Abstract
A novel Pseudomonas, designated strain BBB001T, an aerobic, rod-shaped bacterium, was isolated from the rhizosphere of Nicotiana glauca in Las Palmas Gran Canaria, Spain. Genomic analysis revealed that it could not be assigned to any known species of Pseudomonas, so the name Pseudomonas palmensis sp. nov. was proposed. A 16S rRNA gene phylogenetic analysis suggested affiliation to the Pseudomonas brassicae group, being P. brassicae MAFF212427 T the closest related type strain. Upon genomic comparisons of both strains, all values were below thresholds established for differentiation: average nucleotide identity (ANI, 88.29%), average amino acid identity (AAI, 84.53%), digital DNA-DNA hybridization (dDDH, 35.4%), and TETRA values (0.98). When comparing complete genomes, a total of 96 genes present exclusively in BBB001T were identified, 80 of which appear associated with specific subsystems. Phenotypic analysis has shown its ability to assimilate glucose, potassium gluconate, capric acid malate, trisodium citrate, and phenylacetic acid; it was oxidase positive. It is able to produce auxins and siderophores in vitro; its metabolic profile based on BIOLOG Eco has shown a high catabolic capacity. The major fatty acids accounting for 81.17% of the total fatty acids were as follows: C16:0 (33.29%), summed feature 3 (22.80%) comprising C16:1 ω7c and C16:1 ω6c, summed feature 8 (13.66%) comprising C18:1 ω7c, and C18:1ω6c and C17:0 cyclo (11.42%). The ability of this strain to improve plant fitness was tested on tomato and olive trees, demonstrating a great potential for agriculture as it is able to trigger herbaceous and woody species. First, it was able to improve iron nutrition and growth on iron-starved tomatoes, demonstrating its nutrient mobilization capacity; this effect is related to its unique genes related to iron metabolism. Second, it increased olive and oil yield up to 30% on intensive olive orchards under water-limiting conditions, demonstrating its capacity to improve adaptation to adverse conditions. Results from genomic analysis together with differences in phenotypic features and chemotaxonomic analysis support the proposal of strain BBB001T (=LMG 31775T = NCTC 14418T) as the type strain of a novel species for which the name P. palmensis sp. nov is proposed.
Keywords: Pseudomonas palmensis, rhizobacteria, Nicotiana glauca, PGPB, iron mobilization, plant adaptation, drought, biostimulant
Introduction
Plant growth–promoting bacteria (PGPBs) are bacteria that colonize plant roots and promote plant growth. These bacteria live in the rhizosphere and play a very important role in plant fitness; in some species, they live in symbiosis with the plant, whereas in most of them, they establish a loose relationship colonizing the root surface. Plants often depend on these PGPBs to acquire nutrients, whereas in other cases, PGPB provides protection against certain pathogens, produces phytohormones such as auxins improving plant development, or helps the plant to get adapted to biotic and abiotic stresses such as herbivores, pathogenic microorganisms, or salinity (Barriuso et al., 2005; Delfin et al., 2015; Bano and Muqarab, 2017).
Nicotiana glauca is a plant species from the Solanaceae family, native from South America and naturalized in the Mediterranean area and in the Canary Islands. Able to colonize poor soils, N. glauca contains anabasine, an alkaloid that confers some toxicity, and indicates the existence of a complex secondary metabolism, inducible and probably related to defense and plant adaptation (Sinclair et al., 2004; DeBoer et al., 2009). Therefore, wild populations of N. glauca colonizing harsh environments will select the best candidates for adaptation and would be a good source for putative PGPBs (Ramos-Solano et al., 2010). Many different bacterial genera have been reported to be PGPBs, and among them, Pseudomonas strains are very abundant. They are Gram-negative flagellated bacteria, often known because of the pigment production; they play crucial roles in soil health and plant development (Kloepper, 1992) and affect plant growth (Van Loon, 2007). Among the mechanisms frequently used by Pseudomonas species to benefit plant growth are siderophore production (Mavrodi et al., 2001; Rasouli-Sadaghiani et al., 2014), phosphate solubilization (Anzuay et al., 2013), or stimulation of plant protection triggering induced systemic resistance (Bakker et al., 2007; Fatima and Anjum, 2017).
As part of an ongoing research, bacteria from the rhizosphere of N. glauca were isolated, and strain BBB001T was characterized. In this study, a full genetic analysis was conducted to investigate the phylogenetic relationship of strain BBB001T within the Pseudomonas genus by sequencing the whole genome and comparing with those of the most similar genomes within this genus. A polyphasic characterization was also carried out. In addition, two biological assays were performed to support its potential for agriculture, one to demonstrate its ability to improve iron nutrition in tomato and another one to increase olive production under water-limiting conditions.
Results
Phenotypic Characterization
Strain BBB001T isolated from the rhizosphere of N. glauca was classed as a Gram-negative, non–endospore-forming rod. Strain BBB001T grows on plate count agar (PCA) at 28°C forming circular colonies < 1 mm ∅, with smooth borders, opaque, yellowish, and creamy texture. In liquid culture (nutrient broth), color changes from pale yellow in the exponential growth phase to intense yellow on the stationary phase after 24 h at 28°C.
Temperature growth test shows that the strain BBB001T grows in 24 h from 4 to 40°C, although at low temperatures (4°C) it takes up to 72 h. As regards pH, strain BBB001T grows in a pH range from 6 to 10 and with a salt concentration from 2 to 6%. The optimal temperature, pH, and salinity of growth for strain BBB001T are as follows: 28°C, pH 8, and 0% salinity.
The API20 NE reveals its ability to assimilate several substrates, namely, glucose, potassium gluconate, capric acid, malate, trisodium citrate, and phenylacetic acid; it is oxidase positive (Table 1). Compared to Pseudomonas brassicae MAFF212427T and Pseudomonas laurentiana GSL-010T, strain BBB001T is able to degrade CAP, whereas P. brassicaeT cannot, and it is oxidase positive, whereas P. laurentianaT is not, sharing all other metabolic capabilities with the two type strains.
TABLE 1.
Characteristics that differentiate Pseudomonas palmensis sp. nov. from the type strains of the most closely related Pseudomonas species: Pseudomonas brassicae MAFF212427 T (Sawada et al., 2020) and Pseudomonas laurentiana GSL-010 T (Wright et al., 2018).
| Strain BBB001T | Pseudomonas brassicae MAFF 212427T | Pseudomonas laurentiana GSL-010T | |
| Cell size (μm) | 0.5−0.8 × 1.5−3.8 | 2.1 ± 0.4 × 1.2 ± 0.1 | 1.75−2.2 × 0.5−0.7 |
| Isolation source | Rhizosphere (Spain) | Diseased brocoli | Seawater (Canada) |
| Temperature range (optimal) (°C) | 4–40 (28) | 4–35 (27) | 10–37 (30) |
| NaCl optimal (%, w/v) | 0 | ND | 0.5 |
| Maximum NaCl (%, w/v) | 6 | ND | 3 |
| pH range | 6–10 | ND | 5–10 |
| pH optimum | 8 | ND | 7–7.5 |
| API20 NE | |||
| [CAP] | + | − | + |
| OX | + | + | − |
| Saturated fatty acid | |||
| C10:0 | − | − | 0.11 |
| C12:0 | 4.02 | 4.00 | 2.66 |
| C14:0 | 0.94 | − | 1.76 |
| C15:0 | − | _ | 0.42 |
| C16:0 | 33.29 | 28.10 | 32.29 |
| C17:0 | − | _ | 0.13 |
| C18:0 | 0.52 | _ | 0.18 |
| Unsaturated fatty acid | |||
| C16:1 ω 5c | − | _ | 0.11 |
| C17:1, ω 8c | − | _ | 0.10 |
| C18:1, ω 7c | − | _ | 8.30 |
| Branched fatty acid | |||
| C17:0 cyclo | 11.42 | 7.80 | 1.13 |
| C18:1 11 methyl ω7C | − | − | 0.35 |
| C19:0 cyclo ω8c | 0.81 | − | − |
| Hydroxy fatty acid | |||
| C8:0 3-OH | − | − | − |
| C10:0 3-OH | 9.96 | 4.00 | 3.83 |
| C11:0 3-OH | − | − | − |
| C12:0 2-OH | 4.23 | 2.60 | 4.18 |
| C12:0 3-OH | 4.35 | 3.90 | 3.66 |
| C12:1 3-OH | − | − | 0.07 |
| Summed Features* | |||
| 2 | − | − | − |
| 3 | 22.80 | 29.00 | 40.43 |
| 7 | − | − | 0.17 |
| 8 | 13.66 | 17.5 | 0 |
| Utilization of: | |||
| L-Phenylalanine | + | Unkown | − |
For API20 NE results obtained in this study, all three strains were negative for NO3, TRP, GLU, ADH, URE, ESC, GEL, PNPG, GLU, ARA, MNE, MAN, NAG, MAL, GNT, and ADI and positive for GNT, GLU, MLT, CIT, and PAC. Characteristics for API20 NE and biolog plates are scored as follows: − negative reaction; + positive reaction. Fatty acid profile expressed as% (FAME): − indicates absence of the fatty acid. Summed feature* represents two or three fatty acids that cannot be separated using the MIDI system. Summed feature 3 comprises C16:1 ω7c/C16:1 ω 6c; summed feature 8 comprises C18:1 ω 7c/C18:1 ω 6c.
According to BIOLOG ECO, strain BBB001T is able to degrade several carboxylic acids, among which it is worth noting malic acid, hydroxybutyric acid and glucosaminic acid, the latter can also be consumed as a source of nitrogen. Among sugars, glucose-1-phosphate, cellobiose, lactose, and N-acetyl glucosamine stand out. As a nitrogen source, strain BBB001T catabolizes the two amines (putrescine and phenyl ethylamine), although the consumption of some amino acids, such as serine and glycol-L-glutamic acid, is also remarkable (Supplementary Table 2).
The major fatty acids (>81.17% of the total fatty acids) are C16:0 (33.29%), summed feature 3 (22.80%) comprising C16:1 ω7c and C16:1 ω6c, summed feature 8 (13.66%) comprising C18:1 ω7c, and C18:1 ω6c and C17:0 cyclo (11.42%).
In addition, this strain resulted positive for auxins as for siderophores synthesis and negative for chitinases production.
In addition to API20 information, Table 1 summarizes available information from the most closely related Pseudomonas species, confirming differences for strain BBB001T. Interestingly, and different from BBB001T, “Pseudomonas qingdaonensis” strain JJ3T was not able to degrade D-galacturonic acid, and its temperature range runs from 4 to 37°C, whereas BBB001T runs from 4 to 40°C (Sawada et al., 2020).
Phylogenetic Analysis
The genome of the BBB001T strain was analyzed with the tools available at EzBioCloud. First, EzBioCloud’s identification service provides proven similarity-based searches against quality-controlled databases of 16S rRNA sequences. After that, the ANI (average nucleotide identity), AAI (average amino acid identity), dDDH, and TETRA analysis were performed. Type (Strain) Genome Server (TYGS) was used to analyze the complete genome.
Estimates of the Degree of Similarity Between Genomes
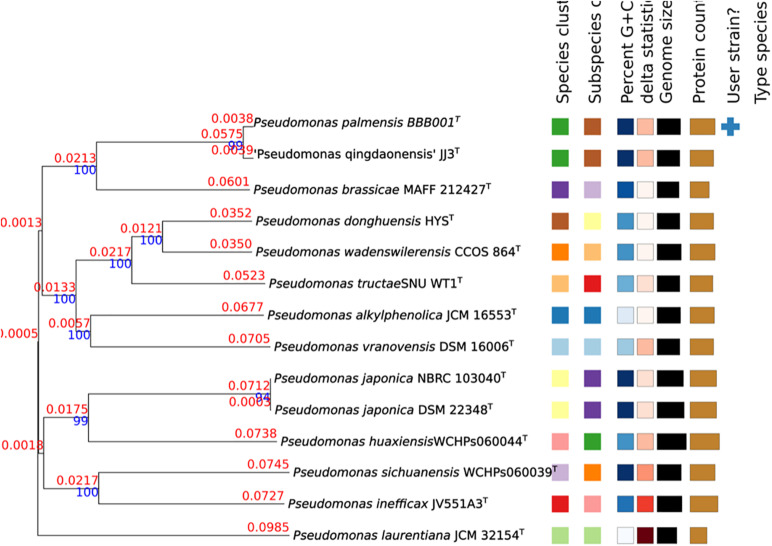
(a) Phylogenetic analysis and delimitation of species based on the analysis of 16S. The size of 16S rRNA gene was 1,541 bp (MW009702). The analysis based on 16S rRNA gene indicated that the strain BBB001T shared the highest similarity with P. qingdaonensis JJ3T (100%, non-validated taxon), P. brassicae MAFF212427T (99.85%), and Pseudomonas defluvii WCHP16T (99.31%), whereas P. laurentiana GSL-010T, Pseudomonas japonica NBRC 103040T, and Pseudomonas huaxiensis WCHPS060044T were between 99.00 and 99.1% (Table 2). A phylogenetic tree was constructed based on comparison of the most similar 16S rRNA gene sequences available in the TYGS database (Figure 1). Three different groups appeared in the tree: group 1 in which Pseudomonas japonica NBRC 103040T, Pseudomonas sinwaenesis WCHPs060039T, and Pseudomonas ineficax JV551A3T were located; group 2, represented by Pseudomonas alkylphenolica KL28T, Pseudomonas huaxinesis WCHPs060044T, Pseudomonas vranoviensis CCM 7279T, Pseudomonas donghuensis HYST, Pseudomonas wandersilveriensis CCOS 864T, and Pseudomonas tructae SNU WT1T; finally group 3, involving P. qingdaonensis JJ3(T), strain BBB001, P. brassicae MAFF212427T, P. laurentiana GSL-010T, and Pseudomonas akapagensis PS24T. In the third group, P. laurentiana GSL-010T and P. akapagensis PS24T separated from the other three, showing that P. qingdaonensis JJ3(T) and strain BBB001 grouped together, separate from P. brassicae MAFF212427T, which is the closest phylogenetically related strain.
TABLE 2.
Similarity of the 16S rRNA of the 30 taxa with valid names and strain BBB001T.
| Rank | Name | Type Strain (T) | Accession | Pairwise Similarity(%) | Authors |
| 1 | Pseudomonas qingdaonensis’ | JJ3 | PHTD01000020 | 100.0 | Wang et al., 2019 |
| 2 | Pseudomonas brassicae | MAFF 212427 | LC514379 | 99.9 | Sawada et al., 2020 |
| 3 | Pseudomonas defluvii | WCHP16 | KY979145 | 99.3 | Qin et al., 2020 |
| 4 | Pseudomonas laurentiana | GSL-010 | KY471137 | 99.1 | Wright et al., 2018 |
| 5 | Pseudomonas japonica | NBRC 103040 | BBIR01000146 | 99.0 | Pungrasmi et al., 2008 |
| 6 | Pseudomonas huaxiensis | WCHPs060044 | MH428812 | 99.0 | Qin et al., 2019a |
| 7 | Pseudomonas alkylphenolica | KL28 | CP009048 | 98.9 | Mulet et al., 2015 |
| 8 | Pseudomonas graminis | DSM 11363 | Y11150 | 98.8 | Behrendt et al., 1999 |
| 9 | Pseudomonas donghuensis | HYS | AJJP01000212 | 98.8 | Gao et al., 2015 |
| 10 | Pseudomonas sichuanensis | WCHPs060039 | QKVM01000121 | 98.8 | Qin et al., 2019b |
| 11 | Pseudomonas rhizosphaerae | DSM 16299 | CP009533 | 98.6 | Peix et al., 2003 |
| 12 | Pseudomonas wadenswilerensi | CCOS 864 | LT009706 | 98.6 | Frasson et al., 2017 |
| 13 | Pseudomonas plecoglossicida | NBRC 103162 | BBIV01000080 | 98.6 | Nishimori et al., 2000 |
| 14 | Pseudomonas wanovensis | CCM 7279 | AY970951 | 98.6 | Tvrzova et al., 2006 |
| 15 | Pseudomonas asiatica | RYU5 | MH517510 | 98.6 | Tohya et al., 2019a |
| 16 | Pseudomonas taiwanensis | BCRC 17751 | EU103629 | 98.6 | Wang et al., 2010 |
| 17 | Pseudomonas putida | NBRC 14164 | AP013070 | 98.5 | Trevisan, 1889; Migula, 1895 |
| 18 | Pseudomonas inefficax | JV551A3 | OPYN01000008 | 98.5 | Keshavarz-Tohid et al., 2019 |
| 19 | Pseudomonas asplenni | ATCC 23835 | LT629777 | 98.4 | Ark and Tompkins, 1946; Savulescu, 1947 |
| 20 | Pseudomonas monteilii | NBRC 103158 | BBIS01000088 | 98.4 | Elomari et al., 1997 |
| 21 | Pseudomonas tructae | SNU WT1 | CP035952 | 98.4 | Oh et al., 2019 |
| 22 | Pseudomonas coleopterorum | Esc2Am | KM888184 | 98.4 | Menéndez et al., 2015 |
| 23 | Pseudomonas reidholzensis | CCOS 865 | LT009707 | 98.4 | Frasson et al., 2017 |
| 24 | Pseudomonas juntendi | BML3 | MK680061 | 98.4 | Tohya et al., 2019b |
| 25 | Pseudomonas cremoricolorata | 1AM 1541 | AB060137 | 98.3 | Uchino et al., 2001 |
| 26 | Pseudomonas entomophila | L48 | CT573326 | 98.3 | Mulet et al., 2012 |
| 27 | Pseudomonas mosselii | CIP 105259 | AF072688 | 98.3 | Dabboussi et al., 2002 |
| 28 | Pseudomonas moorei | RW10 | AM293566 | 98.3 | Cámara et al., 2007 |
| 29 | Pseudomonas lutea | DSM 17257 | JRMB01000004 | 98.2 | Peix et al., 2004 |
| 30 | Pseudomonas helmanticensis | OHA11 | HG940537 | 98.2 | Ramírez-Bahena et al., 2014 |
| 31 | Pseudomonas baetica | a390 | FM201274 | 98.2 | López et al., 2012 |
In column 1, accession number is indicated after the species name.
FIGURE 1.
Tree inferred with FastME 2.1.6.1 (Lefort et al., 2015) from GBDP distances calculated from 16s rRNA gene sequences. The branch lengths are scaled in terms of GBDP distance formula d5. The blue numbers are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 80.9%. The branch length values (in red) represent the evolutionary time between two nodes. Unit: substitutions per sequence site. The tree was rooted at the midpoint (Farris, 1972).
(b) The highest values of ANI and AAI (88.29 and 84.83%, respectively) were obtained with the genome of the species P. brassicae MAFF212427T. The highest value of dDDH (35.4%) and the lower intergenomic distance (0.11) was obtained when comparing strain BBB001T and P. brassicae MAFF212427T (Table 1). G + C content shows a difference of 0.65% between genomes of these two strains. TETRA values were between 0.86 and 0.988, not showing a significant resemblance between the genome of strain BBB001T and the other 24 genomes (Table 3). Comparison with the genome of P. qingdaonensis JJ3(T) (non-validated taxon) showed the highest values (Table 3).
TABLE 3.
Similarity of the 24 genomes with strain BBB001T genome.
| Rank | Name | genome accesion number | ANI% | AAI% | dDDH | Intergenomic distance | Difference G + C (%) | TETRA |
| 1 | Pseudomonas qingdaonensis’ JJ3(T) | PHTD00000000.1 | 99.2 | 95.0 | 93.4 | 0.0855 | 0.14 | 0.9998 |
| 2 | Pseudomonas brassicae MAFF 212427(T) | LC514391.1 | 88.3 | 84.5 | 35.4 | 0.1167 | 0.65 | 0.9880 |
| 3 | Pseudomonas laurentiana JCM 32154(T) | JAAHBT000000000.1 | 81.6 | 81.0 | 25.1 | 0.1732 | 4.35 | 0.8680 |
| 4 | Pseudomonas japonica NBRC 103040(T) | NZ BBIR00000000.1 | 83.6 | 78.0 | 26.7 | 0.1618 | 0.10 | 0.9600 |
| 5 | Pseudomonas alkylphenolica KL28(T) | NZ CP009048.1 | 83.4 | 80.0 | 27.1 | 0.1597 | 3.43 | 0.9384 |
| 6 | Pseudomonas graminis DSM 11363(T) | NZ FOHW00000000.1 | 77.9 | 72.5 | 22.5 | 0.1951 | 3.8 | 0.9040 |
| 7 | Pseudomonas donghuensis HYS(T) | NZ AJJP00000000.1 | 84.2 | 78.0 | 27.7 | 0.1554 | 1.64 | 0.9699 |
| 8 | Pseudomonas sichuanensis WCHPs060039(T) | NZ QKVM00000000.1 | 82.0 | 76.8 | 25.3 | 0.1716 | 0.29 | 0.9840 |
| 9 | Pseudomonas rhizosphaerae DSM 16299 (T) | CP009533.1 | 79.7 | 72.6 | 23.5 | 0.1859 | 2.06 | 0.9550 |
| 10 | Pseudomonas wadenswilerensis CCOS 864(T) | NZ UIDD00000000.1 | 84.1 | 78.8 | 27.7 | 0.1556 | 1.67 | 0.9680 |
| 11 | Pseudomonas plecoglossicida NBRC 103162(T) | NZ BBIV00000000.1 | 81.9 | 74.9 | 25.1 | 0.1732 | 1.26 | 0.9820 |
| 12 | Pseudomonas vranovensis CCM 7279(T) | NZ AUED00000000.1 | 83.3 | 81.1 | 26.6 | 0.1630 | 2.53 | 0.9610 |
| 13 | Pseudomonas asiatica RYU5(T) | NZ BUF01000001.1 | 81.9 | 77.0 | 25.3 | 0.1718 | 1.24 | 0.9760 |
| 14 | Pseudomonas taiwanensis BCRC 17751(T) | NZ AUEC00000000.1 | 81.0 | 77.9 | 24,00 | 0.1823 | 2.19 | 0.9610 |
| 15 | Pseudomonas putida NBRC 14164(T) | NC 021505.1 | 81.4 | 76.3 | 25,00 | 0.1745 | 1.72 | 0.9700 |
| 16 | Pseudomonas inefficax JV551A3(T) | OPYN00000000.1 | 81.8 | 75.8 | 25.5 | 0.1706 | 1.21 | 0.9718 |
| 17 | Pseudomonas monteilii NBRC 103158(T) | NZ BBIS00000000.1 | 81.5 | 77.1 | 24.7 | 0.1768 | 2.37 | 0.9670 |
| 18 | Pseudomonas coleopterorum LMG 28558(T) | FNTZ00000000.1 | 79.6 | 74.3 | 23.4 | 0.1866 | 2.17 | 0.9542 |
| 19 | Pseudomonas reidholzensis CCOS 865(T) | UNOZ00000000 | 82.1 | 76.2 | 25,00 | 0.1741 | 0.03 | 0.9700 |
| 20 | Pseudomonas cremoricolorata 1AM 1541(T) | NZ AUEA00000000.1 | 80.7 | 73.9 | 23.6 | 0.1856 | 0.55 | 0.9531 |
| 21 | Pseudomonas mosselii CIP 105259(T) | NZ MRVJ00000000.1 | 82.3 | 77.4 | 25.4 | 0.1715 | 0.38 | 0.9810 |
| 22 | Pseudomonas moorei DSM 12647(T) | NZ FNKJ01000003.1 | 79.4 | 72.8 | 23.6 | 0.1850 | 4.40 | 0.9120 |
| 23 | Pseudomonas lutea DSM 17257(T) | JRMB00000000.1 | 77.4 | 74.4 | 22.5 | 0.1946 | 3.91 | 0.8920 |
| 24 | Pseudomonas baetica a390(T) | PKLC00000000.1 | 78.8 | 73.4 | 22.9 | 0.1916 | 5.30 | 0.8681 |
For each genome, its length [in base pairs (bp)], number of annotated genes, and the values of ANI, AAI, dDDH, intergenomic distance, G + C difference, and TETRA index estimated in the comparison with the genome of Pseudomonas BBB001T. In column 1, the genome accession number is indicated after the species name in RefSeq.
Phylogenetic Analyses Based on Complete Genome Sequences
The genome of strain BBB001T 1 (WGS: JAFKEC000000000) was compared against all type strain genomes available in the TYGS database via the MASH algorithm, a fast approximation of intergenomic relatedness (Ondov et al., 2016), and the 10 type strains with the smallest MASH distances chosen per user genome resulting in a phylogenetic tree (Figure 2).
FIGURE 2.
Phylogenetic tree constructed in TYGS. Tree inferred with FastME 2.1.6.1 (Lefort et al., 2015) from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5. The numbers above the branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 94.3%. The branch length values represent the evolutionary time between two nodes. Unit: substitutions per sequence site. The tree was rooted at the midpoint (Farris, 1972).
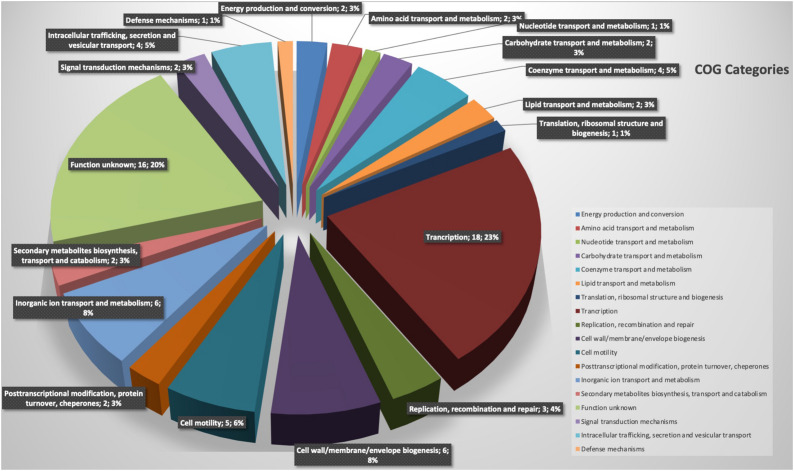
The comparative analysis of the complete genomes identified a total of 79 genes present exclusively in strain BBB001T (Supplementary Table 2). Among these genes (Figure 3), transcription-related genes were the most abundant (18) representing 23% or these unique genes; 16 genes were not associated with specific subsystems (20%); 6 were associated with cell wall, membrane, and envelope synthesis (8%); and 6 (8%) were related to inorganic transport and metabolism (eggnog-emapper). Further analysis was carried out on genes related to inorganic transport and metabolism, and two genes related to the uptake and iron metabolism were identified by blast in antiSMASH 5.02 secondary metabolism database; this is consistent with data from phenotypic characterization.
FIGURE 3.
Representation of functional categories of novel regions of strain BBB001T.
Biological Assays
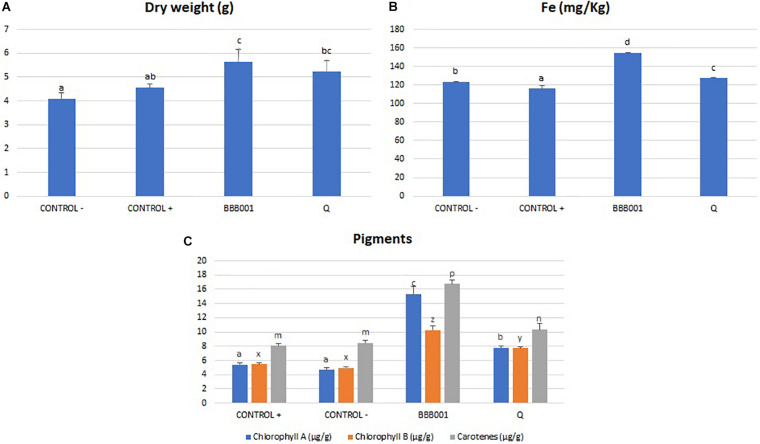
The biological assay on iron-starved tomato was conducted to proof the biological effect associated with iron absorption (Fe3+) and metabolism unique genes of this strain (Figure 3). Data showed that both strain BBB001T and its culture media free of bacteria (chelate) supplemented with non-soluble iron were able to revert chlorosis due to iron deficiency, according to visual evaluation (data not shown). Strain BBB001T significantly increased dry weight (Figure 4A), Fe concentration (Figure 4B), and photosynthetic pigments (chlorophylls and carotenes, Figure 4C), against the positive control (Fe-EDTA) and the negative control. The chelate produced by the bacteria significantly increased all parameters, except the increase in dry weight, which was non-significant; increases were not as marked as with the bacteria (Figure 4).
FIGURE 4.
(A) Dry weight of tomato plants (g). (B) Iron plant content (mg/kg). (C) Pigment concentration (mg/g) on tomato leaves. Different letters denote statistically significant differences according to LSD test (p < 0.05) for chlorophyl a (a, b, and c), chlorophyl b (x, y, and z), and carotenes (m, n, and p).
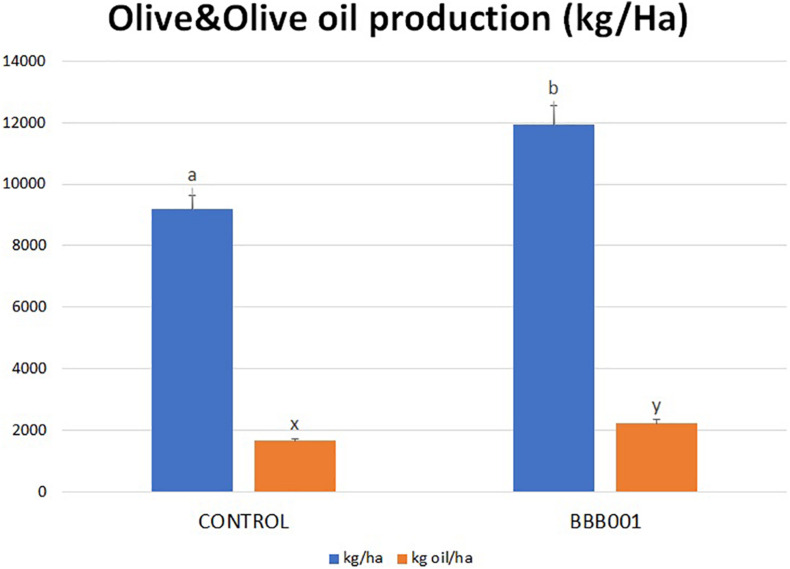
The biological assay in the intensive olive orchard was conducted aiming to demonstrate the ability of the strain to improve plant adaptation to stress situations, decreasing water input by 25% of regular watering. Results showed a statistically significant increase in olive yield up to 30% (kg/ha), as well as in oil production (Figure 5). Interestingly, the production achieved with strain BBB001T under water-limiting conditions (12,000 kg/ha) reached similar values to controls under regular water regime (data not shown).
FIGURE 5.
Olive and olive oil production of Olea europaea trees (kg/ha). Different letters denote statistically significant differences according to LSD test (p < 0.05) for olive yield (a, b), oil yield (x, y).
Discussion
Strain BBB001T isolated from the rhizosphere of N. glauca is able to improve plant yield in olive trees under water-limiting conditions and to efficiently supply iron to tomato under iron-limiting conditions, demonstrating its good potential for agriculture. Although the beneficial effects in plants have been described before for other Pseudomonas (Kloepper, 1992; Mavrodi et al., 2001; Bakker et al., 2007; Van Loon, 2007; Anzuay et al., 2013; Rasouli-Sadaghiani et al., 2014; Fatima and Anjum, 2017), the multiphasic genomic analysis has evidenced that this is a new strain with beneficial properties for agriculture, for which the name Pseudomonas palmensis sp. nov. has been proposed.
The ability of N. glauca to select the best strains to improve adaptation to harsh environments has been evidenced consistent with previous studies (Ramos-Solano et al., 2010). Its complex secondary metabolism, inducible and probably related to defense and plant adaptation (Sinclair et al., 2004; DeBoer et al., 2009), together with scarcity of nutrients, is key to select strains that play crucial roles in soil health and plant development. In line with this statement, strain BBB001T was positive for auxins and for siderophore production. The ability to produce auxins is very common and is related to increases in root surface, which provide plants with an enhanced capacity to absorb water and nutrients and therefore increased growth potential (Gutierrez Mañero et al., 1996).
The carbon source utilization fingerprint characteristic of strain BBB001T indicates a great adaptive capacity to survive in the rhizosphere of plants, quickly metabolizing organic acids and carbohydrates released by roots, which makes this strain a good root colonizer, one of the traits necessary for effective PGPBs (Posada et al., 2018). This is consistent with bioassay results, as field-grown olives evidence an increase in yield due to bacterial effects, probably due to an efficient colonization.
As there is an increasing number of PGPBs being described, many of which belong to Pseudomonas, a multiphasic approach to study its genome was taken to unravel the hidden information of this strain with such a good performance. First, analysis of the 16S rRNA revealed a high similarity (>99%) to five species with validated names. Sequencing of the 16S ribosomal gene (16S rRNA) is the basic tool in the current system classification of bacterial species. Generally, similarity values for the 16S rRNA gene less than 98.65 to 99% are accepted for different species (Kim et al., 2014). However, the 16S rRNA gene may offer limited resolution power as it shows a low capacity for discrimination between closely related species. Furthermore, some species may show very similar sequences of 16S rRNA (>99%), despite being clearly different according to DDH values (Ash et al., 1991; Rosselló-Móra and Amann, 2001). Consistently, ANI and AAI values confirmed differences to existing validated genomes. The values of ANI and AAI represent a robust measure of the evolutionary distance between genomes, as it has been empirically verified that ANI and AAI values of 95 to 96% equals a DDH value (DNA–DNA hybridization) of 70%, commonly used to delimit prokaryotic species (Konstantinidis and Tiedje, 2005; Richter and Rosselló-Móra, 2009; Kim et al., 2014). The closer related strain genomes will show similar frequencies of tetranucleotide use, with correlation indexes (TETRA) equal to or greater than 0.99 (Teeling et al., 2004; Richter and Rosselló-Móra, 2009). The highest values of ANI and AAI obtained in the comparison with the genome of the species P. brassicae were well below the 95% threshold established for considering that both genomes belong to the same species. The highest value of dDDH and the lower intergenomic distance were obtained when comparing strain BBB001T and P. brassicae MAFF212427T (Table 3), being the value of dDDH much lower than the 70% necessary to be able to assign strain BBB001T to this species. This result is also corroborated by TETRA values, which did not show a significant resemblance between the genome of strain BBB001T and the other genomes with values less than 0.99%. In the phylogenetic tree constructed in TYGS with complete genome data (Figure 2), strain BBB001T is closely related with P. brassicae MAFF212427T and P. qingdaonensis JJ3T. In this case, the analysis establishes a closer relationship between strain BBB001T and P. qingdaonensis JJ3T, which taxonomic name is not validated yet3. It is likely therefore that the repertoire of accessory genes of strain BBB001T is more like that of P. qingdaonensis JJ3T than that of P. brassicae MAFF212427T.
Comparison of the most similar complete genomes at Pan-seq revealed novel regions of this strain, a total of 96 genes were present exclusively in strain BBB001T, and 80 of these genes appear associated with specific subsystems. Among these genes (Figure 3), those related to the uptake and iron metabolism were found (Coulton et al., 1983; Braun, 2003; Miethke and Marahiel, 2007) and correlate with data from phenotypic characterization.
Biological assays were designed to evidence the potential of the strain for agriculture based on its unique genes related to iron metabolism on the one hand and of its general performance. The biological assay on iron-starved tomato was conducted based on information about unique genes of this strain related to iron absorption (Fe3+) and metabolism (Figure 3). Data showed that both BBB001T and its culture media free of bacteria (chelate) supplemented with non-soluble iron were able to revert chlorosis due to iron deficiency, according to visual evaluation, significantly increasing dry weight, iron concentration and photosynthetic pigments in iron-starved tomatoes, being the bacteria much more efficient than the chelate (Figure 4).
The differences between the bacteria and the culture media suggest that the increased iron mobilization is strongly improved when the bacteria are present, consistent with the multitarget response proposed for beneficial bacteria (Ilangumaran and Smith, 2017). In this type of response, bacteria improve iron mobilization probably supported by an increased root development that allows an increase in absorption surface (Gutierrez Mañero et al., 1996), a response attributed to auxin concentration, also present in the unique gene set of the strain. The beneficial effect on iron nutrition also shows an increase of photosynthetic pigments on the shoot, being especially striking the increase on chlorophyll a, the key player on light reactions as it is mainly located on photosystems. The increase in photosynthetic potential is evidenced on growth (dry weight), with the highest increase in dry weight on the bacteria treated plants, confirming that not only iron limits plant growth and supporting the better performance of bacteria in agriculture, where many environmental factors change along plant growth cycle.
The biological assay in the intensive olive orchard was conducted aiming to demonstrate the ability of the strain to improve plant adaptation to stress situations, especially the lack of water, as this situation becomes more and more frequent nowadays. The mechanisms involved in plant adaptation to stress are common to many other stressful situations such as salinity or biotic stress, through common signaling pathways (Martin-Rivilla et al., 2020). Plant adaptive mechanisms involve ionic homeostasis and modulation of redox balance among others (Kangasjärvi et al., 2014; Kollist et al., 2019). Olive yield (kg/ha) and oil production increased significantly (up to 30%) despite the 25% decrease of water input (Figure 4). The registered increase in oil was due to two factors: the increment in olive production and the increase in fat yield, reaching similar values to controls under regular water regimen (data not shown).
These results support the suitability of this strain to activate adaptive mechanisms in different plant species, from herbaceous to woody plant species, as well as its versatility to different environmental stress factors, evidencing its multitarget power (Ilangumaran and Smith, 2017).
Genomic analysis on strain BBB001 revealed that it could not be assigned to any known species of Pseudomonas, so the name P. palmensis sp. nov. was proposed for strain BBB001T. A 16S rRNA gene sequence–based phylogenetic analysis suggested that the strain BBB001T was affiliated with the P. brassicae group with P. brassicae MAFF212427T as the most closely related type strain (99.85% similarity). Genomic comparisons of strain BBB001T with the type strain species P. brassicae MAFF212427T were all below thresholds established for differentiation. In the comparative analysis of complete genomes, a total of 96 genes present exclusively in strain BBB001T were identified, 80 of which appear associated with specific subsystems.
Also, the novel taxon possesses several phenotypic traits related to beneficial plant traits, like auxin production or iron mobilization. Results obtained in iron-starved tomato and water-limited olive orchards confirm the potential of this strain for agriculture. On the one hand, it is efficient for iron mobilization under nutrient stress and adverse conditions, consistent with the specific and unique set of genes associated with iron metabolism, so it can be used to develop organic Fe supplies for sustainable agriculture, both the strain and its metabolites. On the other hand, it can improve adaptation to water-limiting conditions while keeping high productivity, so it is an excellent candidate to develop biotechnological products for agriculture under harsh conditions. Finally, the effects recorded in herbaceous species such as tomato, as well as in a woody species such as olive, confirm its wide scope capability to succeed in most agronomic crops, as this strain can trigger plant metabolism in different species.
In summary, all phenotypic and genomic characterization of the novel strain BBB001T reveals a distinct and well-differentiated species from all Pseudomonas species with validated reference taxons and, more precisely, different from P. brassicae MAFF212427T.
Thus, based on the polyphasic approach, we describe a novel Pseudomonas species, for which the name P. palmensis sp. nov. is proposed, based on the geographical origin of isolation, Las Palmas de Gran Canarias (Islas Canarias, Spain). The type strain is strain BBB001T (= LMG 31775 = NCTC 14418).
Description of Pseudomonas palmensis sp. nov
Pseudomonas palmensis (palm.en’sis), N.L. masc./fem. adj. palmensis, belonging to Las Palmas de Gran Canaria, Spain, from which the type strain was isolated.
Cells are Gram-reaction negative, aerobic, rod-shaped bacteria ranging between 0.5 and 0.8 μm × 1.5–3.8 μm long, non–endospore-forming rods. Colonies grow on PCA at 28°C forming circular colonies < 1 mm ∅, with smooth borders, opaque, yellowish, and creamy texture. In liquid culture (nutrient broth), color changes from pale yellow in the exponential growth phase to intense yellow on the stationary phase after 24 h at 28°C. Growth occurs at 4 to 40°C in 24 h but not at 42°C. As regards pH, strain BBB001T grows in a pH range from 6 to 10 and with a salt concentration from 2 to 6%. The optimal temperature, pH, and salinity of growth for BBB001T are 28°C, pH 8, and 0% salinity. It is able to assimilate glucose, potassium gluconate, capric acid malate, trisodium citrate, and phenylacetic acid; it is oxidase positive. Its metabolic profile based on BIOLOG Eco shows a high catabolic capacity. It is able to produce auxins and siderophores, but not chitinases. The major fatty acids (> 81.17% of the total fatty acids) are C16:0 > summed feature 3 (C16:1ω7c/C16:1 ω6c), > summed feature 8 (C18:1ω7c/C18:1ω6c) > C17:0 cyclo.
The strain BBB001T was isolated form the rhizosphere of N. glauca L. in Las Palmas de Gran Canaria Spain, in 2017. The genomic DNA G + C content of the type strain is 64.05%. The draft genome and 16S rRNA gene sequences of the strain BBB001T have been deposited at the NCBI GenBank under accession numbers MW009702 and JAFKEC000000000, respectively. The type strain is BBB001T (= LMG 31775 = NCTC 14418).
Materials and Methods
Origin of Bacteria
The bacterial screening was carried out over the rhizosphere of wild populations of N. glauca in Las Palmas de Gran Canaria (Islas Canarias, Spain; Coordinates UTM: 28°01′36.8″ N 15°23′19.7W) in December 2017. The soil intimately adhered to roots, and the thinner roots (diameter 1–2 mm) of four plants were pooled at random and constituted a replicate; four replicates were sampled. All materials were brought to the laboratory in plastic bags at 4°C. One gram of rhizosphere soil and thinner roots were suspended in 10 mL sterile distilled water and homogenized for 1 min in an Omni Mixer. One hundred microliters of the soil suspension was used to prepare serial 10-fold dilutions in a final volume of 1 mL; 100 μL was plated on Standard Methods Agar (Pronadisa Spain) and incubated for 4 days at 28°C. Individual colonies were selected after 36 h. To avoid duplication, isolated colonies were marked on the plate after selection. Eighty colony-forming units (cfu) were selected from each serial-dilution series, that is, from each replicate (four), constituting 320 cfu. All were isolated and grouped according to Gram staining, morphological characteristics, and sporulating capacity into four parataxonomic groups: Gram-positive endospore-forming bacilli, Gram-positive non–endospore-forming bacilli, Gram-negative bacilli, and other morphologies were grouped under “Others.” All isolates were kept at −20°C on glycerol: water (1:4) (Ramos-Solano et al., 2010). Our bacterium was isolated from these dilutions and was kept at −20°C on glycerol: water (1:4).
Phenotypic Characterization
Our bacterium was tested for in vitro production of auxins, (Benizri et al., 1998), siderophores (Alexander and Zuberer, 1991), and chitinases (Rodríguez-Kábana et al., 1983; Frändberg and Schnürer, 1998) using specific culture media in which a color change reveals the biochemical trait.
To test pH and salt tolerance rates of BBB001T, the following procedure was followed. Growth was tested in presence of 0 to 8% (wt/vol) NaCl (at intervals of 1%) and at pH 4 to 10 (at intervals of 0.5 pH units) by supplementing TSB with appropriate buffer systems (pH 4–5.5, 0.1 M citric acid/0.1 M sodium citrate; pH 6–9, 0.1 M KH2PO4/0.1 M NaOH; pH 8.5–10, 0.1 M NaHCO3/0.1 M Na2CO3 (Xu et al., 2005). After autoclaving, pH medium was adjusted by the addition of NaOH/HCl 1 M before sterile filtration (pore size 0.22 μm). Analyses were carried out at 28°C for 48 h by shaking at 140 revolutions/min (rpm) in an orbital shaker and measuring OD600 nm every 2 h. Each well contained 200 μL of the selected medium and was inoculated at a dilution of 1:1,000 with an overnight culture of cells grown in unmodified TSB at 28°C for 24 h. Periphery pH and NaCl growth conditions previously obtained were also studied, using 5-mL tubes at 28°C for 1 week. Growth at 4 to 42°C was tested in tubes containing 7 mL TSB with shaking at 140 rpm for 8 days. All strains were precultured overnight at 28°C with TSB before suspending one loop of cells in 1 mL TSB as inoculation culture. The 7-mL test tubes were inoculated with 10 μL of the inoculation culture. The optimal growth temperature was determined using the SPECTROstar nano (BMG Labtech) and TSB under the same conditions as used for testing growth at different NaCl concentrations and pH. The range for optimal growth was determined by the length of the lag phase until the cultures achieved an increase in OD600 of 0.2.
The metabolic capabilities were evaluated by API 20NE and BIOLOG ECO according to manufacturer instructions. The bacterial suspension was prepared from bacterial biomass grown for 24 h in PCA resuspended in MgSO4 10 mM to achieve 95% of transmittance at 620 nm. Biolog ECO plates were then inoculated with 150 μL per well. Plates were incubated at 25°C in darkness. Each well contained a substrate and tetrazolium salts, which turn violet when reduced by the activity of microorganisms. Three replicates per treatment and sampling time were performed. Kinetics of average well color development (AWCD) were used to perform a curve and to determine bacterial speed using the 31 provided substrates. From this curve, 7-day incubation time was chosen to represent the catabolic profile of the strain.
Fatty acid composition was determined according to MIDI microbial identification system (Sasser, 1990) on an Agilent 6850 gas chromatograph, provided with the MIDI microbial identification system with method TSBA6 (MIDI, 2008). This fatty acid profile was obtained from strain BBB001 grown in nutrient agar, at 28°C, for 24 h.
Phylogenetic Analysis
To investigate the phylogenetic relationship of strain BBB001T with other species of Pseudomonas genus and define the taxonomic allocation, three main strategies were used:
-
•
Estimate the similarity between genomes: (a) phylogenetic analysis and delimitation of species based on the analysis of 16S rRNA and (b) ANI, AAI, hybridization DNA–DNA in silico or dDDH (digital DNA–DNA hybridization), difference in the content of guanine-cytosine (G + C), and frequency of tetranucleotide use.
-
•
Phylogenetic analysis at complete genome level.
-
•
Characterization of novel regions.
Libraries Preparation
The DNA extraction from the lyophilized culture was performed with the REAL MicroSpin DNA Isolation kit (Durviz), strictly following the manufacturer’s instructions. A sample that did not contain a bacterial culture was processed in parallel with the other samples to verify the absence of cross contamination during DNA extraction. DNA was quantified using fluorometric methods (Qubit, Thermo Fisher Scientific).
Nextera XT Library Prep kit (Illumina) was used to prepare the library, following the manufacturer’s instructions. The fragment size distribution was then checked on an Agilent 2100 Bioanalyzer, using the Agilent DNA 1000 kit. Qubit dsDNA HS kit was used to check library concentration.
Based on these concentration data, the library was equimolarly mixed and loaded in a fraction of a MiSeq Paired-End 300 (Illumina) run.
Quality Analysis and Sequencing Data
The FastQC 0.11.3 program was used to analyze the quality of the sequencing. Trimmomatic 0.36 (Bolger et al., 2014) was used to remove the adapters (ILLUMINACLIP option) and poor-quality regions (SLIDINGWINDOW: 5: 30). All sequences smaller than 80 base pairs were also discarded.
De novo Assembly and Quality Analysis
Short sequences were assembled de novo using SPAdes 3.10 (Bankevich et al., 2012). SPAdes is based on the creation of graphs using the algorithm known as de Bruijin using different kmers. As recommended in the manual, kmers 21, 33, 55, 77, 99, and 127 were used; the –cov-cutoff option was set to auto and the –careful option was enabled. SPAdes use the information from the two pairs of files generated in the sequencing to transform contigs into scaffolds by introducing Ns between the contigs that can be joined.
The quality of the assemblies was analyzed using the QUAST 4.4 program (Gurevich et al., 2013).
The sequences of each sample were mapped in the assembly generated by SPAdes, to determine the coverage of each genomic region using BWA 0.7.12 (Li and Durbin, 2010). SAMtools 0.1.19 (Li et al., 2009) was used to determine the depth of sequencing and eliminate secondary or poor-quality mappings.
Qualimap 2.2.1 (Okonechnikov et al., 2015) was used to observe how the coverage is distributed throughout the genome and, in addition, to extract the specific coverage of each scaffold. To avoid possible inconsistencies or the presence of contaminating sequences, those scaffolds with less than 10 X coverage were discarded from the successive analyses. In addition, those scaffolds with a length less than 1,000 base pairs were also eliminated.
The sequencing of the complete genome on the Illumina platform MiSeq PE300 generated 986,724 sequences and a total of 290 megabases (Mb). The sequences obtained after applying the quality filters were assembled de novo to create larger sequences (scaffolds). Strain BBB001T genome assembly generated 128 scaffolds with a total length of 5.91 Mb. The mapping of the sequences against the assembled genome confirmed that 99.94% of the sequences obtained for strain BBB001T could be used for the generation of the assembly.
Estimates of the Degree of Similarity Between Genomes
Phylogenetic Analysis and Delimitation of Species Based on the Analysis of 16S
16S rRNA sequences were BLAST-queried against all strains with a validated name in the reference database at the Ezbiocloud website.
ANI and AAI
The first stage of this analysis consists in the comparison at the nucleotide (ANI) and amino acid (AAI) level of strain BBB001T genome with all complete genomes available in the EzBiocloud database for similar genomes according to the 16S analysis previously performed. The estimates of the nucleotide identity index (ANI) were calculated using4. The amino acid content (AAI) was calculated at http://enve-omics.ce.gatech.edu/aai/. Values below 95% indicate a new species (Richter and Rosselló-Móra, 2009; Kim et al., 2014).
dDDH and Content in G + C
Based on the results of the previous section, the 26 genomes located in the fourth quartile of the distribution of the ANI values were selected (i.e., predictably, the most closely related to strain BBB001T), to calculate the intergenomic distances and the dDDH indexes, using the GGDC web server (Genome-to-Genome Distance Calculator). This server also calculates the difference in G + C content of genomes analyzed. Values less than 70% are indicative of significant differences (Richter and Rosselló-Móra, 2009; Kim et al., 2014).
Frequencies of Use of Tetranucleotides
The JSpecies software (Richter and Rosselló-Móra, 2009) was used to calculate the differences in tetranucleotide usage profiles between the genome of strain BBB001T and the 26 most related genomes. Values greater than 99% are indicative of significant differences (Teeling et al., 2004; Richter and Rosselló-Móra, 2009).
Phylogenetic Analyses Based on Complete Genome Sequences
The genome sequence of BB001 was uploaded to the TYGS, a free bioinformatics platform available under https://tygs.dsmz.de, for a whole genome-based taxonomic analysis (Meier Kolthoff and Göker, 2019).
First, BB001 genome was compared against all type strain genomes available in the TYGS database via the MASH algorithm, a fast approximation of intergenomic relatedness (Ondov et al., 2016), and the 10 type strains with the smallest MASH distances chosen per user genome. This was used as a proxy to find the best 50 matching type strains (according to the bitscore) for each user genome and to subsequently calculate precise distances using the Genome BLAST Distance Phylogeny (GBDP) approach under the algorithm “coverage” and distance formula d5 (Meier-Kolthoff et al., 2013). These distances were finally used to determine the 10 closest type strain genomes for each of the user genomes.
For the phylogenomic inference, all pairwise comparisons among the set of genomes were conducted using GBDP and accurate intergenomic distances inferred under the algorithm “trimming” and distance formula d5 (Meier-Kolthoff et al., 2013). One hundred distance replicates were calculated each. Digital DDH values and confidence intervals were calculated using the recommended settings of the GGDC 2.1 (Meier-Kolthoff et al., 2013).
The resulting intergenomic distances were used to infer a balanced minimum evolution tree with branch support via FASTME 2.1.6.1 including SPR postprocessing (Lefort et al., 2015). Branch support was inferred from 100 pseudo-bootstrap replicates each. The trees were rooted at the midpoint (Farris, 1972) and visualized with PhyD3 (Kreft et al., 2017).
Characterization of Pseudomonas BBB001T Novel Regions
The novel regions were determined using the web server Pan-seq5. The Novel Region Finder of this software currently identifies any genomic regions present in any of the “Selected Query” sequences (genomes of the 26 species closer in EzBiocloud) that are not present in any of the “Selected Reference” sequences (genome of BB001) and returns these regions in multi-fasta format. These comparisons are done using the nucmer program from MUMmer v3. Annotations of novel sequences were computed using eggnog-mapper (Huerta-Cepas et al., 2017) based on eggNOG 4.5 orthology data (Huerta-Cepas et al., 2018).
Biological Assays Experimental Set Up
Iron Mobilization on Tomato
Tomato (Solanum lycopersicum L.) seeds were germinated in cocopeat with 24°C/19°C (day/night), and 15-h/9-h light–dark and kept in the greenhouse under controlled conditions throughout the 12 weeks of the experiment. Four treatments were used in this experiment: (1) control + (Fe-EDTA), (2) control− (no iron, no bacterial treatment), (3) strain BBB001T, and (4) the culture media free of bacteria [chelate (Q) produced by BBB001T]; to obtain treatments 3 and 4, strain was inoculated on nutrient broth and was grown for 24 h at 28°C under shaking, and then the culture media was centrifuged at 4,000 rpm, and the supernatant was treatment 4 (chelate), and treatment 3 was made by resuspending cells in the same volume of MgSO4 10 mM solution. A total of 96 plants arranged in three replicates with eight plants each per treatment. Plants were grown under iron deficiency conditions achieved by washing substrate with bicarbonate buffer pH 8.5 to bring substrate to basic pH preventing iron absorption. After sowing, plants were watered twice a week with bicarbonate buffer pH 8.5 to maintain a high pH and with Hoagland without iron once a week. When chlorosis appeared (6 weeks after sowing), bacteria were delivered to plant roots by soil drench supplemented with iron (FeCl3). Three doses were delivered every 2 weeks; each dose consisted of 10 mL of strain BBB001T (or chelate), at 1 × 108 cfu/mL per plant supplemented with FeCl3 in an equivalent concentration of iron per plant to even that of Fe-EDTA. Two days after the last inoculation, plants were harvested to analyze dry weight, iron content, and photosynthetic pigments.
After 12 weeks of experiment, plants were harvested and dried, and plant weight was recorded. Leaves from plants in a replicate were pooled, and chlorophylls and carotenes were analyzed according to Lichtenthaler and Hartmut (1987), and iron was determined by atomic absorption spectrometry (method PNT.08.01).
Olea europaea Production in Intensive Orchards Under Water Limitation
A 12-year-old intensive orchard of O. europaea var. Arbequina in Toledo (Spain) (Coordinates UTM 393782–4343510) was used for this experiment. Two lines of 100 m each were selected, and treatments were marked within the two lines on a random block design. Treatments consisted in three replicates of seven trees; BBB001T treatment was root inoculated (500 mL per tree with a bacterial concentration 1 × 108 cfu/mL) every 2 weeks from April to October, and controls were mock inoculated with water. Regular watering consists of four times a week, each 2 L/h for 3 h; in experimental trees, one time was skipped to decrease water input by 25%. Olives were harvested at the end of October, collecting all olives from the seven trees in the replicate, which were weighed, and fat content determined with Soxhlet. Production (kg/ha) was estimated calculating production per plant and considering orchard’s plant density (1,600/ha).
Statistical Analysis
To evaluate treatment effects on all variables, one-way analysis of variance was performed. When significant differences appeared (p < 0.05), LSD test (least significant difference) Fisher was used. Statgraphics Plus for Windows was the program used.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA705568.
Author Contributions
FJGM conceptualized the manuscript. EG-A and IH carried on the bacterial isolation. EG-A analyzed the genomic data. JL, AG-V, and EG-A developed tomato bioassay and drafted the manuscript. EG-A, BR-S, and FJGM developed olive assay. FJGM and BR-S wrote and supervised the final version. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Antonio Marin for providing olive orchards and manipulation.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.672751/full#supplementary-material
References
- Alexander D. B., Zuberer D. A. (1991). Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12 39–45. 10.1007/bf00369386 [DOI] [Google Scholar]
- Anzuay M. S., Frola O., Angelini J. G., Ludueña L. M., Fabra A., Taurian T. (2013). Genetic diversity of phosphate-solubilizing peanut (Arachis Hypogaea L.) associated bacteria and mechanisms involved in this ability. Symbiosis 60 143–154. 10.1007/s13199-013-0250-2 [DOI] [Google Scholar]
- Ark P. A., Tompkins C. M. (1946). Bacterial leaf blight of bird’s-nest fern. Phytopathology 36, 758–761. [Google Scholar]
- Ash C., Farrow J. A. E., Dorsch M., Stackebrandt E., Collins M. D. (1991). Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16s Rrna. Int. J. Syst. Evol. Microbiol. 41 343–346. 10.1099/00207713-41-3-343 [DOI] [PubMed] [Google Scholar]
- Bakker P. A. H. M., Pieterse C. M. J., Van Loon L. C. (2007). Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathol 97 239–243. 10.1094/phyto-97-2-0239 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano A., Muqarab R. (2017). Plant defence induced by PGPR against Spodoptera litura in tomato (Solanum lycopersicum L.). Plant Biol. 19 406–412. 10.1111/plb.12535 [DOI] [PubMed] [Google Scholar]
- Barriuso J., Pereyra M. T., Lucas J. A., Megias M., Manero F. J. G., Ramos B. (2005). Screening for putative PGPR to improve establishment of the symbiosis Lactarius deliciosus-Pinus Sp. Microb. Ecol. 50 82–89. 10.1007/s00248-004-0112-9 [DOI] [PubMed] [Google Scholar]
- Behrendt U., Ulrich A., Schumann P., Erler W., Burghardt J., Seyfarth W. (1999). A taxonomic study of bacteria isolated from grasses: a proposed new species Pseudomonas graminis sp. nov. Int. J. Syst. Bacteriol. 49, 297–308. [DOI] [PubMed] [Google Scholar]
- Benizri E., Courtade A., Picard C., Guckert A. (1998). Role of maize root exudates in the production of auxins by Pseudomonas fluorescens M.3.1. Soil. Biol. Biochem 30 1481–1484. 10.1016/s0038-0717(98)00006-6 [DOI] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 30 2114–2120. [DOI] [PubMed] [Google Scholar]
- Braun V. (2003). Iron uptake by Escherichia coli. Front. Biosci. 8:1409–1421. 10.2741/1232 [DOI] [PubMed] [Google Scholar]
- Cámara B., Strömpl C., Verbarg S., Spröer C., Pieper D. H., Tindall B. J. (2007). Pseudomonas reinekei sp. nov., Pseudomonas moorei sp. nov. and Pseudomonas mohnii sp. nov., novel species capable of degrading chlorosalicylates or isopimaric acid. Int. J. Syst. Evol. Microbiol. 57, 923–931. [DOI] [PubMed] [Google Scholar]
- Coulton J. W., Mason P., Dubow M. S. (1983). Molecular cloning of the ferrichrome-iron receptor of Escherichia coli K-12. J. Bacteriol. 156 1315–1321. 10.1128/jb.156.3.1315-1321.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabboussi F., Hamze M., Singer E., Geoffroy V., Meyer J. M., Izard D. (2002). Pseudomonas mosselii sp. nov., a novel species isolated from clinical specimens. Int. J. Syst. Evol. Microbiol. 52(Pt 2), 363–376. [DOI] [PubMed] [Google Scholar]
- DeBoer K. D., Lye J. C., Campbell D., Aitken A., Su K.-K., Hamill J. D. (2009). The A622 gene in Nicotiana glauca (Tree Tobacco): evidence for a functional role in pyridine alkaloid synthesis. Plant Molec. Biol. 69:299. 10.1007/s11103-008-9425-2 [DOI] [PubMed] [Google Scholar]
- Delfin E. F., Rodriguez F. M., Paterno E. S. (2015). Biomass partitioning, yield, nitrogen and phosphorus uptake of PGPR inoculated tomato (Lycopersicum esculentum L) under field condition. Philipp. J. Crop. Sci. 40 59–65. [Google Scholar]
- Elomari M., Coroler L., Verhille S., Izard D., Leclerc H. (1997). Pseudomonas monteilii sp. nov., isolated from clinical specimens. Int. J. Syst. Bacteriol. 47, 846–852. [DOI] [PubMed] [Google Scholar]
- Farris J. S. (1972). Estimating phylogenetic trees from distance matrices. Am. Nat. 106 645–667. 10.1086/282802 [DOI] [Google Scholar]
- Frasson D., Opoku M., Picozzi T., Torossi T., Balada S., Smits T. H. M., et al. (2017). Pseudomonas wadenswilerensis sp. nov. and Pseudomonas reidholzensis sp. nov., two novel species within the Pseudomonas putida group isolated from forest soil. Int. J. Syst. Evol. Microbiol. 67, 2853–2861. [DOI] [PubMed] [Google Scholar]
- Fatima S., Anjum T. (2017). Identification of a potential ISR determinant from Pseudomonas aeruginosa pm12 against fusarium wilt in tomato. Front. Plant Sci. 8:848. 10.3389/fpls.2017.00848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frändberg E., Schnürer J. (1998). Antifungal activity of chitinolytic bacteria isolated from airtight stored cereal grain. Can. J. Microbiol. 44 121–127. 10.1139/w97-141 33356898 [DOI] [Google Scholar]
- Gao J., Xie G., Peng F., Xie Z. (2015). Pseudomonas donghuensis sp. nov., exhibiting high-yields of siderophore. Antonie Van Leeuwenhoek 107, 83–94. [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesleret G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez Mañero F. J., Acero N., Lucas J. A., Probanza A. (1996). The influence of native rhizobacteria on European alder [Alnus glutinosa (L.) Gaertn.] growth. II. Characterization and biological assays of metabolites from growth promoting and growth inhibiting bacteria. Plant Soil 182 67–74. 10.1007/BF00010996 [DOI] [Google Scholar]
- Huerta-Cepas J., Forslund K., Coelho L. P., Szklarczyk D., Jensen L. J., von Mering C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34 2115–2122. 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., Hernandez-Plaza A., Forslund S. M. K., Cook H., et al. (2018). EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology prediction resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47 D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilangumaran G., Smith D. L. (2017). Plant growth promoting rhizobacteria in amelioration of salinity stress: a systems biology perspective. Front. Plant Sci. 8:1768. 10.3389/fpls.2017.01768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangasjärvi S., Tikkanen M., Durian G., Aro E. M. (2014). Photosynthetic light reactions. An adjustable hub in basic production and plant immunity signaling. Plant Physiol. Biochem. 81 128–134. 10.1016/j.plaphy.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Keshavarz-Tohid V., Vacheron J., Dubost A., Prigent-Combaret C., Taheri P., Tarighi S., et al. (2019). Genomic, phylogenetic and catabolic re-assessment of the Pseudomonas putida clade supports the delineation of Pseudomonas alloputida sp. nov., Pseudomonas inefficax sp. nov., Pseudomonas persica sp. nov., and Pseudomonas shirazica sp. nov. Syst. Appl. Microbiol. 42, 468–480. [DOI] [PubMed] [Google Scholar]
- Kim M., Oh S. H., Park S. C., Chun J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16s Rrna gene sequence similarity for species demarcation of Prokaryotes. Intl. J. System. Evol. Microbiol. 64 346–351. 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]
- Kloepper J. W. (1992). Plant growth-promoting rhizobacteria as biological control agents. Soil microbial ecology applications in agricultural and environmental management. pp.255-274 ref.6 pp. ISBN : 0824787374 ed Metting F.B., Junior. Publisher. Newyork, NY: Marcel Dekker Inc. [Google Scholar]
- Kollist H., Zandalinas S. I., Sengupta S., Nuhkat M., Kangasjärvi J., Mittler R. (2019). Rapid responses to abiotic stress: Priming the landscape for the signal transduction network. Trends Plant. Sci. 24 25–37. 10.1016/j.tplants.2018.10.003 [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T., Tiedje J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Nat. Acad. Sci. 102 2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft L., Botzki A., Coppens F., Vandepoele K., Van Bel M. (2017). PhyD3: A phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics. 33 2946–2947. 10.1093/bioinformatics/btx324 [DOI] [PubMed] [Google Scholar]
- Lefort V., Desper R., Gascuel O. (2015). FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32 2798–2800. 10.1093/molbev/msv150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010). Fast and accurate long-read alignment with burrows–wheeler transform. Bioinformatics 26 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler, Hartmut K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Met. Enzymol. 148 350–382. 10.1016/0076-6879(87)48036-1 [DOI] [Google Scholar]
- López J. R., Diéguez A. L., Doce A., De la Roca E., De la Herran R., Navas J. I.1, et al. (2012). Pseudomonas baetica sp. nov., a fish pathogen isolated from wedge sole, Dicologlossa cuneata (Moreau). Int. J. Syst. Evol. Microbiol. 62, 874–882. [DOI] [PubMed] [Google Scholar]
- Martin-Rivilla H., Garcia-Villaraco A., Ramos-Solano B., Gutierrez-Mañero F. J., Lucas J. A. (2020). Bioeffectors as biotechnological tools to boost plant innate immunity: signal transduction pathways involved. Plants 9:1731. 10.3390/plants9121731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi D. V., Bonsall R. F., Delaney S. M., Soule M. J., Phillips G., Thomashow L. S. (2001). Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa Pao1. J. Bacteriol. 183 6454–6465. 10.1128/jb.183.21.6454-6465.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier Kolthoff J. P., Göker M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome- based taxonomy. Nat. Commun. 10:2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Auch A. F., Klenk H. P., Goker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez E., Ramírez-Bahena M. H., Fabryová A., Igual J. M., Benada O., Mateos P. F., et al. (2015). Pseudomonas coleopterorum sp. nov., a cellulase-producing bacterium isolated from the bark beetle Hylesinus fraxini. Int. J. Syst. Evol. Microbiol. 65, 2852–2858. [DOI] [PubMed] [Google Scholar]
- MIDI (2008). Sherlock microbial identification system operating manual, version 6.1. Newark, DE: MIDI Inc. [Google Scholar]
- Miethke M., Marahiel M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71 413–451. 10.1128/mmbr.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migula W. (1895). “Schizomycetes (Bacteria, Bacterien),” in Die Natürlichen Pfanzenfamilien Teil I, Abteilung Ia, eds Engler A., Prantl K. (Leipzig: Wilhelm Engelmann; ), 1–44. [Google Scholar]
- Mulet M., Gomila M., Lemaitre B., Lalucat J., García-Valdés E. (2012). Taxonomic characterisation of Pseudomonas strain L48 and formal proposal of Pseudomonas entomophila sp. nov. Syst. Appl. Microbiol. 35, 145–149. [DOI] [PubMed] [Google Scholar]
- Mulet M., Sánchez D., Lalucat J., Lee K., García-Valdés E. (2015). Pseudomonas alkylphenolica sp. nov., a bacterial species able to form special aerial structures when grown on p-cresol. Int. J. Syst. Evol. Microbiol. 65, 4013–4018. [DOI] [PubMed] [Google Scholar]
- Nishimori E., Kita-Tsukamoto K., Wakabayashi H. (2000). Pseudomonas plecoglossicida sp. nov., the causative agent of bacterial haemorrhagic ascites of ayu, Plecoglossus altivelis. Int. J. Syst. Evol. Microbiol. 50, 83–89. [DOI] [PubMed] [Google Scholar]
- Oh W. T., Jun J. W., Giri S. S., Yun S., Kim H. J., Kim S. G., et al. (2019). Pseudomonas tructae sp. nov., novel species isolated from rainbow trout kidney. Int. J. Syst. Evol. Microbiol. 69, 3851–3856. [DOI] [PubMed] [Google Scholar]
- Okonechnikov K., Conesa A., García-Alcalde F. (2015). Qualimap 2:advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32 292–294. 10.1093/bioinformatics/btv566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov B. D., Treangen T. J., Melsted P. (2016). Mash: Fast genome and metagenome distance estimation using MinHash. Genome. Biol. 17 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peix A., Rivas R., Mateos P. F., Martínez-Molina E., Rodríguez-Barrueco C., Velázquez E. (2003). Pseudomonas rhizosphaerae sp. nov., a novel species that actively solubilizes phosphate in vitro. Int. J. Syst. Evol. Microbiol. 53, 2067–2072. [DOI] [PubMed] [Google Scholar]
- Peix A., Rivas R., Santa-Regina I., Mateos P. F., Martínez-Molina E., Rodríguez-Barrueco C., et al. (2004). Pseudomonas lutea sp. nov., a novel phosphate-solubilizing bacterium isolated from the rhizosphere of grasses. Int. J. Syst. Evol. Microbiol. 54, 847–850. [DOI] [PubMed] [Google Scholar]
- Posada L., Álvarez J. C., Romero-Tabarez M., Luz de-Bashan, Villegas-Escobar V. (2018). Enhanced molecular visualization of root colonization and growth promotion by Bacillus subtilis Ea-Cb0575 in different growth systems. Microbiol. Res. 217 69–80. 10.1016/j.micres.2018.08.017 [DOI] [PubMed] [Google Scholar]
- Pungrasmi W., Lee H. S., Yokota A., Ohta A. (2008). Pseudomonas japonica sp. nov., a novel species that assimilates straight chain alkylphenols. J. Gen. Appl. Microbiol. 54, 61–69. [DOI] [PubMed] [Google Scholar]
- Qin J., Feng Y., Lü X., Zong Z. (2019a). Pseudomonas huaxiensis sp. nov., isolated from hospital sewage. Int. J. Syst. Evol. Microbiol. 69, 3281–3286. [DOI] [PubMed] [Google Scholar]
- Qin J., Hu Y., Feng Y., Xaioju L., Zong Z. (2019b). Pseudomonas sichuanensis sp. nov., isolated from hospital sewage. Int. J. Syst. Evol. Microbiol. 69, 517–522. [DOI] [PubMed] [Google Scholar]
- Qin J., Hu Y., Wu W., Feng Y., Zong Z. (2020). Pseudomonas defluvii sp. nov., isolated from hospital sewage. Int. J. Syst. Evol. Microbiol. 70, 4199–4203. [DOI] [PubMed] [Google Scholar]
- Ramírez-Bahena M. H., Cuesta M. J., Flores-Félix J. D., Mulas R., Rivas R., Castro-Pinto J., et al. (2014). Pseudomonas helmanticensis sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 64, 2338–2345. [DOI] [PubMed] [Google Scholar]
- Ramos-Solano B., Lucas Garcia J. A., Garcia-Villaraco A., Algar E., Garcia-Cristobal J., Gutierrez Manero F. J. (2010). Siderophore and chitinase producing isolates from the rhizosphere of Nicotiana glauca graham enhance growth and induce systemic resistance in Solanum lycopersicum L. Plant Soil 334 189–197. 10.1007/s11104-010-0371-9 [DOI] [Google Scholar]
- Rasouli-Sadaghiani M. H., Malakouti M. J., Khavazi K., Miransari M. (2014). “Siderophore efficacy of fluorescent Pseudomonades affecting labeled iron (59 Fe) uptake by wheat (Triticum aestivum L.) genotypes differing in Fe efficiency”. In: Use of Microbes for the Alleviation of Soil Stresses. ed Miransari M. Springer: New York, NY. [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Nat. Acad. Sci. 106 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Kábana R., Godoy G., Morgan-Jones G., Shelby R. A. (1983). The determination of soil chitinase activity: conditions for assay and ecological studies. Plant Soil. 75, 95–106. 10.1007/BF02178617 [DOI] [Google Scholar]
- Rosselló-Móra R., Amann R. (2001). The species concept for prokaryotes. FEMS Microbiol. Rev. 25 39–67. 10.1016/s0168-6445(00)00040-1 [DOI] [PubMed] [Google Scholar]
- Sasser M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids, MIDI. Technical note 101. Newark, DE: MIDI. [Google Scholar]
- Savulescu T. (1947). Contribution a la classification des bacteriacees phytopathogenes. Anal. Acad. Romane Ser. III Tom 22. Memoire 4, 1–26. [Google Scholar]
- Sawada H., Fujikawa T., Horita H. (2020). Pseudomonas brassicae sp. nov., a pathogen causing head rot of broccoli in Japan. Int. J. Syst. Evol. Microbiol. 70 5319–5329. 10.1099/ijsem.0.004412 [DOI] [PubMed] [Google Scholar]
- Sinclair S. J., Johnson R., Hamill J. D. (2004). Analysis of wound-induced gene expression in nicotiana species with contrasting alkaloid profiles. Funct. Plant Biol. 31 721–729. 10.1071/fp03242 [DOI] [PubMed] [Google Scholar]
- Teeling H., Meyerdierks A., Bauer M., Amann R., Glöckner F. O. (2004). Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 6 938–947. 10.1111/j.1462-2920.2004.00624.x [DOI] [PubMed] [Google Scholar]
- Tohya M., Watanabe S., Teramoto K., Uechi K., Tada T., Kuwahara-Arai K., et al. (2019a). Pseudomonas asiatica sp. nov., isolated from hospitalized patients in Japan and Myanmar. Int. J. Syst. Evol. Microbiol. 69, 1361–1368. [DOI] [PubMed] [Google Scholar]
- Tohya M., Watanabe S., Teramoto K., Shimojima M., Tada T., Kuwahara-Arai K., et al. (2019b). Pseudomonas juntendi sp. nov., isolated from patients in Japan and Myanmar. Int. J. Syst. Evol. Microbiol. 69, 3377–3384. [DOI] [PubMed] [Google Scholar]
- Trevisan V. B. (1889). I Generi e le Specie delle Batteriacee. L. Zanaboni e Gabuzzi. Available online at: http://scholar.google.com/scholar?cluster=12213764842208350008&hl=en&oi=scholarr [Google Scholar]
- Tvrzova L., Schumann P., Sproer C., Sedlacek I., Pacova Z., Sedo O., et al. (2006). Pseudomonas moraviensis sp. nov. and Pseudomonas vranovensis sp. nov., soil bacteria isolated on nitroaromatic compounds, and emended description of Pseudomonas asplenii. Int. J. Syst. Evol. Microbiol. 56, 2657–2663. [DOI] [PubMed] [Google Scholar]
- Uchino M., Shida O., Uchimura T., Komagata K. (2001). Recharacterization of Pseudomonas fulva Iizuka and Komagata 1963, and proposals of Pseudomonas parafulva sp. nov. and Pseudomonas cremoricolorata sp. nov. J. Gen. Appl. Microbiol. 46, 247–261. [DOI] [PubMed] [Google Scholar]
- Van Loon L. C. (2007). “Plant responses to plant growth-promoting rhizobacteria”. In: New Perspectives and Approaches in Plant Growth-Promoting Rhizobacteria Research, eds Raaijmakers P. A. H. M., Bloemberh G., Hofte M., Lemanceau P., Cooke B.M. New York, NY: Springer [Google Scholar]
- Wang M. Q., Wang Z., Yu L. N., Zhang C. S., Bi J., Sun J. (2019). Pseudomonas qingdaonensis sp. nov., an aflatoxin-degrading bacterium, isolated from peanut rhizospheric soil. Arch. Microbiol. 201, 673–678. [DOI] [PubMed] [Google Scholar]
- Wang L. T., Tai C. J., Wu Y. C., Chen Y. B., Lee F. L., Wang S. L. (2010). Pseudomonas taiwanensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 60, 2094–2098. [DOI] [PubMed] [Google Scholar]
- Wright M. H., Hanna J. G., Ii D. A., Tebo B. M. (2018). Pseudomonas laurentiana sp. nov., an Mn(III)-oxidizing Bacterium Isolated from the St. Lawrence Estuary. Pharmacogn. Commn. 8 153–157. 10.5530/pc.2018.4.32 [DOI] [Google Scholar]
- Xu P., Li W. J., Tang S. K., Zhang Y. Q., Chen G. Z., Chen H. H., et al. (2005). Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int. J. Sys. Evo. Microbiol. 55 1149–1153. 10.1099/ijs.0.63407-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA705568.