Abstract
Plant mitochondrial genomes sometimes carry cytoplasmic male sterility (CMS)-associated genes. These genes have been harnessed in various crops to produce high-yielding F1 hybrid seeds. The gene open reading frame 352 (orf352) was reported to be an RT102-type CMS gene in rice (Oryza sativa), although the mechanism underlying its role in CMS is unknown. Here, we employed mitochondrion-targeted transcription activator-like effector nucleases (mitoTALENs) to knockout orf352 from the mitochondrial genome in the CMS rice RT102A. We isolated 18 independent transformation events in RT102A that resulted in genome editing of orf352, including its complete removal from the mitochondrial genome in several plants. Sequence analysis around the mitoTALEN target sites revealed their induced double-strand breaks were repaired via homologous recombination. Near the 5ʹ-target site, repair involved sequences identical to orf284, while repair of the 3ʹ-target site yielded various new sequences that generated chimeric genes consisting of orf352 fragments. Plants with a chimeric mitochondrial gene encoding amino acids 179–352 of ORF352 exhibited the same shrunken pollen grain phenotype as RT102A, whereas plants either lacking orf352 or harboring a chimeric gene encoding amino acids 211–352 of ORF352 exhibited partial rescue of pollen viability and germination, although these plants failed to set seed. These results demonstrated that disruption of orf352 partially restored pollen development, indicating that amino acids 179–210 from ORF352 may contribute to pollen abortion.
Recent development of a method to edit the mitochondrial genome has revealed one of the causative genes of male sterility in rice, a phenotype that increases seed production.
Introduction
Cytoplasmic male sterility (CMS) was first reported in maize (Zea mays; Rhoades, 1931). In rice (Oryza sativa), the first report of a cytoplasmic effect on male reproductive function was reduction of seed fertility in the progeny of the first backcross between wild rice (Oryza rufipogon) and O. sativa (Katsuo and Mizushima, 1958). In agriculture, CMS is harnessed to produce high-yielding F1 hybrid seeds in various crops. Several genes in the mitochondrial genome cause dysfunction of pollen development, resulting in male sterility. These genes are ordinarily named as open reading frame (orf) followed by a unique number indicating the number of amino acids in encoded proteins, and they are associated with CMS. CMS-causative genes in the mitochondrial genome are broadly referred to as CMS-associated genes because a direct causal relationship between orf and CMS is, for the most part, technically out of reach at present. CMS-associated genes have been reported in many plants (Hanson and Bentolila, 2004). Examples of such genes in rice include orf79 in BT-type CMS (Iwabuchi et al., 1993; Akagi et al., 1994), orfH79 in HL-type CMS (Yi, 2002), orf352 in WA-type CMS (Bentolila and Stefanov, 2012; Luo et al., 2013), and CW-orf307 in CW-type CMS (Fujii et al., 2010). Similarly, in Brassicaceae, cybridization between rapeseed (Brassica napus) and radish (Raphanus sativus) led to the identification of orf125 in Kosena CMS (Iwabuchi et al., 1999), and orf138 in Ogura CMS (Grelon et al., 1994).
We recently employed transcription activator-like effector nucleases (TALENs) to delete the mitochondrial CMS-associated gene orf79 in BT-type CMS rice (Kazama et al., 2019). TALENs, which make genome editing possible, usually comprise a left TALEN and a right TALEN, each containing a designer DNA-binding domain and a nuclease domain. TALENs typically function as obligate heterodimers and are often used to induce double-strand breaks (DSBs) at or near the region recognized by the DNA-binding domain. These are then repaired by imperfect nonhomologous end-joining repair in the nuclear genome, introducing small deletions or insertions in the process. We designed a mitoTALEN vector to add a mitochondrial targeting signal to a TALEN. The mitoTALEN construct is integrated into the nuclear genome via Agrobacterium (Agrobacterium tumefaciens)-mediated transformation, thereby allowing the mitochondrial genome to be edited without the need for direct transformation of the organellar genome. Using this method, we successfully deleted orf79 in BT-type CMS rice and restored fertility, illustrating the power of mitoTALENs as tools to reveal the role of a CMS-associated gene.
Traditionally, the presence of a CMS-causative gene is revealed by outcrossing a line to separate the mitochondrial CMS locus and the nuclear restorer of fertility (Rf) gene that normally suppresses the effects of the CMS-causative gene. In this study, we used RT102-type CMS rice, which is derived from the cytoplasm of the wild rice O. rufipogon Griff., accession W1125 (Motomura et al., 2003). The CMS line RT102A carries the cytoplasm of RT102 and the nucleus of the fertile O. sativa japonica cultivar, Taichung 65 (T65). Pollen grains in RT102A are sterile, with two different morphologies: ∼83% are shrunken and cannot be stained by Lugol’s iodine, while the remaining 17% are spherical and stain strongly (Okazaki et al., 2013). The shrunken pollen phenotype seen in RT102A is like that of WA-CMS, in which microspores abort at the early uninucleate microspore stage, right after meiosis (Li et al., 2007). We previously sequenced and assembled the RT102-type mitochondrial genome into a single circular molecule of 502,250 bp and identified the new chimeric gene, orf352, as a CMS-associated gene (Okazaki et al., 2013). The protein encoded by orf352 was almost identical to WA352, a CMS-associated protein in WA-type CMS (Bentolila and Stefanov, 2012; Luo et al., 2013; Tang et al., 2017), with only 5 nt differences resulting in four amino acid substitutions. The orf352 gene appeared to comprise a fragment identical to a part of orf284, and a fragment with similarity to orf288, both of which encode hypothetical proteins. Previously, orf284 and orf288 have been reported to be involved in the generation of CMS-associated genes (Tang et al., 2017). Functional evidence for the role of orf352/WA352 in RT102-/WA-type CMS as a CMS-causative gene has not yet been described.
Here, we used a loss-of-function analysis approach to assess the contribution of the mitochondrial chimeric gene orf352 to CMS in the CMS line RT102A. We generated two mitoTALEN vectors to target orf352 and introduced them into the CMS line RT102A. We describe the characterization of the genotypes and phenotypes associated with genome editing of orf352 via mitoTALENs and suggest that disruption of orf352 partially restores pollen development.
Results
Nuclear transformation of RT102-type CMS rice with TALENs targeting mitochondrial orf352
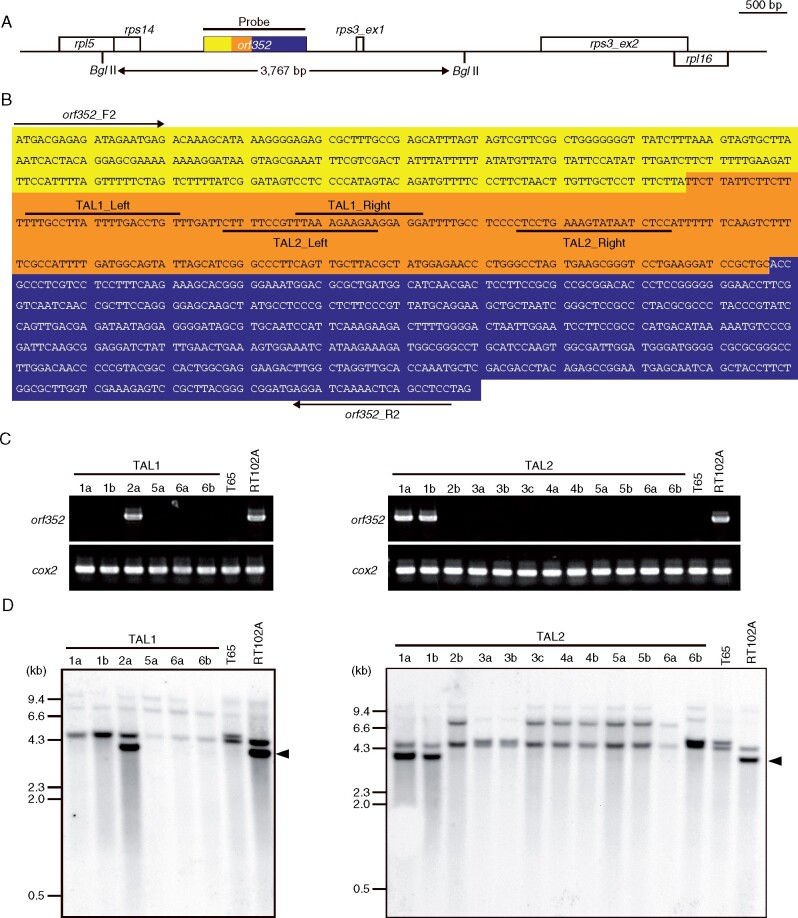
Mitochondrial orf352 was previously reported to be a CMS-associated gene in the RT102-type CMS rice, RT102A (Okazaki et al., 2013). To determine what contribution, if any, orf352 might make to the male sterile phenotype displayed in RT102A, we searched for a sequence unique to orf352 before designing TALENs. The orf352 coding region comprises 1,059 nt and is divided into three fragments based on their homology to other mitochondrial genes. The first orf352 fragment is 297 nt in length and is identical to orf284, while the last 562 nt of orf352 (from 498 to 1,059 nt, where the first adenine from the translation start codon is counted as nt 1 has 97% sequence identity with orf288 (Figure 1, A and B). The middle orf352 fragment (298–497 nt) is specific to orf352. We therefore selected this region as a target sequence for mitoTALENs (Supplemental Figure S1A). We generated two mitoTALEN vectors, pTAL1 and pTAL2, each harboring the left and right part of the TALEN. Each mitoTALEN vector was introduced into RT102A rice calli via Agrobacterium (A. tumefaciens)-mediated transformation. We confirmed stable insertion of the transgene into the plant genome by PCR amplification of an introduced hygromycin resistance gene (HPT) present on the T-DNA (Supplemental Figure S1B). Six primary (T0) transgenic plants harboring pTAL1, and 12 harboring pTAL2, were generated.
Figure 1.
Introduction of the mitoTALENs TAL1 and TAL2, targeting orf352, into the RT102-type CMS rice (RT102A). A, Schematic illustration of the target gene orf352 and the neighboring region in the RT102-type mitochondrial genome. Yellow, region identical to orf284; navy blue, region homologous to orf288; orf352-specific sequences are shown in orange. rpl5, ribosomal protein L5; ψrps14, pseudo ribosomal protein S14; rps3_ex1, exon 1 of ribosomal protein S3; rps3_ex2, exon 2 of rps3; ψrpl16, pseudo ribosomal protein L16. B, Nucleotide sequences of the orf352 coding region. Background colors are as in (A). Primers for PCR analysis (orf352_F2, orf352_R2) are shown, as well as each TAL binding site. C, Genotyping of transgenic plants by PCR over the orf352 region. T65 is a fertile japonica rice cultivar that lacks orf352 and served as a negative control. Cytochrome c oxidase subunit 2 (cox2) is a positive control for amplification. The same images of orf352 are used in Supplemental Figure S3. D, Southern blot analysis of transgenic plants with TAL1 and TAL2-specific probes. Arrowheads indicate signals corresponding to a 3.8-kb orf352 fragment. The data in (C and D) are representative of three independent experiments.
The introduction of mitoTALEN targeting orf79 has been reported to be accompanied by the disappearance of orf79 from the mitochondrial genome of transformants (Kazama et al., 2019). To determine whether orf352 might be similarly deleted by mitoTALENs, we tested genomic DNA from transformants for the presence of orf352 by PCR and Southern blot analysis (Figure 1, C and D). The PCR primers orf352_F2 and orf352_R2 should amplify the full-length orf352 coding region from RT102A genomic DNA, as well as from any transformants retaining the entire orf352 sequence. However, with the exception of TAL1-2a, we failed to amplify orf352 from most plants transformed with pTAL1. Likewise, except for plants TAL2-1a and TAL2-1b, most plants carrying the T-DNA derived from pTAL2 were negative for orf352, as tested by PCR amplification (Figure 1C). We amplified full-length orf352 from RT102A genomic DNA; since RT102A and the transformants TAL1-2a, TAL2-1a, and TAL2-1b plants shared the same band pattern for the PCR products after electrophoresis on agarose gel, we hypothesized that the orf352 PCR-positive plants might not carry any alteration at orf352. To investigate this possibility, we sequenced the PCR products obtained above and compared their sequences with the RT102A mitochondrial genome. The sequences were identical, confirming the absence of rearrangements around the mitoTALEN target sites. We also verified the presence or absence of the orf352 by Southern blot analysis using total genomic DNA extracted from leaf blades and a probe spanning the orf352 region, as indicated in Figure 1A. We detected a 3.8-kb band in RT102A, as expected for the intact orf352 genomic fragment. However, we did not detect a band of the same size or intensity in the fertile japonica cultivar, Taichung 65 (T65), which does not carry orf352 in its mitochondrial genome. Five of the TAL1 transgenic plants (TAL1-1a, 1b, 5a, 6a, and 6b) and 10 of the TAL2 transgenic plants (TAL2-2b, 3a, 3b, 3c, 4a, 4b, 5a, 5b, 6b, and 6c) similarly showed no signal specific for orf352 (Figure 1D). However, the three orf352-positive transgenic plants identified by PCR (TAL1-2a, TAL2-1a, and TAL2-1b) had a strong hybridization signal at the same size as that of RT102A. These results indicate that orf352 is intact in the mitochondrial genome in the transgenic plants.
Aberrant homologous recombination events in mitochondrial genomes were edited at orf352
Previous analysis of mitoTALEN-mediated genome editing of the mitochondrial gene orf79 revealed that the DSB introduced by mitoTALENs is repaired by two independent homologous recombination events at recombination sites A and B on either side of the target site (Kazama et al., 2019; Supplemental Figure S2). Indeed, the DSB is repaired via recombination between site A (or B) and site Aʹ (or Bʹ), which refers to a region anywhere in the mitochondrial genome with high sequence similarity to (site A or B; Supplemental Figure S2). During DSB repair, the region between recombination sites A and B, including the target site itself, is deleted from the mitochondrial genome. The homologous recombination occurs nonreciprocally and is accompanied by replication, so finally, DNA molecules containing the site A/Aʹ and B/Bʹ will be duplicated (Supplemental Figure S2).
To determine the extent of the deletion around orf352, we designed specific primer pairs to PCR-amplify eight fragments mapping up to 1.4-kb upstream and up to 3.8-kb downstream of orf352 (Supplemental Figure S3A). We successfully amplified the four amplicons upstream of orf352 (regions 1–4), as well as the two downstream amplicons (regions 7 and 8), in all transgenic plants. We could not amplify just downstream of orf352 (region 6) in transgenic plants TAL1-1a, TAL1-1b, and TAL1-5a. This indicated that these three transformants carry larger deletions around orf352 than any of the other transgenic plants (Supplemental Figure S3B). To rule out the possibility that DSBs have been repaired by end-joining rather than homologous recombination, we attempted to amplify across the entire region around orf352 (labeled as region 9 in Supplemental Figure S3A) with the forward primer from region 3 and the reverse primer from region 7. End-joining repair would be expected to cause a deletion or an insertion, which would be apparent on an agarose gel when compared with the product from RT102A. However, we failed to obtain a PCR amplicon from orf352-edited plants, even though all orf352-positive plants exhibited an amplicon of the same size as RT102A (Supplemental Figure S3B). This result indicates that the DSBs were not repaired by end-joining.
We have noticed previously that mitoTALEN-induced DSBs are repaired via homologous recombination at an arbitrary recombination site at an arbitrary position from the mitoTALEN target site (Kazama et al., 2019). To better understand the recombination events that occurred in each transformant, we turned to fusion primer and nested integrated PCR (FPNI-PCR), which is a modified method of thermal asymmetric interlaced (TAIL)-PCR (Wang et al., 2011). We then sequenced the FPNI-PCR amplicons using primers flanking the recombination sites and compared the resulting nucleotide sequences to the RT102-type mitochondrial genome using BLAST to identify the template used during repair (Figure 2). Our query sequences sometimes showed high sequence similarity to two regions in the single master circle of the RT102-type mitochondrial genome, likely because of the large duplication the master circle carries. We identified six distinct types of recombination on the 3ʹ-mitoTALEN-target site (Figure 2A; Supplemental Figure S4). In Type 1 (TAL1-1a), Type 2 (TAL1-1b), and Type 3 (TAL1-5a) recombination events, the entire orf352 coding region was lost. Based on BLAST results, the DSB from the Type 1 recombination event was repaired via homologous recombination between a 17-bp region downstream of orf352 (267,450 nt to 267,466 nt in the RT102-type mitochondrial genome, GenBank AP012528), and a distant 17-bp region (221,704–221,719 nt; Figure 2B). In the Type 2 recombination event (TAL1-1b), the DSB was repaired between a 17-bp region (267,510–267,526 nt) downstream of orf352, and one of the two possible distant 17-bp regions (110,198–110,214 nt and 441,826–441,842 nt). Homologous recombination in Type 3 plants occurred at a 100-bp region downstream of orf352 (267,776–267,845 nt) and a distant 96-bp region (209,174–209,269 nt) as template for repair. In plants with Type 4 (TAL1-6a and Type1-6b), Type 5 (TAL2-2b, TAL2-3c, TAL2-4a, TAL2-4b, TAL2-5a, TAL2-5b, and TAL2-6b), and Type 6 (TAL2-3a, TAL2-3b, and TAL2-6c) repair, the recombination sites were located within orf352 (268,339–268,354 nt) and a distant 16-bp region (206,014–206,029 nt). The recombination sites for Types 5 and 6 repair are identical to each other, but Type 5 repair appears to include an additional recombination event 95-bp upstream of the common recombination site for Types 5 and 6 repair (Supplemental Figure S4).
Figure 2.
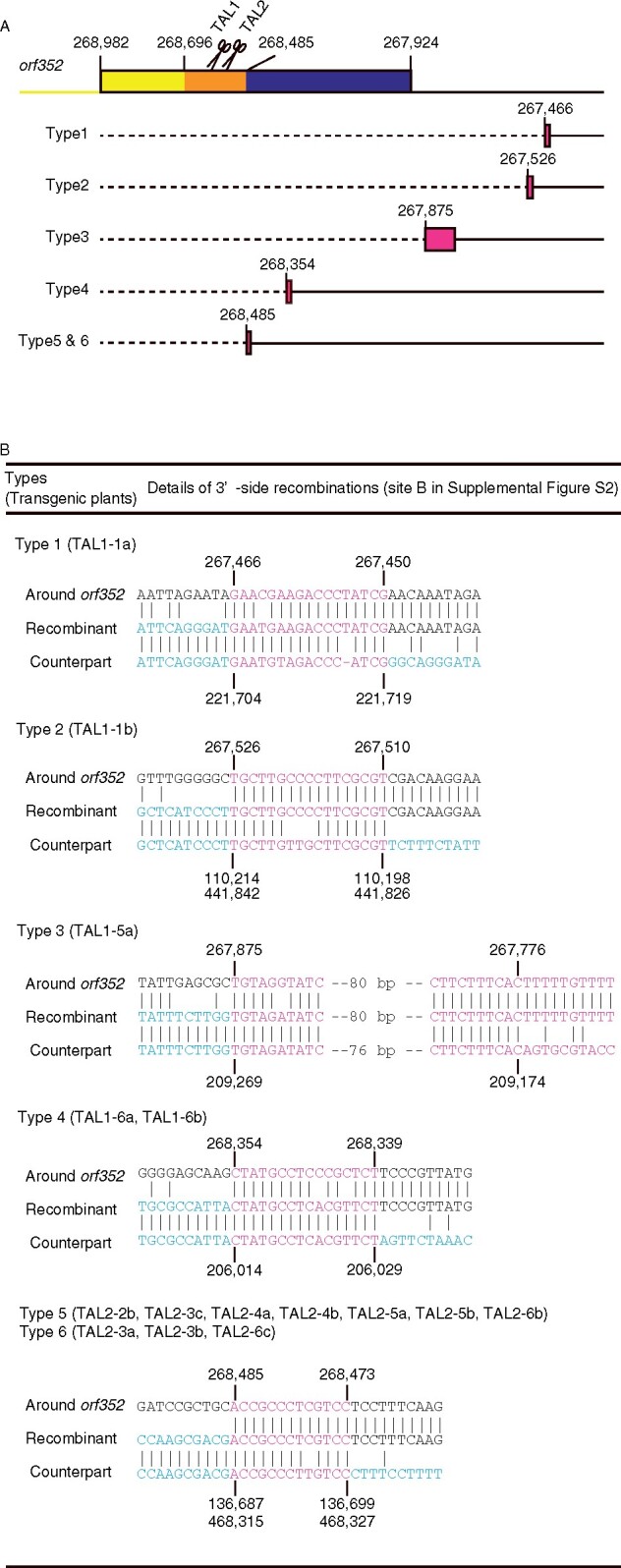
DSBs induced by mitoTALENs are repaired via homologous recombination. A, Schematic diagram of the various recombination types observed at site B (the principle behind recombination is illustrated in Supplemental Figure S2). Top, genetic structure around the orf352 coding region. Yellow, region identical to orf284; navy blue, region homologous to orf288; orange, orf352-specific sequences. Scissors indicate mitoTALENs (TAL1 [left] and TAL2 [right]). Bottom, position (in bp) and structure of the recombination event at site B in each type of transformant. Magenta filled boxes indicate the recombination sites B. The dashed lines indicate the deleted regions. B, Sequence comparisons between the RT102A mitochondrial genome and each recombinant type. Sequence labeled “Around orf352” are sequences near the mitoTALEN targets. “Recombinant” sequences are sequences that arose via homologous recombination. “Counterpart” sequences are sequences distant from the target and near the recombination sites B′. The magenta sequences in (B) correspond to the magenta boxes in (A). The blue sequences are sequences near the recombination sites B′.
Although repair on the 3ʹ-side of the mitoTALEN target site varied across all transgenic plants, recombination at the 5ʹ-site (recombination site A, as illustrated in Supplemental Figure S2) made use of the same site in all orf352-edited transgenic plants (Figure 3; Supplemental Figure S5). Results from BLAST searched against the RT102-type mitochondrial genome indicated that repair took place between the first half of orf352 (268,696–269,254 nt, with homology to orf284 and its promoter region) and a region 5ʹ of orf284 in the mitochondrial genome (115,639–116,493 nt, and 447,267–448,121 nt; Figure 3; Supplemental Figure S5). The identical sequence of 559 bp is illustrated in Supplemental Figure S2 as the recombination sites A and Aʹ. Repair via homologous recombination at this site resulted in the duplication of orf284, with each orf284 having a distinct upstream sequence, which we confirmed by PCR, in all orf352-edited plants. We also independently confirmed the duplication by Southern blot analysis, which showed two bands when using orf284 (amplified with P4 and P5) as a probe (Figure 3). Like the orf79 in BT-type CMS, DSBs in orf352 induced by mitoTALENs were repaired via homologous recombination, and we successfully obtained orf352-edited plants.
Figure 3.
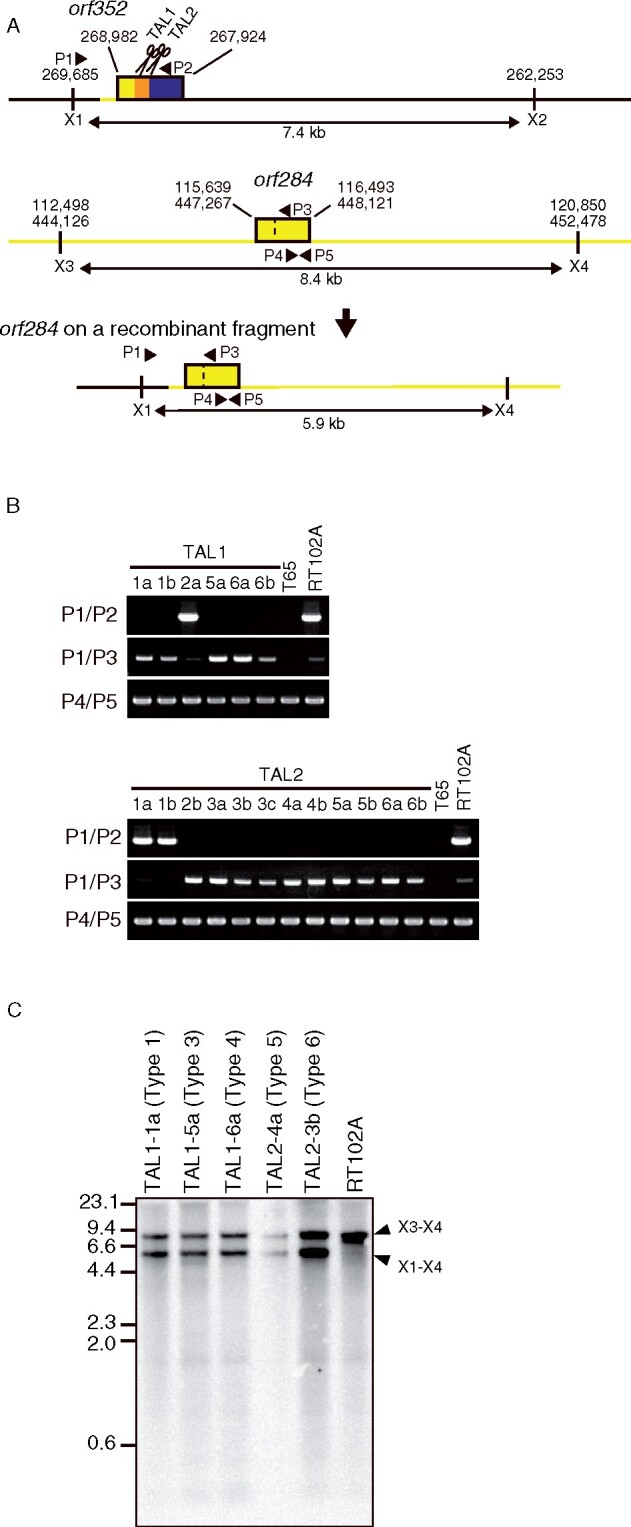
DSBs induced by mitoTALENs are repaired by identical homologous recombination in all orf352-edited plants. A, Schematic diagram of DSB repair at the 5ʹ-side of the target site. Top, genomic structure around the orf352. Yellow, region identical to orf284; navy blue, region homologous to orf288; orange, orf352-specific sequences. Scissors indicate mitoTALENs (TAL1 and TAL2). Middle, genomic structure around orf284. Bottom, genomic structure of new recombinants. Black line, region originating around orf352. P1–P5, primer positions for genotyping PCR; X1–X4, positions of XhoI restriction sites. B, PCR analysis of new recombinants. T65 is a fertile japonica rice that lacks orf352, which serves as a negative control. RT102A serves as a positive control for the presence of orf352. C, Duplication of orf284 during repair via homologous recombination, as detected by Southern blot analysis hybridizing on genomic DNA digested with XhoI. The probe was synthesized using primers P4 and P5. Arrowheads indicate signals corresponding to 8.4-kb orf284 fragment (X3-X4) and 5.9-kb recombinant fragment (X1–X4). The data in (B and C) are representative of three independent experiments.
Partial restoration of pollen development, but not seed set, via genome editing of orf352
Genomic editing of orf79 has been shown to restore fertility of BT-type CMS rice (Kazama et al., 2019). To test the effects of orf352 editing on fertility, we calculated the seed setting rates for all transgenic plants (Table 1). The seed setting rate for the fertile cultivar T65 was close to 90%, whereas the CMS line RT102A was completely sterile, as expected (Okazaki et al., 2013). Contrary to our expectations, all orf352-edited plants were also completely sterile (Table 1), indicating that orf352 is unlikely to be the cause of RT102-type CMS. We expanded our analysis of the consequences of orf352 genome editing to pollen viability and pollen tube elongation. Most pollen grains from cultivar T65 were stained by Lugol’s iodide solution, demonstrating their viability, whereas most RT102A pollen grains were shrunken, and few were stained by Lugol’s (Okazaki et al., 2013; Figure 4). The few spherical pollen grains in RT102A appeared normal but did not germinate when placed on stigmas. In contrast, pollen grains from cultivar T65 germinated on stigmas and their tubes elongated a clear sign of viable and fertile pollen (Figure 4).
Table 1.
Seed setting rate for orf352-edited plants
| Line | No. of spikelets/panicles | No. of filled grains | Seed setting rate (%) | orf352 |
|---|---|---|---|---|
| T65 | 355/6 | 308 | 87.0 ± 2.9 | Absent |
| RT102A | 154/3 | 0 | 0.0 ± 0.0 | Present |
| TAL1-1a (Type 1) | 226/10 | 0 | 0.0 ± 0.0 | Deleted |
| TAL1-1b (Type 2) | 154/3 | 0 | 0.0 ± 0.0 | Deleted |
| TAL1-5a (Type 3) | 291/3 | 0 | 0.0 ± 0.0 | Deleted |
| TAL1-6a (Type 4) | 542/10 | 0 | 0.0 ± 0.0 | Disrupted |
| TAL2-4a (Type 5) | 112/2 | 0 | 0.0 ± 0.0 | Disrupted |
| TAL2-3b (Type 6) | 330/8 | 0 | 0.0 ± 0.0 | Disrupted |
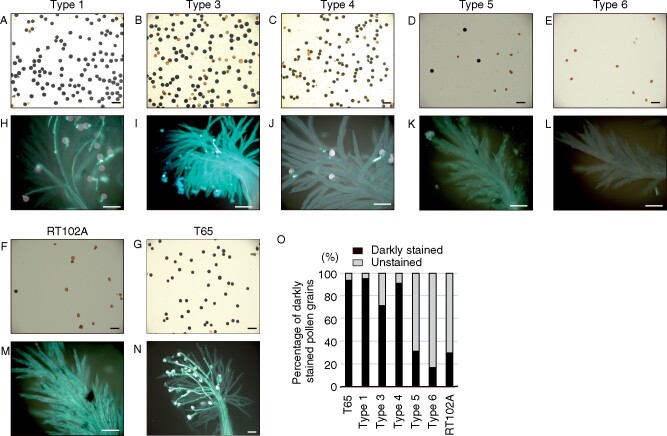
Figure 4.
Disruption of orf352 partially restores pollen development but does not restore pollen fertility. A–G, Pollen grain viability in the various recombinant types, as determined by staining with Lugol’s iodine (1% I2-KI). H–N, Pollen germination and pollen tube elongation on stigmas, observed by staining pollen tubes with aniline blue. O, Percentage of darkly stained pollen grains (viable) in each recombinant type. Scale bars = 200 μm (A–G) and 100 μm (H–N). The data are representative of six independent experiments.
Having established that the control cultivars displayed their expected fertility phenotypes, we next checked the pollen phenotypes for all but one orf352-edited plant (one Type 2 plant became blighted before anthesis). In Type 1 (TAL1-1a), Type 3 (TAL1-5a), and Type 4 (TAL1-6a) plants, the percentage of darkly stained pollen grains was similar to that seen in the fertile cultivar T65 (Figure 4, A–C, and G), with values of 95%, 71%, and 91%, respectively (Figure 4O). In contrast, Types 5 and 6 plants showed a severe reduction in pollen viability (Figure 4, D and E), with percentages of dark pollen grains of 31% and 17%, respectively—lower than or similar to the 30% viable pollen seen in RT102A (Figure 4, F and O). In all transgenic plants, far fewer pollen grains germinated on stigmas relative to pollen grains from T65 (Figure 4N). Although some pollen tubes did start to elongate, they quickly stopped progressing along the stigma, either just after germination or after reaching about halfway down the stigma (Figure 4, H–J). Almost all pollen grains from Types 5 to Type 6 plants were empty and shrunken, similar to RT012A pollen, and none germinated as predicted (Figure 4, K–M). To qualify the pollen phenotypes of orf352-edited plants, we counted the number of darkly stained pollen grains. Over 60% of pollen grains from Types 1, 3, and 4 plants were stained, indicative of starch reserves (Figure 4O). In these transgenic plants, pollen viability therefore partially recovered relative to that observed in Type 5 or Type 6 plants, or in the CMS line RT102A. Types 5 and 6 plants, however, showed no restoration of pollen viability. These data indicate that orf352 is involved in inhibition of pollen development, and other unidentified gene(s) are involved in inducing male sterility in RT102A.
Prediction of genes near recombination sites
Targeting orf352 with mitoTALENs induced the formation of DSBs, which were repaired via homologous recombination between homologous sequences near the target and elsewhere in the mitochondrial genome (Figures 2 and 3; Supplemental Figure S2). New genes may thus emerge at the recombination sites. To explore this possibility, we PCR-amplified the recombined DNA fragments containing either recombination site A/Aʹ or B/Bʹ (Supplemental Figure S2) and determined their sequences. A new gene was defined as the longest gene not previously present in the mitochondrial genome. At the 5′-recombination site (indicated as A/A′ in Supplemental Figure S2), all orf352-edited plants shared the same duplicated gene, orf284. We predicted several new genes at the 3′-recombination sites, which we previously showed to be associated with homologous recombination events (Supplemental Figure S4; Table 2). One new gene was observed in each of Types 1 and 2 recombination repair events, encoding 77 and 47 amino acids, respectively, which we named as orf77 (Type 1) and orf47 (Type 2). Both are completely new orf with no similarity to other genes or known genes; however, the mitochondrial genome of Type 3 plants harbored a new gene encoding a protein of 108 amino acids, which included the entire ATP synthase subunit 9 protein (ATP9). Recombination in Type 4 plants created an orf encoding a 142-aa protein, comprising two new amino acids added to the N terminus of a fragment of ORF352 (from 215 to 352 aa). We named this gene as orf142. Recombination events in Types 5 and 6 plants created the same gene, orf174, encoding a protein comprising aa 179–352 of ORF352. Additional recombination upstream of orf174 occurred in Type 5, so Types 5 and 6 are distinguished from each other (Supplemental Figure S4). While pollen viability was not restored in Type 5 or 6 plants, it occurred in other transgenic plants, raising the possibility that a fragment comprising aa 179–210 of ORF352 is responsible for pollen development inhibition.
Table 2.
Chimeric loci resulting from recombination at the 3ʹ-site
| Recombination type | New gene | Notes |
|---|---|---|
| Type 1 | orf77 | NA |
| Type 2 | orf47 | NA |
| Type 3 | orf108 | Contains full-length atp9 ORF |
| Type 4 | orf142 | Contains partial sequence of orf352 (orf288) |
| Type 5 | orf174 | Contains partial sequence of orf352 (orf288) |
| Type 6 | orf174 | Contains partial sequence of orf352 (orf288) with a different promoter than that seen for Type 5 |
Discussion
Hybrid vigor confers improved performance and increased yield to F1 hybrid plants over either parent. Global seed markets for most vegetables and major crops such as maize (Zea mays), sorghum (Sorghum bicolor), and rice, are thus dominated by F1 hybrid seeds (Hochholdinger and Baldauf, 2018). To avoid self-pollination that would lead to the production of nonhybrid seed, male-sterile plants are routinely employed as the female parent in crosses; thus, CMS has considerable agronomic importance. In rice, wild abortive (WA)-type CMS is mainly used to produce hybrid seed of indica varieties (Huang et al., 2014), and at the flowering stage, it is associated with shrunken pollen grains that cannot be stained with Lugol’s iodine solution. The fertility of this CMS can be restored by the nucleus-encoded fertility restorer genes, Rf3 and Rf4. Sequencing the mitochondrial genome in WA-type CMS plants identified orf352 as a putative CMS-causative gene named as WA352. The WA352 protein has been reported to accumulate preferentially in the anther tapetum and to interact with the cytochrome c oxidase assembly protein 11 (COX11) subunit of respiration complex IV, resulting in ROS production and programmed cell death, which eventually leads to microspore abortion at the early microspore stage (Luo et al., 2013). The Rf proteins RF4 and RF3 are thought to suppress the accumulation of WA352 at the post-transcriptional and post-translational level, respectively. The cloning of Rf4 revealed that RF4 is a pentatricopeptide repeat-containing protein (Kazama and Toriyama, 2014; Tang et al., 2014). The cloning of Rf3 has not been reported.
In this study, we used an RT102-type CMS derived from wild rice (O. rufipogon; Motomura et al., 2003). The mitochondrial genome of the RT102-type CMS (RT102A) harbors orf352, a sequence variant of WA352 whose encoded protein differs from WA352 by four amino acid substitutions. The surrounding genomic structure at orf352 in RT102A is also different from that of WA-type CMS. In WA-type CMS, the mitochondrial genome downstream of WA352 contains exons 4 and 5 of nad5, encoding NADH dehydrogenase, whereas the corresponding sequence in RT102A originates from exons 1 and 2 of rps3, encoding ribosome small subunit 3. The shrunken pollen phenotype we observed in RT102-type CMS is similar to that of the WA-type CMS, although some spherical pollen grains are evident in RT102A (Okazaki et al., 2013). These signs of inhibition of pollen development are likely to be associated with microspore abortion at the early microspore stage, possibly via an identical mechanism in the RT102-type CMS and WA-type CMS. Genomic manipulation of orf352 would validate its role as a CMS-causative gene, but the direct transformation of the rice mitochondrial genome has not been reported. As an alternative technique, we recently developed mitoTALENs; these are genome editors targeted to mitochondria, but expressed from a transgene in the nuclear genome. We used mitoTALENs to edit the mitochondrial gene orf79 to demonstrate that disruption of orf79 restores fertility of BT-type CMS rice (Kazama et al., 2019). Here, we tested the contribution of orf352 to RT102-type CMS using two mitoTALEN constructs targeting orf352, culminating in the isolation of orf352-edited plants (Figure 1, B and C).
We previously showed that DSBs introduced by mitoTALENs around orf79 in the mitochondrial genome are repaired via ectopic homologous recombination (Kazama et al., 2019). Next, using FPNI-PCR, we determined that DSBs introduced by orf352-targeted mitoTALENs were repaired similarly. FPNI-PCR also allowed us to characterize the recombination sites in detail (Figure 2; Supplemental Figure S5). In all transformants, DSBs at the 5ʹ-site involved the same homologous recombination event at the orf352 sequence, which is identical to orf284 and orf284 itself. Repair of DSBs at the 3ʹ-site was more variable and used various sequences of variable length, from 13 to 100 bp. Interestingly, the length of the 5ʹ-recombination repair tract was the greatest, at 559 bp, and was common to all transformants. This observation suggests that the longer and highly similar stretches of sequences make for efficient templates for repair via homologous recombination. We classified all transformants into six types based on recombination at the 3ʹ-site. Recombination resulted in complete loss of orf352 in Types 1, 2, and 3 plants, but only partial loss, with retention of the portion of orf352 that is homologous to orf288, in Types 4, 5, and 6 plants. Genome editing of the orf352 partially rescued the pollen defects observed in RT102A, as evidenced by spherical pollen grains rich in starch seen in Types 1, 3, and 4 plants that can germinate on stigmas. Seed set, however, was not restored. We hypothesized that early microspore development is affected by orf352, while pollen and pollen tube growth may be influenced by new genes such as orf284, and which are now duplicated in the mitochondrial genomes of our orf352-edited plants, or by other unidentified gene(s) in the RT102-type mitochondrial genome.
The partial restoration of pollen viability and development in Types 1 and 3 plants, as demonstrated by the accumulation of mostly spherical and darkly stained pollen grains with some germination potential, may be associated with the loss of orf352. Supporting this hypothesis, Types 5 and 6 plants showed no recovery of pollen development and retained a large segment of the orf352 sequence (encoding aa 179–352 of ORF352, which was homologous to orf288) to yield the new mitochondrial gene orf174. This new gene may be expressed in the anther within spikelets at the meiotic stage. The mitochondrial genome of Type 4 plants harbored the new gene orf142, which encodes a protein comprising aa 211–352 of ORF352. Because these plants showed partial restoration of pollen development, unlike Types 5 and 6 plants, we propose that the 32 amino acids from ORF352, which are included in ORF174 but absent from ORF142 (Figure 5), might be crucial for inhibition of pollen development, resulting in the production of shrunken and unstained pollen grains. A previous study of the function of WA352 in WA-type CMS reported that WA352 contains two COX11-interacting domains, from 218 to 292 aa and from 294 to 352 aa; in addition, interaction between COX11 and WA352 was required to establish CMS (Luo et al., 2013). The region spanning aa 169–199, named the cs2 region, was also proposed to be crucial in regulating the interaction between WA352 and COX11 (Tang et al., 2017). This region overlaps with the 32-aa segment we identified here (Figure 5). The deletion of this region in ORF142 (encoded by the mitochondrial genome of Type 4 plants) might therefore prevent the interaction between ORF142 and COX11, leading to partial recovery of pollen development, although ORF142 is thought to still contain one COX11-interacting region. Further investigation of the function of these truncated ORF352 variants in the context of their interaction with COX11 is required. This report suggests that multiple genes in the mitochondrial genome may be involved in the induction of male sterility in rice. Our study also provides insights into the function of CMS-causative genes and illustrates the usefulness of mitoTALENs for functional research of plant mitochondrial genes.
Figure 5.
Alignment of ORF352, ORF174, and ORF142 protein sequences identified a 32-amino acid stretch that is critical to inhibition of pollen development. Brown, CS3 domain; blue, CS2 domain; orange, COX11-interaction region (Tang et al., 2017); magenta, stretch of 32 amino acids critical for ORF352 function. Boxes indicate predicted transmembrane domains.
Materials and methods
Plant materials
The RT102-type CMS line (RT102A) was developed via multiple backcrosses between O. rufipogon Griff. accession W1125 (National Institute of Genetics, Mishima, Japan) and O. sativa L. cv Taichung 65, as previously described (Okazaki et al., 2013). This RT102-type CMS comprises the nucleus of O. sativa L. cv Taichung 65 and the cytoplasm of O. rufipogon Griff., accession W1125.
Construction of mitoTALEN vectors
MitoTALEN binary vectors used here were constructed as described previously (Kazama et al., 2019). Briefly, DNA recognition motifs for the selected target sequences were assembled via the Golden Gate cloning, using methods with the Platinum Gate TALEN kit (Sakuma et al., 2013), into the multisite-entry vectors for the right or the left TALEN ORFs. ORFs in the entry vectors were then recombined by multi-LR reactions into Gateway destination vectors harboring the cauliflower mosaic virus 35S promoter and the mitochondrial localization signal (MLS) from Arabidopsis ATPase deltaʹ subunit (encoded by At5g47030). The destination vector is based on the binary vector pH7WG (Karimi et al., 2002). An additional Gateway Entry vector, containing a transcription termination sequence from Arabidopsis heat-shock protein (HSP18.2; encoded by At5g59720) and one copy of the 35S promoter followed by one copy of the MLS, was recombined into the destination vector between the right and left region of the TALEN ORFs to provide the transcript terminator for the TALEN right transgene, and the promoter and MLS for TALEN left (Supplemental Figure S1).
Generation of transgenic plants with mitoTALEN vectors
Plants were planted in plastic pots filled with rice culture soil and grown at 30°C/25°C (10-h d/14-h night) in a biotron (Nippon Medical & Chemical Instruments Co. Ltd, Osaka, Japan). Transformation methods were described previously (Kazama et al., 2008). Primary (T0) transformants were confirmed by PCR using a primer pair that amplifing a fragment of Hygromycin Phosphotransferase (HPT; Supplemental Table S1). HPT-positive T0 plants were transplanted to soil and transferred to a biotron. We obtained six transgenic plants with the TAL1 construct (1a, 1b, 2a, 5a, 6a, and 6b) and 12 with the TAL2 construct (1a, 1b, 2b, 3a, 3b, 3c, 4a, 4b, 5a, 5b, 6b, and 6c). The number of each plant reflects the petri plate from which the original T0 seedlings were selected. Plants with the same number but a different letter originated from the same petri plate. When T0 plants started to set seeds, heading panicles were bagged to avoid cross-pollination. At maturity, filled and unfilled spikelets were counted and seed setting rate was calculated. Spikelets were harvested 1 d before anthesis to stain pollen grains with a 1% (w/v) I2-KI Lugol solution to determine pollen viability. Pollen germination was observed on stigmas via aniline blue staining, as previously described (Fujii and Toriyama, 2005).
Isolation of genomic DNA and Southern blot analysis
Total DNA was extracted from green leaf blades using the DNeasy Plant Mini Kit (Qiagen). The structure of the mitochondrial genome around the mitoTALEN target sequences was determined by PCR using the primers listed in Supplemental Table S1, followed by sequencing to define the exact recombination site. Briefly, FPNI-PCR was performed with the primers listed in Supplemental Table S1. The resulting PCR amplicons were purified after gel electrophoresis and sequenced. Recombination sites in the mitochondrial genome obtained by this method were confirmed by PCR and by Southern blot analysis, as described previously (Kazama et al., 2016).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers_LC631941 (orf77), LC631942 (orf47), LC631943 (orf108), LC631944 (orf142), and LC631945 (orf174).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Introduction and confirmation of the orf352-targeting mitoTALEN vectors, pTAL1 and pTAL2.
Supplemental Figure S2. Repair principle of the double-strand break introduced by mitoTALENs in the mitochondrial genome.
Supplemental Figure S3. Detection of the deleted region around the orf352 after homologous recombination by PCR.
Supplemental Figure S4. Schematic diagram of double-strand break repair in each type of transgenic plants.
Supplemental Figure S5. Double-strand breaks induced by mitoTALENs are repaired by the identical homologous recombination at the 5ʹ-side of the target site.
Supplemental Table S1. Primer list used in this study.
Supplementary Material
Acknowledgments
We thank M. Ito (Tohoku University) for technical assistance with TAL transformation.
Funding
This research was partly supported by grants from the Japanese Science and Technology Agency (PRESTO to S.A.) and the Japan Society for the Promotion of Science (grant numbers JP16H06182, JP18H02172, JP20H05680, and JP21H02169 to T.K., and JP19H02927 to S.A.).
Conflict of interest statement. The authors declare no potential competing interests.
T.K., S.A., and K.T. initiated and designed the project. S.A. constructed the vectors. S.O. and T.K. performed experiments. S.O., S.A., K.T., and T.K. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Tomohiko Kazama (tomo-kazama@agr.kyushu-u.ac.jp).
References
- Akagi H, Sakamoto M, Shinjyo C, Shimada H, Fujimura T (1994) A unique sequence located downstream from the rice mitochondrial atp6 may cause male sterility. Curr Genet 25: 52–58 [DOI] [PubMed] [Google Scholar]
- Bentolila S, Stefanov S (2012) A reevaluation of rice mitochondrial evolution based on the complete sequence of male-fertile and male-sterile mitochondrial genomes. Plant Physiol 158: 996–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, , Kazama T, , Yamada M, , Toriyama K ( 2010) Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. BMC Genomic 11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Toriyama K (2005) Molecular mapping of the fertility restorer gene for ms-CW-type cytoplasmic male sterility of rice. Theor Appl Genet 111: 696–701 [DOI] [PubMed] [Google Scholar]
- Grelon M, Budar F, Bonhomme S, Pelletier G (1994) Ogura cytoplasmic male-sterility (CMS)-associated orf138 is translated into a mitochondrial membrane polypeptide in male-sterile Brassica cybrids. Mol Gen Genet 243: 540–547 [DOI] [PubMed] [Google Scholar]
- Hanson MR, Bentolila S (2004) Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16 (Suppl): S154–S169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Baldauf JA (2018) Heterosis in plants. Curr Biol 28: R1089–R1092 [DOI] [PubMed] [Google Scholar]
- Huang JZ, Zg E, Zhang HL, Shu QY (2014) Workable male sterility systems for hybrid rice: Genetics, biochemistry, molecular biology, and utilization. Rice (N Y) 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi M, Koizuka N, Fujimoto H, Sakai T, Imamura J (1999) Identification and expression of the kosena radish (Raphanus sativus cv Kosena) homologue of the ogura radish CMS-associated gene, orf138. Plant Mol Biol 39: 183–188 [DOI] [PubMed] [Google Scholar]
- Iwabuchi M, Kyozuka J, Shimamoto K (1993) Processing followed by complete editing of an altered mitochondrial atp6 RNA restores fertility of cytoplasmic male sterile rice. EMBO J 12: 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Katsuo K, Mizushima U (1958) Studies on the cytoplasmic difference among rice varieties, Oryza sativa L. 1. On the fertility of hybrids obtained reciprocally between cultivated and wild varieties. Jpn J Breed 8: 1–5 [Google Scholar]
- Kazama T, Itabashi E, Fujii S, Nakamura T, Toriyama K (2016) Mitochondrial ORF79 levels determine pollen abortion in cytoplasmic male sterile rice. Plant J 85: 707–716 [DOI] [PubMed] [Google Scholar]
- Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K (2008) Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J 55: 619–628 [DOI] [PubMed] [Google Scholar]
- Kazama T, Okuno M, Watari Y, Yanase S, Koizuka C, Tsuruta Y, Sugaya H, Toyoda A, Itoh T, Tsutsumi N, et al. (2019). Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat Plants 5: 722–730 [DOI] [PubMed] [Google Scholar]
- Kazama T, Toriyama K (2014) A fertility restorer gene, Rf4, widely used for hybrid rice breeding encodes a pentatricopeptide repeat protein. Rice (N Y) 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yang D, Zhu Y (2007) Characterization and use of male sterility in hybrid rice breeding. J Integr Plant Biol 49: 791–804 [Google Scholar]
- Luo D, Xu H, Liu Z, Guo J, Li H, Chen L, Fang C, Bai Q, Zhang M, Yao N, et al. (2013) A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet 45: 573–577 [DOI] [PubMed] [Google Scholar]
- Motomura K, Moromizato Z, Adaniya S (2003) Inheritance of cytoplasmic male sterility and restoration of fertility in rice line, RT102C, derived from Oryza rufipogon. Jpn J Trop Agr 47: 70–76 [Google Scholar]
- Okazaki M, Kazama T, Murata H, Motomura K, Toriyama K (2013) Whole mitochondrial genome sequencing and transcriptional analysis to uncover an RT102-type cytoplasmic male sterility-associated candidate Gene Derived from Oryza rufipogon. Plant Cell Physiol 54: 1560–1568 [DOI] [PubMed] [Google Scholar]
- Rhoades MM (1931) Cytoplasmic inheritance of male sterility in Zea Mays. Science 73: 340–341 [DOI] [PubMed] [Google Scholar]
- Sakuma T, Ochiai H, Kaneko T, Mashimo T, Tokumasu D, Sakane Y, Suzuki K-I, Miyamoto T, Sakamoto N, Matsuura S, et al. (2013) Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci Rep 3: 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Luo D, Zhou D, Zhang Q, Tian D, Zheng X, Chen L, Liu Y-G (2014) The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol Plant 7: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Tang H, Zheng X, Li C, Xie X, Chen Y, Chen L, Zhao X, Zheng H, Zhou J, Ye S, et al. (2017) Multi-step formation, evolution, and functionalization of new cytoplasmic male sterility genes in the plant mitochondrial genomes. Cell Res 27: 130–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ye S, Li J, Zheng B, Bao M, Ning G (2011) Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol 11: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Wang L, Sun Q, Zhu Y (2002) Discovery of mitochondrial chimeric-gene associated with cytoplasmic male sterility of HL-rice. Chin Sci Bull 47: 744–747 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.