Abstract
It is well established that memory CD8 T cells protect susceptible strains of mice from mousepox, a lethal viral disease caused by ectromelia virus (ECTV), the murine counterpart to human variola virus. While mRNA vaccines induce protective antibody (Ab) responses, it is unknown whether they also induce protective memory CD8 T cells. We now show that immunization with different doses of unmodified or N(1)-methylpseudouridine-modified mRNA (modified mRNA) in lipid nanoparticles (LNP) encoding the ECTV gene EVM158 induced similarly strong CD8 T cell responses to the epitope TSYKFESV, albeit unmodified mRNA-LNP had adverse effects at the inoculation site. A single immunization with 10 μg modified mRNA-LNP protected most susceptible mice from mousepox, and booster vaccination increased the memory CD8 T cell pool, providing full protection. Moreover, modified mRNA-LNP encoding TSYKFESV appended to green fluorescent protein (GFP) protected against wild-type ECTV infection while lymphocytic choriomeningitis virus glycoprotein (GP) modified mRNA-LNP protected against ECTV expressing GP epitopes. Thus, modified mRNA-LNP can be used to create protective CD8 T cell-based vaccines against viral infections.
Keywords: mRNA vaccine, modified mRNA, lipid nanoparticle, virus, CD8 T cells, poxvirus
Graphical abstract
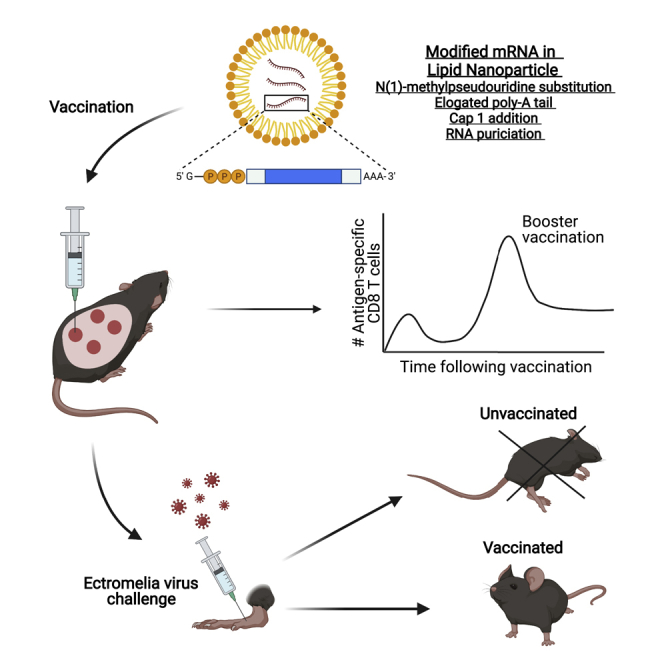
It is known that lipid nanoparticle (LNP)-encapsulated mRNA vaccines induce protective anti-viral antibodies, but whether they can also induce protective memory CD8 T cells is unknown. Sigal and colleagues show that mRNA-LNP vaccines induce memory CD8 T cells that protect from mousepox, a highly lethal viral disease.
Introduction
CD8 T cells play a major role in reducing the pathogen burden by controlling viral infection. CD8 T cells can also complement the antibody (Ab) response by targeting antigens that are not exposed on the surface of pathogens and directly eliminate infected or cancerous cells. While most vaccines are evaluated by their capacity to elicit antibody responses, Abs may not be sufficient or effective for protection. In some cases, such as dengue virus, Abs can also be detrimental, resulting in antibody-dependent enhancement.1 Furthermore, for many pathogens, the dominant Ab epitopes are highly mutable, leaving them ineffective against new strains.2, 3, 4 In principle, the best vaccines should induce both protective Abs and CD8 T cells.
mRNA encapsulated in lipid nanoparticles (LNP) can potentially be used to produce novel CD8 T cell vaccines. Compared to traditional vaccine platforms, immunization with mRNA has multiple advantages, including rapid translation of the gene sequence upon cell entry, no integration into the genome, lack of immunity to an extraneous component of the vaccine, and simple vaccine design and manufacture.5, 6, 7 N(1)-methylpseudouridine modification to mRNA (herein modified mRNA) followed by liquid chromatography purification has been shown to improve mRNA stability and translation.8, 9, 10 Encapsulation of the mRNA into LNP enhances host cell uptake and efficient antigen expression in vivo.11 These recent improvements to RNA-based vaccine formulation and delivery systems have drastically increased the potential of mRNA vaccines for the prevention of infectious diseases.9
Previous studies have demonstrated that modified mRNA delivered by LNP (mRNA-LNP) induces potent and often protective immune responses to a number of pathogens in animal models, including Zika virus,12,13 HIV-1,14 herpes simplex virus (HSV),15,16 Ebola virus,17 influenza virus,18, 19, 20 and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).21, 22, 23 Modified mRNA-LNP has also shown promise in two phase I human clinical trials for influenza virus (NCT03076385 and NCT03345043) and induced higher than 90% protection in recent phase III trials for SARS-CoV-2 (NCT04368728).20,24,25 Indeed, modified mRNA-LNP is currently being used as the main vaccines to combat COVID19 in the United States as well as other countries. While Abs induced by mRNA vaccination are thought to be the major source of protection, CD8 T cell responses have been identified in multiple vaccination studies using mRNA.19,21,23,26,27 However, the role of these CD8 T cells in vaccine-mediated protection is unclear.
We have previously shown that memory CD8 T cells induced by peptide-dendritic cell (DC) immunization protected mice against lethal ectromelia virus (ECTV) challenge.28 Here we used the ECTV model to demonstrate that modified mRNA vaccines encapsulated in LNP induce memory CD8 T cell responses that protect susceptible mice from lethal ECTV infection. Our results indicate that modified mRNA-LNP is an excellent platform to produce anti-viral vaccines that necessitate memory CD8 T cells to induce protection.
Results
Nucleoside-modified mRNA-LNP vaccination elicits virus-specific CD8 T cell responses without skin pathology
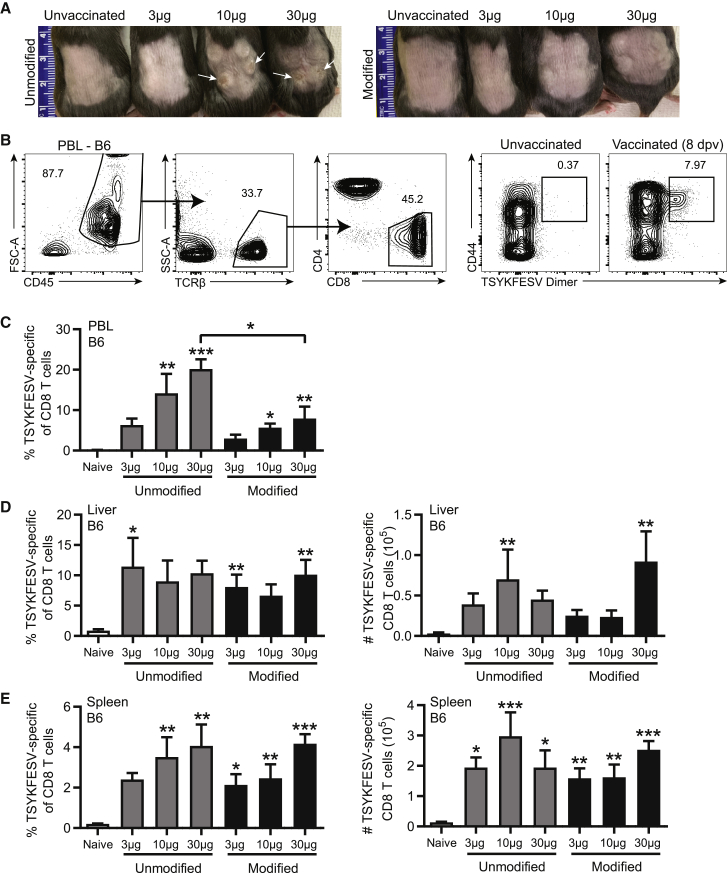
To compare the efficacy of unmodified and modified double-stranded RNA (dsRNA)-depleted mRNA-LNP vaccines at eliciting CD8 T cell responses, we immunized C57BL/6 (B6) mice with 3, 10, or 30 μg of dsRNA-depleted, unmodified or modified mRNA encapsulated in LNP composed of a cationic lipid (ALC0307), phosphatidylcholine, cholesterol, and polyethylene glycol (PEG)-lipid (APCP LNP, herein LNP).29 The encapsulated mRNA corresponded to the ECTV immune evasion gene EVM158, which encodes a secreted interferon (IFN)-γ decoy receptor that harbors the CD8 T cell immunodominant epitope TSYKFESV. This murine CD8 T cell epitope binds to the mouse MHC-I H-2Kb (Kb) and is shared by the poxviruses vaccinia virus (VACV) and ECTV.30,31 Mice immunized with unmodified mRNA at 10 or 30 μg suffered skin inflammation and wounds at 8 days post-vaccination (dpv) (Figure 1A). Immunization with modified mRNA did not result in any adverse effects at 3 or 10 μg. The 30 μg dose resulted in slight inflammation without any wounds, which resolved within several days (Figure 1A). At 8 dpv, mice immunized with 10 or 30 μg unmodified or modified mRNA had high frequencies of Kb-TSYKFESV+ CD8 T cells within the peripheral blood leukocytes (PBL) as determined with Kb-TSYKFESV dimers (DimerX, BD Biosciences) (Figures 1B and 1C). With the 30 μg dose, unmodified mRNA elicited a greater frequency of TSYKFESV-specific CD8 T cells in PBL than the modified version, as determined by ANOVA (Figure 1C). However, unmodified and modified mRNA induced comparable frequencies and total numbers of Kb-TSYKFESV+ CD8 T cells in the liver and spleen (Figures 1D and 1E), two common sites of viral replication following ECTV infection. Interestingly, mRNA dose had a relatively minor impact on the magnitude of the CD8 T cell response in the liver and spleen (Figures 1D and 1E).
Figure 1.
Nucleoside-modified mRNA-LNP vaccination elicits virus-specific CD8 T cell responses without skin pathology
(A–E) B6 mice were immunized i.d. with 3, 10, or 30 μg of unmodified or modified mRNA-LNP encoding EVM158. TSYKFESV-specific CD8 T cells were determined by Kb-TSYKFESV dimer staining. (A) Image of hind back of mice vaccinated with unmodified (top) or modified (bottom) mRNA-LNP at 8 dpv. White arrows indicate areas of lesions. Ruler units are in centimeters. (B) Representative flow plot of gating strategy to identify TSYKFESV-specific CD8 T cells using DimerX complexes. (C) Frequency of TSYKFESV-specific of CD8 T cells in PBL. (D and E) Frequency and total number of TSYKFESV-specific CD8 T cells in (D) liver and (E) spleen at 8 dpv. (C–E) Groups were compared to each other by one-way ANOVA analysis with Tukey’s post-tests. All statistical differences indicated are compared to unvaccinated group unless otherwise designated; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (C–E n = 6).
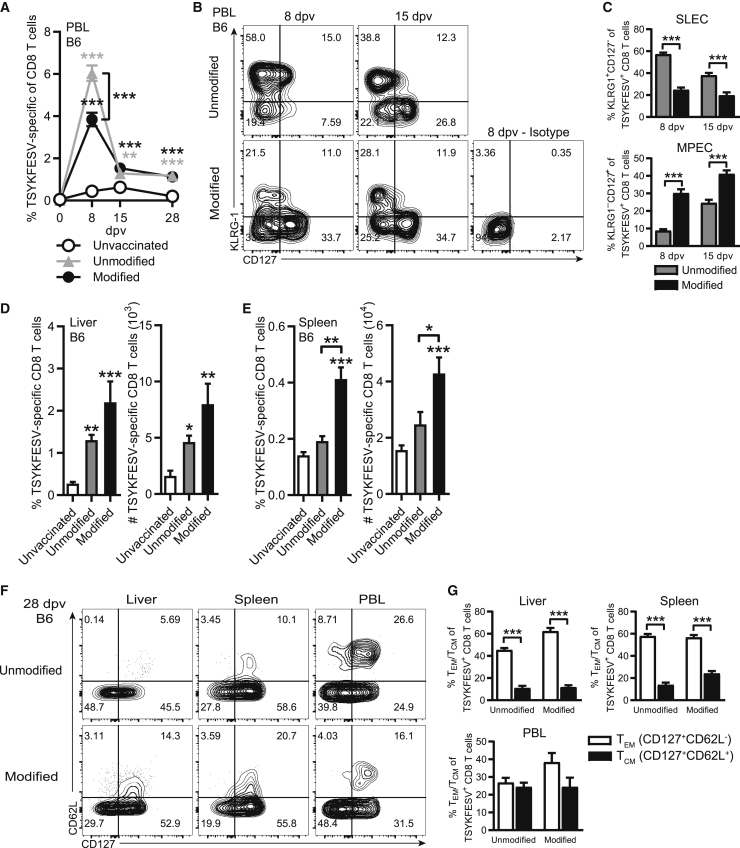
Modified mRNA-LNP induces greater memory precursor effector cell (MPEC) formation and primarily effector memory CD8 T cells
We next analyzed the TSYKFESV-specific response in the blood over a longer period (Figure 2A). For this we used the 10 μg dose, as it was effective, and because the unmodified form had adverse effects. At 8 dpv, the frequency of TSYKFESV-specific CD8 T cells in PBL was significantly higher in B6 mice immunized with unmodified as compared to modified mRNA. However, by 28 dpv, the frequency of TSYKFESV-specific CD8 T cells was similar in the unmodified and modified mRNA groups. These results suggested that modified mRNA vaccination may induce higher frequencies of CD8 T MPECs. In support, modified mRNA induced a greater frequency of TSYKFESV-specific CD8 T cell MPECs based on phenotypic marker expression (CD127+KLRG-1−)32,33 (Figures 2B and 2C), while unmodified mRNA elicited a greater proportion of short-lived effector cells (SLEC; CD127−KLRG-1+) (Figures 2B and 2C) in the PBL at 8 and 15 dpv. The frequency and total numbers of TSYKFESV-specific CD8 T cells in the liver were significantly increased over the unvaccinated control group at 28 dpv (Figure 2D), and they were significantly higher in the spleen of the modified mRNA group compared to the unmodified mRNA group (Figure 2E). With both vaccines, the majority of TSYKFESV-specific CD8 T cells in the liver and spleen were CD127+CD62L− at 28 dpv, indicating that they had become effector memory CD8 T cells34,35 (TEM; Figures 2F and 2G). Taken together, these data indicate that both unmodified and modified mRNA-LNP induce strong CD8 T cell responses. However, modified mRNA is superior because it does not cause any visible adverse effects at the inoculation site with 3 and 10 μg of mRNA-LNP while inducing greater frequencies of MPECs with 10 μg of mRNA-LNP. Based on these results, we used a 10 μg dosage of modified mRNA-LNP in all subsequent experiments.
Figure 2.
Modified mRNA-LNP induces greater MPEC formation and primarily effector memory CD8 T cells
(A) Percentage of TSYKFESV-specific CD8 T cells in PBL over time following immunization of B6 mice with 10 μg mRNA-LNP. Statistical differences are compared to unvaccinated group unless otherwise designated. (B) Concatenated flow plot of KLRG-1 by CD127 staining of TSYKFESV-specific CD8 T cells at 8 and 15 dpv in the PBL of B6 mice following immunization with unmodified or modified mRNA-LNP. (C) Frequency of short-lived effector cells (SLEC; KLRG-1+CD127−) or memory precursor (MPEC; KLRG-1−CD127+) TSYKFESV-specific CD8 T cells in the PBL at 8 and 15 dpv of B6 mice immunized with unmodified or modified mRNA-LNP. (D and E) Frequency and total number of TSYKFESV-specific CD8 T cells in the (D) liver and (E) spleen at 28 dpv in B6 mice. (F) Concatenated flow plots of CD62L by CD127 staining on TSYKFESV-specific CD8 T cells at 28 dpv in B6 mice. (G) Frequency of T effector memory (TEM; CD127+CD62L-) or T central memory (TCM; CD127+CD62L+) TSYKFESV-specific CD8 T cells at 28 dpv in B6 mice. (C and G) Groups were compared using t tests, and (A), (D), and (E) were compared using one-way ANOVA; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (A, D, and E, n = 9; C and G, n = 10).
mRNA-LNP vaccination protects susceptible strains of mice against lethal ECTV challenge
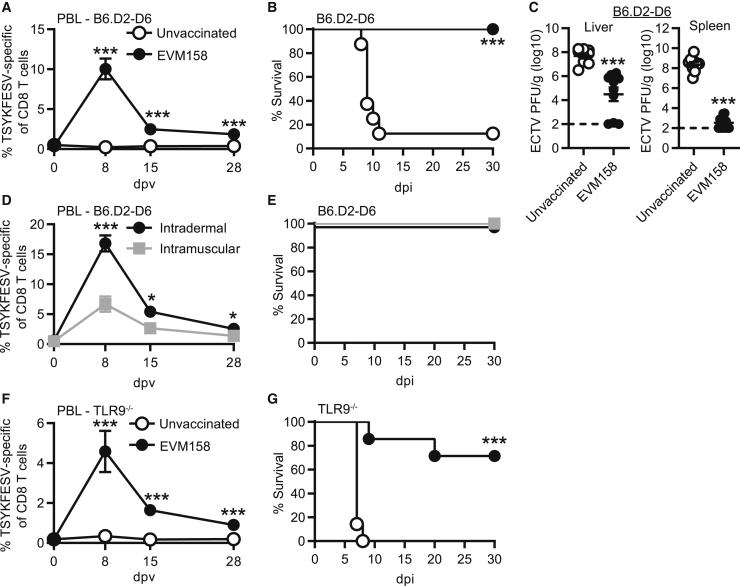
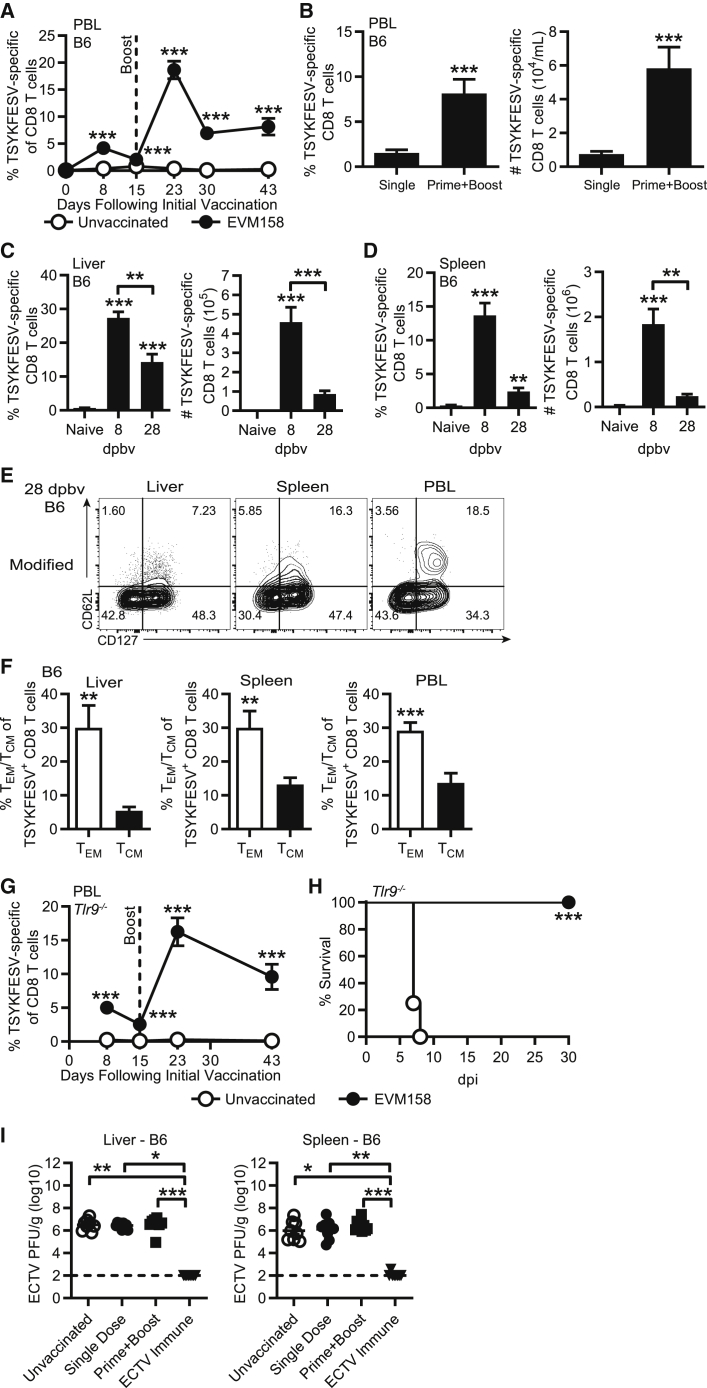
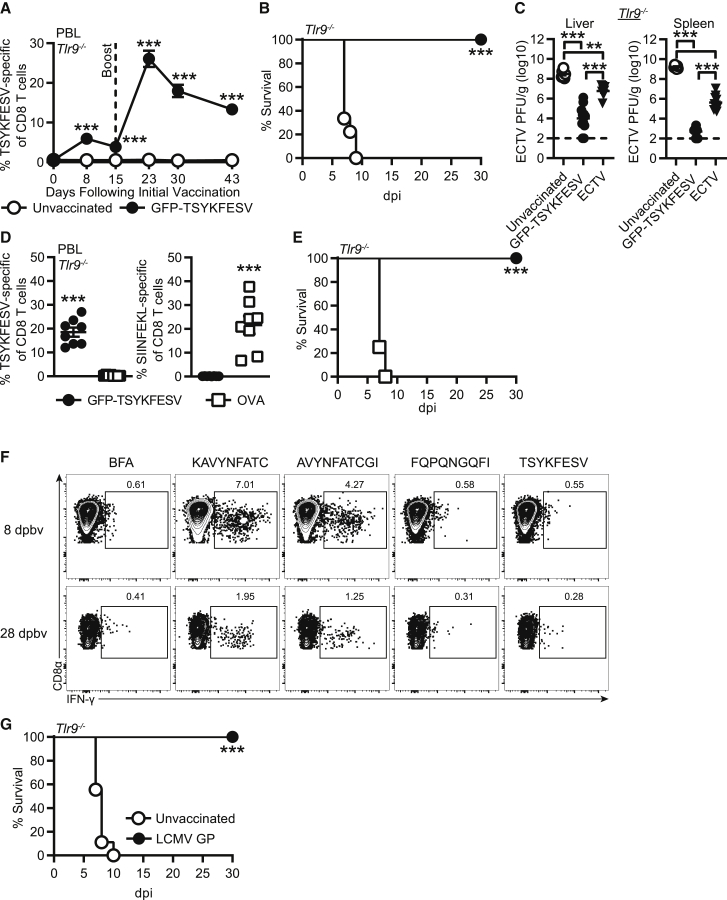
We next sought to determine whether the memory CD8 T cell responses induced with modified EVM158 mRNA-LNP vaccine were sufficient to protect mice from lethal ECTV infection. While B6 mice are naturally resistant to lethal mousepox, other strains of mice, such as B6.D2-(D6Mit149-D6Mit15)/LusJ (B6.D2-D6), are highly susceptible.36 B6.D2-D6 mice are a B6 congenic strain carrying the distal portion of chromosome 6 from DBA/2J mice.37 This chromosomal fragment makes B6.D2-D6 susceptible to lethal mousepox due to an impaired natural killer (NK) cell response.36 Immunization of B6.D2-D6 mice with 10 μg of modified EVM158 mRNA induced a TSYKFESV-specific response that settled to 1%–2% of CD8 T cells at 28 dpv in the PBL (Figure 3A). While ECTV challenge of unvaccinated B6.D2-D6 mice resulted in 88% mortality rate, all immunized mice were protected and survived infection (Figure 3B). Furthermore, virus titers were significantly reduced in the liver and spleen at 7 days post-infection (dpi) (Figure 3C). This suggests that the CD8 T cell response induced by mRNA vaccination protected B6.D2-D6 mice from lethal ECTV infection by promoting viral clearance.
Figure 3.
mRNA-LNP vaccination protects susceptible strains of mice against lethal ECTV challenge
(A–C) B6.D2-D6 mice were immunized i.d. with 10 μg modified mRNA-LNP encoding EVM158. TSYKFESV-specific CD8 T cells were identified by Kb-TSYKFESV dimer staining. (A) Frequency of TSYKFESV-specific CD8 T cells in PBL following mRNA-LNP vaccination. (B) Survival curve of B6.D2-D6 mice following ECTV infection. (C) Virus titers of immunized B6.D2-D6 mice by plaque assay in the liver and spleen at 7 dpi. (D) Percentage of TSYKFESV-specific CD8 T cells in PBL of B6.D2-D6 mice following either i.d. or i.m. immunization with modified mRNA-LNP. (E) Survival curve for B6.D2-D6 mice immunized with mRNA-LNP i.d. or i.m. (F) Frequency of TSYKFESV-specific CD8 T cells in PBL following modified mRNA vaccination of TLR9-deficient mice. (G) Survival curve of immunized TLR9-deficient mice. (A, C, D, and F) Groups were compared by t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (A and B, n = 8–9; C, n = 9–10; D and E, n = 7–8; F and G, n = 7).
We also investigated whether changing the route of immunization to intramuscular (i.m.) altered vaccine efficacy. B6.D2-D6 mice immunized i.m. had a significantly lower frequency of TSYKFESV-specific CD8 T cells in the PBL compared to those immunized intradermally (i.d.) (Figure 3D). However, both routes of immunization induced responses that were sufficient to protect B6.D2-D6 mice from mousepox (Figure 3E). In addition, we evaluated the protective capacity of modified mRNA-LNP vaccination in mice deficient in the DNA CpG sensor TLR9 (Tlr9−/−). ECTV is a DNA poxvirus to which Tlr9−/− mice are highly susceptible.38,39 However, absence of TLR9 should not impact CD8 T cell responses induced by mRNA-LNP vaccination. Modified mRNA-LNP immunization induced memory TSYKFESV-specific CD8 T cell responses in the PBL of Tlr9−/− mice (Figure 3F) that were comparable to what was observed with B6 and B6.D2-D6 mice in previous figures. In contrast to unvaccinated B6.D2-D6 mice, which mostly died between 9–11 dpi (Figure 3B), all unvaccinated Tlr9−/− mice succumbed to ECTV infection at 7–8 dpi (Figure 3G). On the other hand, only 30% of the immunized Tlr9−/− mice died, indicating relatively strong but still incomplete protection (Figure 3G). These results suggested that compared to B6.D2-D6 mice, the number of memory CD8 T cells induced in Tlr9−/− mice by a single immunization may not be sufficient to fully protect this highly susceptible strain.
Prime-boost mRNA-LNP immunization increases the magnitude of the memory CD8 T cell pool and protects Tlr9−/−, but sera from immunized mice are not protective
We next tested whether a booster immunization with mRNA-LNP at 15 dpv could increase the number of TSYKFESV-specific CD8 T cells as we previously demonstrated for DC-peptide immunization.28 This boosting regimen strongly increased the magnitude of the TSYKFESV-specific CD8 T cell response in the PBL of B6 mice at 23, 30, and 43 dpv (8, 15, and 28 days post booster vaccination [dpbv]) (Figure 4A). The prime-boost immunization regimen induced a greater frequency and number of TSYKFESV-specific CD8 T cells in the PBL at 28 dpbv as compared to a single immunization at 28 dpv (Figure 4B). In addition, prime-boost immunization induced high frequencies and total numbers of TSYKFESV-specific CD8 T cells in the liver (Figure 4C) and spleen (Figure 4D) at 8 and 28 dpbv. The frequency and number of TSYKFESV-specific CD8 T cells present in the liver and after prime-boost were higher than those observed after single immunization (compare Figures 4C and 4D with 2D and 2E). Similar to a single immunization, TSYKFESV-specific CD8 T cells at 28 dpbv in the PBL, liver, and spleen were largely effector memory cells (TEM) (Figures 4E and 4F).
Figure 4.
Prime-boost mRNA-LNP immunization increases magnitude of CD8 T cell response and improves survival in TLR9-deficient mice, but sera from immunized mice are not protective
(A–D) Previously immunized B6 mice were i.d. vaccinated with 10 μg modified mRNA-LNP 15 days following the first immunization. (A) Frequency of TSYKFESV-specific CD8 T cells in the PBL following prime-boost immunization. (B) Frequency and number of TSYKFESV-specific CD8 T cells in the PBL 28 days following a single or booster immunization. (C and D) Percentage and total number of TSYKFESV-specific CD8 T cells in the (C) liver and (D) spleen at 8 and 28 dpbv. Statistical differences are compared to naive mice unless otherwise designated. (E) Concatenated flow plots of CD62L and CD127 on TSYKFESV-specific CD8 T cells in the PBL of B6 mice following prime-boost immunization at 28 dpbv. (F) Percent TEM or T central memory (TCM) of TSYKFESV-specific CD8 T cells in the PBL of B6 mice at 28 dpbv following prime-boost immunization with modified mRNA-LNP. (G) Frequency of TSYKFESV-specific CD8 T cells following prime-boost immunization in Tlr9−/− mice. (H) Survival curve in prime-boost-vaccinated Tlr9−/− mice. (I) WT B6 mice received 200 μL of serum from unvaccinated, EVM158-modified mRNA-LNP-immunized, or previously ECTV-infected B6 mice 1 day prior to ECTV challenge. Viral titers of liver and spleens from ECTV-infected, serum-recipient mice at 5 dpi. Groups indicate source of sera. (A, B, F, and G) Groups were compared using t tests; data in (C), (D), and (I) were compared using one-way ANOVA; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗0.001 (A–D, n = 9; E and F, n = 9; G–I, n = 8–9).
Given the increased CD8 T cell responses following booster vaccination in B6 mice, we next evaluated prime-boost immunization in Tlr9−/− mice. Like B6 mice, approximately 10% of the CD8 T cells in the PBL of Tlr9−/− mice receiving a booster immunization were TSYKFESV-specific at 28 dpbv (43 dpv) (Figure 4G). While the unvaccinated Tlr9−/− mice succumbed to ECTV infection, all mice receiving prime-boost mRNA vaccination were protected upon challenge (Figure 4H). These data indicate that prime-boost immunization with mRNA-LNP vaccines can be used to increase the magnitude of the CD8 T cell response and achieve a higher level of protection.
Since EVM158 encodes a non-structural, secreted IFN-γ decoy receptor that is not required for ECTV pathogenesis,40 the data above suggested that the mechanism of protection induced by EVM158 mRNA-LNP vaccination was due to CD8 T cells and not Abs. However, this needed to be ruled out, as we assumed anti-EVM158 Abs were generated given the efficacy of mRNA-LNP at inducing Ab responses.9 Compared to sera from unvaccinated mice, passive immunization of B6 mice with immune sera obtained from mice at 28 dpv or dpbv with EVM158 mRNA-LNP did not reduce virus loads in the liver or spleen at 5 dpi (Figure 4I). In contrast, sera from mice that had recovered from ECTV infection significantly reduced viral titers in both organs of recipient mice at 5 dpi (Figure 4I). Thus, even if anti-EVM158 protein Abs were present in EVM158 mRNA-LNP-vaccinated mice, they were insufficient to protect mice from ECTV infection.
CD8 T cell response induced by modified mRNA-LNP vaccination protects mice against ECTV infection
While there are no known murine CD4 epitopes against the EVM158 protein, it was important to further confirm that the protection induced by mRNA-LNP vaccination was mediated by anti-TSYKFESV memory CD8 T cells and independent of memory CD4 T cells or Abs. To this end, we produced a modified mRNA-LNP vaccine expressing TSYKFESV (the minimal CD8 T cell epitope) attached to the C terminus of green fluorescent protein (GFP; GFP-TSYKFESV mRNA). Approximately 13% of the CD8 T cells in the PBL of Tlr9−/− mice prime-boosted with GFP-TSYKFESV mRNA-LNP were TSYKFESV-specific at 28 dpbv (Figure 5A). Similar to immunization with mRNA encoding EVM158, vaccination with GFP-TSYKFESV protected all Tlr9−/− mice from lethal mousepox (Figure 5B). Furthermore, virus titers at 6 dpi in the livers and spleens of GFP-TSYKFESV-mRNA-vaccinated mice were significantly reduced compared to the unvaccinated group (Figure 5C). The virus titers of immunized Tlr9−/−mice were even lower than those of unvaccinated, mousepox-resistant B6 mice infected with ECTV (Figure 5C). These results indicate that mRNA encoding just the specific epitope attached to GFP can induce protective CD8 T cell memory.
Figure 5.
CD8 T cell response induced by modified mRNA-LNP vaccination protects mice against ECTV infection
(A–C) Tlr9−/− mice were immunized with 10 μg modified mRNA-LNP encoding GFP-TSYKFESV. (A) Frequency of TSYKFESV-specific CD8 T cells in the PBL of Tlr9−/− mice following immunization with GFP-TSYKFESV mRNA. (B) Survival curve of Tlr9−/−mice immunized with GFP-TSYKFESV mRNA following ECTV infection. (C) Virus titers in liver and spleen at 6 dpi in unvaccinated and GFP-TSYKFESV-immunized Tlr9−/− mice or unvaccinated B6 mice infected with ECTV. (D) Frequency of TSYKFESV or SIINFEKL-specific CD8 T cells in the PBL at 28 dpbv of Tlr9−/− mice immunized with mRNA-LNP encoding either GFP-TSYKFESV or chicken OVA. (E) Survival curve of Tlr9−/− mice immunized with GFP-TSYKFESV or OVA mRNA-LNP following ECTV infection. (F) B6 mice were immunized with mRNA-LNP encoding LCMV GP. Representative flow plot of IFN-γ+ CD8 T cells from the spleen following peptide restimulation for 5 h at 8 and 28 dpbv. (G) Survival curve of Tlr9−/− mice immunized with LCMV GP-encoding mRNA and infected by a recombinant ECTV-MEE strain expressing LCMV epitopes KAVYNFATC and AVYNFATCGI. (A and D) Comparisons were done using t tests, whereas in (C), groups were compared using one-way ANOVA; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (A and B, n = 9–10; C, n = 7–11; D and E, n = 8; G, n = 9–10).
As a control to ensure that protection was not mediated by the innate immune response or bystander CD8 T cell activity, we immunized Tlr9−/− mice with mRNA-LNP encoding chicken ovalbumin (OVA). Tlr9−/− mice immunized with GFP-TSYKFESV mounted robust TSYKFESV-specific memory CD8 T cell response and no detectable SIINFEKL-specific CD8 T cells in the PBL at 28 dpbv (Figure 5D). As anticipated, OVA mRNA-LNP immunization induced ∼21% SIINFEKL-specific memory CD8 T cells at 28 dpbv in the PBL with no TSYKFESV-specific CD8 T cells (Figure 5D). However, only Tlr9−/− mice vaccinated with GFP-TSYKFESV were protected upon ECTV challenge, whereas all OVA-immunized mice rapidly succumbed to infection (Figure 5E). This demonstrates that the virus-specific CD8 T cell memory response, and not an innate immune response induced by the mRNA-LNP vaccination, was responsible for the resistance of mRNA-LNP immunized mice to lethal mousepox.
Last, to test whether CD8 T cells to epitopes other than TSYKFESV could protect from ECTV and to further confirm that protection can be mediated by memory CD8 T cells independent of circulating Abs or memory CD4 T cells, we immunized B6 mice with a modified mRNA-LNP vaccine encoding lymphocytic choriomeningitis virus (LCMV) glycoprotein (GP). The GP contains overlapping CD8 T cell epitopes KAVYNFATC and AVYNFATCGI, which respectively bind to H-2Db (Db) and Kb. We assessed GP-specific CD8 T cell responses at 8 and 28 dpbv by determining IFN-γ production following in vitro peptide restimulation of splenocytes from LCMV-GP-immunized B6 mice. Relatively large numbers of CD8 T cells produced IFN-γ after restimulation with the GP-specific peptides KAVYNFATC or AVYNFATCGI (Figure 5F). As expected, the negative control peptides LCMV nucleoprotein-derived FQPQNGQFI and ECTV-derived TSYKFESV did not induce IFN-γ in CD8 T cells, as they were not expressed by the LCMV GP mRNA-LNP vaccine (Figure 5F). Unvaccinated and modified GP mRNA-LNP-immunized Tlr9−/− mice were infected with a recombinant ECTV strain (multiple epitopes with enhanced GFP; ECTV-MEE) expressing an engineered construct containing KAVYNFATCGI (covering both the KAVYNFATC and AVYNFATCGI epitopes) and other irrelevant CD8 T cell epitopes. Similar to previous experiments, the unvaccinated group succumbed to the infection, while all mice immunized with LCMV GP mRNA-LNP survived (Figure 5G). These data indicate that the protection induced by modified mRNA-LNP vaccination is not limited to a specific CD8 T cell epitope and confirm that it is mediated by memory CD8 T cells.
Discussion
Our work demonstrates that nucleoside-modified mRNA encapsulated in APCP LNP (which are quite similar to those in the current Pfizer/BioNTech BNT162b2 vaccine, which contains ALC3015 instead of ALC309) can induce very robust epitope-specific CD8 T cell responses that transition to an expanded population of memory CD8 T cells following i.d. and i.m. immunization. Of note, the mice that we used were sufficient for the type I interferon (IFN-I) receptor IFNAR. Our results were surprising, as they contrast with previous reports that mRNA vaccines encapsulated in 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP)/dioleoylphosphatidylethanolamine (DOPE) require the inhibition of type I interferon (IFN-I) signaling in order to induce CD8 T cell responses after subcutaneous, but not intravenous, inoculation.41,42 Our results suggest that the composition of the LNP may profoundly impact the effectiveness of an mRNA-LNP vaccine at inducing CD8 T cell responses in individuals with an intact IFN-I response. Given its translational importance, this issue should be studied in more detail. Similarly, additional studies are required to determine whether IFN-I signaling is necessary or dispensable for the induction of memory CD8 T cell responses by mRNA encapsulated in APCP or similar LNP.
While unmodified mRNA-LNP also induced CD8 T cell responses that may protect mice from lethal mousepox, following i.d. immunization it caused prominent skin inflammation and wounds, particularly at the higher doses. These adverse effects could be due to elevated TLR stimulation and induction of type I interferons by the unmodified mRNA.8,43 Interestingly, modified mRNA-LNP induced more TSYKFESV-specific memory CD8 T cells in the spleen than unmodified mRNA-LNP. This is likely a result of the higher frequency of MPECs induced by modified versus unmodified mRNA-LNP at earlier time points. Booster immunization with modified mRNA-LNP significantly increased the memory CD8 T cell pool as compared to a single vaccination, which was required for the complete protection of Tlr9−/− mice. It should be noted that the early booster immunization at 15 dpv could result in a similar peak magnitude but a less stable memory CD8 T cell pool as compared to boosting at later times.44 Determining the optimal prime-boost regimen that provides rapid onset but also long-lasting protection should be a goal of future studies.
Vaccination with just the TSYKFESV epitope appended to GFP still induced memory CD8 T cells that protected from lethal mousepox. Interestingly, the magnitude of the CD8 T cell response induced by GFP-TSYKFESV was comparable to that with mRNA encoding the entire EVM158 gene. Thus, it may be beneficial to hone the immune response to known CD8 T cell epitopes that are protective or conserved across multiple virus strains. It has been previously demonstrated that antigen-specific CD4 T cells can be induced by mRNA vaccination.9,19,21,23,27,45 It would be interesting to determine if individual CD4 T cell epitopes could also be expressed by modified mRNA-LNP and whether they could contribute to anti-viral immunity. We did not observe a significant contribution from potential antibodies induced by EVM158 mRNA immunization at reducing virus loads. This was expected because EVM158 is an IFN-γ decoy receptor that is not expressed by virions but rather produced and secreted after ECTV infects host cells. Moreover, EVM158 is not required for ECTV pathogenesis.40,46,47 Thus, EVM158-specific antibodies would be unable to inhibit ECTV attachment and infection or to induce Ab-mediated cell or viral lysis.
Together, our data indicate that memory CD8 T cells induced by mRNA-LNP vaccination were sufficient to protect mice from lethal viral challenge. These results suggest that mRNA vaccines can be designed to synthesize engineered proteins tailored to include Ab and multiple CD8 T cell epitopes restricted to different human MHC-I molecules, as well as to target conserved regions of pathogens. Of note, a major advantage of mRNA vaccination as compared to other methods is the ability to focus the immune response to the antigen of choice and, as we show here, even minimal CD8 T cell epitopes. This offers the possibility of avoiding pre-existing immunity to components of the vaccine that could inhibit efficacy, such as may occur with vectored vaccines. Therefore, the same mRNA-LNP platform can potentially be used to immunize against multiple pathogens by simply changing the identity of the mRNA. Vaccination with mRNA to elicit CD8 T cell immunity should also be useful in other areas of medicine, such as vaccines and immunotherapies for cancer. Overall, our findings further highlight the potential of nucleoside-modified mRNA-LNP as an excellent platform for future vaccine development.
Materials and methods
Mice
All experiments were approved by the Thomas Jefferson University Institutional Animal Care and Use Committee (IACUC). Wild-type C57BL/6N mice were purchased from Charles River. B6.D2-(D6Mit149-D6Mit15)/LusJ (B6.D2-D6) mice were purchased from The Jackson Laboratory. B6.129-Tlr9tm1Aki/Obs (Tlr9−/−) mice were produced by Dr. S. Akira (Osaka University, Japan) and generously provided by Dr. Robert Finberg (University of Massachusetts, Worcester, MA, USA).48 All mice were bred and maintained in-house. For the B6.D2-D6 strain, only male mice were used, because they are more susceptible to lethal mousepox than females (∼90% mortality for males versus 50% for females). For all other strains, both males and females were used, as there were no gender-biased results observed. All mice were 6–12 weeks of age when used in experiments.
Viruses and infection
ECTV strain Moscow was obtained from ATCC (VR-1374) and propagated as previously described.49 For specific experiments, we used a recombinant ECTV-MEE strain expressing an engineered protein containing multiple epitopes, including KAVYNFATC and AVYNFATCGI, as well as enhanced GFP (eGFP). Briefly, we introduced MEE downstream of position 189897 of ECTV, which lies in a long intergenic region and where introduction of foreign sequences does not affect viral pathogenicity. We generated a construct by recombinant PCR containing the following segments in order: ECTV Moscow fragment 189543–189897 (5′ homology recombination arm), the vaccinia virus early/late promoter p7.5, a Kozac sequence, a fragment encoding amino acids 214–265 of chicken ovalbumin (contains SIINFEKL), LCMV GP amino acid fragment 29–45 (contains KAVYNFATCGI), LCMV NP fragment 114–130, LCMV NP fragment 392–408 (contains FQPQNGQFI), the sequence for eGFP, and the ECTV Moscow fragment 189950–190297 (3′ homology recombination arm). The construct was cloned into plasmid Bluescript II SK+ to generate the targeting vector pBS-MEE. This targeting vector was used to transfect mouse A9 cells using Lipofectamine 2000 as per manufacturer instructions (Invitrogen). The transfected cells were infected with wild-type ECTV Moscow strain at 0.3 plaque-forming units (PFU) per cell in 6-well plates. Two days later, transfected/infected A9 cells were harvested using a scraper, frozen, and thawed, and different dilutions of cell lysates were used to infect BS-C-1 cells in 6-well plates. Two hours after infection, the cells were overlaid with media containing 0.5% agarose. Four days later, green-fluorescent plaques were extracted with a pipette tip and used to infect a new set of cells. The purification procedure was iterated 5 times, until all plaques were fluorescent. The resulting ECTV-MEE was expanded similar to the wild-type virus. Mice were infected with 30 μL in the right hind footpad with 3,000 PFU wild-type ECTV or 10,000 PFU recombinant strain ECTV-MEE diluted in PBS.
Serum transfer
B6 mice were immunized with a single dose or prime-boosted using modified mRNA-LNP encoding gene EVM158. At 28 dpv or 28 dpbv, immunized mice were terminally bled via cardiac puncture. Serum was collected, pooled by similar vaccination group, and 200 μL of serum was adoptively transferred to naive B6 mice. As controls, serum from unvaccinated and ECTV immune mice, 30 days following infection, were also administered to naive B6 mice. One day following serum transfer, recipient mice were infected with 3,000 PFU of ECTV. Livers and spleens were collected at 5 dpi for plaque assay.
Plaque assay
Titers of ECTV were determined by plaque assay as previously outlined,50 with slight modifications. Briefly, BS-C-1 cells (ATCC CCL-26) were grown in 24-well tissue culture plates to 80%–90% confluency in DMEM tissue culture medium (Invitrogen Life Technologies) supplemented with 10% fetal calf serum (FCS; Sigma-Aldrich), 4.5 g/L glucose, 4.5 g/L L-glutamine, 4.5 g/L sodium pyruvate, 1× non-essential amino acids, and 100 IU/mL penicillin and streptomycin (complete DMEM). BS-C-1 monolayers were infected with 10-fold dilutions of organ homogenate for 1.5 h at 37°C with 5% CO2 in complete DMEM medium containing 2% FCS. Organ homogenate was obtained from processing samples for flow cytometry or by whole-organ mechanical disruption using a TissueLyser (QIAGEN) on a frequency of 30 iterations/s for 2 min. Following incubation, virus was removed, and monolayers were overlaid with 1:1 mixture of 2% carboxymethyl cellulose and complete DMEM medium containing 5% FCS. After 4–5 days of incubation at 37°C with 5% CO2, monolayers were fixed for 20 min at room temperature in 1% crystal violet in 20% ethanol solution containing 4% paraformaldehyde. Excess crystal violet was washed off in a pool of water and plaques were quantified. Virus titers were assessed at 6 dpi in TLR9-deficient mice, as unvaccinated mice succumbed to infection 1 day later.
mRNA in LNP production and vaccination
Codon-optimized sequences for ECTV EVM158, GFP-TSYKFESV, chicken OVA, and LCMV GP were codon-optimized, synthesized (GenScript), and cloned into an mRNA production plasmid as previously described.19,21 Briefly, plasmids were linearized, and mRNAs generated using MEGAscript T7 RNA polymerase (Ambion). mRNAs were transcribed to contain poly(A) tails of 101 nucleotides in length. Uridine 5′-triphosphates were substituted for N(1)-methylpseudouridine 5′-triphosphates (TriLink), and cap1 structure was generated using CleanCap (TriLink). mRNA was purified by cellulose purification as previously described51 and analyzed by agarose gel electrophoresis. Purified mRNAs were encapsulated in LNP using a self-assembly process by rapidly mixing an aqueous solution of mRNA at pH 4.0 with a solution of lipids dissolved in ethanol; LNP were similar in composition to those described previously,29,52 which contain a cationic lipid (ALC0307, proprietary to Acuitas Therapeutics)/phosphatidylcholine/cholesterol/PEG-lipid.29 The proprietary lipid and LNP composition are described in US patent US10221127. They had a diameter of ∼80 nm as measured by dynamic light scattering using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) instrument. For vaccinations, the hind backs of mice were shaved, and mRNA was injected i.d. at 4 distinct locations approximately 1 cm apart with 20 μL of mRNA (80 μL total) using insulin syringes (28–29 gauge). For i.m. injection, mice were administered 40 μL mRNA in both hind thigh areas (80 μL total). Graphical abstract depicting mRNA-LNP vaccination was generated using BioRender.
Flow cytometry
Blood was collected in hematocrit capillary tubes containing heparin (Fisher Scientific). When enumerating cell numbers in the PBL, 50 μL of blood was harvested and counted. Spleens were processed into single-cell suspensions by gentle tissue dissociation using frosted microscope slides (Fisher Scientific). Livers were carefully manipulated through stainless steel wire mesh (88 T316 0.0035-inch diameter; TWC) in a 1.5 × 1.25-in polyvinyl chloride (PVC) female trap adaptor (#4804; Nibco). Hepatocytes were removed following re-suspension in 37% percoll (GE Healthcare Life Sciences) and centrifugation for 20 min at 930 relative centrifugal force at room temperature. The resulting liver cell pellet, splenocytes, or blood was treated with ammonium chloride potassium (ACK) buffer (155 mM NH4Cl, 1 mM KHCO3, 0.1 mM EDTA) for 5 min to lyse red blood cells and washed with RPMI 1640 medium. For T cell re-stimulation experiments, up to 2 × 106 cells were incubated with brefeldin A (BFA; 10 μg/mL) and peptide (1 μM; Genscript) or BFA alone as a control for 5 h at 37°C and 5% CO2 in complete RPMI 1640 medium. To prevent non-specific Fc receptor binding to Abs, cells were stained with anti-CD16/32 (Fc-Block; 2.4G2 ATCC). For extracellular staining of surface molecules, single-cell suspensions were incubated with Abs in FACS buffer for 30 min at 4°C. To detect TSYKFESV or SIINFEKL-specific CD8 T cells, BD DimerX Kb (BD Biosciences) molecules were incubated with TSYKFESV or SIINFEKL peptide and PBS (0.2:0.075:0.725 volume ratio) overnight at 37°C. DimerX Kb-TSYKFESV/SIINFEKL complexes were conjugated with anti-mouse IgG1 at a volume ratio of 4:1 (clone RMG1-1; PE) for 1 h at room temperature. Cells were incubated with 1 μL DimerX complexes for 30 min at 4°C prior to staining with other extracellular Abs. For intracellular staining, samples were stained as above and then fixed for 10 min in 1% paraformaldehyde in PBS. Cells were then incubated in 1× Perm/Wash buffer (BD Biosciences) for 5 min at 4°C and stained for 30 min with Abs in 1× Perm/Wash buffer. Data were acquired using the BD LSRFortessa or BD LSR II cytometers (BD Biosciences) and analyzed with FlowJo software (BD Biosciences).
Abs targeting the following molecules were used in this study: CD4 (clone M4-5; BV785), CD8a (clone 53-6.7; BV711), CD44 (clone IM7; BV421 BioLegend, BUV395 BD Biosciences), CD45 (clone 30-F11; PerCP/Cy5.5), CD62L (clone MEL-14; fluorescein isothiocyanate [FITC]), CD90.2 (clone 53-2.1; BV605, Pacific Blue), CD127 (clone SB/199; APC), IFN-γ (clone XMG1.2; PE/Cy7), KLRG-1 (clone 2F1/KLRG1; PE/Cy7), and TCRβ (clone H57-597; Pacific Blue). All Abs were purchased from BioLegend unless otherwise stated. Data were acquired using the BD LSRFortessa or BD LSR II cytometers (BD Biosciences) and analyzed with FlowJo software (BD Biosciences).
Statistical analysis
Data were analyzed using Prism (GraphPad) software. Groups were assessed for normal distribution using both Anderson-Darling and D’Agostino and Pearson tests. If all groups passed normality tests (p > 0.05), two groups were analyzed using unpaired t test or one-way ANOVA with Tukey’s multiple comparisons test for more than two groups. If any groups failed to pass one of the normality tests, we compared two groups using non-parametric Mann-Whitney test and more than two groups using Kruskal-Wallis with Dunn’s multiple comparisons test. Survival curves were analyzed using the log-rank Mantel-Cox test. For tracking TSYKFESV-specific CD8 T cells in the PBL, comparisons were always done between groups for each day using the appropriate t test or Mann-Whitney test. For most experiments with more than two groups, highlighted statistical differences were compared with the naive or unvaccinated group unless indicated otherwise. All experiments were done a minimum of 2 times, and, where possible, experiments have been compiled and displayed as mean ± SEM. For all figures, p values are represented by the following symbols: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Acknowledgments
This work was supported by grants R01AI110457 and R01AI065544 from the National Institute of Allergy and Infectious Diseases (NIAID) and AG048602 from the National Institute on Aging (NIA) to L.J.S. N.P. was supported by grants R01AI146101 and R01AI153064 from NIAID. D.W. was supported by a sponsored research agreement from BioNTech. C.J.K. was supported by T32 AI134646 from NIAID. Research reported in this publication utilized the Flow Cytometry and Laboratory Animal facilities at the Sidney Kimmel Cancer Center at Jefferson Health and was supported by the National Cancer Institute of the National Institutes of Health under award number P30CA056036.
Author contributions
C.J.K., P.A.P., and L.J.S. designed the experiments, analyzed the results, and wrote the manuscript. D.W., N.P., and H.M. provided the resources, developed the methodology, and helped with the review and editing process. C.J.K. and P.A.P. performed most experiments. C.S. and L.T. helped with some experiments. P.J.C.L. and Y.K.T. designed and prepared the mRNA LNP. L.J.S. conceived the initial idea and supervised the work.
Declaration of interests
N.P. and D.W. are named on patents that describe the use of nucleoside-modified mRNA as a platform to deliver therapeutic proteins and vaccines. They have disclosed those interests fully to the University of Pennsylvania and have in place an approved plan for managing any potential conflicts arising from licensing of the patents. P.J.C.L. and Y.K.T. are employees of Acuitas Therapeutics, a company focused on the development of lipid nanoparticulate nucleic acid delivery systems for therapeutic applications and are named on patent applications describing lipid nanoparticles for delivery of mRNA.
References
- 1.Halstead S.B., Rojanasuphot S., Sangkawibha N. Original antigenic sin in dengue. Am. J. Trop. Med. Hyg. 1983;32:154–156. doi: 10.4269/ajtmh.1983.32.154. [DOI] [PubMed] [Google Scholar]
- 2.Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plotkin S.A., Plotkin S.L. The development of vaccines: how the past led to the future. Nat. Rev. Microbiol. 2011;9:889–893. doi: 10.1038/nrmicro2668. [DOI] [PubMed] [Google Scholar]
- 4.Trovato M., Sartorius R., D’Apice L., Manco R., De Berardinis P. Viral Emerging Diseases: Challenges in Developing Vaccination Strategies. Front. Immunol. 2020;11:2130. doi: 10.3389/fimmu.2020.02130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9:1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pascolo S. Vaccination with messenger RNA. Methods Mol. Med. 2006;127:23–40. doi: 10.1385/1-59745-168-1:23. [DOI] [PubMed] [Google Scholar]
- 7.Scorza F.B., Pardi N. New Kids on the Block: RNA-Based Influenza Virus Vaccines. Vaccines (Basel) 2018;6:20. doi: 10.3390/vaccines6020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 9.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;169:176. doi: 10.1016/j.cell.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Awasthi S., Hook L.M., Pardi N., Wang F., Myles A., Cancro M.P., Cohen G.H., Weissman D., Friedman H.M. Nucleoside-modified mRNA encoding HSV-2 glycoproteins C, D, and E prevents clinical and subclinical genital herpes. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw7083. eaaw7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaTourette P.C., 2nd, Awasthi S., Desmond A., Pardi N., Cohen G.H., Weissman D., Friedman H.M. Protection against herpes simplex virus type 2 infection in a neonatal murine model using a trivalent nucleoside-modified mRNA in lipid nanoparticle vaccine. Vaccine. 2020;38:7409–7413. doi: 10.1016/j.vaccine.2020.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer M., Huang E., Yuzhakov O., Ramanathan P., Ciaramella G., Bukreyev A. Modified mRNA-Based Vaccines Elicit Robust Immune Responses and Protect Guinea Pigs From Ebola Virus Disease. J. Infect. Dis. 2018;217:451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., Tam Y.K., Karikó K., Barbosa C.J., Madden T.D. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freyn A.W., Ramos da Silva J., Rosado V.C., Bliss C.M., Pine M., Mui B.L., Tam Y.K., Madden T.D., de Souza Ferreira L.C., Weissman D. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., 3rd A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman R.A., Fuhr R., Smolenov I., Mick Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson Ӧ., Pujar H.S. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 25.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc J., Moreira E.D., Zerbini C., Baily R. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., mRNA-1273 Study Group An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 28.Remakus S., Rubio D., Ma X., Sette A., Sigal L.J. Memory CD8+ T cells specific for a single immunodominant or subdominant determinant induced by peptide-dendritic cell immunization protect from an acute lethal viral disease. J. Virol. 2012;86:9748–9759. doi: 10.1128/JVI.00981-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tscharke D.C., Karupiah G., Zhou J., Palmore T., Irvine K.R., Haeryfar S.M., Williams S., Sidney J., Sette A., Bennink J.R., Yewdell J.W. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alcamí A., Smith G.L. Soluble interferon-gamma receptors encoded by poxviruses. Comp. Immunol. Microbiol. Infect. Dis. 1996;19:305–317. doi: 10.1016/0147-9571(96)00013-6. [DOI] [PubMed] [Google Scholar]
- 32.Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., Kaech S.M. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaech S.M., Tan J.T., Wherry E.J., Konieczny B.T., Surh C.D., Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 34.Flynn K.J., Belz G.T., Altman J.D., Ahmed R., Woodland D.L., Doherty P.C. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 35.Usherwood E.J., Hogan R.J., Crowther G., Surman S.L., Hogg T.L., Altman J.D., Woodland D.L. Functionally heterogeneous CD8(+) T-cell memory is induced by Sendai virus infection of mice. J. Virol. 1999;73:7278–7286. doi: 10.1128/jvi.73.9.7278-7286.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang M., Orr M.T., Spee P., Egebjerg T., Lanier L.L., Sigal L.J. CD94 is essential for NK cell-mediated resistance to a lethal viral disease. Immunity. 2011;34:579–589. doi: 10.1016/j.immuni.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis R.C., Schadt E.E., Smith D.J., Hsieh E.W., Cervino A.C., van Nas A., Rosales M., Doss S., Meng H., Allayee H., Lusis A.J. A genome-wide set of congenic mouse strains derived from DBA/2J on a C57BL/6J background. Genomics. 2005;86:259–270. doi: 10.1016/j.ygeno.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Samuelsson C., Hausmann J., Lauterbach H., Schmidt M., Akira S., Wagner H., Chaplin P., Suter M., O’Keeffe M., Hochrein H. Survival of lethal poxvirus infection in mice depends on TLR9, and therapeutic vaccination provides protection. J. Clin. Invest. 2008;118:1776–1784. doi: 10.1172/JCI33940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubio D., Xu R.H., Remakus S., Krouse T.E., Truckenmiller M.E., Thapa R.J., Balachandran S., Alcamí A., Norbury C.C., Sigal L.J. Crosstalk between the type 1 interferon and nuclear factor kappa B pathways confers resistance to a lethal virus infection. Cell Host Microbe. 2013;13:701–710. doi: 10.1016/j.chom.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remakus S., Ma X., Tang L., Xu R.-H., Knudson C., Melo-Silva C.R., Rubio D., Kuo Y.M., Andrews A., Sigal L.J. Cutting Edge: Protection by Antiviral Memory CD8 T Cells Requires Rapidly Produced Antigen in Large Amounts. J. Immunol. 2018;200:3347–3352. doi: 10.4049/jimmunol.1701568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Hoecke L., Roose K., Ballegeer M., Zhong Z., Sanders N.N., De Koker S., Saelens X., Van Lint S. The Opposing Effect of Type I IFN on the T Cell Response by Non-modified mRNA-Lipoplex Vaccines Is Determined by the Route of Administration. Mol. Ther. Nucleic Acids. 2020;22:373–381. doi: 10.1016/j.omtn.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Beuckelaer A., Pollard C., Van Lint S., Roose K., Van Hoecke L., Naessens T., Udhayakumar V.K., Smet M., Sanders N., Lienenklaus S. Type I Interferons Interfere with the Capacity of mRNA Lipoplex Vaccines to Elicit Cytolytic T Cell Responses. Mol. Ther. 2016;24:2012–2020. doi: 10.1038/mt.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson E.A., Beura L.K., Nelson C.E., Anderson K.G., Vezys V. Shortened Intervals during Heterologous Boosting Preserve Memory CD8 T Cell Function but Compromise Longevity. J. Immunol. 2016;196:3054–3063. doi: 10.4049/jimmunol.1501797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182:1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai H., Buller R.M., Chen N., Green M., Nuara A.A. Biosynthesis of the IFN-gamma binding protein of ectromelia virus, the causative agent of mousepox. Virology. 2005;334:41–50. doi: 10.1016/j.virol.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 47.Chen N., Buller R.M., Wall E.M., Upton C. Analysis of host response modifier ORFs of ectromelia virus, the causative agent of mousepox. Virus Res. 2000;66:155–173. doi: 10.1016/s0168-1702(99)00135-5. [DOI] [PubMed] [Google Scholar]
- 48.Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 49.Fang M., Roscoe F., Sigal L.J. Age-dependent susceptibility to a viral disease due to decreased natural killer cell numbers and trafficking. J. Exp. Med. 2010;207:2369–2381. doi: 10.1084/jem.20100282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu R.H., Cohen M., Tang Y., Lazear E., Whitbeck J.C., Eisenberg R.J., Cohen G.H., Sigal L.J. The orthopoxvirus type I IFN binding protein is essential for virulence and an effective target for vaccination. J. Exp. Med. 2008;205:981–992. doi: 10.1084/jem.20071854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]