ABSTRACT
Background
Healthy plant-based diet index (hPDI) is associated with a lower risk of cardiometabolic conditions, but its association as well as interactions with microbiome have not been elucidated.
Objectives
We aimed to investigate the interrelations between hPDI, gut microbiome, and cardiometabolic risk markers.
Methods
hPDI was derived from dietary assessments by a validated FFQ and was examined in relation to metagenomic profiles of 911 fecal samples collected from 303 men aged 71 ± 4 y with an average BMI (in kg/m2) of 25.2 ± 3.6 in the Men's Lifestyle Validation Study. Principal coordinate (PCo) analysis based on Bray–Curtis dissimilarity was conducted, and interactions between hPDI and PCo were examined by using a metabolic risk score composed of blood lipids, BMI, and glycated hemoglobin.
Results
After multivariable adjustment, hPDI was significantly associated with the relative abundance of 7 species and 9 pathways. In particular, higher hPDI was significantly associated with a higher relative abundance of Bacteroides cellulosilyticus and Eubacterium eligens, amino acid biosynthesis pathways (l-isoleucine biosynthesis I and III and l-valine biosynthesis), and the pathway of pyruvate fermentation to isobutanol. A favorable association between hPDI and the metabolic risk score was more pronounced among men with a higher PCo characterized by higher abundance of Bacteroides uniformis and lower abundance of Prevotella copri. At the individual species level, a similar interaction was also observed between hPDI and P. copri, as well as with Clostridium clostridioforme or Blautia hydrogenotrophica (all P-interaction < 0.01).
Conclusion
A greater adherence to a healthy plant-based diet by older men was associated with a microbial profile characterized by a higher abundance of multiple species, including B. cellulosilyticus and E. eligens, as well as pathways in amino acid metabolism and pyruvate fermentation. In addition, inverse associations between healthy plant-based diet and human metabolic risk may partially depend on microbial compositions.
Keywords: plant-based diet index, microbiome, metabolic, species, diet
Introduction
There is a rapidly growing body of evidence suggesting that diet plays a lifelong role in the mutualistic relation between humans and their gastrointestinal microbiota (1). The gastrointestinal microbiota harvest energy and micronutrients through fermentation of dietary fiber and other constituents from the host's diet. The host may subsequently also absorb energy and beneficial compounds (e.g., SCFAs and enterolignans) that are otherwise inaccessible to humans (2). Conversely, the microbiota also generate bioactive compounds that may adversely influence human health, such as trimethylamine N-oxide from animal products (3). This complex, mutual relation of human diet with microbial composition and functionality requires a deep understanding (4, 5).
In this regard, several studies have demonstrated that a variety of healthy dietary patterns can be metabolized by the microbiota to exert beneficial effects on host health, while the dietary patterns may also gradually influence microbial composition over time (e.g., via selection for resistant starch or carbohydrate fermentation) (6–9). Our previous analyses showed that greater adherence to healthy plant-based diets that do not completely exclude animal proteins was significantly associated with a lower risk of cardiometabolic conditions (10), and the diets could be as beneficial as the strict vegan or vegetarian diets with regard to reducing diabetes risk (11, 12). In contrast to the vegan/vegetarian diets that are consumed by a small proportion of populations, plant-based diets are much more commonly practiced globally. Indeed, plant-based diets are gaining in popularity because of their potentially beneficial effects to both human and planetary health (10–13). In addition, there is a potentially intrinsic relation between these diets and microbiota because the diets emphasize healthy plant-based foods, which are primary contributors of fiber, lignans, and other healthy prebiotics. However, the relation between plant-based diets and human gut microbiota remains to be defined.
To fill this important knowledge gap, in the current analysis, we focused on the adherence to healthy plant-based diets as reflected by a healthy plant-based diet index (hPDI) score and evaluated its relation with microbial composition and functionality and metabolic risk. Our central hypothesis is that hPDI score is associated with a beneficial profile of microbial compositions and pathways. We also explored potential interactions between hPDI and microbiome composition on the profile of metabolic risk.
Methods
Study population
The Men's Lifestyle Validation Study (MLVS) is a substudy within the Health Professionals Follow-Up Study (HPFS), which was established in 1986 when 51,529 male US health professionals aged 40–75 y completed a mailed questionnaire about their medical history and lifestyle at baseline, with follow-up questionnaires being administered subsequently in order to assess and update lifestyle, diet, and medical history (12, 14). The current ad hoc analysis is based on existing data collected through the MLVS.
The study protocol was approved by the Harvard T.H. Chan School of Public Health Institutional Review Board, and written informed consent was obtained from all participants.
Dietary assessments
In the MLVS, diet was assessed repeatedly at the beginning and the end of the study (1 y apart) using a validated semiquantitative FFQ (15, 16). The FFQ consisted of >130 questions inquiring how often, on average, participants consumed a prespecified amount of the foods during the previous year. The questions had 9 possible responses, ranging from never or <1 time per month to ≥6 times per day. The reproducibility and validity of the FFQ in measuring food intake have been documented in previous studies (17, 18). For example, the average deattenuated Pearson correlation coefficients between FFQ assessments and those by multiple 7-d diet records ranged from 0.45 for nuts to 0.85 for tea/coffee for the 18 food groups considered in the hPDI in a previous validation study in the HPFS (18). Daily intake of energy and fiber was calculated by multiplying the frequency of consumption of each food item by its nutrient content and summing the nutrient contributions of all foods. Nutritional composition data were from Harvard food composition databases.
hPDI
Based on the frequency and prespecified serving size, we calculated servings of intake per day for each food item listed in the FFQ. We then grouped the food items into 18 groups based on nutritional and culinary similarities of foods within the broad categories of plant-based foods and animal products by summing intakes of all food items in each food group. We considered whole grains, fruits, vegetables, nuts, legumes, vegetable oils, and tea/coffee as healthy plant food groups; fruit juices, sugar-sweetened beverages, refined grains, potatoes, and sweets/desserts were considered unhealthy plant food groups. Animal food groups included animal fats, dairy, eggs, fish/seafood, meat (poultry and red meat), and miscellaneous animal-based foods. Next, we categorized these 18 food groups (servings/day) into quintiles, and each quintile was assigned a score between 1 and 5. Participants receive a score of 1–5 (1 for lowest quintile and 5 for highest quintile) for each healthy plant food group; for animal food groups and unhealthy plant food groups, we reversed the score so that higher intake received a lower score (1 for highest quintile and 5 for lowest quintile). We then summed the scores across the 18 food groups to derive the hPDI score, which has a theoretical range of 18 (lowest possible score) to 90 (highest possible score). Overall, a higher hPDI score reflects higher intake of healthy plant-based foods and lower intake of animal products and unhealthy plant-based food (12, 13).
We considered both FFQ surveys in the MLVS and dietary assessments collected using similar FFQs quadrennially from 1986 to 2010 in the HPFS and calculated the cumulative average of hPDI scores based on all FFQ assessments (up to 9 assessments) to measure habitual long-term adherence to the healthy plant-based diet.
In secondary analyses, we also derived an overall PDI score without differentiating between healthy and unhealthy plant-based food groups (i.e., foods in both categories were positively ranked corresponding to quintiles), as well as unhealthy PDI (uPDI) score by assigning positive scores corresponding to the quintile distribution of unhealthy plant-based food intake and reversely assigning scores to the intake of other foods (12, 13).
Fecal sample collection
The MLVS comprised 308 participants who underwent repeated examinations of diet and lifestyle and multiple collections of blood and fecal samples in 2012–2013 (19). All examinations were conducted within a 1-y period, and fecal samples were self-collected by participants as previously described (19). Briefly, participants provided up to 2 pairs of fecal samples (6 mo apart) from 2 adjacent bowel movements. A short questionnaire was administered for the fecal sample collections and inquired about fecal consistency as indicated through the Bristol Stool Chart, questions regarding the use of acid-lowering and antibiotic medications, antibiotic usage, and other information regarding gastrointestinal health. At approximately the same time as fecal sample collection, a fasting blood sample was also collected (twice, 6 mo apart). Body weight and other anthropometric and lifestyle variables were also assessed at these time points. Participants reported date of birth, geographic location, ethnicity, alcohol consumption, and smoking status at baseline examination when they were enrolled in the MLVS. All MLVS participants were free of a history of coronary heart disease, stroke, cancer, or major neurological disease.
Taxonomic and functional profiling of metagenomic samples
Shotgun metagenomes were generated from study specimens using paired-end 100nt Illumina HiSeq shotgun sequencing as previously described (19, 20). DNA from fecal samples was extracted using standard protocols and subsequentially prepared for sequencing using the Nextera XT DNA Library Preparation Kit and sequenced to a target depth of 1–2 Gnt each. Taxonomic and functional profiles were generated using the bioBakery workflow (21). Briefly, quality controls included the removal of human sequences, quality trimming, and depletion of duplicate reads using KneadData (http://huttenhower.sph.harvard.edu/kneaddata), taxonomic profiling by MetaPhlAn2 (22), and functional profiling by HUMAnN2 (23). Microbiome profiles of the MLVS men have been previously described (19, 20).
We excluded participants with missing values of plasma metabolic marker measurements [including blood lipids and glycated hemoglobin (HbA1c)] or hPDI score, which resulted in 911 metagenomes from 303 participants included in the current analysis (Supplemental Figure 1). We further filtered all taxonomic features with a relative abundance <10−4 in >10% of all samples. Similarly, we filtered all pathways with a relative abundance <10−5 in >10% of all samples.
Assessment of metabolic risk factors
Fasting blood samples were collected through venipuncture into sodium heparin tubes and shipped by overnight mail with an ice pack. Plasma levels of HDL cholesterol and triacylglycerol were assayed using enzymatic methods. HbA1c levels were measured using turbidimetric immunoinhibition (Roche Diagnostics). Blind quality control samples (10%) were randomly interspersed in the assay batches, based on which we estimated coefficients of variation <7% for all plasma assays.
To build the metabolic risk score, we first divided triacylglycerol, HDL cholesterol, BMI (in kg/m2), and HbA1c into quintiles, and each quintile was assigned a score between 1 and 5. Participants received a score of 1–5 (1 for the lowest quintile and 5 for the highest quintile) for triacylglycerol, BMI, and HbA1c; they received a reverse score for HDL cholesterol (1 for the highest quintile and 5 for the lowest quintile). We then summed the scores across the 4 variables to derive a metabolic score that has a theoretical range of 5 (lowest possible score, low risk) to 20 (highest possible score, high risk).
Statistical analysis
All species/pathways data were normalized via arc-sin square root transformation. MaAslin2 (https://huttenhower.sph.harvard.edu/maaslin2) was used to examine the associations between the hPDI score and the relative abundance of taxonomy and pathways, with a random effect to account for within-person correlations between the 4 time points. Visualizations were constructed using Graphical Phylogenetic Analysis (https://huttenhower.sph.harvard.edu/graphlan). In secondary analyses, we also examined the association between the 18 individual food groups, as well as dietary fiber, and microbial features. Analyses were based on per SD of hPDI and other dietary variables. In multivariable analyses, we adjusted for time-varying covariates assessed proximately to each fecal/blood sample collection, including age, total energy intake, physical activity, smoking, alcohol consumption, Bristol categories, use of antibiotics in past year, and consumption of any probiotics (except yogurt) in the past 2 mo. The analyses of associations between hPDI score and metabolic risk factors were based on 466 plasma measurements of the 303 participants, of whom 163 had repeated measurements. Linear mixed-effect models (PROC MIXED, SAS version 9.4; SAS Institute) were used to examine the associations between hPDI and metabolic risk factors, with an unstructured covariance matrix specified to account for within-person correlations between the 2 time points.
To derive microbial patterns at the species level, we used principal coordinate (PCo) analysis based on Bray–Curtis dissimilarity. Permutational multivariate analysis of variance (PERMANOVA) test was applied to assess the association of the overall microbial community with PDI scores. To alleviate concerns of multiple comparisons, we examined interactions between PDI scores and PCo on the metabolic score, and in secondary analyses we also examined interactions with individual species that were associated with hPDI. Test for interaction was conducted by including an interaction term between PDIs and PCo/species (high compared with low) in the multivariable-adjusted linear mixed model and examining the significance of the interaction term. We used the detection rate to guide the categorization of individuals by the abundance of individual species in the interaction tests: for species that were detected in ≥800 (of the 916) samples, we classified the participants into high-abundance or low-abundance category based on the median value of the relative abundance of the species. For species with lower detection rate, we defined the low-abundance group as the absence of the species and high-abundance group as the presence of the species.
False discovery rate (FDR) values <0.05 after FDR correction following the Benjamini–Hochberg method were considered statistically significant.
Results
Table 1 presents baseline characteristics of participants according to quintiles of hPDI. The average hPDI score ranged from 46.5 in the lowest quintile to 64.1 in the highest quintile. A high hPDI was associated with a higher intake of fiber, plant proteins, whole grains, fruits, vegetables, nuts, and legumes and a lower intake of energy, animal proteins, refined grains, potatoes, sweets, animal fat, egg, dairy, and meats (Table 1).
TABLE 1.
Characteristics of 303 older men in the MLVS by quantile of hPDI1
| hPDI | ||||||
|---|---|---|---|---|---|---|
| Quintile 1 (n = 59) | Quintile 2 (n = 62) | Quintile 3 (n = 60) | Quintile 4 (n = 62) | Quintile 5 (n = 60) | P-trend | |
| Age,2 y | 70.1 ± 3.5 | 70.6 ± 4.4 | 71.7 ± 4.4 | 70.9 ± 4.3 | 71.3 ± 4.6 | 0.09 |
| BMI, kg/m2 | 26.2 ± 4.0 | 25.6 ± 3.4 | 24.5 ± 3.6 | 24.8 ± 2.5 | 23.9 ± 3.1 | 0.001 |
| Total activity,3 MET-h/wk | 117 ± 56 | 116 ± 46 | 108 ± 48 | 115 ± 59 | 114 ± 51 | 0.42 |
| Current smoking, % | 0.0 | 1.0 | 1.9 | 4.9 | 5.0 | 0.49 |
| Using of antibiotics in past 12 mo, % | 18.4 | 37.2 | 23.8 | 31.8 | 23.8 | 0.82 |
| Consumed any probiotics in past 2 mo, % | 6.4 | 5.5 | 2.6 | 6.1 | 5.0 | 0.98 |
| PDI score | 54.0 ± 3.9 | 56.3 ± 4.8 | 56.0 ± 4.5 | 55.9 ± 4.7 | 58.9 ± 4.4 | <0.0001 |
| uPDI score | 56.2 ± 5.1 | 54.6 ± 6.0 | 53.5 ± 6.2 | 51.5 ± 5.8 | 50.5 ± 4.5 | <0.0001 |
| hPDI score | 46.5 ± 2.6 | 51.3 ± 0.9 | 54.4 ± 0.9 | 58.0 ± 1.2 | 64.1 ± 2.8 | <0.0001 |
| Dietary intakes | ||||||
| Energy, kcal/d | 2353 ± 411 | 2254 ± 512 | 2069 ± 481 | 1987 ± 510 | 1921 ± 459 | <0.0001 |
| Protein, g/d | 96.0 ± 17.5 | 92.7 ± 23.9 | 90.2 ± 23.7 | 85.5 ± 20.8 | 81.0 ± 18.7 | 0.0001 |
| Animal protein, g/d | 67.5 ± 14.5 | 62.3 ± 17.4 | 60.0 ± 17.3 | 55.5 ± 14.1 | 46.0 ± 16.2 | <0.0001 |
| Plant protein, g/d | 28.6 ± 6.1 | 30.3 ± 8.6 | 30.1 ± 8.9 | 30.0 ± 8.9 | 35.0 ± 12.7 | 0.003 |
| Carbohydrates, g/d | 275 ± 56.8 | 273 ± 70.7 | 251 ± 64.5 | 241 ± 69.2 | 254 ± 76.7 | 0.01 |
| Fiber, g/d | 21.8 ± 5.5 | 24.5 ± 7.4 | 24.1 ± 7.4 | 25.2 ± 7.9 | 30.4 ± 10.6 | <0.0001 |
| Alcohol, g/d | 12.3 ± 12.0 | 16.6 ± 13.7 | 12.2 ± 10.6 | 15.7 ± 13.1 | 12.2 ± 11.6 | 0.83 |
| Whole grains, serving/d | 1.6 ± 0.7 | 1.7 ± 0.9 | 2.1 ± 1.2 | 1.9 ± 0.9 | 2.5 ± 1.7 | <0.0001 |
| Fruits, serving/d | 1.3 ± 0.8 | 1.6 ± 0.6 | 1.7 ± 0.8 | 1.9 ± 1.1 | 2.6 ± 1.4 | <0.0001 |
| Vegetables, serving/d | 3.2 ± 1.0 | 3.4 ± 1.4 | 3.5 ± 1.7 | 3.9 ± 1.7 | 4.6 ± 1.6 | <0.0001 |
| Nuts, serving/d | 0.6 ± 0.4 | 0.7 ± 0.5 | 0.8 ± 0.6 | 0.7 ± 0.5 | 1.0 ± 0.6 | 0.0005 |
| Legumes, serving/d | 0.4 ± 0.2 | 0.5 ± 0.2 | 0.4 ± 0.2 | 0.4 ± 0.2 | 0.7 ± 0.5 | <0.0001 |
| Vegetable oils, serving/d | 0.5 ± 0.4 | 0.4 ± 0.3 | 0.4 ± 0.4 | 0.6 ± 0.5 | 0.6 ± 0.5 | 0.01 |
| Tea and coffee, serving/d | 1.3 ± 1.2 | 2.4 ± 1.5 | 2.2 ± 1.5 | 2.7 ± 1.5 | 2.2 ± 1.2 | 0.001 |
| Fruit juices, serving/d | 1.1 ± 0.9 | 0.9 ± 0.7 | 0.8 ± 0.6 | 0.5 ± 0.4 | 0.6 ± 0.5 | <0.0001 |
| Refined grains, serving/d | 1.8 ± 0.6 | 1.8 ± 0.9 | 1.4 ± 0.7 | 1.5 ± 0.9 | 1.2 ± 0.5 | <0.0001 |
| Potatoes, serving/d | 0.8 ± 0.3 | 0.6 ± 0.3 | 0.6 ± 0.3 | 0.4 ± 0.2 | 0.3 ± 0.2 | <0.0001 |
| Sugar-sweetened beverages, serving/d | 0.6 ± 0.6 | 0.4 ± 0.4 | 0.2 ± 0.2 | 0.1 ± 0.2 | 0.1 ± 0.1 | <0.0001 |
| Sweets and desserts, serving/d | 1.8 ± 0.9 | 1.8 ± 0.9 | 1.4 ± 0.9 | 1.2 ± 0.7 | 1.0 ± 0.7 | <0.0001 |
| Animal fat, serving/d | 0.8 ± 0.7 | 0.3 ± 0.3 | 0.4 ± 0.5 | 0.3 ± 0.4 | 0.2 ± 0.2 | <0.0001 |
| Dairy, serving/d | 2.6 ± 1.2 | 2.1 ± 1.0 | 2.1 ± 0.9 | 1.8 ± 0.9 | 1.6 ± 0.8 | <0.0001 |
| Egg, serving/d | 0.4 ± 0.3 | 0.3 ± 0.2 | 0.3 ± 0.3 | 0.3 ± 0.3 | 0.2 ± 0.2 | 0.0001 |
| Fish or seafood, serving/d | 0.3 ± 0.1 | 0.4 ± 0.2 | 0.3 ± 0.2 | 0.4 ± 0.2 | 0.4 ± 0.2 | 0.02 |
| Meat, serving/d | 1.6 ± 0.5 | 1.4 ± 0.5 | 1.3 ± 0.6 | 1.1 ± 0.4 | 0.7 ± 0.4 | <0.0001 |
| Miscellaneous animal-based foods,serving/d | 0.4 ± 0.5 | 0.3 ± 0.4 | 0.3 ± 0.5 | 0.3 ± 0.4 | 0.2 ± 0.4 | 0.046 |
Values are age-standardized means ± SDs or percentages. hPDI, healthy plant-based diet index; MET, metabolic equivalents; MLVS, Men's Lifestyle Validation Study; PDI, pland-based diet index; uPDI, unhealthy plant-based diet index.
Not age-adjusted.
Metabolic equivalent hours per week from recreational and leisure-time activities.
TABLE 2.
Main effects of PDI and cardiometabolic risk markers based on 466 plasma measurements of the 303 participants1
| PDI | |||
|---|---|---|---|
| hPDI | PDI | uPDI | |
| HDL cholesterol, mg/dL | 2.160 (0.637)1 | 1.003 (0.702) | –1.863 (0.719)1 |
| Triglycerides, mg/dL | –8.879 (2.657)1 | –4.199 (2.928) | 4.617 (3.010) |
| HbA1c, % | –0.057 (0.018)1 | –0.011 (0.020) | 0.043 (0.020)1 |
| BMI, kg/m2 | –0.918 (0.176)1 | –0.988 (0.192)1 | 0.225 (0.203) |
| Metabolic score2 | –0.979 (0.177)1 | –0.471 (0.198)1 | 0.557 (0.203)1 |
The analysis was based on 466 plasma measurements of the 303 participants, 163 of whom had repeated measurements. P < 0.05, main effects were expressed as the β coefficients (SEM) of the differential metabolic risk factors associated with per SD changes of PDIs. Multivariable adjustment considered age, energy intake, alcohol, smoking, physical activity, using of antibiotics, consumed any probiotics, and fecal sample characteristics as well as the repeated measurements (participant's ID as random intercept). HbA1c, glycated hemoglobin; hPDI, healthy plant-based diet index; PDI, plant-based diet index; uPDI, unhealthy plant-based diet index.
The metabolic score (range: 5–20) was calculated as a sum of quintile scores of triglyceride, HDL cholesterol, BMI, and HbA1c, where 1 was assigned for the lowest quintiles and 5 for the highest quintiles of triglyceride, BMI, and HbA1c. The score was reversed for HDL cholesterol (1 for the highest quintile and 5 for the lowest quintile).
Taxonomies associated with PDI scores, fiber, and individual foods
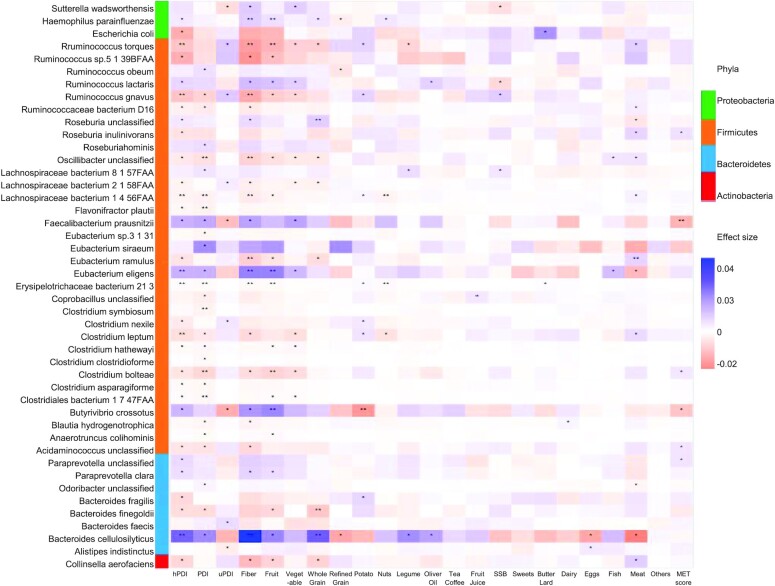
A higher hPDI score was significantly associated with 7 species at the FDR < 0.05 level (Figure 1), including a higher relative abundance (%) ofBacteroides cellulosilyticus (2.58%; 95% CI: 1.39, 3.77) and Eubacterium eligens (1.37%; 95% CI: 0.55, 2.20) and a lower abundance of Ruminococcus torques (–1.09%; 95% CI: –1.67, –0.50), Ruminococcus gnavus (–1.10%; 95% CI: –1.69, –0.52), Clostridium leptum (–0.66%; 95% CI: –1.03, –0.30), Lachnospiraceae bacterium 1_4_56faa (–0.29%; 95% CI: –0.45, –0.12), and Erysipelotrichaceae bacterium 21_3 (–0.12%; 95% CI: –0.18, –0.05). At the 0.05 < FDR < 0.25 level, a higher hPDI score was positively associated with relative abundance of Butyrivibrio crossotus, Faecalibacterium prausnitzii, Ruminococcus lactaris, Roseburia hominis, Paraprevotella clara, and Haemophilus parainfluenzae (Supplemental Figure 2). The majority of species (96/138, 69.6%) were associated with both hPDI and the overall PDI score in the same direction, whereas the majority of species (107/138, 77.5%) were associated with hPDI and uPDI in the opposite direction (Supplemental Figure 2).
FIGURE 1.
Heatmap of the species that significantly associated with PDIs, fiber, and individual food groups based on 911 repeated measurements of the 303 participants. Effect size colors are based on the relative abundance (arc sine of square root of the normalized abundance values) changes with per SD increasing of PDIs, foods, fiber, or MET score. Filtering criteria: relative abundance <0.001% at 10%+ samples. Generalized linear mixed-effects regressions implemented in MaAsLin2 were adjusted for repeated measurements (participant's ID as random intercept), age, energy intake, alcohol, smoking, physical activity, using of antibiotics, consumed any probiotics, and fecal sample characteristics). hPDI, healthy plant-based diet index; MET, metabolic risk; PDI, plant-based diet index; SSB, sugar-sweetened beverage; uPDI, unhealthy plant-based diet index.
Six of the 7 hPDI species were also significantly associated with dietary fiber intake in the same direction (Figure 1), including B. cellulosilyticus, E. eligens, R. torques, R. gnavus, L. bacterium 1_4_56faa, and E. bacterium 21_3. The associations between these species and individual food components of the hPDI score, especially fruits and whole grains, largely mirrored the associations with hPDI score. Of the 7 species that were significantly associated with hPDI, E. eligens, R. torques, and E. bacterium 21_3 were also significantly associated with fruits; B. cellulosilyticus was also significantly associated with whole grains; and L. bacterium 1_4_56faa and E. bacterium 21_3 were also significantly associated with the frequency of nut consumption (Figure 1). In contrast, associations in the opposite direction were observed between these species and intake of animal products, such as meats, fish, and dairy products (Figure 1). Only a few species were significantly associated with individual foods but not the PDI scores, such as red meat intake in relation to Coprococcus comes and Bacteroides nordii (Supplemental Figure 2).
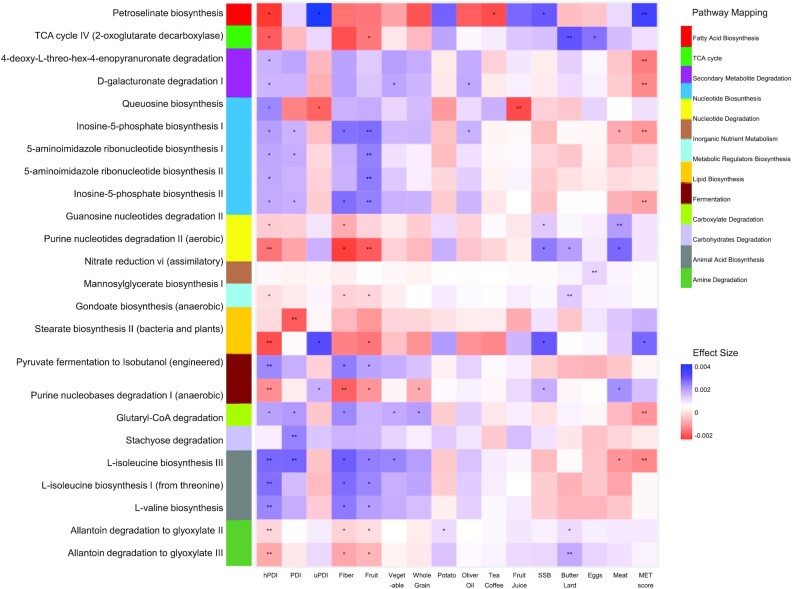
Pathways associated with PDI scores, fiber, and individual foods
Of the 239 pathway features that passed the filtering criteria, 4 pathways were significantly enriched and 5 pathways significantly depleted with increasing values of the hPDI score (FDR < 0.05) (Figure 2). At the enzyme level, hPDI score was significantly associated with the relative abundances of 155 enzymes (FDR < 0.05; Supplemental Table 1). The 4 enriched pathways included 3 branched-chain amino acid (BCAA) biosynthesis pathways (l-isoleucine biosynthesis I and III and l-valine biosynthesis) and 1 fermentation pathway (pyruvate fermentation to isobutanol). Consistently, relative abundance of the BCAA transaminase was also significantly enriched with increasing hPDI (FDR < 0.001; Supplemental Table 1). Relative abundance of amino acid biosynthesis pathways was also positively associated with fiber and fruits and negatively associated with meats (Figure 2).
FIGURE 2.
Heatmap of the pathway (DNA) that significantly associated with PDIs, fiber, and individual food groups based on 911 repeated measurements of the 303 participants. Effect size colors are based on the relative abundance (arc sine of square root of the normalized abundance values) changes with per SD increasing of PDIs, foods, fiber, or MET score. Filtering criteria: relative abundance <0.0001% at 10%+ samples. Generalized linear mixed-effects regressions implemented in MaAsLin2 were adjusted for repeated measurements (participant's ID as random intercept), age, energy intake, alcohol, smoking, physical activity, using of antibiotics, consumed any probiotics, and fecal sample characteristics). hPDI, healthy plant-based diet index; MET, metabolic risk; PDI, plant-based diet index; SSB, sugar-sweetened beverage; TCA, tricarboxylic acid; uPDI, unhealthy plant-based diet index.
We also detected 5 pathways that were inversely associated with hPDI (Figure 2). A low hPDI score, reflecting a high intake of refined grains and animal foods, was associated with an enrichment of purine nucleobases degradation pathway (Figure 2) and related formate dehydrogenase (EC 1.2.1.2), glycine reductase (EC 1.21.4.2), guanine deaminase (EC 3.5.4.3), and methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9) (all FDRs < 0.05; Supplemental Table 1). Other low hPDI-related pathways included the lipid biosynthesis pathway of stearate biosynthesis II and related 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100), as well as amine degradation pathways of allantoin degradation to glyoxylate II and III and related allantoate deiminase (EC 3.5.3.9) (all FDRs < 0.05; Supplemental Table 1). The majority of the pathways that were inversely associated with hPDI were positively associated with increasing intake of eggs, meats, butter, or lard, whereas the relative abundance of lipid biosynthesis pathway was positively associated with uPDI and sugar-sweetened beverages (Figure 2).
Microbial composition, hPDI, and their interactions on metabolic risk
The first 2 PCos of species derived from the principal coordinates analysis captured 9.5% and 8.8% of the overall variations and largely reflected compositions of Firmicutes and Bacteroidetes, respectively (Supplemental Figure 3D). In particular, the abundance of Bacteroides uniformis positively (R2 = 0.64) and P. copri inversely (R2 = 0.61) accounted for the PCo1 loading. The abundance of Eubacterium rectale (R2 = 0.76) accounted for the PCo2 loading. The overall microbial communities were significantly associated with the PDI scores (both R2 for hPDI and PDI = 0.0069, R2 = 0.0053 for uPDI, all P < 0.001; PERMANOVA with Bray–Curtis distances) (Supplemental Figure 3A–C).
Per SD increment of hPDI score was significantly associated with a 2.2 mg/dL (SE = 0.6) higher HDL cholesterol, –8.9 mg/dL (SE = 2.7) lower triacylglycerol, 0.06% (SE = 0.02) lower HbA1c, 0.9 kg/m2 (SE = 0.2) lower BMI, and a 1.0 unit (SE = 0.2) lower value of the metabolic score (Table 2). On the contrary, increasing uPDI was significantly associated with lower HDL cholesterol but higher triacylglycerol and a higher metabolic score. The overall PDI was associated with lower BMI and metabolic score (Table 2).
The association of hPDI with metabolic score was significantly modified by the overall microbial composition pattern in that the inverse association between hPDI and metabolic score was more pronounced in participants with a higher PCo1 than in participants with a lower PCo1 (P-interaction = 0.004, FDR = 0.01; Figure 3A). This interaction was mainly driven by the interaction between hPDI and PCo1 on HbA1c (P-interaction = 0.00004, FDR = 0.002; Figure 3A). Consistently, when we further examined the interaction between hPDI and the presence of P. copri, per SD increment of hPDI score was significantly associated with lower levels of triacylglycerol, HbA1c, and metabolic score among individuals without P. copri than among P. copri carriers (Figure 3B). Bacteroides uniformis primarily modulated the association between hPDI and HbA1c; per SD hPDI score was significantly associated with a 0.25 SD less HbA1c among carries of B. uniformis only (Figure 3C). We did not find significant interactions between PCo2 and hPDI or between E. rectale and hPDI on metabolic risks.
FIGURE 3.
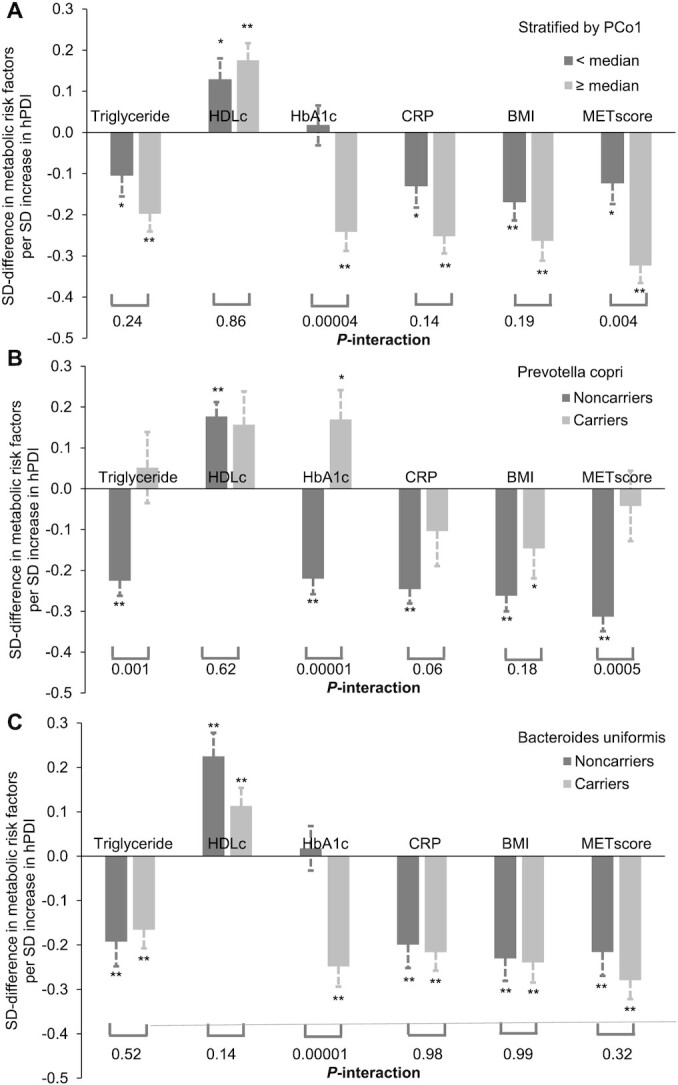
Interactions between microbiome and hPDI on metabolic risk factors based on 911 repeated measurements of the 303 participants. *0.01 < P < 0.05; **P < 0.01; associations between hPDI and metabolic risk factors were calculated from a linear mixed model that included participant's identifier as random effects and the hPDI, PCo1 score, or Prevotella copri/Bacteroides uniformis carriages, as well as simultaneously adjusting for age, energy intake, alcohol, smoking, physical activity, using of antibiotics, consumed any probiotics, and fecal sample characteristics. P-interactions between hPDI and the first PCo (A), the P. copri carriage (B), and B. uniformis (C) on individual and/or overall metabolic risk were calculated from the same models by further including the product term. Triglyceride, HDLc, HbA1,c and CRP were plasma concentrations. CRP, C-reactive protein; HbA1c, glycated hemoglobin; HDLc, HDL concentration; hPDI, healthy plant-based diet index; MET, metabolic risk; PCo; principal coordinate.
Interactions between hPDI and individual species on metabolic risk
We found that a few other species potentially modulated the associations between hPDI and metabolic risk score. For example, hPDI score was more strongly associated with lower metabolic score in the presence of Clostridium clostridioforme than in the absence of this species: the βs were –1.58 and –0.70 per SD hPDI, respectively (P-interaction = 0.0003, FDR = 0.01; Supplemental Figure 4A). In contrast, the presence of Blautia hydrogenotrophica significantly attenuated the associations between hPDI and metabolic score, with the βs of –0.28 and –1.16 per SD hPDI with high and low abundance of this species, respectively (P-interaction = 0.002, FDR = 0.036; Supplemental Figure 4B).
Other findings
The relative abundance of Coprococcus sp. ART55/1 and that of F. prausnitzii were significantly associated with a lower MET score (FDR < 0.05; Supplemental Table 2). The relative abundances of d-galacturonate degradation I, inosine-5-phosphate biosynthesis I, 4-deoxy-l-threo-hex-4-enopyranuronate degradation, glutaryl-CoA degradation, petroselinate biosynthesis, l-isoleucine biosynthesis III, and inosine-5′-phosphate biosynthesis II were significantly associated with a lower metabolic score, all in a direction opposite their associations with hPDI (Figure 2).
Discussion
In this study of free-living healthy men, a higher hPDI score that reflects a higher intake of fresh fruits, vegetables, whole grains, and other healthy plant-based foods and a lower intake of animal products was significantly associated with the overall gut microbiome composition, as well as multiple individual species, such as B. cellulosilyticus and E. eligens. hPDI was also associated with the enrichment of pathways involved in fermentation from pyruvate to isobutanol and biosynthesis pathways of BCAAs, such as l-isoleucine and l-valine, as well as depletion of fatty acid biosynthesis, lipid biosynthesis, and amine degradation. Last, the overall microbial composition significantly modulated the association between hPDI and metabolic risk profile in that the favorable association was more pronounced among men with a higher PCo characterized by higher B. uniformis and lower P. copri.
The fermentation of glycans, especially complex carbohydrates, such as fiber, that are not digestible by the host's own digestive enzymes, represents one of the dominant microbial metabolic activities in the gut that produce butyrate and other SCFAs and may exert subsequent favorable physiological effects to the host (24–26). Our findings regarding individual species are broadly consistent with prior evidence linking fiber intake and microbes in feeding studies or in vitro experiments (27–30). For example, our study showed that better adherence of hPDI was associated with the enrichment of E. eligens, R. lactaris, R. hominis, and B. cellulosilyticus. In vivo, the abundance of E. eligens was significantly enriched by nondigestible polysaccharides (27). Similarly, R. lactaris was significantly enriched following intake of resistant starch, and this enrichment was also significantly associated with the production of SCFAs (30). Last, durum wheat flour and whole-grain barley pasta in healthy subjects significantly increased the abundance of R. hominis (28). Bacteroides cellulosilyticus is known to be involved in fiber metabolism (31, 32). Studies examining habitual vegetarian and vegan diets have consistently shown that these diets are linked to the enrichment of fiber-degrading bacteria in the gut (5). Our study findings are also broadly consistent with those from studies that examined other healthful dietary indices. For example, a recent study (33) found that both the Healthy Eating Index and Mediterranean diet score were positively associated with the abundance of F. prausnitzii, whereas the abundance of Lactobacillus was inversely associated with both dietary indices.
In the current study, we observed that a high hPDI was significantly associated with the enrichment of pathways involved in amino acid biosynthesis and a low hPDI associated with enrichment of pathways involved in the metabolism of nutrients rich in animal products. Previous studies indicated that vegan diet differed from animal-based diets with respect to several microbe-dependent metabolic pathways, including increased metabolism of fiber and polyphenols, enrichment of BCAA synthesis and decreased metabolism of bile acids, choline, l-carnitine, and amino acids (34–36).
Our study also identified several species and functional pathways that were significantly associated with a lower metabolic risk, such as abundance of F. prausnitzii as well as pathways of d-galacturonate degradation I and 4-deoxy-l-threo-hex-4-enopyranuronate degradation. A recent study indicated that an isocaloric Mediterranean diet intervention led to increased levels of the relative abundance of F. prausnitzii and genes for microbial carbohydrate degradation linked to butyrate metabolism (37). The pathways of d-galacturonate degradation I and 4-deoxy-l-threo-hex-4-enopyranuronate degradation were positively associated with fruit intake and inversely associated with metabolic risk score in the Lifelines DEEP study (38). Collectively, these findings provided further evidence that gut microbiome may function as a mediator of dietary factors on the host metabolic status (39).
The interplay between hPDI and microbiome profile and individual species on the metabolic risk markers is worth discussing. Of note, similar microbiome–diet interactions were also detected in previous studies. For instance, the health effect of adherence to the Mediterranean diet was enhanced at the enrichment of F. prausnitzii (33) or reduced at a higher abundance of P. copri (40). Existing evidence indicates that the production of bioactive microbiota metabolites following the intake of dietary precursors is highly variable between individuals. For example, the same dose of flaxseed supplementation could result in a 0.9- to 43.8-fold increase in urinary excretion of enterolignans (41). Experiments revealed that this between-person variability was largely due to the variability in the capacity of microbiota in digesting the complex carbohydrate (42, 43). Collectively, both our data and existing data indicate that microbiota may significantly modulate the effects of diet on the production of bioactive compounds and subsequently cardiometabolic health and thus, to a certain extent, account for the individualized response to the same dietary interventions observed in clinical studies.
The current analysis leveraged repeated assessments of long-term diet, gut microbiome, and cardiometabolic risk markers, which allowed us to systematically account for within- and between-person variability. Another strength is that all participants were healthy men free of major chronic diseases, and thus the strong influence of diseases on the basic biochemical associations of interest is small.
The primary limitation of this study is the cross-sectional design, which cannot help establish temporal relations between diet and microbial compositions and functionalities. However, the use of repeated assessments of diet many years before the collection of fecal samples renders it possible that detected associations—often significant but of low effect size—are the result of gradual, long-term selective pressures on the gut microbiome. However, they could also be the joint result of external factors (e.g., a generally healthier or more active lifestyle corresponding with a higher hPDI and with microbiome effects). The measurement errors in dietary assessments are inevitable. Although we calculated and used the cumulative average of hPDI scores, we cannot exclude the possibility of misclassification of adherence to hPDI. Our multivariable-adjusted analysis controlled for multiple established and potential confounders, although residual or unmeasured confounding may still exist, such as that by the use of different types of probiotics. Our study participants were exclusively healthy male health professionals of largely European ancestries. Although the homogeneity may help alleviate confounding by socioeconomic factors, the findings may lack generalizability to other populations, such as women or minorities. Previous studies indicated similar associations of PDI scores with risk of diabetes and coronary heart disease between men and women (12, 13, 20), but the interplay between PDIs and microbiota on health might be modified by hormonal or other differences between gender and warrants further investigations. Furthermore, the relation between diet and the human gut microbiome is complex, with vastly different effects over the short term, long term, at different life stages, under varying health and disease conditions, and across ethnogeographical environments (44). This complexity further limits the generalizability of the current findings. Also, we were not able to assess metabolites that are produced by microbiome, especially SCFAs, and thus cannot evaluate which small molecules might explain interactions between diet, the microbiome, and systemic host effects such as cardiometabolic risk markers. Last, we examined the habitual adherence to hPDI in relation to microbiome in the current analysis, in light of the notion that long-term dietary habits play an important role in shaping composition and function of the gut microbiome (45) and in affecting the risk of developing chronic diseases. As such, our findings cannot be extrapolated to reflect acute effects of hPDI on microbiome. Nonetheless, the microbial composition and functional potential have been shown to be largely stable in the MLVS, suggesting that the impact of short-term variations of diet on the microbiome is likely to be relatively minor in this population (20).
In conclusion, a greater adherence to a healthy plant-based diet was associated with an overall gut microbiome profile, primarily with the enrichment of B. cellulosilyticus and E. eligens as well as enriched pathways of amino acid biosynthesis, and the depletion of pathways involved in processing constituents from animal products. Both the overall microbial composition and some individual species that were associated with the hPDI also modulated the beneficial association between hPDI and metabolic risk. Although these findings warrant replication and further evaluation in future studies, they indicate that healthy plant-based diets are associated with a unique microbial composition, which may jointly determine the effects of the diet on metabolic health. Further studies are warranted to examine the generalizability of these findings to populations of different biological or demographic characteristics.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—YL, DDW, and QS: designed the analysis; YL, AS, and DDW: analyzed the data; JW and RL: provided statistical support; YL and QS: wrote the manuscript; DDW, KLI, JL, JEW, MB, ATC, CH, EBR, FBH, and QS: provided critical editorial comments; YL and QS: had primary responsibility for the final content; and all authors: read and approved the final manuscript.
Notes
Supported by research grants R01HL035464 (EBR, QS), R01DK120870 and R01DK126698 (QS), HL060712 (FBH), R01CA202704 (ATC, CH), K24DK098311 (ATC), U54DE023798 (CH), K99DK119412 (DDW), and K99DK122128 (JL) from the NIH. The Men's Lifestyle Validation Study was supported by grant U01CA152904 from the National Cancer Institute (NCI). The Health Professionals Follow-Up Study was supported by grant U01CA167552 from NCI and grant R01HL35464 from the National Heart, Lung, and Blood Institute.
Author disclosures: YL was supported by grants from the California Walnut Commission and Swiss Reinsurance Company. All other authors report no conflicts of interest.
The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and the decision to submit the manuscript for publication.
Supplemental Figures 1–4 and Supplemental Tables 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
YL, DDW, and AS contributed equally to this work.
Abbreviations used: BCAA, branched-chain amino acid; FDR, false discovery rate; HbA1c, glycated hemoglobin; hPDI, healthy plant-based diet index; HPFS, Health Professionals Follow-Up Study; MLVS, Men's Lifestyle Validation Study; PCo, principal coordinate; PERMANOVA, permutational multivariate ANOVA; uPDI, unhealthy plant-based diet index.
Contributor Information
Yanping Li, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
Dong D Wang, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
Ambika Satija, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
Kerry L Ivey, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Microbiome and Host Health Programme, South Australian Health and Medical Research Institute, North Terrace, Adelaide, Australia; Department of Nutrition and Dietetics, College of Nursing and Health Sciences, Flinders University, Adelaide, Australia.
Jun Li, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
Jeremy E Wilkinson, Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
Ruifeng Li, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
Megu Baden, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
Andrew T Chan, Clinical and Translational Epidemiology Unit, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
Curtis Huttenhower, Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Eric B Rimm, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA.
Frank B Hu, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA.
Qi Sun, Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA; Joslin Diabetes Center, Boston, MA, USA.
Data Availability
All relevant data are available upon approval. Access is restricted due to participant confidentiality and privacy concerns. Further information, including the procedures to obtain and access data from the Health Professionals Follow-Up Study, is provided at https://sites.sph.harvard.edu/hpfs/resources-for-hpfs-investigators/.
References
- 1.Hills RD Jr, Pontefract BA, Mishcon HR, Black CA, Sutton SC, Theberge CR. Gut microbiome: profound implications for diet and disease. Nutrients. 2019;11(7):1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57(1):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang WH, Hazen SL. Microbiome, trimethylamine N-oxide, and cardiometabolic disease. Transl Res. 2017;179:108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowyer RCE, Jackson MA, Pallister T, Skinner J, Spector TD, Welch AA, Steves CJ. Use of dietary indices to control for diet in human gut microbiota studies. Microbiome. 2018;6(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi Cet al. . High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65(11):1812–21. [DOI] [PubMed] [Google Scholar]
- 6.De Filippis F, Parente E, Ercolini D. Metagenomics insights into food fermentations. Microb Biotechnol. 2017;10(1):91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graf D, Di Cagno R, Fak F, Flint HJ, Nyman M, Saarela M, Watzl B. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis. 2015;26:26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vital M, Howe A, Bergeron N, Krauss RM, Jansson JK, Tiedje JM. Metagenomic insights into the degradation of resistant starch by human gut microbiota. Appl Environ Microbiol. 2018;84(23):e01562–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Filippis F, Pellegrini N, Laghi L, Gobbetti M, Ercolini D. Unusual sub-genus associations of faecal Prevotella and Bacteroides with specific dietary patterns. Microbiome. 2016;4(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satija A, Hu FB. Plant-based diets and cardiovascular health. Trends Cardiovasc Med. 2018;28(7):437–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian F, Liu G, Hu FB, Bhupathiraju SN, Sun Q. Association between plant-based dietary patterns and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA Intern Med. 2019;179(10):1335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satija A, Bhupathiraju SN, Rimm EB, Spiegelman D, Chiuve SE, Borgi L, Willett WC, Manson JE, Sun Q, Hu FB. Plant-based dietary patterns and incidence of type 2 diabetes in US men and women: results from three prospective cohort studies. PLoS Med. 2016;13(6):e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willett W, Rockström J, Loken B, Springmann M, Lang T, Vermeulen S, Garnett T, Tilman D, DeClerck F, Wood Aet al. . Food in the Anthropocene: the EAT-Lancet Commission on healthy diets from sustainable food systems. Lancet North Am Ed. 2019;393(10170):447–92. [DOI] [PubMed] [Google Scholar]
- 14.Satija A, Bhupathiraju SN, Spiegelman D, Chiuve SE, Manson JE, Willett W, Rexrode KM, Rimm EB, Hu FB. Healthful and unhealthful plant-based diets and the risk of coronary heart disease in U.S. adults. J Am Coll Cardiol. 2017;70(4):411–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willett WC, Sampson L, Stampfer MJ, Rosner B, Bain C, Witschi J, Hennekens CH, Speizer FE. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol. 1985;122:51–65. [DOI] [PubMed] [Google Scholar]
- 16.Al-Shaar L, Yuan C, Rosner B, Dean SB, Ivey KL, Clowry CM, Sampson LA, Barnett JB, Rood J, Harnack LJet al. . Reproducibility and validity of a semi-quantitative food frequency questionnaire in men assessed by multiple methods. Am J Epidemiol. 2020:kwaa280. doi: 10.1093/aje/kwaa280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salvini S, Hunter DJ, Sampson L, Stampfer MJ, Colditz GA, Rosner B, Willett WC. Food-based validation of a dietary questionnaire: the effects of week-to-week variation in food consumption. Int J Epidemiol. 1989;18:858–67. [DOI] [PubMed] [Google Scholar]
- 18.Feskanich D, Rimm EB, Giovannucci EL, Colditz GA, Stampfer MJ, Litin LB, Willett WC. Reproducibility and validity of food intake measurements from a semiquantitative food frequency questionnaire. J Am Diet Assoc. 1993;93:790–6. [DOI] [PubMed] [Google Scholar]
- 19.Abu-Ali GS, Mehta RS, Lloyd-Price J, Mallick H, Branck T, Ivey KL, Drew DA, DuLong C, Rimm E, Izard Jet al. . Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat Microbiol. 2018;3(3):356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehta RS, Abu-Ali GS, Drew DA, Lloyd-Price J, Subramanian A, Lochhead P, Joshi AD, Ivey KL, Khalili H, Brown GTet al. . Stability of the human faecal microbiome in a cohort of adult men. Nat Microbiol. 2018;3(3):347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIver LJ, Abu-Ali G, Franzosa EA, Schwager R, Morgan XC, Waldron L, Segata N, Huttenhower C. bioBakery: a meta'omic analysis environment. Bioinformatics. 2018;34(7):1235–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12(10):902–3. [DOI] [PubMed] [Google Scholar]
- 23.Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, Huttenhower C. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15(11):962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macfarlane GT, Macfarlane S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol. 2011;45(Suppl):S120–7. [DOI] [PubMed] [Google Scholar]
- 25.Russell WR, Hoyles L, Flint HJ, Dumas ME. Colonic bacterial metabolites and human health. Curr Opin Microbiol. 2013;16(3):246–54. [DOI] [PubMed] [Google Scholar]
- 26.Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, Date P, Farquharson F, Johnstone AM, Lobley GEet al. . Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8(11):2218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung WS, Walker AW, Louis P, Parkhill J, Vermeiren J, Bosscher D, Duncan SH, Flint HJ. Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol. 2016;14:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Angelis M, Montemurno E, Vannini L, Cosola C, Cavallo N, Gozzi G, Maranzano V, Di Cagno R, Gobbetti M, Gesualdo L. Effect of whole-grain barley on the human fecal microbiota and metabolome. Appl Environ Microbiol. 2015;81:7945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3(4):289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Upadhyaya B, McCormack L, Fardin-Kia AR, Juenemann R, Nichenametla S, Clapper J, Specker B, Dey M. Impact of dietary resistant starch type 4 on human gut microbiota and immunometabolic functions. Sci Rep. 2016;6:28797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stender EGP, Dybdahl Andersen C, Fredslund F, Holck J, Solberg A, Teze D, Peters GHJ, Christensen BE, Aachmann FL, Welner DHet al. . Structural and functional aspects of mannuronic acid-specific PL6 alginate lyase from the human gut microbe Bacteroides cellulosilyticus. J Biol Chem. 2019;294(47):17915–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RLet al. . Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013;11(8):e1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz-Saavedra S, Salazar N, Suárez A, de Los Reyes-Gavilán CG, Gueimonde M, González S. Comparison of different dietary indices as predictors of inflammation, oxidative stress and intestinal microbiota in middle-aged and elderly subjects. Nutrients. 2020;12:3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaut M, Clavel T. Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr. 2007;137(3):751S–5S. [DOI] [PubMed] [Google Scholar]
- 35.Glick-Bauer M, Yeh MC. The health advantage of a vegan diet: exploring the gut microbiota connection. Nutrients. 2014;6(11):4822–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hooper LV, Midtvedt T, Gordon JI. How host–microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. [DOI] [PubMed] [Google Scholar]
- 37.Meslier V, Laiola M, Roager HM, De Filippis F, Roume H, Quinquis B, Giacco R, Mennella I, Ferracane R, Pons Net al. . Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut. 2020;69(7):1258–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurilshikov A, van den Munckhof ICL, Chen L, Bonder MJ, Schraa K, Rutten JHW, Riksen NP, de Graaf J, Oosting M, Sanna Set al. . Gut microbial associations to plasma metabolites linked to cardiovascular phenotypes and risk. Circ Res. 2019;124(12):1808–20. [DOI] [PubMed] [Google Scholar]
- 39.Sonnenburg JL, Backhed F. Diet–microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang DD, Nguyen LH, Li Y, Yan Y, Ma W, Rinott E, Ivey KL, Shai I, Willett WC, Hu FBet al. . The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat Med. 2021;27:333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lampe JW, Martini MC, Kurzer MS, Adlercreutz H, Slavin JL. Urinary lignan and isoflavonoid excretion in premenopausal women consuming flaxseed powder. Am J Clin Nutr. 1994;60(1):122–8. [DOI] [PubMed] [Google Scholar]
- 42.Clavel T, Henderson G, Alpert CA, Philippe C, Rigottier-Gois L, Doré J, Blaut M. Intestinal bacterial communities that produce active estrogen-like compounds enterodiol and enterolactone in humans. Appl Environ Microbiol. 2005;71(10):6077–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Possemiers S, Bolca S, Eeckhaut E, Depypere H, Verstraete W. Metabolism of isoflavones, lignans and prenylflavonoids by intestinal bacteria: producer phenotyping and relation with intestinal community. FEMS Microbiol Ecol. 2007;61(2):372–83. [DOI] [PubMed] [Google Scholar]
- 44.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10(5):323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight Ret al. . Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available upon approval. Access is restricted due to participant confidentiality and privacy concerns. Further information, including the procedures to obtain and access data from the Health Professionals Follow-Up Study, is provided at https://sites.sph.harvard.edu/hpfs/resources-for-hpfs-investigators/.