Abstract
V(D)J recombination occurs at recombination signal sequences (RSSs) containing conserved heptamer and nonamer elements. RAG-1 and RAG-2 initiate recombination by cleaving DNA between heptamers and antigen receptor coding segments. RAG-1 alone contacts the nonamer but interacts weakly, if at all, with the heptamer. RAG-2 by itself has no DNA-binding activity but promotes heptamer occupancy in the presence of RAG-1; how RAG-2 collaborates with RAG-1 has been poorly understood. Here we examine the composition of RAG-RSS complexes and the relative contributions of RAG-1 and RAG-2 to heptamer binding. RAG-1 exists as a dimer in complexes with an isolated RSS bearing a 12-bp spacer, regardless of whether RAG-2 is present; only a single subunit of RAG-1, however, participates in nonamer binding. In contrast, multimeric RAG-2 is not detectable by electrophoretic mobility shift assays in complexes containing both RAG proteins. DNA-protein photo-cross-linking demonstrates that heptamer contacts, while enhanced by RAG-2, are mediated primarily by RAG-1. RAG-2 cross-linking, while less efficient than that of RAG-1, is detectable near the heptamer-coding junction. These observations provide evidence that RAG-2 alters the conformation or orientation of RAG-1, thereby stabilizing interactions of RAG-1 with the heptamer, and suggest that both proteins interact with the RSS near the site of cleavage.
Immunoglobulin and T-cell receptor genes are assembled by rearrangement of antigen receptor gene segments during lymphocyte development. This process, termed V(D)J recombination, is mediated by recombination signal sequences (RSSs) composed of conserved heptamer and nonamer elements, separated by spacers of 12 or 23 bp (12-RSSs and 23-RSSs, respectively); recombination normally occurs between gene segments whose RSSs bear spacers of different length (the 12/23 rule). DNA rearrangement is initiated by the recombination activating proteins RAG-1 and RAG-2 (21, 30), which act in concert to introduce a double-strand break (DSB) at the junction between the RSS and the adjacent coding DNA (14, 38). This reaction proceeds in two steps: in the first, a nick is introduced at the 5′ end of the heptamer element flanking the coding DNA; in the second, the resulting 3′ hydroxyl on the coding end attacks a phosphodiester on the opposite strand (14). As a result, two DNA ends are produced: a signal end, terminating in a blunt, 5′-phosphorylated DSB, and a coding end, terminating in a DNA hairpin (14, 23, 25, 31, 38, 39).
Several lines of evidence indicate that V(D)J recombination is a specialized form of DNA transposition (24). These include (i) chemical similarity between RAG-mediated DSB formation and Mu transposition (39), (ii) an analogy between hybrid joint formation and the retroviral disintegration reaction (16), (iii) the ability of the RAG proteins to catalyze integration of signal ends into nonhomologous DNA (1, 9), and (iv) the involvement of hairpin intermediates in the transposition of Tn10 (10). A deeper appreciation of the similarity between V(D)J rearrangement and other types of transposition will require detailed understanding of the form and function of RAG-RSS complexes at different stages of recombination.
At present, however, even RAG-RSS complexes formed prior to initiation of V(D)J recombination are incompletely defined. For example, RAG-1 binds the RSS nonamer through interactions that resemble those of the bacterial invertase Hin with its target site hixL (4, 19, 34, 36) but the stoichiometry of RAG-1-RSS association and its relevance to assembly of the V(D)J preinitiation complex has remained unclear. Likewise, while heptamer contacts exhibit strong dependence on the presence of both RAG-1 and RAG-2 (36), the composition of this complex and the relative roles of RAG-1 and RAG-2 in mediating heptamer contact have not been determined.
Here we define the stoichiometry of RAG proteins in 12-RSS complexes containing RAG-1 with or without RAG-2 and probe the association of RAG-1 and RAG-2 with the RSS heptamer. In complexes with a 12-RSS substrate in the presence or absence of RAG-2, RAG-1 exists as a dimer, although only a single subunit of RAG-1 participates in nonamer binding. In contrast, multimeric RAG-2 was undetectable in electrophoretic mobility shift complexes containing both RAG proteins. By photo-cross-linking to aryl azide-substituted substrate DNA, we show that RAG-2 promotes direct contact between RAG-1 and the heptamer. Cross-linking of RAG-2, while less efficient than that of RAG-1, is most evident near the heptamer-coding junction. These observations indicate that RAG-2 alters the conformation or orientation of RSS-bound RAG-1, thereby stabilizing interactions of RAG-1 with the heptamer, and suggest that both proteins interact with the RSS near the site of cleavage.
MATERIALS AND METHODS
DNA constructs and protein purification.
Expression constructs encoding core fragments of RAG-1 or RAG-2, fused at the amino terminus to a single copy of the maltose binding protein (MBP) and possessing or lacking a carboxy-terminal myc epitope, have been previously described (36). Versions of these vectors encoding two tandemly arrayed copies of MBP fused to RAG-1 and RAG-2 were also constructed. Briefly, an MBP fragment was amplified by PCR from pBSR1MBP(+) (36), by using the reverse primer (5′-GGAAACAGCTATGACCATG) and a primer specific for the MBP–RAG-1 junction, into which an XbaI site had been introduced (5′-GGATCTCTAGAAGAGTCTGACGACCGCTGG). The MBP fragment was subcloned by TA cloning (Invitrogen) to produce the plasmid pCRMBP, and its nucleotide sequence was verified. A 1.2-kb, MBP-encoding fragment was obtained from pCRMBP by digestion with XbaI and cloned into the unique XbaI site upstream of the MBP-RAG coding sequence in pBSR1(MBP+) or pBSR2(MBP+) (36). BamHI-NotI fragments from the resulting pBSR1(MBP2) or pBSR2(MBP2) plasmids were cloned into pcDNA1 as described previously (36).
Single or double MBP-RAG fusion proteins were expressed individually or coexpressed (where noted) in 293 cells and purified by amylose affinity chromatography as described previously (13, 36). For photo-cross-linking experiments, RAG-1 and RAG-2 were coexpressed in 293 cells (10 μg of each plasmid per 10-cm-diameter plate) and purified according to a protocol previously used for RAG-1 (13).
Oligonucleotide substrates for binding, cleavage, and photo-cross-linking.
The standard substrate used in binding and cleavage assays was a 50-bp duplex containing a single 12-RSS, formed by annealing two oligonucleotides, DAR39 and DAR40 (14). Derivatives of DAR39 and DAR40 (SD2504 and SD2505, respectively) containing mutant heptamer and nonamer sequences have been described previously (36). Where indicated, pairs of phosphorothioate linkages were introduced at specific sites within DAR39 or SD2504 during chemical synthesis by using a sulfurizing reagent (Glen Research). Aryl azides were coupled to the phosphorothioate positions within the substrate DNA following a procedure adapted from that of Yang and Nash (42). Briefly, the phosphorothioate-containing oligonucleotides were 5′-end labeled with 32P by using T4 polynucleotide kinase and annealed to a fivefold excess of its unlabeled complement. The DNA was exchanged into 40 mM sodium bicarbonate (pH 9.0) by gel filtration over Sephadex G-50 (NICK column; Pharmacia), and an equal volume (400 μl) of 10 mM 4-azidophenacyl bromide (Fluka) in 100% dimethyl sulfoxide was added. After 1 h at room temperature, the mixture was extracted with isobutanol and the DNA was precipitated in ethanol with linear polyacrylamide as a carrier. The duplex DNA was purified by native polyacrylamide gel electrophoresis (PAGE) as previously described (13).
EMSA.
Binding reactions containing single or double MBP-RAG fusion proteins (alone or coexpressed) were assembled, incubated, and analyzed by electrophoretic mobility shift assay (EMSA) as described previously (36).
DNA cleavage assay.
Cleavage reactions containing RAG-1 and/or RAG-2 were assembled as described previously (13), except that DNA and Me2+ were omitted and incubated at 25°C for various times. Subsequently, the reaction mixtures were supplemented with MgCl2 to a 1 mM final concentration, 32P-labeled substrate DNA, and the omitted RAG protein, where appropriate. Samples were transferred to 37°C and incubated for 20 min. Reaction products were fractionated by denaturing PAGE; 32P was detected by autoradiography and quantified by phosphorimager analysis (Molecular Dynamics). In control reactions, cleavage buffer was incubated at 25°C for the times noted above. Subsequently, the remaining components (RAG proteins, MgCl2, and 32P-labeled DNA) were added simultaneously; samples were immediately transferred to 37°C and incubated for an additional 20 min.
Photo-cross-linking.
RAG-1 or coexpressed RAG-1 and RAG-2 chimeras (∼75 ng each), myc tagged or untagged as described in the text, were incubated with 32P-labeled, aryl azide-derivatized substrate DNA (1 nM) in a 96-well U-bottom tissue culture plate (Becton-Dickinson) under the DNA-binding conditions described previously (36), except that a single-stranded oligonucleotide (DAR81 [5 μM]) was added as a nonspecific competitor. Samples (30 μl) were incubated in the dark for 20 min in a 37°C water bath, placed in an ice-water bath for 5 min, and irradiated through polystyrene with 250,000 μJ of 312-nm UV light (Stratalinker 1800; Stratagene) per cm2. After irradiation, samples were supplemented with sodium dodecyl sulfate (SDS) (2% final concentration), incubated for 10 min at 37°C to disrupt RAG protein complexes and combined with 900 μl of IP buffer (150 mM NaCl, 25 mM TrisCl [pH 8.0], 1% Nonidet P-40, 0.5% deoxycholate) containing 1 mM phenylmethylsulfonyl fluoride. Protein A-purified anti-myc antibody 9E10 (10 μg) was added to each sample, and the mixture was incubated on ice for 1.5 h. Complexes were recovered by immunoprecipitation using protein A/G agarose (30 μl; Protein A/G Plus; Santa Cruz Biotechnology); beads were collected by centrifugation and washed three times with IP buffer containing 0.1% SDS. Radioactivity was detected in samples by Cerenkov counting. Immunoprecipitates were fractionated by SDS-PAGE; RAG proteins were detected by immunoblotting with affinity-purified rabbit polyclonal antibodies to MBP (probe C-18; Santa Cruz Biotechnology).
RESULTS
Stoichiometry of RAG-1 and RAG-2 in RAG-RSS complexes.
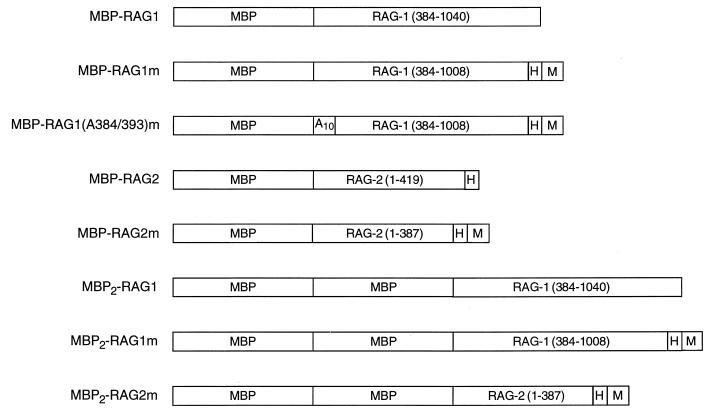
In previous studies using an EMSA, RAG-1 was found to support the formation of distinct DNA-protein complexes in the absence and presence of RAG-2 (36). To probe the stoichiometry of the RAG molecules within these two complexes, chimeric core RAG-1 (amino acids 384 to 1008) and RAG-2 (amino acids 1 to 387) proteins, fused at the amino terminus to one or two copies of the MBP and at the carboxyl terminus to a myc epitope and a polyhistidine tag, were constructed (Fig. 1) (15, 36, 38).
FIG. 1.
RAG-1 and RAG-2 fusion proteins used in this study. MBP, myc (M), and polyhistidine (H) sequences are indicated. RAG-1 and RAG-2 residues are indicated and numbered.
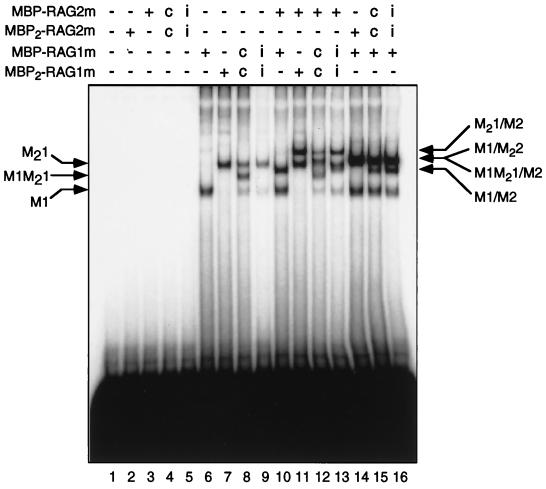
Single or double MBP fusions of RAG-1 or RAG-2 were expressed individually or coexpressed in 293 cells, purified by amylose affinity chromatography, and examined by EMSA for their ability to bind a 12-RSS substrate (Fig. 2). Consistent with previous results (36), no binding was seen in reactions lacking protein (Fig. 2, lane 1), or containing RAG-2 fusion proteins alone (Fig. 2, lanes 2 to 5). Species with retarded mobility (M1 and M21) were detected in reactions containing MBP–RAG-1m or MBP2–RAG-1m alone (Fig. 2, lanes 6 and 7). As expected, the mobilities of these complexes differed, reflecting the presence of one (M1) or two (M21) copies of MBP in the fusion protein. When MBP–RAG-1m and MBP2–RAG-1m were coexpressed, a species whose mobility was intermediate between those of the individually expressed proteins was detected (M1M21) (Fig. 2, lane 8); the mobility of this species, as expected, was shifted by an anti-myc antibody (data not shown). The ratio of the complexes observed in this reaction, as assessed by a phosphorimager, was approximately 1:2:1 (M21:M1M21:M1), as expected if dimerization is random. Dimer formation appears to precede DNA binding, as M1M21 complexes were not detected when individually expressed MBP–RAG-1m and MBP2–RAG-1m were combined and incubated with substrate DNA (Fig. 2, lane 9). This observation is consistent with the predominance of dimeric RAG-1 in the absence of RAG-2 and DNA, as assessed by native PAGE (27). Two observations indicate that formation of the M1M21 complex is not an artifact of the composition of the RAG-1 fusion proteins. First, the M1M21 complex can be formed by RAG-1 chimeras lacking carboxy-terminal tags (data not shown); second, MBP does not mediate dimerization, as indicated by the inability of MBP itself to dimerize (20) and the existence of RAG-2 as a monomer in complex with the 12-RSS and RAG-1 (see below). Thus, we infer that RAG-1 exists as a dimer in the 12-RSS complex.
FIG. 2.
Stoichiometry of RAG proteins in RAG-RSS complexes. A labeled DNA probe bearing a canonical 12-RSS was incubated without (−) protein (lane 1) or with (+) combinations of RAG fusion proteins, as defined in Fig. 1 and indicated at the top, and analyzed by EMSA. Positions of DNA-protein complexes containing only MBP–RAG-1m (M1), only MBP2–RAG-1m (M21), or both (M1M21) are indicated at the left. Positions of DNA-protein complexes containing forms of RAG-1 with either MBP–RAG-2m (M1/M2, M1M21/M2, and M21/M2) or MBP2–RAG-2m (M1/M22) are designated at the right. i, protein obtained from cells expressing single or double MBP-RAG fusions individually; c, protein obtained from cells coexpressing single and double RAG fusions.
When the RSS substrate was incubated with MBP–RAG-2m and MBP–RAG-1m or MBP2–RAG-1m, two species were observed: a faster-migrating complex which comigrated with the species formed in the presence of RAG-1 alone (Fig. 2, compare lanes 6 and 7 to lanes 10 and 11) and an additional complex with retarded mobility (M1-M2 and M21-M2, respectively; Fig. 2, lanes 10 and 11). When individually expressed MBP–RAG-1m and MBP2–RAG-1m were combined and incubated with the RSS substrate and MBP–RAG-2m, four species were distinguishable (Fig. 2, lane 13). These corresponded in mobility to the pairs of species present in reactions containing a single RAG-2 chimera and MBP–RAG-1m or MBP2–RAG-1m (compare lane 13 to lanes 10 and 11). Coexpressed MBP–RAG-1m and MBP2–RAG-1m, in contrast, formed more than four species in the presence of MBP–RAG-2m and the RSS substrate (Fig. 2, lane 12). Several of these were easily resolved and comigrated with M1, M1M21, and M21-M2. Complexes corresponding to M21 and M1-M2 were faint or poorly resolved. However, a new species with mobility intermediate between that of M21 and M21-M2 was clearly visible (compare lanes 12 and 13), and is consistent with formation of an M1M21-M2 complex. These data suggest that RAG-1 retains its dimer configuration when associated with the 12-RSS substrate and RAG-2.
In agreement with an earlier study (36), two RAG-RSS complexes were detected in reactions containing MBP–RAG-1m and MBP2–RAG-2m: a slower species (M1/M22) and a species that comigrated with the M1 complex (Fig. 2, lane 14). When MBP–RAG-2m and MBP2–RAG-2m were expressed individually, combined and incubated with MBP–RAG-1m, the same qualitative and quantitative pattern of RAG-RSS complexes was observed as in reactions containing the coexpressed RAG-2 chimeras (Fig. 2, compare lanes 15 and 16). Notably, these complexes comigrated with species formed in reactions containing MBP–RAG-1m and individually expressed MBP–RAG-2m or MBP2–RAG-2m (compare lanes 10 and 14 to lanes 15 and 16), even at lower exposures (data not shown). Taken together, these data suggest that the 12-RSS precleavage complex contains dimeric RAG-1 and monomeric RAG-2.
A single functional subunit is sufficient for binding of RAG-1 to an isolated RSS nonamer.
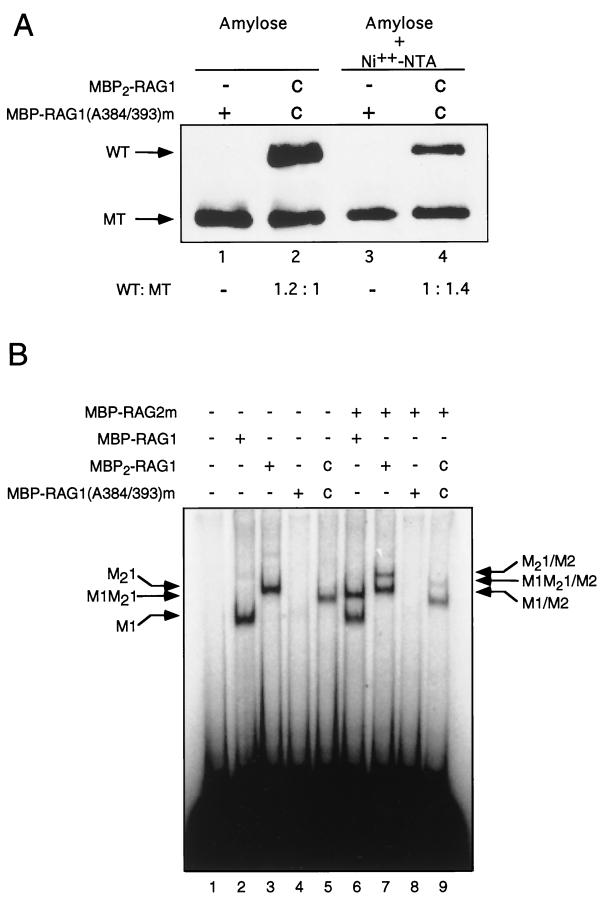
We next asked whether one or both subunits of the RAG-1 dimer participated in nonamer binding. To address this, a binding-deficient RAG-1 mutant chimera, containing amino-terminal MBP and a carboxy-terminal polyhistidine tag, was expressed alone or together with a wild-type (WT) RAG-1 chimera containing two tandem, amino-terminal copies of MBP but lacking a polyhistidine tag (MBP2-RAG1) (Fig. 1). The mutant (MT) protein used in this experiment, MBP–RAG-1(A384/393)m (Fig. 1), carries an alanine substitution that impairs specific binding to the RSS (36). This mutation lies in a region of the protein previously shown to be essential for nonamer binding (4, 34). The expressed proteins were purified by amylose affinity chromatography, followed by affinity chromatography over Ni2+-nitrilotriacetic (Ni2+-NTA) resin. From cells expressing both proteins, the amylose column was expected to retain WT or MT homodimers and the heterodimer. Only the MT homodimer and heterodimer were expected to be retained on the Ni2+-NTA column, as the WT chimera lacks the polyhistidine tag. RAG-1 chimeras retained at the first and second chromatographic steps were examined by immunoblotting (Fig. 3A). Although the amount of MBP2–RAG-1 eluted from the amylose resin was present at a 1.2-fold excess over MBP–RAG-1(A384/393)m (Fig. 3A, lanes 1 and 2), the ratio of WT to MT RAG-1 chimera following elution from Ni2+-NTA was 1:1.4 (Fig. 3A, lanes 3 and 4), consistent with loss of WT dimers. DNA cleavage activity was detectable in the Ni2+-NTA eluate containing heterodimeric RAG-1, but not in the eluate containing homomeric mutant RAG-1 alone (data not shown).
FIG. 3.
Nonamer binding in a 12-RSS substrate is supported by a single subunit of the RAG-1 dimer. (A) MBP–RAG-1(A384/393)m was expressed alone or coexpressed (c) with MBP2–RAG-1. After amylose affinity chromatography (lanes 1 and 2) and Ni2+-chelate affinity chromatography (lanes 3 and 4), the levels of WT and MT RAG-1 fusion proteins were determined by immunoblotting with anti-RAG-1 antibody Ab307 (14). The positions of WT and MT RAG-1 fusion proteins are designated at left. (B) The radiolabeled 12-RSS probe was incubated without (−) protein (lane 1) or with (+) RAG-1 fusion proteins (indicated above and defined in Fig. 1) which had been purified by amylose affinity chromatography (lanes 2, 3, 6, and 7) or consecutive rounds of amylose and Ni2+ affinity chromatography (lanes 4, 5, 8, and 9). Reactions were carried out in the absence (lanes 2 to 5) or presence (lanes 6 to 9) of RAG-2. The positions of DNA-protein complexes are indicated as described in the legend to Fig. 2.
Purified RAG-1 chimeras were then assayed for RSS binding by EMSA in the absence (Fig. 3B, lanes 2 to 5) or presence (Fig. 3B, lanes 6 to 9) of RAG-2. As expected, MBP–RAG-1 and MBP2–RAG-1 formed RSS complexes in the absence of RAG-2 (M1 and M21, respectively; Fig. 3B, lanes 2 and 3). As expected, the MT chimera did not detectably bind substrate DNA in the absence or presence of RAG-2 (Fig. 3B, lanes 4 and 8). When MBP2–RAG-1 and MBP–RAG-1(A384/393)m were coexpressed and copurified, they yielded a species of intermediate mobility (M1M21), consistent with the formation of a binding-competent heterodimer containing WT and MT subunits (Fig. 3B, lane 5). No species comigrating with M21 was detected, indicating that the WT homodimer was efficiently removed by the purification regimen and that subunit exchange occurs inefficiently under these conditions. In the presence of RAG-2, additional species with retarded mobility were observed in reactions containing MBP–RAG-1 or MBP2–RAG-1 (M1-M2 and M21-M2, respectively; Fig. 3B, lanes 6 and 7), consistent with observations presented above (Fig. 2). In the reaction containing MBP2–RAG-1 and MBP–RAG-1(A384/393)m, a species (M1M21-M2) was detected whose mobility was lower than that of M1M21 and intermediate between those of M1-M2 and M21-M2 (Fig. 3B, lane 9). Taken together these observations indicate that (i) in the single RSS precleavage complexes examined here, only a single subunit of the RAG-1 dimer participates in nonamer binding and (ii) a RAG-1 dimer containing only one binding-competent subunit can associate with RAG-2 in an RSS complex. Moreover, because mutant homodimers failed to form stable complexes, while heterodimers containing MT and WT subunits were able to bind DNA, we infer, assuming one DNA binding site per RAG-1 subunit, that a single DNA fragment is present in the shifted species observed in Fig. 2 and Fig. 3B.
Prior interaction of RAG-1 with RAG-2 in the absence of DNA enhances cleavage activity.
Several laboratories have described an association of RAG-1 with RAG-2 in the absence of DNA (12, 15, 27, 33), but the relationship of this association to RSS recognition or cleavage has remained unclear. Indeed, because RAG-1 alone can interact with an RSS it has been suggested that RAG-1 recruits RAG-2 to the RSS (4). We wished to address whether the efficiency of DNA cleavage at the RSS is affected by prior interaction of RAG-1 with RAG-2 in the absence of substrate DNA.
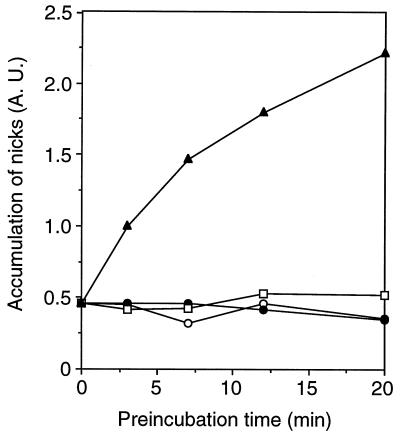
RAG-1 and RAG-2, alone or in combination, were incubated at 25°C for times ranging from 0 to 20 min. Mg2+ and radiolabeled RSS substrate were then added, the samples were transferred to 37°C and the amount of nicked substrate was assessed 20 min thereafter (Fig. 4). In reactions containing a single RSS, Mg2+ supports nicking but not hairpin formation, simplifying assessment of RAG activity (14). The amount of nicked-product formation was unaltered by preincubation of RAG-1 or RAG-2 alone over the period examined. In these instances the yields of nicked product were similar to those obtained from matched control reactions in which RAG-1, RAG-2, Mg2+, and DNA were combined simultaneously just prior to incubation at 37°C. Thus, the individual RAG proteins are stable under these conditions for at least 20 min in the absence of DNA and Mg2+. When RAG-1 and RAG-2 were preincubated together, however, the yield of nicked product increased sharply with increasing time of preincubation (Fig. 4). These results provide evidence that RAG-1 and RAG-2 interact prior to substrate recognition in a manner that increases their activity. This interaction may reflect formation of a stable complex between the two proteins, as RAG-1 and RAG-2 can be coimmunoprecipitated from these preincubation reactions (data not shown), consistent with previous results.
FIG. 4.
DNA cleavage activity is enhanced by preincubation of RAG-1 with RAG-2 in the absence of substrate. RAG-1 and RAG-2 were incubated separately or together at 25°C in cleavage buffer lacking DNA and Me2+ for times ranging from 0 to 20 min. The reaction mixtures were then supplemented with 32P-labeled substrate DNA, MgCl2 to a concentration of 1 mM, and the omitted RAG protein where appropriate. Samples were transferred to 37°C and incubated for 20 min. In control reactions, cleavage buffer was incubated at 25°C for the times noted above. Subsequently, the remaining components (RAG proteins, MgCl2, and 32P-labeled DNA) were added simultaneously; samples were immediately transferred to 37°C and incubated for an additional 20 min. Reaction products were fractionated by denaturing PAGE and quantified with a phosphorimager. Accumulation of nicked product (measured in arbitrary absorption units [A.U.]) is plotted as a function of preincubation time for reactions in which RAG-1 or RAG-2 was preincubated separately (closed circles and open squares, respectively), preincubated together (filled triangles), or combined simultaneously with MgCl2 and labeled substrate (open circles).
RAG-2 induces RAG-1 to contact the RSS heptamer region.
In RSS complexes containing RAG-1 alone, DNA contacts are centered on the nonamer, while in complexes containing RAG-1 and RAG-2, protein-RSS interactions extend through the spacer and into the heptamer (36). While RAG-1 and RAG-2 collaborate in heptamer recognition, the nature of this collaboration has remained unknown. Extension of DNA-protein contacts into the heptamer could reflect binding of RAG-2 at a site adjacent to RAG-1; alternatively or in addition, RAG-2 might alter the conformation or orientation of RAG-1 so as to promote contact between RAG-1 and the heptamer. We addressed this question by covalent trapping (42). Photoreactive aryl azides were coupled to phosphorothioates that had been introduced at specific positions in or around the heptamer during chemical synthesis of substrate oligonucleotides. The placement of phosphorothioates was guided by the pattern of heptamer phosphate contacts previously identified by ethylation interference (36). Three pairs of backbone positions were chosen (Fig. 5): one pair (S15-S16) was placed at the coding end, adjacent to the heptamer; the other two pairs were placed on either side of the region of RAG-2-dependent ethylation interference (S20-S21 and S26-S27). Denaturing gel electrophoresis of derivatized substrates bearing canonical (WT) or MT heptamer and nonamer elements showed a reduction in the amount of uncoupled oligonucleotide and the appearance of two slower-migrating species, reflecting the coupling of an aryl azide to one or both phosphorothioates (data not shown). Coupling efficiency was similar for both WT and MT substrates, with at least 80% of the input oligonucleotide derivatized in each instance (data not shown). All three derivatized wild-type substrates were bound similarly by the RAG proteins, as assessed by EMSA (data not shown).
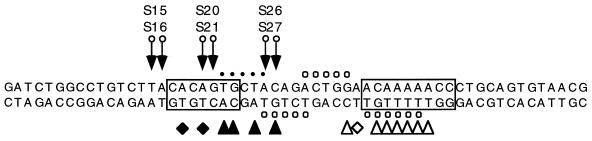
FIG. 5.
Aryl azide derivatization of 12-RSS substrates. Aryl azide moieties (arrows) were coupled to one of three pairs of phosphorothioates (S15-S16, S20-S21, and S26-S27) in the heptamer regions of WT or MT 12-RSS substrates, as defined in Materials and Methods (the wild-type substrate is depicted here). Specific RAG-DNA contacts, as defined by ethylation interference (circles) and methylation or KMnO4 interference (triangles), are marked. Putative RAG-induced structural perturbations in substrate DNA, as defined by overrepresentation of KMnO4 modification, are indicated by diamonds. Open symbols represent interactions detected in 12-RSS complexes containing RAG-1 alone; closed and open symbols, taken together, denote interactions detected in complexes containing RAG-1 and RAG-2.
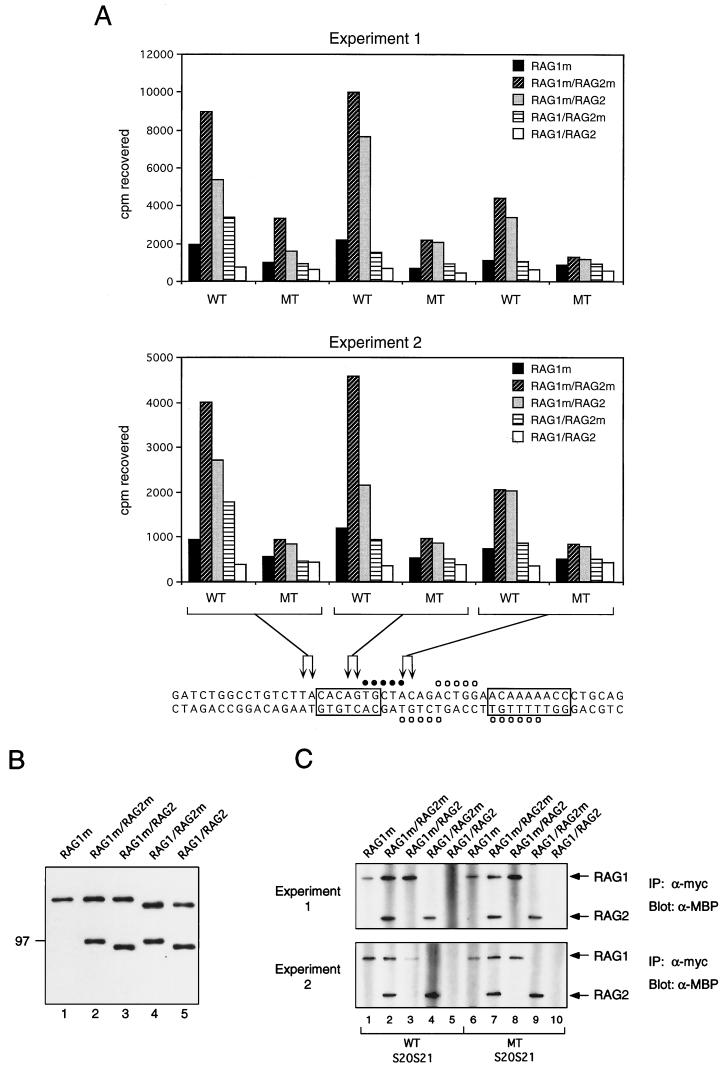
MBP fusions of RAG-1 and RAG-2 containing or lacking the myc epitope (Fig. 1) were incubated in various combinations with radiolabeled, derivatized RSS substrates. The reaction mixes were irradiated at 312 nm, noncovalent associations were disrupted by detergent, and protein(s) was immunoprecipitated with an anti-myc antibody. Coexpressed RAG-1 and RAG-2 proteins were used in this experiment, as they are more active than their individually expressed counterparts (8). The yields of singly expressed and coexpressed RAG-1 chimeras were similar to each other and to the yields of RAG-2, as assessed by immunoblotting (Fig. 6B). The RSS binding and cleavage activities of coexpressed proteins were similar within a factor of 2 (data not shown).
FIG. 6.
Photo-cross-linking of RAG proteins to a 12-RSS substrate. (A) Aryl azide-derivatized, radiolabeled WT or MT substrates were incubated in two independent experiments with myc-tagged RAG-1 (RAG1m), or with pairwise combinations of RAG chimeras containing (RAG1m or RAG2m) or lacking (RAG1 or RAG2) epitope tags, as indicated. All fusion proteins contained one copy of MBP at the amino terminus. After binding, samples were irradiated with 312-nm UV light. Noncovalent associations were disrupted by treatment with detergent, and radioactivity was immunoprecipitated with an anti-myc antibody. Samples are grouped according to the positions of aryl azide derivatives, as indicated. Sites of ethylation interference in complexes containing RAG-1 alone (open circles) or RAG-1 and RAG-2 (open and closed circles) are shown. (B) Purified fusion proteins, described in panel A, fractionated by SDS-PAGE, and detected by immunoblotting with anti-MBP antibody. The purity of the proteins was judged to be greater than 90% by silver staining. (C) Anti-myc immunoprecipitates (IP: α-myc) from cross-linking reactions in panel A were fractionated by SDS-PAGE, and RAG fusion proteins were detected by immunoblotting with an anti-MBP antibody (α-MBP). A representative immunoblot from each experiment, comparing RAG protein levels in immunoprecipitates from the S20-S21 sample group, is shown. The positions of RAG-1 and RAG-2 fusion proteins are indicated at right.
In two independent experiments, the highest levels of protein-DNA cross-linking were observed when myc-tagged RAG-1 was incubated with WT substrate in the presence of tagged or untagged RAG-2; the most efficient cross-linking occurred when the substrate was derivatized at positions S20-S21, in the middle of the heptamer, although specific cross-linking was observed at the other positions as well (Fig. 6A). In contrast, substantially less radioactivity was precipitated from reactions containing tagged RAG-1 in the absence of RAG-2 or untagged RAG-1 in the presence of tagged RAG-2 (Fig. 6A). In the presence of RAG-2, cross-linking of RAG-1 was RSS specific, as mutation of the heptamer and nonamer greatly reduced the amount of radiolabeled DNA precipitated (Fig. 6A). Very little radioactivity was recovered from reactions in which neither RAG protein was tagged, demonstrating dependence of precipitation on the presence of the myc epitope (Fig. 6A). Moreover, the amount of radioactivity recovered from reactions containing underivatized, phosphorothioate-containing oligonucleotides was at least 10-fold lower than that from reactions containing derivatized substrates (data not shown), indicating that irradiation at 312 nm specifically cross-linked DNA to protein through the aryl azide moieties.
Immunoblotting of anti-myc immunoprecipitates with anti-MBP antibodies demonstrated that detergent treatment of samples after cross-linking efficiently disrupted RAG-1–RAG-2 interactions (Fig. 6C); moreover, differential recovery of these proteins does not account for differences in radioactivity precipitated with RAG-1 as opposed to RAG-2 (compare Fig. 6A and C). (The relatively low yield of radioactivity seen in the reaction of experiment 2 containing RAG-1m, RAG-2, and the WT S20-S21 substrate [Fig. 6A] may be explained in part by the poor recovery of RAG-1m in the immunoprecipitate [Fig. 6C, lower panel, lane 3].) Although RAG-1 exhibited a low level of specific cross-linking to the heptamer region in the absence of RAG-2, the degree of discrimination between MT and WT substrates was two- to threefold lower than in reactions containing both proteins (Fig. 6A), consistent with the relative nonspecificity of DNA binding by RAG-1 in isolation (3, 36). In reactions containing both RAG proteins, RAG-2 exhibited a low level of specific cross-linking to the substrate (Fig. 6A). Specific RAG-2 cross-linking showed relatively little dependence on the positions of aryl azides but was most evident at the coding base pairs abutting the heptamer (S15-S16). From these results we infer that heptamer contacts, while enhanced by the presence of RAG-2, are mediated primarily by RAG-1. These observations provide strong evidence that RAG-2 alters the conformation or orientation of RAG-1 so that the latter is more able to engage the heptamer and suggest that both proteins interact with the RSS near the site of cleavage.
DISCUSSION
Collaboration between RAG-2 and RAG-1.
While RAG-1 are RAG-2 are both necessary and sufficient for the initiation of V(D)J recombination, the relative roles of the two proteins in mediating RSS recognition have been unclear. In the absence of RAG-2, RAG-1 exhibits relatively poor binding specificity for RSS sequences (3, 36), although interactions with the nonamer are evident from one-hybrid assays (4), surface plasmon resonance (34), modification interference, and DNA footprinting (19, 36). These interactions are impaired by mutations in a region of purported homology between RAG-1 and the prokaryotic invertase Hin, whose recognition site hix resembles the RSS nonamer (4, 34). Modification interference and direct footprinting analysis support an analogy between Hin-hix and RAG-1–nonamer interactions (19, 36). Nonetheless, in the absence of RAG-2 the ability of RAG-1 to discriminate between specific and nonspecific DNA is weak.
Several lines of evidence have indicated that RAG-2 collaborates with RAG-1 to achieve recognition of the heptamer: (i) in one-hybrid assays, transactivation in the presence of RAG-1 and RAG-2 is more heptamer dependent than in the presence of RAG-1 alone (4); (ii) protein-DNA complexes containing RAG-1 and RAG-2, as detected by EMSA, are more sensitive to heptamer mutation than complexes containing RAG-1 alone (3, 8, 36); and (iii) heptamer contacts are observed in protein-DNA complexes containing both RAG-1 and RAG-2, but not in those containing RAG-1 alone (36). In principle, RAG-2 could exert its effect on RSS recognition directly, by binding the heptamer, or indirectly, by modifying the conformation or orientation of RAG-1. These possibilities are not mutually exclusive.
The photo-cross-linking experiments presented here demonstrate that RAG-1 makes direct, RAG-2-dependent contacts with the RSS in the vicinity of the heptamer. This result is consistent with the observation that certain RAG-1 mutations impart sensitivity to changes in the coding sequence flanking the RSS, providing indirect evidence for the interaction of RAG-1 with the heptamer-coding junction (22, 26). The distances between RAG-1 and its points of contact with the heptamer region are likely to be on the order of 11 Å or less, which is the distance between a backbone phosphorus atom and the reactive nitrogen of the aryl azide to which it is coupled (42). The corresponding uncertainty in localization of RAG-DNA contacts is on the order of 3.3 to 3.6 bp or less. Specific cross-linking of RAG-2 to the heptamer region is less efficient, consistent with a greater average backbone-to-protein distance than exists for RAG-1. Insofar as it is detectable, specific cross-linking of RAG-2 is most apparent at the coding positions abutting the RSS, while RAG-1 is most efficiently cross-linked to residues in the middle of the heptamer. This observation leaves open the possibility that RAG-2 most closely approaches the RSS near the site of DNA cleavage.
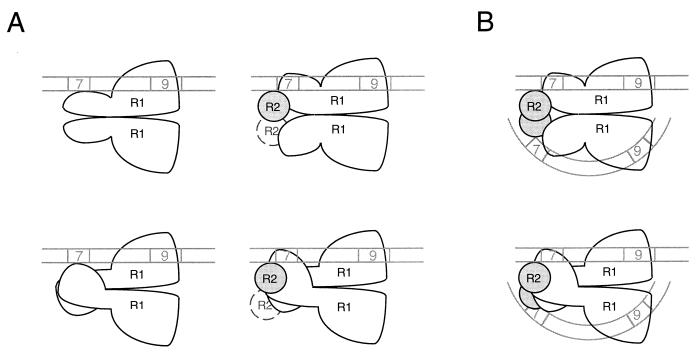
How might RAG-2 promote heptamer occupancy by RAG-1? In principle, RAG-2 might alter the stoichiometry of RAG-1 in RSS complexes, but this possibility is eliminated by the observation that RAG-1 exists as a dimer in RSS complexes regardless of the presence of RAG-2 (Fig. 2) (27). We favor, rather, a mechanism in which RAG-2 alters the conformation of one or both subunits of the RAG-1 dimer. This conformational change need not occur after association with DNA. Indeed, the enhancement of RSS cleavage activity observed upon preincubation of the RAG proteins in the absence of DNA (Fig. 4) could be explained if association of RAG-1 with RAG-2, or a subsequent, DNA-independent isomerization of the RAG-1–RAG-2 complex, were rate limiting for DNA cleavage. The available data are consistent with the following interpretation (Fig. 7). On an isolated 12-RSS substrate in the absence of RAG-2, a single subunit of dimeric RAG-1 participates in nonamer binding; heptamer interactions are undetectable by EMSA or modification interference (36) (Fig. 7A). Inclusion of RAG-2 with RAG-1 in the 12-RSS binding reaction permits formation of a complex in which one or both RAG-1 subunits have undergone a RAG-2-dependent conformational change that enforces occupancy of the heptamer region. Heptamer binding by RAG-1 could, in principle, be mediated by either subunit of the RAG-1 dimer. The data do not imply that either of these single RSS-containing complexes is an intermediate in physiologic V(D)J recombination.
FIG. 7.
Models of RAG-RSS interactions. (A) Models for RAG interactions with an isolated 12-RSS. 12-RSS complexes in the absence (left) and presence (right) of RAG-2 are depicted. A single subunit mediates binding of the RAG-1 (R1) dimer to the nonamer; RAG-1 interacts weakly, if at all, with the heptamer in the absence of RAG-2. RAG-2 (R2), by effecting a change in the conformation of RAG-1, induces the latter protein to contact the heptamer. The available data do not distinguish between models in which RAG-1 contacts the heptamer in cis (upper) or trans (lower). Mobility shift experiments suggest that RAG-2 exists as a monomer in the single-site complex, but the presence of an additional RAG-2 subunit (dashed circle) cannot be rigorously excluded. The diagram is not meant to imply that RAG-2 is recruited to a prestablished RAG-1–DNA complex; rather, RAG-1 and RAG-2 may associate before interacting with the RSS. Moreover, these models are not meant to represent intermediates in the formation of synaptic complexes. (B) Speculative views of the synaptic complex. 12- and 23-RSSs are depicted by straight and curved double lines, respectively. Each subunit of the RAG-1 dimer interacts with a separate nonamer element; RAG-1 interactions with the heptamer could occur in cis (upper) or trans (lower). RSS orientation is based on the observation that the minimum intersignal distance required for efficient coupled cleavage (5) or recombination (32) is shortest when signals are oriented heptamer to heptamer. DNA bending proteins (e.g., HMG-1 or HMG-2) that may facilitate synaptic complex assembly by enhancing RAG binding to 23-RSSs (37) are not shown. For details, see the text.
Implications for synaptic complex formation.
The results of stoichiometric measurements and cross-linking studies presented here, although carried out with an isolated RSS substrate, may provide insight into the composition of synaptic complexes containing 12- and 23-RSSs. Because a single functional subunit of the RAG-1 dimer can support nonamer binding (Fig. 3B), a dimer of RAG-1 could in principle bind two RSSs. Thus, in one simple model for a synaptic complex, each subunit of a RAG-1 dimer would bind a nonamer; the associated heptamer could, in principle, be bound by the same or the opposite subunit (Fig. 7B). Such a complex could be assembled by any of several pathways, including (i) stepwise capture of each RSS, (ii) simultaneous capture of both RSSs, or (iii) formation of RAG-RSS complexes at separate sites, followed by synapsis formation through protein-protein interactions. The available data do not distinguish among these possibilities, although the existence of RAG-1 as a stable dimer would be most consistent with the first two.
Within the limitations of the mobility shift assay employed here, only monomeric RAG-2 was detectable in 12-RSS complexes containing RAG-1 (Fig. 2; Fig. 7B). It remains possible, however, that single RSS complexes containing RAG-2 dimers are short-lived and thus undetectable by our assay. Moreover, the stoichiometry of RAG-2 in synaptic complexes is not known; heptamer recognition by a second RAG-1 subunit could require the participation of an additional RAG-2 monomer, as drawn here (Fig. 7B). Other proteins, such as HMG-1, are likely to be required for efficient assembly of synaptic complexes (2, 7, 37). The clustering of RAG-RSS backbone contacts on one side of the helix (36) could, in principle, leave the opposite face of the DNA free to contact accessory proteins such as HMG-1. Because HMG-1 is expected to bend DNA away from itself (41), this arrangement might facilitate simultaneous engagement of the 23-spacer nonamer and heptamer by RAG-1. The dimerization of RAG-1 and its ability, in the presence of RAG-2, to contact both the heptamer and the nonamer, suggest that one RAG-1 subunit may transmit information concerning the length of its associated spacer to the other subunit, thereby assisting enforcement of the 12/23 rule (5, 7, 35, 40).
RAG-RSS recognition and formation of DNA transposition complexes.
V(D)J recombination is a specialized form of transposition, specifically related to the cut-and-paste reactions employed by Tn7 and Tn10 (1, 9, 10, 39). In addition, similarities between nonamer recognition by RAG-1 and hix recognition by Hin (4, 19, 34, 36) have suggested that V(D)J recombination and Hin-mediated inversion share additional features.
The synaptic complexes associated with bacteriophage Mu transposition and Hin-mediated DNA inversion have been characterized in some detail, and it may be useful to compare these systems with the model for RAG-RSS recognition presented here. In the case of phage Mu transposition, stable synaptic complex formation, strand cleavage, and strand transfer occur within a protein-DNA complex termed the transpososome; within this complex, four Mu A monomers are tightly bound to the ends of the Mu genome (11). Assembly of the transpososome is supported by Ca2+, but strand cleavage does not occur unless Ca2+ is replaced with Mn2+ or Mg2+ (18). Notably, the electrophoretic mobility of the stable synaptic complex is not affected when Ca2+ is substituted for Mg2+ (29). In these respects, the Mu transpososome resembles a RAG-RSS complex (8, 14).
The model of RAG synaptic complex formation presented in Fig. 7B would also resemble the Mu and Hin systems in the use of a four-subunit core, except that the RAG system would utilize two distinct proteins, rather than a Mu tetramer or a pair of Hin dimers (6, 11). The use of two gene products to initiate strand cleavage and transfer has precedence in the Tn7 transposition system (28). Unlike Tn7 transposition, however, in which TnsA and TnsB catalyze distinct DNA processing reactions at the ends of the transposon (28), no such distinction exists between the sites of DNA cleavage in V(D)J recombination, as isolated 12- and 23-RSSs can each serve as substrates for the recombinase in vitro (14). A closer analogy to the action of RAG-2 may be provided by the mechanism by which the protein Fis stimulates Hin-catalyzed inversion. A dimer of Fis, bound to a recombinational enhancer element, is proposed to induce a conformational change in Hin; this would place the Hin active sites close to the scissile phosphodiester bonds at hixL and hixR, thereby facilitating double strand cleavage (17). Whether RAG-2 plays an analogous role in the initiation of V(D)J recombination awaits determination of the relative roles of the RAG proteins in catalysis of DNA cleavage.
ACKNOWLEDGMENTS
This work was supported by the Howard Hughes Medical Institute and by grant CA16519 from the National Cancer Institute. P.C.S. is an associate of the Howard Hughes Medical Institute.
Footnotes
We dedicate this paper to the memory of our friend and colleague Eugenia Spanopoulou.
REFERENCES
- 1.Agrawal A, Eastman Q M, Schatz D G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal A, Schatz D G. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 3.Akamatsu Y, Oettinger M A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequence. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 5.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 6.Heichman K A, Johnson R C. The Hin invertasome: protein-mediated joining of distant recombination sites at the enhancer. Science. 1990;249:511–517. doi: 10.1126/science.2166334. [DOI] [PubMed] [Google Scholar]
- 7.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 8.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 9.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy A K, Guhathakurta A, Kleckner N, Haniford D B. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 11.Lavoie B D, Chan B S, Allison R G, Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu Type 1 transpososome. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leu T M J, Schatz D G. rag-1 and rag-2 are components of a high-molecular-weight complex, and association of rag-2 with this complex is rag-1 dependent. Mol Cell Biol. 1995;15:5657–5670. doi: 10.1128/mcb.15.10.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Swanson P, Desiderio S. RAG-1- and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol Cell Biol. 1997;17:6932–6939. doi: 10.1128/mcb.17.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 15.McMahan C J, Sadofsky M J, Schatz D G. Definition of a large region of RAG1 that is important for coimmunoprecipitation of RAG2. J Immunol. 1997;158:2202–2210. [PubMed] [Google Scholar]
- 16.Melek M, Gellert M, van Gent D C. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 1998;280:301–303. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- 17.Merickel S K, Haykinson M J, Johnson R C. Communication between Hin recombinase and Fis regulatory subunits during coordinate activation of Hin-catalyzed site-specific DNA inversion. Genes Dev. 1998;12:2803–2816. doi: 10.1101/gad.12.17.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuuchi M, Baker T A, Mizuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992;70:303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- 19.Nagawa F, Ishiguro K-I, Tsuboi A, Yoshida T, Ishikawa A, Takemori T, Otsuka A J, Sakano H. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol Cell Biol. 1998;18:655–663. doi: 10.1128/mcb.18.1.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.New England Biolabs. Unpublished data.
- 21.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 22.Roman C, Baltimore D. Genetic evidence that the RAG1 protein directly participates in V(D)J recombination through substrate recognition. Proc Natl Acad Sci USA. 1996;93:2333–2338. doi: 10.1073/pnas.93.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth D, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth D B, Craig N L. VDJ recombination: a transposase goes to work. Cell. 1998;94:411–414. doi: 10.1016/s0092-8674(00)81580-9. [DOI] [PubMed] [Google Scholar]
- 25.Roth D B, Menetski J P, Nakajima P B, Bosma M J, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 26.Sadofsky M J, Hesse J E, van Gent D C, Gellert M. RAG-1 mutations that affect the target specificity of V(D)J recombination: a possible direct role of RAG-1 in site recognition. Genes Dev. 1995;9:2193–2199. doi: 10.1101/gad.9.17.2193. [DOI] [PubMed] [Google Scholar]
- 27.Santagata S, Aidinis V, Spanopoulou E. The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J Biol Chem. 1998;273:16325–16331. doi: 10.1074/jbc.273.26.16325. [DOI] [PubMed] [Google Scholar]
- 28.Sarnovsky R J, May E W, Craig N L. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 29.Savilahti H, Rice P A, Mizuuchi K. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 31.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 32.Sheehan K M, Lieber M R. V(D)J recombination: signal and coding joint resolution are upcoupled and depend on parallel synapsis of the sites. Mol Cell Biol. 1993;13:1363–1370. doi: 10.1128/mcb.13.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spanopoulou E, Cortes P, Shih C, Huang C-M, Silver D P, Svec P, Baltimore D. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 1995;3:715–726. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 34.Spanopoulou E, Zaitseva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 35.Steen S B, Gomelsky L, Roth D B. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 36.Swanson P C, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 37.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 39.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 40.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 41.Werner M H, Hugh J R, Gronenborn A M, Clore G M. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 42.Yang S-W, Nash H A. Specific photocrosslinking of DNA-protein complexes: identification of contacts between integration host factor and its target DNA. Proc Natl Acad Sci USA. 1994;91:12183–12187. doi: 10.1073/pnas.91.25.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]