Carrera et al. describe the first genome-wide association study on ischaemic stroke patients treated with pharmacological thrombolysis. They show that polymorphisms in the ZBTB46 gene are associated with the risk of parenchymal haematoma, and replicate the finding in an independent cohort.
Keywords: GWAS, ischaemic stroke, pharmacogenetics, parenchymal haematoma, thrombolysis
Abstract
Haemorrhagic transformation is a complication of recombinant tissue-plasminogen activator treatment. The most severe form, parenchymal haematoma, can result in neurological deterioration, disability, and death. Our objective was to identify single nucleotide variations associated with a risk of parenchymal haematoma following thrombolytic therapy in patients with acute ischaemic stroke. A fixed-effect genome-wide meta-analysis was performed combining two-stage genome-wide association studies (n = 1904). The discovery stage (three cohorts) comprised 1324 ischaemic stroke individuals, 5.4% of whom had a parenchymal haematoma. Genetic variants yielding a P-value < 0.05 1 × 10−5 were analysed in the validation stage (six cohorts), formed by 580 ischaemic stroke patients with 12.1% haemorrhagic events. All participants received recombinant tissue-plasminogen activator; cases were parenchymal haematoma type 1 or 2 as defined by the European Cooperative Acute Stroke Study (ECASS) criteria. Genome-wide significant findings (P < 5 × 10−8) were characterized by in silico functional annotation, gene expression, and DNA regulatory elements. We analysed 7 989 272 single nucleotide polymorphisms and identified a genome-wide association locus on chromosome 20 in the discovery cohort; functional annotation indicated that the ZBTB46 gene was driving the association for chromosome 20. The top single nucleotide polymorphism was rs76484331 in the ZBTB46 gene [P = 2.49 × 10−8; odds ratio (OR): 11.21; 95% confidence interval (CI): 4.82–26.55]. In the replication cohort (n = 580), the rs76484331 polymorphism was associated with parenchymal haematoma (P = 0.01), and the overall association after meta-analysis increased (P = 1.61 × 10−8; OR: 5.84; 95% CI: 3.16–10.76). ZBTB46 codes the zinc finger and BTB domain-containing protein 46 that acts as a transcription factor. In silico studies indicated that ZBTB46 is expressed in brain tissue by neurons and endothelial cells. Moreover, rs76484331 interacts with the promoter sites located at 20q13. In conclusion, we identified single nucleotide variants in the ZBTB46 gene associated with a higher risk of parenchymal haematoma following recombinant tissue-plasminogen activator treatment.
Introduction
Despite the effectiveness of thrombolytic recombinant tissue-plasminogen activator (rtPA) treatment for acute ischaemic stroke (AIS), a 6- to 7-fold increased risk of intracerebral haemorrhage (ICH) remains a serious therapeutic limitation.1 The most severe presentation of rtPA-induced intracerebral bleeding—parenchymal haematoma—worsens the patient’s outcome and increases the risk of 3-month mortality from 2.4 to 4.5 times.2
The damage on the blood vessel caused by ischaemia and reperfusion,3 the breakdown of the blood–brain barrier,4 and the coagulopathy produced by rtPA5 result in blood extravasation within the ischaemic core. Despite the efforts to identify risk factors, such as older age, stroke severity, high blood pressure, circulating glucose levels, leukoaraiosis, and prediction scores,2,6,7 6% of patients still suffer a parenchymal haematoma after thrombolysis.8
Part of the inter-individual variability in the response to AIS treatment could be explained by the genetic architecture of the patients. Previous results used a candidate gene approach that recognized the polymorphisms involved in the risk of parenchymal haematoma, which included, but were not limited to: alpha-2-macroglobulin (A2M), coagulation factor XII (FXII, F12),6 and survivin (BIRC5).9 However, the results of some of these findings need to be confirmed by replication in larger populations. To date, no genome-wide association studies (GWAS) have been published in this field. The hypothesis-free method has been able to find and replicate genetic variants and biological pathways associated with the risk of stroke,10 and a similar success is expected when studying AIS phenotypes, such as parenchymal haematoma.
This study aimed to identify polymorphisms associated with parenchymal haematoma in AIS patients undergoing thrombolytic therapy through a GWAS approach.
Materials and methods
Study population
The study consisted of AIS patients admitted to an Emergency Department and treated with intravenous rtPA. Thrombolysis was performed within 4.5 h of symptom onset at a standard dose of 0.9 mg/kg/dose (10% bolus and 90% continuous 1-h infusion). Eligible participants were at least 18 years old, who suffered a neurological deficit, diagnosed by an expert neurologist, and confirmed by neuroimaging. The exclusion criteria were: patients with remote parenchymal haematoma (as different physiological mechanisms may be involved in this condition, e.g. cerebral microbleeds, amyloid angiopathy); cases in which information on the presence of haemorrhagic transformation was not available; onset to treatment time >4.5 h; and patients who had undergone endovascular therapy. Furthermore, an extreme phenotype approach was implemented, then we excluded haemorrhagic infarctions as previous studies did not show an association of haemorrhagic infarction types 1 and 2 with worsened outcomes.11
The Discovery Cohort (n = 3217) consisted of AIS patients recruited via hospital-based studies between 2003 and 2017. The participants were part of the Genetics of Early Neurological Instability After Ischaemic Stroke (GENISIS), Genetic contribution to Functional Outcome and Disability after Stroke (GODS) and the Genotyping Recurrence Risk of Stroke (GRECOS) studies. Participants in the Replication Stage (n = 1172) were enrolled through the effort of collaborative networks: The International Stroke Genetics Consortium (ISGC), the Spanish Stroke Genetics Consortium (Genestroke), the Spanish Stroke Research Network (INVICTUS plus), the Stroke Genetics Network (SiGN), the Genetic Study in Ischaemic Stroke Patients treated with tPA (Geno-tPA), BAse de Datos de ICtus del hospital del MAR (BASICMAR) (Stroke database of the Hospital del Mar), Leuven Stroke Genetics Study (LSGS), Helsinki 2000 Ischaemic Stroke Genetics Study and GENISIS study. In particular, the Replication Study was intentionally strengthened with parenchymal haematoma cases to ensure a sufficient number for conducting studies (see Supplementary material for more detailed information on these studies and patient recruitment).
Clinical protocol
Demographic data, past medical history, cardiovascular risk factors, clinical examination, stroke severity assessed with the National Institutes of Health Stroke Score (NIHSS) at initial evaluation, and treatment decisions were retrieved from the medical records. CT scans were obtained prior to thrombolytic administration (baseline), and 24 h after symptom onset (follow-up) or whenever a neurological deterioration was detected. Neurological deterioration was defined as an increase of at least 4 points in the NIHSS score. All brain images were reviewed by a radiologist or neuro-radiologist. The radiological, clinical and genetic evaluations were mutually blinded.
Phenotype definition
The presence of haemorrhagic transformation was assessed in the follow-up CT scan and radiologically-classified according to the ECASS criteria12 into haemorrhagic infarctions types 1 and 2 and parenchymal haematoma types 1 and 2. Parenchymal haematoma was defined as bleeding in <30% of the infarcted area with mild space-occupying effect (PH-1) or haematoma >30% of this area and a significant mass effect (PH-2). Symptomatic ICH was defined according to the SITS-MOST criteria as local PH-2 on the CT brain scan combined with an increase of ≥4 points in the NIHSS score within 22–36 h of treatment.13
Standard protocol approvals, registrations and patient consents
An Institutional Review Board or Ethics Committee approved the study at each participating site. All patients or their relatives provided written informed consent.
Genome-wide genotyping and imputation
DNA samples were genotyped on commercial arrays from Illumina (Table 1). Stringent quality controls were performed (Supplementary material), such as removing genetic variants based on the genotyping call rate (<97%), minor allele frequency (MAF <1%), significant deviations from the Hardy-Weinberg equilibrium (P = 1 × 10−6), and identity by state to analyse relatedness among subjects. Studies genotyped on the same platforms were combined with the exception of the Finnish participants, resulting in one stratum for the Discovery and three strata for the Replication stages (Supplementary Fig. 1). Quality controls were applied again,14 then phase haplotypes by stratum were estimated and imputation was performed using the Michigan Imputation Server Portal15 based on the 1000 Genomes Phase 3v5 panel. The Finnish cohort was imputed separately due to their genetic background, which differs from other European populations.
Table 1.
Cohorts included in the meta-analysis
| Stage | Cohort | n | PH (%) | Location | Array |
|---|---|---|---|---|---|
| Discovery | GENISIS | 1062 | 52 (4.9) | Spain | HumanCore ExomeChip |
| GODS | 234 | 14 (6) | Spain | HumanCore ExomeChip | |
| GRECOS | 28 | 5 (17.9) | Spain | HumanCore ExomeChip | |
| Overall | 1324 | 71 (5.4) | – | – | |
| Replicationa | Geno-tPAb | 157 | 36 (22.9) | Spain | Omni 1.5 M |
| BASICMARb | 91 | 8 (8.8) | Spain | Omni 5 M | |
| LSGSb | 45 | 8 (17.8) | Belgium | Omni 5 M | |
| HELSINKI 2000c | 164 | 12 (7.3) | Finland | HumanCore ExomeChip | |
| GENISISc | 70 | 2 (2.9) | Finland | HumanCore ExomeChip | |
| GENISIS | 53 | 4 (7.5) | Poland | HumanCore ExomeChip | |
| Overall | 580 | 70 (12.1) | – | – | |
| Overall | – | 1904 | 141 (7.4) | – | – |
BASICMAR = Base de Datos de Ictus del Hospital del Mar (Hospital del Mar Stroke Database); GENISIS = Genetics of Early Neurological Instability after Ischaemic Stroke study; Geno-PA = Genetic study in ischaemic stroke patients treated with tPA; GODS = Genetic contribution to Functional Outcome and Disability after Stroke study; GRECOS = Genotyping Recurrence Risk of Stroke; HELSINKI 2000 = Helsinki 2000 Ischaemic Stroke Genetics Study; LSGS = Leuven Stroke Genetics Study; PH = parenchymal haematoma.
Contributing cohorts were not analysed by ancestry stratum due to the limited samples available.
In the replication stage, in these cohorts, groups were pooled and imputed based on genotyping array.
Association analysis and meta-analysis
We performed a Discovery case-control association analysis. Frequentist association test for each allele were performed in SNPTEST v2.5.216, under an additive genetic model adjusted for age, sex, three principal components (Supplementary Fig. 2), and clinical variables associated with parenchymal haematoma after a logistic regression: baseline NIHSS and diabetes (P < 0.05). Polymorphisms that achieved genome-wide significance and nominal significance, defined as P < 5 × 10−8 and P < 1 × 10−5, respectively, were selected for replication in new independent cohorts. Furthermore, to obtain enough cases, all participants of the replication study were combined into a single dataset. Any possible bias created by population substructure was then checked through analysis with and without adjustment for the principal components.
Subsequently, we examined the single nucleotide polymorphisms (SNPs) identified during the first stage using an association study adjusted for age, sex, baseline NIHSS, genotyping platform and 10 principal components (Supplementary Fig. 3). A different number of principal components compared with the discovery cohort was required due to the different genetic background of the populations studied. To homogenize the replication cohort (Supplementary Fig. 3) we used this different number of principal components. This difference was mainly due to the Finnish population. Lastly, we conducted a fixed effect inverse-variance meta-analysis using METAL.17 Standard error and genomic control options were applied. The genome-wide significance was set at P < 5 × 10−8. In addition, we performed gene-based analysis using the Multi-marker Analysis of GenoMic Annotation (MAGMA)18; the study included variants located within 2 kb of the gene at 3′ and 5′UTR. Additionally we ran S-MultiXcan,19 the multi-tissue tool of the S-PrediXcan software to confirm the MAGMA findings.
We performed GNOVA20 analysis to evaluate the genetic architecture shared with other diseases. We tested genetic correlation using MEGASTROKE European analysis data10 for the ischaemic stroke phenotypes. The sample sizes of the stroke subtypes were: for ischaemic stroke (n = 440 328), large artery atherosclerosis stroke (n = 301 663), cardioembolic stroke (n = 362 661), and small vessel stroke (n = 348 946). For ICH we used the summary statistics available from the ICH GWAS 201421 (n = 6965) and its lobar (n = 1148) and deep (n = 2075) ICH subtypes. Additionally, we evaluated genetic correlation for the white matter hyperintensity (WMH) volume phenotype (n = 11 226) using published data.22 All datasets were obtained from http://cerebrovascularportal.org, accessed 6 November 2020.
We generated a polygenic risk score (PRS) with the data from our study. We used the PRSice-2 software that runs logistic regressions to determine the P-threshold with the largest variance explained by the PRS, assessed as the increment in Nagelkerke’s pseudo-R2.
Bioinformatic functional analysis
After meta-analysis, novel SNPs significantly associated with parenchymal haematoma were examined to identify their biological function using publicly available online bioinformatic tools. We tested tissue-specific expression for the polymorphism-containing gene where the polymorphism is located through the Genotype-Tissue Expression (GTEx) Project,23 BRAINEAC dataset24 and by mapping the gene expression in brain tissue using the single nuclei RNA sequencing expression browser.25 In addition, we checked the effect of the top genetic variant over the gene expression levels in a 1 Mb window (eQTL, expression quantitative trait loci) in different tissues using the summary data-based Mendelian randomization and the heterogeneity in dependent instruments test (SMR and HEIDI),26 the eQTL consortium,27 and the GTEx project.23 An extended eQTL analysis in vascular and brain tissues was performed as well. Interactions between the top SNPs with distal DNA regulatory elements and gene promoter sites were evaluated by Capture HiC Plotter28 in the lymphoblastoid cell line GM12878 and by the RegulomeDB database.29 Furthermore, previous associations of the most significant SNP and proxies (r2 > 0.6) with stroke, cardiovascular traits, and any genome-wide association studies were investigated with the cerebrovascular disease knowledge portal30 and the PhenoScanner31 web tool.
Statistical analyses
Statistical analyses for clinical variables were performed using SPSS statistical package version 17.0 (IBM, Chicago, USA). Univariate analysis for case-controls was evaluated by χ2 or Fisher’s exact test. T-tests, Mann-Whitney U-tests or Kruskal-Wallis tests were used for continuous variables. Logistic regression was conducted with the forward stepwise method to select clinical variables, such as covariates in the association studies. Meta-analysis heterogeneity was calculated using Cochran’s Q-test.
Data availability
Any qualified investigator may request the summary statistics and dataset. Detailed methods are available in the Supplementary material.
Results
Genome-wide association analysis and meta-analysis
Following quality controls and imputation, 1324 AIS patients treated with rt-PA met the inclusion criteria and 7 989 272 polymorphisms were tested (Supplementary Fig. 1). Parenchymal haematoma occurred in 5.4% (n = 71) (Table 1), 2.4% (n = 32) had PH-1 and 2.9% (n = 39) had PH-2. Moreover, the incidence of symptomatic ICH was 1.4% (n = 18). The symptomatic ICH was associated with in-hospital mortality, disability and mortality at 3 months (P < 0.001); significantly, PH-1 and PH-2 haemorrhages excluding symptomatic ICH were also associated with disability [P = 0.007; odds ratio (OR): 7.19; 95% confidence interval (CI): 3.18–16.25] and mortality at 3 months (P = 4.48 × 10−8; OR: 6.05; 95% CI: 3.22–11.36) (Supplementary Table 1). The univariate analysis showed that diabetes, cardioembolism aetiology and baseline NIHSS appeared to be associated with parenchymal haematoma risk (P = 0.02; P = 0.04; P = 1.79 × 10−10). After logistic regression, only diabetes and baseline NIHSS were significantly associated with parenchymal haematoma and used for the GWAS adjustment (Supplementary Table 2). The clinical characteristics are summarized in Table 2.
Table 2.
Clinical findings and univariate analysis of the Discovery Study subjects
| PH |
|||||
|---|---|---|---|---|---|
| Total (n = 1324) | Absence (n = 1253) | Presence (n = 71) | P | OR (95% CI) | |
| Sex, male (%) | 733 (55.4) | 692 (55.2) | 41 (57.7) | 0.7 | 1.11 (0.68–1.79) |
| AF (%) | 361 (27.3) | 336 (26.9) | 25 (35.2) | 0.13 | 1.48 (0.89–2.44) |
| Diabetes (%) | 329 (24.8) | 303 (24.2) | 26 (36.6) | 0.02* | 1.81 (1.10–2.99) |
| HTN (%) | 867 (65.8) | 817 (65.5) | 50 (71.4) | 0.31 | 1.32 (0.78–2.24) |
| ST (%) | 374 (34.5) | 356 (34.7) | 18 (31) | 0.57 | 0.85 (0.48–1.50) |
| TOAST (%) | |||||
| CE | 556 (43.4) | 518 (42.6) | 38 (55.1) | 0.04* | 1.65 (1.02–2.69) |
| LAA | 219 (17.1) | 215 (17.7) | 4 (5.9) | 0.01* | 0.29 (0.07–0.79) |
| SVO | 56 (4.4) | 55 (4.5) | 1 (1.5) | 0.36 | 0.31 (0.01–1.89) |
| OT | 24 (1.9) | 23 (1.9) | 1 (1.5) | 1 | 0.77 (0.02–4.91) |
| UND | 426 (33.3) | 402 (33.1) | 24 (35.3) | 0.71 | 1.1 (0.6–1.8) |
| Age, years (IQR) | 76 (65–82) | 75 (65–82) | 77 (70–82) | 0.28 | |
| Baseline NIHSS (IQR) | 12 (7–18) | 11 (7–18) | 18 (14–22) | <0.001* | |
| Glucose, mg/dl (IQR) | 120 (103–147) | 119 (103–146) | 130 (105–168) | 0.18 | |
| OTT, min (IQR)a | 130 (90–180) | 128 (90–180) | 140 (90–180) | 0.54 | |
| SBP, mmHg (IQR) | 155 (138–172) | 154 (138–172) | 158 (144–173) | 0.36 | |
| DBP, mmHg (IQR) | 82 (71–98) | 81 (71–97) | 85 (70–112) | 0.34 | |
For categorical variables, frequencies were described as percentages. For continuous variables, the median values and interquartile range (IQR) were calculated. AF = atrial fibrillation; CE = cardioembolism; DBP = diastolic blood pressure; HTN = hypertension; LAA = large artery atherosclerosis; OT = other aetiology; OTT = time from onset to treatment; PH = parenchymal haematoma; SBP = systolic blood pressure; ST = statins; SVO = small vessel occlusion; TOAST = Trial of Org 1072 in Acute Stroke Treatment; UND = undetermined aetiology.
*P-value < 0.05. P-values after logistic regression: diabetes: P = 0.04; cardioembolism aetiology: P = 0.29; baseline NIHSS: P = 4.8 × 10−9.
Time from onset to treatment was available in 788 participants.
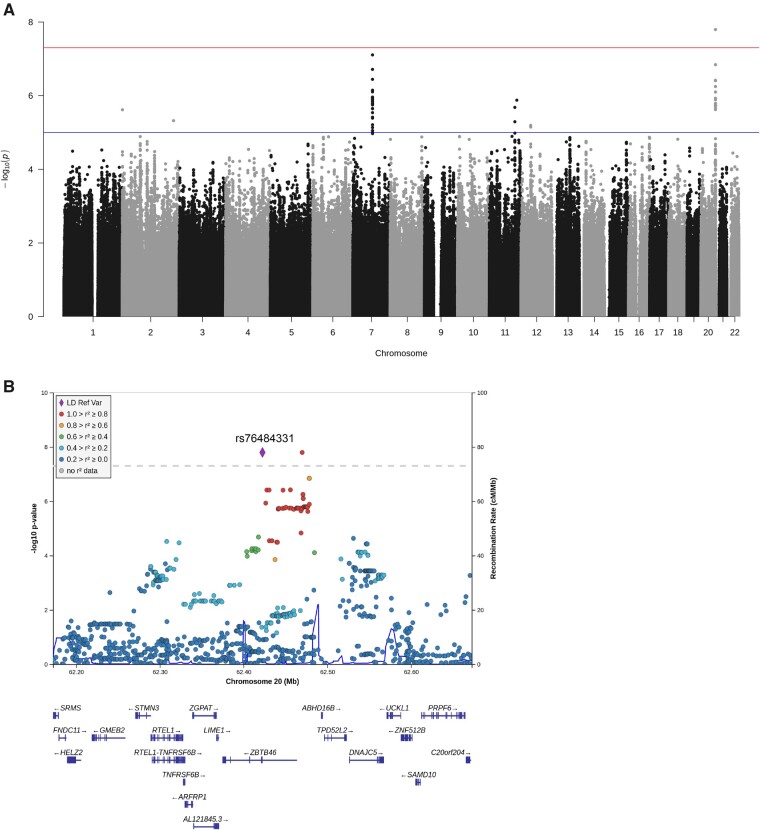
The Manhattan plot obtained is presented in Supplementary Fig. 4. We did not observe overall inflation of P-values (genomic inflation factor λ = 1.018) (Supplementary Fig. 4). The discovery analysis identified one polymorphism that reached genome-wide significance: rs76484331 located on chromosome 20, in intron 1 of ZBTB46, (P = 2.49 × 10−8; OR: 11.31; 95% CI: 4.82–26.55). Furthermore, 182 SNPs were selected for the follow-up stage based on the P-value cut-off 1 × 10−5, including 43 polymorphisms at the ZBTB46 gene (Supplementary Table 3).
Replication was performed in 580 AIS patients. The prevalence of parenchymal haematoma was 12.1% (n = 70); PH-1 developed in 6.4% (n = 37) and PH-2 in 5.7% (n = 33). The higher occurrence of bleeding events can be attributed to the Genot-PA project, a study designed to understand the genetic variability of thrombolytic response. The number of parenchymal haematoma samples included in this study was larger than expected. Age, sex and NIHSS were required as minimum clinical covariates (Table 3). Cardioembolism aetiology stroke was not included as a covariate because it was not significant after logistic regression. Moreover, 10 principal components accounted for the GWAS population substructure adjustment. The genome-wide significant SNP rs76484331 (P < 5 × 10−8) selected from the discovery phase was significant in the validation cohort (P = 0.01). When we included cardioembolism aetiology stroke as a covariable the results did not change significantly (Supplementary Table 4). Additionally, we analysed 182 polymorphisms with nominally significant P-values (P < 1 × 10−5), obtaining the lowest P-value at rs1962779 on chromosome 7 (P = 3.34 × 10−3). Three of these SNPs could not be included in the analysis because of failed imputation quality controls (Supplementary Table 3).
Table 3.
Clinical characteristics of the replication cohort
| PH (%) |
|||||
|---|---|---|---|---|---|
| Total (n = 580) | Absence (n = 510) | Presence (n = 70) | P | OR (95% CI) | |
| Sex, male (%) | 287 (49.5) | 256 (50.2) | 31 (44.3) | 0.35 | 0.79 (0.48–1.30) |
| CE (%) | 286 (49.8) | 239 (47.3) | 47 (68.1) | 0.001* | 2.38 (1.39–4.06) |
| Age, years (IQR) | 71 (60–78) | 70 (59–78) | 74 (68–80) | 0.004* | |
| Diabetes, yes (%) | 42 (12.9) | 35 (12.7) | 7 (13.7) | 0.84 | 1.09 (0.46–2.61) |
| Baseline NIHSS (IQR) | 10 (6–17) | 9 (5–15) | 16 (11–19) | <0.001* | |
For categorical variables, frequencies were described as percentages. For continuous variables, median values and interquartile range (IQR) were calculated. CE = cardioembolism aetiology by trial of Org 1072 in Acute Stroke Treatment classification; PH = parenchymal haematoma.
*P-value < 0.05.
The meta-analysis of both stages revealed one locus on chromosome 20q13 that reached genome-wide significance in association with parenchymal haematoma: rs76484331 (P = 1.61 × 10−8; OR: 5.84, 95% CI: 3.16–10.76; heterogeneity P = 0.03), formed by 43 polymorphisms that exceeded the nominal significance cut-off (P < 1 × 10−5) (Fig. 1). Moreover, we found five loci (overall 45 SNPs) in syntrophin gamma 2 (SNTG2), RUN and FYVE domain-containing protein 4 (RUFY4), semaphorin-3A (SEMA3A), Down syndrome cell adhesion molecule-like protein 1 (DSCAML1), and PDZ domain-containing ring finger 4 (PDZRN4) that reached P-values < 1 × 10−5. All the top SNPs had a consistent direction of effect (Table 4); the best association obtained was for SEMA3A, which addressed a P-value close to the significance cut-off point (P = 7.85 × 10−8; OR: 2.43; 95% CI: 1.76–3.37; heterogeneity P = 0.25).
Figure 1.
Genome-wide association meta-analysis of parenchymal haematoma. SNPs were represented by dots and were plotted based on their GWAS meta-analysis P-values. (A) Manhattan plot of genome-wide association meta-analysis. The red line shows genome-wide significance (P < 5 × 10−8) and the blue line represents the suggestive association significance threshold (P < 1 × 10−5). Results were adjusted for age, sex, baseline NIHSS, diabetes, and principal components. (B) Regional association plot centred on rs76484331 (ZBTB46). The regional plot was drawn using LocusZoom software v0.4.8. Linkage disequilibrium with the top SNP is represented by the coloured dots. Genes were characterized by horizontal lines. The ZNF512B gene was omitted.
Table 4.
Independent leading SNPs and the most significant associations with parenchymal haematoma in the meta-analysis
| SNP | CHR | Position (bp) | Location | Gene | EA/NEA | No. variants | EAF | Stage | OR (95% CI) | P |
|---|---|---|---|---|---|---|---|---|---|---|
| rs77557904 | 2 | 1047076 | Intronic | SNTG2 | G/C | 1 | 0.06 | Meta-analysis | 3.82 (2.19–6.68) | 2.43 × 10−6 |
| Discovery | 6.56 (3.10–13.89) | 8.62 × 10−7 | ||||||||
| Replication | 2.01 (0.88–4.59) | 0.1 | ||||||||
| rs112541215 | 2 | 218916291 | Intronic | RUFY4 | A/T | 1 | 0.08 | Meta-analysis | 3.01 (1.88–4.84) | 4.79 × 10−6§ |
| Discovery | 4.67 (2.43–8.96) | 3.76 × 10−6 | ||||||||
| Replication | 1.90 (0.96–3.74) | 0.07 | ||||||||
| rs1962779 | 7 | 83837734 | Intronic | SEMA3A | C/G | 40 | 0.19 | Meta-analysis | 2.43 (1.76–3.37) | 7.85 × 10−8§ |
| Discovery | 2.94 (1.87–4.61) | 2.77 × 10−6 | ||||||||
| Replication | 2.00 (1.26–3.18) | 3.34 × 10−3 | ||||||||
| rs4356265 | 11 | 117301818 | Intronic | DSCAML1 | T/C | 1 | 0.11 | Meta-analysis | 2.75 (1.83–4.15) | 1.33 × 10−6§ |
| Discovery | 3.92 (2.23–6.88) | 1.89 × 10−6 | ||||||||
| Replication | 1.87 (1.03–3.38) | 0.04 | ||||||||
| rs564865745 | 12 | 41626444 | Intronic | PDZRN4 | G/A | 57 | 0.05 | Meta-analysis | 4.23 (2.37–7.54) | 1.03 × 10−6 |
| Discovery | 9.89 (4.20–23.28) | 1.56 × 10−7 | ||||||||
| Replication | 2.12 (0.97–4.62) | 0.06 | ||||||||
| rs76484331 | 20 | 62422504 | Intronic | ZBTB46 | A/C | 43 | 0.1 | Meta-analysis | 5.84 (3.16–10.76) | 1.61 × 10−8* |
| Discovery | 11.31 (4.82–26.55) | 2.49 × 10−8* | ||||||||
| Replication | 2.97 (1.24–7.09) | 0.01 |
CHR = chromosome; EA = effect allele; EAF = effect allele frequency; No. variants = number of variants reaching P > 1 × 10−5; NEA = non-effect allele. Only independent SNPs (r2 < 0.1, within a 1 Mb window) with a P-value < 1 × 10−5 are shown. Alleles and chromosomal positions were identified on the basis of the 1000 Genomes Phase 3 Project. Location was described following the ANNOVAR system.
*P-value < 5 × 10−8.
Heterogeneity P-value > 0.05.
On the other hand, the gene-based analysis computed one statistically significant gene associated with our phenotype: chromosome 20 open reading frame 181 (C20orf181; P = 0.05/18647; P < 2.68 × 10−6) (Supplementary Table 5), located at 20q13.33. In addition, ZBTB46 showed a P-value of 0.0035. S-MultiXcan using GTEx 23 v8 data confirmed the results for ZBTB46 (P = 3.16 × 10−4); however, the C20orf181 transcript was not covered by the 22 313 transcripts evaluated with S-MultiXcan and it could not be analysed (Supplementary Table 6).
Genetic correlation analysis revealed a shared genetic background of parenchymal haematoma and deep ICH, lobar ICH and WMH (P < 9.2 × 10−3) (Supplementary Table 7). After Bonferroni correction, only lobar ICH was significantly associated with parenchymal haematoma.
The best-fit for the PRS estimation of disability after 3 months using the genetic data from the discovery cohort was observed for a threshold of P = 0.00530005, pseudo-R2 = 0.0684721, and composed of 3506 SNPs. A significant association was observed with parenchymal haematoma and mortality after 3 months, but no association was found with in-hospital mortality (Supplementary Tables 8 and 9). The most significant association was with disability after 3 months [modified Rankin Score (mRS) 0–2 versus 3–6; P = 1.5 × 10−6]. Interestingly, in a multivariable logistic regression for disability after 3 months the PRS remained significant in the logistic regression after inclusion of the clinical variables: baseline NIHSS, sex and age (Supplementary Table 10).
Functional analysis
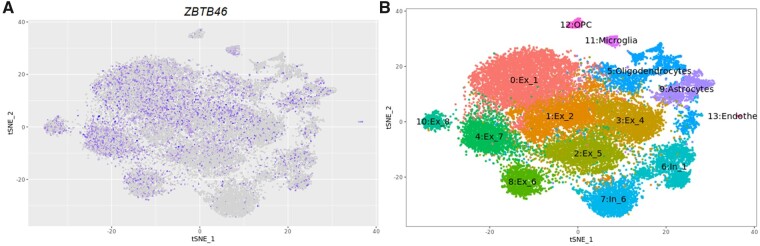
The GTEx and BRAINEAC portal revealed gene expression of ZBTB46 in different tissues, with the highest expression detected in the brain, specifically in the region of the cerebellum (Supplementary Figs 5 and 6). Besides, single nuclei RNA sequencing confirmed ZBTB46 RNA expression in neurons and endothelial cells (Fig. 2). Furthermore, the eQTL study showed a nominal association of rs76484331 with expression levels of ZBTB46 in blood tissue (P = 2.2 × 10−6) (Supplementary Table 11); similarly, interactions between rs76484331 and promoter sites at the ZBTB46 gene were evidenced by Capture HiC Plotter (Supplementary Fig. 7). The RegulomeDB classified rs76484331 as transcription factor-binding or DNase peak, chromatin states included quiescent action in blood cells and weak transcription in the brain (Supplementary Table 12). However, rs76484331 did not affect a single gene exclusively. We observed additional cis-eQTL and promoter sites located in the near window of 1 Mb associated with the presence of the polymorphism including Lck interacting transmembrane adaptor 1 (LIME1) and DnaJ heat shock protein family (Hsp40) member C5 (DNAJC5), further interactions are detailed in Supplementary Table 13 and Supplementary Fig. 7.
Figure 2.
RNA-seq expression of ZBTB46 in the brain. (A) The T-distributed stochastic neighbour embedding (tSNE) plots represent 26 331 nuclei and clusters. ZBTB46 gene expression levels are represented by colour: high expression is presented in purple and low expression in grey. (B) Clusters are identified by cell type: excitatory neurons (Ex), inhibitory neurons (In), oligodendrocytes, microglia, oligodendrocyte precursor cells (OPC), and endothelial cells. Data were extracted from the web-based application http://ngi.pub/snuclRNA-seq/.
Traits that have been reported as being associated with rs76484331 include cardiomyopathy, blood pressure, and coronary atherosclerosis. Moreover, using the cerebrovascular disease knowledge portal (https://cerebrovascularportal.org/), we could not find any association with haemorrhagic stroke and ischaemic stroke subtypes (Supplementary Table 14). Additionally, we did not find evidence of association within rs76484331 and hypertension or blood pressure levels in our cohort (Supplementary Tables 15 and 16).
Discussion
This study found a genome-wide polymorphism associated with parenchymal haematoma in an intron of ZBTB46. The SNP, rs7648433, was initially identified in 1324 patients and further validation was sought in 580 participants; both stages included AIS treated with intravenous thrombolysis.
ZBTB46 is a member of the Poxvirus and Zinc Finger and Krüppel-type (POK) protein family that contains a POZ domain for protein-protein interactions and zinc fingers for DNA binding.32 The POK proteins act as transcription factors, facilitating the recruitment of co-repressors to promoter regions33 and play a role in the development of haematopoietic,34 dendritic,35 and endothelial cells,35 and in lymphocyte differentiation during the immune response.36 No studies have associated ZBTB46 with ischaemic or haemorrhagic stroke; however, involvement in stroke mechanisms, such as shear stress and atherosclerosis, has been reported previously.37 Moreover, ZBTB46 has also been linked with prostate cancer38 and multiple sclerosis.39
The polymorphisms identified lay within intronic regions, portions of gene that are spliced out prior to protein translation. Introns are part of the non-coding variants that represent >90% of all the hits identified by GWAS in complex diseases. Finding the causality of these SNPs could be challenging; however, these genome regions may regulate gene expression.40 We tested whether the GWAS significant polymorphism could influence other neighbouring genes using in silico analysis tools. Considering the importance of the integrity of the blood–brain barrier in the pathophysiology of haemorrhagic events after thrombolysis, we emphasized the SNP effect on gene expression in brain, blood and blood vessel tissues.
We found a nominal association between rs7648433 and ZBTB46 gene expression reported in arterial vessel tissue; in addition, ZBTB46 RNA expression was identified in endothelial cells and neurons in brain tissue. Moreover, cis-eQTL of rs7648433 within a neighbouring region of 1 Mb identified six differential gene expressions in blood tissue. None of the cis-eQTL genes identified have been previously associated with cerebrovascular traits.
Regulation of gene expression by ZBTB46 is mediated through binding of the zinc finger domain to a chromatin complex.41 The POK proteins act over a large and assorted group of genes, which includes transcription factors, RNA processing factors, chromatin regulators, kinases, peptidases, ubiquitin ligases, and phosphatases.42 The current study exhibited an interaction among rs7648433, and several promoter sites located in chromosome 20, including ZBTB46. Moreover, the gene-based analysis identified the open read frame C20orf181, located at 231 bp from the upstream region of ZBTB46; open read frames can mediate the protein synthesis through the translation regulation of genes.43
Our results suggest the modulation of genetic targets affects the parenchymal haematoma risk and could influence the response to rtPA therapy. On the other hand, rs76484331 has been associated with cardiomyopathies and systolic blood pressure levels in the UK Biobank (UKBB) project. Polymorphism pleiotropy could highlight the biological effect of the variant; however, replications should be performed in independent cohorts and a secondary analysis is needed in order to clarify this. In addition, ZBTB46 plays a role in keeping cells in a quiescent state (G0–G1)37 and could be activated through several proteins or events like the Toll-like receptor proteins (TLR)42 or disturbed vessel flow.37 Little is known about how ZBTB46 responds under ischaemic conditions and with the data available, we are not able to determine which molecular process is modified in the parenchymal haematoma risk by rs76484331 or ZBTB46.
Our study also revealed five loci that reached a nominal association with parenchymal haematoma; the second most significant locus, SEMA3A, is related to the vascular permeability of the blood–brain barrier and brain damage after cerebral ischaemia in murine models.44 Moreover, SEMA3A is expressed by the ischaemic brain and the ischaemic core during reperfusion.45 It seems that the genetics of SEMA3A could modulate the response to ischaemia and promote the onset of bleeding events. Studies with larger sample sizes are required to determine whether this GWAS hit is associated with the onset of parenchymal haematoma.
Interestingly, genetic correlation analysis revealed a shared genetic correlation of parenchymal haematoma with several traits, such as lobar and deep ICH or WMH. These findings indicate that ICH genetic risk factors could play a role in the risk of parenchymal haematoma; however, further studies are needed to confirm this hypothesis.
The consequences of symptomatic ICH on the stroke outcome at 3 months and in-hospital mortality13 are well established. In our cohort the occurrence of PH-1 and non-symptomatic PH-2 also had a damaging effect on disability after stroke. In view of this, efforts to translate causal variants and genes to clinical practice should be encouraged. This is supported by our results with the PRS generated using the data from the genetic analysis of parenchymal haematoma. The PRS was associated in the independent replication cohort with parenchymal haematoma, disability and mortality after 3 months. In the case of disability, it remains significantly associated after logistic regression indicating a potential use in clinical practice as a possible biomarker or as something to be explored in further studies looking for drug targets.
The GWAS findings could feasibly allow the identification of new drug targets or the repurposing of existing drug components used for other diseases that address the newly-identified causal genes or pathways; for example, the identification of IL23R polymorphisms associated with ankylosing spondylitis46 generated the repositioning of secukinumab, an anti-IL-17A monoclonal antibody involved in the IL23 pathway. Secukinumab is widely used in the treatment of psoriasis and psoriatic arthritis and is currently used successfully in ankylosing spondylitis too.47 Another strategy for translating discoveries from GWAS includes the development of tools (e.g. risk scores) for measuring the individual predisposition to a trait. The Geno-tPA score, based on clinical characteristics and two polymorphisms, demonstrated its prediction capability of bleeding events in patients undergoing thrombolysis alone or in combination with mechanical thrombectomy.6,48 The implementation of predictive scores could help physicians in decision-making, avoiding treatment delays, adjusting the drug dose, or implementing additional therapies.
We performed the first GWAS of the haemorrhagic transformation risk after thrombolysis, based on a relatively small sample size in comparison with large-scale GWAS. To address this issue, petechial bleeding cases were excluded, thus through an extreme phenotyping study approach, we aimed at increasing the variant effect sizes49 to detect parenchymal haematoma-associated polymorphisms. Furthermore, to reduce false-positive results due to low-frequency variants, we removed extremely large beta estimates,50 applied a stringent filter of minor allele frequency and evaluated the presence of alleles in order to guarantee a total of at least 25 alleles51 in each stage.
The limitations of the study included not being able to analyse the Finnish cohort because of the small number of cases available. The Finnish population is considered genetically isolated52 and its genetic population structure could generate a bias; therefore, we attempted to reduce this limitation by including a sufficient number of principal components in the association study.53 Based on isolation by distance, we used 10 eigenvectors in our analysis. Another limitation is the absence of patients treated with mechanical thrombectomy, a treatment that is currently available for certain AIS patients.54 Moreover, our results cannot be extrapolated to non-European populations. Further studies are required to validate our findings in diverse populations and patients who underwent mechanical recanalization.
In summary, our findings identified a previously unreported polymorphism in the ZBTB46 gene associated with the development of parenchymal haematoma following rtPA treatment. Besides, suggestive loci were identified, which require confirmation in future independent studies. Further functional studies are required to clarify the genetic mechanism involved in the bleeding risk of thrombolysis.
Web resources
The data used for the analyses described in this manuscript were obtained from the GTEx Portal (gtexportal.org) on 20 June 201928; the CHiCP online tool (https://www.chicp.org) on 29 October 2019; and the Cerebrovascular Disease Knowledge Portal (cerebrovascularportal.org) on 20 June 2019.
Supplementary Material
Acknowledgements
We are grateful to Lucía Muñoz (Hospital Germans Trias i Pujol), Anna Penalba (Vall d’Hebron Research Institute), Uxue Lazkano (IMIM-Hospital del Mar), Carmen Jimenez (Hospital Universitari Son Espases), Elisa Cortijo (Hospital Clínico Universitario), Rebeca Marín Bueno (Hospital de la Santa Creu i Sant Pau, IIB-Sant Pau), Esther Sarasola Diez (Hospital de Basurto), Carmen Gubern (Doctor Josep Trueta University Hospital), Aki Havulinna (Institute for Molecular Medicine Finland), Veikko Salomaa (Institute for Molecular Medicine Finland), and Antoni Ferens (Jagiellonian University), for their contribution to patient recruitment; and to Agustin Ruiz and Oscar Sotolongo (Fundació ACE) for their technical support.
Funding
The GENISIS study is funded by the National Institute of Health (K23 NS099487, and R01NIH NS085419). The Neurovascular Research Laboratory is supported by the Spanish stroke research network (INVICTUS plus). The Stroke Pharmacogenomics and Genetics Laboratory is supported by the Spanish stroke research network (INVICTUS plus); the Generation Project (PI15/01978) and the Pre-Test Stroke Project (PMP15/00022) are funded by the Instituto de Salud Carlos III and Fondo Europeo de Desarrollo Regional (ISCIII-FEDER). The Neurovascular Research Group, IMIM is supported by INVICTUS plus (RD16/0019/0002). The Biomedical Research Institute Hospital de la Santa Creu i Sant Pau, IIB Sant Pau is supported by INVICTUS plus (RD16/0019/0010). Ibiostroke (AC19/00106), Maestro Project (PI18/01338) is supported by the Instituto de Salud Carlos III and Fondo Europeo de Desarrollo Regional (ISCIII-FEDER) and the Epigenisis project is supported by Marató TV3 (201711.30). I.F.-C. (CPII17/00021), T.S. (CPII17/00027) and F.C. (CPII19/00020) are recipients of a research contract from the Miguel Servet Program from the Instituto de Salud Carlos III. A.B. is supported by a Juan Rodes research contract from the Carlos III Health Institute (JR16/00008). J.C.-M. is supported by an AGAUR Contract (agència de gestió d'ajuts universitaris i de recerca; FI_DGR 2019, grant number 2019_FI_B 00853) co-financed with Fons Social Europeu (FSE). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- AIS
acute ischaemic stroke
- GWAS
genome-wide association studies
- ICH
intracerebral haemorrhages; NIHSS = National Institutes of Health Stroke Score
- PRS
polygenic risk score
- rtPA
recombinant tissue-plasminogen activator
- SNP
single nucleotide polymorphism
References
- 1.Whiteley WN, Emberson J, Lees KR, et al. ; Stroke Thrombolysis Trialists' Collaboration. Risk of intracerebral haemorrhage with alteplase after acute ischaemic stroke: A secondary analysis of an individual patient data meta-analysis. Lancet Neurol. 2016;15(9):925–933. [DOI] [PubMed] [Google Scholar]
- 2.Strbian D, Sairanen T, Meretoja A, et al. Helsinki Stroke Thrombolysis Registry Group. Patient outcomes from symptomatic intracerebral haemorrhage after stroke thrombolysis. Neurology. 2011;77(4):341–348. [DOI] [PubMed] [Google Scholar]
- 3.Khatri R, McKinney AM, Swenson B, Janardhan V.. Blood-brain barrier, reperfusion injury, and haemorrhagic transformation in acute ischaemic stroke. Neurology. 2012;79:S52–S57. [DOI] [PubMed] [Google Scholar]
- 4.Richard Leigh M, Shyian S, Jen M, et al. on behalf of the STIR and VISTA Imaging Investigators. Pretreatment blood brain barrier damage and post treatment intracranial haemorrhage in patients receiving IV tPA. Bone. 2014;23(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panni P, Gory B, Xie Y, et al. ; on behalf of the ETIS (Endovascular Treatment in Ischaemic Stroke) Investigators. Acute stroke with large ischaemic core treated by thrombectomy: Predictors of good outcome and mortality. Stroke. 2019;50(5):1164–1171. [DOI] [PubMed] [Google Scholar]
- 6.Del Río-Espínola A, Fernández-Cadenas I, Giralt D, et al. ; the GRECOS Investigators. A predictive clinical-genetic model of tissue plasminogen activator response in acute ischaemic stroke. Ann Neurol. 2012;72(5):716–729. [DOI] [PubMed] [Google Scholar]
- 7.Lou M, Safdar A, Mehdiratta M, et al. The HAT Score: A simple grading scale for predicting haemorrhage after thrombolysis. Neurology. 2008;71(18):1417–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed N, Audebert H, Turc G, et al. Consensus statements and recommendations from the ESO-Karolinska Stroke Update Conference, Stockholm 11-13 November 2018. Eur Stroke J. 2019;4(4):307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallolas J, Rodríguez R, Gubern C, Camós S, Serena J, Castellanos M.. A polymorphism in the promoter region of the survivin gene is related to haemorrhagic transformation in patients with acute ischaemic stroke. NeuroMol Med. 2014;16(4):856–861. [DOI] [PubMed] [Google Scholar]
- 10.Malik R, Chauhan G, Traylor M, et al. ; MEGASTROKE Consortium. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina CA, Alvarez-Sabín J, Montaner J, et al. Thrombolysis-related haemorrhagic infarction: A marker of early reperfusion, reduced infarct size, and improved outcome in patients with proximal middle cerebral artery occlusion. Stroke. 2002;33(6):1551–1556. [DOI] [PubMed] [Google Scholar]
- 12.Larrue V, Von Kummer R, Del Zoppo G, Bluhmki E.. Hemorrhagic transformation in acute ischaemic stroke: Potential contributing factors in the European Cooperative Acute Stroke Study. Stroke. 1997;28(5):957–960. [DOI] [PubMed] [Google Scholar]
- 13.Wahlgren N, Ahmed N, Dávalos A, et al. ; SITS-MOST investigators. Thrombolysis with alteplase for acute ischaemic stroke in the Safe Implementation of Thrombolysis in Stroke-Monitoring Study (SITS-MOST): An observational study. Lancet. 2007;369(9558):275–282. [DOI] [PubMed] [Google Scholar]
- 14.Zuvich RL, Armstrong LL, Bielinski SJ, et al. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet Epidemiol. 2011;35(8):887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchini J, Howie B, Myers S, McVean G, Donnelly P.. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. [DOI] [PubMed] [Google Scholar]
- 17.Willer CJ, Li Y, Abecasis GR.. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Leeuw CA, Mooij JM, Heskes T, Posthuma D.. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbeira AN, Pividori MD, Zheng J, Wheeler HE, Nicolae DL, Im HK.. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 2019;15(1):e1007889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Q, Li B, Ou D, et al. A powerful approach to estimating annotation-stratified genetic covariance via GWAS summary statistics. Am J Hum Genet. 2017;101(6):939–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo D, Falcone GJ, Devan WJ, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral haemorrhage. Am J Hum Genet. 2014;94(4):511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Traylor M, Tozer DJ, Croall ID, et al. ; International Stroke Genetics Consortium. Genetic variation in PLEKHG1 is associated with white matter hyperintensities (n = 11,226). Neurology. 2019;92(8):E749–E757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battle A, Brown CD, Engelhardt BE, et al. ; eQTL manuscript working group. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramasamy A, Trabzuni D, Guelfi S, et al. North American Brain Expression Consortium. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del-Aguila JL, Li Z, Dube U, et al. A single-nuclei RNA sequencing study of Mendelian and sporadic AD in the human brain. Alzheimers Res Ther. 2019;11(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. [DOI] [PubMed] [Google Scholar]
- 27.Võsa U, Claringbould A, Westra H-J, et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv. [Preprint] doi:10.1101/447367 [Google Scholar]
- 28.Schofield EC, Carver T, Achuthan P, et al. CHiCP: A web-based tool for the integrative and interactive visualization of promoter capture Hi-C datasets. Bioinformatics. 2016;32(16):2511–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawford KM, Gallego-Fabrega C, Kourkoulis C, et al. ; International Stroke Genetics Consortium. Cerebrovascular disease knowledge portal an open-access data resource to accelerate genomic discoveries in stroke. Stroke. 2018;49(2):470–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: An expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35(22):4851–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SU, Maeda T.. POK/ZBTB proteins: An emerging family of proteins that regulate lymphoid development and function. Immunol Rev. 2012;247(1):107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perissi V, Jepsen K, Glass CK, Rosenfeld MG.. Deconstructing repression: Evolving models of co-repressor action. Nat Rev Genet. 2010;11(2):109–123. [DOI] [PubMed] [Google Scholar]
- 34.Bilic I, Ellmeier W.. The role of BTB domain-containing zinc finger proteins in T cell development and function. Immunol Lett. 2007;108(1):1–9. [DOI] [PubMed] [Google Scholar]
- 35.Satpathy AT, Kc W, Albring JC, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012;209(6):1135–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chevrier S, Corcoran LM.. BTB-ZF transcription factors, a growing family of regulators of early and late B-cell development. Immunol Cell Biol. 2014;92(6):481–488. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Sun HY, Kumar S, Del Mar Puerta M, Jo H, Rezvan A.. ZBTB46 is a shear-sensitive transcription factor inhibiting endothelial cell proliferation via gene expression regulation of cell cycle proteins. Lab Investig. 2019;99(3):305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fararjeh AFS, Liu YN.. ZBTB46, SPDEF, and ETV6: Novel potential biomarkers and therapeutic targets in castration-resistant prostate cancer. Int J Mol Sci. 2019;20(11):2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lill CM, Schjeide BMM, Graetz C, et al. ; International Multiple Sclerosis Genetics Consortium. MANBA, CXCR5, SOX8, RPS6KB1 and ZBTB46 are genetic risk loci for multiple sclerosis. Brain. 2013;136(6):1778–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishizaki SS, Boyle AP.. Mining the unknown: Assigning function to noncoding single nucleotide polymorphisms toward the goal of understanding variation. Trends Genet. 2017;33(1):34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beaulieu AM, Sant'Angelo DB.. The BTB-ZF family of transcription factors: Key Regulators of lineage commitment and effector function development in the immune system. J Immunol. 2011;187(6):2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meredith MM, Liu K, Darrasse-Jeze G, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209(6):1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leppek K, Das R, Barna M.. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19(3):158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou ST, Nilchi L, Li X, et al. Semaphorin3A elevates vascular permeability and contributes to cerebral ischemia-induced brain damage. Sci Rep. 2015;5:7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujita H, Zhang B, Sato K, Tanaka J, Sakanaka M.. Expressions of neuropilin-1, neuropilin-2 and semaphorin 3A mRNA in the rat brain after middle cerebral artery occlusion. Brain Res. 2001;914(1-2):1–14. [DOI] [PubMed] [Google Scholar]
- 46.Burton PR, Clayton DG, Cardon LR, et al. ; Breast Cancer Susceptibility Collaboration (UK). Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39(11):1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sieper J, Deodhar A, Marzo-Ortega H, et al. Secukinumab efficacy in anti-TNF-naive and anti-TNF-experienced subjects with active ankylosing spondylitis: Results from the MEASURE 2 Study. Ann Rheum Dis. 2017;76(3):571–575. [DOI] [PubMed] [Google Scholar]
- 48.Carrera C, Cullell N, Torres-Águila N, et al. ; Spanish Stroke Genetic Consortium. Validation of a clinical-genetics score to predict haemorrhagic transformations after rtPA. Neurology. 2019;93(9):e851–e863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tesi N, van der Lee SJ, Hulsman M, et al. Centenarian controls increase variant effect sizes by an average twofold in an extreme case–extreme control analysis of Alzheimer’s disease. Eur J Hum Genet. 2019;27(2):244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler TW, Day FR, Croteau-Chonka DC, et al. ; Genetic Investigation of Anthropometric Traits (GIANT) Consortium. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9(5):1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howrigan D. Details and considerations of the UK Biobank GWAS. Neale lab blog. 20 September 2017. Accessed 11 February 2021. http://www.nealelab.is/blog/2017/9/11/details-and-considerations-of-the-uk-biobank-gwas
- 52.Lim ET, Würtz P, Havulinna AS, et al. ; for the Sequencing Initiative Suomi (SISu) Project. Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population. PLoS Genet. 2014;10(7):e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crosslin DR, Tromp G, Burt A, et al. Controlling for population structure and genotyping platform bias in the eMERGE multi-institutional biobank linked to electronic health records. Front Genet. 2014;5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Powers WJ, Rabinstein AA, Ackerson T, et al. ; American Heart Association Stroke Council. 2018 guidelines for the early management of patients with acute ischaemic stroke: A guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2018;49(3):e46–e110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any qualified investigator may request the summary statistics and dataset. Detailed methods are available in the Supplementary material.