Abstract
Random homozygous knockout (RHKO) is an antisense RNA strategy capable of identifying genes whose homozygous functional inactivation yields a selectable phenotype in cells growing in culture. Using this approach, we isolated NIH 3T3 fibroblast clones that showed the ability to form colonies on 0.5% agar and tumors in nude mice. The gene inactivated in one of these clones was found to encode VASP (vasodilator-stimulated phosphoprotein), a previously identified protein that binds to components of the cadherin-catenin junctional complex and has been implicated in cell-cell interactions, the formation of actin filaments, and the transmission of signals at the cytoskeleton-membrane interface. Fibroblasts made deficient in VASP by RHKO showed loss of contact inhibition, and consequently, continued cell division past confluence. Restoration of VASP function by reversal of RHKO yielded cells that had lost the neoplastic capabilities acquired during RHKO. Overproduction of VASP mRNA in the sense or antisense orientation from expression constructs introduced by transfection into naive NIH 3T3 fibroblasts also resulted in neoplastic transformation, implying that normal cell growth may require the maintenance of VASP expression within a narrow range. Our results implicate VASP in tumorigenesis and/or cancer progression.
Vasodilator-stimulated phosphoprotein (VASP), a proline-rich founding member of the Ena-VASP protein family, was discovered both as a substrate of cyclic AMP- and cyclic GMP-dependent protein kinases and a component of the actin-based cytoskeleton (10, 25–27, 65). VASP is associated with focal adhesions, actin filaments, and highly dynamic membrane regions (50) and has been shown to be a multiligand protein that binds to actin, profilin, ActA, zyxin, and vinculin (9, 12, 51, 52). Vinculin in turn associates with tensin, α-actinin, α-catenin, and talin (4, 31, 63, 66, 68), and zyxin and profilin are involved in actin filament assembly and organization (3, 14, 43, 56, 60). In vivo evidence indicates that VASP is a crucial factor in the formation of actin filaments and the integration of signals transmitted between the cytoskeleton, the cytoskeleton-membrane interface, and two cyclic nucleotide signal transduction pathways (2, 25, 65). Additionally, recent data indicate that VASP is functionally homologous to the Drosophila melanogaster Enabled (Ena) locus, which was identified initially as a dominant genetic suppressor of mutations in the Abelson tyrosine kinase (1). However, the biological role of VASP has remained largely unknown.
Recently, a novel gene discovery strategy termed random homozygous knockout (RHKO) was shown to be capable of identifying genes whose functional inactivation in murine fibroblasts leads to reversible cellular transformation (40). This approach uses a promoter within a randomly inserted, chromosomally integrated gene search vector (GSV) to produce antisense transcripts complementary to those originating in the chromosomal gene containing the GSV, and consequently, also complementary to transcripts from other copies of that gene. Use of a β-geo reporter gene within the GSV allows selection of integration events in transcriptionally active genes and also enables the monitoring of antisense effects. Reversal of antisense inhibition is accomplished by Cre- and lox-mediated deletion of a gene encoding an activator of the antisense promoter (40, 69).
Using the RHKO strategy, we generated multiple NIH 3T3 cell clones able to form colonies on 0.5% agar and metastatic tumors in nude mice. We report here that the gene affected by RHKO in one of these clones is the previously identified cell adhesion protein and signal transducer, VASP. We further show that the tumorigenic capabilities associated with VASP deficiency are reversible and that overexpression of VASP, as well as underproduction, can lead to neoplastic transformation. Our results suggest that VASP has a previously unsuspected role in tumorigenesis and/or cancer progression.
MATERIALS AND METHODS
Cell culture and transformation.
NIH 3T3 cells (American Type Culture Collection) were grown in Dulbecco modified Eagle medium supplemented with 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 10% calf serum (Life Technologies, Inc., Rockville, Md.). RHKO was done as previously described (40). Briefly, pLLGSV, a Moloney murine leukemia virus-derived retroviral GSV containing the β-geo reporter gene, was introduced into NIH 3T3 cells, where it integrated at multiple chromosomal sites. Infected cells were selected by using 800 μg of G418 per ml for 2 to 3 weeks. Suspensions of G418-resistant NIH 3T3 cells were transfected with pLLTX DNA by electroporation. Following selection in hygromycin (500 μg/ml) for 2 to 3 weeks, hygromycin-resistant clones were plated onto 0.5% agar (11, 13, 39) and the colonies that formed after 4 to 5 weeks were isolated and expanded to cell lines. Cells transfected with pRSV-Cre for removal of the transactivator were selected by using 1 μM gancyclovir for 2 to 3 weeks. Resistant clones were isolated and expanded individually, and the status of cellular transformation was confirmed by soft agar assays as described above.
Tumorigenicity assays.
Tumorigenicity was assayed by injection of 105 cells into athymic nude mice (NIH nu/nu, female, and 6 weeks of age) subcutaneously over the lateral thorax. Mice were examined twice weekly and sacrificed 32 days later (40).
In vitro cell proliferation.
Cells were seeded at a density of 5 × 105 in 10-cm-diameter tissue culture dishes. The live cells were counted 24, 48, 72, and 96 h later with a hemacytometer.
Identification of fusion transcripts.
Polyadenylated mRNA was isolated from cells from which the transactivator gene had been removed and a cDNA library was constructed with the GIBCO BRL SuperScript plasmid system (Life Technologies). Oligo(dT) was used to prime the mRNA for synthesis of the first cDNA strand, and cDNAs were cloned into pCMVSPORT 3.0. Fusion transcripts containing the chromosomal vasp gene and the gene search vector were identified by using the GeneTrapper cDNA positive selection system (Life Technologies). To confirm that fusion transcripts contained both vasp and GSV sequences, reverse transcription-PCR (RT-PCR) was done by using a specific forward primer for a mouse vasp sequence (5′-ACA GTA GTA AGA GTA ACC GCG-3′), and a specific reverse primer for the gene search vector sequence (5′-GAT CCG CCA TGT CAC AGA TC-3′).
Isolation of cDNA clones.
cDNAs corresponding to fusion transcripts were prepared by RT-PCR with poly(A) RNA as an initial template. The vasp-specific reverse primer was 5′-ACT CCA GAG ACC TTT GCA TT-3′, and the forward primer was 5′-TGG AAC GAA ATG TAG CAA AGA GAG-3′. PCR was performed as follows: 48°C for 45 min; 94°C for 2 min; 40 cycles, with 1 cycle consisting of 94°C for 30 s, 60°C for 1 min, and 68°C for 2 min; and 68°C for 7 min. PCR products were resolved in 1.5% agarose gels stained with ethidium bromide, and DNA fragments were cut from gels and purified with a Qiagen gel extraction kit. The sequences of the purified DNA were determined with an Applied Biosystems model 310 genetic analyzer.
Full-length vasp cDNA was cloned into the pCR3.1 vector by using a eukaryotic TA cloning kit (Invitrogen). The mouse vasp cDNA was inserted between the cytomegalovirus (CMV) promoter and polyadenylation site in either the sense or antisense direction. A mutation was introduced into the construct containing vasp cDNA in the sense orientation by a 4-bp insertion at codon 38 (BglII digestion and Klenow fill-in), which produces a frameshift and a translation stop signal 7 codons later.
Western blot analysis.
Cells were grown to 90% confluence, and whole-cell lysates were prepared in RIPA buffer (1× phosphate-buffered saline, 1% Nonidet P-40 or lgepa CA-630, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate). Fifty-microgram amounts of these lysates were separated by electrophoresis on sodium dodecyl sulfate–10% polyacrylamide gels, and electroblotted onto nitrocellulose membranes. The blots were probed with anti-VASP monoclonal antibody (a generous gift of Michael Zimmer and Ulrich Walter) and detected by an enhanced chemiluminescence procedure (Amersham). The same blots were stripped and reprobed with monoclonal antibody to α-tubulin (clone DM 1A; Sigma) as a loading control. Protein levels were determined by scanning autoradiograms of Western blots with Scanmaster 3 (Howtek), which were analyzed by the Quality One program (pdi, Huntington Station, N.Y.).
Southern blot analysis.
Genomic DNA was isolated from VK cells by standard procedures, and 25-μg samples were digested with restriction enzymes, subjected to electrophoresis on a 1% agarose gel, blotted onto Hybond N nylon membrane (Amersham), and fixed to the membrane by UV cross-linking. The blots were probed with a 1.3-kb β-geo fragment labelled by the random priming method (18).
RESULTS
Isolation of cells in which homozygous gene inactivation produces colony formation on 0.5% agar and tumor formation in nude mice.
We employed the RHKO procedure as previously described (40) to isolate clones of G418-resistant NIH 3T3 cells able to form colonies on 0.5% agar, which has commonly been used to select neoplastically transformed cells that have high metastatic potential (13, 21, 39). One of these clones (Fig. 1A) was expanded into the cell line designated VK; Southern blot analysis showed that VK cells contain the provirus form of the pLLGSV retroviral gene search vector integrated as a head-to-tail tandem repeat at a single chromosomal site (Fig. 2).
FIG. 1.
Transactivation of antisense promoter leads to cell transformation and tumorigenesis. (A) Effect of transactivation of antisense promoter on cell growth. Cell line VK contained the transactivator vector, pLLTX, introduced as previously described (40). The LAP 348 transactivator gene has been deleted in cell line VKΔT2 by introducing pRSV-Cre and selecting cells resistant to ganciclovir. All cells were incubated for 3 weeks in 0.5% agar. Bar, 100 μm. (B) Assay for tumorigenicity of the two cell lines shown in panel A. A total of 105 cells of each cell line were injected subcutaneously over the lateral thorax into four animals as described in Materials and Methods; a representative mouse receiving each cell line is shown 32 days after injection.
FIG. 2.
Structure of integrated provirus and results of Southern blot analysis. (A) Map of integrated provirus. RP, regulated antisense promoter (arrow indicates direction of transcription). 3′dLTR or 5′dLTR, defective 3′ or 5′ retroviral long terminal repeat lacking sequences required for production of virions; SA, splice acceptor site; β-geo, reporter gene fusion of Escherichia coli lacZ and neo (aph) genes (arrow indicates sense direction of transcription into the fusion gene). Relevant restriction enzyme sites are shown, along with the regions used as probes for Southern blots. (B) Southern blot. Genomic DNA (25 μg) used for each restriction enzyme digestion was separated by electrophoresis and analyzed as described in Materials and Methods. Blots were probed with a 1.3-kb DNA probe complementary to the region of the provirus. The positions of size markers are indicated to the right of the blot.
To establish that the transformed phenotype observed for VK cells is dependent on expression of antisense RNA from the pLLGSV-derived chromosomally integrated provirus, the antisense promoter in the provirus was turned off. This was accomplished by transfecting VK cells with pRSV-Cre, which deletes the LAP 348 (42) transactivator gene and an adjacent fusion of hygromycin resistance and thymidine kinase genes by site-specific recombination at lox sites bracketing a segment containing these genes (40). Cells losing thymidine kinase activity were identified by their consequent resistance to ganciclovir (40). Examination of 19 cell clones from a population of ganciclovir-resistant, hygromycin-sensitive cells indicated that 7 had lost the ability to form colonies on agar. The reversion of phenotype of VKΔT2, one of these clones that was chosen for further study, is shown in Fig. 1A. None of 20 individual subclones of VK cells, which retain the transactivator, lost their ability to produce colonies on 0.5% agar.
Properties of VK and VKΔT2 cell lines.
Subcutaneous injection of VK cells into nude mice resulted in tumors at the site of injection in all animals (Fig. 1B and Table 1). The tumor in one of these mice was observed at autopsy to have metastasized spontaneously to the lungs. The VKΔT2 cell line, which had undergone reversal of the ability to form colonies in agar following excision of the transactivator, showed no tumorigenic capabilities following injection into nude mice, and no tumors were observed in any of the four animals receiving these cells (Fig. 1).
TABLE 1.
Properties of cell line VK and its derivative
Presence (+) or absence (−) of combined ganciclovir sensitivity and hygromycin resistance.
Number of colonies per 104 cells plated. Colonies were counted 20 days following seeding in 0.5% agar, as described by Li and Cohen (40).
Number of animals showing tumors 32 days following subcutaneous injection of 105 cells of four animals tested.
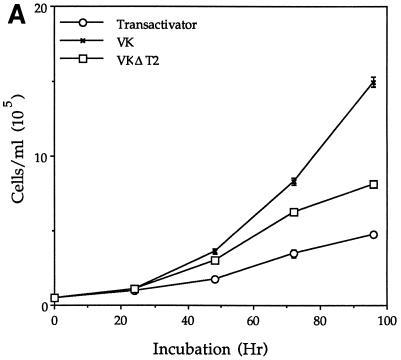
Comparison of the morphology and growth properties of VK, VKΔT2, and control cells (Fig. 3) showed that the tumorigenic capabilities observed for VK were accompanied by marked differences in morphology and growth properties during culture. VK cells were much smaller, as determined visually, than control NIH 3T3 cells containing the transactivator vector only and failed to show the contact inhibition at confluence characteristic of NIH 3T3 cells. They also grew to a cell density at 96 h of culture that was two to three times the maximum density attained by NIH 3T3 cells (Fig. 3A). VKΔT2 cells resembled the control and showed normal contact inhibition, consistent with their inability to form colonies on agar or tumors in nude mice. VK cells, which contain an active promoter producing antisense RNA complementary to VASP transcripts, assumed a dramatically round shape for the first 24 h after plating (Fig. 3B), as has been observed also for cells treated by antisense oligonucleotides to some cell adhesion proteins (44, 46, 61), but became flatter after 2 days in culture. VKΔT2 cells displayed the characteristically flat shape characteristic of fibroblasts in addition to showing contact inhibition at confluence (Fig. 3B), demonstrating reversibility of abnormal morphology upon excision of the transactivator.
FIG. 3.
Characteristics of VK and VKΔT2 cell lines compared with NIH 3T3 cells containing only the pLLTX transactivator vector in VASP function. (A) Growth properties of cells in vitro. Cells (5 × 105) were plated into 10-cm-diameter culture dishes, and at the indicated times, cells were removed and counted. Values indicate averages of three independent experiments, each done in triplicate (mean ± standard error). (B) Morphology of cells in vitro 24 h following the plating of cultures. All fields were examined at the same magnification. Bar, 100 μm.
cDNA cloning and characterization of the fusion transcript.
To identify the chromosomal gene initiating transcripts fused to the β-geo reporter gene in VK cells, a cDNA library was constructed by using poly(A) RNA isolated from cells from which the transactivator gene had been removed. cDNA cloning and sequencing led to identification of a cDNA clone containing 313 bp of a chromosomally encoded transcript sequence fused in frame to the splice acceptor site 5′ to β-geo (Fig. 4). A database search using the BLAST program showed that the segment 5′ to β-geo corresponded to the 5′ untranslated region of the VASP gene plus the first five nucleotides of the VASP protein-coding sequence (70).
FIG. 4.
Murine vasp cDNA sequence fused to the GSV. The nucleotides in uppercase indicate the nucleotides in the vasp sequence which match those of the published vasp sequence from nucleotides 23 to 335 (70). The nucleotides in lowercase are those in the sequence at the splice acceptor site of the GSV. The splice junction 5′ to β-geo is underlined. The nucleotides shown in bold type are the first five nucleotides of the coding sequence of exon 1 of vasp.
Independent evidence that transcripts fusing the VASP sequence to β-geo are made in VK cells was obtained by RT of poly(A) RNA from VKΔT2 and amplification of the resulting cDNA by PCR, using primers corresponding to chromosomal and vector sequences, as described in Materials and Methods. Such PCR amplification yielded cDNA corresponding to the transcript predicted for the VASP–β-geo fusion segment identified by gene trapping, as demonstrated by agarose gel electrophoresis and Southern blotting. Sequencing of the PCR product confirmed that fusion of VASP and β-geo sequences had occurred at the expected splice acceptor site.
VASP expression is reduced in VKΔT2 cells. As seen in Fig. 5 and Table 2, VKΔT2 cells, which contain a GSV insertion in one chromosomal copy of VASP but lack the transactivator that turns on the GSV antisense promoter, maintained a steady-state level of VASP protein at approximately 58% of the level observed in control NIH 3T3 cells. Western blots of protein isolated from VK cells showed 28% of the wild-type level of VASP protein, indicating that total ablation of VASP production is not required for neoplastic transformation of NIH 3T3 cells.
FIG. 5.
Western blot analysis of VASP. Whole-cell lysate (50 μg) in each lane was separated by electrophoresis on a sodium dodecyl sulfate–10% polyacrylamide gel. Anti-VASP monoclonal antibody was used to detect the protein levels. Anti-α-tubulin antibody was used as a loading control. Blots were prepared as described in Materials and Methods.
TABLE 2.
Effect of RHKO on VASP protein level
| Cell line | VASPa (mean ± SE) | α-Tubulina (mean ± SE) | VASP/α-tubulinb
|
|
|---|---|---|---|---|
| Calculated | Normalized | |||
| NIH 3T3 | 6.79 ± 0.48 | 4.12 ± 0.63 | 1.65 | 1.00 |
| VK | 1.99 ± 0.51 | 4.28 ± 0.42 | 0.46 | 0.28 |
| VKΔT2 | 5.83 ± 0.12 | 6.10 ± 0.47 | 0.96 | 0.58 |
Density trace of autoradiograms. Autoradiograms of Western blots were scanned with Scanmaster 3 (Howtek) and analyzed with the Quality One program (pdi). Data represent averages of density traces of autoradiograms from three independent experiments.
Relative density units.
Abnormal VASP expression transforms naive NIH 3T3 fibroblasts.
Transcription of chromosomally integrated VASP cDNA in an antisense orientation in stably transfected naive NIH 3T3 cells under control of the CMV early promoter resulted in the ability of 0.51% of 105 plated cells to form colonies in 0.5% agar (Fig. 6B), providing independent confirmation of the effects of VASP inactivation in these cell lines. Additionally, overexpression of chromosomally integrated VASP cDNA from the CMV promoter in the sense direction surprisingly also resulted in colony formation by NIH 3T3 cells (frequency, 0.48% of plated cells) (Fig. 6C). This was abolished by a 4-bp insertion in codon 38 of the VASP protein-coding sequence that causes premature termination of the VASP protein (Fig. 6D), indicating specifically that VASP protein overexpression is responsible for the observed effect. Cells receiving vectors lacking a VASP insert also showed no evidence of cellular transformation (Fig. 6A). Examination of transfected populations of NIH 3T3 cells expressing VASP cDNA in either the sense or antisense direction indicated that during the first 24 h after plating, approximately 10% of the cells had an abnormal round shape similar to that observed for the VK cell line. The lower frequency of morphologically abnormal cells in these populations than that in VK cells, which are clonal, correlates with the lower frequency of cellular transformation by these VASP cDNA transfectants (Table 3). We speculate that VASP cDNA may be expressed to different extents at different chromosomal insertion sites in cells of the population of cDNA transfectants.
FIG. 6.
Colony growth in 0.5% agar by NIH 3T3 cells transfected with vasp cDNA and control constructs. Representative results are shown for soft agar assays measuring anchorage-independent growth of NIH 3T3 fibroblasts expressing different forms of vasp cDNA and control constructs. Transfected cells were selected for growth in 800 μg of G418 per ml for 18 days, and 105 resistant cells were plated in 0.5% agar and incubated for 3 weeks. Cells were transfected with different vectors. Vector control lacking vasp cDNA (A), vectors containing full-length vasp cDNA in antisense (B) or sense (C) orientation, and vector containing vasp cDNA in sense orientation but containing a protein-terminating insertion as described in Materials and Methods (D) were used. Bar, 100 μm.
TABLE 3.
Colony formation by NIH 3T3 cells transfected with vasp cDNA constructs
| vasp cDNA construct | No. of coloniesa (mean ± SE) | Efficiency of colony formation (%) |
|---|---|---|
| Vector only | 0 | 0 |
| Antisense | 510 ± 41 | 0.51 |
| Sense | 478 ± 36 | 0.48 |
| Sense mutant | 0 | 0 |
Number of colonies formed per 105 cells in 0.5% agar. Colonies were counted 21 days after plating. The data represent averages of three independent experiments, each done in triplicate.
DISCUSSION
The discovery that the VASP gene, which was identified previously in another context (29, 65), is implicated in the pathogenesis and/or progression of cancer is an unexpected outcome of the use of an RHKO screen to isolate candidate tumor suppresser genes. However, given the evidence that a variety of genes having a role in focal adhesions and cytoskeleton interactions have a role in neoplasia, it is perhaps not surprising to have detected VASP, which is a component of the actin-based cytoskeleton (10, 25–27, 65) and has been demonstrated to affect actin filaments and the transmission of signals at the cytoskeleton-membrane interface, in this screen. Consistent with indications that cytoskeletal alterations may be primary events in tumorigenesis (6, 20, 32, 49, 57) are indications of tumor suppressor function of actin regulatory proteins such as α-actinin, tropomyosin 1, gelsolin, merlin, DCC, tensin, and vinculin (16, 17, 19, 22, 41, 48, 58, 59, 66).
The neoplastic transformation, lack of contact inhibition at confluence, and morphological alterations that resulted from the production of antisense RNA complementary to VASP transcripts were reversed in 7 of 19 transformed NIH 3T3 cell clones upon inactivation of the GSV antisense promoter, whereas no spontaneous reversion was observed in any of 20 VK cell subclones retaining the transactivator. Of 19 clones tested following restoration of VASP activity, 12 retained abnormal growth properties and the capacity for tumorigenesis, suggesting that permanent genetic alterations may have occurred in these cells during VASP inactivation. Analogous observations have been made following restoration of function of the tsg101 gene (40, 69), whose role in tumorigenesis was also discovered by an RHKO screen, for retinoblastoma (Rb) (8, 30) and for tumor suppressor genes that are believed to function as caretakers (28, 34).
The human VASP gene maps to chromosome 19q13.2-13.3 (70), a site suspected to harbor tumor suppressor gene(s) involved in a variety of human cancers, and loss of heterozygosity in this region has been identified in 35 to 81% of gliomas (5, 53–55) and 53% of ovarian cancers (7, 47); however, currently there is no evidence of genomic abnormalities at the VASP locus in human malignancies.
NIH 3T3 cells in which VASP protein was reduced to 58% of the wild-type level by haploid insufficiency resulting from a GSV insert in one chromosomal copy of the VASP gene grew normally and showed no tumorigenic capabilities. However, abnormal cell morphology and the ability to produce metastatic cancer in nude mice were observed when VASP protein was reduced by antisense interference to 28% of the wild-type level, as assayed by Western blotting. Cells in which the TSG101 protein was diminished but not absent are also tumorigenic (69), and transforming growth factor β1 and murine p27kip1 recently have been reported to show true haploid insufficiency in their ability to protect against cancer development (45, 62). Moreover, Venkatachalam et al. (64) have demonstrated that loss of both p53 alleles is not a prerequisite for neoplasia and that simple reduction in p53 levels may be sufficient to promote tumorigenesis. These examples of tumorigenesis associated with reduction in gene expression appear to be exceptions to the traditional view that neoplasia related to dysfunction of tumor suppressor genes requires genomic mutations that ablate gene activity (24, 34–36, 67). Whereas such loss-of-function mutations classically have provided the framework for defining, and the basis for the discovery of, tumor suppressor genes, the RHKO procedure now makes practical the identification of additional classes of genes whose abnormal regulation may lead to tumorigenesis. Because RHKO selects for GSV insertions at sites that yield phenotypic effects and such effects may result from less than total ablation of protein, RHKO provides a tool for discovering loci implicated in tumorigenesis by mechanisms such as alterations in transcript splicing or decay or events that affect proteins at the translational or posttranslational level (15, 33).
Stable expression of VASP cDNA in naive NIH 3T3 cells in an antisense orientation resulted in neoplastic transformation, consistent with results obtained by RHKO. Additionally, expression of chromosomally inserted VASP cDNA in NIH 3T3 cells also resulted in cellular transformation, and this was reversed by an insertion mutation that leads to premature termination of the VASP protein. Sequencing of the VASP cDNA derived from VKΔT2 cells shows two differences from the sequence reported by Zimmer et al. (70) (i.e., A · T-to-G · C and C · G-to-G · C transitions at 955 and 1192 bp, respectively, and an AGG deletion at 1193 to 1195 bp). However, the sequences of RT-PCR products generated from total RNA isolated from VK, VKΔT2, and parental NIH 3T3 cells were identical, making it likely that the transformation we observed in cells overexpressing VASP cDNA in the sense direction is caused by excess wild-type protein rather than from dominant-negative (38) or gain-of-function (23) mutations such as those reported for p53 (37). Together, these results suggest that VASP expression outside of a narrow range can perturb cell growth, perhaps by shifting the dynamic equilibrium between pools of assembled and unassembled cytoskeletal proteins (50). Analogous effects of over- and underproduction have been observed also for tsg101, whose excess or deficiency in NIH 3T3 fibroblasts can lead to tumorigenesis (40).
ACKNOWLEDGMENTS
We thank Michael Zimmer and Ulrich Walter for anti-VASP antibody and for helpful discussions.
This study was supported in part by funds from the 1993 Helmut Horten Foundation Research Award to S.N.C. and by a gift from the Chiron Corporation. K.L. is a recipient of NIH postdoctoral fellowship awards from the National Cancer Institute (PHS NRSA CA09302) and Human Genome Training Program (HG 00044-04).
REFERENCES
- 1.Ahern-Djamali S M, Comer A R, Bachmann C, Kastenmeier A S, Reddy S K, Beckerle M C, Walter U, Hoffmann F M. Mutations in Drosophila enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains. Mol Biol Cell. 1998;9:2157–2171. doi: 10.1091/mbc.9.8.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aszodi A, Pfeifer A, Ahmad M, Glauner M, Zhou X H, Ny L, Andersson K E, Kehrel B, Offermanns S, Fassler R. The vasodilator-stimulated phosphoprotein (VASP) is involved in cGMP- and cAMP-mediated inhibition of agonist-induced platelet aggregation, but is dispensable for smooth muscle function. EMBO J. 1999;18:37–48. doi: 10.1093/emboj/18.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckerle M C. Zyxin: zinc fingers at sites of cell adhesion. Bioessays. 1997;19:949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- 4.Belkin A M, Koteliansky V E. Interaction of iodinated vinculin, metavinculin and α-actinin with cytoskeletal proteins. FEBS Lett. 1987;220:291–294. doi: 10.1016/0014-5793(87)80832-3. [DOI] [PubMed] [Google Scholar]
- 5.Bello M J, Leone P E, Nebreda P, Kusak M E, Decampos J M, Vaquero J, Sarasa J L, Pestana A, Rey J A. Molecular abnormalities of chromosome 19 in malignant gliomas: preferential involvement of the 19q13.2-19q13.4 region. Int J Oncol. 1995;6:655–658. doi: 10.3892/ijo.6.3.655. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Ze’ev A. The cytoskeleton in cancer cells. Biochim Biophys Acta. 1985;780:197–212. doi: 10.1016/0304-419x(85)90003-4. [DOI] [PubMed] [Google Scholar]
- 7.Bicher A, Ault K, Kimmelman A, Gershenson D, Reed E, Liang B. Loss of heterozygosity in human ovarian cancer on chromosome 19q. Gynecol Oncol. 1997;66:36–40. doi: 10.1006/gyno.1997.4709. [DOI] [PubMed] [Google Scholar]
- 8.Bookstein R, Shew J Y, Chen P L, Scully P, Lee W H. Suppression of tumorigenicity of human prostate carcinoma cells by replacing a mutated RB gene. Science. 1990;247:712–715. doi: 10.1126/science.2300823. [DOI] [PubMed] [Google Scholar]
- 9.Brindle N P, Holt M R, Davies J E, Price L J, Critchley D R. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem J. 1996;318:753–757. doi: 10.1042/bj3180753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butt E, Abel K, Krieger M, Palm D, Hoppe V, Hoppe J, Walter U. cAMP- and cCMP-dependent protein kinase phophorylation sites of the focal adhesion vasodilator-stimulated phosphoprotein (VASP) in vitro and in intact human platelets. J Biol Chem. 1994;269:14509–14517. [PubMed] [Google Scholar]
- 11.Cartwright C A, Eckhart W, Simon S, Kaplan P L. Cell transformation by pp60c-src mutated in the carboxy-terminal regulatory domain. Cell. 1987;49:83–91. doi: 10.1016/0092-8674(87)90758-6. [DOI] [PubMed] [Google Scholar]
- 12.Chakraborty T, Ebel F, Domann E, Niebuhr K, Gerstel B, Pistor S, Temm-Grove C J, Jockusch B M, Reinhard M, Walter U, Wehland J. A novel focal adhesion factor directly linking the intracellularly motile Listeria monocytogenes and Listeria ivanovii to the actin-based cytoskeleton of mammalian cells. EMBO J. 1995;14:1314–1321. doi: 10.1002/j.1460-2075.1995.tb07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cifone M A, Fidler I J. Correlation of patterns of anchorage-independent growth with in vivo behavior of cells from a murine fibrosarcoma. Proc Natl Acad Sci USA. 1980;77:1039–1043. doi: 10.1073/pnas.77.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford A W, Beckerle M C. Purification and characterization of zyxin, and 82,000-dalton component of adherens junctions. J Biol Chem. 1991;266:5847–5853. [PubMed] [Google Scholar]
- 15.Cui W, Fowlis D J, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst R J. TGF-β1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 16.Dosaka-Akita H, Hommura F, Fujita H, Kinoshita I, Nishi M, Morikawa T, Katoh H, Kawakami Y, Kuzumaki N. Frequent loss of gelsolin expression in no-small cell lung cancers of heavy smokers. Cancer Res. 1998;15:322–327. [PubMed] [Google Scholar]
- 17.Fearon E, Cho K, Nigro J, Kern S, Simon J, Ruppert J, Hamilton S, Preisinger A, Thomas G, Kinsler K, Vogelstein B. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 19.Fernadez J L R, Geiger B, Salomon D, Sabanay M Z, Ben-Ze’ev A. Suppression of tumorigenicity in transformed cells after transfection with vinculin cDNA. J Cell Biol. 1992;119:427–438. doi: 10.1083/jcb.119.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fidler I J, Hart I R. Biological diversity in metastatic neoplasms: origins and implications. Science. 1982;217:998–1003. doi: 10.1126/science.7112116. [DOI] [PubMed] [Google Scholar]
- 21.Freedman V H, Shin S. Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell. 1974;3:355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- 22.Gluck U, Kwiatkowski D J, Ben-Ze’ev A. Suppression of tumorigenicity in simian virus 40-transformed 3T3 cells transfected with α-actinin cDNA. Proc Natl Acad Sci USA. 1993;90:383–387. doi: 10.1073/pnas.90.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gualberto A, Aldape K, Kozakiewicz K, Tlsty T D. An oncogenic form of p53 confers a dominant, gain of function phenotype that disrupts spindle checkpoint control. Proc Natl Acad Sci USA. 1998;95:5166–5171. doi: 10.1073/pnas.95.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haber D, Harlow E. Tumour-suppressor genes: evolving definitions in the genomic age. Nat Genet. 1997;16:320–322. doi: 10.1038/ng0897-320. [DOI] [PubMed] [Google Scholar]
- 25.Haffner C, Jarchau T, Reinhard M, Hoppe J, Lohmann S M, Walter U. Molecular cloning, structural analysis and functional expression of the proline-rich focal adhesion and microfilament-associated protein VASP. EMBO J. 1995;14:19–27. doi: 10.1002/j.1460-2075.1995.tb06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halbrugge M, Walter U. Purification of a vasodilator-regulated phosphoprotein from human platelets. Eur J Biochem. 1989;185:41–50. doi: 10.1111/j.1432-1033.1989.tb15079.x. [DOI] [PubMed] [Google Scholar]
- 27.Halbrugge M, Friedrich C, Eigenthaler M, Schanzenbacher P, Walter U. Stoichiometric and reversible phosphorylation of a 46-kDa protein in human platelets in response to cGMP- and cAMP-elevating vasodilators. J Biol Chem. 1990;265:3088–3093. [PubMed] [Google Scholar]
- 28.Hartwell L H, Weiner T A, Kadyk L, Garvik B. Cell cycle checkpoints, genomic integrity, and cancer. Cold Spring Harbor Symp Quant Biol. 1994;49:259–263. doi: 10.1101/sqb.1994.059.01.030. [DOI] [PubMed] [Google Scholar]
- 29.Holt M R, Critchley D R, Brindle N P. The focal adhesion phosphoprotein, VASP. Int J Biochem Cell Biol. 1998;30:307–311. doi: 10.1016/s1357-2725(97)00101-5. [DOI] [PubMed] [Google Scholar]
- 30.Huang H J, Yee J K, Shew J Y, Chen P L, Bookstein R, Friedmann T, Lee E Y, Lee W H. Suppression of the neoplastic phenotype by replacement of the RB gene in human cancer cells. Science. 1988;242:1563–1566. doi: 10.1126/science.3201247. [DOI] [PubMed] [Google Scholar]
- 31.Huttelmaier S, Mayboroda O, Harbeck B, Jarchau T, Jockusch B M, Rudiger M. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4,5-bisphosphate. Curr Biol. 1998;8:479–488. doi: 10.1016/s0960-9822(98)70199-x. [DOI] [PubMed] [Google Scholar]
- 32.Keely P, Parise L, Juliano R. Integrins and GTPases in tumour cell growth, motility and invasion. Trends Cell Biol. 1998;8:101–106. doi: 10.1016/s0962-8924(97)01219-1. [DOI] [PubMed] [Google Scholar]
- 33.Kimura Y, Koga H, Araki N, Mugita N, Fujita N, Takeshima H, Nishi T, Yamashima T, Saido T, Yamasaki T, Moritake K, Saya H, Nakao M. The involvement of calpain-dependent proteolysis of the tumor suppressor NF2 (merlin) in schwannomas and meningiomas. Nat Med. 1998;4:915–922. doi: 10.1038/nm0898-915. [DOI] [PubMed] [Google Scholar]
- 34.Kinzler K W, Vogelstein B. Cancer-susceptibility genes: gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 35.Klein G. The approaching era of the tumor suppressor genes. Science. 1987;238:1539–1545. doi: 10.1126/science.3317834. [DOI] [PubMed] [Google Scholar]
- 36.Knudson A J. Antioncogenes and human cancer. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine A. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 38.Levine A J, Momand J, Finlay C A. The p53 tumor suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Price J E, Fan D, Zhang R D, Bucana C D, Fidler I J. Correlation of growth capacity of human tumor cells in hard agarose with their in vivo proliferative capacity at specific metastatic sites. J Natl Cancer Inst. 1989;81:1406–1412. doi: 10.1093/jnci/81.18.1406. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Cohen S N. tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 41.Lo S H, Weisberg E, Chen L B. Tensin: a potential link between the cytoskeleton and signal transduction. Bioessays. 1994;16:817–823. doi: 10.1002/bies.950161108. [DOI] [PubMed] [Google Scholar]
- 42.Lupton S D, Brunton L L, Kalberg V A, Overell R W. Dominant positive and negative selection using a hygromycin phosphotransferase-thymidine kinase fusion gene. Mol Cell Biol. 1991;11:3374–3378. doi: 10.1128/mcb.11.6.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machesky L M, Pollard T D. Profilin as a potential mediator of membrane-cytoskeleton communication. Trends Cell Biol. 1993;3:381–385. doi: 10.1016/0962-8924(93)90087-h. [DOI] [PubMed] [Google Scholar]
- 44.Martin M, Andeoli C, Sahuquet A, Montcourrier P, Algrain M, Mangeat P. Ezrin NH2-terminal domain inhibits the cell extension activity of the COOH-terminal domain. J Cell Biol. 1995;128:1081–1093. doi: 10.1083/jcb.128.6.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthew L F, Randel E, Gurley K E, Roberts J M, Kemp C J. The murine gene p27KIP1 is haplo-insufficient for tumour suppression. Nature. 1998;396:177–180. doi: 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narayanan R, Lawlor K G, Schaapveld R Q, Cho K R, Vogelstein B, Bui-Vinh Tran P, Osborne M P, Telang N T. Antisense RNA to the putative tumor-suppressor gene DCC transforms Rat-1 fibroblasts. Oncogene. 1992;7:553–561. [PubMed] [Google Scholar]
- 47.Pejovic T. Genetic changes in ovarian cancer. Ann Med. 1995;27:73–78. doi: 10.3109/07853899509031940. [DOI] [PubMed] [Google Scholar]
- 48.Prasad G L, Fuldner R A, Cooper H L. Expression of transduced tropomyosin 1 cDNA suppresses neoplastic growth of cells transformed by the ras oncogene. Proc Natl Acad Sci USA. 1993;90:7039–7043. doi: 10.1073/pnas.90.15.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raz A, Geiger B. Altered organization of cell-substrate contacts and membrane-associated cytoskeleton in tumor cell variants exhibiting different metastatic capabilities. Cancer Res. 1982;42:5183–5190. [PubMed] [Google Scholar]
- 50.Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch B M, Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch B, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reinhard M, Jouvenal K, Tripier D, Walter U. Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phophoprotein) Proc Natl Acad Sci USA. 1995;74:5463–5467. doi: 10.1073/pnas.92.17.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritland S R, Ganju V, Jenkins R B. Region-specific loss of heterozygosity on chromosome 19 is related to the morphologic type of human glioma. Genes Chromosomes Cancer. 1995;12:277–282. doi: 10.1002/gcc.2870120407. [DOI] [PubMed] [Google Scholar]
- 54.Rosenberg J E, Lisle D K, Burwick J A, Ueki K, vonDeimling A, Mohrenweiser H W, Louis D N. Refined deletion mapping of the chromosome 19q glioma tumor suppressor gene to the D19S412-STD interval. Oncogene. 1996;13:2483–2485. [PubMed] [Google Scholar]
- 55.Rubio M P, Correa K M, Ueki K, Mohrenweiser H W, Gusella J F, vonDeimling A, Louis D N. The putative glioma tumor suppressor gene on chromosome 19q maps between APOC2 and HRC. Cancer Res. 1994;54:4760–4763. [PubMed] [Google Scholar]
- 56.Sadler I, Crawford A W, Michelsen J W, Beckerle M C. Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J Cell Biol. 1992;119:1573–1587. doi: 10.1083/jcb.119.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz M A. Integrins, oncogenes, and anchorage independence. J Cell Biol. 1997;39:575–578. doi: 10.1083/jcb.139.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scoles D R, Huynh D P, Morcos P A, Coulsell E R, Robinson N G G, Tamanoi F, Pilst S T. Neurofibromatosis 2 tumor suppressor schwannomin interacts with βII-spectrin. Nat Genet. 1998;18:354–359. doi: 10.1038/ng0498-354. [DOI] [PubMed] [Google Scholar]
- 59.Shaw R J, McClatchey A I, Jacks T. Regulation of the neurofibromatosis type 2 tumor suppressor protein, Merlin, by adhesion and growth arrest stimuli. J Biol Chem. 1998;273:7757–7764. doi: 10.1074/jbc.273.13.7757. [DOI] [PubMed] [Google Scholar]
- 60.Suetsugu S, Miki H, Takenawa T. The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 1998;17:6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeuchi K, Sato N, Kasahara H, Funayama N, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Perturbation of cell adhesion and microvilli formation by antisense oligonucleotides to ERM family members. J Cell Biol. 1994;125:1371–1384. doi: 10.1083/jcb.125.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang B, Bottinger E P, Jakowlew S B, Bagnall K M, Mariano J, Anver M R, Jetterio J J, Wakefield L M. Transforming growth factor-β1 is a new form of tumor suppressor with true haploid insufficiency. Nat Med. 1998;4:802–807. doi: 10.1038/nm0798-802. [DOI] [PubMed] [Google Scholar]
- 63.Turner C E, Glenney J R, Burridge K. Paxillin: a new vinculin binding protein present in focal adhesions. J Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venkatachalam S, Shi Y, Jones S N, Vogel H, Bradley A, Pinkeland D, Donehower L A. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waldmann R, Nieberding M, Walter U. Vasodilator-stimulated protein phophorylation in platelets is mediated by cAMP- and cGMP-dependent protein kinases. Eur J Biochem. 1987;167:441–448. doi: 10.1111/j.1432-1033.1987.tb13357.x. [DOI] [PubMed] [Google Scholar]
- 66.Weiss E E, Kroemker M, Rudiger A, Jockusch B M, Rudiger M. Vinculin is part of the cadherin-catenin junctional complex: complex formation between α-catenin and vinculin. J Cell Biol. 1998;141:755–764. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White R L. Tumor suppressing pathways. Cell. 1998;92:591–592. doi: 10.1016/s0092-8674(00)81124-1. [DOI] [PubMed] [Google Scholar]
- 68.Wilkins J A, Risinger M A, Lin S. Studies of proteins that co-purify with smooth muscle vinculin: identification of immunologically related species in focal adhesions of nonmuscle and Z-lines of muscle cells. J Cell Biol. 1986;102:1483–1494. doi: 10.1083/jcb.103.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie W, Li L, Cohen S N. Cell cycle-dependent subcellular localization of the tsg101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc Natl Acad Sci USA. 1998;95:1595–1600. doi: 10.1073/pnas.95.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zimmer M, Fink T, Fiselore L, Hauser W, Scherer K, Lichter P, Walter U. Cloning of the VASP (vasodilator-stimulated phosphoprotein) genes in human and mouse: structure, sequence, and chromosomal localization. Genomics. 1996;36:227–233. doi: 10.1006/geno.1996.0457. [DOI] [PubMed] [Google Scholar]