Abstract
Phytopathogenic members of the Sclerotinia genus cause widespread disease across a broad range of economically important crops. In particular, Sclerotinia sclerotiorum is considered one of the most destructive and cosmopolitan of plant pathogens. Here, were review the epidemiology of the pathogen, its economic impact on agricultural production, and measures employed toward control of disease. We review the broad approaches required to tackle Sclerotinia diseases and include cultural practices, crop genetic resistance, chemical fungicides, and biological controls. We highlight the benefits and drawbacks of each approach along with recent advances within these controls and future strategies.
Keywords: sclerotia, necrotroph, fungal pathogen, biocontrol, biopesticide, fungicide, horticulture, row crops
Introduction
Sclerotinia rot, also referred to as white mould on some crops, is a widespread fungal disease caused by phytopathogenic members of the Sclerotinia genus. In particular, S. sclerotiorum is considered one of the most destructive and cosmopolitan of plant pathogens (Bolton et al., 2006; Saharan and Mehta, 2008; Smolinska and Kowalska, 2018). Sclerotinia is widely distributed throughout temperate regions but also occurs in more arid areas. A lack of adequate host genetic resistance, the wide host range of the pathogen, and the general difficulty, both culturally and chemically, in managing the disease are the main drivers for Sclerotinia species causing extensive crop damage within both broad acre and horticultural farming sectors. Economic losses result from collapsed vegetable crops that are completely unmarketable, and in grain or oilseed crops as a reduction in seed number, weight, or quality (Bolton et al., 2006; Saharan and Mehta, 2008; Peltier et al., 2012; Derbyshire and Denton-Giles, 2016). Sclerotia, the resting stage of the fungus, can also contaminate harvested seed, reducing seed price because of the detection of foreign material in the product (Peltier et al., 2012).
Sclerotinia rots can be caused by three closely related species: S. sclerotiorum, Sclerotinia Trifoliorum, and Sclerotinia minor. Combined, they are known to infect over 500 plant species, mostly from Dicotyledonae but a few are from Monocotyledonae such as onion and garlic (Laemmlen, 2001; Saharan and Mehta, 2008; Willbur et al., 2019). S. sclerotiorum has a wide host range, and S. minor affects similar hosts but within a reduced range. The host range of S. trifoliorum is narrower, usually, only forage legumes such as clover and lucerne/alfalfa. Numerous broadleaf weeds such as wild clover, dandelion, capeweed, and wild radish are also hosts and play a role in carry-over of the disease and inoculum between crops. Although Sclerotinia diseases are typically a sporadic production problem, the disease has become a consistent issue in intensive cropping systems with short rotations. The drivers for increased disease emergence include favourable environmental/seasonal conditions such as reliable rainfall or irrigation, and the frequency of susceptible hosts in the cropping rotation, which build up levels of soil-borne sclerotia.
Economically important broad acre crops commonly affected by S. sclerotiorum include the oilseed crops canola and sunflower, and the legumes soybean, peanuts, chickpea, and several bean species, and its infection often leads to a significant loss in crop production (Agrios, 2005; Link and Johnson, 2007; Saharan and Mehta, 2008; Derbyshire and Denton-Giles, 2016). The incidence of Sclerotinia stem rot on canola is problematic worldwide and reported in most canola-producing regions of the world such as China, India, Europe, Australia, and North and South America (reviewed in Alkooranee et al., 2017; Zheng et al., 2020). In the United States, collective crop losses to S. sclerotiorum exceed US$200 million annually (Bolton et al., 2006). In 2010, the disease cost Canada an estimated US$600M in lost canola production (Dupont Pioneer Report, 2012). In smaller production areas such as Australia, losses estimated over AUS$59M have been recorded (Khangura et al., 2014). In canola, diseases can be widespread but highly sporadic, requiring specific environmental conditions to develop. As such, disease incidence can vary greatly from year to year, but is most damaging under prolonged wet conditions, particularly leading up to and during flowering and where the crop is grown intensively, resulting in a build-up of pathogen load, in the form of sclerotia, within the cropping system (Khangura et al., 2014, 2015; Derbyshire and Denton-Giles, 2016). Under conducive conditions, yield losses typically exceed 20–35%, but incidences of over 50 and up to 80–100% have been reported in some global markets (Markell et al., 2009; Dokken-Bouchard et al., 2010; Murray and Brennan, 2012; reviewed in Alkooranee et al., 2017). In Canada, it is predicted that for every 1% of canola crop infections, there is a 0.5% loss in potential yield results (Derbyshire and Denton-Giles, 2016).
In legumes, years that favour the development of Sclerotinia diseases are a significant cause of economic losses. The disease is considered “chronic to epidemic” on soybean throughout the world with the worst losses occurring in North and South America (Grau et al., 2004). For example, in a particularly bad year for Sclerotinia on soybean in the United States in which it ranked second out of 23 diseases, an estimated cost of US$560 million was reported (Peltier et al., 2012). It has been estimated that, in bad years, losses in peanut yield can reach US$1 million in North Carolina alone (Smith et al., 2008).
Despite the regular application of fungicides, Sclerotinia diseases are one of the major causes of losses in horticultural crops (Vallalta and Porter, 2004; Saharan and Mehta, 2008). This includes both leafy vegetables, such as vegetable brassicas, and root vegetables, such as potatoes and carrots. The fungus can cause losses in both the field and under storage conditions, although most plants become infected in the field (Dillard, 1987). Lettuce is reported as one of the crops that are most susceptible to Sclerotinia. Losses in field-grown lettuce in the United Kingdom are commonly 10%, but greater losses of up to 50% can occur under wet conditions (Young et al., 2004). S. Minor-induced losses in lettuce are reported to range from 10 to 45% in intensive lettuce-growing regions of Australia, despite the use of fungicide in spray programs (Vallalta and Porter, 2004).
Sclerotinia minor is less common than S. sclerotiorum but has a somewhat narrower host range (Willetts and Wong, 1980). It is a major concern for peanuts as the cause of Sclerotinia blight (Chenault Chamberlin et al., 2010). Losses in peanut crops in North Carolina alone due to Sclerotinia blight have been estimated at US$1–4 million/year (Smith et al., 2008). Other crops impacted by S. minor include soybean and common beans (Grau et al., 2004).
Of the Sclerotinia species, Sclerotinia trifoliorum has the narrowest host range, infecting mostly legumes (Willetts and Wong, 1980). It causes white mold, crown, and stem rot of forage legumes, such as lucerne/alfalfa and clovers, and cool season grain legumes (e.g., chickpea, lentil, faba beans) (Jellis et al., 1990; Njambere et al., 2014). All species that are infected by S. trifoliorum are also susceptible to S. sclerotiorum (Willetts and Wong, 1980; Njambere et al., 2014).
Pathogen Epidemiology and Disease Cycle
Sclerotinia Disease Development and Symptoms
As a necrotrophic fungal pathogen, the Sclerotinia fungus needs to kill plant cells in order to establish disease and obtain nutrients. It is such a destructive pathogen in agriculture, because it is able to infect plants at any growth stage including young seedlings, mature plants, and fruits in the field or in storage. In addition, when not infecting plants, the fungus may spend more than 90% of its life as sclerotia, its primary resting stage. Sclerotia are hard, melanised survival structures (Figure 1) that are resistant to desiccation and act as food storage reserves, allowing the fungus to survive in soil or stubble for up to 5 years or more (Agrios, 2005; Peltier et al., 2012; Young and Werner, 2012; Smolinska and Kowalska, 2018). These resting structures allow the pathogen to survive in the absence of a plant host and serve as a source of infection in subsequent crops (Agrios, 2005; Young and Werner, 2012). The sclerotia of S. sclerotiorum and S. trifoliorum are typically 2–5 mm in diameter but sometimes greater than 10 mm, while S. minor produces small sclerotia of 0.5–2 mm in diameter.
FIGURE 1.
Generalised Sclerotinia disease cycle. Image credits: Smith, 2018 CSIRO and Tian, 2020 CSIRO.
Both Sclerotinia sclerotiorum and Sclerotinia trifoliorum reproduce carpogenically by producing small (5–10 mm in diameter), tan, cup-shaped mushroom-like structures called apothecia (Figure 1) which contain sexual spores called ascospores (Willetts and Wong, 1980). Sclerotial germination and ascospore production are favoured by moist, cool soils with temperatures around 4–18°C; however, temperature requirements are variable depending on the origin of the Sclerotinia isolates (Willetts and Wong, 1980; Peltier et al., 2012; Smolinska and Kowalska, 2018). Cold-conditioned sclerotia within the top centimetres of soil typically begin to produce apothecia after 2–6 weeks at 15°C (Willetts and Wong, 1980; Young et al., 2004; Peltier et al., 2012; Clarkson et al., 2014). The apothecia-fruiting bodies burst, releasing thousands of ascospores that are readily spread by the wind throughout plant canopies and surrounding areas. S. sclerotiorum ascospores require certain environmental conditions in order to germinate and infect plants. Temperatures around 15–25°C and wetness or high humidity are required for >48 h (Young et al., 2004; Clarkson et al., 2014; Derbyshire and Denton-Giles, 2016; Willbur et al., 2019). Susceptible plant tissues such as flower petals and senescing leaves are typically infected first (Link and Johnson, 2007), serving as the nutrient source for Sclerotinia mycelium to grow and subsequently infect other plant tissues. In canola, for example, infected senescent petals are the predominant source of infection. They drop into the canopy and lodge onto leaf axils or stem branches, or stick to leaves and stems (reviewed in Derbyshire and Denton-Giles, 2016; Thatcher et al., 2017). Surveillance for infested petals is commonly performed to measure pathogen load in the field (reviewed in Derbyshire and Denton-Giles, 2016).
Infection can also occur myceliogenically by contact with hyphae from sclerotia onto plant tissues, or directly from contact with other diseased plants. This is a common infection route for S. minor, which rarely produces apothecia, but is relatively rare in S. sclerotiorum and S. Trifoliorum (Link and Johnson, 2007). The environmental drivers for differential carpogenic or mycelial germination of S. sclerotiorum or S. trifoliorum sclerotia are not well understood. A review of abiotic conditions promoting myceliogenic germination of S. sclerotiorum highlighted moisture availability and extremes in temperatures as key factors influencing myceliogenic germination (Lane et al., 2019).
Upon infection, typical Sclerotinia lesions first appear on the leaves and leaf axils as water-soaked spots or lesions with a pale greyish white or brownish white appearance caused by cell death and the action of pathogen pectolytic enzymes. The water-soaked lesions, particularly at nodes, can rapidly progress along and around the stem above and below the infected nodes. Infected leaves may also fall and lodge further down the canopy and spread the infection to other plants. Plants infested with Sclerotinia develop a white, cottony growth on the stems followed by the formation of sclerotia (Figure 1), which can occur inside or outside of stems, and pods of legumes such as soybean, or on canola pods if the weather is favourable (Fernando et al., 2004; Bolton et al., 2006; Kamal et al., 2016; Willbur et al., 2019). The infected part of the stem turns soft and greyish-white in colour. The lesions girdle the stem, cutting off vascular transport, causing infected stems to become bleached and stringy or shred, and plant tissues above the lesion wilt and die (Grau et al., 2004; Derbyshire and Denton-Giles, 2016; Willbur et al., 2019). Legume pods on infected plants may appear white in colour, smaller, and contain fewer seeds. They may also contain sclerotia. Other common symptoms include stunting, premature ripening, and lodging of plants (Willetts and Wong, 1980; Peltier et al., 2012). Sclerotia produced within infected stems can be distributed after harvest by equipment or may be harvested with the seed, causing contamination of seed lots. Therefore, it is important to screen seeds for contamination.
Infection by Sclerotinia sclerotiorum is often patchy across a field, and symptoms on row or broad acre grain and oilseed crops are generally only evident late in the season after the flowering stage (Grau et al., 2004; Derbyshire and Denton-Giles, 2016). Plants can be attacked at any growth stage, but infection by S. sclerotiorum generally occurs during flowering, because favourable weather conditions tend to coincide with a fully developed crop canopy supporting cool, moist, shaded conditions in soils, which encourage the development of apothecia and the release of ascospores (Willbur et al., 2019). Mist, dew, and fog are all potential sources of moisture. Disease development is dependent on sufficient moisture in the canopy and typically extended periods of leaf wetness. Losses tend to be greatest when cool (15–25°C), wet, humid conditions, combine with management practices that favour high yield potential such as high planting density, narrow row spacing, and high plant nutrition that support the development of dense canopies that can contain high humidity (Grau et al., 2004; Link and Johnson, 2007; Peltier et al., 2012). Crops grown by overhead spray irrigation will also favour disease development.
Sclerotinia sclerotiorum can infect vegetable crops at any growth stage. Infections typically occur on the stem or leaves at the base or from infection events on the top of densely grown crops (Dillard, 1987). Blossoms from weeds are a common source of nutrients for ascospores and, consequently, a source of fungal inoculum on nearby vegetable crops. Infection starts with water-soaked lesions that become infested with fungal mycelium. The host tissue becomes soft and watery, ultimately leading to complete crop failure, and sclerotes produced with the diseased tissues.
Sclerotinia minor infects through eruptive myceliogenic germination; therefore, infection below the canopy at roots, crowns, and leaves is observed. Disease symptoms of S. minor are similar to those of S. sclerotiorum but not necessarily coincide with flowering. Initially, water-soaked lesions appear on the stems or leaves. As the disease progresses, fluffy white mycelia may become visible, and then the lesions become bleached, and necrotic and infected stems become shredded and die (Smith et al., 2008).
Rots of forage legumes caused by Sclerotinia trifoliorum manifest as a soft rot of the crown and roots, which begins with the appearance of brown leaf lesions that then spread to stems and shoots. New growth on the infected plants begins to wilt, then dies. The dead tissues are often covered with white mycelium, particularly in wet, cool weather (Barbetti and You, 2014). While plants can be infected at all growth stages, losses tend to be most severe when infection occurs at the seedling stage (Kanbe et al., 2002).
Pathogenicity
An understanding of the cytological and molecular mechanisms of Sclerotinia colonisation and infection has increased in recent years thanks to advances in genomic sequencing, transcriptomic analysis, and functional gene characterisation. Recent findings show that the interactions between Sclerotinia and its wide range of hosts may be significantly more sophisticated than previously assumed, with the molecular mechanisms of infection and pathogenicity recently well reviewed (Mbengue et al., 2016; Liang and Rollins, 2018; Xia et al., 2020). Here, we summarise these findings and provide an overview of Sclerotinia pathogenicity, and in subsequent sections, we will detail how this knowledge could be exploited in new controls for disease suppression.
Host Infection
As a canonical necrotrophic fungus, Sclerotinia attacks and kills the cells of its host by secreting an arsenal of cell wall-degrading enzymes and toxins, and it consumes dead cells for energy (Mbengue et al., 2016; Xu et al., 2018; McCaghey et al., 2019; Wang et al., 2019a). It has been proposed that this pathogen exhibits a dynamic two-phased infection model in which it first evades or counteracts host defence, colonising the host and growing in apoplastic spaces (Kabbage et al., 2015; Liang and Rollins, 2018; Ding et al., 2021). Through the production of cell-degrading enzymes and toxins, the pathogen then progressively kills and degrades the host cells. It is further proposed that this phased infection process is not just defined at the temporal or spatial level, but rather the possibility that it isdefined through different sectors or developmental stages of the advancing fungal colony (Kabbage et al., 2015).
The initial colonisation of the host by Sclerotinia occurs via a compound appressoria (a modified hyphal tip), which penetrates the cuticle of the host via a penetration peg. Following initial penetration, the colonisation phase involves the growth and branching of sub-cuticular hyphae (Liang and Rollins, 2018). There is evidence that these subcuticular hyphae can spread several cell layers ahead of killed epidermal cells, in which the hyphae are interacting with healthy cells (Kabbage et al., 2015). Once the subcuticular hyphae are established, they branch into smaller, ramifying hyphae that grow inter- and intra-cellularly into the epidermal and mesophyll cells. Liang and Rollins (2018) noted that subcuticular and ramifying hyphae have distinct morphology and colonisation patterns, suggesting potential functional specialisation, and that subcuticular hyphae are likely to be more important in defence suppression and infection establishment, while ramifying hyphae are more important in initiating cell death and cell wall degradation.
Further evidence for the two-phase infection model in Sclerotinia was found in a study analysing sequential gene expression patterns during infection of Brassica napus (Seifbarghi et al., 2017). During the first 12 to 24 h post-infection period the expression of Sclerotinia effector-like genes (e.g., genes encoding the LysM domain protein and a cysteine rich protein) associated with suppression of the pathogen recognition and defence systems in the host was upregulated. At the later stage of infection (after 24 h) Sclerotinia genes associated with the induction of necrosis and programmed cell death were upregulated.
The secretion of a wide array of cell-wall-degrading enzymes facilitates the degradation of host cell walls, as well as numerous proteases and hydrolases, to macerate tissues and release nutrient sources (Mbengue et al., 2016; Xu et al., 2018; McCaghey et al., 2019; Wang et al., 2019a). These include proteinases, cutinase, cellulases, polygalacturonases, glucanases, and xylanases. It has been suggested that the diversity of enzymes produced by Sclerotinia may enhance its adaptability and therefore contribute to its wide host range (Ding et al., 2021).
Secreted Proteins and Host Defence Suppression
Important virulence components of Sclerotinia include secreted and effector-like proteins and are reviewed in detail by Derbyshire et al. (2017), Xu et al. (2018), McCaghey et al. (2019), Xia et al. (2020), and Shao et al. (2021). Historically, fungal effectors have been defined as small, secreted proteins that modulate the host cell to facilitate infection. More broadly, they encompass a wider array of proteins or molecules that function to establish and progress disease.
Bioinformatics approaches applied to the full genome sequence of S. sclerotiorum have identified a large number of genes encoding a range of virulence-related secretory effector proteins (Amselem et al., 2011). Secretome analyses have shown that S. sclerotiorum has the potential to produce 400 secreted proteins including nearly 80 virulence factor candidates (Guyon et al., 2014; Heard et al., 2015; Derbyshire et al., 2017). Of those characterised to date, manipulation of host salicylic acid, jasmonic acid, and ROS signalling are common themes to suppress or interfere with host defense responses (McCaghey et al., 2019; Wang et al., 2019a; Ding et al., 2021). For example, the S. sclerotiorum-secreted integrin-like protein SsITL suppresses host immunity at the early stage of infection (Mbengue et al., 2016; Tang et al., 2020). It interacts with the plant chloroplast-localised CAS (chloroplast-localised calcium-sensing receptor) protein that is a positive regulator of salicylic acid signalling and resistance against Sclerotinia. Manipulators of host cell death include the Sclerotinia Cu/Zn superoxide dismutase SsSOD1 and the Ca-binding SsCAF1 protein also involved in Sclerotinia development (reviewed in McCaghey et al., 2019; Xia et al., 2020). The chorismate mutase enzyme Ss-Cmu1 is predicted to be secreted and dampen host salicylic acid synthesis (Liang and Rollins, 2018). In another example, the S. sclerotiorum small, cysteine-rich secreted protein SsSSVP1 induces plant cell death and interacts with the plant QCR8, a subunit of the cytochrome b-c1 complex of mitochondrial respiratory chain, and disturbs the localisation of QCR8 in mitochondria (Lyu et al., 2016). Another strategy employed by S. sclerotiorum has been recently shown in the Sclerotinia-Brassica pathosystem. Chen et al. (2020) demonstrated that S. sclerotiorum was able to metabolise, via a hydrolase, toxic isothiocyanates produced by the plant and therefore was able to promote its growth and contribute to its virulence on glucosinolate-producing plants. Together, these examples highlight the multiple strategies employed by the Sclerotinia secretome to manipulate or avoid host cell processes and likely contribute to its broad host range and effectiveness as a destructive plant pathogen. Further functional characterisation of these and other Sclerotinia virulence-related secreted proteins may provide new targets for disease control targeting either the pathogen directly or through the modification of its host target.
The Roles of Oxalic Acid, ROS, and Host Cell Death
Sclerotinia also produces the non-selective phytotoxin, oxalic acid, that contributes to pathogenesis through acidification and suppression of host defence responses by manipulating the host redox environment (Williams et al., 2011; Smolinska and Kowalska, 2018). Evidence for the involvement of oxalic acid in plant infection has been demonstrated by the recovery of oxalate from infected tissues (Bateman and Beer, 1965; Maxwell and Lumsden, 1970; Marciano et al., 1983; Godoy et al., 1990) or by injection of oxalate into plants followed by the development of Sclerotinia disease-like symptoms (Bateman and Beer, 1965; Noyes and Hancock, 1981). In addition, it could be demonstrated that Sclerotinia mutant strains that were deficient in oxalate synthesis were non-pathogenic but regained normal virulence upon regaining their oxalate synthesis capacities (Godoy et al., 1990). Recent studies proposed that it is not oxalic acid specifically that is necessary for the ability of Sclerotinia to cause disease but rather its acidic pH (reviewed in Xu et al., 2018).
In neutral or alkaline environments, Sclerotinia grows slowly and produces acids such as oxalic acid to acidify its environment (Tourneau, 1979). The fungus increases its growth rate under acidic conditions (Maxwell and Lumsden, 1970) and initiates the production of sclerotia (Rollins and Dickman, 2001; Chen et al., 2004). Lytic and hydrolytic enzymes are expressed and secreted under acidic conditions, which are needed for optimal enzyme activity and are necessary for disease development (Cotton et al., 2003; Girard et al., 2004; Kasza et al., 2004; Li et al., 2004; Seifbarghi et al., 2017). If the ambient pH gets too low, Sclerotinia will be able to produce ammonia or oxalate decarboxylase that degrades oxalate, thereby raising the pH (Vega et al., 1970; Magro et al., 1988; Billon-Grand et al., 2012; Liang et al., 2015). A recent study compared the pathogenicity of different Sclerotinia isolates in canola and observed a correlation between pathogen aggressiveness and its ability to acidify its environment (Denton-Giles et al., 2018). The ability to sense and change pH in its environment enables Sclerotinia to survive under many different conditions and thereby makes it one of the most successful pathogens of plants.
In addition to its direct effect on cellular pH, oxalic acid has been shown to induce programmed cell death, and it is thought that the manipulation of host programmed cell death plays a crucial role in the pathogenicity of Sclerotinia (Kim et al., 2008; Mbengue et al., 2016). Oxalic acid induces increased levels of ROS in the host, which is also associated with programmed cell death. When ROS production is inhibited, apoptotic-like cell death due to oxalic acid is also inhibited (Kim et al., 2008). Kim et al. (2008) suggested that additional roles of oxalic acid include acting as a signalling molecule that induces programmed cell death. Sclerotinia also produces a range of other factors that induce the necrosis of host cells. Most are proteins or peptides such as ethylene-inducing peptides, endopolygalacturonases, and a cutinase (Liang and Rollins, 2018). Two small secretory proteins, Ss-SSVP1 and Ss-CP1, have been shown to induce cell death (Lyu et al., 2016; Yang et al., 2018).
Gene Silencing and sRNAs
A recent study by Derbyshire et al. (2019) showed that during infection, Sclerotinia produces at least 374 distinct small RNAs, many of which downregulate functional domains associated with plant immunity. These small RNAs induce gene silencing through RNA interference (RNAi) pathways (Mbengue et al., 2016).
A genome wide association (GWAS) study has shown that bioinformatically predicted targets of Sclerotinia small RNAs included functional domains associated with plant immunity and quantitative disease resistance (Derbyshire et al., 2019). Mutants of both plant and fungal RNAi components were found to have reduced silencing of host immunity genes and therefore reduced disease symptoms (Weiberg et al., 2013).
Foe to Most, Friend to Some?
While Sclerotinia sclerotiorum predominantly infects dicots, in a recent study, it was found to grow endophytically within several monocots including wheat, barley, oat, rice and maize (Tian et al., 2020). While it is not uncommon for fungal pathogens of a plant species to colonise another plant species and not cause disease, in the study by Tian, they found that S. sclerotiorum colonisation actually had a positive influence on the host. Endophytic growth within wheat reduced disease severity incited by the phytopathogenic fungal pathogens that cause Fusarium head blight and stripe rust, likely through manipulation of defence responses and hormone signalling. It is interesting that such a broad host range and highly destructive pathogen of dicots had a beneficial effect on monocots. These results have implications in the cultural management of Sclerotinia in farming systems where susceptible break crops such as canola and legumes are grown in rotation or in close proximity to cereals. Further studies are needed to determine if mycelium carry-over between seasons on monocot stubble can serve as an inoculum source for the following year.
Disease Control
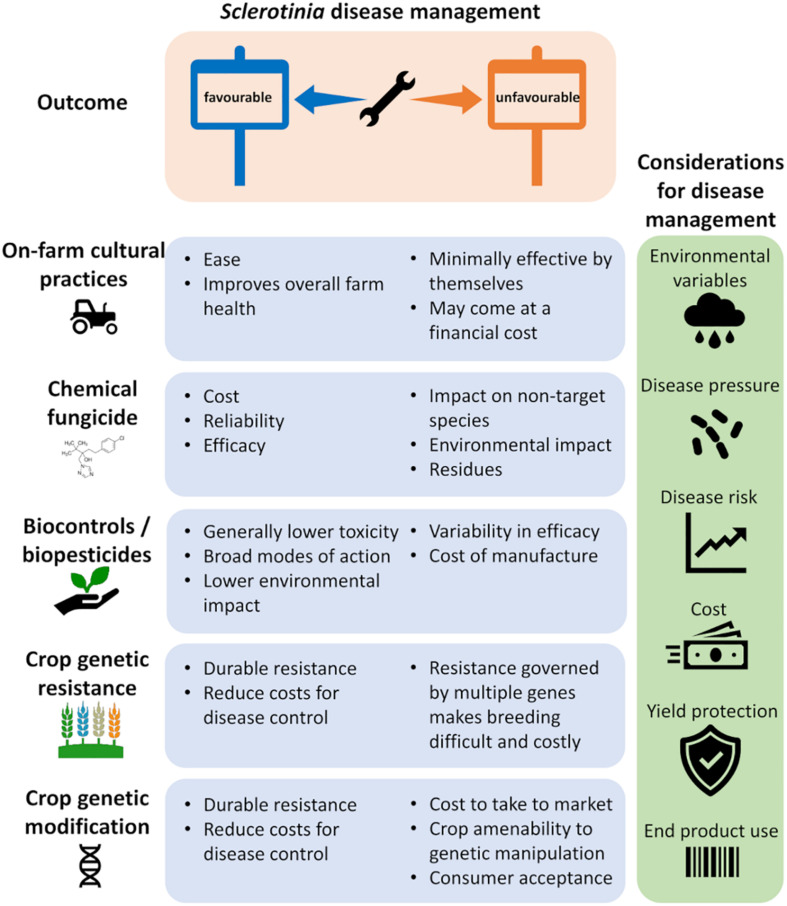
As with many diseases, there is no single treatment that can completely control Sclerotinia, so most growers employ an integrated management approach to reduce their risk of losses from disease. This involves combining several approaches including cultural practices, variety selection, and chemical and biological controls (Peltier et al., 2012). With limited genetically resistant cultivars available to growers and the broad host range of the pathogen, cultural and management practices, such as fungicide use, are the predominant approaches for control but provide variable protection depending on the timing of application. Growers, therefore, need to consider environmental variables, disease pressure, and risks when planning their management strategy (Figure 2).
FIGURE 2.
Sclerotinia disease management. Current strategies to control Sclerotinia diseases rely on integrated disease management, each with its pros and cons (blue panels) toward the desired outcome. This may involve a combination of management strategies suited to the host crop and requires consideration and monitoring of multiple factors to guide decision on best suited disease control tools (green panel).
Cultural Practices
Cultural and agronomic measures are an important part of disease management and can reduce disease severity but are not effective by themselves in controlling the disease. Cultural practices include crop rotation, reducing plant density, and practices to reduce ascospore production and release. Because Sclerotinia has a wide host range, many broad leaf crops and weeds support the disease, so weed management and careful selection of crops to rotate are required, as repeated cropping of susceptible crops increases sclerotial numbers in each subsequent crop. Small grain crops (maize, wheat, barley, oats, sorghum) are not susceptible to infection by Sclerotinia spp. and so are suitable rotation crops but a break of 2–3 years may be required to decrease the number of sclerotia in soil (Peltier et al., 2012). However, in light of the potential for these small grain crops to still be colonised by Sclerotinia (Tian et al., 2020), avoidance of infested or adjacent fields for 1–4 years could be considered the best management option if economically viable.
Canopy management, including wider row spacing or lower seeding rates, can be used to increase air flow and decrease humidity in the crop canopy. Studies on a range of different crops have shown that this can reduce numbers of apothecia and disease incidence (McDonald et al., 2013). Reducing fertilisers and delaying planting to manage vegetative growth should also be considered, as over-fertilising and early planting can result in tall, bulky crops at the time of flowering that will create denser canopies, and, with high rainfall, will put crops at higher risk of disease.
Stubble management practices to minimise the carry-over of viable sclerotia between seasons have demonstrated efficacy. These include burning crop residues or using irrigation to increase rotting of sclerotia. Other studies have suggested that there may be lower disease incidence in no-till systems (Kurle et al., 2001; Simpfendorfer et al., 2004) but these results are inconsistent (Peltier et al., 2012). It is speculated that sclerotia may be more likely to degrade in no-till systems because they are not buried and are more prone to attack by predators, desiccation, or exposure to UV in the upper soil layers (Peltier et al., 2012). It is also advisable not to keep seeds from infected crops for replanting, as this will also increase the risk of spreading infection into potentially previously clean areas.
Crop Resistance
For all crops that are susceptible to Sclerotinia species, it is generally advised that where partially resistant cultivars are available, they should be used as part of an integrated management approach to reduce the risk of yield losses and to decrease the load of inoculum in future seasons (Kurle et al., 2001; Grau et al., 2004; Smith et al., 2008).
There are ongoing efforts to breed Sclerotinia-resistant cultivars of a range of broad acre and horticultural crops (Kim and Diers, 2000; Arahana et al., 2001; Cruickshank et al., 2002; Lithourgidis et al., 2005; Vuong et al., 2008; Chenault Chamberlin et al., 2010; Barbetti and You, 2014; Taylor et al., 2015; Derbyshire and Denton-Giles, 2016; Bennett et al., 2018). In all cases, it is important to consider that the disease screening method is reflective of conditions experienced by crops grown under field conditions. For example, in canola, one of the most significant limitations in high throughput seedling assays is that plant disease resistance is not measured at the growth stage during which the disease is most likely to occur in the field, which means that plants should be grown at the flowering developmental stage (Derbyshire and Denton-Giles, 2016; Thatcher et al., 2017).
Molecular breeding is an important strategy in several crops for improving host resistance as an avenue for disease control. There is no genetic source of complete plant resistance to these broad-host range pathogens known to date, but partial resistance has been identified in several economically important crops (e.g., canola and soybean) (Mbengue et al., 2016; McCaghey et al., 2019). Resistance to Sclerotinia is polygenetic and made up of the complex interaction of numerous minor effect genes (Wang et al., 2019a). This results in quantitative disease resistance, in which a natural population of plants will display a continuum of resistance phenotypes from highly susceptible to partially resistant.
The current status of resistance mapping and breeding is well reviewed by Wang et al. (2019a) and Ding et al. (2021). Early studies screening for resistance loci in B. napus used quantitative trait loci (QTL) mapping. More recently, alternative approaches that include genome-wide association mapping studies using natural plant populations have been applied. This, paired with high-throughput sequencing, omics, and improved data analytics combined with an increased understanding of molecular mechanisms of plant defense against Sclerotinia have improved the identification of quantitative resistance gene/loci and of host genes or processes that can be targeted and manipulated for enhancing resistance or reducing susceptibility. For example, Ding et al. (2021) broadly categorised resistance responses into three types: (1) the activation of defence signaling pathways, (2) the production of secondary metabolites that kill Sclerotinia or inhibit growth and infection processes, and (3) the production of activating enzymes, proteins, or antimicrobial peptides that affect Sclerotinia cell wall integrity or block its pathogenicity factors. Wang et al. (2019a) noted that the GWAS studies on Sclerotinia resistance in B. napus predominantly linked loci-containing genes involved in downstream defence responses, ROS production, detoxification, oxidative protection, and secondary metabolite enzymes. These findings suggest that Sclerotinia resistance is likely a function of a variety of cellular processes and that this needs to be taken into account when breeding for crop varieties with enhanced resistance.
Natural Sources of Resistance or Non-GM Approaches
The fact that Sclerotinia tolerance in several species is controlled by multiple minor genes makes breeding resistant varieties difficult and costly, and raises the risk that undesirable traits can be inadvertently introgressed through the process of linkage drag (Wang et al., 2019a). In some cases, QTLs that are associated with physiological resistance to S. sclerotiorum have been reported (Grau et al., 2004). It is thought that the severity of disease caused by S. sclerotiorum is governed by a combination of physiological resistance (e.g., phytoalexin production) and disease escape mechanisms (e.g., open canopy architecture, lodging resistance, and timing of maturity) (Grau et al., 2004). This further complicates efforts to breed Sclerotinia-resistant varieties, as the disease tolerance phenotypes may not be suitable as breeding material.
There is a relatively large body of information on quantitative trait locus mapping data available for resistance to Sclerotinia in various hosts, but relatively little is known about the molecular basis for the quantitative trait loci. The advances in molecular breeding are likely to be made possible because of the work being done to increase the understanding of the molecular basis of host genetic resistance (Wang et al., 2019a). Using a GWAS approach analysing global B. napus germplasm, Gyawali et al. (2016) identified 669 polymorphic loci that were associated with Sclerotinia resistance including 21 alleles related to resistance and 13 related to susceptibility. A map of Sclerotinia resistance has been constructed in B. napus using 347 markers and integrated 35 QTLs that have been linked to disease resistance (Li et al., 2015). Similar studies in soybean have identified 103 QTLs that have been mapped to 16 and 11 loci that were significantly correlated with resistance in field and greenhouse by GWAS (Wen et al., 2018). It has been noted in both canola and soybean that resistance to Sclerotinia is conferred by minor QTLs, but each only makes a small contribution (typically less than 10%) to phenotypic variance (Wen et al., 2018; McCaghey et al., 2019).
In canola, the lack of complete genetic resistance has prompted researchers to screen close relatives of Brassica napus such as Brassica oleracea and also wild crucifers that lie outside the Brassica genus (reviewed in Derbyshire and Denton-Giles, 2016). Partial stem resistance has been identified through a variety of screening methods (Garg et al., 2008, 2010; Denton-Giles et al., 2018). The challenge will be to breed this material into elite cultivars. Avoiding petal infection is another option that has been explored. Apetalous B. napus plants develop less disease but are still equally infected by S. sclerotiorum as full-petalled cultivars (Young and Werner, 2012).
In soybean, there are no cultivars with complete resistance to Sclerotinia sclerotiorum, but there are partially resistant varieties available (Grau et al., 2004; McCaghey et al., 2019; Willbur et al., 2019). There are commercial varieties of peanut available that are resistant to S. minor and S. sclerotiorum. It has been estimated that the resistant peanut cultivars, Toalson, Tamspan 90, and Tamrun 98, save the United States peanut industry from ∼US$5 million in losses due to Sclerotinia annually (Bennett et al., 2018). There are ongoing efforts to breed elite varieties with S. sclerotiorum and S. minor resistance that are adapted to a wider range of environments (Cruickshank et al., 2002). There is evidence that resistance to S. trifoliorum exists within several forage and grain legumes such as alfalfa, clover, and faba beans, but there are currently no commercially available resistant cultivars, and breeding efforts are ongoing (Halimi et al., 1998; Kanbe et al., 2002; Lithourgidis et al., 2005; Mikaliuniene et al., 2015). Screening for S. sclerotiorum resistance in wild Cicer populations, with the aim to adopt this into commercial chickpea varieties, identified partial stem resistance (Mwape et al., 2021).
Genetic Modification
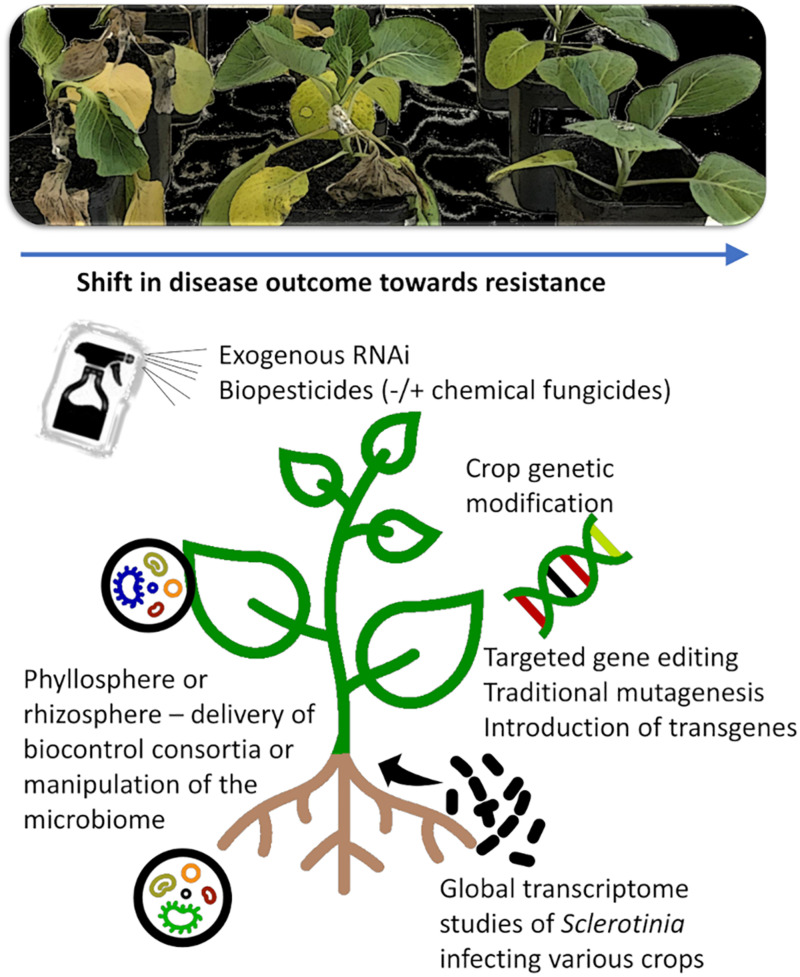
The lack of strong, major effect resistance in most crops makes classical breeding a challenging task. In crops where genetically modified (GM) varieties are already available for other traits, such as canola or soybean, transgenic or targeted genetic modification approaches for Sclerotinia resistance may be an economical option (Figure 3).
FIGURE 3.
New opportunities and tools for Sclerotinia control. New tools exist or are emerging that allow targeted manipulation of pathogen, host, or beneficial microbial populations that facilitate a reduction in Sclerotinia disease symptom development. These includes exogenous controls such as RNAi or biochemicals and biopesticides that can act directly on the pathogen (e.g., antifungal activity, fungal development) or indirectly by enhancing plant defense responses. Powerful endogenous controls such as crop genetic modification can provide durable resistance, but the process is costly, and not all crops are readily transformable or afford public acceptance. New knowledge of the pathogenicity and virulence of Sclerotinia species can be acquired through global omics studies on the pathogen infecting diverse crops of varying disease resistance, and this knowledge can be used to identify essential pathogen processes or weaknesses that can be targeted in new management tools.
The overexpression of oxalic acid degrading oxalate oxidase genes in a number of crops has enhanced resistance to S. Sclerotiorum (Lithourgidis et al., 2005; Yang et al., 2019; van Esse et al., 2020). In most cases, complete resistance is not observed but rather a delay in disease progression. This may, however, be sufficient in the field to provide a yield benefit or reduce the requirement for fungicide applications.
A general strategy to enhance plant resistance to pathogens is to upregulate the expression of key or master switches of defence responses such as transcription factors or mitogen-activated protein kinases (MPKs) (Thatcher et al., 2005; Amorim et al., 2017). The overexpression of specific WRKY transcription factors or MPKs in Arabidopsis and/or canola has been demonstrated to provide increased resistance against Sclerotinia (Chen et al., 2013; Liu et al., 2018; Wang et al., 2019b). Other approaches taken include the overexpression of downstream pathogen inhibiting defence genes, such as chitinase genes or polygalacturonase-inhibiting proteins, to slow down disease progression (Zarinpanjeh et al., 2016; Wang et al., 2018; Yang et al., 2020). More recently, plant-derived antifungal metabolites have shown a potential to inhibit Sclerotinia, and their expression could be manipulated for enhanced resistance. For example, Ranjan et al. (2019) demonstrated that metabolite extracts from the stems of enhanced resistance soybean lines had an antifungal activity and targeted pathogen ergosterol biosynthesis, disrupting enzymes involved in fungal lipid and sterol biosynthesis.
More recently, targeted gene editing of a Sclerotinia effector host target has shown potential as a new strategy to deliver increased resistance. In the study by Zhang et al. (2021), they found that Brassica napus has eight homologous copies of BnQCR8, the plant target of the Sclerotinia effector-like protein SsSSVP1 (Lyu et al., 2016). Using CRISPR/Cas9 to reduce the BnQCR8 copy number, they found that B. napus mutants with one or more edited copies displayed stronger resistance against S. sclerotiorum. The mutants also showed increased resistance to the related necrotrophic fungal pathogen Botrytis cinerea, which contains an SsSSVP1 homologue. This study highlights the power of editing host targets shared by pathogen effectors.
Numerous other genes have been implied in resistance responses against Sclerotinia, highlighting the arsenal of attack strategies employed by this pathogen over a diverse array of hosts (reviewed in Wang et al., 2019a). The challenge, therefore, if a targeted genetic manipulation approach is taken, is what host gene(s) will be the best targets, or, if the transgenic approach is undertaken, what transgenes will be the most effective. This may include the expression of antimicrobial proteins, the overexpression of specific defence pathways or key regulators, or employing RNA interference (RNAi) approaches to target key host or Sclerotinia processes (Andrade et al., 2016; McCaghey et al., 2019; van Esse et al., 2020; Xia et al., 2020).
Chemical Fungicides
In addition to cultural control, fungicides are widely used to control Sclerotinia. A range of fungicides are commercially available to prevent disease losses; however, the environmental drivers of disease can make the timing of application decisions difficult. The decision to apply a fungicide should also not only be made with the current harvest in mind but also aimed at reducing sclerotia load in the soil for future crops. The decision of whether to apply fungicides often becomes an economic trade-off between the cost of fungicide applications and the risk of disease (Figure 2).
Several different classes of fungicides with diverse modes of action are used globally to treat Sclerotinia on different crops. This includes anilinopyrimidines (inhibit methionine biosynthesis), methyl benzimidazole carbamates (MBCs) (inhibit cell division by disrupting microtubule formation), dicarboxamides (thought to inhibit osmotic signal transduction), demethylation inhibitors (DMIs) (inhibit membrane sterol biosynthesis and the development of functional cell walls), quinone outside Inhibitors (QoIs) or strobilurins (inhibit mitochondrial respiration), and succinate dehydrogenase inhibitors (SDHIs) (inhibit mitochondrial respiration) (reviewed in Peltier et al., 2012; Derbyshire and Denton-Giles, 2016). Not all are registered for use on all crops, and in some cases there are restrictions on the number of applications of certain fungicides per crop, per season. Several chemical classes have limited systemic movement in the plant, meaning they are not functional on non-treated plant tissues (Peltier et al., 2012).
In broad acre or flowering row crops such as canola or soybean, fungicides can reduce disease incidence, but the timing of application is critical and can be difficult to achieve. Plants typically become susceptible to infection once flowering commences. In most years, fungicides are targeted at the early flowering stage, but optimal timing varies depending on the season and is only economical when there is a moderate to high risk for Sclerotinia infection (Mueller et al., 2002; Peltier et al., 2012; Derbyshire and Denton-Giles, 2016; Willbur et al., 2019). The efficacy of fungicides greatly decreases if they are applied after symptoms become visible. Foliar fungicides applied during early flowering can provide some protection but are typically preventative rather than curative, and the decision to apply must be made early, before symptoms have developed (Peltier et al., 2012). The goals of fungicide application are to protect early petals, achieve maximum penetration of the product into the crop canopy, and protect potential infection sites. For regions with high disease levels or long-flowering periods, a second spray is often targeted at later flowering stages (Paulitz et al., 2015). Fungicide application during flowering constitutes a significant monetary investment, and because of the sporadic nature of the disease (variability in disease incidence from year-to-year, region-to-region, and field-to-field), routine prophylactic application of fungicides is uneconomical and undesirable. The long flowering time of canola in some growing regions such as Australia (up to 6 weeks) and the effective activity of preventative fungicides (up to 3 weeks) often necessitate more than one fungicide application. In some instances, high infection levels are observed in canola even after two foliar fungicide applications. This indicates the difficulty in controlling the disease and the importance of understanding the environmental triggers for more targeted controls.
Fungicides recommended for control of S. minor on peanut, for example, include aminopyridines, dicarboxamides, and SDHIs (Ryley et al., 2000; Smith et al., 2008). All the three compounds are considered preventive and therefore must be applied before the onset of disease. As with the control of S. sclerotiorum, several fungicide applications may be needed to provide continuous protection in years with conditions highly favourable to disease development (Ryley et al., 2000). For many growers, this is not economical, and most will make a maximum of two applications (Smith et al., 2008).
Several fungicides have been trialled for their ability to protect against Sclerotinia trifoliorum in pasture legumes but, similar to treatments for Sclerotinia sclerotiorum and Sclerotinia minor, they are often not considered cost-effective (Kandula et al., 2015). Controlling S. trifoliorum in alfalfa is problematic. Several studies have suggested that it can be controlled with monthly sprays of dicarboxamides (Hoveland et al., 1996; Sulc and Rhodes, 1997). More recently Frate and Long (2005) found that dicarboxamides reduced disease symptoms and increased biomass yields of alfalfa in fields with known disease risk.
In leafy vegetable crops such as lettuce, the accurate timing of fungicide application is needed for efficacious control. This has prompted the development of disease forecasting models (Clarkson et al., 2014). Strategic plant-targeted sprays of dicarboxamides can be effective in Sclerotinia control, resulting in 80–96% reduction in Sclerotinia lettuce drop disease (Vallalta and Porter, 2004). Drenching of plants at transplanting and fungicide applications at the early crop stage before canopy closure are critical for controlling mycelial growth and preventing lower leaf infections. However, in some countries, permitted fungicide options are limited, and the increasing pressure to produce vegetable crops with low pesticide inputs necessitates alternate options to be implemented. In lettuce, most of the other registered fungicide classes (MBCs, strobilurins, aminopyridines, DMIs) were found to be less effective than dicarboxamides under high disease pressure (Pung and O’Brien, 2001; Vallalta and Porter, 2004).
Because most major fungicide classes rely on a single mode of action, the chance for the development of resistance in pathogen populations might be considered high (Derbyshire and Denton-Giles, 2016). However, Sclerotinia appears to have a low propensity for resistance, as it is considered homothallic (the ability of a single spore to produce a sexually reproducing colony) or undergoes asexual reproduction (Aldrich-Wolfe et al., 2015). However, instances of resistance to MBCs and dicarboxamides have been reported globally, and strains with decreased sensitivity to SDHIs have been reported in France (Ma et al., 2009; Wang et al., 2015; Derbyshire and Denton-Giles, 2016; reviewed in Liu et al., 2021). It is, therefore, important to not rely on a single mode of action to control Sclerotinia. Furthermore, the selective pressure caused by the overuse of fungicides may create resistance in other non-target pathogen populations. Recently, Liu et al. (2021) assessed the DMI fungicide metconazole for its potential usefulness in Sclerotinia control. They found no cross-resistance between metconazole and representative fungicides of the MBC, dicarboxamide, and SDHI classes over the 22 S. sclerotiorum isolates they tested, concluding that metconazole, if registered, could be used in spray rotations.
Biological Control, Biopesticides, and Microbiome Manipulation
The high risk of losses as well as difficulty controlling Sclerotinia using traditional methods has driven research into alternative approaches. There is a significant body of research into potential biocontrols for Sclerotinia spp. Several different species of fungi, such as Coniothyriumminitans and Trichoderma spp., and bacteria, such as Pseudomonas fluorescens, Bacillus spp., and Streptomyces spp. are known to be antagonistic to Sclerotinia spp. (Whipps and Gerlagh, 1992; De La Fuente et al., 2004; Gorgen et al., 2009; Luduena et al., 2012; Zeng et al., 2012b; Chen et al., 2016b; Smolinska and Kowalska, 2018). Importantly, biocontrols offer new modes of action and can be interchanged with chemical fungicides, thereby reducing the reliance on chemicals whilst also aiding to prolong chemical modes of action and reducing the development of resistant pathogen populations (Figure 2).
While many studies show the antagonism of Sclerotinia spp. in vitro with a range of fungi and bacteria, the efficacy needs to be proven in-planta and, critically, in field trials where performance is strongly influenced by environmental factors (Boland, 1997). Many biocontrol candidates that appear promising in the lab fail to show sufficient efficacy in the field, as their efficacy can be highly influenced by environmental conditions (Chitrampalam et al., 2008). The key to their success will depend on correct delivery and ensuring durability under multiple environments. This was clearly articulated in a study of a commercial product for the biocontrol of S. sclerotinia. Nicot et al. (2019) found differences in the susceptibly of 75 S. sclerotiorum isolates, highlighting the need to test biocontrols against pathogen species diversity panels and the possibility of biocontrol selection pressure on pathogen populations, as commercial biocontrol products become more widely adopted by growers. For Sclerotinia control, several different biocontrol species have been commercialised, and there are ongoing efforts to develop other promising candidates. Several commercial examples are discussed below.
Beneficial Fungi
Of the range of potential biocontrol agents for Sclerotinia, the fungal parasite Coniothyrium minitans is the best studied and commercially established. C. minitans was first isolated from infected sclerotia of S. sclerotiorum in 1947 (Tribe, 1957). The fungal predator attacks and degrades sclerotia in the soil (de Vrije et al., 2001). The commercially available biocontrol formulation of C. minitans is Contans WG (Bayer CropScience, Cambridge, United Kingdom). Many other Coniothyrium spp. are necrotrophic plant pathogens, but C. minitans appears to have lost the ability to infect plants and become specialised in the infection of sclerotia in the soil as food source (Whipps and Gerlagh, 1992). Sclerotia attacked by C. minitans will not produce apothecia and, therefore, will not produce ascospores that initiate the onset of disease symptoms (de Vrije et al., 2001). The efficacy of C. minitans in controlling Sclerotinia disease has mostly been shown on high value crops such as canola, sunflower, lettuce, cucumber, beans, and peanuts (de Vrije et al., 2001; Jones and Whipps, 2002; Chitrampalam et al., 2008; McQuilken and Chalton, 2009; Ojaghian, 2010). In field trials of C. minitans to control S. sclerotiorum across multiple crops, reductions of 10–70% in disease symptoms have been reported (Whipps and Gerlagh, 1992; Zeng et al., 2012a; reviewed in Derbyshire and Denton-Giles, 2016; Smolinska and Kowalska, 2018). Reductions in sclerotia development by as much as 95% have also been reported [soybean (Zeng et al., 2012a)]. Glasshouse trials for the protective effect of Contans WG in combination with low doses of a range of fungicides on bean (Phaseolus vulgaris, L.) showed that Contans WG applied with a low dose of a dicarboximide fungicide completely eliminated the formation of symptoms in infected plants (Elsheshtawi et al., 2017), demonstrating the potential role of Contans WG in an integrated pest management system.
There is sufficient evidence supporting the efficacy of C. minitans in supressing Sclerotinia spp. that the commercially available formulation of Contans WG is recommended as part of an integrated pest management approach in some parts of the world. It can be applied as a soil drench or applied to heavily diseased crop debris post-harvest before they are ploughed into the soil to destroy sclerotia and reduce the risk of infestation in future crops (de Vrije et al., 2001; Yang et al., 2010). Despite several reports of C. minitans suppressing Sclerotinia, its efficacy in the field is also reported as inconsistent and may be linked to varying susceptibility of different Sclerotinia strains (reviewed in Derbyshire and Denton-Giles, 2016; Kamal et al., 2016; Nicot et al., 2019).
Another fungal biocontrol example is the use of Trichoderma strains where many studies have shown that several Trichoderma spp. interfere with hyphal growth of Sclerotinia spp., parasitise sclerotia, and reduce apothecia abundance in laboratory studies (Woo et al., 2014). There are over 100 commercially registered biofungicide and/or growth-promoting products based on various Trichoderma spp. (reviewed in Woo et al., 2014), and there is still a considerable research activity in biocontrol using Trichoderma spp. Reports on Sclerotinia field efficacy are, however, limited (Smolinska and Kowalska, 2018). Positive examples include disease control on cabbage and legumes with reductions in sclerotia levels and/or up to a 50–65% decrease in disease symptoms (Zeng et al., 2012a; Geraldine et al., 2013; Jones et al., 2014).
Beneficial Bacteria
It is known that several different species of bacteria, many commonly isolated from healthy plant tissues, rhizospheres or bulk soils, suppress pathogens and promote plant growth. Most of these bacteria directly antagonise pathogens through the production of antimicrobial compounds or lytic enzymes, directly out-compete pathogens or sequester nutrients from them, or induce systemic disease resistance within the plant, helping it to fight off pathogen attacks. Some beneficial bacteria also promote plant fitness by producing plant hormone-mimics and antioxidant compounds (Berg and Hallmann, 2006; Narayanasamy, 2013; Rey and Dumas, 2017).
There are numerous studies demonstrating the suppression of Sclerotinia by Bacillus in vitro and exploring the mechanism of suppression (Li et al., 2009; Tonelli et al., 2010; Luduena et al., 2012; Figueredo et al., 2014; Vinodkumar et al., 2017; Ansary et al., 2018; Lopes et al., 2018; Massawe et al., 2018). The efficacy of Bacillus spp. in preventing and treating Sclerotinia disease in planta and in the field is not as well demonstrated. Examples of field evaluation include B. subtilis (BY2) and B. megaterium (A6) protecting canola in field studies, leading to statistically significant increases in grain yield (Hu et al., 2013, 2014). In three field trials run over two seasons, Kamal et al. (2015) showed the protection of canola against S. sclerotiorum with a control efficacy of 71–80% with two foliar applications of B. cereus (SC1-1). Treatment of soybeans with B. subtilis strains SB01 or SB24 in field trials produced significant decreases in disease severity (Zhang et al., 2011). Vinodkumar et al. (2017) showed that several strains of B. amyloliquefaciens and B. cereus could decrease symptoms of root rot in carnation by up to 88% when applied as a root dip prior to transplanting. In pot studies, several Bacillus spp. significantly reduced the disease severity of S. sclerotiorum in common beans and mustard plants (Sabate et al., 2018). However, more field scale studies to prove the efficacy of Bacillus-based products are needed to support recommendations of commercial use. There are several Bacillus amyloliquefaciens/subtilis strains marketed as biofungicide treatments for a range of fungal diseases, such as Sclerotinia [e.g., Serenade Optimum, (Bayer CropScience, St. Louis, MO, United States), Cease, (Bioworks Inc., Victor, NY, United States), and Amplitude, (Marrone Bio Innovations Inc., Davis, CA, United States)]. These products are either formulations of Bacillus cells or Bacillus cells and their spent fermentation media, or a complex mixture of plant-supportive biochemicals that the bacteria produce. Together, these products are registered for applications of Sclerotinia control across leafy and root vegetables, legumes, and oilseed crops.
Actinobacteria, including Streptomyces spp., are known to produce a range of antimicrobial secondary metabolites, such as many antibiotics used in human and veterinary medicine, and play important roles in soil and rhizosphere ecology (Demain, 2000). Many Streptomyces spp. form associations with plants, either by living as endophytes in the tissues of the plant or closely associating within the rhizosphere. This makes them promising targets for the development of agricultural products for disease control and plant growth promotion (Palaniyandi et al., 2013; Rey and Dumas, 2017). The commercial products Actinovate (Novozymes BioAg Ltd. Saskatoon, Canada) and Mycostop (Verdera Oy, Espoo, Finland) are formulations of Streptomyces lydicus WYEC 108 and Streptomyces griseoviridis K61, respectively. Applications for the Actinovate control of S. sclerotiorum or S. minor include soil applications for disease control on Brassica head and stem vegetables, leafy vegetables, and legume vegetables. Mycostop is not registered for Sclerotinia control, but studies suggest that it may offer some protection. In a field trial testing for the efficacy of several different biocontrol products in controlling S. sclerotiorumin soybean, Zeng et al. (2012a) showed that S. lydicus (applied as a soil drench of Actinovate) reduced the disease severity index by 30.8% and t the number of sclerotia in harvested soybeans by 93.8% relative to diseased control plots. Chen et al. (2016b) compared the efficacy of two Streptomyces strains, S. exfoliatusFT05W and S. cyaneus ZEA17I against Streptomyces lydicusWYEC108, isolated from Actinovate, for the control of S. sclerotiorum in lettuce. Under growth chamber conditions, they reported that all three strains provided some level of protection against disease, with S. lydicus outperforming the others. However, under field conditions, S. exfoliates FT05W and S. cyaneus ZEA17I reduced the disease incidence, whereas S. lydicusWYEC108 did not. This highlights the need to conduct disease assays under field conditions but also highlights that several other Streptomyces strains may have a potential for commercialisation as biocontrol products for the control of Sclerotinia disease.
Mycoviruses
Mycoviruses, or viruses of fungi, also show potential as new biocontrol agents for crop fungal pathogens, such as Sclerotinia (Xie and Jiang, 2014; Zhang et al., 2020). Sclerotinia can host various mycoviruses, such as ssRNA, dsRNA, and single-stranded circular DNA viruses (reviewed in Xie and Jiang, 2014). The DNA mycovirus S. sclerotiorum hypovirulence-associated DNA virus 1 (SsHADV-1) can infect and confer hypovirulence on S. sclerotorium (Yu et al., 2013). When leaves of Arabidopsis thaliana or Brassica napus were sprayed with SsHADV-1 viral particles followed by inoculation with S. sclerotiorum, the leaves displayed reduced lesion development. Further research on this mycovirus found that it could convert pathogenic S. sclerotiorum into a beneficial plant endophyte by downregulating the expression of key S. sclerotinia virulence genes such as those encoding cell-wall degrading enzymes or effector-like genes, for example, Ss-Cmu1, SsITL, and SsSSVP1 (Zhang et al., 2020). Interestingly, the SsHADV-1 infection of S. sclerotiorum not only affected the expression of Sclerotinia genes, but when the viral-infected Sclerotinia colonised plant tissues it also caused increased the expression of B. napus defence- and hormone-associated genes, coupled with enhanced plant growth (Zhang et al., 2020). The viral infection system was also tested in the field. Spraying hyphal fragments of a SsHADV-1 infected S. sclerotiorum strain in B. napus plants during early flowering reduced Sclerotinia stem rot disease severity by 30–67% (Zhang et al., 2020).
Biocontrol Consortia and Microbiome Management
Biocontrols are typically sold and applied as single microbial inoculants. There is, however, an increasing interest in the delivery of consortiums of beneficial microbes to promote plant health (Czajkowski et al., 2020). Studies on Sclerotinia are scarce, but there is evidence of the potential for this approach in disease control. For example, in pea, the co-delivery of T. harzianum, B. subtilis, and Pseudomonas aeruginosa resulted in reduced mortality when challenged with S. sclerotiorum (Jain et al., 2015). In another example, Bahkali et al. (2014) found the C. minitans, T. viride, and T. hamatum, applied as soil drenches, were more effective than S. griseoviridis, B. subtilis, or Pseudomonas fluorescens alone in S. sclerotiorum disease suppression in beans. However, the best observed results (100% disease suppression) were achieved with the combination of either C. minitans + S griseoviridis or T. hamatum + S. griseoviridis. This highlights the possibility that combinations of biocontrol agents that have differing modes of action may be more effective than any single biocontrol applied alone. While promising, this is an area that requires more study, as there are currently few studies that show the impact of combinations of biocontrol organisms.
The concept of delivering microbial consortia for plant health is moving beyond applying a handful of microbes and toward the delivery of optimised synthetic communities (French et al., 2021; Trivedi et al., 2021). This research area is still in its early stages, but several studies show promise for disease control. The challenge for application to control Sclerotinia diseases will be the establishment of consortia, potentially over a range of plant tissues, and their persistence over sometimes long crop growing periods, as is the case for many row crops. An alternate approach is to manipulate the crop microbiome toward one that supports plant health. There is evidence this can be achieved by modifying farm management practices (e.g., reducing agrichemical use, increasing crop diversity) (French et al., 2021). Again, this area of research is also in its infancy, and significant research gaps are needed to be addressed before this approach becomes a reality.
Biochemicals and Biologically Derived Pesticide Products
There is a significant and growing body of research exploring the ability of biologically derived pesticide products to inhibit the growth of plant pathogens, such as Sclerotinia. This includes, for example, the fermentation and production or extraction of antimicrobial compounds or lytic enzymes, plant growth promoters, or induced systemic resistance (ISR) inducing compounds (Figure 3; Trejo-Estrada et al., 1998; El-Tarabily et al., 2000; Conn et al., 2008; Prapagdee et al., 2008; Baharlouei et al., 2010; Alam et al., 2012; Froes et al., 2012; Mathys et al., 2012; Wu et al., 2015; Chen et al., 2016a; Vergnes et al., 2020). At this time, the vast majority of these studies are in the in-vitro stage, with few examples of pot-scale, in-planta studies, and even fewer progressing to field studies. However, most of these reports describe promising candidates that have shown the ability to inhibit the germination and growth of Sclerotinia species.
Another promising area of research is ribonucleic acid interference approaches for the exogenous application of ribonucleic acid molecules for fungal pathogen control (Das and Sherif, 2020; Kuo and Falk, 2020). The foliar application of double-stranded RNAs to the leaf surface of canola or Arabidopsis significantly decreased S. sclerotiorum disease development (McLoughlin et al., 2018). There were, however, differences in the efficacy of some molecules over the two plant species. The authors suggest that the discrepancies may be due to differences in leaf morphology or cell structure characteristics.
Future Perspectives
The severity of disease and crop losses across a broad range of horticultural and broad-acre farming sectors suggests that new and effective control measures need to be developed against Sclerotinia pathogens. This could include registration and/or development of fungicide chemistries with an increased window of preventative activity to minimise yield loss, and new modes of action to delay/prevent fungicide resistance. Increasing the understanding of how biocontrols work will facilitate optimisation of their formulation and delivery and provide insight into screening for more efficient/potent strains of commercial potential. Further, opportunities exist to combine chemical fungicides and biocontrols, or multiple biocontrols, to afford multiple modes of action and/or synergistic or additive effects. Together, these approaches can be incorporated with farming cultural practices in a coordinated attack on Sclerotinia. An understanding of how these approaches function together will be critical for durable disease control. Lastly, opportunities exist for the genetic modification of certain crops to attain complete disease resistance against Sclerotinia.
Author Contributions
CO’S and LT conceived the review topic and outline. CO’S, KB, and LT wrote the review. All the authors listed have made a substantial, direct, and intellectual contribution to the study and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Elaine Smith and Jonathan Anderson for their helpful discussions.
Footnotes
Funding. KB was supported by CSIRO Research Office Postdoctoral Fellowship.
References
- Agrios G. N. (2005). “chapter eleven - Plant diseases caused by fungi,” in Plant Pathology, Fifth Edn, ed. Agrios G. N. (San Diego: Academic Press; ), 385–614. 10.1016/b978-0-08-047378-9.50017-8 [DOI] [Google Scholar]
- Alam M., Dharni S., Abdul K., Srivastava S. K., Smad A., Gupta M. K. (2012). A promising strain of Streptomyces sp with agricultural traits for growth promotion and disease management. Indian J. Exper. Biol. 50 559–568. [PubMed] [Google Scholar]
- Aldrich-Wolfe L., Travers S., Nelson B. D., Jr. (2015). Genetic variation of Sclerotinia sclerotiorum from multiple crops in the North Central United States. PLoS One 10:e0139188. 10.1371/journal.pone.0139188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkooranee J. T., Aledan T. R., Ali A. K., Lu G., Zhang X., Wu J., et al. (2017). Detecting the hormonal pathways in oilseed rape behind induced systemic resistance by Trichoderma harzianum TH12 to Sclerotinia sclerotiorum. PLoS One 12:e0168850. 10.1371/journal.pone.0168850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim L., da Fonseca dos Santos R., Neto J. P. B., Guida-Santos M., Crovella S., Benko-Iseppon A. M. (2017). Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 18 335–351. 10.2174/1389203717666160619185308 [DOI] [PubMed] [Google Scholar]
- Amselem J., Cuomo J. C. A., van Kan J. A. L., Viaud M., Benito E. P., Couloux A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genetics 7:e1002230. 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade C. M., Tinoco M. L. P., Rieth A. F., Maia F. C. O., Aragão F. J. L. (2016). Host-induced gene silencing in the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Pathol. 65 626–632. 10.1111/ppa.12447 [DOI] [Google Scholar]
- Ansary M. W. R., Prince M., Haque E., Sultana F., West H. M., Rahman M. M., et al. (2018). Endophytic Bacillus spp. from medicinal plants inhibit mycelial growth of Sclerotinia sclerotiorum and promote plant growth. Z. Naturforsch. C J. Biosci. 73 247–256. 10.1515/znc-2018-0002 [DOI] [PubMed] [Google Scholar]
- Arahana V. S., Graef G. L., Specht J. E., Steadman J. R., Eskridge K. M. (2001). Identification of QTLs for resistance to Sclerotinia sclerotiorum in Soybean. Crop Sci. 41 180–188. 10.2135/cropsci2001.411180x [DOI] [Google Scholar]
- Baharlouei A., Sharifi-Sirchi G. R., Bonjar G. H. S. (2010). Identification of an antifungal chitinase from a potential biocontrol agent, Streptomyces plicatus Strain 101, and its new antagonistic spectrum of activity. Philippine Agric. Sci. 93 439–445. [Google Scholar]
- Bahkali A. H., Elsheshtawi M., Mousa R. A., Elgorban A. M., Alzaroaa A. A. (2014). Biological control of Sclerotinia sclerotiorum in beans with antagonistic microorganisms under greenhouse conditions. Res. Crops 1 884–892. 10.5958/2348-7542.2014.01426.0 [DOI] [Google Scholar]
- Barbetti M. J., You M. P. (2014). Opportunities and challenges for improved management of foliar pathogens in annual clover pastures across southern Australia. Crop Pasture Sci. 65 1249–1266. 10.1071/cp14117 [DOI] [Google Scholar]
- Bateman D. F., Beer S. V. (1965). Simultaneous production and synergistic action of oxalic acid and polygalacturonase during pathogenesis by Sclerotium rolfsii. Phytopathology 55 204–211. [PubMed] [Google Scholar]
- Bennett R. S., Chamberlin K. D., Damicone J. P. (2018). Sclerotinia blight resistance in the US peanut mini-core collection. Crop Sci. 58 1306–1317. 10.2135/cropsci2017.09.0591 [DOI] [Google Scholar]
- Berg G., Hallmann J. (2006). “Control of plant pathogenic fungi with bacterial endophytes,” in Soil Biology, eds Schulz B., Boyle C., Sieber T. N. (Berlin Heidelberg: Springer-Verlag; ), 53–69. 10.1007/3-540-33526-9_4 [DOI] [Google Scholar]
- Billon-Grand G., Rascle C., Droux M., Rollins J. A., Poussereau N. (2012). pH modulation differs during sunflower cotyledon colonization by the two closely related necrotrophic fungi Botrytis cinerea and Sclerotinia sclerotiorum. Mol. Plant Pathol. 13 568–578. 10.1111/j.1364-3703.2011.00772.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland G. J. (1997). Stability analysis for evaluating the influence of environment on chemical and biological control of white mold (Sclerotinia sclerotiorum) of bean. Biol. Control 9 7–14. 10.1006/bcon.1997.0515 [DOI] [Google Scholar]
- Bolton M. D., Thomma B. P., Nelson B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7 1–16. 10.1111/j.1364-3703.2005.00316.x [DOI] [PubMed] [Google Scholar]
- Chen C., Harel A., Gorovoits R., Yarden O., Dickman M. B. (2004). MAPK regulation of sclerotial development in Sclerotinia sclerotiorum is linked with pH and cAMP sensing. Mol. Plant Microbe Interact. 17 404–413. 10.1094/mpmi.2004.17.4.404 [DOI] [PubMed] [Google Scholar]
- Chen J., Ullah C., Reichelt M., Beran F., Yang Z. L., Gershenzon J., et al. (2020). The phytopathogenic fungus Sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat. Commun. 11:3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Bonaldi M., Erlacher A., Kunova A., Pizzatti C., Saracchi M., et al. (2016a). Biocontrol of two Streptomyces spp. strains against lettuce basal drop caused by Sclerotinia sclerotiorum. Phytopathology 106:7. [Google Scholar]
- Chen X., Pizzatti C., Bonaldi M., Saracchi M., Erlacher A., Kunova A., et al. (2016b). Biological control of lettuce drop and host plant colonization by rhizospheric and endophytic Streptomycetes. Front. Microbiol. 7:714. 10.3389/fmicb.2016.00714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu J., Lin G., Wang A., Wang Z., Lu G. (2013). Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Rep. 32 1589–1599. 10.1007/s00299-013-1469-3 [DOI] [PubMed] [Google Scholar]
- Chenault Chamberlin K. D., Melouk H. A., Payton M. E. (2010). Evaluation of the U.S. peanut mini core collection using a molecular marker for resistance to Sclerotinia minor Jagger. Euphytica 172 109–115. 10.1007/s10681-009-0065-7 [DOI] [Google Scholar]
- Chitrampalam P., Figuili P. J., Matheron M. E., Subbarao K. V., Pryor B. M. (2008). Biocontrol of ylettuce drop caused b Sclerotinia sclerotiorum and S. minor in Desert Agroecosystems. Plant Dis. 92 1625–1634. 10.1094/pdis-92-12-1625 [DOI] [PubMed] [Google Scholar]
- Clarkson J. P., Fawcett L., Anthony S. G., Young C. (2014). A Model for Sclerotinia sclerotiorum infection and disease development in lettuce, based on the effects of temperature, relative humidity and ascospore density. PLoS One 9:e94049. 10.1371/journal.pone.0094049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn V. M., Walker A. R., Franco C. M. M. (2008). Endophytic actinobacteria induce defense pathways in arabidopsis thaliana. Mol. Plant Microbe Interact. 21 208–218. 10.1094/mpmi-21-2-0208 [DOI] [PubMed] [Google Scholar]
- Cotton P., Kasza Z., Bruel C., Rascle C., Fèvre M. (2003). Ambient pH controls the expression of endopolygalacturonase genes in the necrotrophic fungus Sclerotinia sclerotiorum. FEMS Microbiol. Lett. 227 163–169. 10.1016/s0378-1097(03)00582-2 [DOI] [PubMed] [Google Scholar]
- Cruickshank A. W., Cooper M., Ryley M. J. (2002). Peanut resistance to Sclerotinia minor and S. sclerotiorum. Aust. J. Agric. Res. 53 1105–1110. 10.1071/ar02013 [DOI] [Google Scholar]
- Czajkowski R., Maciag T., Krzyzanowska D. M., Jafra S. (2020). “Biological Control Based on Microbial Consortia – From Theory to Commercial Products,” in How Research Can Stimulate the Development of Commercial Biological Control Against Plant Diseases, eds De Cal A., Melgarejo P., Magan N. (Cham: Springer International Publishing; ), 183–202. 10.1007/978-3-030-53238-3_12 [DOI] [Google Scholar]
- Das P. R., Sherif S. M. (2020). Application of exogenous dsRNAs-induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 11:946. 10.3389/fpls.2020.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente L., Thomashow L., Weller D., Bajsa N., Quagliotto L., Chernin L., et al. (2004). Pseudomonas fluorescens UP61 isolated from birdsfoot trefoil rhizosphere produces multiple antibiotics and exerts a broad spectrum of biocontrol activity. Eur. J. Plant Pathol. 110 671–681. 10.1023/b:ejpp.0000041569.35143.22 [DOI] [Google Scholar]
- de Vrije T., Antoine N., Buitelaar R. M., Bruckner S., Dissevelt M., Durand A., et al. (2001). The fungal biocontrol agent Coniothyrium minitans: production by solid-state fermentation, application and marketing. Appl. Microbiol. Biotechnol. 56 58–68. 10.1007/s002530100678 [DOI] [PubMed] [Google Scholar]
- Demain A. L. (2000). Small bugs, big business: The economic power of the microbe. Biotechnol. Adv. 18 499–514. 10.1016/s0734-9750(00)00049-5 [DOI] [PubMed] [Google Scholar]
- Denton-Giles M., Derbyshire M. C., Khentry Y., Buchwaldt L., Kamphuis L. G. (2018). Partial stem resistance in Brassica napus to highly aggressive and genetically diverse Sclerotinia sclerotiorum isolates from Australia. Can. J. Plant Pathol. 40 551–561. 10.1080/07060661.2018.1516699 [DOI] [Google Scholar]
- Derbyshire M., Denton-Giles M., Hegedus D., Seifbarghy S., Rollins J., Van Kan J., et al. (2017). The complete genome sequence of the Phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 9 593–618. 10.1093/gbe/evx030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire M., Mbengue M., Barascud M., Navaud O., Raffaele S. (2019). Small RNAs from the plant pathogenic fungus Sclerotinia sclerotiorum highlight host candidate genes associated with quantitative disease resistance. Mol. Plant Pathol. 20 1279–1297. 10.1111/mpp.12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire M. C., Denton-Giles M. (2016). The control of sclerotinia stem rot on oilseed rape (Brassica napus): current practices and future opportunities. Plant Pathol. 65 859–877. 10.1111/ppa.12517 [DOI] [Google Scholar]
- Dillard H. R. (1987). Sclerotinia rot of cabbage in: Cornell University, Fact sheet 730.30. Ithaca, NY: Cornell University. [Google Scholar]
- Ding L. N., Li T., Guo X. J., Li M., Liu X. Y., Cao J., et al. (2021). Sclerotinia stem rot resistance in rapeseed: recent progress and future prospects. J. Agric. Food Chem. 69 2965–2978. 10.1021/acs.jafc.0c07351 [DOI] [PubMed] [Google Scholar]
- Dokken-Bouchard F. L., Bassendowski K. A., Boyle T., Cowell L. E., Gugel R. K., Ippolito J., et al. (2010). Survey of canola diseases in Saskatchewan, 2009. Can. Plant Dis. Surv. 90 127–129. [Google Scholar]
- Dupont Pioneer Report (2012). “Economic impact of sclerotinia stem rot,” in Agronomy Sciences Research Summary 2012, Canadian Edition, eds Hacault K., Faust R., Butzen S., Jeschke M. (Johnston, IA: Dupont Pioneer; ), 34–37. [Google Scholar]
- Elsheshtawi M., Elkhaky M. T., Sayed S. R., Bahkali A. H., Mohammed A. A., Gambhir D., et al. (2017). Integrated control of white rot disease on beans caused by Sclerotinia sclerotiorum using Contans (R) and reduced fungicides application. Saudi J. Biol. Sci. 24 405–409. 10.1016/j.sjbs.2016.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Tarabily K. A., Soliman M. H., Nassar A. H., Al-Hassani H. A., Sivasithamparam K., Mckenna F., et al. (2000). Biological control of Sclerotinia minor using a chitinolytic bacterium and actinomycetes. Plant Pathol. 49 573–583. 10.1046/j.1365-3059.2000.00494.x [DOI] [Google Scholar]
- Fernando W. G. D., Nakkeeran S., Zhang Y. (2004). Ecofriendly methods in combating Sclerotinia sclerotiorum (Lib.) de Bary. Recent Res. Devel. Environ. Biol. 1 329–347. [Google Scholar]
- Figueredo M. S., Tonelli M. L., Taurian T., Angelini J., Ibanez F., Valetti L., et al. (2014). Interrelationships between Bacillus sp CHEP5 and Bradyrhizobium sp SEMIA6144 in the induced systemic resistance against Sclerotium rolfsii and symbiosis on peanut plants. J. Biosci. 39 877–885. 10.1007/s12038-014-9470-8 [DOI] [PubMed] [Google Scholar]
- Frate C. A., Long R. F. (2005). Sclerotinia in alfalfa: Biology and control in the Central Valley”, in 35th California Alfalfa Symposium. Visalia, California, USA: University of California, Davis. [Google Scholar]
- French E., Kaplan I., Iyer-Pascuzzi A., Nakatsu C. H., Enders L. (2021). Emerging strategies for precision microbiome management in diverse agroecosystems. Nat. Plants 7 256–267. 10.1038/s41477-020-00830-9 [DOI] [PubMed] [Google Scholar]
- Froes A., Macrae A., Rosa J., Franco M., Souza R., Soares R., et al. (2012). Selection of a Streptomyces strain able to produce cell wall degrading enzymes and active against Sclerotinia sclerotiorum. J. Microbiol. 50 798–806. 10.1007/s12275-012-2060-2 [DOI] [PubMed] [Google Scholar]
- Garg H., Sivasithamparam K., Banga S. S., Barbetti M. J. (2008). Cotyledon assay as a rapid and reliable method of screening for resistance against Sclerotinia sclerotiorum in Brassica napus genotypes. Austr. Plant Pathol. 37, 106–111. 10.1071/AP08002 [DOI] [Google Scholar]
- Garg H., Atri C., Sandhu P. S.1, Kaur B., Renton B., Banga S. K., et al. (2010). High level of resistance to Sclerotinia sclerotiorum in introgression lines derived from hybridization between wild crucifers and the crop Brassica species B. napus and B. juncea. Field Crops Res. 117, 51–58. 10.1016/j.fcr.2010.01.013 [DOI] [Google Scholar]
- Geraldine A. M., Lopes F. A. C., Carvalho D. D. C., Barbosa E. T., Rodrigues A. R., Brandão R. S., et al. (2013). Cell wall-degrading enzymes and parasitism of sclerotia are key factors on field biocontrol of white mold by Trichoderma spp. Biol. Control 67 308–316. 10.1016/j.biocontrol.2013.09.013 [DOI] [Google Scholar]
- Girard V., Fèvre M., Bruel C. (2004). Involvement of cyclic AMP in the production of the acid protease Acp1 by Sclerotinia sclerotiorum. FEMS Microbiol. Lett. 237 227–233. 10.1111/j.1574-6968.2004.tb09700.x [DOI] [PubMed] [Google Scholar]
- Godoy G., Steadman J. R., Dickman M. B., Dam R. (1990). Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 37 179–191. 10.1016/0885-5765(90)90010-u [DOI] [Google Scholar]
- Gorgen C. A., Da Silveira A. N., Carneiro L. C., Ragagnin V., Lobo M. (2009). White mold control with mulch and Trichoderma harzianum 1306 on soybean. Pesquisa Agropecuaria Brasileira 44 1583–1590. [Google Scholar]
- Grau C. R., Dorrance A. E., Bond J., Russin J. S. (2004). “Fungal Diseases,” in Soybeans: Improvement, production and uses, 3 Edn, eds Boerma H. R., Specht J. E. (Madison, WI, USA: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; ), 679–763. [Google Scholar]
- Guyon K., Balague C., Roby D., Raffaele S. (2014). Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15:336. 10.1186/1471-2164-15-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyawali S., Harrington M., Durkin J., Horner K., Parkin I. A. P., Hegedus D. D., et al. (2016). Microsatellite markers used for genome-wide association mapping of partial resistance to Sclerotinia sclerotiorum in a world collection of Brassica napus. Mol. Breed. 36:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halimi E. S., Rowe D. E., Pratt R. G. (1998). Responses of alfalfa to stem-tip inoculations with five isolates of Sclerotinia trifoliorum. Crop Sci. 38 1179–1182. 10.2135/cropsci1998.0011183x003800050011x [DOI] [Google Scholar]
- Heard S., Brown N. A., Hammond-Kosack K. (2015). An interspecies comparative analysis of the predicted secretomes of the necrotrophic plant pathogens Sclerotinia sclerotiorum and Botrytis cinerea. Plos One 10:e0130534. 10.1371/journal.pone.0130534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoveland C. S., Durham R. G., Bouton J. H. (1996). No-till seeding of grazing-tolerant alfalfa as influenced by grass suppression, fungicide, and insecticide. J. Produc. Agric. 9 410–414. 10.2134/jpa1996.0410 [DOI] [Google Scholar]
- Hu X. J., Roberts D. P., Xie L. H., Maul J. E., Yu C. B., Li Y. S., et al. (2013). Bacillus megaterium A6 suppresses Sclerotinia sclerotiorum on oilseed rape in the field and promotes oilseed rape growth. Crop Protec. 52 151–158. 10.1016/j.cropro.2013.05.018 [DOI] [Google Scholar]
- Hu X. J., Roberts D. P., Xie L. H., Maul J. E., Yu C. B., Li Y. S., et al. (2014). Formulations of Bacillus subtilis BY-2 suppress Sclerotinia sclerotiorum on oilseed rape in the field. Biol. Control 70 54–64. 10.1016/j.biocontrol.2013.12.005 [DOI] [Google Scholar]
- Jain A., Singh A., Singh S., Singh H. B. (2015). Biological management of Sclerotinia sclerotiorum in pea using plant growth promoting microbial consortium. J. Basic Microbiol. 55 961–972. 10.1002/jobm.201400628 [DOI] [PubMed] [Google Scholar]
- Jellis G. J., Smith D. B., Scott E. S. (1990). Identification of Sclerotinia spp. on Vicia faba. Mycol. Res. 94 407–409. 10.1016/s0953-7562(09)80366-3 [DOI] [Google Scholar]
- Jones E. E., Rabeendran N., Stewart A. (2014). Biocontrol of Sclerotinia sclerotiorum infection of cabbage by Coniothyrium minitans and Trichoderma spp. Biocontrol Sci. Technol. 24 1363–1382. [Google Scholar]
- Jones E. E., Whipps J. M. (2002). Effect of inoculum rates and sources of Coniothyrium minitans on control of Sclerotinia sclerotiorum disease in glasshouse lettuce. Eur. J. Plant Pathol. 108 527–538. [Google Scholar]
- Kabbage M., Yarden O., Dickman M. B. (2015). Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233 53–60. 10.1016/j.plantsci.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Kamal M. M., Lindbeck K. D., Savocchia S., Ash G. J. (2015). Biological control of sclerotinia stem rot of canola using antagonistic bacteria. Plant Pathol. 64 1375–1384. 10.1111/ppa.12369 [DOI] [Google Scholar]
- Kamal M. M., Savocchia S., Lindbeck K. D., Ash G. J. (2016). Biology and biocontrol of Sclerotinia sclerotiorum (Lib.) de Bary in oilseed Brassicas. Aust. Plant Pathol. 45 1–14. 10.1007/s13313-015-0391-2 [DOI] [Google Scholar]
- Kanbe M., Mizukami Y., Fujimoto F. (2002). Improvement of resistance to Sclerotinia crown and stem rot of alfalfa through phenotypic recurrent selection. Jarq-Japn. Agric. Res. Q. 36 1–5. 10.6090/jarq.36.1_1 [DOI] [Google Scholar]
- Kandula D. R. W., Jones E. E., Stewart A., Mclean K. L., Hampton J. G. (2015). Trichoderma species for biocontrol of soil-borne plant pathogens of pasture species. Biocontrol Sci. Technol. 25 1052–1069. 10.1080/09583157.2015.1028892 [DOI] [Google Scholar]
- Kasza Z., Vagvölgyi C., Févre M., Cotton P. (2004). Molecular characterization and in planta detection of Sclerotinia sclerotiorum endopolygalacturonase genes. Curr. Microbiol. 48 208–213. 10.1007/s00284-003-4166-6 [DOI] [PubMed] [Google Scholar]
- Khangura R., Beard C., Hills A. (2015). Managing sclerotinia stem rot in canola. South Perth: Department of Agriculture and Food, Government of Western Australia. [Google Scholar]
- Khangura R., Van Burgel A., Salam M., Aberra M., Wj Macleod W. (2014). Why Sclerotinia was so bad in 2013? Understanding the disease and management options. Bentley, Australia: Grains Industry Association of Western Australia Inc. [Google Scholar]
- Kim H. S., Diers B. W. (2000). Inheritance of partial resistance to Sclerotinia stem rot in soybean. Crop Sci. 40 55–61. 10.2135/cropsci2000.40155x [DOI] [Google Scholar]
- Kim K. S., Min J. Y., Dickman M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant-Microbe Interac. 21 605–612. 10.1094/mpmi-21-5-0605 [DOI] [PubMed] [Google Scholar]
- Kuo Y.-W., Falk B. W. (2020). RNA interference approaches for plant disease control. BioTechniques 69 469–477. 10.2144/btn-2020-0098 [DOI] [PubMed] [Google Scholar]
- Kurle J. E., Gran C. R., Oplinger E. S., Mengistu A. (2001). Tillage, crop sequence, and cultivar effects on Sclerotinia stem rot incidence and yield in soybean. Agron. J. 93 973–982. 10.2134/agronj2001.935973x [DOI] [Google Scholar]
- Laemmlen F. (2001). ”Sclerotinia Diseases”, in Publication 8042 University of California Agriculture and Natural Resources Communication Services. Oakland, CA: University of California. [Google Scholar]
- Lane D., Denton-Giles M., Derbyshire M., Kamphuis L. G. (2019). Abiotic conditions governing the myceliogenic germination of Sclerotinia sclerotiorum allowing the basal infection of Brassica napus. Aust. Plant Pathol. 48 85–91. 10.1007/s13313-019-0613-0 [DOI] [Google Scholar]
- Li J., Yang Q., Zhao L. H., Zhang S. M., Wang Y. X., Zhao X. Y. (2009). Purification and characterization of a novel antifungal protein from Bacillus subtilis strain B29. J. Zhejiang Univ. Sci. B 10 264–272. 10.1631/jzus.b0820341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Q., Zhao Z. K., Hayward A., Cheng H. Y., Fu D. H. (2015). Integration analysis of quantitative trait loci for resistance to Sclerotinia sclerotiorum in Brassica napus. Euphytica 205 483–489. 10.1007/s10681-015-1417-0 [DOI] [Google Scholar]
- Li R., Rimmer R., Buchwaldt L., Sharpe A. G., Séguin-Swartz G., Hegedus D. D. (2004). Interaction of Sclerotinia sclerotiorum with Brassica napus: cloning and characterization of endo- and exo-polygalacturonases expressed during saprophytic and parasitic modes. Fungal Genet. Biol. 41 754–765. 10.1016/j.fgb.2004.03.002 [DOI] [PubMed] [Google Scholar]
- Liang X., Moomaw E. W., Rollins J. A. (2015). Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol. Plant Pathol. 16 825–836. 10.1111/mpp.12239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. F., Rollins J. A. (2018). Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 108 1128–1140. 10.1094/phyto-06-18-0197-rvw [DOI] [PubMed] [Google Scholar]
- Link V. H., Johnson K. B. (2007). White mold (Sclerotinia). St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Lithourgidis A. S., Roupakias D. G., Damalas C. A. (2005). Inheritance of resistance to Sclerotinia stem rot (Sclerotinia trifoliorum) in faba beans (Vicia faba L.). Field Crops Res. 91 125–130. 10.1016/j.fcr.2004.06.007 [DOI] [Google Scholar]
- Liu F., Li X., Wang M., Wen J., Yi B., Shen J., et al. (2018). Interactions of WRKY15 and WRKY33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnol. J. 16 911–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Fu L., Chen J., Wang S., Liu J., Jiang J., et al. (2021). Baseline sensitivity of Sclerotinia sclerotiorum to metconazole and the analysis of cross-resistance with carbendazim, dimethachlone, boscalid, fluazinam, and fludioxonil. Phytoparasitica 49 123–130. 10.1007/s12600-020-00867-8 [DOI] [Google Scholar]
- Lopes K. B. D., Carpentieri-Pipolo V., Fira D., Balatti P. A., Lopez S. M. Y., Oro T. H., et al. (2018). Screening of bacterial endophytes as potential biocontrol agents against soybean diseases. J. Appl. Microbiol. 125 1466–1481. 10.1111/jam.14041 [DOI] [PubMed] [Google Scholar]
- Luduena L. M., Taurian T., Tonelli M. L., Angelini J. G., Anzuay M. S., Valetti L., et al. (2012). Biocontrol bacterial communities associated with diseased peanut (Arachis hypogaea L.) plants. Eur. J. Soil Biol. 53 48–55. 10.1016/j.ejsobi.2012.08.002 [DOI] [Google Scholar]
- Lyu X., Shen C., Fu Y., Xie J., Jiang D., Li G., et al. (2016). A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotiniasclerotiorum with its host plants. PLoS Pathog. 12:e1005435. 10.1371/journal.ppat.1005435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. X., Chen Y., Wang J. X., Yu W. Y., Tang Z. H., Chen C. J., et al. (2009). Activity of carbendazim, dimethachlon, iprodione, procymidone and boscalid against Sclerotinia stem rot in Jiangsu Province of China. Phytoparasitica 37:421. 10.1007/s12600-009-0057-0 [DOI] [Google Scholar]
- Magro P., Marciano P., Di Lenna P. (1988). Enzymatic oxalate decarboxylation in isolates ofSclerotinia sclerotiorum. FEMS Microbiol. Lett. 49 49–52. 10.1111/j.1574-6968.1988.tb02680.x [DOI] [Google Scholar]
- Marciano P., Di Lenna P., Magro P. (1983). Oxalic acid, cell wall-degrading enzymes and pH in pathogenesis and their significance in the virulence of two Sclerotinia sclerotiorum isolates on sunflower. Physiol. Plant Pathol. 22 339–345. 10.1016/s0048-4059(83)81021-2 [DOI] [Google Scholar]
- Markell S., Kandel H., Rio L. D., Halley S., Olson L., Mathew F., et al. (2009). Sclerotinia of Canola. Madison: North Dakota State University Extension Publication. [Google Scholar]
- Massawe V. C., Hanif A., Farzand A., Mburu D. K., Ochola S. O., Wu L. M., et al. (2018). Volatile compounds of endophytic Bacillus spp. have biocontrol activity against Sclerotinia sclerotiorum. Phytopathology 108 1373–1385. 10.1094/phyto-04-18-0118-r [DOI] [PubMed] [Google Scholar]
- Mathys J., De Cremer K., Timmermans P., Van Kerkhove S., Lievens B., Vanhaecke M., et al. (2012). Genome-wide characterization of ISR Induced in Arabidopsis thaliana by Trichoderma hamatum T382 against botrytis cinerea infection. Front. Plant Sci. 3:108. 10.3389/fpls.2012.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell D. P., Lumsden R. D. (1970). Oxalic acid production by Sclerotinia sclerotiorum in Infected bean and in culture. Phytopathology 60 1395–1398. 10.1094/phyto-60-1395 [DOI] [Google Scholar]
- Mbengue M., Navaud O., Peyraud R., Barascud M., Badet T., Vincent R., et al. (2016). Emerging trends in molecular interactions between plants and the broad host range fungal pathogens Botrytis cinerea and Sclerotinia sclerotiorum. Front. Plant Sci. 7:422. 10.3389/fpls.2016.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaghey M., Willbur J., Smith D. L., Kabbage M. (2019). The complexity of the Sclerotinia sclerotiorum pathosystem in soybean: virulence factors, resistance mechanisms, and their exploitation to control Sclerotinia stem rot. Trop. Plant Pathol. 44 12–22. 10.1007/s40858-018-0259-4 [DOI] [Google Scholar]
- McDonald M. R., Gossen B. D., Kora C., Parker M., Boland G. (2013). Using crop canopy modification to manage plant diseases. Eur. J. Plant Pathol. 135 581–593. 10.1007/s10658-012-0133-z [DOI] [Google Scholar]
- McLoughlin A. G., Wytinck N., Walker P. L., Girard I. J., Rashid K. Y., De Kievit T., et al. (2018). Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci. Rep. 8:7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilken M. P., Chalton D. (2009). Potential for biocontrol of Sclerotinia rot of carrot with foliar sprays of Contans WG (Coniothyrium minitans). Biocontrol Sci. Technol. 19 229–235. 10.1080/09583150802635549 [DOI] [Google Scholar]
- Mikaliuniene J., Lemeziene N., Danyte V., Suproniene S. (2015). Evaluation of red clover (Trifolium pratense L.) resistance to Sclerotinia crown and root rot (Sclerotinia trifoliorum) in the laboratory and field conditions. Zemdirbyste-Agric. 102 167–176. 10.13080/z-a.2015.102.022 [DOI] [Google Scholar]
- Mueller D. S., Hartman G. L., Pedersen W. L. (2002). Effect of crop rotation and tillage system on Sclerotinia stem rot on soybean. Can. J. Plant Pathol. 24 450–456. 10.1080/07060660209507033 [DOI] [Google Scholar]
- Murray G., Brennan J. (2012). The current and potential costs from diseases of oilseed crops in Australia. Kingston, Australia: Grains Research & Development Corporation. [Google Scholar]
- Mwape V. W., Khentry Y., Newman T. E., Denton-Giles M., Derbyshire M., Chen K., et al. (2021). Identification of sources of Sclerotinia sclerotiorum resistance in a collection of wild Cicer germplasm. Plant Dis. 10.1094/PDIS-02-21-0367-RE [Online ahead of print]. [DOI] [PubMed] [Google Scholar]
- Narayanasamy P. (2013). Biological management of diseases of crops. Volume 1 Characteristics of biological control agents. Dordrecht, NY: Springer. [Google Scholar]
- Nicot P. C., Avril F., Duffaud M., Leyronas C., Troulet C., Villeneuve F., et al. (2019). Differential susceptibility to the mycoparasite Paraphaeosphaeria minitans among Sclerotinia sclerotiorum isolates. Trop. Plant Pathol. 44 82–93. 10.1007/s40858-018-0256-7 [DOI] [Google Scholar]
- Njambere E. N., Peever T. L., Vandemark G., Chen W. (2014). Genotypic variation and population structure of Sclerotinia trifoliorum infecting chickpea in California. Plant Pathol. 63 994–1004. 10.1111/ppa.12176 [DOI] [Google Scholar]
- Noyes R. D., Hancock J. G. (1981). Role of oxalic acid in the Sclerotinia wilt of sunflower. Physiol. Plant Pathol. 18 123–132. 10.1016/s0048-4059(81)80033-1 [DOI] [Google Scholar]
- Ojaghian M. R. (2010). Biocontrol of potato white mold using coniothyrium minitans and resistance of potato cultivars to Sclerotinia sclerotiorum. Plant Pathol. J. 26 346–352. 10.5423/ppj.2010.26.4.346 [DOI] [Google Scholar]
- Palaniyandi S. A., Yang S. H., Zhang L. X., Suh J. W. (2013). Effects of actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 97 9621–9636. 10.1007/s00253-013-5206-1 [DOI] [PubMed] [Google Scholar]
- Paulitz T., Schroeder K., Beard T. L. (2015). Sclerotinia stem rot or white mold of canola. Pullman, WA: Washington State University Extension. [Google Scholar]
- Peltier A. J., Bradley C. A., Chilvers M. I., Malvick D. K., Mueller D. S., Wise K. A., et al. (2012). Biology, yield loss and control of Sclerotinia stem rot of soybean. J. Integ. Pest Manage. 3 B1–B7. [Google Scholar]
- Prapagdee B., Kuekulvong C., Mongkolsuk S. (2008). Antifungal potential of extracellular metabolites produced by Streptomyces hygroscopicus against phytopathogenic fungi. Int. J. Biol. Sci. 4 330–337. 10.7150/ijbs.4.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pung H., O’Brien R. (2001). Integrated management of Sclerotinia disease in beans. Australia: Horticultural Australia Ltd. [Google Scholar]
- Ranjan A., Westrick N. M., Jain S., Piotrowski J. S., Ranjan M., Kessens R., et al. (2019). Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 17 1567–1581. 10.1111/pbi.13082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey T., Dumas B. (2017). Plenty is no plague: Streptomyces symbiosis with Crops. Trends Plant Sci. 22 30–37. 10.1016/j.tplants.2016.10.008 [DOI] [PubMed] [Google Scholar]
- Rollins J. A., Dickman M. B. (2001). pH signaling in Sclerotinia sclerotiorum: identification of a pacC/RIM1 homolog. Appl. Environ. Microbiol. 67 75–81. 10.1128/aem.67.1.75-81.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryley M. J., Kyei N. A., Tatnell J. R. (2000). Evaluation of fungicides for the management of sclerotinia blight of peanut. Aust. J. Agric. Res. 51 917–924. 10.1071/ar97123 [DOI] [Google Scholar]
- Sabate D. C., Brandan C. P., Petroselli G., Erra-Balsells R., Audisio M. C. (2018). Biocontrol of Sclerotinia sclerotiorum (Lib.) de Bary on common bean by native lipopeptide-producer Bacillus strains. Microbiol. Res. 211 21–30. 10.1016/j.micres.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Saharan G. S., Mehta N. (2008). Sclerotinia Diseases of Crop Plants: Biology, Ecology and Disease Management. Berlin: Springer. [Google Scholar]
- Seifbarghi S., Borhan M. H., Wei Y., Coutu C., Robinson S. J., Hegedus D. D. (2017). Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genom. 18:266. 10.1186/s12864-017-3642-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D., Smith D. L., Kabbage M., Roth M. G. (2021). Effectors of plant necrotrophic fungi. Front. Plant Sci. 12:687713. 10.3389/fpls.2021.687713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpfendorfer S., Heenan D. P., Kirkegaard J. A., Lindbeck K. D., Murray G. M. (2004). Impact of tillage on lupin growth and the incidence of pathogenic fungi in southern New South Wales. Aust. J. Exper. Agric. 44 53–56. 10.1071/ea03140 [DOI] [Google Scholar]
- Smith D. L., Garrison M. C., Hollowell J. E., Isleib T. G., Shew B. B. (2008). Evaluation of application timing and efficacy of the fungicides fluazinam and boscalid for control of Sclerotinia blight of peanut. Crop Protec. 27 823–833. 10.1016/j.cropro.2007.11.010 [DOI] [Google Scholar]
- Smolinska U., Kowalska B. (2018). Biological control of the soil-borne fungal pathogen Sclerotinia sclerotiorum - a review. J. Plant Pathol. 100 1–12. 10.1007/s42161-018-0023-0 [DOI] [Google Scholar]
- Sulc R. M., Rhodes L. H. (1997). Planting date, fungicide, and cultivar effects on Sclerotinia crown and stem rot severity in alfalfa. Plant Dis. 81 13–17. 10.1094/pdis.1997.81.1.13 [DOI] [PubMed] [Google Scholar]
- Tang L., Yang G., Ma M., Liu X., Li B., Xie J., et al. (2020). An effector of a necrotrophic fungal pathogen targets the calcium sensing receptor in chloroplasts to inhibit host resistance. Mol. Plant Pathol. 21 686–701. 10.1111/mpp.12922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A., Coventry E., Jones J. E., Clarkson J. P. (2015). Resistance to a highly aggressive isolate of Sclerotinia sclerotiorum in a Brassica napus diversity set. Plant Pathol. 64 932–940. [Google Scholar]
- Thatcher L. F., Anderson J. P., Singh K. B. (2005). Plant defence responses: what have we learnt from Arabidopsis? Funct. Plant Biol. 32 1–19. 10.1071/fp04135 [DOI] [PubMed] [Google Scholar]
- Thatcher L. F., Myers C. A., Pain N., O’sullivan C. A., Roper M. M. (2017). A Sclerotinia disease assay for screening flowering canola plants in controlled environments. Aust. Plant Pathol. 46 333–338. 10.1007/s13313-017-0486-z [DOI] [Google Scholar]
- Tian B., Xie J., Fu Y., Cheng J., Li B., Chen T., et al. (2020). A cosmopolitan fungal pathogen of dicots adopts an endophytic lifestyle on cereal crops and protects them from major fungal diseases. ISME J. 14 3120–3135. 10.1038/s41396-020-00744-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli M. L., Taurian T., Ibanez F., Angelini J., Fabra A. (2010). Selection and in vitro characterization of biocontrol agents with potential to protect peanut plants against fungal pathogens. J. Plant Pathol. 92 73–82. [Google Scholar]
- Tourneau D. L. (1979). Morphology, cytology, and physiology of Sclerotinia species in culture. Phytopathology 69:887. 10.1094/phyto-69-887 [DOI] [Google Scholar]
- Trejo-Estrada S. R., Sepulveda I. R., Crawford D. L. (1998). In vitro and in vivo antagonism of Streptomyces violaceusniger YCED9 against fungal pathogens of turfgrass. World J. Microbiol. Biotechnol. 14 865–872. [Google Scholar]
- Tribe H. T. (1957). On the parasitism of Sclerotinia trifoliorum by Coniothyrium minitans. Trans. Br. Mycol. Soc. 40 489–499. 10.1016/s0007-1536(57)80055-2 [DOI] [Google Scholar]
- Trivedi P., Mattupalli C., Eversole K., Leach J. E. (2021). Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol. 230 2129–2147. 10.1111/nph.17319 [DOI] [PubMed] [Google Scholar]
- Vallalta O., Porter I. J. (2004). Development of biological controls for Sclerotinia diseases of horticultural crops in Australasia. Australia: Horticultural Australia Ltd. [Google Scholar]
- van Esse H. P., Reuber T. L., Van Der Does D. (2020). Genetic modification to improve disease resistance in crops. New Phytol. 225 70–86. 10.1111/nph.15967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega R. R., Corsini D., Le Tourneau D. (1970). Nonvolatile organic acids produced by Sclerotinia sclerotiorum in synthetic liquid media. Mycologia 62 332–338. 10.2307/3757591 [DOI] [PubMed] [Google Scholar]
- Vergnes S., Gayrard D., Veyssière M., Toulotte J., Martinez Y., Dumont V., et al. (2020). Phyllosphere colonization by a soil Streptomyces sp. Promotes plant defense responses against fungal infection. Mol. Plant Microbe Interact. 33 223–234. 10.1094/mpmi-05-19-0142-r [DOI] [PubMed] [Google Scholar]
- Vinodkumar S., Nakkeeran S., Renukadevi P., Malathi V. G. (2017). Biocontrol potentials of antimicrobial peptide producing BaIcillus Species: multifaceted antagonists for the management of stem rot of carnation caused by Sclerotinia sclerotiorum. Front. Microbiol. 8:446. 10.3389/fmicb.2017.00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong T. D., Diers B. W., Hartman G. L. (2008). Identification of QTL for resistance to Sclerotinia stem rot in soybean plant introduction 194639. Crop Sci. 48 2209–2214. 10.2135/cropsci2008.01.0019 [DOI] [Google Scholar]
- Wang Y., Duan Y.-B., Zhou M.-G. (2015). Molecular and biochemical characterization of boscalid resistance in laboratory mutants of Sclerotinia sclerotiorum. Plant Pathol. 64 101–108. 10.1111/ppa.12246 [DOI] [Google Scholar]
- Wang Z., Ma L.-Y., Cao J., Li Y.-L., Ding L.-N., Zhu K.-M., et al. (2019a). Recent advances in mechanisms of plant defense to Sclerotinia sclerotiorum. Front. Plant Sci. 10:1314. 10.3389/fpls.2019.01314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Bao L. L., Zhao F. Y., Tang M. Q., Chen T., Li Y., et al. (2019b). BnaMPK3 is a key regulator of defense responses to the devastating plant pathogen Sclerotinia sclerotiorum in Oilseed Rape. Front. Plant Sci. 10:91. 10.3389/fpls.2019.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wan L., Xin Q., Chen Y., Zhang X., Dong F., et al. (2018). Overexpression of OsPGIP2 confers Sclerotinia sclerotiorum resistance in Brassica napus through increased activation of defense mechanisms. J. Exper. Bot. 69 3141–3155. 10.1093/jxb/ery138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F. M., Zhao H., Zhang Z., Kaloshian I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342 118–123. 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. X., Tan R. J., Zhang S. C., Collins P. J., Yuan J. Z., Du W. Y., et al. (2018). Integrating GWAS and gene expression data for functional characterization of resistance to white mould in soya bean. Plant Biotechnol. J. 16 1825–1835. 10.1111/pbi.12918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipps J. M., Gerlagh M. (1992). Biology of coniothyrium-minitans and its potential for use in disease biocontrol. Mycol. Res. 96 897–907. 10.1016/s0953-7562(09)80588-1 [DOI] [Google Scholar]
- Willbur J., Mccaghey M., Kabbage M., Smith D. L. (2019). An overview of the Sclerotinia sclerotiorum pathosystem in soybean: impact, fungal biology, and current management strategies. Trop. Plant Pathol. 44 3–11. 10.1007/s40858-018-0250-0 [DOI] [Google Scholar]
- Willetts H. J., Wong J. A. L. (1980). The biology of Sclerotinia sclerotiorum, S. trifoliorum, and S. minor with emphasis on specific nomenclature. Bot. Rev. 46 101–165. 10.1007/bf02860868 [DOI] [Google Scholar]
- Williams B., Kabbage M., Kim H.-J., Britt R., Dickman M. B. (2011). Tipping the Balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7:e1002107. 10.1371/journal.ppat.1002107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S. L., Ruocco M., Vinale F., Nigro M., Marra R., Nadia Lombardi N., et al. (2014). Trichoderma-based products and their wdespread use in agriculture. Open Mycol. J. 8 71–126. 10.2174/1874437001408010071 [DOI] [Google Scholar]
- Wu Y. C., Yuan J., Yaoyao E., Raza W., Shen Q. R., Huang Q. W. (2015). Effects of volatile organic compounds from Streptomyces albulus NJZJSA2 on growth of two fungal pathogens. J. Basic Microbiol. 55 1104–1117. 10.1002/jobm.201400906 [DOI] [PubMed] [Google Scholar]
- Xia S., Xu Y., Hoy R., Zhang J., Qin L., Li X. (2020). The notorious soilborne pathogenic fungus Sclerotinia sclerotiorum: An update on genes studied with mutant analysis. Pathogens 9:27. 10.3390/pathogens9010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Jiang D. (2014). New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 52 45–68. 10.1146/annurev-phyto-102313-050222 [DOI] [PubMed] [Google Scholar]
- Xu L., Li G., Jiang D., Chen W. (2018). Sclerotinia sclerotiorum: An evaluation of virulence theories. Annu. Rev. Phytopathol. 56 311–338. 10.1146/annurev-phyto-080417-050052 [DOI] [PubMed] [Google Scholar]
- Yang G., Tang L., Gong Y., Xie J., Fu Y., Jiang D., et al. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217 739–755. 10.1111/nph.14842 [DOI] [PubMed] [Google Scholar]
- Yang L., Li G. Q., Long Y. Q., Hong G. P., Jiang D. H., Huang H. C. (2010). Effects of soil temperature and moisture on survival of Coniothyrium minitans conidia in central China. Biol. Control 55 27–33. 10.1016/j.biocontrol.2010.06.010 [DOI] [Google Scholar]
- Yang X., Yang J., Li H., Niu L., Xing G., Zhang Y., et al. (2020). Overexpression of the chitinase gene CmCH1 from Coniothyrium minitans renders enhanced resistance to Sclerotinia sclerotiorum in soybean. Transgen. Res. 29 187–198. 10.1007/s11248-020-00190-2 [DOI] [PubMed] [Google Scholar]
- Yang X., Yang J., Wang Y., He H., Niu L., Guo D., et al. (2019). Enhanced resistance to sclerotinia stem rot in transgenic soybean that overexpresses a wheat oxalate oxidase. Transgen. Res. 28 103–114. 10.1007/s11248-018-0106-x [DOI] [PubMed] [Google Scholar]
- Young C. S., Clarkson J. P., Smith J. A., Watling M., Phelps K., Whipps J. M. (2004). Environmental conditions influencing Sclerotinia sclerotiorum infection and disease development in lettuce. Plant Pathol. 53 387–397. 10.1111/j.1365-3059.2004.01018.x [DOI] [Google Scholar]
- Young C. S., Werner C. P. (2012). Infection routes for Sclerotinia sclerotiorum in apetalous and fully petalled winter oilseed rape. Plant Pathol. 61 730–738. 10.1111/j.1365-3059.2011.02556.x [DOI] [Google Scholar]
- Yu X., Li B., Fu Y., Xie J., Cheng J., Ghabrial S. A., et al. (2013). Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc. Natl. Acad. Sci. USA. 110 1452–1457. 10.1073/pnas.1213755110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarinpanjeh N., Motallebi M., Zamani M. R., Ziaei M. (2016). Enhanced resistance to Sclerotinia sclerotiorum in Brassica napus by co-expression of defensin and chimeric chitinase genes. J. Appl. Genet. 57 417–425. 10.1007/s13353-016-0340-y [DOI] [PubMed] [Google Scholar]
- Zeng W., Kirk W., Hao J. (2012a). Field management of Sclerotinia stem rot of soybean using biological control agents. Biol. Control 60 141–147. 10.1016/j.biocontrol.2011.09.012 [DOI] [Google Scholar]
- Zeng W., Wang D., Kirk W., Hao J. (2012b). Use of Coniothyrium minitans and other microorganisms for reducing Sclerotinia sclerotiorum. Biol. Control 60 225–232. 10.1016/j.biocontrol.2011.10.009 [DOI] [Google Scholar]
- Zhang H., Xie J., Fu Y., Cheng J., Qu Z., Zhao Z., et al. (2020). A 2-kb mycovirus converts a pathogenic fungus into a beneficial endophyte for Brassica protection and yield enhancement. Mol. Plant 13 1420–1433. 10.1016/j.molp.2020.08.016 [DOI] [PubMed] [Google Scholar]
- Zhang J., Xue A. G., Morrison M. J., Meng Y. (2011). Impact of time between field application of Bacillus subtilis strains SB01 and SB24 and inoculation with Sclerotinia sclerotiorum on the suppression of Sclerotinia stem rot in soybean. Eur. J. Plant Pathol. 131 95–102. 10.1007/s10658-011-9790-6 [DOI] [Google Scholar]
- Zhang X., Cheng J., Lin Y., Fu Y., Xie J., Li B., et al. (2021). Editing homologous copies of an essential gene affords crop resistance against two cosmopolitan necrotrophic pathogens. Plant Biotechnol. J. 2021:13667. 10.1111/pbi.13667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Koopmann B., Ulber B., Von Tiedemann A. (2020). A global survey on diseases and pests in oilseed rape—current challenges and innovative strategies of control. Front. Agron. 2:590908. 10.3389/fagro.2020.590908 [DOI] [Google Scholar]