Abstract
The reproductive transition is an important event that is crucial for plant survival and reproduction. Relative to the thorough understanding of the vegetative phase transition in angiosperms, a little is known about this process in perennial conifers. To gain insight into the molecular basis of the regulatory mechanism in conifers, we used temporal dynamic transcriptome analysis with samples from seven different ages of Pinus tabuliformis to identify a gene module substantially associated with aging. The results first demonstrated that the phase change in P. tabuliformis occurred as an unexpectedly rapid transition rather than a slow, gradual progression. The age-related gene module contains 33 transcription factors and was enriched in genes that belong to the MADS (MCMl, AGAMOUS, DEFICIENS, SRF)-box family, including six SOC1-like genes and DAL1 and DAL10. Expression analysis in P. tabuliformis and a late-cone-setting P. bungeana mutant showed a tight association between PtMADS11 and reproductive competence. We then confirmed that MADS11 and DAL1 coordinate the aging pathway through physical interaction. Overexpression of PtMADS11 and PtDAL1 partially rescued the flowering of 35S::miR156A and spl1,2,3,4,5,6 mutants in Arabidopsis (Arabidopsis thaliana), but only PtMADS11 could rescue the flowering of the ft-10 mutant, suggesting PtMADS11 and PtDAL1 play different roles in flowering regulatory networks in Arabidopsis. The PtMADS11 could not alter the flowering phenotype of soc1-1-2, indicating it may function differently from AtSOC1 in Arabidopsis. In this study, we identified the MADS11 gene in pine as a regulatory mediator of the juvenile-to-adult transition with functions differentiated from the angiosperm SOC1.
As an age biomarker, the MADS transcription factor DAL1 interacts with a strict maturity-related MADS transcription factor to mediate the vegetative-to-reproductive transition in pine.
Introduction
The irreversible transformation of plants from the vegetative to reproductive phase is referred to as the vegetative phase change (Poethig, 2013) and is a crucial stage in the lifecycle of plants. Prior to beginning reproduction during their long-life cycle, many perennial woody plants experience a prolonged juvenile phase. In the conifer Norway spruce (Picea abies), about 20–25 years are required to reach reproductive age (Chalupka and Cecich, 1997), while Pinus tabuliformis needs 5–7 years to complete its juvenile phase. Despite considerable progress in the understanding of flowering time control in model plants, such as Arabidopsis thaliana (from now on Arabidopsis) and economically important corps, the mechanism underlying the aging pathway in perennial trees, especially conifers, remains largely unclear. Therefore, to explore the effects of aging on the processes of reproductive development in forest trees is of great theoretical and practical importance.
In angiosperms, studies have revealed that an aging pathway mediated mainly by microRNAs (miRNAs) affects the juvenile-to-adult phase transition (Khan et al., 2014); among them, miR156 and miR172 have been identified as key regulatory factors (Lauter, 2005; Wu, 2006;Wu et al., 2009). miR156 and miR172 show evolutionary and expression conservation in annual and perennial angiosperm plants including Arabidopsis (Wu et al., 2009), Oryza sativa (Xie, 2006), Zea mays (Chucka, 2011), and Populus × canadensis (Wang et al., 2011). The miR156 accumulates to high levels in seedlings and its expression level gradually decreases during the development of Arabidopsis (Wu, 2006), while miR172 shows the opposite expression pattern (Wu et al., 2009). The expression level of miR156 gradually decreases, resulting in a gradual increase in SQUAMOSA-PROMOTER-BINDING PROTEIN-LIKE (SPL) proteins, after which SPL9 activates miR172 expression (Wang, 2009; Wu et al., 2009). The miR172 continuously inhibits the expression of a small group of APETALA2-like (AP2-like) genes, including AP2, TARGET OF EAT1,3 (TOE1 and TOE3), and SCHLAFMÜTZE (SMZ), eliminating AP2-like protein inhibition over FLOWERING LOCUS T (FT), SUPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), APETALA1 (AP1), and other MADS-box genes, allowing plants to shift from vegetative development to reproductive growth (Wang, 2009; Yant et al., 2010; Zhu and Helliwell, 2011). The expression patterns of miR156 and miR172, which strongly correlated with age, are similar in perennial broadleaf trees (Wang et al., 2011), indicating the conservation of this signaling pathway in angiosperms.
The same transcriptional regulation models between miR156 with SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-box genes and miR172 with AP2 homologous genes have been observed in gymnosperms, such as Ginkgo biloba (Zhang et al., 2015), Norway spruce, and Picea glauca (Shigyo et al., 2006; Nilsson et al., 2007; Guo et al., 2008; Zhang et al., 2019). In P. tabuliformis, we confirmed that miR156 and miR172 specifically cut mRNA target sites for SPL1,2,3 and AP2 homologous genes; however, the expression pattern of miR156 and its target genes showed no correlation with age (Niu et al., 2015, 2016). In addition to reproductive regulation, miR156 and miR172 are also involved in the regulation of various developmental processes and stress responses, and whether these functions are conserved evolutionarily has not been elucidated. Thus, determining whether miR156 and miR172 play key roles in the vegetative phase transition in gymnosperms is essential.
MADS-box family genes, such as AP1 and SOC1, play critical roles in the aging pathway (Wu et al., 2009; Yant et al., 2010; Zhu and Helliwell, 2011). The SOC1 gene is a flowering pathway integrator that is widespread among plants and in Arabidopsis connects many flowering signals related to photoperiod, temperature, hormone, and age (Hepworth et al., 2002; Moon et al., 2003; Jungeun, 2010; Jung, 2012). Although SOC1-like genes have been isolated from some gymnosperms, including Gnetum gnemon (Winter et al., 1999), P. abies (Tandre et al., 1995; Uddenberg et al., 2013), and Pinus radiata (Walden et al., 1998), and a SOC1-like member in Cryptomeria japonica functions in accelerating flowering (Katahata et al., 2014), the regulatory mechanism of these genes in gymnosperms remains unclear.
Among MADS-box genes, AGAMOUS-LIKE 6 (AGL6)-like genes belong to one of the most mysterious and elusive subfamilies, with a variety of expression patterns and functions in different plants (Rijpkema et al., 2009; Li et al., 2010; DuAn et al., 2012; Dreni and Zhang, 2016; Ma et al., 2019). The AGL6 subfamily, present in both gymnosperms and angiosperms, plays important roles in control of the floral meristem and the production of floral organs and seeds in angiosperms (Rijpkema et al., 2009; Li et al., 2010; Shen et al., 2019). Ectopic expression of AGL6-like genes of orchid (Oncidium Gower; Hsu et al., 2014) and rice (Li et al., 2010) promoted flowering by activating the FT gene and negatively regulating a subset of FLOWERING LOCUS C (FLC)/MADS AFFECTING FLOWERING (FAM) clade genes in Arabidopsis (Yoo et al., 2011; Hsu et al., 2014). Moreover, studies in Petunia hybrida, rice, and maize indicated that AGL6 have redundant functions with SEPALATTA (SEP) (Rijpkema et al., 2009; Thompson et al., 2009; DuAn et al., 2012). Nevertheless, the functions and regulatory mechanism of the AGL6 subfamily have not yet been fully elucidated in either angiosperms or gymnosperms.
The spruce DEFICIENS-AGAMOUS-LIKE 1 (DAL1) gene is a homolog of the AGL6 subfamily isolated from conifers (Tandre et al., 1995). DAL1 has been proposed as a possible mediator of phase change, as its expression is stimulated in the mid-juvenile phase and increases with age in Norway spruce (Carlsbecker et al., 2004). The expression of DAL1 in conifers rises gradually with age, similar to that of miR172 in angiosperms (Carlsbecker et al., 2004; Xiang et al., 2019). In addition, heterologous expression of conifer DAL1 in Arabidopsis has led to an obvious early flowering phenotypic change, along with adult features such as the development of leaves with predominantly abaxial trichomes (Carlsbecker et al., 2004; Katahata et al., 2014). The processes triggered by age-related genes have been characterized in conifers mainly based on studies of homologous genes of angiosperms, and the molecular landscape of the aging pathway in conifers has remained largely unexplored.
With long-growth cycles and highly heterozygous genetic backgrounds, the juvenile-to-adult transition in gymnosperms shows much greater complexity than that in angiosperms. To gain further insights into aging pathways in conifers, we screened for a gene module that was significantly associated with aging in P. tabuliformis using samples of seven different age classes for temporal dynamic transcriptome analysis. Further analysis of the age-related gene module revealed that the phase change in P. tabuliformis did not occur gradually with age, but instead was more rapid than the expected trend. Moreover, MADS-box family genes, especially SOC1-like genes in the age-related gene module play substantial roles in the juvenile-to-adult transition. Expression–pattern analysis in P. tabuliformis and a super-late cone-setting P. bungeana mutant confirmed the tight connection between PtMADS11 and reproductive competence. Genetic assays conducted in wild-type and mutant Arabidopsis verified the differing functions and active sites of PtDAL1 and PtMADS11 in regulating floral initiation. In this study, we further elucidated the regulatory mechanism of the reproductive transformation in pine.
Results
A gene module significantly associated with aging in P. tabuliformis
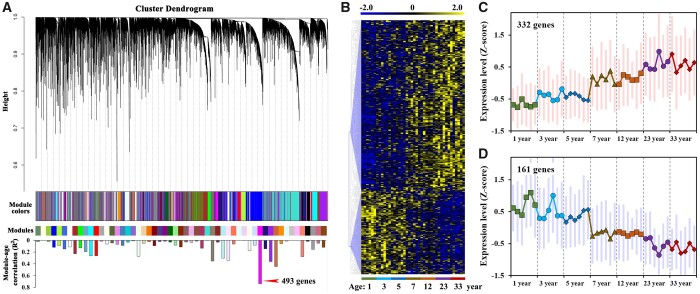
Conifers typically have a very long-life cycle in contrast to other model plants, such as Arabidopsis with short lifespans. In conifers, reproductive development occurs after several years. To explore the mystery of the conifer phase transition and reproductive development, we collected samples of P. tabuliformis from plants at seven different age stages for temporal dynamic transcriptome sequencing. Through weighted gene coexpression network analysis (WGCNA), we identified a module containing 493 differentially expressed genes that were significantly correlated with age (R2 = 0.74; Figure 1A and Table S1). The expression patterns of these genes could be divided into two age-related submodules (Figure 1B), including one that was upregulated with age, containing 332 genes (Figure 1C), and the other one consisting of 161 genes that showed age-related decline (Figure 1D). The expression profiles of genes in these two submodules were divided into seven distinct age groups from juvenile to adult. At 5–7 years, a growth phase change occurred, which is consistent with the timing of the reproductive phase in P. tabuliformis. The seventh year is when the transformation from juvenile growth to maturity occurs in P. tabuliformis. The polynomial model and general linear model were conducted to match the ages of P. tabuliformis and gene expressions (Supplemental Figure S1). The result showed that a simple linear model has poor fit over the time in which reproductive competency is obtained, but is better fit by a polynomial model (Supplemental Figure S1). This suggests an attainment of the reproductive phase change that is more rapid than gradual, contrary to what was previously expected (Nilsson and Brunner, 2004; García-López et al., 2014; Khan et al., 2014). The submodule of genes that are downregulated with age likely plays important roles in maintenance of juvenility and vegetative growth, whereas the upregulated submodule is likely a regulatory component of the juvenile-to-adult phase transition in conifers.
Figure 1.
WGCNA and expression patterns of differentially expressed genes from seven different ages. A, WGCNA was conducted on a total of 13,607 genes that were differentially expressed between any two sample groups of the seven different age stages (P < 0.01). Six biological replicates (trees) were sequenced for each age group. Based on topological overlap, a cluster dendrogram was produced using gene average linkage hierarchical clustering. Modules of coexpressed genes are shown with different color bars beneath the dendrograms. B, Heatmap of 493 differentially expressed genes that were significantly correlated with age. C, Expression profiles of genes that were upregulated with age. D, Expression patterns of genes that were downregulated with age. The pointes with same shape and color indicate samples in same age group. Each age group represent by six biological replicates (six different trees). The red and blue shading in (C) and (D) represents the maximum and minimum values of all genes in each sample.
The MADS-domain transcription factors PtDAL1 and SOC1-like are key players in the aging module of P. tabuliformis
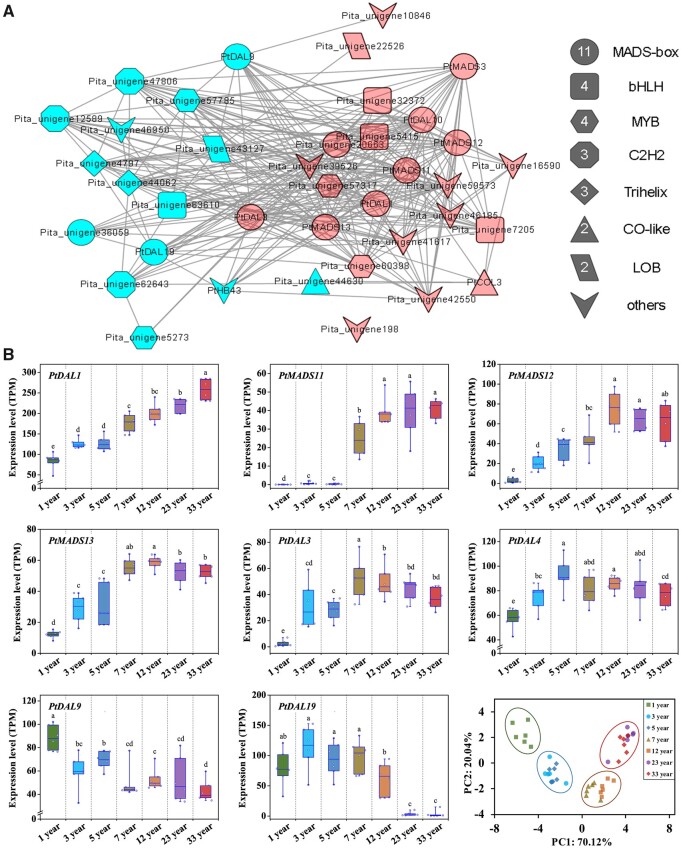
In the age-related module, 33 transcription factors (TFs) were identified that tightly integrated a coexpression network (Figure 2A). Of note, 11 of these 33 TFs belonged to the MADS-box family, making it the most abundant family, including PtDAL1 and PtDAL10 (Carlsbecker et al., 2004; Carlsbecker et al., 2003), which have previously been found to play important roles in the reproductive development of conifers (Figure 2A). The DAL1, a potential mediator of the juvenile-to-adult transition in Norway spruce, also showed the highest correlation coefficient with age in P. tabuliformis (R2 = 0.86; Supplemental Figure S2). These results indicate that the MADS-domain TFs are key players in the vegetative phase transition of conifers.
Figure 2.
MADS-box TFs are enriched among differentially expressed genes associated with age in P. tabuliformis. The coexpression network of 33 tightly integrated TFs. The red shapes represent the genes with expression increase with age, and the blue shapes represent a decrease. B, Expression pattern analysis of PtDAL1 and seven SOC1-like genes in different age groups and PCA for different age groups. The box lower and upper boundaries indicate the 25th and 75th percentiles for each data set. The horizontal lines inside the boxes represent the median values. Whiskers indicate the maximum and minimum values. Different lowercase letters above the plots indicate significant differences at the 0.05 level (ANOVA multiple range test), n = 6.
Moreover, six out of the seven SOC1-like genes (except PtDAL4) in P. tabuliformis were found in the age-related coexpression module (Supplemental Figure S3), implying that SOC1-like genes may play a dominant role in the vegetative to reproductive transformation, which is mediated by an age-related pathway (Figure 2A; Supplemental Figure S3). The expression levels of PtMADS11,12,13, PtDAL3, and PtDAL4 were upregulated in older trees as compared to seedlings, while PtDAL9 and PtDAL19 showed a decreasing trend. PtDAL19 was undetectable in 23- and 33-year-old trees. In addition, principal component analysis (PCA) based on the expression of PtDAL1 and SOC1-like genes clearly divided the samples into four age groups (Figure 2B), showing that PtDAL1 and SOC1-like genes could serve as aging marker genes in P. tabuliformis.
Among seven SOC1-like genes, PtMADS11 showed a unique expression pattern and the correlation coefficient with age was R2 = 0.70 (Supplemental Figure S2). PtMADS11 remained at undetectable levels in juvenile samples without reproductive development, but was suddenly upregulated at 7 years of age and increased with time and developmental status (Figure 2B). The ability to produce cones in P. tabuliformis is normally acquired at the age of 7 years, implying that PtMADS11 may be a primary TF that is closely associated with the age-related initiation of reproductive growth.
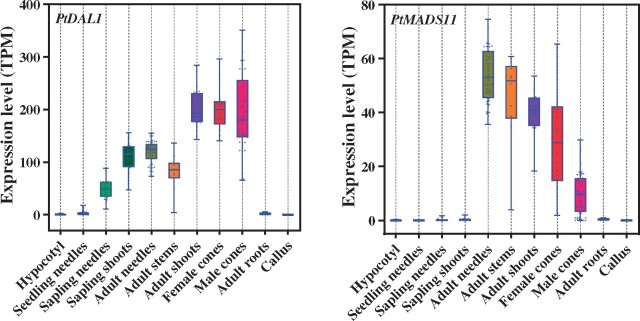
To gain further insights into the functions of the seven SOC1-like and PtDAL1 genes, their detailed expression profiles were analyzed in 347 samples from various tissues and age groups of P. tabuliformis. As shown in Figure 3 and Supplemental Figure S4, the expression patterns of the seven SOC1-like and PtDAL1 genes were not specific to reproductive development, but also generally accumulated in vegetative organs. High expression of PtDAL1 was primarily detected in adult shoots as well as female and male cones, whereas low expression was observed in seedling needles, adult roots, and callus (Figure 3). PtMADS12 showed a similar expression pattern, with high expression levels in adult shoots and female cones, but low levels in juvenile shoots, adult needles and stems, and negligible expression in seedling needles and callus (Supplemental Figure S4). Similarly, PtMADS13 was expressed at higher levels in adult needles, stems, and shoots. PtDAL3 was detected primarily in adult stems, with low levels in juvenile shoots, adult needles, adult stems and shoots, male and female cones, and almost undetectable levels in seedling needles, adult roots and callus, in accordance with the general expression profile of SOC1-like genes (Supplemental Figure S4). The expression of PtDAL4 was apparently low in needles but high in shoots of both juvenile and adult trees, but this gene was weakly detected in other tissues (Supplemental Figure S4). Of note, PtDAL9 was preferentially expressed in seedling needles, but was detected at low levels (barely above background levels) in other samples, and PtDAL19 was upregulated specifically in juvenile shoots and adult roots (Supplemental Figure S4). PtDAL9 and PtDAL19 have been identified in tender and undifferentiated tissues, and the expression profiles of these two genes were distinct from those of the other five SOC1-like genes (Supplemental Figure S5). The functions of SOC1-like gene family members have been reported to be broad and diversified. The expression of PtMADS11 followed a strict maturity-related pattern; it was preferentially induced in mature tissues other than roots, and undetectable in any immature organs. Notably, roots are generally considered to maintain a state of continuous division and have a young physiological age (George et al., 2008).
Figure 3.
PtDAL1 and PtMADS11 expression profiles in various tissues of P. tabuliformis. The expression levels of PtDAL1 and PtMADS11 in 347 total samples of P. tabuliformis were analyzed through RNA-seq, including samples of the hypocotyl (2 months of age, n = 36), seedling needles (2 months of age, n = 90), sapling needles (3–5 years of age, n = 18), sapling shoots (3–5 years of age, n = 18), adult needles (33 years of age, n = 48), adult stems (33 years of age, n = 10), adult shoots (22–33 years of age, n = 30), female cones (33 years of age, n = 18), male cones (33 years of age, n = 57), adult roots (33 years of age, n = 4), and callus (induced from the cotyledon, n = 18). The lower box and upper boundaries indicate the 25th and 75th percentiles for each data set. The horizontal lines inside the boxes represent the median values. Whiskers indicate the maximum and minimum values.
MADS11 is a potential mediator of reproductive ability in conifers
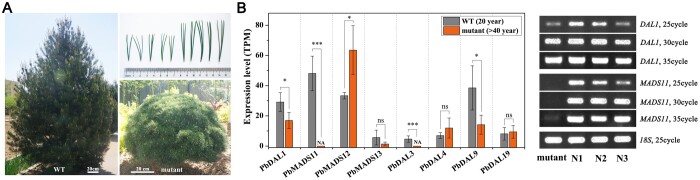
Transformation of conifer species is not possible due to the limited genetic tools available for this group, and functional verification of promising candidate aging pathway genes is extremely difficult. In lieu of such tests, we identified a dwarfed and super-late cone-setting mutant of Pinus bungeana from a common garden. The mutant displayed a bushy appearance and shorter needles, a reduced apical dominance and lack of reproductive ability compared to normal P. bungeana (Figure 4A; Supplemental Figure S6). Pinus bungeana generally transitions to reproductive growth after 10–15 years of vegetative growth, but the mutant had never produced male or female cones at the age of 40 years. We analyzed the expression levels of PbDAL1 and SOC1-like homologs in normal trees and this mutant using RNA sequencing (RNA-seq) and reverse transcription-PCR (RT-PCR) techniques (Table S2). The results showed that PbMADS11 and PbDAL3 were not detectable in the mutant, and that the expression of PbDAL1 and PbDAL9 was downregulated compared with younger normal trees (Figure 4B). The lack of PbMADS11 in the P. bungeana mutant indicates that MADS11 may be a mediator of reproductive ability.
Figure 4.
MADS11 is a potential mediator of reproductive ability in conifers. A, Phenotype of the late-cone-setting P. bungeana mutant. Normal P. bungeana trees grow 3–5 m tall. The P. bungeana mutant displayed a dwarfed and bushy appearance. Needle phenotypes of normal (N1–N3) and mutant P. bungeana. B, Expression levels of PbDAL1 and SOC1-like homologs in normal and mutant P. bungeana trees. The mean of three biological replicates is plotted with se. t test was used to analyze the differences between the two groups of samples. *P≤0.05 and ***P≤0.001 between the two groups.
In general, coniferous maturation and reproductive ability are assumed to occur in an apical–basal gradient along the tree stem (Hartmann et al., 1997; Carlsbecker et al., 2004). In contrast to basal branches, upper-layer branches generally have greater reproductive ability and typically hold more cones. To further investigate the relationship among PtDAL1, SOC1-like genes, and reproductive ability, we detected the activities of those genes in six branch whorls from the top toward the base of 15-year-old P. tabuliformis through RNA-seq. The apical shoots showed substantially higher expression levels of PtMADS11, and expression levels decreased toward the basal part of the tree. The PtDAL3 also showed spatial patterns along the axis of the tree, but the decreasing trend was not as obvious as that of PtMADS11 (Supplemental Figure S7). Based on these results, the PtMADS11 showed a striking relationship with reproductive capacity.
MADS11 and DAL1 coordinate the aging pathway through physical interaction
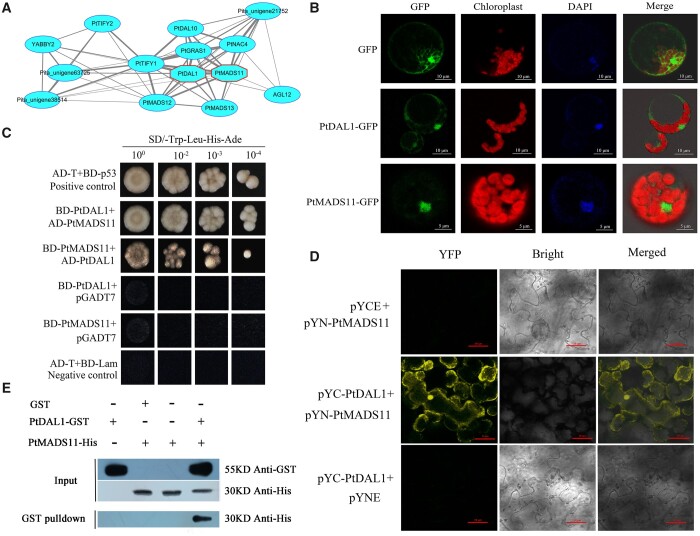
To investigate the molecular basis of the vegetative to reproductive stage transition regulatory process, a TF collaborative network (TFCN) using an algorithm based on a shared coexpression connectivity matrix (SCCM; Niu et al., 2019) was constructed to screen for hub-coordinated TFs associated with the phase change in conifers. PtMADS11 and PtDAL1 showed the highest coordination of TFs among MADS-box genes, implying functional coordination (Figure 5A). PtMADS11 and PtDAL1 were both observed in the nucleus and cytoplasm of Arabidopsis protoplasts (Figure 5B). To identify the interaction between PtDAL1 and PtMADS11, yeast two-hybrid (Y2H) assays were performed. Both AD-PtDAL1/BD-PtMADS11 and AD-PtMADS11/BD-PtDAL1 cotransformed yeast strains grew in all dilutions, supporting the role of PtMADS11 as a strong PtDAL1-interacting protein in yeast cells (Figures 5C). Bimolecular fluorescence complementation (BiFC) assays were conducted in Nicotiana benthamiana leaves. PtDAL1 was fused with the split N terminus of a yellow fluorescent protein (YFP, namely PYN), and PtMADS11 was fused with the split C-terminus of YFP (PYC). When we transiently expressed PYC-PtDAL1 and PYN-PtMADS11, strong YFP signals were observed not only in the nucleus but also in the cytoplasm of leaf epidermal cells (Figure 5D). Moreover, glutathione S-transferase (GST) pull-down assays demonstrated that PtDAL1-GST bound to PtMADS11-His (Figure 5E). Together, these results clearly demonstrate that PtDAL1 and PtMADS11 can form heterodimers through direct protein–protein interaction.
Figure 5.
PtMADS11 physically interacts with PtDAL1. A, TFCN based on the SCCM among age-related MADS-box TFs. The red octagons indicate the PtDAL1 and PtMADS11. B, Subcellular localization analysis of PtDAL1 and PtMADS11 performed in Arabidopsis protoplasts. Scale bars: 10 μm (upper two rows) and 5 μm (bottom row). C, Y2H assay of interactions between PtDAL1 and PtMADS11. AD-T and BD-p53 were used as positive control; AD-T and BD-Lam were used as negative control. D, BiFC assay of interactions between PtDAL1 and PtMADS11 in N. benthamiana leaf epidermis. YFP, YFP fluorescence; Bright, bright field image; Merge, merged image of YFP and Bright; pYN-PtMADS11, expression of pYN-PtMADS11 alone; pYC-PtDAL1, expression of pYC-PtDAL1 alone; pYN-PtMADS11+pYC-PtDAL1, coexpression of pYN-PtMADS11 and pYC-PtDAL1. Scale bars: 20 μm. E, GST pull-down analysis of the protein interaction between PtDAL1 and PtMADS11.
PtMADS11 and PtDAL1 induce early flowering through different mechanisms in Arabidopsis
To elucidate how the PtDAL1 and PtMADS11 function in the vegetative phase-change process, PtDAL1- and PtMADS11-overexpressing transgenic Arabidopsis lines were generated. The transgenic plants displayed visible alterations in morphology from the wild-type, and these improved phenotypic properties were inherited over successive generations. Ectopic expression of the PtDAL1 gene in wild-type Arabidopsis significantly advanced the phase transition, and the inflorescences reached a height of only 7–9 cm, compared with 30 cm or more for the wild-type (Figure 6A(a); Roberts and Shirsat, 2006). The reproduction of most transgenic plants was initiated considerably after the formation of four rosette leaves bearing abaxial trichomes (Supplemental Figure S8, Aa–d). The geminiflorous organ formed from the fusing of two small carpels and multiple stamens, and the petals were loosely enveloped by four sepals at the top of the inflorescence in transgenic lines (Supplemental Figure S8, Ae–l). The structure formed by the fusion of carpels further developed into two siliques within one carpopodium (Supplemental Figure S8, C(c)–(e)). In addition to severe phenotypic aberrations in the flowers, oval-shaped cotyledons and curled rosette leaves were produced in the transformants (Supplemental Figure S8Ca). The transgenic plants produced shorter siliques that were often curled and contained fewer seeds compared with those of wild-type plants (Supplemental Figure S8, C(b) and C(f)). These findings confirm the pivotal role of PtDAL1 in reproductive initiation and control of floral organ identity.
Figure 6.
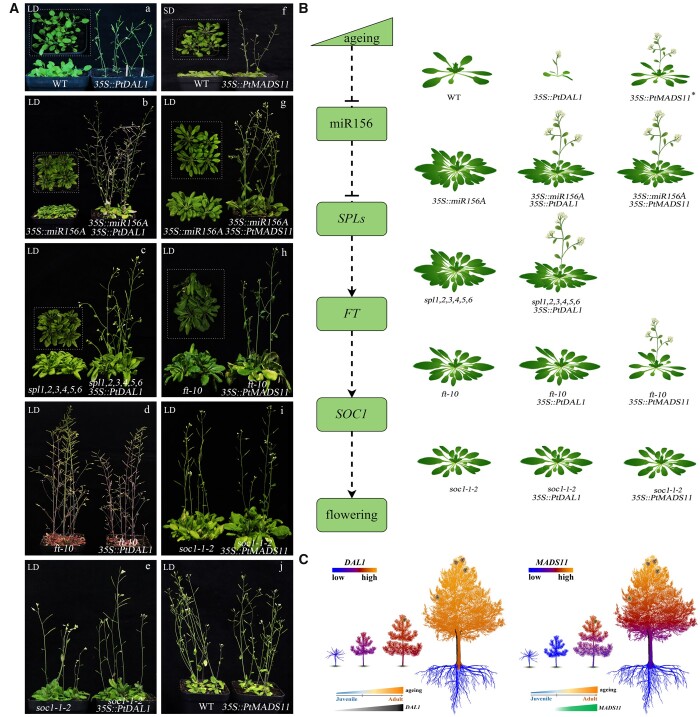
The function of the PtDAL1 and PtMADS11 in the flowering regulatory network in Arabidopsis and pine. Transgenic phenotypes of 35S::DAL1 in wild-type and loss-of-function mutant Arabidopsis. a, Ectopic expression of the PtDAL1 gene significantly advanced the flowering of wild-type Arabidopsis; b, overexpression of PtDAL1 rescued the late flowering of 35S::miR156A Arabidopsis lines; c, the late-flowering phenotype of the spl1,2,3,4,5,6 mutant was restored by overexpression of PtDAL1; d, the early flowering phenotype of the PtDAL1 overexpression lines was suppressed by ft-10 mutation; e, the overexpression of 35S::PtDAL1 in soc1-1-2 mutant. f, 35S::PtMADS11 transgenic Arabidopsis exhibited early flowering under SD conditions; g, heterotopic expression of PtMADS11 restored the severe late-flowering phenotype of 35S::miR156A Arabidopsis; h, overexpression of PtMADS11 accelerated flowering with ft-10 mutation; i, transgenic PtMADS11 in the soc1-1-2 mutant caused no apparent alteration in morphology; and j, no obvious phenotypes of overexpression of 35S::MADS11 in wild-type Arabidopsis under LD conditions. The parts of the figure were assembled from more than one photo, and the outlines of each original photo are indicated by dotted lines. B, The model of the PtDAL1 and PtMADS11 in the flowering regulatory network of Arabidopsis. *, under SD conditions. C, The expression patterns of PtDAL1 and PtMADS11 in P. tabuliformis growth process. The red and blue indicate high- and low-expression level, respectively. PtDAL1 was expressed at low levels in seedlings, and gradually accumulated with age (black triangle). And PtMADS11 was suddenly activated in tree at 7 years, and then the expression increased with age (green quadrilateral). In adult trees, PtMADS11 had a downward trend of top-down expression, while PtDAL1 is relatively consistently expressed between different layer branches. The cones in the trees indicate the reproductive ability of these trees.
The 35S::PtMADS11 transgenic plants showed early flowering and prematurely terminated inflorescence structures under short-day (SD) conditions, but this flowering promotion effect was greatly attenuated under long-day (LD) conditions (Figure 6, A(f) and A(j)). The loss of inflorescence indeterminacy resulted in production of terminal flowers at the end of the main inflorescence, which then shifted from continuous growth to finite growth (Supplemental Figure S8, B(a)–(h)). Taken together, these results indicate that PtMADS11 functions as a positive regulator of flowering and a determinant of meristem fate within an inflorescence.
Phenotypic rescue tests were conducted using Arabidopsis mutants defective in key genes of flowering regulatory networks to determine the regulatory mechanism of PtDAL1 and PtMADS11 in the phase-transition regulation pathway, as well as their functions in the promotion of flowering in Arabidopsis. Plants overexpressing miR156 and hexamutants of its targets spl1,2,3,4,5,6 show an extremely severe phenotype of delayed flowering, but the late-flowering phenotype can be restored with overexpression of PtDAL1, which allows the transgenic plants to transform normally into the reproductive growth phase (Figure 6, A(b) and A(c)). However, in this study, we found that the early flowering phenotype of PtDAL1 overexpression lines was suppressed by ft loss-of-function mutation (Figure 6A(d)). Similarly, PtDAL1 did not normalize the phenotype of soc1-1-2 mutants (Figure 6A(e)). These findings show that the regulatory roles of PtDAL1 in the reproductive transition are compromised in the absence of either FT or SOC1. Of note, fd-3 mutations introduced into the PtDAL1 gene triggered premature floral development and produced several curly rosettes and cauline leaves (Supplemental Figure S8, D(a)–(c)). Thus, the function of PtDAL1 in promoting the vegetative phase change in Arabidopsis may be independent of the FLOWERING LOCUS D (FD) gene. However, heterotopic expression of PtMADS11 restored the severely late-flowering phenotype of both 35S::miR156A and ft-10, indicating that PtMADS11 acts independently of miR156 and FT in the regulation of flowering (Figure 6, A(g) and A(h)). Similar to PtDAL1, PtMADS11 accumulation had no obvious effect when SOC1 was absent (Figure 6A(i)). The rosette statistical analysis of transgenic lines was showed in Supplemental Figure S9. Although PtMADS11 is a SOC1-like gene, its function does not overlap with that of AtSOC1 in Arabidopsis. These results imply that PtMADS11 and PtDAL1 play different roles in flowering regulatory networks.
Discussion
There has been considerable progress in understanding flowering time control in model plants such as Arabidopsis (Blumel et al., 2015) and economically important crops such as rice (Yano et al., 2001). However, there are major differences in the regulatory mechanisms of the phase change between conifers and angiosperms (Tandre et al., 1995), and the mechanism underlying the aging pathway in perennial trees, especially conifers, has remained elusive.
In the present study, we identified a gene module that was significantly correlated with aging (Figure 1A). Previous studies suggested that the phase transition in trees occurs gradually (Nilsson and Brunner, 2004; García-López et al., 2014; Khan et al., 2014), but we observed an unexpectedly rapid transition in gene expression from vegetative growth to reproductive development in P. tabuliformis. The expression patterns of differentially expressed genes within the age-related module were divided into two age-related submodules (Figure 1B; Supplemental Table S1). The polynomial model and general linear model were further conducted to match the ages with gene expressions (Supplemental Figure S1), which supported our opinion that the phase change in P. tabuliformis was a more drastic change than the expected trend. The gene module that was highly correlated with age included two subsets of genes, one of which was upregulated and the other was downregulated as age increased (Figure 1, C and D). In the processes of attaining reproductive competence and reproductive bud differentiation in conifers, up and downregulated gene sets may regulate the balance between vegetative and reproductive growth. A clear understanding of the transcriptional regulation of these up- and downregulated gene sets in the aging pathway will require further research.
In the age-related module, 11 TFs in the MADS-box family accounted for the largest group of the 33 total TFs that tightly integrate a coexpression network (Figure 2A), implying an important role in the reproductive development of conifers. In addition to the MADS-box family, TFs such as MYB, CO-like, and bHLH, which could play roles in the conifer reproductive transformation, were also identified (Figure 2A). In angiosperms, the CONSTANS (CO) gene functions upstream of the photoperiod pathway and directly regulates the expression of the downstream FT gene in leaves to induce flowering (Priyanka and Kishore, 2015; Shim et al., 2017). Several studies on the expression patterns and functions of CO-like genes in gymnosperms have suggested that it functions as an inducer that responds to light signals to initiate reproductive growth in gymnosperms (Holefors et al., 2009; Nilsson, 2009; Yan et al., 2017). miR156-targeted SPLs negatively regulate the R2R3-MYB–bHLH–WD40 complex to inhibit the expression of anthocyanin biosynthesis genes (An et al., 2012). MYB TFs regulate floral organ differentiation and development in Arabidopsis (Baumann et al., 2007; Zhang et al., 2009). Thus, MYB and bHLH TFs may be involved in the vegetative growth transition in conifers under the control of the aging pathway.
Phylogenetic analyses imply that SOC1-like genes in conifers are not orthologs of angiosperm SOC1-like genes (Supplemental Figure S3), which derived from the same ancestor and diverged through evolution. After independent functional differentiation of gymnosperm and angiosperm SOC1-like genes, a modified cluster consisting of SOC1-like genes may act as an integral dominant module that regulates reproduction in gymnosperms, as expression of six SOC1-like members of P. tabuliformis was tightly correlated with age (Figure 2A). In angiosperms, the CO gene positively regulates SOC1 expression via the FT gene in the upstream flowering pathway (Böhlenius, 2006), while FLC inhibits SOC1 expression in the upstream vernalization pathway (Hepworth et al., 2002; Helliwell et al., 2006). The function and mode of action of the CO gene in gymnosperms are conserved (Holefors et al., 2009; Nilsson, 2009), but no FLC homologs have been found in gymnosperms (Nystedt et al., 2013; Chen et al., 2017). The functions of SOC1-like genes in P. tabuliformis appeared differentiated within the subcluster due to the presence of two different expression patterns (Supplemental Figure S5). DAL19 in Norway spruce was specifically upregulated in cone-setting shoots and maintained a high-expression level in adult buds (Uddenberg et al., 2013; Akhter et al., 2018). However, in the present study, PtDAL19 was undetectable in adult needles, shoots, female cones, and male cones. The opposite expression profiles of DAL19 between Norway spruce and P. tabuliformis reveal the functional differentiation of DAL19 between Norway spruce and P. tabuliformis. Based on large-scale expression data, we propose that PtMADS11, PtDAL3, PtDAL4, PtMADS13, and PtMADS12 predominately promote reproductive growth in the aging pathway, while PtDAL19 and PtDAL9 likely act as negative regulators of the juvenile-to-adult phase transition process in P. tabuliformis. SOC1-like members with opposite functions might share comparable regulatory mechanisms related to age, thereby balancing vegetative growth and reproductive development in P. tabuliformis.
Although the expression patterns of PtDAL1 and SOC1-like homologs were strongly correlated with age, expression of these genes was not limited to reproductive organs, and high-expression levels were also detected in vegetative organs (Figure 3; Supplemental Figure S4). Genes such as FT, AP1, and LEAFY (LFY), which activate reproductive growth, tend to be activated in vegetative organs, and this feature is conserved among angiosperms (Mandel and Yanofsky, 1995; Moyroud et al., 2010). Moreover, emerging studies have shown that SOC1-like genes are involved in a broader range of functions related to enhancing plant tolerance to high temperature and inducing chloroplast biogenesis in petals than previously expected (Wang et al., 2019; Ning et al., 2020). PtDAL1 and SOC1-like homologs may directly sense or transmit signals upstream of age-regulated pathways, with crucial effects on broader network function and reproductive development. The age-mediated response to environmental factors in conifers is worthy of further in-depth investigation.
Expression of PtMADS11 was almost undetectable in young plants during vegetative growth (1–5 years), and the expression increases after 7 years of growth (Figures 2, B and 6, C). These results imply that PtMADS11 plays a role in the transformation from vegetative to reproductive growth, and that the seventh year is the end of juvenility in P. tabuliformis. The lack of PbMADS11 in the super-late cone-setting P. bungeana mutant supports the close relationship between MADS11 and reproductive capacity (Figure 4B).
We identified the DAL1 gene as a primary age biomarker, and it showed the highest correlation coefficient with age (R2 = 0.86) among the genes investigated in P. tabuliformis (Supplemental Figure S2). A transcriptome-wide analysis in Larix kaempferi supports the conservation of DAL1 expression patterns in conifers (Xiang et al., 2019). In contrast to the undetectable expression of DAL1 in Norway spruce before 5 years (Tandre et al., 1995; Carlsbecker et al., 2004), expression of PtDAL1 in P. tabuliformis was detectable at the seedling stage (Figure 6C). This difference could arise due to the different lengths of the vegetative growth stages of Norway spruce and P. tabuliformis. Norway spruce requires 20–25 years before reproducing, while the juvenile phase for P. tabuliformis is 5–7 years. PtDAL1 may function as an activator for shoots to initiate reproduction, but this process may require other coregulators.
In the age-related TFCN module (Figure 5A), beyond DAL1 and MADS11, the DAL10 gene, which is specifically active in seed cones and pollen cones in Norway spruce, was closely related to reproductive development (Carlsbecker et al., 2003), and AGL12 was reported to regulate flowering transition in Arabidopsis (Tapia-Lopez et al., 2008). The GRAS members functioned on the regulation of phase transition process in barley and rice (Curaba et al., 2013; Fan et al., 2015), and showed an age-related expression pattern in L. kaempferi (Xiang et al., 2019). It was also reported that the putative GA-NAC-LFY regulator module might play roles in accelerating reproductive development (Fang et al., 2021). These results confirmed the accuracy of the predicted gene module. The expression of PtDAL1 increased gradually during the stage of undetectable PtMADS11 expression (1–5 years) and maintained a higher level than that of PtMADS11 during the reproductive growth phase (Supplemental Figure S5). In addition, PtDAL1 had a wider expression range than that of PtMADS11 among different tissues (Figure 3). The subcellular localization of PtMADS11 protein in cytoplasm and nucleus reasonably explains the interaction between PtDAL1 and PtMADS11 in cytoplasm and nucleus (Figure 5, B and D), which was consistent with MADS-domain protein–protein interactions in Arabidopsis (Immink et al., 2002). These results imply a tight functional coordination between PtDAL1 and PMADS11 in an age-mediated module that links the previously expressed regulator PtDAL1 to the direct interaction factor PtMADS11 in P. tabuliformis. Nevertheless, further evidence is needed to verify the direct regulatory relationship between PtDAL1 and PtMADS11.
PtDAL1 is a homolog of the AGL6 subfamily in conifers. The AGL6 subfamily is widespread in plants and has a variety of functions that have been poorly characterized, along with diverse and divergent expression patterns (Reinheimer and Kellogg, 2009 ; Dreni and Zhang, 2016; Ma et al., 2019). The AtAGL6 is expressed in floral organs and stems in Arabidopsis (Yoo et al., 2011), and two Gnetum gnemon AGL6 subfamily genes (GGM9 and GGM11) are expressed in both male and female cones, but absent in leaves (Winter et al., 1999; Shen et al., 2019), while PtDAL1 was detected in the stems, needles, and reproductive cones of P. tabuliformis (Figure 3). Despite the divergent expression patterns of the AGL6 subfamily in plants with distant evolutionary relationships, AGL6 is generally associated with reproduction (Koo et al., 2010; Callens et al., 2018) and cone formation (Winter et al., 1999). The orchid AGL6 homolog OMDS1, when overexpressed in Arabidopsis, causes an early flowering phenotype by inducing expression of SOC1, FT, LFY, and AP1 genes (Hsu et al., 2003). This phenotypic similarity in transgenic plants indicates that DAL1 may have conserved regulatory mechanisms. As a member of the MADS-domain family, AGL6 appears to form multimers with other TFs, which have various interactions with SQUA/AP1-like and AGL proteins (Ma et al., 2019), and to integrate signals at the core of the regulatory network. In conifers, DAL1 is a key node of the aging pathway that can form heterodimers with MADS11 and integrates signals to initiate the juvenile-to-adult transition process.
Ectopic expression of PtDAL1 led to an extremely early flowering in Arabidopsis (Figure 6A(a)), indicating that this gene plays a dominant role in the initiation of reproductive growth. These results are consistent with those reported in Norway spruce (Carlsbecker et al., 2004). However, the phenotypic changes caused by transformation of genes from conifers into Arabidopsis did not elucidate the underlying mechanism. High expression levels of a number of other MADS-box family genes caused early flowering, including DAL2 and DAL10 from spruce (Tandre et al., 1998; Carlsbecker et al., 2003), but the effects of DAL1 on flowering time were more severe than those of other MADS-box genes (Carlsbecker et al., 2004), and its effect on the growth of Arabidopsis was particularly strong. The formation of the geminiflorous organ from two short carpels in the 35S::PtDAL1 transgenic lines implied its broader functional roles in floral organ identity and flower development.
During the transition from vegetative growth to flowering in Arabidopsis, SOC1 had upregulated expression in the apical meristem, which effectively promoted early flowering (Lee et al., 2000; Samach, 2000). The expression level of CO increased under LD conditions, which promoted expression of the FT gene and then the SOC1 gene (Yoo et al., 2005). Transgenic PtMADS11 plants showed no apparent alteration under LD conditions, but exhibited early flowering along with premature termination of inflorescence under SD conditions (Figure 6, A(f) and A(j)). PtMADS11 may require a positive factor to promote its function in Arabidopsis under SD conditions. Under SD conditions, the Arabidopsis growth cycle was prolonged and the wild-type Arabidopsis contained about 26 rosette leaves compared with 14 rosette leaves under LD conditions (Supplemental Figure S9). The SD conditions extended the period in which PtMADS11 could play its role and thereby intensified its flowering promotion.
Genetic assays were conducted to verify the functional difference between PtDAL1 and PtMADS11 (Figure 6B), as the flowering regulatory network in Arabidopsis has been extensively studied (Blumel et al., 2015). The extremely severe delayed flowering phenotype of 35S::miR156A lines could be rescued with constitutive expression of PtDAL1 or PtMADS11, implying that the target loci of both transgenic PtDAL1 and PtMADS11 were downstream of miR156. The early flowering phenotype of PtDAL1-overexpression lines was suppressed by the ft loss-of-function mutation, while ft-10 mutant lines that expressed 35S::PtMADS11 had accelerated flowering (Figure 6, A(d) and A(h)). Theoretically, as an upstream regulator of FT, PtDAL1, and PtMADS11 functioned on different positions of the regulatory network in Arabidopsis. Of note, fd-3 mutations introduced into the PtDAL1 gene triggered premature floral development, which revealed that the function of PtDAL1 in promoting flowering in Arabidopsis may be independent of the FD gene. In Arabidopsis, 35S::FT partially rescued the late-flowering phenotype of fd-1 mutants (Abe et al., 2005), indicating that an FD-independent flowering time regulation pathway exists. Notably, all of the gymnosperm SOC1-like proteins queried nested in a clade outside of the Arabidopsis SOC1 and FLC clade proteins (Supplemental Figure S3), indicating their functions may have differentiated during the evolution (Uddenberg et al., 2013). In the present study, we revealed that PtMADS11 cannot restore the late-flowering phenotype of soc1-1-2 mutants; this result suggests that the gymnosperm SOC1-like genes probably cannot replace the SOC1 in Arabidopsis. The differentiated functions of PtMADS11 support a specific regulatory role in the conifer aging pathway.
In this study, we determined that the phase change in pine was a more rapid change than the expected trend. In total, 33 tightly coordinated TFs were identified from the age-related gene module, among which SOC1-like genes were enriched. Among SOC1-like homologs, two distinct expression patterns represented two internal submodules with opposite functions, which together could balance vegetative growth and reproductive growth. In this study, we confirmed the function of PtDAL1 as a regulator in phase transition and identified an interacting gene, PtMADS11, which may function as a positive regulator of reproductive capacity in P. tabuliformis. The differing active sites of PtDAL1 and PtMADS11 were validated through genetic analysis. The results confirm that the functions of PtDAL1 and PtMADS11 differ, and that PtMADS11 may play a unique role in the aging pathway in conifers. Our findings provide a basis for further in-depth studies into the specific regulatory relationships between DAL1 and MADS11 in conifers using more advanced plant genetic engineering tools, such as carbon nanotube carriers.
Materials and methods
Plant materials
For the different ages of P. tabuliformis Carr., plants were selected from a primary clonal seed orchard located in Pingquan City, Hebei Province, China (118°44.6758′ E, 40°98.8784′ N, 560–580 m above sea level). Vegetative shoot apexes were collected from the top branches of the trees after bud-break on 10 May 2016. Six different trees were used as biological replicates for RNA-seq analysis. For tissue specific expression analysis, we collected the needles and hypocotyl from P. tabuliformis seedlings that were cultured in control conditions (24°C, 16 h for light and 8 h for dark), and the sapling needles and shoots were sampled from 3-year-old trees. The callus was induced from seedling hypocotyls following the published method, and propagated for several generations (Liu et al., 2020). Adult tissue samples, including needles, shoots, stems, cones, and roots, were collected from the same primary clonal seed orchard on 10 May 2016. The mutant of P. bungeana was found in a common garden of Beijing Ming Tombs Forest Farm, Beijing, China. Three normal P. bungeana trees were selected as controls for gene expression analysis. From six apical to base whorls of branches of three 15-year-old trees in the same primary seed orchard, vegetative shoots of each whorl have been collected on 11 May 2019 and stored at −80 °C for total RNA extraction. All the samples were collected between 12:00 and 13:00 to control the influence of photoperiod during sampling.
All the mutants and transgenic lines of Arabidopsis used in this study were obtained from The Arabidopsis Information Resource (http://arabidopsis.org). The 35::miR156A transgenic lines prolonged the expression of juvenile vegetative traits and delayed flowering (Wu and Poethig, 2006). The spl1,2,3,4,5,6 mutant that functioned in delaying vegetative phase change and increasing rate of leaf initiation delayed flowering time equal to 35S::miR156A (Xu et al., 2016). The ft-10 mutant displayed a late flowering phenotype under LD conditions (Yoo et al., 2005). The fd-3 mutant displayed a late flowering phenotype that was unaffected by vernalization, and an increased number of rosette leaves (Koornneef et al., 1991). And the soc1-1-2 mutant delayed flowering both in LD and SD growth conditions (Lee et al., 2000).
RNA-seq analysis
Total RNA from different samples of P. tabuliformis were extracted by the Trizol method (Invitrogen, CA, USA). The cleaved RNA fragments were then reverse-transcribed to create the final complementary DNA (cDNA) libraries using the mRNA-Seq sample preparation kit (Illumina, Inc., San Diego, CA, USA). The cDNA libraries were sequenced on the Illumina NovaSeq platform by using the paired-end module (2 × 150 bp). Clean reads for each sample were aligned to the P. tabuliformis reference transcriptome (Niu et al., 2013). Sleuth software was used to analyze differential expression of the genes (Pimentel et al., 2017). The heatmap of the expression cluster of differential genes was analyzed and displayed by Multiple array viewer software. The gene expression patterns of the samples from different conditions were calculated and normalized by the Z-score transformation (Cheadle et al., 2003). The coding sequences of SOC1-like homologs and PtDAL1 were available in Supplemental Data Set S1.
Phylogenetic analysis
Multiple alignments of protein sequences were obtained by MAFFT tool (Katoh and Standley, 2013) and the maximum likelihood tree based on the JTT model was obtained using MEGA software (Sohpal et al., 2010). Bootstrap values were obtained by 1,000 bootstrap replicates.
Network construction
The WGCNA was performed on all the samples using the standard method (Zhang and Horvath, 2005; Horvath, 2008). On the basis of topological overlap, a cluster dendrogram was produced by evaluating the average linkage hierarchical clustering of genes. The correlation between module eigengenes and the aging was analyzed. We then chose the module that was most significantly correlated with age for further analysis in the coexpression network.
The TFCN was further constructed using an algorithm based on the SCCM (Nie et al., 2011; Niu et al., 2019). We considered two TFs to be collaborative and connected in the TFCN only if they shared at least 40 coexpressed genes in common from the top 200 most tightly coexpressed genes for each TF.
Subcellular localization analysis
For subcellular localization analysis, 35S::PtDAL1–GFP and 35S::PtMADS11–GFP fusions were constructed under the control of the constitutive 35S promoter with the green fluorescent protein (GFP) coding region. The rosette leaves of Arabidopsis growing for 3–4 weeks under LD conditions (16-h light/8-h dark, 23°C) were cutoff for enzymatic hydrolysate at 22°C for 2 h in dark (Wu et al., 2009). An equal volume W5 solution was added, shaken gently, and the protoplasts were filtered into the new tube by 100 mesh cell sieves. The protoplasts were collected after centrifugation at 150 rpm at room temperature for 2 min. The supernatant was carefully removed and the protoplast was resuspended by adding an appropriate amount of precooled MMG solution. The 35S::PtDAL1-GFP and 35S::PtMADS11-GFP plasmids were each added into 100 μL protoplasts and lightly mixed. Then 110 μL PEG4000 solution was added and incubated at room temperature for 15 min. A 440-μL W5 solution was added to gently terminate the transfection process. The protoplasts were collected after centrifugation at 150 rpm for 2 min and 400-μL WI solution was added. The solution was incubated at 22°C in dark for 16–24 h. After removing the WI solution, DAPI staining solution (5 μg/mL) was added to cover the protoplasts for 10 min and phosphate-buffered saline (PBS) solution was used to wash the protoplasts three times. The GFP signal was observed by confocal microscope (Leica SP8), the excitation wavelengths and emission filters sets were as 488 nm (Ex)/BP505 to 530 nm (Em). Image analysis was conducted with the Leica LAS AF software.
BiFC assay
For BiFC assays, full-length PtDAL1 and PtMADS11 were cloned into pYCE and pYNE vectors containing the C- or N-terminal end of YFP. The constructs (pYC-PtDAL1 and pYN-PtMADS11) and corresponding empty vectors were each transformed into Agrobacterium strain GV3101-psoup, then the culture was shaken in a constant temperature shaker for 12–18 h until OD600 = 0.6. The bacterial liquid was collected after centrifugation at a room temperature and resuspended with infection solution (10 mM MES, 150 μM AS, 10 mM MgCl2) keeping OD600 = 1.0 and pH = 5.6. Then we added 20 μL AS to the bacteria solution and let it stay in the dark for 2–3 h at a room temperature. The two strains of bacteria liquid were mixed and injected into N. benthamiana leaves by sterile syringes with the needle removed. During the injection, the blade face of leaves was held with fingers to help the bacteria liquid insinuate the leaves slowly. A small wound could be made by the needle hub to ensure the injection would go smoothly. The inoculated N. benthamiana was treated under dark conditions for 24 h, and the fluorescence signal was observed after growth under normal conditions for 48 h (16-h light/8-h dark, 23°C). Fluorescence was then observed by confocal microscope (Leica-SP8), the excitation wavelengths and emission filters sets were 514 nm (Ex)/BP525 to 550 nm (Em). Image analysis was conducted with the Leica LAS AF software.
Y2H assay
Y2H assay was performed to investigate the protein–protein interactions following the Yeast Protocols Handbook (Clontech). The full coding regions of PtDAL1 and PtMADS11 were fused with pGADT7 and pGBKT7 vectors. These constructs and empty vectors as control were then transformed into the Y2H gold yeast strain by the lithium acetate method. The yeast cells cotransformed with plasmids encoding two tested proteins were plated onto selective medium (lacking leucine, tryptophan, histidine, and adenine), and supplemented with aureobasidin A for screening of possible interactions.
GST pull down
Bacteria with overexpressed PtDAL1-GST and PtMADS11-His proteins were resuspended in 20 mM Tris–HCl buffer (pH = 7.5) that contained 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.15 M NaCl, 4 mM benzamidine, and 0.02% NaN3, and ruptured using a French press. The GST and PtDAL1-GST proteins were adsorbed onto the completely resuspended immobilized glutathione during incubation for 4 h with rotation. After adding PtMADS11-His protein to the glutathione-Sepharose beads, the GST–fusion protein complex was rotated during an overnight incubation under 4°C conditions. The protein complex was centrifuged, the supernatant discarded, and the proteins resuspended in 500 μL PBS (1,250 rpm, 5 min). Then, 100 μL of eluent buffer (20 mM Tris–HCl, pH = 7.5) was used to separate the supernatant from the protein precipitates (30 min, 4°C). The precipitates were analyzed through western blotting. Western blot detection was performed as described previously with minor modifications (Gibbons, 2014).
Plasmid construction and transformation of A. thaliana
The PtDAL1 and PtMADS11 genes were amplified from the cDNA of P. tabuliformis new shoot by PCR using primers that introduced XbaI and XhoI restriction sites (Supplemental Table S2). The CDS of PtDAL1 and PtMADS11 each recombined into the plant expression vector pBI121, which contained the nptII gene as a selectable marker, with the 35S promoter. Constructs were transformed into Agrobacterium strain GV3101, and 35S::PtDAL1 and 35S::PtMADS11 overexpression transgenic Arabidopsis lines were generated by the floral-dip process (Clough and Bent, 1998). All the experiments on Arabidopsis mutants, transgenic lines, and wild-type were conducted with transgenic 35S::PtDAL1 and 35S::PtMADS11 to determine their functions in the flowering regulatory network. The harvested seeds were screened on MS mediums containing Kanamycin antibiotic (50 mg·mL−1) until homozygous lines were obtained. All of the Arabidopsis plants were grown in an artificial climate box under LD conditions (16-h light/8-h dark, 23°C) or SD conditions (8-h light/16-h dark, 23°C). The transgenic Arabidopsis lines used in this study were shown in Supplemental Table S3. Nine transgenic lines for each experiment were randomly selected, and at least 30 events were counted for rosette leaf number statistics. The significance of differences between transgenic lines and mutants or wild-type Arabidopsis was analyzed by t test.
Data availability
The authors declare that all data supporting the findings of this study are available within the article and Supplementary Material online or are available upon request from the corresponding author. The RNA-seq data that support the findings of this study have been deposited in the CNSA (https://db.cngb.org/cnsa/) of China National GeneBank Database with accession number CNP0001648.
Accession numbers
The accession numbers of Arabidopsis mutants mentioned in this article are as follows: CS69799 (spl10-3; spl11-1; spl13-1; spl15-1; spl2-1; spl9-4); CS67849 (35S::miR156A); SALK_054421C (fd-3); SALK_006054C (soc1-1-2); and CS9869 (ft-10).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The polynomial model (R2 = 0.86) and general linear model (R2 = 0.74) of the expression level of 493 differential genes matching age stages in P. tabuliformis.
Supplemental Figure S2. The correlation coefficient of age with the expression level of PtDAL1 and PtMADS11.
Supplemental Figure S3. The phylogenetic tree of SOC1-like proteins in Arabidopsis and P. tabuliformis.
Supplemental Figure S4. The expression patterns of SOC1-like genes except PtMADS11 expression during aging in P. tabuliformis.
Supplemental Figure S5. The PtDAL1 and SOC1-like genes expression pattern during aging in P. tabuliformis.
Supplemental Figure S6. The silhouettes of dwarf mutant and normal P. bungeana at the same scale.
Supplemental Figure S7. Expression profiles of PtDAL1 and seven SOC1-like homologs in six branch whorls from top to base of 15-year-old P. tabuliformis.
Supplemental Figure S8. The phenotype of 35S::PtDAL1 transgenic Arabidopsis lines.
Supplemental Figure S9. Analysis of rosette leaf number at flowering of Arabidopsis transgenic and mutant lines.
Supplemental Table S1. A list of all 493 differentially expressed genes of P. tabuliformis among different ages.
Supplemental Table S2. Primers used for plasmid construction RT-PCR analysis.
Supplemental Table S3. The transgenic Arabidopsis lines used in this study.
Supplemental Dataset S1. The coding sequences of SOC1-like homologs and PtDAL1 in P. tabuliformis.
Supplementary Material
Acknowledgments
We would like to thank Meiqin Liu and the Testing and Analysis Center of Beijing Forestry University for the help with confocal microscopy. We would like to thank Dongmei Hu for her help in drawing the diagram.
Funding
This work was supported by The National Natural Science Foundation of China (grant no. 31870651), National Key R&D Program of China (grant no. 2017YFD0600500), and The Program of Introducing Talents of Discipline to Universities (111 project, B13007).
Conflict of interest statement. The authors declare that they have no conflicts of interest.
J.J.M. acquired, analyzed, interpreted data, and drafted the manuscript. J.J.M., X.C. and Y.T.S. participated in the sample preparation and performed experiments. G.F.Z., J.X.L., and T.P. provided valuable discussions and helped to refine the draft. X.Q.Z. provided the cultivation and conservation the different age P. tabuliformis materials. S.P.Q. and F.M. discovered and identified the Pinus bungeana mutant. S.H.N., W.L., Y.L., and H.X.W. participated in the design of the study and performed the statistical analysis. S.H.N. conceived the study and participated in the funding acquisition and project administration, and helped to draft the manuscript. All authors read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Shi-Hui Niu (arrennew@bjfu.edu.cn).
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, et al. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Akhter S, Kretzschmar WW, Nordal V, Delhomme N, Street NR, Nilsson O, et al. (2018) Integrative analysis of three RNA sequencing methods identifies mutually exclusive exons of MADS-box isoforms during early bud development in Picea abies. Front Plant Sci 9: 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An X, Tian Y, Chen K, Wang X, Hao Y (2012) The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J Plant Physiol 169: 710–717 [DOI] [PubMed] [Google Scholar]
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, et al. (2007) Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134: 1691–1701 [DOI] [PubMed] [Google Scholar]
- Blumel M, Dally N, Jung C (2015) Flowering time regulation in crops-what did we learn from Arabidopsis? Curr Opin Biotechnol 32: 121–129 [DOI] [PubMed] [Google Scholar]
- Callens C, Tucker MR, Zhang D, Wilson ZA (2018) Dissecting the role of MADS-box genes in monocot floral development and diversity. J Exp Bot 69: 2435–2459 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Sundstrom J, Tandre K, Englund M, Kvarnheden A, Johanson U, et al. (2003) The DAL10 gene from Norway spruce (Picea abies) belongs to a potentially gymnosperm-specific subclass of MADS-box genes and is specifically active in seed cones and pollen cones. Evol Dev 5: 551–561 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Tandre K, Johanson U, Englund M, Engström P (2004) The MADS-box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies). Plant J 40: 546–557. [DOI] [PubMed] [Google Scholar]
- Chalupka W and , Cecich RA ( 1997) Control of the first flowering in forest trees. Scand J For Res 12: 102–111 [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG (2003) Analysis of microarray data using Z score transformation. J Mol Diagn 5: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang X, Liu X, Zhang L (2017) Evolutionary analysis of MIKCc-Type MADS-Box genes in gymnosperms and angiosperms. Front Plant Sci 8: 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D (2011) Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc Natl Acad Sci USA 42: 17550–17555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curaba J, Talbot M, Li Z, Helliwell C (2013) Over-expression of microrna171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol 13: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L, Zhang D (2016) Flower development: the evolutionary history and functions of the AGL6 subfamily MADS-box genes. J Exp Bot 67: 1625–1638 [DOI] [PubMed] [Google Scholar]
- Duan Y, Xing Z, Diao Z, Xu W, Li S, Du X, et al. (2012) Characterization of Osmads6-5, a null allele, reveals that OsMADS6 is a critical regulator for early flower development in rice (Oryza sativa L.). Plant Mol Biol 80: 429–442 [DOI] [PubMed] [Google Scholar]
- Fan T, Li X, Wu Y, Xia K, Ouyang J, Zhang M, et al. (2015) Rice Osa-miR171c mediates phase change from vegetative to reproductive development and shoot apical meristem maintenance by repressing four OsHAM transcription factors. PLos One 10: e0125833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Chai Z, Yao W, Chen B, Zhang M (2021) Interactions between ScNAC23 and ScGAI regulate GA-mediated flowering and senescence in sugarcane. Plant Sci 304: 110806. [DOI] [PubMed] [Google Scholar]
- García-López M, C, Vidoy I, Jiménez-Ruiz J, Muñoz-Mérida A, Fernández-Ocaña A, de la Rosa R, et al. (2014) Genetic changes involved in the juvenile-to-adult transition in the shoot apex of Olea europaea L. occur years before the first flowering. Tree Genet Genomes 10: 585–603 [Google Scholar]
- George EF, Hall MA, De Klerk G, editors (2008) Stock plant physiological factors affecting growth and morphogenesis . In Plant Propagation by Tissue Culture, Ed 3, Springer, Dordrecht, pp 205–226 [Google Scholar]
- Gibbons J ( 2014) Western blot: Protein transfer overview. N Am J Med Sci 6: 158–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Zhu Q, Gu X, Ge S, Yang J, Luo J (2008) Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418: 1–8 [DOI] [PubMed] [Google Scholar]
- Ning G, Yan X, Chen H, Dong R, Zhang W, Ruan Y, et al. (2020) Genetic manipulation of Soc1-like genes promotes photosynthesis in flowers and leaves and enhances plant tolerance to high temperature. Plant Biotechnol J 19: 8–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann HT, Kester DE, Davies FT Jr,, Geneve RL (1997) Plant propagation: principles and practices. Am Midl Nat 63: 253 [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directlyin vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Böhlenius H (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. Embo J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holefors A, Opseth L, Ree Rosnes A, K, Ripel L, Snipen L, Fossdal G, et al. (2009) Identification of PaCOL1 and PaCOL2, two CONSTANS-like genes showing decreased transcript levels preceding short day induced growth cessation in Norway spruce. Plant Physiol Biochem 47: 105–115 [DOI] [PubMed] [Google Scholar]
- Horvath P (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HF, Huang CH, Chou LT, Yang CH (2003) Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol 44: 783–794 [DOI] [PubMed] [Google Scholar]
- Hsu W, Yeh T, Huang K, Li J, Chen H, Yang C (2014) AGAMOUS-LIKE13, a putative ancestor for the E functional genes, specifies male and female gametophyte morphogenesis. Plant J 77: 1–15 [DOI] [PubMed] [Google Scholar]
- Immink RGH, Gadella TWJ, Ferrario S, Busscher M, Angenent GC (2002) Analysis of Mads-box protein-protein interactions in living plant cells. Proc Natl Acad Sci USA 99: 2416–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J-H, Ju Y, Pil J, et al. (2012) The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J 69: 577–588 [DOI] [PubMed] [Google Scholar]
- Jungeun L (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61: 2247–2254 [DOI] [PubMed] [Google Scholar]
- Katahata S, Futamura N, Igasaki T, Shinohara K (2014) Functional analysis of SOC1-like and AGL6-like MADS-box genes of the gymnosperm Cryptomeria japonica. Tree Genet Genomes 10: 317–327 [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MRG, Ai X, Zhang J (2014) Genetic regulation of flowering time in annual and perennial plants. RNA 5: 347–359 [DOI] [PubMed] [Google Scholar]
- Koo SC, Bracko O, Park MS, Schwab R, Chun HJ, Park KM, et al. (2010) Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J 62: 807–816 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Veen VDJH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose S (2005) microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA 26: 9412–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, et al. (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liang W, Jia R, Yin C, Zong J, Kong H, et al. (2010) The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Liao S, Cui K, Wan Y, Zhou Z, Li Z, Cui Y, Zhang P (2014) Reproductive biology of the endangered cypress Calocedrus macrolepis. Nord J Bot 32: 98–105 [Google Scholar]
- Liu S, , Ma J, , Liu H, , Guo Y, , Li W, , Niu S ( 2020) An efficient system for Agrobacterium-mediated transient transformation in Pinus tabuliformis. Plant Methods 16: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Deng S, Chen L, Jia Z, Sang Z, Zhu Z, et al. (2019) Gene duplication led to divergence of expression patterns, protein-protein interaction patterns and floral development functions of AGL6-like genes in the basal angiosperm Magnolia wufengensis (Magnoliaceae). Tree Physiol 39: 861–876 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377: 522–524 [DOI] [PubMed] [Google Scholar]
- Moon J, Suh S, Lee H, Choi K, Hong CB, Paek N, et al. (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Moyroud E, Kusters E, Monniaux M, Koes R, Parcy F (2010) LEAFY blossoms. Trends Plant Sci 15: 346–352 [DOI] [PubMed] [Google Scholar]
- Nie J, Stewart H, Zhang JA, Thomson F, Ruan X, Cui, Wei H (2011). TF-Cluster: a pipeline for identifying functionally coordinated transcription factors via network decomposition of the shared coexpression connectivity matrix (SCCM). BMC Syst Biol 5: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O, Brunner AM (2004) Revisiting tree maturation and floral initiation in the poplar functional genomics era. New Phytol 164: 43–51 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Carlsbecker A, Sundås-Larsson A, Vahala T (2007) APETALA2 like genes from Picea abies show functional similarities to their Arabidopsis homologues. Planta 225: 589–602 [DOI] [PubMed] [Google Scholar]
- Nilsson O (2009) Plant evolution: measuring the length of the day. Curr Biol 19: R302–R303 [DOI] [PubMed] [Google Scholar]
- Niu SH, Liu SW, Ma JJ, Han FX, Li Y, Li W (2019) The transcriptional activity of a temperature-sensitive transcription factor module is associated with pollen shedding time in pine. Tree Physiol 39: 1173–1186 [DOI] [PubMed] [Google Scholar]
- Niu S, Huwei Y, Xinrui S, Ilga PYL, Yousry A (2016) A transcriptomics investigation into pine reproductive organ development. New Phytol 209: 1278–1289 [DOI] [PubMed] [Google Scholar]
- Niu S, Li Z, Yuan H, Chen X, Li Y, Li W (2013) Transcriptome characterization of Pinus tabuliformis and evolution of genes in the Pinus phylogeny. BMC Genomics 14: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S, Liu C, Yuan H, Li P, Li Y, Li W (2015) Identification and expression profiles of sRNAs and their biogenesis and action-related genes in male and female cones of Pinus tabuliformis. BMC Genomics 16: 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin Y, Scofield DG, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L (2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690 [DOI] [PubMed] [Google Scholar]
- Poethig RS (2013) Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol 105: 125–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyanka M, Kishore CP (2015) GIGANTEA—an emerging story. Front Plant Sci 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheimer R, Kellogg EA (2009) Evolution of AGL6-like MADS-box genes in Grasses (Poaceae): ovule expression is ancient and palea expression is new. Plant Cell 21: 2591–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema A S, Zethof J, Gerats T, Vandenbussche M (2009) The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J 60: 1–9 [DOI] [PubMed] [Google Scholar]
- Roberts K, Shirsat AH (2006). Increased extensin levels in Arabidopsis affect inflorescence stem thickening and height. J Exp Bot 57: 537–545 [DOI] [PubMed] [Google Scholar]
- Samach A (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Shen G, Yang C, Shen C, Huang K (2019) Origination and selection of ABCDE and AGL6 subfamily MADS-box genes in gymnosperms and angiosperms. Biol Res 52: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigyo M, Hasebe M, Ito M (2006) Molecular evolution of the AP2 subfamily. Gene 366: 256–265 [DOI] [PubMed] [Google Scholar]
- Shim JS, Kubota A, Imaizumi T (2017) Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol 173: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohpal VK, Dey A, Singh A (2010) MEGA biocentric software for sequence and phylogenetic analysis: a review. Int J Bioinform Res Appl 6: 230–240 [DOI] [PubMed] [Google Scholar]
- Tandre K, Albert V A, Sundas A, Engstrom P (1995) Conifer homologues to genes that control floral development in angiosperms. Plant Mol Biol 27: 69–78 [DOI] [PubMed] [Google Scholar]
- Tandre K, Svenson M, Svensson ME, Engstrom P (1998) Conservation of gene structure and activity in the regulation of reproductive organ development of conifers and angiosperms. Plant J 15: 615–623 [DOI] [PubMed] [Google Scholar]
- Tapia-Lopez R, Garcia-Ponce B, Dubrovsky JG, Garay-Arroyo A, Perez-Ruiz RV, Kim SH, et al. (2008) An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol 146: 1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B E, Bartling L, Whipple C, Hall DH, Sakai H, Schmidt R, et al. (2009) bearded-ear encodes a MADS-box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddenberg D, Reimegård J, Clapham D, Almqvist C, von Arnold S, Emanuelsson O, et al. (2013) Early cone setting in Picea abies acrocona is associated with increased transcriptional activity of a MADS-box transcription factor. Plant Physiol 161: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden AR, Wang DY, Walter C, Gardner RC (1998) A large family of TM3 MADS-box cDNAs in Pinus radiata includes two members with deletions of the conserved K domain. Plant Sci 138: 167–176 [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-Regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, et al. (2011. ) miRNA control of vegetative phase change in trees. Plos Genet 7: e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter KU, Becker A, Munster T, Kim JT, Saedler H, Theissen G (1999) MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc Natl Acad Sci USA 96: 7342–7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Shen S, Lee L, Lee S, Chan M, Lin C (2009) Tape-Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang J, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang W, Li W, Zhang S, Qi L (2019) Transcriptome-wide analysis to dissect the transcription factors orchestrating the phase change from vegetative to reproductive development in Larix kaempferi. Tree Genet Genomes 15:68 [Google Scholar]
- Xie K (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, Zhao J, Park M-Y, Earley KW, Wu G, Yang L, Poethig RS (2016) Developmental functions of miR156-regulated squamosa promoter binding protein-like (SPL) genes in Arabidopsis thaliana. Plos Genet 12: e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Mao D, Liu X, Wang L, Xu F, Wang G, et al. (2017) Isolation and functional characterization of a circadian-regulated CONSTANS homolog (GbCO) from Ginkgo biloba. Plant Cell Rep 36: 1387–1399 [DOI] [PubMed] [Google Scholar]
- Yano M, Kojima S, Takahashi Y, Lin H, Sasaki T (2001) Genetic control of flowering time in rice, a short-day plant. Plant Physiol 127: 1425–1429 [PMC free article] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, et al. (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, et al. (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Wu X, Lee JS, Ahn JH (2011) AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J 65: 62–76 [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: e17. [DOI] [PubMed] [Google Scholar]
- Zhang M, Chen Y, Jin X, Cai Y, Yuan Y, Fu C, et al. (2019) New different origins and evolutionary processes of AP2/EREBP transcription factors in Taxus chinensis. BMC Plant Biol 19: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li J, Sang Y, Xing S, Wu Q, Liu X (2015) Identification and characterization of microRNAs in Ginkgo biloba var. epiphylla Mak. Plos One 10: e127184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cao G, Qu LJ, Gu H (2009) Characterization of Arabidopsis MYB transcription factor gene AtMYB17 and its possible regulation by LEAFY and AGL15. J Genet Genomics 36: 99–107 [DOI] [PubMed] [Google Scholar]
- Wang Z, Shen Y, Yang X, Pan Q, Ma G, Bao M (2019) Overexpression of particular MADS-Box transcription factors in heat stressed plants induces chloroplast biogenesis in petals: chloroplasts biogenesis in petals. Plant Cell Environ 42: 1545–1560 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62: 487–495 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and Supplementary Material online or are available upon request from the corresponding author. The RNA-seq data that support the findings of this study have been deposited in the CNSA (https://db.cngb.org/cnsa/) of China National GeneBank Database with accession number CNP0001648.