat (STR) profiling (Supplementary materials), and confirmed free from mycoplasma. MDA-MB-231 was cultured in L15 (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin/streptomycin (Sigma-Aldrich, USA) at 37°C in a humidified incubator without CO2. Other cell lines were cultured in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified incubator with 5% CO2.
2.6. Cell transfection
To knock down gene expression, small interfering RNAs (siRNAs) were transfected into cells with jetPRIME® (Polyplus-transfection Inc, France) according to the manufacturer's instructions. The sequences of siRNAs were as follows: siACSL4, 5′-GGGAGUGAUGAUGCAUCAUAGCAAU-3′; siGPX4, 5′-ACGAAGAGAUCAAAGAGUU-3′. Negative control was a scrambled siRNA that targets unknown gene sequence. The transfection efficiency was verified by real-time quantitative polymerase chain reaction (RT-qPCR) and western blot (WB) analysis after 48 h.
2.7. RNA extraction and RT-qPCR
Total RNA of cells was extracted using Simply P Total RNA Extraction kit (Bioer Technology, China) and reverse transcribed to complementary deoxyribonucleic acids (cDNAs) using HiScript® II Q RT SuperMix (Vazyme Biotech co., ltd, China). Obtained cDNAs were quantified by RT-qPCR assay using ChamQ SYBR Color qPCR Master Mix (Vazyme Biotech co., ltd, China) on LightCycler® 96 (Roche, Germany) following the manufacturer's instructions. The gene-specific primers used were listed in Supplementary Table 1. Gene expression levels were normalized to GAPDH expression using the 2−△CT method. Each cDNA sample was triplicated in 96-microwell plates.
2.8. WB
To isolate protein, cells were lysed with RIPA Lysis Buffer (Merck Millipore, Germany) and protease inhibitor (Merck Millipore, Germany). Protein concentration was measured with BCA Protein Assay Kit (Beyotime Biotechnology, China). Equal amount of protein was loaded and separated by 10% SDS-PAGE gels and then transferred to PVDF membranes (Merck Millipore, Germany). After blocked in 5% skimmed milk, the membranes were incubated with primary antibodies at 4°C overnight and then incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (1:3000; RRID: AB_2099233; Cat#7074, Cell Signaling Technology, USA) at room temperature for 1 h. Enhanced chemiluminescence was performed with Immobilon Western Chemiluminescent HRP Substrate (Millipore, USA) and detected by Chemidoc Touching Imaging System (BIO-RAD, USA). The primary antibodies used were shown as follows: ACSL4 (1:10000; RRID: AB_2714020; Cat#ab155282, Abcam, UK), GPX4 (1:1000; RRID: AB_10973901; Cat#ab125066, Abcam, UK), and β-actin (1:1000; RRID: AB_2242334; Cat#3700, Cell Signaling Technology, USA).
2.9. Bioinformatics analysis
The Cancer Genome Atlas (TCGA) datasets were employed to calculate the associations of ACSL4 and GPX4 expression with pathological tumor features [32]. Correlations of gene expression were calculated using TCGA breast cancer datasets downloaded by University of California Santa CRUZ (UCSC) Xena [33]. The Kaplan-Meier Plotter database (http://kmplot.com/analysis/) was used to perform survival analysis of distant metastasis-free survival (DMFS) and overall survival (OS), detected by log-rank test as well as Cox proportional hazard regression [34]. The gene profiles GSE40968 and GSE162069 were downloaded from Geno Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) datasets to derive the differentially expressed genes (DEGs), which were then analyzed with gene ontology (GO) enrichment and kyoto encyclopedia of genes and genomes (KEGG) pathway by Database for Annotation, Visualization and Integrated Discovery (DAVID) [35,36]. Pathway interaction analyses were drawn by the R package “clusterProfiler” [37]. The StarBase database (http://starbase.sysu.edu.cn/) was used to predict RNA binding proteins of ACSL4 and GPX4 [38], which were utilized to conduct protein-protein association networks by STRING database [39].
2.10. Statistical analysis
The mean density of ACSL4 and GPX4 was shown as average ± standard deviation (range). The combination status of ACSL4 with GPX4 expression levels was categorized into ACSL4high/GPX4low, ACSL4high/GPX4high, ACSL4low/GPX4low, and ACSL4low/GPX4high. The relationships between ACSL4 or GPX4 expression levels and clinicopathological characteristics (age, menopausal status, clinical T stage, clinical nodal status, ER, PR, HER2, Ki-67, molecular subtype, BMI, and family history) were analyzed as categorical variables by Chi-squared test or Fisher's exact test and as continuous variables by Spearman's rank correlation test. Student's t test was performed for comparing continuous variables following normal distribution between two groups. Patients were divided into the pCR group and the non-pCR group according to treatment outcomes. Univariate and multivariate logistic regression analyses were performed to derive odds ratios (ORs) and 95% confidence intervals (CIs) when analyzing the predictive value of ACSL4 or GPX4 for pCR in whole group and subgroups, and the interactions between ACSL4 or GPX4 and clinicopathological variables. The reverse Kaplan-Meier method was used to calculate the estimated median follow-up period [40,41]. Survival curves were compared by Kaplan-Meier method, and survival rates were compared by log-rank test. Cox proportional hazard regressions were used to calculate hazard ratios (HRs) with 95% CIs for DFS, with baseline age, menopausal status, clinical T stage, clinical nodal status, ER status, HER2 status, and BMI as adjustment factors. Gene correlation analysis was performed by Pearson correlation method. All statistical analyses were performed by R language v3.6.3 (http://www.R-project.org). The P value < 0.05 was considered significant.
2.11. Ethics approval and consent to participate
Both trials were ethically approved by the Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University (SHPD001, approval ID [2014]14K; SHPD002, approval ID [2017]088). All the participants signed written informed consents.
2.12. Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The authors had full access to all the data in the study and accept responsibility to submit for publication.
3. Results
3.1. Baseline features
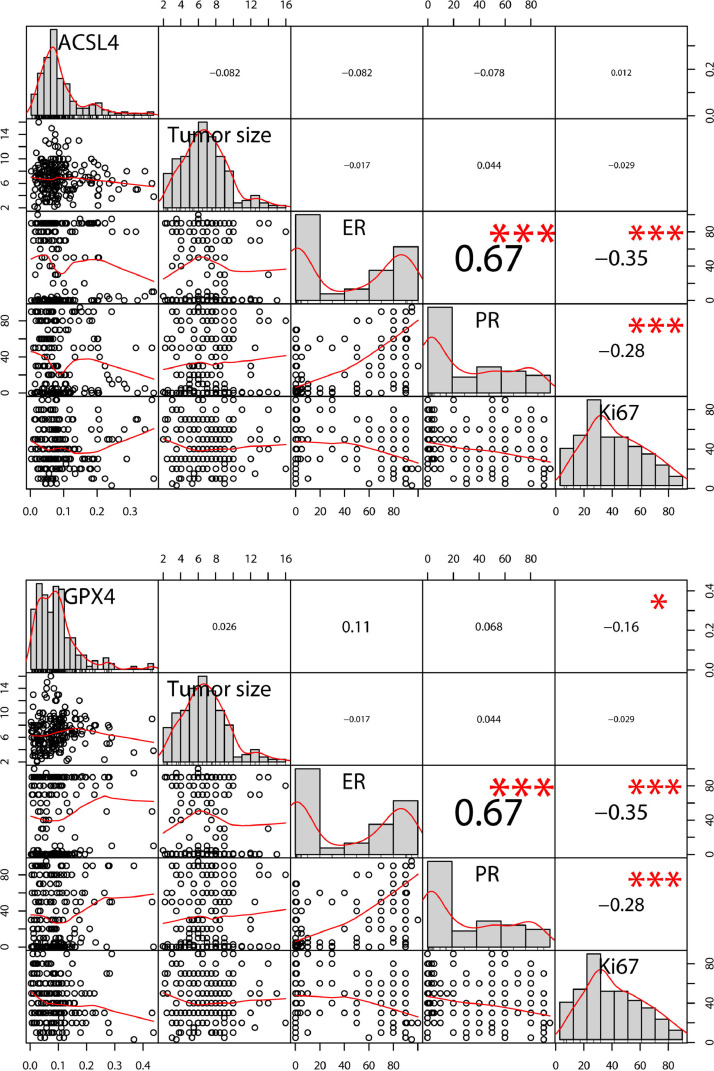
Both ACSL4 and GPX4 were expressed mainly in the cytoplasm and occasionally in the interstitial of breast cancer cells. The representative IHC staining images are shown in Fig. 1. The mean density of ACSL4 and GPX4 was 0.092 ± 0.070 (range, 0.002–0.370) and 0.092 ± 0.072 (range, 0.003–0.436), respectively (Table 1). As categorical variables, GPX4 was positively associated with ER (P=0.004) and luminal-like subtype (P=0.025; Chi-squared test; Table 2). As continuous variables, GPX4 was inversely related to Ki-67 (Spearman's r=-0.16, P=0.026; Fig. 2).
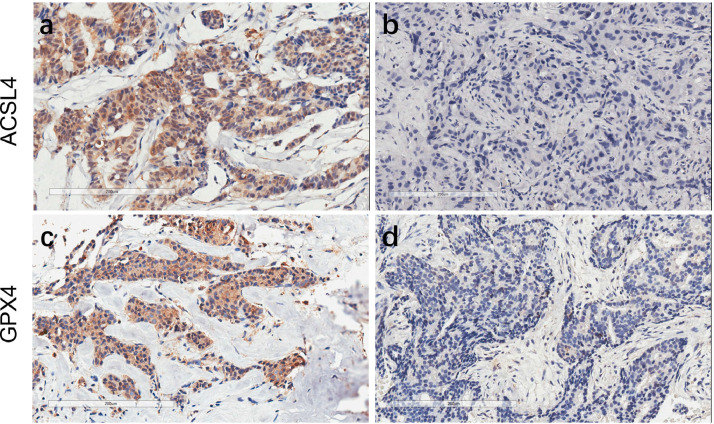
Fig. 1.
Expression of ACSL4 and GPX4 in pre-NAC breast cancer specimens. Notes: Representative immunohistochemistry pictures of high ACSL4 expression (a), low ACSL4 expression (b), high GPX4 expression (c), and low GPX4 expression (d) at ×200 magnification. Scale bar = 200 μm.
Table 1.
Mean density of ACSL4 and GPX4.
| Staining | Density | ||
| Mean | Standard Deviation | Range | |
| ACSL4 | 0.092 | 0.070 | 0.002-0.370 |
| GPX4 | 0.092 | 0.072 | 0.003-0.436 |
Table 2.
Relationships of ACSL4 and GPX4 expression with clinicopathological factors.
| Factors | Total | ACSL4 |
GPX4 |
||||
|---|---|---|---|---|---|---|---|
| (N=199) | Low (N=146) | High (N=53) | P | Low (N=175) | High (N=24) | P | |
| Age | 0.542 | 0.505 | |||||
| <35 | 15 (7.5%) | 10 (6.8%) | 5 (9.4%) | 14 (8.0%) | 1 (4.2%) | ||
| ⩾35 | 184 (92.5%) | 136 (93.2%) | 48 (90.6%) | 161 (92.0%) | 23 (95.8%) | ||
| Menopausal status | 0.112 | 0.783 | |||||
| Premenopausal | 86 (43.2%) | 68 (46.6%) | 18 (34.0%) | 75 (42.9%) | 11 (45.8%) | ||
| Postmenopausal | 113 (56.8%) | 78 (53.4%) | 35 (66.0%) | 100 (57.1%) | 13 (54.2%) | ||
| Clinical T stage | 0.695 | 0.306 | |||||
| T1 | 1 (0.5%) | 1 (0.7%) | 0 | 1 (0.6%) | 0 | ||
| T2 | 46 (23.1%) | 31 (21.2%) | 15 (28.3%) | 41 (23.4%) | 5 (20.8%) | ||
| T3 | 91 (45.7%) | 69 (47.3%) | 22 (41.5%) | 76 (43.4%) | 15 (62.5%) | ||
| T4 | 61 (30.7%) | 45 (30.8%) | 16 (30.2%) | 57 (32.6%) | 4 (16.7%) | ||
| Clinical nodal status | 0.572 | 0.404 | |||||
| Node negative | 23 (11.6%) | 18 (12.3%) | 5 (9.4%) | 19 (10.9%) | 4 (16.7%) | ||
| Node positive | 176 (88.4%) | 128 (87.7%) | 48 (90.6%) | 156 (89.1%) | 20 (83.3%) | ||
| ER status | 0.643 | 0.004 | |||||
| ER negative | 69 (34.7%) | 52 (35.6%) | 17 (32.1%) | 67 (38.3%) | 2 (8.3%) | ||
| ER positive | 130 (65.3%) | 94 (64.4%) | 36 (67.9%) | 108 (61.7%) | 22 (91.7%) | ||
| PR status | 0.223 | 0.086 | |||||
| PR negative | 54 (27.1%) | 43 (29.4%) | 11 (20.8%) | 51 (29.1%) | 3 (12.5%) | ||
| PR positive | 145 (72.9%) | 103 (70.6%) | 42 (79.2%) | 124 (70.9%) | 21 (87.5%) | ||
| HER2 status | 0.549 | 0.403 | |||||
| HER2 negative | 117 (58.8%) | 84 (57.5%) | 33 (62.3%) | 101 (57.7%) | 16 (66.7%) | ||
| HER2 positive | 82 (41.2%) | 62 (42.5%) | 20 (37.7%) | 74 (42.3%) | 8 (33.3%) | ||
| Ki-67 | 0.935 | 0.261 | |||||
| <30% | 48 (24.1%) | 35 (24.0%) | 13 (24.5%) | 40 (22.9%) | 8 (33.3%) | ||
| ⩾30% | 151 (75.9%) | 111 (76.0%) | 40 (75.5%) | 135 (77.1%) | 16 (66.7%) | ||
| Molecular subtype | 0.407 | 0.025 | |||||
| Luminal A-like | 15 (7.5%) | 12 (8.2%) | 3 (5.7%) | 12 (6.9%) | 3 (12.5%) | ||
| Luminal B-like | 140 (70.3%) | 98 (67.1%) | 42 (79.3%) | 119 (68.0%) | 21 (87.5%) | ||
| HER2-enriched | 19 (9.6%) | 15 (10.3%) | 4 (7.5%) | 19 (10.9%) | 0 | ||
| Basal-like | 25 (12.6%) | 21 (14.4%) | 4 (7.5%) | 25 (14.2%) | 0 | ||
| Family history | 0.824 | 0.802 | |||||
| Negative | 145 (72.9%) | 107 (72.3%) | 38 (71.7%) | 127 (72.6%) | 18 (75.0%) | ||
| Positive | 54 (27.1%) | 39 (26.7%) | 15 (28.3%) | 48 (27.4%) | 6 (25.0%) | ||
Abbreviations: T, tumor; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Fig. 2.
The correlations of ACSL4 and GPX4 density with tumor size, ER, PR, and Ki-67. Notes: Distribution of the data is present along the diagonals. The upper part exhibits correlation coefficients and P values calculated by Spearman's rank correlation analysis (* P < 0.05, ** P < 0.01, and *** P < 0.001). The lower part shows scatter plots with smooth fitting curves. There was a significant correlation between GPX4 and Ki-67 (r=-0.16, P=0.026), ER and PR (r=0.67, P < 0.001), ER and Ki-67 (r=-0.35, P < 0.001), and PR and Ki-67 (r=-0.28, P < 0.001). Abbreviations: ER, estrogen receptor; PR, progesterone receptor.
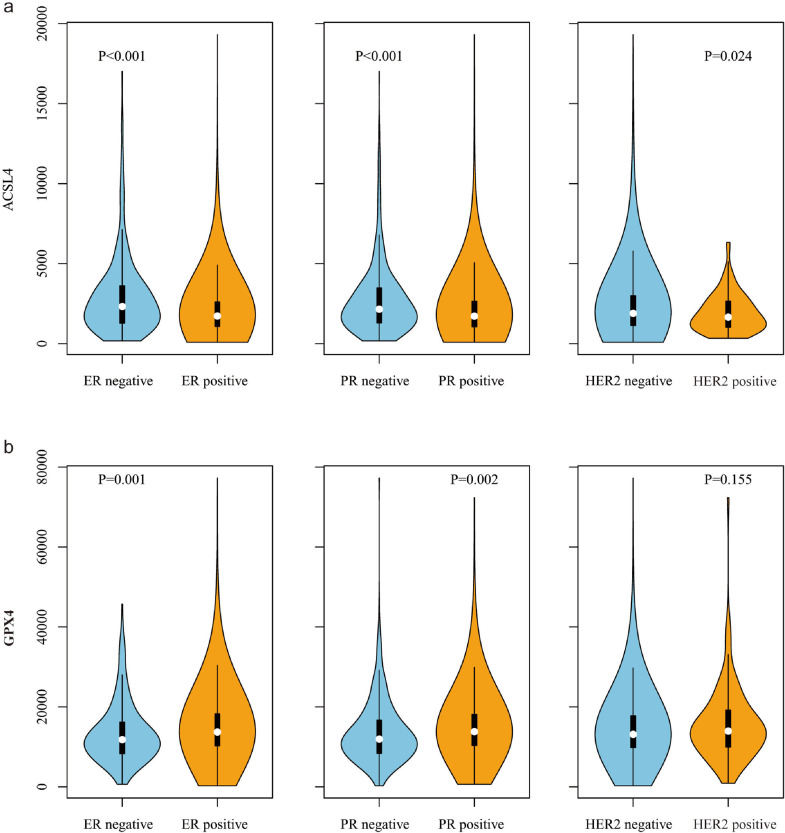
In the TCGA datasets, we observed an inverse correlation of ACSL4 expression with ER (P < 0.001), PR (P < 0.001), and HER2 (P=0.024; Student's t test; Fig. 3a), and a positive correlation of GPX4 expression with ER (P=0.001) and PR (P=0.002; Student's t test; Fig. 3b).
Fig. 3.
The correlations of ACSL4 (a) and GPX4 (b) expression with ER, PR, and HER2 in TCGA datasets (n=1070). Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
3.2. Relationships of ACSL4 and GPX4 levels with therapeutic effect
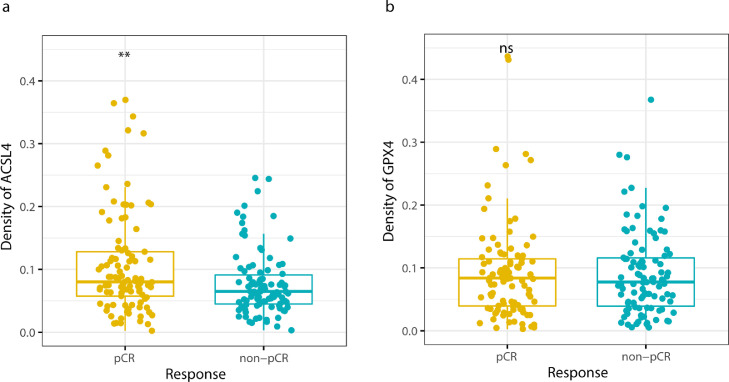
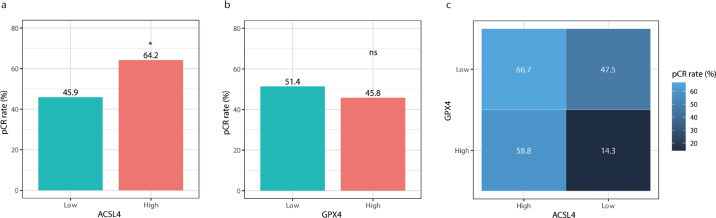
Among 199 patients, 101 (50.8%) achieved pCR while 98 (49.2%) failed to. ACSL4 expression was significantly higher in the pCR group than in the non-pCR group (P=0.002; Student's t test; Fig. 4a). However, there was no significant difference of GPX4 expression between the pCR group and non-pCR group (P=0.657; Student's t test; Fig. 4b). On the other hand, the pCR rate was 64.2% in ACSL4 high-expression group, much higher than 45.9% in ACSL4 low-expression group (P=0.023; Chi-squared test; Fig. 5a), while the pCR rates showed no significant difference according to GPX4 expression levels (51.4% vs. 45.8%, P=0.607; Chi-squared test; Fig. 5b). According to the combination status of the two proteins, patients with the highest pCR rate appeared to be ACSL4high/GPX4low group (66.7%), followed by ACSL4high/GPX4high group (58.8%), ACSL4low/GPX4low group (47.5%), and ACSL4low/GPX4high group (14.3%; P=0.038; Chi-squared test; Fig. 5c).
Fig. 4.
ACSL4 and GPX4 expression between pCR and non-pCR groups. Notes: (a) ACSL4 density was higher in pCR group than non-pCR group (P=0.002; Student's t test). (b) No difference of GPX4 density was detected between pCR and non-pCR group (P=0.657; Student's t test). Abbreviations: pCR, pathological complete response; ns, not significant.
Fig. 5.
The pCR rates of patients with different ACSL4 or GPX4 levels. Notes: (a) The pCR rate was higher in ACSL4-high group than ACSL4-low group (P=0.023; Chi-squared test). (b) No difference was detected in pCR rates between GPX4-high and GPX4-low groups (P=0.607; Chi-squared test). (c) The pCR rates in the four subgroups were as follows: 66.7% in ACSL4high/GPX4low, 58.8% in ACSL4high/GPX4high, 47.5% in ACSL4low/GPX4low and 14.3% in ACSL4low/GPX4high (P=0.038; Chi-squared test). Abbreviations: pCR, pathological complete response; ns, not significant.
In the univariate logistic regression analysis, ACSL4 level (OR=2.110, 95% CI 1.103-4.037, P=0.024) and the combination of ACSL4 with GPX4 levels (OR=2.234, 95% CI 1.047–4.767, P=0.038) were predictors of pCR. However, GPX4 level alone was not capable of predicting pCR (OR=0.799, 95% CI 0.340–1.881, P=0.608; likelihood ratio test; Table 3), which remained non-significant after adjustment (OR=1.573, 95% CI 0.572–4.325, P=0.380; likelihood ratio test; Table 4). The multivariate logistic regression analysis suggested that both ACSL4 expression level (OR=3.487, 95% CI 1.505-8.077, P=0.004; likelihood ratio test; Table 5) and the combination of ACSL4 with GPX4 expression levels (OR=2.708, 95% CI 1.035–7.086, P=0.042; likelihood ratio test; Table 6) could serve as independent predictors of pCR, along with age, clinical T stage, ER status, HER2 status, and Ki-67 (P < 0.05; likelihood ratio test; Tables 5, 6).
Table 3.
Univariate analyses of the predictive markers of pCR.
| Characteristics | Comparison for OR | Univariate OR | 95% CI | P | |
| GPX4 | High vs. low | 0.799 | 0.340 | 1.881 | 0.608 |
| ACSL4 | High vs. low | 2.110 | 1.103 | 4.037 | 0.024 |
| ACSL4/GPX4 combination | ASCL4high/GPX4low vs. others | 2.234 | 1.047 | 4.767 | 0.038 |
| Age (years) | ⩾35 vs. <35 | 0.348 | 0.107 | 1.133 | 0.080 |
| Menopausal status | Post- vs. pre-menopausal | 1.145 | 0.653 | 2.006 | 0.637 |
| Clinical T stage | T4 vs. T1-3 | 0.466 | 0.251 | 0.863 | 0.015 |
| ER status | Positive vs. negative | 0.177 | 0.091 | 0.343 | <0.001 |
| HER2 status | Positive vs. negative | 4.159 | 2.268 | 7.626 | <0.001 |
| Ki-67 | ⩾30% vs. <30% | 3.761 | 1.841 | 7.680 | <0.001 |
Abbreviations: pCR, pathological complete response; OR, odds ratio; CI, confidence interval; T, tumor; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2.
Table 4.
Multivariate analysis of the predictive markers of pCR using GPX4 expression level.
| Characteristics | Comparison for OR | Multivariate OR | 95% CI | P | |
| GPX4 | High vs. low | 1.573 | 0.572 | 4.325 | 0.380 |
| Age (years) | ⩾35 vs. <35 | 0.219 | 0.055 | 0.880 | 0.032 |
| Menopausal status | Post- vs. pre-menopausal | 1.108 | 0.534 | 2.301 | 0.782 |
| Clinical T stage | T4 vs. T1-3 | 0.445 | 0.205 | 0.969 | 0.041 |
| ER status | Positive vs. negative | 0.169 | 0.079 | 0.362 | <0.001 |
| HER2 status | Positive vs. negative | 4.463 | 2.216 | 8.985 | <0.001 |
| Ki-67 | ⩾30% vs. <30% | 2.530 | 1.088 | 5.881 | 0.031 |
Abbreviations: pCR, pathological complete response; OR, odds ratio; CI, confidence interval; T, tumor; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2.
Table 5.
Multivariate analysis of the predictive markers of pCR using ACSL4 expression level.
| Characteristics | Comparison for OR | Multivariate OR | 95% CI | P | |
| ACSL4 | High vs. low | 3.487 | 1.505 | 8.077 | 0.004 |
| Age (years) | ⩾35 vs. <35 | 0.234 | 0.056 | 0.973 | 0.046 |
| Menopausal status | Post- vs. pre-menopausal | 0.908 | 0.424 | 1.942 | 0.803 |
| Clinical T stage | T4 vs. T1-3 | 0.444 | 0.201 | 0.982 | 0.045 |
| ER status | Positive vs. negative | 0.154 | 0.071 | 0.335 | <0.001 |
| HER2 status | Positive vs. negative | 5.082 | 2.444 | 10.566 | <0.001 |
| Ki-67 | ⩾30% vs. <30% | 2.561 | 1.087 | 6.033 | 0.031 |
Abbreviations: pCR, pathological complete response; OR, odds ratio; CI, confidence interval; T, tumor; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2.
Table 6.
Multivariate analysis of the predictive markers of pCR using combination of ACSL4 and GPX4 expression levels.
| Characteristics | Comparison for OR | Multivariate OR | 95% CI | P | |
| ACSL4/GPX4 combination | High vs. low | 2.708 | 1.035 | 7.086 | 0.042 |
| Age (years) | ⩾35 vs. <35 | 0.239 | 0.059 | 0.968 | 0.045 |
| Menopausal status | Post- vs. pre-menopausal | 1.011 | 0.481 | 2.125 | 0.977 |
| Clinical T stage | T4 vs. T1-3 | 0.420 | 0.193 | 0.913 | <0.029 |
| ER status | Positive vs. negative | 0.184 | 0.087 | 0.389 | <0.001 |
| HER2 status | Positive vs. negative | 4.688 | 2.301 | 9.549 | <0.001 |
| Ki-67 | ⩾30% vs. <30% | 2.501 | 1.073 | 5.825 | 0.034 |
Abbreviations: pCR, pathological complete response; OR, odds ratio; CI, confidence interval; T, tumor; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2.
3.3. Subgroup analysis
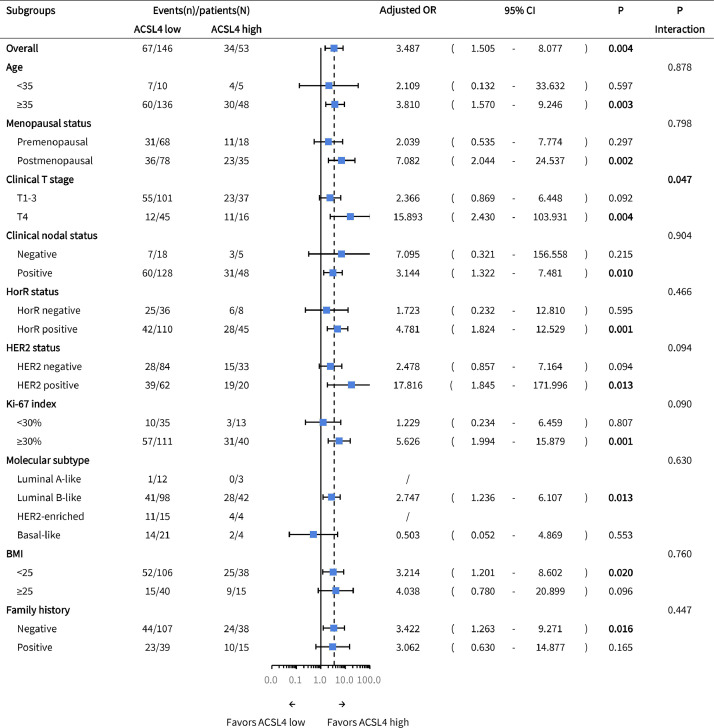
Patients with higher ACSL4 expression achieved higher pCR rates than the low-expression group in those aged ⩾ 35 years (adjusted OR=3.810, 95% CI 1.570–9.246, P=0.003) and postmenopausal (adjusted OR=7.082, 95% CI 2.044–24.537, P=0.002), clinical T4 (adjusted OR=15.893, 95% CI 2.430–103.931, P=0.004), clinical node-positive (adjusted OR=3.144, 95% CI 1.322–7.481, P=0.010), HorR-positive (adjusted OR=4.781, 95% CI 1.824–12.529, P=0.001), HER2-positive (adjusted OR=17.816, 95% CI 1.845-171.996, P=0.013), Ki-67 ⩾ 30% (adjusted OR=5.626, 95% CI 1.994–15.879, P=0.001), luminal B-like (adjusted OR=2.747, 95% CI 1.236–6.107, P=0.013), BMI < 25 (adjusted OR=3.214, 95% CI 1.201–8.602, P=0.020), and family history-negative subgroups (adjusted OR=3.422, 95% CI 1.263–9.271, P=0.016; likelihood ratio test; Fig. 6). Besides, a significant interaction was observed between ACSL4 expression and clinical T stage (P=0.047; likelihood ratio test; Fig. 6).
Fig. 6.
Subgroup analysis of pCR according to ACSL4 expression levels. Abbreviations: pCR, pathological complete response; OR, odds ratio; CI, confidence interval; T, tumor; HorR, hormone receptor; HER2, human epidermal growth factor receptor 2; BMI, body mass index.
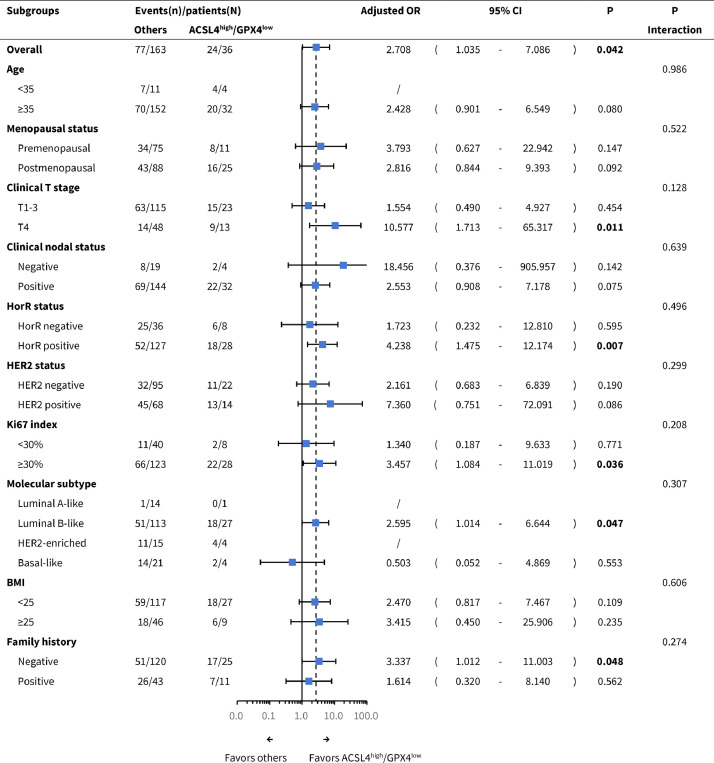
In parallel, pCR was more easily achieved by ACSL4high/GPX4low patients compared with others in clinical T4 (adjusted OR=10.577, 95% CI 1.713-65.317, P=0.011), HorR-positive (adjusted OR=4.238, 95% CI 1.475–12.174, P=0.007), Ki-67 ⩾ 30% (adjusted OR=3.457, 95% CI 1.084–11.019, P=0.036), luminal B-like (adjusted OR=2.595, 95% CI 1.014–6.644, P=0.047), and family history-negative subgroups (adjusted OR=3.337, 95% CI 1.012–11.003, P=0.048; likelihood ratio test; Fig. 7).
Fig. 7.
Subgroup analysis of pCR according to the combination of ACSL4 and GPX4 expression levels. Abbreviations: pCR, pathological complete response; OR, odds ratio; CI, confidence interval; T, tumor; HorR, hormone receptor; HER2, human epidermal growth factor receptor 2; BMI, body mass index.
3.4. Survival analysis
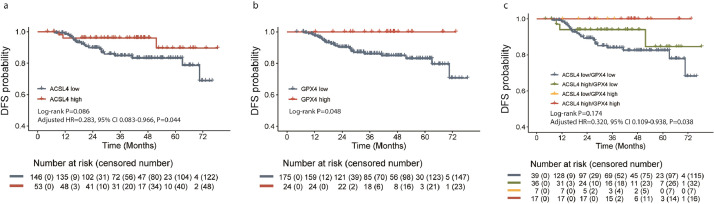
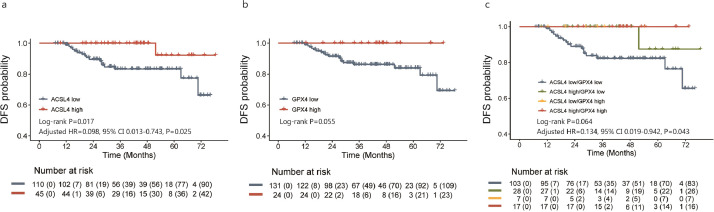
The median follow-up period was 41.2 months (range, 4.2–78.5 months). In all, 24 events were observed in the total patients. There was a marginally significant advantage of DFS in ACSL4 high-expression group over the low-expression group (log-rank P=0.086), while ACSL4 expression could serve as an independent prognostic factor of DFS in the multivariate analysis (adjusted HR=0.283, 95% CI 0.083–0.966, P=0.044; likelihood ratio test; Fig. 8a). Besides, GPX4 high-expression group achieved a longer DFS than those with lower GPX4 expression (log-rank P=0.048; Fig. 8b). When patients were divided into four groups by the combination status of ACSL4 and GPX4 expression levels, the best DFS was observed in ACSL4high/GPX4high group and the worst in ACSL4low/GPX4low group (log-rank P=0.174; adjusted HR=0.320, 95% CI 0.109–0.938, P=0.038; likelihood ratio test; Fig. 8c). In HorR-positive subgroup, better DFS were also observed in those with higher ACSL4 expression (log-rank P=0.017; adjusted HR=0.098, 95% CI 0.013–0.743, P=0.025; likelihood ratio test; Fig. 9a), higher GPX4 expression (log-rank P=0.055; Fig. 9b), and the ACSL4high/GPX4high group women (log-rank P=0.064; adjusted HR=0.134, 95% CI 0.019–0.942, P=0.043; likelihood ratio test; Fig. 9c).
Fig. 8.
Cumulative disease-free survival curves of patients with different ACSL4 or GPX4 expression levels. Notes: Kaplan-Meier plots according to different ACSL4 expression levels (a), GPX4 expression levels (b), and their combination status (c). Abbreviations: HR, hazard ratio; CI, confidence interval.
Fig. 9.
Cumulative disease-free survival curves of patients with different ACSL4 or GPX4 expression levels in HorR-positive subgroup. Notes: Kaplan-Meier plots according to different ACSL4 expression levels (a), GPX4 expression levels (b), and their combination status (c) in HorR-positive subgroup. Abbreviations: HR, hazard ratio; CI, confidence interval; HorR, hormone receptor.
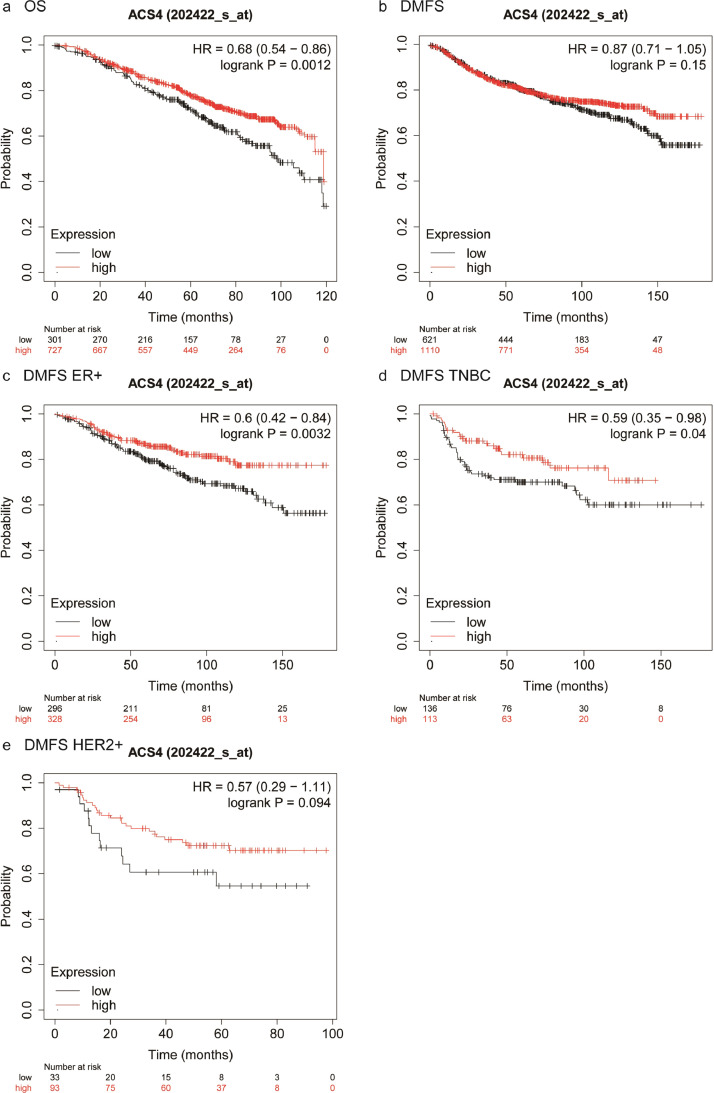
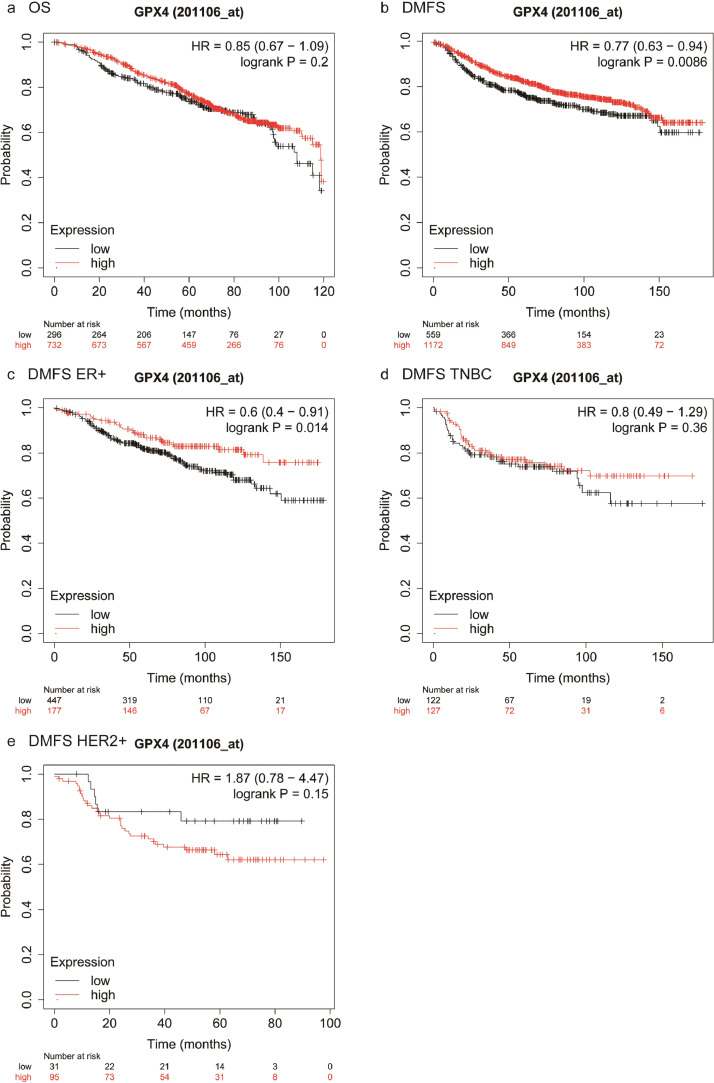
We further performed survival analyses of external breast cancer cohorts based on the Kaplan-Meier Plotter database, a well-known public database that is capable to assess the effect of 54k genes on survival in 21 cancer types. It suggested that patients with higher ACSL4 expression showed better OS compared with those with lower ACSL4 expression (log-rank P=0.0012; HR=0.68, 95% CI 0.54–0.86; Fig. 10a), although there was no significant difference in DMFS (log-rank P=0.15; HR=0.87, 95% CI 0.71–1.05; Fig. 10b). However, the DMFS in patients with higher ACSL4 expression was significantly better than those with lower expression in ER-positive (log-rank P=0.0032; HR=0.6, 95% CI 0.42–0.84; Fig. 10c) and TNBC women (log-rank P=0.04; HR=0.59, 95% CI 0.35–0.98; Fig. 10d), but not in HER2-positive women (log-rank P=0.094; HR=0.57, 95% CI 0.29–1.11; Fig. 10e). In terms of GPX4, there was no statistical difference of OS (log-rank P=0.2; HR=0.85, 95% CI 0.67–1.09; Fig. 11a), whereas GPX4 high-expression group achieved significantly longer DMFS than GPX4 low-expression group (log-rank P=0.0086; HR=0.77, 95% CI 0.63–0.94; Fig. 11b). Consistent results were observed in ER-positive women (log-rank P=0.014; HR=0.6, 95% CI 0.4–0.91; Fig. 11c), while no DMFS difference showed in TNBC patients (log-rank P=0.36; HR=0.8, 95% CI 0.49–1.29; Fig. 11d) or HER2-positive (log-rank P=0.15; HR=1.87, 95% CI 0.78–4.47; Fig. 11e).
Fig. 10.
Kaplan-Meier plots of patients with different ACSL4 expression levels in Kaplan-Meier plotter database. Notes: Cumulative OS curves in the whole group (a) and DMFS curves in the whole group (b), ER-positive subgroup (c), TNBC subgroup (d), and HER2-positive subgroup (e). Abbreviations: OS, overall survival; DMFS, distant metastasis-free survival; HR, hazard ratio; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.
Fig. 11.
Kaplan-Meier plots of patients with different GPX4 expression levels in Kaplan-Meier plotter database. Notes: Cumulative OS curves in the whole group (a) and DMFS curves in the whole group (b), ER-positive subgroup (c), TNBC subgroup (d), and HER2-positive subgroup (e). Abbreviations: OS, overall survival; DMFS, distant metastasis-free survival; HR, hazard ratio; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.
3.5. GO, KEGG, and pathway interaction analysis
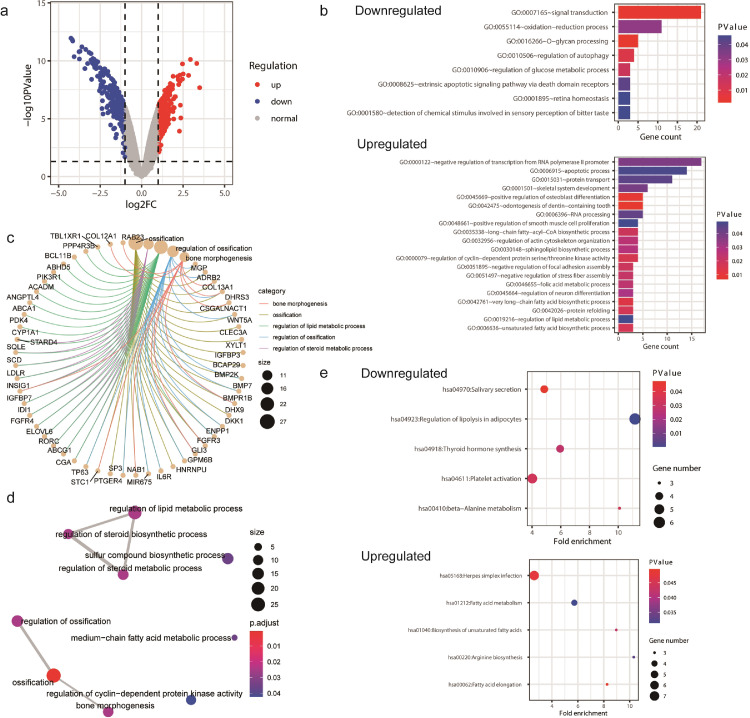
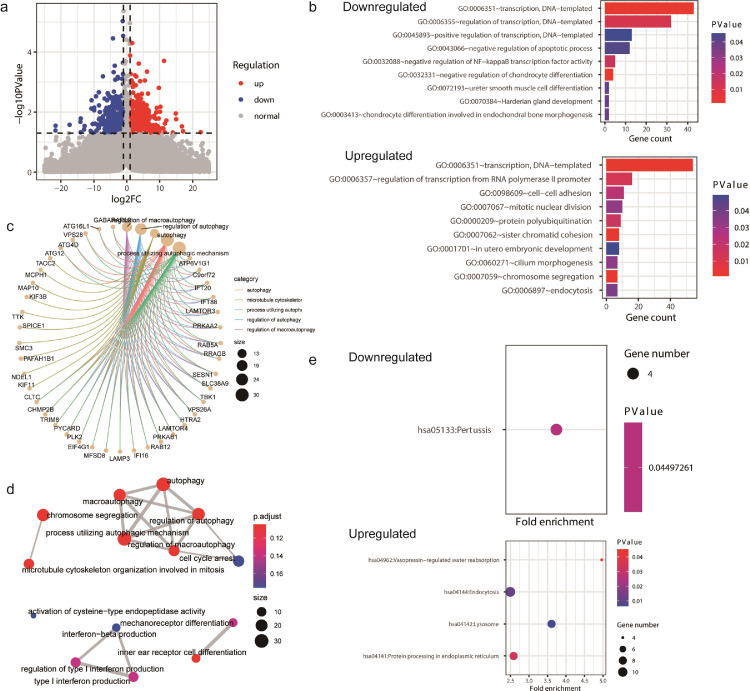
To further investigate the potential roles of ACSL4 and GPX4 in breast cancer, we searched through GEO database and obtained the gene profiles GSE40968 and GSE162069, which respectively revealed the DEGs after overexpressing ACSL4 and knocking down GPX4 in breast cancer cell lines. The gene expression data GSE40968 was applied to bioinformatics analysis to explore the potential function of ACSL4 in breast cancer. As depicted by the volcano plot, a total of 412 and 252 genes were upregulated and downregulated, respectively, after overexpressing ACSL4 in SKBR3 cell line (Fig. 12a). GO enrichment pointed out that the downregulated DEGs might participate in multiple crucial biological processes including signal transduction, autophagy, and glucose metabolic process, while the upregulated DEGs play great roles in negative regulation of transcription, apoptosis, cyclin-dependent protein kinase activity, long-chain fatty-acyl-coA biosynthetic process, and lipid metabolic process (Fig. 12b). The DEGs also take great part in steroid metabolism (Fig. 12c, d). Besides, KEGG analysis highlighted several important pathways such as platelet activation, thyroid hormone synthesis, and fatty acid metabolism (Fig. 12e).
Fig. 12.
Bioinformatics analyses of ACSL4. Notes: (a) Visualization of differentially expressed genes (DEGs) after overexpressing ACSL4 in SKBR3 cell line. (b) Gene ontology (GO) enrichment of the downregulated and upregulated DEGs. (c) The netplot of GO pathways of the DEGs. (d) The mapplot of GO pathways of the DEGs. (e) Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis of the downregulated and upregulated DEGs. Abbreviations: FC, fold change.
The gene profile GSE162069 was analyzed to investigate the potential function of GPX4 in breast cancer. There were 442 genes upregulated and 399 genes downregulated after knocking down GPX4 in MDA-MB-231 cell line (Fig. 13a). GO analysis suggested that the downregulated DEGs might function in apoptosis, and the upregulated DEGs might regulate cell adhesion, mitotic nuclear division, chromosome segregation, etc. (Fig. 13b). The DEGs were also active in cell cycle arrest and autophagy (Fig. 13c, d). In addition, GPX4 was implicated to be involved in lysosome and protein processing in endoplasmic reticulum by KEGG analysis (Fig. 13e).
Fig. 13.
Bioinformatics analyses of GPX4. Notes: (a) Visualization of differentially expressed genes (DEGs) after knocking down GPX4 in MDA-MB-231 cell line. (b) Gene ontology (GO) enrichment of the downregulated and upregulated DEGs. (c) The netplot of GO pathways of the DEGs. (d) The mapplot of GO pathways of the DEGs. (e) Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis of the downregulated and upregulated DEGs. Abbreviations: FC, fold change.
Furthermore, we conducted the protein-protein interaction network for the potential RNA binding proteins of ACSL4 and GPX4 predicted by StarBase database, an open-source platform for studying molecule interactions from CLIP-seq, degradome-seq, and RNA-RNA interactome data (Fig. S1). It implicates the interactions between the potential binding proteins of ACSL4 and GPX4.
3.6. Validation of the DEGs
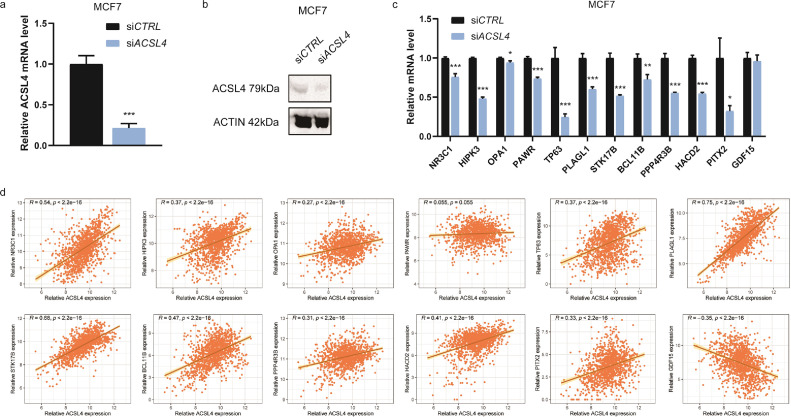
For verification of the DEGs, ACSL4 siRNAs were transfected into MCF7, MDA-MB-231, and Hs578T cell lines and the interfering efficiency was detected by both RT-qPCR (Figs. 14a, S2a) and WB (Figs. 14b, S2b). Interfering ACSL4 significantly decreased the expression of NR3C1, HIPK3, OPA1, PAWR, TP63, PLAGL1, STK17B, all of which take part in the apoptotic process, in MCF7 (Fig. 14c). BCL11B and PPP4R3B, two critical proteins for regulation of lipid metabolic process, and HACD2, which regulates long-chain fatty-acyl-coA biosynthetic process, were also reduced after interfering ACSL4 (Fig. 14c). One out of the genes that negatively regulates transcription from RNA polymerase II promoter, PITX2, was dominantly decreased after knocking down ACSL4 in MCF7 (Fig. 14c), MDA-MB-231 (Fig. S2c), and Hs578T (Fig. S2d). Besides, GDF15, which partakes signal transduction, was significantly increased after knocking down ACSL4 in Hs578T cell line (Fig. S2d). These gene changes were in consistence with those of GSE40968 dataset. The relationships between these genes and ACSL4 were further verified with their expression in breast cancer tissues of public cohorts. Consistently, it revealed a significantly reverse correlation between ACSL4 and GDF15, while other DEGs were positively correlated to ACSL4 in TCGA breast cancer dataset (Fig. 14d).
Fig. 14.
Verification of the differentially expressed genes of ACSL4. Notes: (a) Interfering efficiency in MCF7 cell line detected by RT-qPCR. (b) Interfering efficiency in MCF7 cell line detected by WB. (c) Differentially expressed genes validated by RT-qPCR after transfecting ACSL4 siRNAs into MCF7 cell line. (d) Correlation analyses for the differentially expressed genes and ACSL4 in TCGA breast cancer dataset (n=1247). * P < 0.05, ** P < 0.01, and *** P < 0.001 (Student's t test).
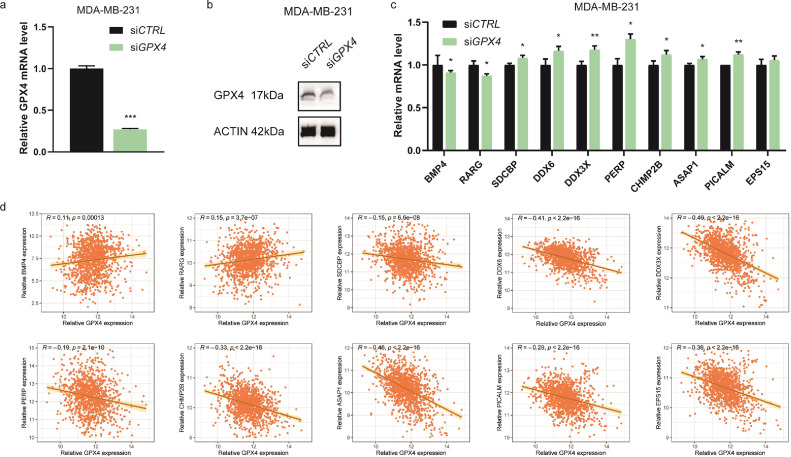
To knock down GPX4, the three breast cancer cell lines were transfected with GPX4 siRNAs and detected for interfering efficiency by RT-qPCR (Figs. 15a, S3a) and WB (Figs. 15b, S3b). As a result, GPX4 knockdown significantly reduced the expression of BMP4 and RARG, which could negatively regulate apoptotic process, in MDA-MB-231 (Fig. 15c) and MCF7 (Fig. S3c). Besides, the expression of genes that play roles in cell adhesion were significantly enhanced after interfering GPX4 in MDA-MB-231 (Fig. 15c) and Hs578T (Fig. S3d), including SDCBP, DDX6, DDX3X, PERP, CHMP2B, ASAP1, and PICALM. These results validated those of the bioinformatics analysis for GSE162069 dataset. In TCGA breast cancer dataset, positive associations were detected between BMP4 as well as RARG and GPX4, whereas reverse associations were seen between SDCBP, DDX6, DDX3X, PERP, CHMP2B, ASAP1, PICALM, EPS15, and GPX4 (Fig. 15d). The public clinical data for gene expression correlation well supports the in vitro experimental results.
Fig. 15.
Verification of the differentially expressed genes of GPX4. Notes: (a) Interfering efficiency in MDA-MB-231 cell line detected by RT-qPCR. (b) Interfering efficiency in MDA-MB-231 cell line detected by WB. (c) Differentially expressed genes validated by RT-qPCR after transfecting GPX4 siRNAs into MDA-MB-231 cell line. (d) Correlation analyses for the differentially expressed genes and GPX4 in TCGA breast cancer dataset (n=1247). * P < 0.05, ** P < 0.01, and *** P < 0.001 (Student's t test).
4. Discussion
To the best of our knowledge, this study for the first time reported not only the predictive value of ferroptosis-related genes ACSL4 and GPX4 for pCR but also their prognostic value for survival outcomes in locally advanced breast cancer treated with neoadjuvant chemotherapy.
Our data first revealed an inverse association between GPX4 expression and Ki-67 index in patients with breast cancer. Besides, there was a positive association observed between GPX4 and ER. The TCGA datasets also indicated that GPX4 expression was positively related to HorR status. Supporting our results, Rusolo et al. reported GPX4 is downregulated in breast cancer MCF7 and MDA-MB-231 cell lines compared with non-cancerous breast MCF10A cell line [42]. Consistently, Cejas et al. demonstrated lower GPX4 expression in breast tumor cells than in benign ductal epithelium [43]. Additionally, they observed a positive correlation of GPX4 expression with ER and PR staining. Moreover, GPX4 expression was repressed in poorly differentiated breast invasive ductal carcinoma compared with the well-differentiated type [43]. Altogether, GPX4 might serve as a tumor suppressor and indicate better differentiated status in breast cancer. On the other hand, the TCGA datasets suggested that ACSL4 expression was inversely associated with HorR and HER2 status. In line with it, Wu et al. summarized the inverse relationships of ACSL4 with ER and HER2 based on multiple public databases [44]. Monaco et al. [45] and Yen et al. [46] also found a negative correlation between expression of ACSL4 and ER. Besides, ACSL4 is overexpressed in breast cancer tissues compared with adjacent normal tissues [47]. In vitro, ACSL4 inhibition significantly suppresses cell proliferation, invasion, and migration in MDA-MB-231 cell line [48,49]. Taking the evidence together, aggressive biological behavior is indicated to ACSL4 overexpressed tumors.
This study demonstrated that ACSL4 expression is an independent predictive factor for pCR, with higher expression contributing to enhanced sensitivity to neoadjuvant chemotherapy in breast cancer. It is supported by the TCGA data that tumors with ACSL4 overexpression are more often ER-negative, PR-negative, and HER2-negative, which is exactly the most sensitive subtype to neoadjuvant cisplatin- and paclitaxel-based treatment [22]. Our results are supported by the findings of basic research as well. Ferroptotic cell death has been recognized as one of the key mechanisms through which chemotherapeutic drugs kill cancer cells [15], [16], [17], [18]. Doll's study found that ferroptosis is much easier to be induced by the ferroptosis inducer RSL3 in ACSL4 high-expressed cell lines MDA-MB-157, MDA-MB-231, and MDA-MB-468 than the low-expressed AU565, MCF7, and T47D [13]. Yuan's study found that ferroptosis-sensitive cell lines HepG2 and HL60 express ACSL4 at relatively high levels, while targeting ACSL4 significantly suppresses ferroptotic cell death induced by erastin [14]. Therefore, we speculated that patients with higher ACSL4 expression might respond better to chemotherapy due to their hypersensitivity to ferroptosis. To further explore the association between chemosensitivity and ACSL4, we performed bioinformatics analysis and validated the DEGs for ACSL4. It highlighted the crucial function of BCL11B, which is increased after ACSL4 overexpression, in lipid metabolism. Yang et al. reported that BCL11B expression greatly sensitizes HepG2 and Huh7 hepatocarcinoma cell lines towards doxorubicin [50]. Besides, the signaling transduction factor GDF15, which is a secreted cytokine of TGF-β superfamily, is decreased after ACSL4 overexpression. Zhao et al. found that high pretreatment serum GDF15 level is correlated with the occurrence of resistance to paclitaxel-carboplatin-based chemotherapy in patients with epithelial ovarian cancer [51]. Therefore, ACSL4 might sensitize breast cancer cells to cytotoxic drugs through promoting BCL11B and inhibiting GDF15, which requires future study to elucidate. Of note, two basic studies found ACSL4 inhibitor PRGL493 slows down breast cancer cell proliferation treated by low-dose chemotherapeutic agents, but the role of ACSL4 in chemotherapy-induced ferroptosis is to be investigated [48,52]. In future, ferroptosis-inducing agents may become a new therapeutic strategy in breast cancer. Moreover, ACSL4 may be a novel predictive biomarker for neoadjuvant chemotherapy.
Additionally, we discovered the combination status of ACSL4 and GPX4 expression could independently predict pCR. The pCR rates elevated step by step with increase of ACSL4 and decrease of GPX4. We postulated that this phenomenon might be associated with the equilibrium state of the two genes. As known about apoptosis, another classical form of cell death that is promoted and inhibited by Bax and Bcl2, respectively, the balance of Bax and Bcl2 will determine the cells’ fate to apoptosis [53], [54]. Similarly, ACSL4 is involved in lipid metabolism to promote ferroptosis [55], while GPX4 takes part in amino acid metabolism to inhibit occurrence of ferroptosis [7,55]. Therefore, the hypersensitivity to chemotherapy of ACSL4high/GPX4low patients might be related to the combined action of ACSL4 and GPX4 in ferroptosis. On the contrary, ACSL4low/GPX4high patients might be resistant to the chemotherapeutic agents that mainly induce ferroptotic cell death. It hints us that ACSL4 inducer and/or GPX4 inhibitor might promote treatment efficacy for these patients. Clinical research will be necessary to unearth the utility of this promising strategy.
Interestingly, we found that ACSL4 expression level and its combination with GPX4 expression level could both predict pCR for family history-negative patients rather than those with family history. We postulated that it might be associated with the different forms of cell death induced by platinum between patients with and without family history. Reportedly, about 20% of breast cancer with family history can be explained by homologous recombination repair gene mutations such as BRCA1/2 mutations [56], while this proportion rises to 55% in patients with family history of both breast and ovarian cancer [57]. Previous research has revealed that platinum-induced double-strand breaks are more easily accumulated in homologous recombination deficient tumors to cause apoptotic cell death [58], [59], [60]. Thus, ferroptotic cell death might be a less important process compared with apoptosis through which platinum kills cancer cells in patients with family history. In contrast, our data suggested the ACSL4high/GPX4low group achieved pCR more easily than others among family history-negative women. Thus, it might implicate ferroptosis as the potentially main pattern of platinum-induced cell death in breast cancer without family history.
Our study showed that ACSL4 and/or GPX4 expression could be independently prognostic for breast cancer treated with neoadjuvant chemotherapy, especially in HorR-positive subgroup. It suggested that pCR benefits with high ACSL4 expression could successfully translate into survival benefits. On the other hand, as a biomarker inversely correlated to Ki-67, GPX4 might prevent pCR and improve prognosis by inhibiting cell proliferation [61,62]. Our results were supported by previous evidence by Chen et al., that patients with higher ACSL4 expression achieved superior OS in the PrognoScan breast cancer cohort [63]. Besides, the prognosis analyses from Kaplan-Meier Plotter datasets were in line with ours. The OS in patients with higher ACSL4 expression was superior to that in the low-expression patients. Patients with higher GPX4 expression obtained longer DMFS than those with lower GPX4 expression. And the subgroup results of ER-positive women were also consistent with ours. Notably, our study is the first to focus on the prognostic values of ACSL4 and GPX4 in patients that receive neoadjuvant chemotherapy. Prolonged follow-up period is necessary in future study.
There are still limitations in this study. First, the sample size was relatively small. Nevertheless, since this is an exploratory study of the prospective trials, it provides intrinsic rules which require validation with enlarged sample size. Second, subgroup analysis was not mature in TNBC. It signifies the necessity to expand the sample size as well. Third, our follow-up time was too limited to perform analysis of OS. However, public databases were utilized to analyze OS with ten-year follow-up period. Prolonged follow-up time are underway for our cohort. In addition, this study focused on two of the ferroptosis-associated genes to evaluate their predictive value of patients’ response to neoadjuvant chemotherapy as well as their prognostic value in the neoadjuvant setting. Actually, ferroptosis is a newly discovered form of programmed cell death regulated by multiple genes. The clinical significance of the remaining genes is still to be explored.
In conclusion, our study for the first time reported that the expression of ferroptosis-related genes ACSL4 and GPX4 could serve as novel predictive and prognostic biomarkers for neoadjuvant chemotherapy for patients with breast cancer. It might help screen candidate responders and determine chemotherapy strategies. Basic research is required to elucidate the mechanism of ferroptosis affecting chemosensitivity.
Contributors
JS Lu and WJ Yin designed and conducted the study. YH Wang, YP Lin, SG Xu, LH Zhou, J Zhang, and WJ Yin collected the clinical data. R Sha, CW Yuan, ZP Wu, and J Peng carried out immunochemistry. YQ Xu performed the in vitro experiments. YQ Xu and XN Sheng performed data analysis. YQ Xu and R Sha drafted the manuscript. WJ Yin and YH Wang verified the underlying data. JS Lu revised the manuscript. All authors have read and approved the final manuscript.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgments
Acknowledgments
We would like to thank all the investigators and patients who participated in the present study. The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Shanghai Natural Science Foundation [Grant No. 19ZR1431100], Clinical Research Plan of Shanghai Hospital Development Center [Grant No. SHDC2020CR3003A, 16CR3065B, and 12016231], Shanghai “Rising Stars of Medical Talent” Youth Development Program for Youth Medical Talents - Specialist Program [Grant No. 2018-15], Shanghai “Rising Stars of Medical Talent” Youth Development Program for Outstanding Youth Medical Talents [Grant No. 2018-16], Shanghai Collaborative Innovation Center for Translational Medicine [Grant No. TM201908], Multidisciplinary Cross Research Foundation of Shanghai Jiao Tong University [Grant No. YG2017QN49, ZH2018QNA42, and YG2019QNA28], Nurturing Fund of Renji Hospital [Grant No. PYMDT-002, PY2018-IIC-01, PY2018-III-15, and PYIII20-09], Science and Technology Commission of Shanghai Municipality [Grant No. 20DZ2201600 and 15JC1402700], and Shanghai Municipal Key Clinical Specialty.
Data sharing statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103560.
Contributor Information
Yaohui Wang, Email: wangyaohui@renji.com.
Wenjin Yin, Email: yinwenjin@renji.com.
Jinsong Lu, Email: lujingsong@renji.com.
Appendix. Supplementary materials
References
- 1.Korde L.A., Somerfield M.R., Carey L.A., Crews J.R., Denduluri N., Hwang E.S. Neoadjuvant chemotherapy, endocrine therapy, and targeted therapy for breast cancer: ASCO guideline. J Clin Oncol. 2021;39(13):1485–1505. doi: 10.1200/JCO.20.03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortazar P., Zhang L., Untch M., Mehta K., Costantino J.P., Wolmark N. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 3.Houssami N., Macaskill P., von Minckwitz G., Marinovich M.L., Mamounas E. Meta-analysis of the association of breast cancer subtype and pathologic complete response to neoadjuvant chemotherapy. Eur J Cancer. 2012;48(18):3342–3354. doi: 10.1016/j.ejca.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 4.Cortazar P., Geyer C.E. Pathological complete response in neoadjuvant treatment of breast cancer. Ann Surg Oncol. 2015;22(5):1441–1446. doi: 10.1245/s10434-015-4404-8. [DOI] [PubMed] [Google Scholar]
- 5.Rastogi P., Anderson S.J., Bear H.D., Geyer C.E., Kahlenberg M.S., Robidoux A. Preoperative chemotherapy: updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J Clin Oncol. 2008;26(5):778–785. doi: 10.1200/JCO.2007.15.0235. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y., Yu C., Luo M., Cen C., Qiu J., Zhang S. Ferroptosis in cancer treatment: another way to Rome. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.571127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1-2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seibt T.M., Proneth B., Conrad M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic Biol Med. 2019;133:144–152. doi: 10.1016/j.freeradbiomed.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hangauer M.J., Viswanathan V.S., Ryan M.J., Bole D., Eaton J.K., Matov A. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551(7679):247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kagan V.E., Mao G., Qu F., Angeli J.P., Doll S., Croix C.S. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol. 2017;13(1):81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W.S., Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016;26(3):165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13(1):91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan H., Li X., Zhang X., Kang R., Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun. 2016;478(3):1338–1343. doi: 10.1016/j.bbrc.2016.08.124. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X., Sui S., Wang L., Li H., Zhang L., Xu S. Inhibition of tumor propellant glutathione peroxidase 4 induces ferroptosis in cancer cells and enhances anticancer effect of cisplatin. J Cell Physiol. 2020;235(4):3425–3437. doi: 10.1002/jcp.29232. [DOI] [PubMed] [Google Scholar]
- 16.Guo J., Xu B., Han Q., Zhou H., Xia Y., Gong C. Ferroptosis: a novel anti-tumor action for Cisplatin. Cancer Res Treat. 2018;50(2):445–460. doi: 10.4143/crt.2016.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv C., Qu H., Zhu W., Xu K., Xu A., Jia B. Low-dose paclitaxel inhibits tumor cell growth by Regulating Glutaminolysis in colorectal carcinoma cells. Front Pharmacol. 2017;8:244. doi: 10.3389/fphar.2017.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giannakakou P., Robey R., Fojo T., Blagosklonny MV. Low concentrations of paclitaxel induce cell type-dependent p53, p21 and G1/G2 arrest instead of mitotic arrest: molecular determinants of paclitaxel-induced cytotoxicity. Oncogene. 2001;20(29):3806–3813. doi: 10.1038/sj.onc.1204487. [DOI] [PubMed] [Google Scholar]
- 19.Ma S., Henson E.S., Chen Y., Gibson SB. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016;7(7):e2307. doi: 10.1038/cddis.2016.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma S., Dielschneider R.F., Henson E.S., Xiao W., Choquette T.R., Blankstein A.R. Ferroptosis and autophagy induced cell death occur independently after siramesine and lapatinib treatment in breast cancer cells. PLoS One. 2017;12(8) doi: 10.1371/journal.pone.0182921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou L., Xu S., Yin W., Lin Y., Du Y., Jiang Y. Weekly paclitaxel and cisplatin as neoadjuvant chemotherapy with locally advanced breast cancer: a prospective, single arm, phase II study. Oncotarget. 2017;8(45):79305–79314. doi: 10.18632/oncotarget.17954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bethea T.N., Rosenberg L., Castro-Webb N., Lunetta K.L., Sucheston-Campbell L.E., Ruiz-Narvaez E.A. Family history of cancer in relation to breast cancer subtypes in African American Women. Cancer Epidemiol Biomark Prev. 2016;25(2):366–373. doi: 10.1158/1055-9965.EPI-15-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff A.C., Hammond M.E., Hicks D.G., Dowsett M., McShane L.M., Allison K.H. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 25.Curigliano G., Burstein H.J., Winer E.P., Gnant M., Dubsky P., Loibl S. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. 2017;28(8):1700–1712. doi: 10.1093/annonc/mdx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodenhuis S., Mandjes I.A.M., Wesseling J., van de Vijver M.J., Peeters M., Sonke G.S. A simple system for grading the response of breast cancer to neoadjuvant chemotherapy. Ann Oncol. 2010;21(3):481–487. doi: 10.1093/annonc/mdp348. [DOI] [PubMed] [Google Scholar]
- 27.Ye Z., Hu Q., Zhuo Q., Zhu Y., Fan G., Liu M. Abrogation of ARF6 promotes RSL3-induced ferroptosis and mitigates gemcitabine resistance in pancreatic cancer cells. Am J Cancer Res. 2020;10(4):1182–1193. [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J., Ding C., Chen Y., Hu W., Lu Y., Wu W. ACSL4 promotes hepatocellular carcinoma progression via c-Myc stability mediated by ERK/FBW7/c-Myc axis. Oncogenesis. 2020;9(4):42. doi: 10.1038/s41389-020-0226-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen G.Q., Benthani F.A., Wu J., Liang D., Bian Z.X., Jiang X. Artemisinin compounds sensitize cancer cells to ferroptosis by regulating iron homeostasis. Cell Death Differ. 2020;27(1):242–254. doi: 10.1038/s41418-019-0352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayr L., Grabherr F., Schwärzler J., Reitmeier I., Sommer F., Gehmacher T. Dietary lipids fuel GPX4-restricted enteritis resembling Crohn's disease. Nat Commun. 2020;11(1):1775. doi: 10.1038/s41467-020-15646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X., Yang J., Ye Y., Chen G., Zhang Y., Wu H. SPTBN1 prevents primary osteoporosis by modulating osteoblasts proliferation and differentiation and blood vessels formation in bone. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.653724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Cancer Genome Atlas Research Network. https://www.cancer.gov/tcga (accessed on 31th December 2019).
- 33.Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 35.Huang da W., Sherman B.T., Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W., Sherman B.T., Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. D607-d13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuster J.J. Median follow-up in clinical trials. J Clin Oncol. 1991;9(1):191–192. doi: 10.1200/JCO.1991.9.1.191. [DOI] [PubMed] [Google Scholar]
- 41.Altman D.G., De Stavola B.L., Love S.B., Stepniewska K.A. Review of survival analyses published in cancer journals. Br J Cancer. 1995;72(2):511–518. doi: 10.1038/bjc.1995.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rusolo F., Capone F., Pasquale R., Angiolillo A., Colonna G., Castello G. Comparison of the seleno-transcriptome expression between human non-cancerous mammary epithelial cells and two human breast cancer cell lines. Oncol Lett. 2017;13(4):2411–2417. doi: 10.3892/ol.2017.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cejas P., Garcia-Cabezas M.A., Casado E., Belda-Iniesta C., De Castro J., Fresno J.A. Phospholipid hydroperoxide glutathione peroxidase (PHGPx) expression is downregulated in poorly differentiated breast invasive ductal carcinoma. Free Radic Res. 2007;41(6):681–687. doi: 10.1080/10715760701286167. [DOI] [PubMed] [Google Scholar]
- 44.Wu X., Li Y., Wang J., Wen X., Marcus M.T., Daniels G. Long chain fatty Acyl-CoA synthetase 4 is a biomarker for and mediator of hormone resistance in human breast cancer. PLoS One. 2013;8(10):e77060. doi: 10.1371/journal.pone.0077060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monaco M.E., Creighton C.J., Lee P., Zou X., Topham M.K., Stafforini DM. Expression of long-chain fatty Acyl-CoA Synthetase 4 in Breast and prostate cancers is associated with sex steroid hormone receptor negativity. Transl Oncol. 2010;3(2):91–98. doi: 10.1593/tlo.09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yen M.C., Kan J.Y., Hsieh C.J., Kuo P.L., Hou M.F., Hsu YL. Association of long-chain acyl-coenzyme A synthetase 5 expression in human breast cancer by estrogen receptor status and its clinical significance. Oncol Rep. 2017;37(6):3253–3260. doi: 10.3892/or.2017.5610. [DOI] [PubMed] [Google Scholar]
- 47.Dinarvand N., Khanahmad H., Hakimian S.M., Sheikhi A., Rashidi B., Pourfarzam M. Evaluation of long-chain acyl-coenzyme A synthetase 4 (ACSL4) expression in human breast cancer. Res Pharm Sci. 2020;15(1):48–56. doi: 10.4103/1735-5362.278714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castillo A.F., Orlando U.D., Maloberti P.M., Prada J.G., Dattilo M.A., Solano A.R. New inhibitor targeting Acyl-CoA synthetase 4 reduces breast and prostate tumor growth, therapeutic resistance and steroidogenesis. Cell Mol Life Sci. 2021;78(6):2893–2910. doi: 10.1007/s00018-020-03679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maloberti P.M., Duarte A.B., Orlando U.D., Pasqualini M.E., Solano A.R., Lopez-Otin C. Functional interaction between acyl-CoA synthetase 4, lipooxygenases and cyclooxygenase-2 in the aggressive phenotype of breast cancer cells. PLoS One. 2010;5(11):e15540. doi: 10.1371/journal.pone.0015540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang W.J., Sun Y.F., Jin A.L., Lv L.H., Zhu J., Wang B.L. BCL11B suppresses tumor progression and stem cell traits in hepatocellular carcinoma by restoring p53 signaling activity. Cell Death Dis. 2020;11(10):895. doi: 10.1038/s41419-020-03115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao D., Wang X., Zhang W. GDF15 predict platinum response during first-line chemotherapy and can act as a complementary diagnostic serum biomarker with CA125 in epithelial ovarian cancer. BMC Cancer. 2018;18(1):328. doi: 10.1186/s12885-018-4246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orlando U.D., Castillo A.F., Medrano M.A.R., Solano A.R., Maloberti P.M., Podesta E.J. Acyl-CoA synthetase-4 is implicated in drug resistance in breast cancer cell lines involving the regulation of energy-dependent transporter expression. Biochem Pharm. 2019;159:52–63. doi: 10.1016/j.bcp.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Edlich F. BCL-2 proteins and apoptosis: recent insights and unknowns. Biochem Biophys Res Commun. 2018;500(1):26–34. doi: 10.1016/j.bbrc.2017.06.190. [DOI] [PubMed] [Google Scholar]
- 54.Rogerio F., Jordao H., Vieira A.S., Maria C.C., Santos de Rezende A.C., Pereira G.A. Bax and Bcl-2 expression and TUNEL labeling in lumbar enlargement of neonatal rats after sciatic axotomy and melatonin treatment. Brain Res. 2006;1112(1):80–90. doi: 10.1016/j.brainres.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 55.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson D., Easton D. The genetic epidemiology of breast cancer genes. J Mammary Gland Biol Neoplas. 2004;9(3):221–236. doi: 10.1023/B:JOMG.0000048770.90334.3b. [DOI] [PubMed] [Google Scholar]
- 57.Martin A.M., Blackwood M.A., Antin-Ozerkis D., Shih H.A., Calzone K., Colligon T.A. Germline mutations in BRCA1 and BRCA2 in breast-ovarian families from a breast cancer risk evaluation clinic. J Clin Oncol. 2001;19(8):2247–2253. doi: 10.1200/JCO.2001.19.8.2247. [DOI] [PubMed] [Google Scholar]
- 58.Jamieson E.R., Lippard S.J. Structure, recognition, and processing of cisplatin-DNA adducts. Chem Reviews. 1999;99(9):2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 59.Heeke A.L., Pishvaian M.J., Lynce F., Xiu J., Brody J.R., Chen W.J. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol. 2018;2018 doi: 10.1200/PO.17.00286. PO.17.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi Y., Zhou F., Jiang F., Lu H., Wang J., Cheng C. PARP inhibitor reduces proliferation and increases apoptosis in breast cancer cells. Chin J Cancer Res. 2014;26(2):142–147. doi: 10.3978/j.issn.1000-9604.2014.02.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 62.Luporsi E., Andre F., Spyratos F., Martin P.M., Jacquemier J., Penault-Llorca F. Ki-67: level of evidence and methodological considerations for its role in the clinical management of breast cancer: analytical and critical review. Breast Cancer Res Treat. 2012;132(3):895–915. doi: 10.1007/s10549-011-1837-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen W.C., Wang C.Y., Hung Y.H., Weng T.Y., Yen M.C., Lai M.D. Systematic analysis of gene expression alterations and clinical outcomes for long-chain acyl-coenzyme a synthetase family in cancer. PloS One. 2016;11(5) doi: 10.1371/journal.pone.0155660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.