Abstract
Seed dormancy and germination are fundamental processes for plant propagation, both of which are tightly regulated by internal and external cues. Phytochrome B (phyB) is a major red/far-red-absorbing photoreceptor that senses light signals that modulate seed dormancy and germination. However, the components that directly transduce that signal downstream of phyB are mostly unknown. Here, we show that the transposase-derived transcription factor FAR-RED ELONGATED HYPOCOTYL3 (FHY3) inhibits seed dormancy and promotes phyB-mediated seed germination in Arabidopsis thaliana. FHY3 physically interacts with phyB in vitro and in vivo. RNA-sequencing and reverse transcription-quantitative polymerase chain reaction analyses showed that FHY3 regulates multiple downstream genes, including REVEILLE2 (RVE2), RVE7, and SPATULA (SPT). Yeast one-hybrid, electrophoresis mobility shift, and chromatin immunoprecipitation assays demonstrated that FHY3 directly binds these genes via a conserved FBS cis-element in their promoters. Furthermore, RVE2, RVE7, and GIBBERELLIN 3-OXIDASE 2 (GA3ox2) genetically act downstream of FHY3. Strikingly, light and phyB promote FHY3 protein accumulation. Our study reveals a transcriptional cascade consisting of phyB-FHY3-RVE2/RVE7/SPT-GA3ox2 that relays environmental light signals and thereby controls seed dormancy and germination.
Introduction
Light is a critical environmental factor for plant growth and developmental processes, such as seed germination, seedling morphogenesis, flowering time, shade avoidance, and leaf senescence. Multiple photoreceptors, including phytochromes (absorbing red and far-red light), cryptochromes and phototropins (absorbing blue and ultraviolet [UV]-A light), and UV RESISTANCE LOCUS 8 (UVR8, perceiving UV-B light), respond to different light qualities and trigger specific light-signaling pathways by relaying the signals to different sets of downstream components, leading to changes in gene expression and, ultimately, various physiological responses (Tilbrook et al., 2013; Legris et al., 2020; Wang and Lin, 2020). Phytochromes are master regulators of light responses and have been extensively studied. Phytochromes exist in either a red (R) light-absorbing, inactive Pr form or a far-red (FR) light-absorbing, bioactive Pfr form. The Arabidopsis (Arabidopsis thaliana) genome encodes five phytochrome apoproteins, namely PHYA–E. Whereas phyA is light labile, the others are light stable (Li et al., 2011a).
Red light converts the phytochrome Pr form to the Pfr form and triggers its translocation from the cytoplasm to the nucleus. There, one of the major functions of phytochromes is that the Pfr form interacts with the basic helix-loop-helix (bHLH) transcription factors PHYTOCHROME-INTERACTING FACTORs (PIFs), promoting their phosphorylation and degradation through the 26S proteasome pathway (Leivar and Quail, 2011; Leivar and Monte, 2014). The transposase-derived transcription factor FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and its homolog FAR-RED IMPAIRED RESPONSE1 (FAR1) play important roles in phytochrome A (phyA)-mediated light responses. Loss-of-function alleles of both genes are associated with elongated hypocotyls under far-red light conditions (Hudson et al., 1999; Wang and Deng, 2002). FHY3 and FAR1 modulate phyA nuclear accumulation and far-red light responses by directly activating FHY1 and FHY1-LIKE expression via their conserved FHY3/FAR1-binding site (FBS) cis-regulatory motif (Lin et al., 2007). A genome-wide chromatin immunoprecipitation sequencing study showed that FHY3 binds more than 1,000 target genes, suggesting that it has broad functions in Arabidopsis (Ouyang et al., 2011). However, the relationship between FHY3 and phyB remains poorly understood.
Seed dormancy and germination are two fundamental processes for plant propagation. Germination initiates a new life cycle, whereas dormancy is a vital fitness trait that allows seeds to withstand unfavorable environmental conditions and prevents undesirable pre-harvest sprouting (Bentsink and Koornneef, 2008; Nonogaki, 2019). Both processes are intimately connected but distinctly controlled by endogenous and exogenous cues. Gibberellin (GA) and abscisic acid (ABA) are two major endogenous phytohormones that antagonistically determine the states of dormancy and germination (Holdsworth et al., 2008; Shu et al., 2016). As an environmental signal, light affects the metabolism and signaling pathways of both GA and ABA and modulates seed germination (Zhao et al., 2007; Seo et al., 2009; Lau and Deng, 2010; de Wit et al., 2016). Accumulating evidence reveals that all phytochromes are involved in light-mediated seed germination; however, phyB plays a key role in this process (Shinomura et al., 1994, 1996, 1998; Botto et al., 1995, 1996; Poppe and Schafer, 1997; Hennig et al., 2002; Donohue et al., 2008; Lee et al., 2012). Notably, PIF1 acts as a central regulator in modulating red/far-red light-reversible seed germination via interaction with phyB. PIF1 directly or indirectly controls the expression of diverse downstream signaling factors, which eventually affect the signaling pathways and/or metabolic levels of GA and ABA (Oh et al., 2004, 2006, 2007; Seo et al., 2009; de Wit et al., 2016). SPATULA (SPT) is another bHLH transcription factor that controls light- and temperature-induced germination of dormant seeds (Penfield et al., 2005). Surprisingly, SPT has opposite roles in two Arabidopsis ecotypes, suppressing dormancy in Landsberg erecta but promoting it in Columbia (Col; Vaistij et al., 2013).
We previously demonstrated that phyB also controls seed dormancy independently of PIF1 (Jiang et al., 2016). Furthermore, two Myb-like transcription factors, REVEILLE1 (RVE1) and RVE2, promote primary seed dormancy maintenance, but repress red/far-red light-reversible germination downstream of phyB. RVE1/2 suppresses the biosynthesis of bioactive GA by directly binding the GIBBERELLIN 3-OXIDASE 2 (GA3ox2) promoter and inhibiting its transcription (Jiang et al., 2016). Thus, phyB-RVE1/2 may represent an uncharacterized pathway with dual roles in regulating seed dormancy and germination (Yang et al., 2020c). Although phyB represses RVE1 and RVE2 expression (Jiang et al., 2016), the underlying mechanism is unknown. In this study, we investigated the role of FHY3 in seed dormancy and germination. We showed that FHY3 is required for the germination of freshly harvested and after-ripened seeds upon treatment with far-red or red light. Moreover, we showed that FHY3 physically interacts with the phyB photoreceptor. Detailed molecular and genetic analyses demonstrated that FHY3 directly regulates RVE2, RVE7, and SPT by binding their promoters and that RVE2 and RVE7 act downstream of FHY3. Furthermore, light was found to promote FHY3 protein accumulation in a partially phyB-dependent manner and FHY3 was found to inhibit phyB expression during seed imbibition. Therefore, our study identifies a transcriptional module through which plants monitor the light environment and decide their germination state.
Results
FHY3 regulates both seed dormancy and germination
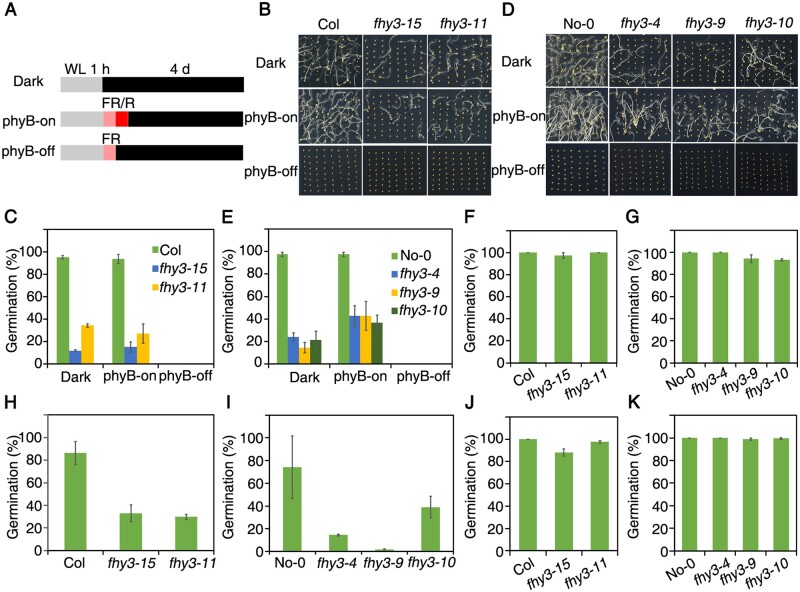
FHY3 is a positive regulator of seedling de-etiolation under far-red light. The fhy3-1 mutant did not show differences in seed germination compared to wild-type when the seeds were cold stratified for 3 d (Yanovsky et al., 2000). Hence, we investigated whether FHY3 was also involved in light-regulated seed germination without cold stratification. We performed germination assays under different light conditions (Figure 1A) using the after-ripened seeds of multiple fhy3 mutant alleles and their corresponding wild-types. In darkness or under phyB-on (a 5-min pulse of far-red light followed by a 5-min pulse of red light to activate phyB), fhy3-11 and fhy3-15 mutants had lower seed germination rates than the Columbia (Col) wild-type. However, the mutant seeds could not germinate under phyB-off conditions (a 5-min pulse of far-red light to inactivate phyB) similar to Col (Figure 1, B and C; Supplemental Figure S1, A–C). Similarly, fhy3-4, fhy3-9, and fhy3-10 had lower seed germination rates than the Nossen (No-0) wild-type in darkness or under phyB-on conditions (Figure 1, D and E). The after-ripened seeds of the fhy3 mutants almost fully germinated under a 16-h light and 8-h dark photoperiodic condition without cold stratification (Figure 1, F and G), suggesting that seed dormancy was released in these after-ripened fhy3 mutants.
Figure 1.
The fhy3 mutants have seed dormancy and germination responses. A, Diagram of light treatments. Seeds were sown on plates under white light (WL) within 1 h and then exposed to far-red (FR) light for 5 min followed by a subsequent 5-min red (R) light pulse (phyB-on) or not (phyB-off). Seeds were incubated in darkness at 22°C for 4 d before recording germination frequency. B and D, Germination phenotype. C and E, Germination frequency of fhy3 mutants and the corresponding wild-type as shown in B and D, respectively. F and G, Germination frequency under a 16-h white light and 8-h dark photoperiodic condition. For B–G, seeds were harvested and stored at room temperature for 9 months and germination was performed without cold stratification. H and I, Germination frequency of freshly harvested seeds without cold treatment. J and K, Germination frequency of freshly harvested seeds after 3 d of 4°C cold treatment. For C, E–K, data are means ± SD, n=3 biological repeats.
To further support this notion, we performed germination assays using different batches of seeds that were stored for different periods. As storage time was extended, the germination rate of Col wild-type gradually increased and reached almost 100% for seeds after-ripened for 9 months and germinated under phyB-on conditions. However, the germination rates of fhy3-11 and fhy3-15 were much lower than that of Col in different batches of seeds (Supplemental Figure S1, D–G). Together, these data indicate that FHY3 is indeed involved in regulating phyB-mediated seed germination. We also assessed phyA-mediated germination by illuminating the seeds with far-red light. The fhy3 mutants had lower germination rates than the wild-type (Supplemental Figure S1H). Thus, FHY3 positively regulates both phyB- and phyA-mediated seed germination.
Primary seed dormancy is developed during seed maturation and is predominantly high when seeds shed from mother plants. We found that FHY3 transcript levels gradually increased in developing siliques and peaked in dry seeds (Supplemental Figure S2; Winter et al., 2007). This expression pattern of FHY3 prompted us to explore whether it is involved in regulating seed dormancy. We examined the seed dormancy response under white light conditions using freshly harvested seeds. The germination rate was much lower in the fhy3 mutants than in the corresponding wild-type seeds (Figure 1, H and I). This decreased germination was not due to developmental defects, because all seeds germinated after 3 d of cold stratification (Figure 1, J and K). These results suggest that FHY3 inhibits seed dormancy.
FHY3 physically interacts with phyB
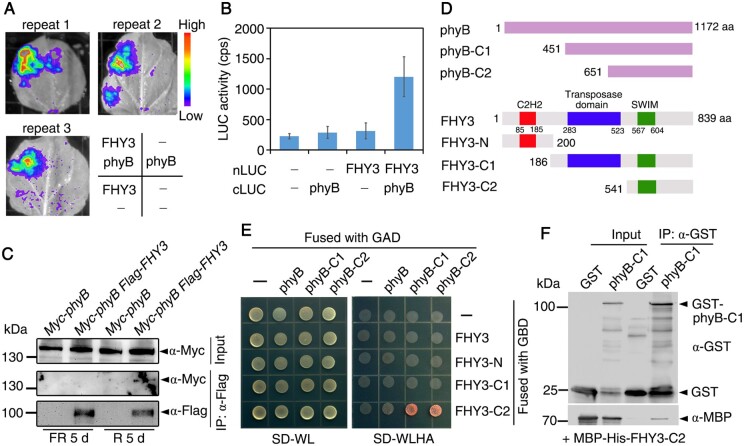
phyB is a main photoreceptor regulating seed dormancy and germination. The involvement of FHY3 in mediating seed dormancy and germination prompted us to explore whether FHY3 could interact with phyB. First, using a luciferase complementation imaging (LCI) assay, we observed strong positive luciferase luminescence signals in Nicotiana benthamiana leaves co-transformed with FHY3-nLUC (fused with the N-terminus of luciferase) and phyB-cLUC (fused with the C-terminus of luciferase; Figure 2, A and B). Second, we grew transgenic plants harboring 35S:Myc-phyB and 35S:Myc-phyB/35S:Flag-FHY3 in far-red or red light for 5 d to generate the Pr form or Pfr form of phyB, respectively, and performed subsequent immunoprecipitation analysis. An anti-Flag antibody was able to pull-down Myc-phyB protein in the double transgenic plants under red light, but not in far-red light (Figure 2C). This result further indicates that FHY3 interacts with the Pfr form of phyB in vivo. Third, we performed a yeast two-hybrid assay to determine the FHY3 and phyB domains that mediate their interaction. The C-terminus regions of FHY3 (FHY3-C2, 541–839 amino acids [aa] containing the SWIM domain) interacted strongly with the C-terminus of phyB (phyB-C1, 451–1,172 aa) or phyB-C2 (651–1,172 aa; Figure 2, D and E). Conversely, the N-terminus (FHY3-N, 1–200 aa containing the C2H2 DNA-binding domain) or the middle and C-terminal parts of FHY3 (FHY3-C1, 186–839 aa containing both the transposase domain and SWIM domain) failed to interact with phyB-C1 (Figure 2, D and E). These data suggest that FHY3 and phyB interact through their C-terminal regions. The SWIM domain of FHY3 likely mediates this interaction, whereas the transposase domain may interfere with the interaction in yeast cells. Finally, we incubated recombinant GST-phyB-C1 (fused with glutathione S-transferase) and MBP-His-FHY3-C2 (fused with maltose-binding protein) proteins in vitro and performed pull-down assays. The anti-GST antibody pulled down MBP-His-FHY3-C2 in samples co-incubated with GST-phyB-C1, but not in those co-incubated with GST alone (Figure 2F). These observations confirm that FHY3 physically interacts with phyB in vivo and in vitro.
Figure 2.
phyB physically interacts with FHY3. A, LCI assay. FHY3 and phyB were fused with either the N- or C-terminus of luciferase (LUC). Different combinations of constructs were co-transformed into N. benthamiana leaves and LUC luminescence was monitored. B, Quantification of relative LUC levels (counts per second, cps) as shown in (A). Data are means ± SD, n=3. C, Co-IP assay. Seedlings were grown in red or far-red light for 5 d. Total proteins were immunoprecipitated with anti-Flag antibody followed by blotting with anti-Flag or anti-Myc. D, Diagram of FHY3 and phyB and their truncations. Numbers indicate positions of the amino acid residues. E, Yeast two-hybrid assay. FHY3 and its fragments were fused with the GAL4 DNA binding domain (GBD), whereas phyB and its fragments were fused with the GAL4 activation domain (GAD). SD-WL indicates synthetic dropout medium lacking Trp and Leu; SD-WLHA denotes synthetic dropout medium lacking Trp, Leu, His, and Ade. F, In vitro pull-down assay. After incubation, the recombinant proteins were immunoprecipitated with anti-GST antibody followed by blotting with anti-GST or anti-MBP.
FHY3 regulates RVE2, RVE7, and SPT expression
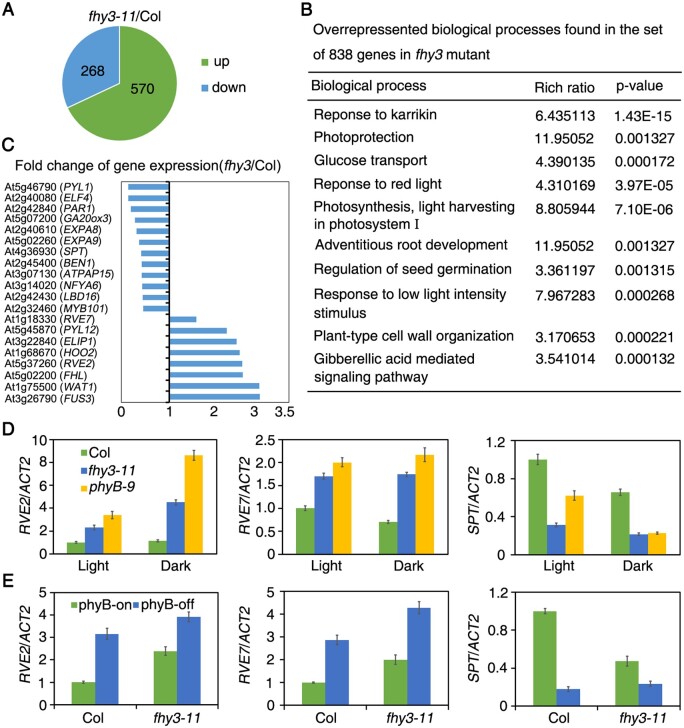
The discovery that FHY3 modulates seed dormancy prompted us to investigate how FHY3 regulates downstream gene expression. Freshly harvested Col and fhy3-11 seeds were imbibed for 24 h under white light and total RNAs were extracted for RNA-seq analysis. We found that 268 and 570 genes were up- or downregulated (1.5-fold changes with P ≤ 0.05) in the fhy3 mutant, respectively (Supplemental Data Set S1). Gene ontology (GO) analysis revealed that these genes are involved in biological processes, including light and hormone responses, photosynthesis, glucose transport, seed germination, root development, and plant-type cell organization (Figure 3B).
Figure 3.
FHY3 regulates downstream gene expression. A, Venn diagram showing the overlap of differentially regulated genes in fhy3-11 compared to Col wild-type. Freshly harvested seeds were imbibed under white light for 24 h before RNA extraction. B, GO analysis of commonly regulated genes by FHY3. C, List of partial FHY3-regulated genes from RNA-seq. D, RT-qPCR analysis of transcript accumulation in Col wild-type, fhy3-11, and phyB-9 mutants. Freshly harvested seeds were imbibed under white light or in darkness for 24 h. E, RT-qPCR analysis of gene expression in Col and fhy3-11 under phyB-on and phyB-off conditions. After-ripened seeds were imbibed for 24 h. For (D) and (E), data are means ± SD, n=3 biological replicates. Relative expression was normalized to the expression level of Actin2.
FHY3 was seen to either positively or negatively regulate genes involved in cell expansion and embryogenesis, which are closely related to seed germination (Figure 3C). To further confirm these expression changes, we performed reverse transcription quantitative polymerase chain reaction (RT-qPCR) using freshly harvested seeds imbibed for 24 h. RVE2 and RVE7 transcript levels were higher in fhy3-11, fhy3-15, and phyB-9 than in Col wild-type seeds. Conversely, SPT expression was lower in all mutants than in Col (Figure 3D; Supplemental Figure S3). Similarly, in fhy3-11 under both phyB-on and -off conditions, RVE2 and RVE7 expression was higher, whereas SPT expression was lower than that in Col (Figure 3E). These results indicate that FHY3 suppresses RVE2 and RVE7 but promotes SPT expression in the Col ecotype. Consistently, the freshly harvested rve7 seeds exhibited higher germination rates than the wild-type, whereas RVE7 overexpression (EPR1-OX; Zhang et al., 2007) led to reduced seed germination (Supplemental Figure S4).
FHY3 directly binds to RVE2, RVE7, and SPT promoters
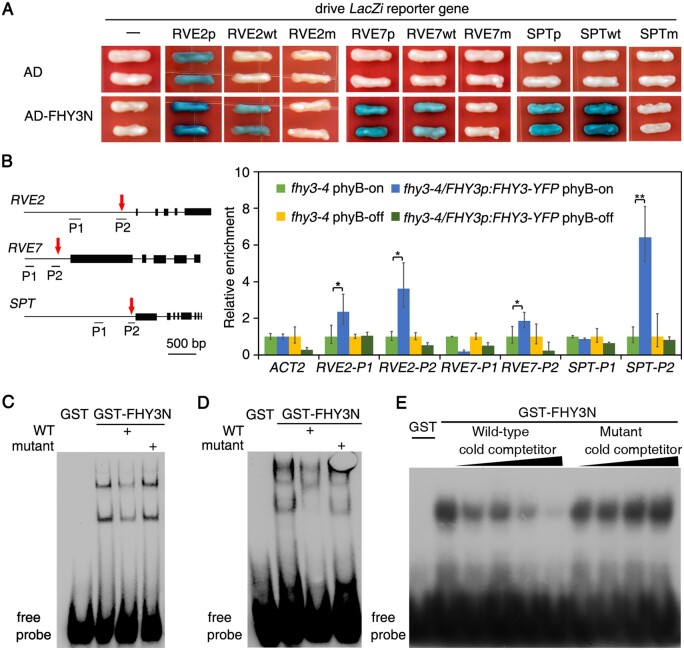
The transcriptome data indicated that FHY3 may directly or indirectly regulate downstream genes. Sequence analysis revealed that a conserved putative FHY3/FAR1-binding site (FBS, CACGCGC, Lin et al., 2007) is present in the RVE2, RVE7, and SPT promoters. We first performed a yeast one-hybrid assay to test the DNA–protein interaction. We generated constructs with the LacZ reporter gene driven by the promoters or promoter fragments containing the wild-type or mutated RVE2, RVE7, and SPT FBS motifs. Incubation of RVE2p:LacZ, RVE7p:LacZ, or SPTp:LacZ with AD-FHY3N (N-terminal 200 aa of FHY3 fused with a B42 activation domain) resulted in strong LacZ expression, although RVE2p:LacZ exhibited an activation background (Figure 4A). Furthermore, AD-FHY3N bound the wild-type oligonucleotide containing the wild-type RVE2, RVE7, and SPT FBS motifs (RVE2wt:LacZ, RVE7wt:LacZ, and SPTwt:LacZ, respectively) and activated reporter expression, but it did not bind the mutant oligonucleotides in the FBS motif (RVE2m:LacZ, RVE7m:LacZ, and SPTm:LacZ; Figure 4A).
Figure 4.
FHY3 directly binds to RVE2, RVE7, and SPT. A, Yeast one-hybrid assay. LacZ reporter gene was driven by promoter (p), promoter fragment containing the FBS motif (wt), or fragment with mutation in the FBS motif (m) of RVE2, RVE7, and SPT. The reporter plasmids were co-transformed with pB42AD or AD-FHY3N into yeast strain EGY48. B, ChIP assay. The after-ripened seeds were imbibed for 24 h under phyB-on or phyB-off conditions. Diagrams of gene structure and primer-binding positions (P1, P2) for PCR are shown in the left panels. Black bars show exons. Red arrows denote the positions of the FBS motif. Data are means ± SD, n=3 replicates. The * and ** indicates significant differences between two columns according to F test (ANOVA) at the levels of 5% or 1%, respectively. C–E, EMSA. FHY3N recombinant proteins were incubated with the biotin-labeled oligonucleotides of RVE2 C, RVE7 D, or 32P-labeled SPT E in the presence of unlabeled wild-type (WT) or mutant cold competitors.
To test if FHY3 binds these regions in vivo during seed germination, we carried out a chromatin immunoprecipitation (ChIP) assay using imbibed fhy3-4 and fhy3-4 FHY3p:FHY3-YFP seeds (Supplemental Figure S5) subjected to phyB-on or phyB-off treatment. DNA fragments (P2) spanning the RVE2, RVE7, and SPT FBS motifs were significantly enriched only in the fhy3-4 FHY3p:FHY3-YFP phyB-on treatment chromatin samples precipitated with anti-GFP mAb-Agarose, but not in the fhy3-4 control or the fhy3-4 FHY3p:FHY3-YFP phyB-off treatment samples (Figure 4B). We did not observe any differences for the ACT2 control (Figure 4B). These results suggest that the interaction between FHY3 and RVE2, RVE7, and SPT promoters is likely dependent on phyB action.
Finally, we performed an electrophoresis mobility shift assay (EMSA) using a GST-FHY3N recombinant protein. Incubation of GST-FHY3N with wild-type RVE2, RVE7, or SPT oligonucleotide probes created upshift bands. It should be noted that there might exist both monomer and dimer forms for binding RVE2 and RVE7. These upshifted bands were abolished or reduced by excess unlabeled wild-type oligonucleotides, but not by unlabeled mutant oligonucleotides (Figure 4, C–E). These results indicate that FHY3 directly binds RVE2, RVE7, and SPT via their specific FBS motifs.
RVE2 and RVE7 act downstream of FHY3
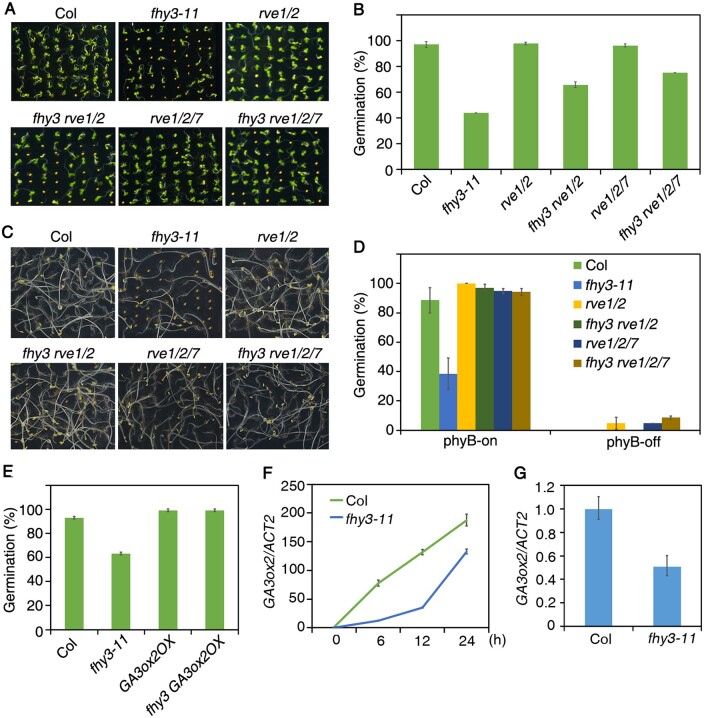
To uncover the genetic relationships between FHY3 and RVE2, RVE7, and SPT, we generated double or triple mutants by crossing. The germination rates of the freshly harvested seeds of fhy3 rve1 rve2 triple mutants and fhy3 rve1 rve2 rve7 quadruple mutants were significantly higher than that of the fhy3 single mutant under white light treatment (Figure 5, A and B). In addition, the after-ripened fhy3 rve1 rve2 and fhy3 rve1 rve2 rve7 seeds basically rescued the reduced germination phenotype of fhy3 under phyB-on conditions (Figure 5, C and D). Besides, these after-ripened seeds could completely germinate under white light (Supplemental Figure S6A). These data indicate that RVE2 and RVE7 (and RVE1) act downstream of FHY3. We were not able to study the relationship between FHY3 and SPT because adult fhy3 spt homozygous plants produced shrunken siliques without seeds (Supplemental Figure S6B), suggesting that FHY3 and SPT are essential for plant reproductive development.
Figure 5.
Genetic interaction between FHY3 and RVE2/RVE7 and GA3ox2. A, Germination phenotype of Col wild-type and various mutants. Freshly harvested seeds were germinated under white light condition for 4 d. B, Germination frequency of seeds as shown in (A). C, Germination phenotype of Col wild-type and various mutants. After-ripened seeds were germinated under phyB-on conditions. D, Germination frequency of seeds as shown in (C). E, Germination frequency of fhy3, GA3ox2-OX, and their double mutants. F and G, GA3ox2 expression in Col versus fhy3-11 mutant in freshly harvested seeds during imbibition for up to 24 h under white light (F) or in post-harvested seeds after imbibition for 24 h under phyB-on condition (G). For (B), (D–G), data are means ± SD, n=3 replicates. Relative expression was normalized to the expression level of Actin2.
Because GA3ox2 acts downstream of RVE2 (Jiang et al., 2016), we introduced a GA3ox2-OX overexpression transgenic line into fhy3. Overexpression of GA3ox2 restored fhy3-11 seed germination to wild-type levels (Figure 5E), suggesting that GA3ox2 is epistatic to FHY3. In agreement, GA3ox2 expression was lower in fhy3 than in Col during imbibition of the freshly harvested and after-ripening seeds under phyB-on conditions (Figure 5, F and G). Besides, we also tested GA3ox2 transcription levels in freshly harvested seeds of the spt-12 mutant and found that GA3ox2 expression was lower in spt-12 than in Col after 24 h imbibition (Supplemental Figure S6C), which suggests that SPT promotes GA3ox2 expression during early imbibition stage.
Light promotes FHY3 protein accumulation, and FHY3 feedback regulates phyB transcription during seed imbibition
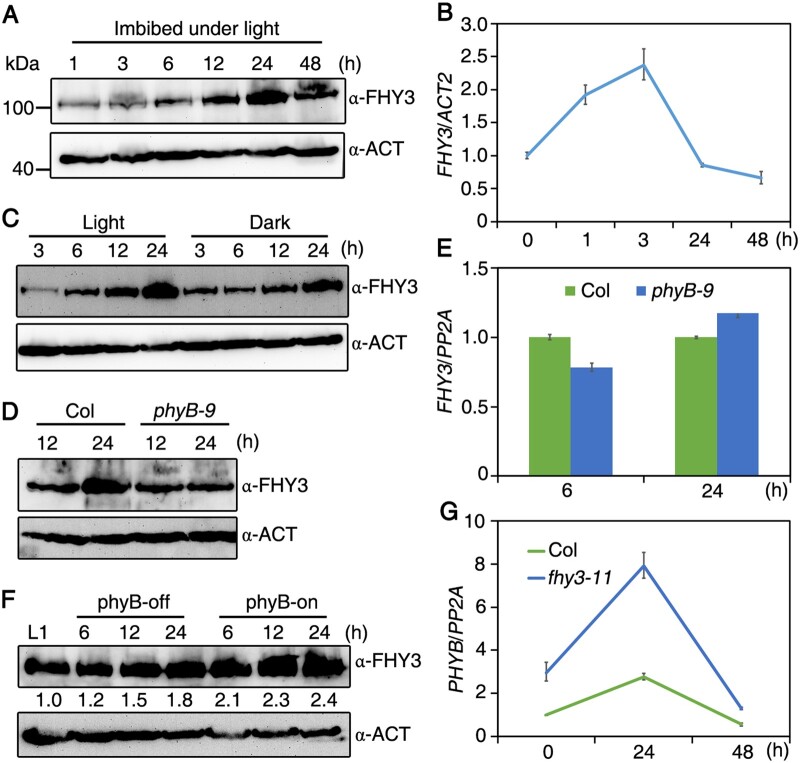
Next, we examined how light and phyB regulate FHY3 expression. We investigated FHY3 protein level in freshly harvested seeds as they were imbibed under white light. Immunoblot analysis revealed that FHY3 accumulated gradually and peaked after 24 h of seed imbibition (Figure 6A). FHY3 transcript levels slightly increased after 3 h of imbibition and subsequently dropped to a steady level (Figure 6B). Furthermore, when the Col seeds were imbibed under white light, the FHY3 protein level was higher than that in darkness (Figure 6C). In addition, the phyB-9 mutants had markedly lower FHY3 level than Col after 24 h of imbibition under white light (Figure 6D). However, FHY3 expression did not drastically differ in phyB-9 compared to Col after seed imbibition (Figure 6E). Furthermore, in the after-ripened seeds, FHY3 was increased under phyB-on but not phyB-off conditions (Figure 6F). Thus, light and phyB promote FHY3 accumulation during seed imbibition.
Figure 6.
phyB positively regulates FHY3 protein accumulation and FHY3 inhibits phyB expression. A, Immunoblot analysis of FHY3. Freshly harvested Col wild-type seeds were imbibed under white light for up to 48 h, and total proteins were blotted with anti-FHY3 or anti-Actin antibody. B, RT-qPCR analysis of FHY3 expression in freshly harvested Col seeds during imbibition. C, Immunoblot analysis of FHY3. Freshly harvested Col wild-type seeds were imbibed under white light or dark for 3, 6, 12, or 24 h, and total proteins were blotted with anti-FHY3 or anti-Actin antibody. D, Immunoblot analysis of FHY3. Freshly harvested seeds of Col and phyB-9 mutants were imbibed under white light for 12 or 24 h. E, RT-qPCR analysis of FHY3 expression in Col and phyB-9 mutants imbibed for 6 or 24 h under white light. F, Immunoblot analysis of FHY3. After-ripened Col seeds (exposure to white light for 1 h) were treated under phyB-on or phyB-off conditions and imbibed for the indicated periods. Values below the bands indicate relative amount of the blotting signals normalized to the Actin controls. G, RT-qPCR analysis of PHYB expression in Col and fhy3-11 mutants imbibed for 24 or 48 h under white light. For (B), (E), and (G), data are means ± SD, n=3 biological replicates. Relative expression was normalized to the level of Actin2 or PP2A.
We previously demonstrated that FHY3 binds the phyB promoter through the FBS motif in yeast cells (Lin et al., 2007). RT-qPCR analysis indicated that phyB mRNA level was increased in the fhy3-11 mutant compared to Col control during seed imbibition (Figure 6G), suggesting that FHY3 feedback represses phyB expression.
Discussion
Seed dormancy and germination are two closely connected but distinct processes (Nonogaki, 2019). Previous studies demonstrated that PIF1 represses phyB-mediated seed germination through directly interacting with phyB and regulating gene expression (reviewed in de Wit et al., 2016; Seo et al., 2009; Yang et al., 2020c). However, phyB controls both seed dormancy and germination, whereas PIF1 only modulates seed germination. The dual function of phyB relies on the RVE1 and RVE2 transcription factors. phyB and RVE1/2 thus define a distinct pathway independent of PIF1 (Jiang et al., 2016; Yang et al., 2020c). We recently also demonstrated that RVE1 and PIF1 interact and form a transcriptional feedback loop that coordinately inhibits seed germination (Yang et al., 2020a). Our current study identifies a role for FHY3 in regulating seed dormancy and germination and establishes a link between phyB and the RVE factors through FHY3.
We showed that FHY3 physically interacts with phyB and that it regulates the transcription of RVE2, RVE7, and SPT, which are also involved in seed germination (Penfield et al., 2005; Jiang et al., 2016). Yeast one-hybrid and ChIP experiments demonstrated that FHY3 directly binds to the specific FBS motifs located in the promoters of RVE2, RVE7, and SPT (Figure 4). Previous ChIP-sequencing data also indicated that RVE2 and SPT are targets of FHY3 in seedlings (Ouyang et al., 2011). Genetic analysis suggested that RVE2 and RVE7 act downstream of FHY3 (Figure 5, A–D). Additionally, FHY3 represses seed dormancy and promotes phyB-mediated seed germination, in a manner opposite to RVE1 and RVE2 (Figure 1; Jiang et al., 2016). RVE2 and SPT directly regulate the expression of GA3ox2, whose encoded enzyme produces bioactive GAs (Penfield et al., 2005; Jiang et al., 2016). Consequently, overexpression of GA3ox2 complements fhy3 mutant phenotypes (Figure 5E). Hence, FHY3 bridges the gap between phyB and RVE2, RVE7, and SPT. This phyB-FHY3-RVE2/RVE7/SPT-GA3ox2 transcriptional cascade perceives and sequentially transduces light signals that regulate endogenous GA biosynthesis and, probably, other metabolic pathways, thus modulating seed dormancy and germination (Figure 7). However, FHY3 may indirectly regulate RVE1, as the RVE1 promoter does not possess a typical FBS motif. RVE proteins also integrate GA signaling in seed germination through regulating protein stability of REPRESSOR OF GA-LIKE2, a DELLA factor in the GA signaling pathway (Yang et al., 2020b). It will be interesting to investigate whether the RVE2, RVE7, and SPT transcription factors coordinately or independently regulate downstream gene expression.
Figure 7.
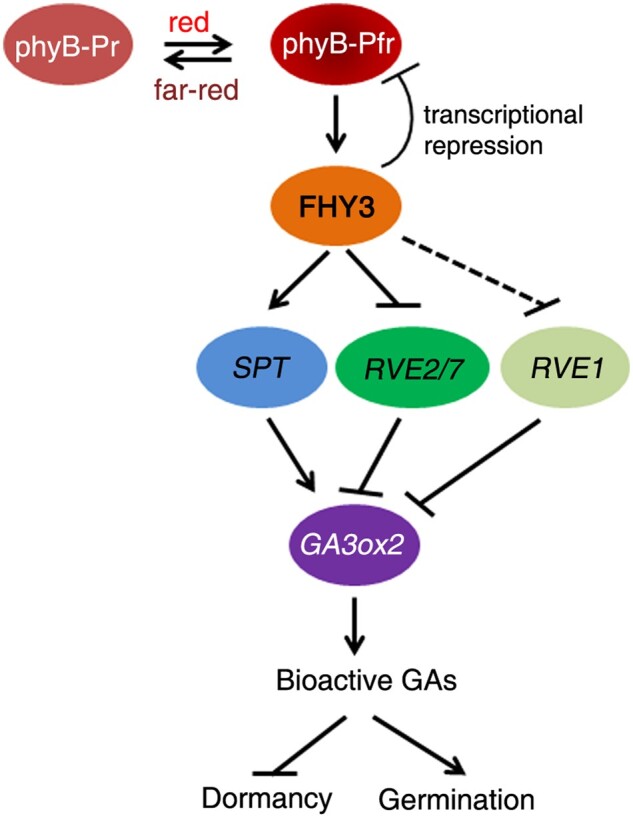
The proposed working model. Light-activated phyB interacts with FHY3 and promotes its protein accumulation, whereas FHY3 negatively feedback inhibits phyB expression. FHY3 directly regulates the expression of SPT (in the Col ecotype), RVE2, and RVE7, but indirectly regulates RVE1 expression. The encoded transcription factors in turn control GA3ox2 transcript levels, resulting in a change of bioactive GA levels and thus seed dormancy or germination.
We propose that the physical interaction between FHY3 and phyB efficiently transduces the light signal to FHY3 and that phyB has at least two biochemical effects on FHY3. Firstly, upon light activation, phyB moves into the nucleus and promotes the FHY3-mediated transcription of downstream gene expression. However, FHY3 only exhibits transcriptional activation activity (Lin et al., 2007). Other transcriptional co-repressor(s) might associate with FHY3 and phyB to inhibit RVE2 and RVE7 expression. PIF1 could function as a co-repressor as it interacts with FHY3 to repress its activation of chlorophyll biosynthesis (Tang et al., 2012). A recent study shows that DET1 is a co-repressor for FHY3 to inhibit ABI5 expression during seeding greening (Xu et al., 2020). DET1 also plays an important role in phyB-mediated seed germination by promoting PIF1 stability and degrading HFR1 protein (Shi et al., 2015). DET1 could act as a co-repressor candidate of FHY3. RVE2 has higher expression in phyB-9 mutants in darkness (Figure 3D) and RVE2 and RVE7 transcripts are increased in fhy3-11 compared to Col under phyB-off conditions (Figure 3E), suggesting that FHY3 is partially independent of phyB in regulating seed germination. Indeed, FHY3 is also involved in regulating phyA-mediated seed germination.
Previous studies also support the transcriptional regulatory activity of phyB. phyB interacts with and regulates the transcriptional or DNA-binding activity of BRASSINAZOLERESISTANT 1 (BZR1) and BZR2, thereby repressing brassinosteroid signaling (Dong et al., 2019; Wu et al., 2019). FHY3 protein accumulation was reduced in the phyB mutant background and under phyB-off conditions compared to under phyB-on (Figure 6). This suggests that red light promotes FHY3 protein accumulation during seed imbibition mostly through phyB. FHY3 protein level is reversely regulated by red and far-red light under photoperiod conditions (Siddiqui et al., 2016), and its abundance is increased by light in seedlings (Liu et al., 2017; Xie et al., 2020a). Therefore, light-triggered FHY3 accumulation promotes seed germination and seedling development at the early stages of a plant’s life cycle. phyB likely regulates FHY3 primarily at the post-translational level, but the underlying mechanism requires further investigation. Furthermore, FHY3 directly and negatively regulates phyB expression (Lin et al., 2007; Figure 6G). The inhibitory effect of FHY3 on phyB may contribute to the phenotype of fhy3 mutants, which, unlike the phyB-9 mutants, germinated under phyB-on conditions. This negative feedback regulation by FHY3 likely slows phyB signaling during seed germination in response to changing light conditions.
These data indicate that the light-triggered phyB-FHY3 interaction promotes FHY3 transcriptional activity. Conversely, phyB-PIF1 interaction results in PIF1 degradation and thus the release of its inhibitory effect on seed germination (Shen et al., 2005). Therefore, the phyB photoreceptor relays light signals to either positive (e.g. FHY3) or negative (e.g. PIF1) regulators using distinct pathways and mechanisms that control seed germination. Alternatively, the FHY3- and PIF1-mediated pathways could be integrated as PIF1 interacts with FHY3 and inhibits the transcriptional activity of FHY3 (Tang et al., 2012). Notably, RVE1 and PIF1 also physically interact and regulate their respective transcriptional activities (Yang et al., 2020a). FHY3 also plays a role in phytochrome-mediated circadian regulation (Allen et al., 2006, Siddiqui et al., 2016). In addition, FHY3 mediates phyA signaling in controlling photomorphogenesis by directly regulating the expression of FHY1 and FHL, which in turn control the nuclear translocation of phyA (Lin et al., 2007). Whether FHY3 interacts with phyA to modulate phyA-mediated seed germination is still an open question. Nevertheless, in addition to controlling phyA signaling, FHY3 regulates the phyB-mediated pathway, revealing its broad roles in relaying phytochrome-derived signals.
Increasing evidence has shown that FHY3 plays essential roles in regulating diverse plant growth and developmental processes, such as chloroplast division, circadian clock entrainment, branching, flowering time, inositol biosynthesis, reactive oxygen species homeostasis, plant immunity, and phosphate starvation (Wang and Wang, 2015; Liu et al., 2017, 2019; Ma et al., 2016, 2019; Xie et al., 2020a, 2020b). These processes are all involved in the vegetative or reproductive growth stages. In this study, we identified a new function of FHY3 in regulating seed germination, pointing out the universal roles of FHY3 in controlling plant development and responses to environmental stimuli. Light quality and quantity both affect seed dormancy establishment during seed maturation (He et al., 2014, 2016). FHY3 transcripts accumulate during seed development and are highly expressed in dry seeds (Supplemental Figure S2), consistent with the negative role of FHY3 in regulating seed dormancy, whereas its biological consequence in this response requires further investigation. Our study supports the biochemical role of FHY3 in directly binding target genes and controlling their expression (Lin et al., 2007) and shows that FHY3 directly transduces signals from a photoreceptor (phytochrome) to control a specific biological process.
Methods
Plant materials and growth conditions
The Arabidopsis (Arabidopsis thaliana) wild-type; the mutants fhy3-11 (Salk_002711; Stirnberg et al., 2012), fhy3-15 (Salk_059264), phyB-9 (Reed et al., 1993), rve2-1 (Salk_051843), rve7-1 (Salk_047716), rve7-2 (SAIL_195_F11), and spt-12 (CS853891, Ichihashi et al., 2010); and the transgenic lines 35S:Myc-phyB (Jiang et al., 2016), GA3ox2-OX (Jiang et al., 2016), and EPR1-OX (RVE7 overexpressing line; Kuno et al., 2003; Zhang et al., 2007) are of the Columbia (Col) ecotype unless otherwise indicated. The mutants fhy3-4, fhy3-9, fhy3-10, fhy3-4 FHY3p:FHY3-YFP and 35S:Flag-FHY3 are of Nossen (No-0) ecotype (Wang and Deng, 2002; Lin et al., 2008; Li et al., 2011b). Double mutants (including those in the background of transgenic plants) were produced by genetic crossing and homozygous lines were used in all experiments. Far-red, red, and white light were supplied by light-emitting diode sources. Plants were grown in a growth chamber with regular irrigation at 22 ± 2°C, 60%–70% humidity, and under long-day (16 h light/8 h dark) conditions. Seeds were harvested at the same developmental stage in each batch.
Dormancy and germination assay
The seed dormancy and germination phenotypes were determined as previously described (Jiang et al., 2016). Briefly, for seed dormancy experiments, freshly harvested seeds were surface sterilized and plated on 0.6% (w/v) agar (pH 5.7) within an hour. Seeds were either stratified or not at 4°C for 3 d, which was then followed by incubation in darkness or white light (80 µmol m−2 s−1) for 4 d and scoring of the germination rate. For phyB-mediated germination experiments, seeds were dry-stored at room temperature for different periods as stated in the text. After sterilization and plating (within 1 h), seeds were incubated in darkness or exposed to far-red light (12 µmol m−2 s−1) for 5 min to inactivate phyB (phyB-off), which was followed by 5 min of red light (10 µmol m−2 s−1) to activate phyB (phyB-on) or not. Seeds were kept in darkness for 4 d and the germination frequency was determined. For phyA-mediated germination assay, post-harvested seeds were imbibed for 1 h and irradiated with far-red light for 5 min. After 48 h of dark incubation, the seeds were irradiated with far-red light for 12 h, and germination frequency was determined after 48 h in darkness. Seeds with a protruded radicle were considered as germinated seeds.
Vector construction
To generate yeast two-hybrid constructs for phyB and FHY3, the fragments of FHY3 (1-839 aa), FHY3-N (1–200 aa), FHY3-C1 (186–839 aa), and FHY3-C2 (541–839 aa) were amplified and cloned into the pEASY-blunt vector (TransGen), resulting in pEASY-FHY3, -FHY3-N, -FHY3-C1, and -FHY3-C2, respectively. FHY3, FHY3-C1, FHY3-C2, and FHY3-N were released from pEASY fusing vector cut with Mfe I (or EcoR I, for FHY3-N) and Sal I, and were ligated into the EcoR I-Sal I sites of pGBKT7 vector (Clontech) to generate GBD-FHY3, -FHY3-N, -FHY3-C1, and -FHY3-C2, respectively. For the GAD-fusion constructs, fragments of phyB-C1 (451–1,172 aa) and phyB-C2 (651–1,172 aa) were amplified and cloned into pEASY-blunt vector, these fragments were cut with Mfe I and Xho I and ligated into EcoR I- Xho I sites of pGADT7 vector (Clontech) to generate GAD-phyB-C1 and -phyB-C2, respectively. For GAD-phyB, the full-length coding sequence of phyB was amplified and cloned into the EcoR I-BamH I sites of pGADT7 vector through recombination (YEASEN, 10911ES25).
To construct vectors for the transient LCI assay, the full-length coding sequence of phyB was amplified and cloned into Kpn I and Sal I sites of pCAMBIA1300-cLUC through recombination, to generate pCAMBIA1300-cLUC-phyB constructs. The coding sequence of FHY3 was amplified and inserted into pEASY vector to get pEASY-FHY3. FHY3 was released from pEASY-FHY3 and ligated into the Kpn I-Sal I sites of pCAMBIA1300-nLUC to generate pCAMBIA1300-FHY3-nLUC.
For pull-down assay, the fragment of phyB-C1 was released from pEASY-phyB-C1 through digesting with Mfe I and Xho I and ligated into the EcoR I-Xho I sites of pGEX-5X-1 (GE Healthcare) to give rise to GST-phyB-C1. For MBP-His-FHY3-C2, the fragment was released from pEASY-FHY3-C2, and ligated into the EcoR I-Sal I sites of pETMALc-H.
To prepare constructs for the yeast one-hybrid assay, the promoter fragments of RVE2, RVE7, and SPT were PCR amplified from Col genomic DNA and ligated into the pEASY vector to give rise to pEASY-RVE2p, pEASY-RVE7p, and pEASY-SPTp, respectively. RVE2p, RVE7p, and SPTp were released from pEASY vectors and inserted into the EcoR I-Xho I sites (for RVE2p) or Kpn I-Sal I sites (for RVE7p and SPTp) of pLacZi2µ (Lin et al., 2007) to generate RVE2p:LacZ, RVE7p:LacZ, and SPTp:LacZ, respectively. To construct LacZ reporter genes driven by the sub-fragments of the promoters (including the wild-type and mutant form of the FBS), about 40-bp oligonucleotides were synthesized as two complementary primers with an EcoR I site overhang at the 5ʹ end and Xho I site overhang at 3ʹ end. The oligo primers were annealed and the double-stranded oligonucleotides were ligated into the EcoR I-XhoI sites of pLacZi2µ to produce RVE2wt:LacZ, RVE7wt:LacZ, SPTwt:LacZ, RVE2m:LacZ, RVE7m:LacZ, and SPTm:LacZ, respectively. To obtain JG-FHY3N, the fragment of about 200 aa was amplified through PCR from the initiation codon ATG and cloned into pEASY vector to give rise to pEASY-FHY3N. FHY3N was released and ligated into the EcoR I-Xho I sites of pB42AD (Clontech).
The oligonucleotide sequences used in vector construction are shown in Supplemental Table S1.
RT-qPCR and RNA-seq assays
Total RNA was isolated from seeds using RNA extraction kit (Biotech, RP3302). The first-strand cDNA was synthesized using reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. RT-qPCR was performed using the TB GreenTM Premix Ex Taq (TaKaRa). Expression level of ACTIN2 or PP2A was used as the internal control and at least three biological replicates were analyzed. The primers used for RT-qPCR are shown in Supplemental Table S1.
For RNA-seq analysis, freshly harvested Col-0 and fhy3-11 seeds were imbibed for 24 h under white light and total RNA was extracted. The experiments were performed in three biological replicates. Data analysis was performed as previously described (Huai et al., 2018).
Purification of recombinant protein
GST, GST-FHY3N, GST-phyB-C1, and His-FHY3-C2 recombinant fusion proteins were expressed in Escherichia coli BL21 (DE3) strain and induced by isopropyl β-D-thiogalactopyranoside at 16°C. These proteins were purified using Glutathione Sepharose 4B beads (GE Healthcare) or Ni-NTA Agarose (QIAGEN) following the respective manufacturer’s instructions.
EMSA
EMSA experiments were performed using LightShift Chemiluminescent EMSA kit (Pierce) according to the manufacturer’s protocol or as previously described (for 32P-labeled probes, Tang et al., 2012). Briefly, 0.5 μg GST or GST-FHY3N was incubated together with biotin-labeled probes (RVE2 and RVE7) or 32P-labeled probes (SPT) in the presence or absence of excess amounts of unlabeled competitors for 20 min at room temperature. The protein–DNA samples were separated on 6% polyacrylamide gels. The oligonucleotide sequences were shown in Supplemental Table S1.
Yeast assays
For yeast one-hybrid, various LacZ reporter plasmids were co-transformed with AD-FHY3N into yeast strain EGY48, and the transformants were grown on SD/-Ura-Trp dropout plates containing X-gal (5-bromo-4-chioro-3-indodyl-β-D-galactopyranoside) for color development. For yeast two-hybrid, different combinations of GAD- and GBD-fusion plasmids were co-transformed into yeast strain Y2H Gold. The transformants were grown on SD/-Trp-Leu-His-Ade dropout plates for interaction analysis.
Protein immunoblotting
Total protein from seeds was isolated as previously described (Oh et al., 2006). The protein extracts were electrophoresed on a 10% SDS-PAGE gel and semi-dry transferred to a polyvinylidene difluoride membrane. The proteins were immunoblotted with anti-FHY3 (Li et al., 2010) or anti-ACT (Cwbio, CW0264M) primary antibodies followed by blotting with horseradish peroxidase-conjugated secondary antibody (Cwbio, CW0102S). The immunoblotting bands were visualized with a chemiluminescence imaging system (Biostep).
Pull-down assay
For in vitro pull-down assay, 2-μg recombinant bait proteins (GST-phyB C1 and GST) and 2-μg recombinant prey protein (MBP-His-FHY3-C2) were added into a new tube containing 1-mL binding buffer (50-mM Tris-HCl pH 7.5, 100-mM NaCl, and 0.6% (v/v) TritonX-100). After incubation at 4°C for 2 h, 20-μL Glutathione Sepharose 4B beads (GE Healthcare) was added and a further 1-h incubation at 4°C was performed. Then, after washing beads 4 times with binding buffer, pull-down proteins were eluted with 2× SDS loading buffer at 95°C for 10 min. The input and IP samples were separated on 10% SDS-PAGE gel, and proteins were detected with anti-GST (Abcam, ab19256; 1:1,000), anti-MBP (Abcam, ab9084; 1:2,000)) antibodies
Co-IP assay
For co-immunoprecipitation, total protein was extracted with lysis buffer containing 50-mM Tris-HCl pH7.5, 100-mM NaCl, 10-mM MgCl2, 5% (v/v) glycerol, 1-mM EDTA, 0.2% (v/v) Tween 20, 10-mM NaF, 2-mM sodium orthovanadate, 1-mM PMSF, and 1× complete protease inhibitor cocktail (Roche), and then centrifuged 3 times for 10 min at 15,000g, 4°C. Cleared extract incubated with anti-Flag antibody (Sigma, F3165) for 2 h at 4°C. About 30-μL protein G-Sepharose beads was added for another 2 h at 4°C. The beads were washed 3 times with lysis buffer, and the precipitated proteins were separated by SDS-PAGE gel. The target protein was detected by immunoblotting with anti-Flag and anti-Myc (TransGen, HT101).
LCI and LUC activity assays
The firefly luciferase complementation imaging (LCI) was performed using the leaves of 7-week-old N. benthamiana plants. Full-length PHYB and FHY3 fused with C-terminal LUC (cLUC) or N-terminal LUC (nLUC) were transformed into Agrobacterium tumefaciens (strain GV3101 or EHA105). The Agrobacterium suspensions were then coinfiltrated into well-grown leaves using a needleless syringe. After injection, these plants were incubated under 16-h light/8-h dark for 3 d before imaging using the NightSHADE LB 985 plant imaging apparatus equipped with a CCD camera (Berthold Technologies). Data was quantified with IndiGo software.
ChIP assay
About 0.5-g post-harvest seeds were used for ChIP assay and the experiments were performed as described previously (Li et al., 2019). Briefly, fhy3-4 and fhy3-4/FHY3p:FHY3-YFP (Lin et al., 2008) seeds were treated under different conditions as described in the text. These samples were fixed in 1% (v/v) formaldehyde for 30 min under vacuum. Glycine was added to a final concentration of 0.125 M and incubated for another 5 min to terminate this reaction. Nuclei were suspended with 600-μL lysis buffer and sonicated to shear DNA into ∼0.5–2-kb fragments. After centrifugation, the supernatant was pre-cleared with salmon sperm shear DNA/protein G agarose beads for 60 min at 4°C. After centrifugation, the supernatant was transferred into a new tube and incubated with 30-μL anti-GFP mAb-Agarose (MBL, D153-8) overnight at 4°C. After washing, the protein–DNA complex was eluted from the beads and reverse crosslinked by incubation with 5-M NaCl at 65°C overnight. The precipitated DNA fragments were quantified by qPCR using the primers shown in Supplemental Table 1.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: FHY3 (AT3G22170), PHYB (AT2G18790), RVE1 (AT5G17300), RVE2 (AT5G37260), RVE7 (AT1G18330), SPT (AT4G36930), and GA3ox2 (AT1G80340).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. FHY3 regulates seed germination.
Supplemental Figure S2. FHY3 expression in siliques and dry seeds.
Supplemental Figure S3. Expression level of RVE2, RVE7, and SPT in the fhy3-15 mutants.
Supplemental Figure S4. Characterization of the rve7 mutant.
Supplemental Figure S5. Phenotype of fhy3-4/FHY3p:FHY3-YFP.
Supplemental Figure S6. Phenotype of the various mutants.
Supplemental Table S1. List of primers used in this study.
Supplemental Data Set S1. List of differentially regulated genes in RNA-seq assay.
Supplementary Material
Acknowledgments
We thank Dr. Lijia Qu for providing the EPR1-ox transgenic seeds, Dr. Xiaodong Xu for gifting the spt mutants, Dr. Gang Li for providing 35S:Flag-FHY3 seeds, and Arabidopsis Biological Research Center for providing the Salk T-DNA lines.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100405, 2017YFA0503802) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27030205) to R.L.
Conflict of interest statement. The authors declare no conflicts of interest.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Rongcheng Lin (rclin@ibcas.ac.cn).
References
- Allen T, Koustenis A, Theodorou G, Somers DE, Kay SA, Whitelam GC, Devlin PF (2006) Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell 18:2506–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Koornneef M (2008) Seed dormancy and germination. The Arabidopsis Book 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto JF, Sánchez Rodolfo A., Casal JJ (1995) Role of phytochrome B in the induction of seed germination by light in Arabidopsis thaliana. J. Plant Physiol 146: 307–312 [Google Scholar]
- Botto JF, Sanchez RA, Whitelam G C, Casal J J (1996) Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol 110: 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M, Galvão VC, Fankhauser C (2016) Light-mediated hormonal regulation of plant growth and development. Annu Rev Plant Biol 67: 513–537 [DOI] [PubMed] [Google Scholar]
- Dong H, Liu J, He G, Liu P, Sun J (2019) Photoexcited phytochrome B interacts with brassinazoleresistant 1 to repress brassinosteroid signaling in Arabidopsis. J Integr Plant Biol 62: 652–667 [DOI] [PubMed] [Google Scholar]
- Donohue K, Heschel MS, Butler CM, Barua D, Sharrock RA, Whitelam GC, Chiang GC (2008) Diversification of phytochrome contributions to germination as a function of seed-maturation environment. New Phytol 177: 367–379 [DOI] [PubMed] [Google Scholar]
- He H, de Souza Vidigal D, Snoek LB, Schnabel S, Nijveen H, Hilhorst H, Bentsink L (2014) Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis. J Exp Bot 65: 6603–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Willems LA, Batushansky A, Fait A, Hanson J, Nijveen H, Hilhorst HW, Bentsink L (2016) Effects of parental temperature and nitrate on seed performance are reflected by partly overlapping genetic and metabolic pathways. Plant Cell Physiol 57: 473–487 [DOI] [PubMed] [Google Scholar]
- Hennig L, Stoddart WM, Dieterle M, Whitelam GC, Schäfer E (2002) Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol 128: 194–200 [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Huai J, Zhang X, Li J, Ma T, Zha P, Jing Y, Lin R (2018) SEUSS and PIF4 coordinately regulate light and temperature signaling pathways to control plant growth. Mol Plant 11: 928–942 [DOI] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13: 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Horiguchi G, Gleissberg S, Tsukaya H (2010) The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol 51: 252–261 [DOI] [PubMed] [Google Scholar]
- Jiang Z, Xu G, Jing Y, Tang W, Lin R (2016) Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat Commun 7: 12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno N, Møller SG, Shinomura T, Xu X, Chua NH, Furuya M (2003) The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15: 2476–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Turečková V, Carat S, Chappuis R, Strnad M, Fankhauser C, Lopez-Molina L (2012) Spatially and genetically distinct control of seed germination by phytochromes A and B. Gene Dev 26: 1984–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Ince YC, Fankhsuser C (2020) Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat Commun 10: 5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Wang Deng X (2011a) Phytochrome signaling mechanisms. Arabidopsis Book 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Siddiqui H, Teng Y, Lin R, Wan XY, Li J, Lau OS, Ouyang X, Dai M, Wan J, Devlin PF, Deng XW, Wang H (2011b) Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol 13: 616–624 [DOI] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y, Wang H, Deng XW (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen T, Li Y, Wang Z, Cao H, Chen F, Li Y, Soppe WJJ, Li W, Liu Y (2019) ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell 31: 832–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Teng Y, Park HJ, Ding L, Black C, Fang P, Wang H (2008) Discrete and essential roles of the multiple domains of Arabidopsis FHY3 in mediating phytochrome A signal transduction. Plant Physiol 148: 981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wei H, Ma M, Li Q, Kong D, Sun J, Ma X, Wang B, Chen C, Xie Y, Wang H (2019) Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell 31: 2089–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xie Y, Wang H, Ma X, Yao W, Wang H (2017) Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 29: 2269–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li Y, Li X, Xu D, Lin X, Liu M, Li G, Qin X (2019) FAR-RED ELONGATED HYPOCOTYLS3 negatively regulates shade avoidance responses in Arabidopsis. Plant Cell Environ 42: 3280–3292 [DOI] [PubMed] [Google Scholar]
- Ma L, Tian T, Lin R, Deng XW, Wang H, Li G (2016) Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol Plant 9: 541–557 [DOI] [PubMed] [Google Scholar]
- Nonogaki H (2019) Seed germination and dormancy: the classic story, new puzzles, and evolution. J Integr Plant Biol 61: 541–563 [DOI] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G (2006) Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J 47: 124–139 [DOI] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X, Li J, Li G, Li B, Chen B, Shen H, Huang X, Mo X, Wan X, Lin R, Li S, Wang H, Deng XW (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA (2005) Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol 15: 1998–2006 [DOI] [PubMed] [Google Scholar]
- Poppe C, Schäfer E (1997) Seed germination of Arabidopsis thaliana phyA/phyB double mutants is under phytochrome control. Plant Physiol 114: 1487–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69: 463. [DOI] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shi H, Wang X, Mo X, Tang C, Zhong S, Deng XW (2015) Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc Natl Acad Sci USA 112: 3817–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Hanzawa H, Schäfer E, Furuya M (1998) Mode of phytochrome B action in the photoregulation of seed germination in Arabidopsis thaliana. Plant J 13: 583–590 [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M (1994) The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol 104: 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M (1996) Action spectra for phytochrome A-and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 8129–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9: 34–45 [DOI] [PubMed] [Google Scholar]
- Siddiqui H, Khan S, Rhodes BM, Devlin PF (2016) FHY3 and FAR1 act downstream of light stable phytochromes. Front Plant Sci 7: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Zhao S, Williamson L, Ward S, Leyser O (2012) FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J 71: 907–920 [DOI] [PubMed] [Google Scholar]
- Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R (2012) Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during de-etiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R (2013) The UVR8 UV-B photoreceptor: perception, signaling and response. Arabidopsis Book 11: e0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Gan Y, Penfield S, Gilday AD, Dave A, He Z, Josse EM, Choi G, Halliday KJ, Graham IA (2013) Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc Natl Acad Sci USA 110: 10866–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang H (2015) Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci 20: 453–461 [DOI] [PubMed] [Google Scholar]
- Wang Q, Lin C (2020) Mechanisms of cryptochrome-mediated photoresponses in plants. Annu Rev Plant Biol 71: 23.1–23.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang W, Xu P, Pan J, Zhang T, Li Y, Li G, Yang H, Lian H (2019) phyB interacts with BES1 to regulate brassinosteroid signaling in Arabidopsis. Plant Cell Physiol 60: 353–366 [DOI] [PubMed] [Google Scholar]
- Xie Y, Liu Y, Ma M, Zhou Q, Zhao Y, Zhao B, Wang B, Wei H, Wang H (2020a) Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat Commun 11: 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zhou Q, Zhao Y, Li Q, Liu Y, Ma M, Wang B, Shen R, Zheng Z, Wang H (2020b) FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate Arabidopsis flowering. Mol Plant 13: 483–498 [DOI] [PubMed] [Google Scholar]
- Xu D, Wu D, Li XH, Jiang Y, Tian T, Chen Q, Ma L, Wang H, Deng XW, Li G (2020) Light and abscisic acid coordinately regulate greening of seedlings. Plant Physiol 183: 1281–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Jiang Z, Jing Y, Lin R (2020a) PIF1 and RVE1 form a transcriptional feedback loop to control light-mediated seed germination in Arabidopsis. J Integ Plant Biol 62: 1372–1384 [DOI] [PubMed] [Google Scholar]
- Yang L, Jiang Z, Liu S, Lin R (2020b) Interplay between REVEILLE1 and RGA-LIKE2 regulates seed dormancy and germination in Arabidopsis. New Phytol 225: 1593–1605 [DOI] [PubMed] [Google Scholar]
- Yang L, Liu S, Lin R (2020c) The role of light in regulating seed dormancy and germination. J Integr Plant Biol 62: 1310–1326 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Whitelam GC, Casal JJ (2000) fhy3-1 retains inductive responses of phytochrome A. Plant Physiol 123: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu X, Liu X, Lin C (2007) Light regulation of gibberellins metabolism in seedling development. J Integr Plant Biol 49: 21–27 [Google Scholar]
- Zhang X, Chen Y, Wang ZY, Chen Z, Gu H, Qu LJ (2007) Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J 51: 512–525 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.