Abstract
Two regions of the gene coding for 16S rRNA in Nocardia species were selected as genus-specific primer sequences for a PCR assay. The PCR protocol was tested with 60 strains of clinically relevant Nocardia isolates and type strains. It gave positive results for all strains tested. Conversely, the PCR assay was negative for all tested species belonging to the most closely related genera, including Dietzia, Gordona, Mycobacterium, Rhodococcus, Streptomyces, and Tsukamurella. Besides, unlike the latter group of isolates, all Nocardia strains exhibited one MlnI recognition site but no SacI restriction site. This assay offers a specific and rapid alternative to chemotaxonomic methods for the identification of Nocardia spp. isolated from pathogenic samples.
Nocardia spp. are gram-positive, aerobic actinomycetes with a worldwide distribution in soil. At least six species are pathogenic for both humans and animals and may enter the body via inhalation of contaminated dust particles or via wounds contaminated with dust or soil (18). They are responsible for several infections including pulmonary, central nervous system, and cutaneous infections (1). An increasing number of cases have been reported since 1980, which supports the view of many investigators that the incidence of nocardiosis is on the rise (2). Nocardiosis is diagnosed by isolation and culture identification. However, colonial characteristics and cellular morphology are variable, and Nocardia spp. may be misidentified and confused with members of closely related genera such as Dietzia (22), Gordona, Mycobacterium, Rhodococcus, Tsukamurella, and even Streptomyces. Conventional identification to the genus level is based on chemotaxonomic characteristics, such as the presence of a major amount of meso-diaminopimelic acid, arabinose, galactose, and mycolic acids with 46 to 60 carbons in cell walls (12). However, these techniques sometimes fail to fully differentiate some of the most closely related genera. Moreover, they are laborious, time-consuming, and expensive and can take 1 to 3 weeks to accomplish.
In this study, we developed an alternative PCR-based identification strategy targeted at the gene coding for 16S rRNA. Although the sequence for this gene is largely phylogenetically conserved, there may be variable sequences characteristic of particular organisms. The detection of these variable regions can therefore allow bacteria to be identified and differentiated from each other (13, 17, 26). The comparative analysis of the sequences of the complete 16S rRNA genes from 85 actinomycetes provided the basis for the design of diagnostic primers with a potential for the genus-specific detection of Nocardia by a PCR assay. We evaluated this approach for the fast, sensitive, and accurate identification of a wide range of Nocardia spp. isolated from pathogenic samples.
MATERIALS AND METHODS
Bacterial strains.
One hundred seventeen clinical strains and eighteen reference strains of actinomycetes have been studied (Table 1). Cultures were obtained from the National Reference Center for Human Mycosis, Antifungal Agents, and Actinomycetes (Institut Pasteur, Paris, France) or from the American Type Culture Collection (ATCC; Rockville, Md.). All isolates were identified by conventional methods including biochemical tests, whole-cell composition, and enzymatic profiles (3). The isolates included 5 strains of Dietzia, 8 strains of Gordona, 17 strains of Mycobacterium, 60 strains of Nocardia belonging to 8 different species, 20 strains of Rhodococcus, 20 strains of Streptomyces, and 5 strains of Tsukamurella. Cultures were maintained on Bennett’s agar medium at room temperature before being treated simultaneously.
TABLE 1.
Strains used in the study
| Genus and species | No. of isolatesa | Reference strains used |
|---|---|---|
| Nocardia asteroides | 17 | ATCC 19247 |
| Nocardia farcinica | 14 | ATCC 3318 |
| Nocardia nova | 11 | ATCC 33727 |
| Nocardia brasiliensis | 7 | ATCC 19296 |
| Nocardia otitidiscaviarum | 5 | ATCC 14629 |
| Nocardia carnea | 2 | ATCC 6847 |
| Nocardia transvalensis | 2 | ATCC 6865 |
| Nocardia brevicatena | 2 | ATCC 15333 |
| Rhodococcus equi | 13 | ATCC 6939 |
| Rhodococcus rhodochrous | 3 | ATCC 13808 |
| Rhodococcus erythropolis | 2 | ATCC 4277 |
| Rhodococcus coprophilus | 2 | ATCC 29080 |
| Streptomyces sp. | 20 | |
| Dietzia marinum | 5 | ATCC 35013 |
| Gordona bronchialis | 2 | ATCC 25592 |
| Gordona sputi | 2 | ATCC 29627 |
| Gordona terrae | 2 | ATCC 25594 |
| Gordona rubropertinctus | 2 | ATCC 14352 |
| Tsukamurella spp. | 5 | |
| Mycobacterium tuberculosis | 9 | ATCC 51910 |
| Mycobacterium spp. | 8 |
National Reference Center for Human Mycosis, Antifungal Agents, and Actinomycetes (Institut Pasteur, Paris, France).
Primer selection.
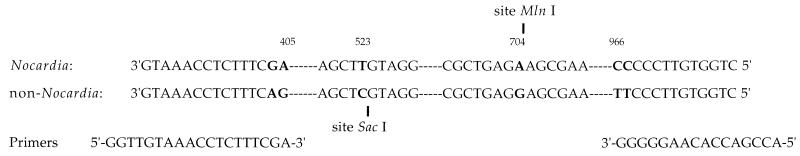
The sequences of the 16S rRNA genes from a large number (n = 85) of isolates belonging to the genera Dietzia, Gordona, Mycobacterium, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella were found in the DDBJ/EMBL/Genbank database. They were analyzed with the PILEUP program (9) of Genetics Computer Group software package (8). The existence of regions exhibiting possible sequence signatures to the genus level was investigated. The results allowed the design of two primers with 3′ extremities specific for the Nocardia genus (Fig. 1): forward primer NG1 (5′-ACCGACCACAAGGGG-3′) was complementary to positions 966 to 982 on the antisense strand, and reverse primer NG2 (5′-GGTTGTAACCTCTTCGA-3′) was complementary to positions 386 to 405 on the sense strand. Both primers were custom synthesized by Appligene (Alkirch, France). Thus, the amplified product was to be 596 bp in length.
FIG. 1.
Comparison of the DNA sequences of the 16S rRNA genes from members of the Nocardia genus and other non-Nocardia genera of aerobic actinomycetes. Differences in base sequences are indicated with boldface characters.
DNA extraction, PCR amplification, and detection and digestion of PCR products.
Isolated colonies were subcultured for 3 to 5 days at 34°C on sterile cellulose acetate membranes (Millipore, Bedford, Mass.) that had been deposited on blood agar (bioMérieux, Marcy l’Etoile, France). DNA was extracted as described previously (21). A colony was suspended in 650 μl of a reaction mixture containing 500 μl of sterile pyrolyzed water and 150 μl of Chelex solution (15% [wt/vol] Chelex 100 resin, 0.1% [wt/vol] sodium dodecyl sulfate, 1% [vol/vol] Nonidet P-40, 1% [vol/vol] Tween 80) and boiled for 30 min at 100°C. The samples were centrifuged for 8 min at 5,000 × g. The final supernatant containing the extracted DNA was recovered, and 15 μl of a 1/20 dilution aliquot was used for amplification. The 100-μl DNA amplification reaction mixture contained 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, each deoxynucleoside triphosphate at a concentration of 200 μM, 1 μM each primer, and 1 U of AmpliTaq Gold (Perkin-Elmer, Norwalk, Conn.). Amplified reactions were carried out in a DNA thermal cycler (Hybaid; Omnigene, Teddington, United Kingdom). After initial denaturation at 94°C for 11 min, the reaction mixture was run through 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 20 s, and extension at 72°C for 1 min. Cycling was followed by a final extension incubation of 10 min at 72°C.
To ensure that no contaminating DNA would give positive results, one sample lacking a template was included in each series of reactions. After amplification, 12-μl samples were analyzed by electrophoresis for 2 h at 100 V in horizontal TAE (40 mM Tris-acetate, 2 mM Na2 EDTA; 2H2O) gels containing 2% agarose (Ultrapure; Bio-Rad Laboratories, Hercules, Calif.) and 0.5 μg of ethidium bromide per ml. The gels were then exposed to UV light to visualize the amplified products and photographed. The reproducibility of the reaction was tested at intervals of different numbers of days with freshly extracted DNA.
The identity of the amplified products was further investigated by restriction fragment length polymorphism (RFLP) analysis with SacI (Boehringer Mannheim GmbH, Mannheim, Germany) and MlnI (New England Biolabs GmbH, Taunus, Germany). Restriction endonucleases were selected by using the DNA Strider program in order to provide differentiation of Nocardia spp. from other aerobic actinomycetes. The digestion of PCR products was carried out for 2 h at 37°C in 20-μl volumes of commercially supplied incubation buffer containing 1 U of restriction enzyme. The volume of the amplimer used in the restriction mixture was adjusted arbitrarily on the basis of the visually observed fluorescence intensity of the amplified DNA fragment in the control gel. Restriction fragment patterns were analyzed by gel electrophoresis of 20 μl of each restriction mixture for 2 h at 100 V in a 3% (wt/vol) Metaphor Agarose gel (FMC BioProducts, Rockland, Maine) containing 0.5 μg of ethidium bromide per ml. A 100-bp ladder was used as a DNA molecular weight marker (Boehringer Mannheim GmbH) to evaluate the sizes of the fragments in each pattern. Fragments smaller than 50 bp were not considered for the interpretation of the restriction pattern because of poor visibility.
RESULTS AND DISCUSSION
Because of the slow growth of Nocardia, we used a direct DNA extraction method from a single colony (isolated in primary culture or from samples received by the reference laboratory) using the Chelex method in less than 40 min. This method was found to be effective in recovering sufficiently purified DNA for amplification even if DNA cannot be visualized. By reducing the time and labor associated with the conventional DNA isolation procedure with the rapid-release method, the entire PCR assay and restriction endonuclease cleavage can be completed within 2 days.
PCR, which allows the specific and sensitive amplification of a preselected DNA region, has been intensively applied to the species identification of numerous organisms (7, 11, 14, 23, 27). Its use requires the detailed examination of the molecular genetics of organisms, especially the identification of the DNA sequences that are specific for the organism tested. The target regions usually are pathogenicity or virulence genes. On the basis of the homologies of a heat shock protein (Hsp65) gene from Nocardia strains and rapidly growing Mycobacterium strains, Lungu et al. (16) developed such an approach: an RFLP analysis of DNA amplified from the Hsp65 gene enabled them to distinguish these two genera. In this case, however, no extensive study concerning other actinomycetes was done. Moreover, because the primers were not specific, only endonuclease digestion gave specificity to this test. Finally, the profiles obtained were sometimes not easy to interpret since the number of patterns was large for rapidly growing Mycobacterium and since fragment sizes were difficult to analyze.
Little is known about DNA from the Nocardia genus. The 16S rRNA gene (rDNA) sequences are the only extensive sequences that have been documented (6, 24). Thus, it is now established that the primary structure of rRNA is composed of regions with highly conserved sequences together with other regions composed of variable or hypervariable sequences, i.e., signatures, that are of special interest for identification to the genus or species level (4, 5, 10). The similarities in nucleotides of rRNA preparations provide a means of establishing relationships among representatives of diverse bacterial taxa. In the late 1970s, Mordarski et al. (19, 20) used DNA-rRNA pairing experiments to show the phylogenetic relationship between a genus belonging to the actinomycetes and the clustering of each genus. Recently, a comprehensive phylogenetic analysis of the genus Nocardia by 16S rRNA gene sequencing (6, 24) confirmed these results and indicated that the genus Nocardia (after several taxonomic modifications) constitutes a homogeneous taxon that can be distinguished from the other mycolic acid-containing organisms. In addition, ribotyping with a probe coding for a part of 16S RNA had demonstrated a species-level specificity for isolates belonging to the N. asteroides complex (15). Therefore, 16S rDNA appeared to be a potential interesting target in the development of new molecular techniques for Nocardia genus identification.
Thus, the presence of regions within 16S rDNA sequences of actinomycetes exhibiting possible sequence signatures at the genus level for Nocardia was investigated. The comparative analysis of the 16S rDNA sequences from 85 strains belonging to actinomycetes (including data for 25 Nocardia strains) revealed a high degree of homology. However, all Nocardia strains examined exhibited the genus-specific GA at nucleotide positions 404 and 405 and CC at nucleotide positions 966 and 967. Primers NG1 and NG2 were constructed by using these sequences containing the Nocardia-specific signature and the corresponding preceding 5′ area (Fig. 1). Although this area was highly conserved among actinomycetes, the sequence of two nucleotides specific for Nocardia was sufficient for the design of diagnostic primers.
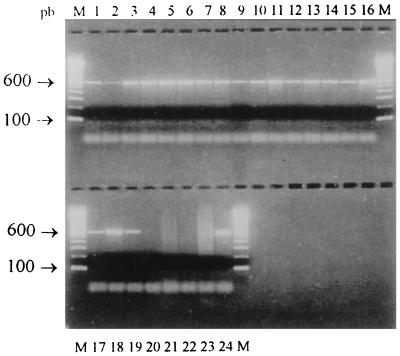
Figure 2 shows an example of the results of a PCR assay with the primers that were designed. The general applicability of the primers was tested against a diverse array of Nocardia strains (including type strains). A 596-bp amplimer was observed in all Nocardia strains, whereas no amplification product was obtained in control experiments with DNA from any of the other actinomycetes strains (Fig. 2). These results demonstrated that all strains of Nocardia spp. tested can be distinguished from representative members of the genera Dietzia, Gordona, Mycobacterium, Rhodococcus, Streptomyces, and Tsukamurella by using the selected primer combination.
FIG. 2.
Agarose gel electrophoresis of PCR products of actinomycetes strains by using primers NG1 and NG2. Lanes: 1 to 4, N. asteroides sensu stricto; 5 to 8, N. farcinica; 9 to 12, N. nova; 13 to 15, N. brasiliensis; 16, N. otitidiscaviarum; 17, N. carnea; 18, N. transvalensis; 19, N. brevicatena; 20, Gordona bronchialis; 21, Streptomyces spp.; 22, Rhodococcus equi; 23, Mycobacterium tuberculosis; 24, N. asteroides ATCC 19247; M, 100-bp DNA ladder (Boehringer Mannheim GmbH). pb, base pairs.
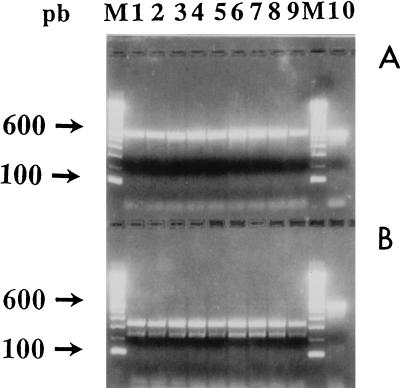
Restriction endonuclease analysis was performed as a second level of control to verify the specificity of the 596-bp amplified fragment. If a non-Nocardia isolate shows an amplified product despite theoretical and observed primer specificity, a second level of check exists. Two recognition sites at nucleotide positions 704 (MlnI) and 523 (SacI) on the amplified fragment allowed the clear differentiation of members of the Nocardia genus from other genera of the aerobic actinomycetes (Fig. 1). As shown in Fig. 3, all Nocardia strains exhibited one MlnI recognition site, producing two DNA fragments of 278 and 318 bp. On the other hand, no SacI restriction site was observed in Nocardia strains, and as expected, no restriction fragments were produced following endonuclease analysis; conversely, a potential SacI restriction site was noted at position 523 of the DNA sequences of non-Nocardia strains. In case of nonspecific amplification of the 16S rRNA gene fragment, the SacI restriction site may produce two fragments of 137 and 459 bp, and these differentiate an isolate from Nocardia species. In fact, even though we never observed nonspecific amplification, we retained the restriction endonuclease analysis step to increase the security of genus identification.
FIG. 3.
RFLP patterns of PCR products from strains belonging to Nocardia genus, obtained by using endonucleases SacI (A) and MlnI (B). Lanes: 1 and 2, N. asteroides sensu stricto; 3, N. farcinica; 4, N. nova; 5, N. brasiliensis; 6, N. otitidiscaviarum; 7, N. carnea; 8, N. transvalensis; 9, N. brevicatena; 10, PCR products of N. asteroides ATCC 19247 without digestion; M, 100-bp DNA ladder (Boehringer Mannheim GmbH). pb, base pairs.
On the basis of our results, we could establish a simple and practical scheme for the identification of genus Nocardia through actinomycetes: (i) 596-bp fragment amplification with an MlnI restriction site but no SacI restriction site indicates the DNA of a strain belonging to the genus Nocardia; (ii) no amplification fragment indicates the DNA of a strain not belonging to the genus Nocardia; and (iii) amplification of a 596-bp fragment with a SacI restriction site and/or without an MlnI restriction site indicates the possible nonspecific amplification, requiring confirmation of the identification by conventional methods.
PCR amplification with two primers (primers TB11 and TB12) enabled amplification of a 440-bp fragment coding for a part of a 65-kDa heat shock protein. This fragment was digested for restriction endonuclease analysis with five enzymes (BstEII, HaeIII, MspI, HinfI, and BsaHI). The results allowed establishment of a practical identification scheme according to the numbers and sizes of the bands generated with each enzyme. It has been used for the identification of different species of aerobic actinomycetes, including the Nocardia species, with an accuracy of 98.6% (25, 28). However, since the scheme described for the identification of actinomycete species presented several subgroups (several profiles with several bands for which sizes were not easy to estimate), the interpretation of the result for a clinical strain was difficult within the actinomycete group. On the contrary, if we knew in advance whether the strains belonged to the Nocardia genus, use of interpretation by the technique of Steingrube et al. (25) made identification of the different species easier. Therefore, the technique described here complements the technique of Steingrube et al. (25). The genus- and species-specific techniques could be combined to accomplish the identification of Nocardia clinical isolates within 48 h after receipt of cultures in the reference laboratories. Further studies are needed to develop different systems of identification of all genera belonging to the pathogenic aerobic actinomycetes after the selection of primers specific for each one.
In the future, the PCR method that we developed could be performed directly with clinical samples such as skin biopsy, blood, sputum, or bronchoalveolar lavage specimens. The direct diagnosis of nocardiosis by PCR of cerebrospinal fluid could replace the use of cerebral biopsy, which requires the use of an aggressive technique. It would improve the speed of diagnosis that is closely related to the favorable course for the patients (in terms of mortality, relapses, and aftereffects).
In short, this study showed that the enzymatic amplification of 16S rDNA combined with restriction analysis of the amplimer can be applied to the identification of strains of the genus Nocardia. The technique, as it is described here, is conceived to enable the rapid identification of isolates from pure cultures. The development of such a rapid, simple, and valid assay for detection of members of the genus Nocardia combined with the technique described by Steingrube et al. (25) for the Nocardia species identification will facilitate rapid diagnosis and prompt the initiation of the appropriate chemotherapy. Moreover, it will facilitate epidemiological studies of the human carriage, environmental contamination, and/or soil distribution of these bacteria.
ACKNOWLEDGMENT
We are grateful to Claude de Bievre for help in using the GCG software package.
REFERENCES
- 1.Beaman B L, Beaman L. Nocardia species: host-parasite relationships. Clin Microbiol Rev. 1994;7:213–264. doi: 10.1128/cmr.7.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boiron P, Provost F, Chevrier G, Dupont B. Review of nocardial infections in France 1987 to 1990. Eur J Clin Microbiol Infect Dis. 1992;11:709–714. doi: 10.1007/BF01989975. [DOI] [PubMed] [Google Scholar]
- 3.Boiron P, Provost F, Dupont B, editors. Laboratory methods for the diagnosis of nocardiosis. Paris, France: Institut Pasteur; 1993. [Google Scholar]
- 4.Borrell N, Acinas S G, Figueras M J, Martinez-Murcia A J. Identification of Aeromonas clinical isolates by restriction fragment length polymorphism of PCR-amplified 16S rRNA genes. J Clin Microbiol. 1997;35:1671–1674. doi: 10.1128/jcm.35.7.1671-1674.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng S, McCleskey F K, Gress M J, Petroziello J M, Liu R, Namdari H, Beninga K, Salmen A, DelVecchio V G. A PCR assay for identification of Enterococcus faecium. J Clin Microbiol. 1997;35:1248–1250. doi: 10.1128/jcm.35.5.1248-1250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun J, Goodfellow M. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol. 1995;45:240–245. doi: 10.1099/00207713-45-2-240. [DOI] [PubMed] [Google Scholar]
- 7.Crotchefelt K A, Welsh L E, DeBonville D, Rosentraus D, Quinn T C. Detection of Neisseria gonorrhoeae and Chlamydia trachomatis in genitourinary specimens from men and women by coamplification PCR assay. J Clin Microbiol. 1997;35:1536–1540. doi: 10.1128/jcm.35.6.1536-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng D, Doolittle R F. Progressive sequence alignment as a prerequisite to phylogenetic trees. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 10.Fox G E, Stackebrandt E. The application of 16S rRNA cataloguing and 5S rRNA sequencing in bacterial systematics. Methods Microbiol. 1991;19:405–458. [Google Scholar]
- 11.Garnier F, Gerbaud G, Courvalin P, Galimand M. Identification of clinically relevant viridans group streptococci to the species level by PCR. J Clin Microbiol. 1997;35:2337–2341. doi: 10.1128/jcm.35.9.2337-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodfellow M, Lechevalier M P. Genus Nocardia Trevisan 1889, 9AL. In: Williams S T, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 4. Baltimore, Md: The Williams & Wilkins, Co.; 1989. pp. 2350–2361. [Google Scholar]
- 13.Hou X G, Kawamura Y, Sultana F, Hirose K, Miyake M, Otsuka Y, Misawa S, Oguri T, Yamamoto H, Ezaki T. Genetic identification of members of the genus Corynebacterium at genus and species levels with 16S rDNA-targeted probes. Microbiol Immunol. 1997;41:453–460. doi: 10.1111/j.1348-0421.1997.tb01878.x. [DOI] [PubMed] [Google Scholar]
- 14.Koide M, Saito A. Diagnosis of Legionella pneumophila infection by polymerase chain reaction. Clin Infect Dis. 1995;21:199–201. doi: 10.1093/clinids/21.1.199. [DOI] [PubMed] [Google Scholar]
- 15.Laurent F, Carlotti A, Boiron P, Villard J, Freney J. Ribotyping: a tool for taxonomy and identification of the Nocardia asteroides complex species. J Clin Microbiol. 1996;34:1079–1082. doi: 10.1128/jcm.34.5.1079-1082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lungu O, Della Latta P, Weitzman I, Silverstein S. Differentiation of Nocardia from rapidly growing Mycobacterium species by PCR-RFLP analysis. Diagn Microbiol Infect Dis. 1994;18:13–18. doi: 10.1016/0732-8893(94)90128-7. [DOI] [PubMed] [Google Scholar]
- 17.Mateicka F, Kozakova D, Rosa P A, Kmety E. Identification of Borrelia burgdorferi sensu lato tick isolates from Slovakia by PCR typing with 16S rRNA primers. Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig. 1997;286:355–361. doi: 10.1016/s0934-8840(97)80092-8. [DOI] [PubMed] [Google Scholar]
- 18.McNeil M M, Brown J M. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin Microbiol Rev. 1994;7:357–417. doi: 10.1128/cmr.7.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mordarski M, Schaal K P, Szyba K, Pulverer G, Tkacz A. Interrelation of Nocardia asteroides and related taxa as indicated by deoxyribonucleic acid reassociation. Int J Syst Bacteriol. 1977;27:66–70. [Google Scholar]
- 20.Mordarski M, Schaal K P, Tkacz G, Pulverer G, Szyba K, Goodfellow M. Deoxyribonucleic acid base composition and homology studies on Nocardia. Zentbl Bakteriol Suppl. 1978;6:91–97. [Google Scholar]
- 21.Provost F, Laurent F, Uzcategui L R, Boiron P. Molecular study of persistence of Nocardia asteroides and Nocardia otitidiscaviarum strains in patients with long-term nocardiosis. J Clin Microbiol. 1997;35:1157–1160. doi: 10.1128/jcm.35.5.1157-1160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rainey F A, Klatte S, Korppenstedt R M, Stackebrandt E. Dietzia, a new genus including Dietzia maris comb. nov., formerly Rhodococcus maris. Int J Syst Bacteriol. 1995;45:32–36. doi: 10.1099/00207713-45-1-32. [DOI] [PubMed] [Google Scholar]
- 23.Ramzan N N. Diagnosis and monitoring of Whipple’s disease by polymerase chain reaction. Ann Intern Med. 1997;126:520–527. doi: 10.7326/0003-4819-126-7-199704010-00004. [DOI] [PubMed] [Google Scholar]
- 24.Ruimy R, Boiron P, Boivin V, Christen R. A phylogeny of the genus Nocardia deduced from the analysis of small-subunit ribosomal DNA sequences, including transfer of Nocardia amarae to the genus Gordona as Gordona amarae comb. nov. FEMS Microbiol Lett. 1994;123:261–268. doi: 10.1111/j.1574-6968.1994.tb07234.x. [DOI] [PubMed] [Google Scholar]
- 25.Steingrube V A, Wilson R W, Brown B A, Kenneth C J, Jr, Blacklock Z, Gibson J L, Wallace R J., Jr Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella isolates by DNA amplification and restriction endonuclease analysis. J Clin Microbiol. 1997;35:817–822. doi: 10.1128/jcm.35.4.817-822.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urakawa H, Kita-Tsukamoto K, Ohwada K. 16S rDNA genotyping using PCR/RFLP (restriction fragment length polymorphism) analysis among the family Vibrionaceae. FEMS Microbiol Lett. 1997;152:125–132. doi: 10.1111/j.1574-6968.1997.tb10418.x. [DOI] [PubMed] [Google Scholar]
- 27.White T, Madej J, R, Persing D H. The polymerase chain reaction for the diagnosis of infectious diseases. Adv Clin Chem. 1992;29:161–196. doi: 10.1016/s0065-2423(08)60224-3. [DOI] [PubMed] [Google Scholar]
- 28.Wilson R W, Steingrube V A, Brown B A, Wallace R J., Jr Clinical application of PCR-restriction enzyme pattern analysis for rapid identification of aerobic actinomycetes isolates. J Clin Microbiol. 1998;36:148–152. doi: 10.1128/jcm.36.1.148-152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]