Abstract
Together with auxin transport, auxin metabolism is a key determinant of auxin signaling output by plant cells. Enzymatic machinery involved in auxin metabolism is subject to regulation based on numerous inputs, including the concentration of auxin itself. Therefore, experiments characterizing altered auxin availability and subsequent changes in auxin metabolism could elucidate the function and regulatory role of individual elements in the auxin metabolic machinery. Here, we studied auxin metabolism in auxin-dependent tobacco BY-2 cells. We revealed that the concentration of N-(2-oxindole-3-acetyl)-l-aspartic acid (oxIAA-Asp), the most abundant auxin metabolite produced in the control culture, dramatically decreased in auxin-starved BY-2 cells. Analysis of the transcriptome and proteome in auxin-starved cells uncovered significant downregulation of all tobacco (Nicotiana tabacum) homologs of Arabidopsis (Arabidopsis thaliana) DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1), at both transcript and protein levels. Auxin metabolism profiling in BY-2 mutants carrying either siRNA-silenced or CRISPR-Cas9-mutated NtDAO1, as well as in dao1-1 Arabidopsis plants, showed not only the expected lower levels of oxIAA, but also significantly lower abundance of oxIAA-Asp. Finally, ability of DAO1 to oxidize IAA-Asp was confirmed by an enzyme assay in AtDAO1-producing bacterial culture. Our results thus represent direct evidence of DAO1 activity on IAA amino acid conjugates.
Introduction
Auxin is one of the most important plant morphogenic compounds since it regulates many processes of plant development at several levels (Paque and Weijers, 2016). Concentration of auxin, mainly represented by indole-3-acetic acid (IAA), is tightly regulated through biosynthesis, transport, degradation, and conjugation; the last involves both reversible and irreversible pathways. Together, these processes maintain auxin homeostasis, which, when disrupted, leads to physiological or morphological changes (Ludwig-Müller, 2014; Casanova-Sáez and Voß, 2019). Transport across the plasma membrane, facilitated mostly through the activities of PIN and AUX/LAX proteins, is probably the most studied auxin regulatory mechanism. Many detailed studies focused on localization, regulation of activity, as well as kinetic parameters, have been published in recent years (Reemmer and Murphy, 2014; Adamowski and Friml, 2015; Sauer and Kleine-Vehn, 2019). However, a plethora of gaps remain in our understanding of auxin metabolism. While several mechanisms of auxin biosynthesis were predicted, only one pathway—wherein intermediate indole-3-pyruvate is catabolized by members of tryptophan aminotransferase and flavin monooxygenase families—was identified in plants (Mashiguchi et al., 2011). Auxin oxidation is a major mechanism of auxin degradation, but the enzyme involved in the formation of 2-oxindole-3-acetic acid (oxIAA), DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1), has been identified only recently (Porco et al., 2016; Zhang et al., 2016). Most of the IAA in plant cells is present in conjugated forms, usually with amino acids or sugars, even though other forms exist as well. Levels of these metabolites depend on cell type as well as on plant species (Ludwig-Müller, 2011). Ester-linked sugar conjugation in plants is catalyzed by specific uridine-diphosphate glycosyltransferases (UGTs). IAA-specific UGTs have been described, for example, in Arabidopsis (Arabidopsis thaliana; Jackson et al., 2001) and maize (Zea mays; Szerszen et al., 1994), but their wider identification and characterization in other plants is difficult since UGTs comprise a large gene family. Amide-bond-linked amino acid conjugates (IAA-aa) are products of the activity of the GRETCHEN-HAGEN 3 (GH3) family of acyl acid-amido synthetases. Some members of this group are known for their promiscuous character due to the range of amino acid donors as well as plant hormone acceptors (Staswick, 2005; Westfall et al., 2016; Aoi et al., 2020). Products of both oxidation and conjugation, for example, 2-oxindole-3-acetic acid-glucosyl ester (oxIAA-GE) or 2-oxindole-3-acetic acid-amino acid conjugates (oxIAA-AA), have been identified in plants, suggesting close coordination of oxidative and conjugating reactions (Zhang and Peer, 2017). In addition, compartmentalization plays an important role in auxin metabolism. Several members of PIN and PIN-likes (PILS) protein families were localized to the membrane of the endoplasmic reticulum (ER; Barbez et al., 2012; Simon et al., 2016), and IAA-aa amidohydrolases IAA-LEUCINE RESISTANT 1, IAA-ALANINE RESISTANT 1, and IAA-LEUCINE RESISTANT (ILR)-LIKE 2 are localized to the ER lumen (Sanchez Carranza et al., 2016).
While a detailed transport-based mechanism of auxin homeostasis in tobacco BY-2 (Nicotiana tabacum L., cv Bright Yellow) cells was recently revealed, no information is known about the natural metabolism of auxin in BY-2 cells. In the present work, we performed a multi-omical characterization of BY-2 cells cultured in the presence (control) or absence (auxin-starved) of synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D). Substantial qualitative and quantitative changes in auxin metabolic profiles in conjunction with changes in transcript and protein levels of auxin metabolic machinery suggested a role of the tobacco homolog of DAO1 in oxidation of IAA amino acid conjugates. This function of DAO1 was confirmed in mutants of DAO1 in tobacco BY-2 cells and Arabidopsis plants.
Results
2,4-D-supplemented and auxin-starved BY-2 cells show the same growth parameters during the first 2 days of cultivation
Tobacco BY-2 cells are typically cultured with synthetic auxin 2,4-D in the cultivation medium. When 2,4-D is not present, auxin starvation leads to an inhibition of cell division, induction of cell elongation, and to other morphological and biochemical changes (Winicur et al., 1998; Sakai et al., 1999). To select the optimal conditions for our analysis of auxin-responsive IAA metabolism, growth parameters (i.e. cell density, weight, and viability of cells) were followed during the growth cycle of BY-2 cells cultured in presence or absence of 2,4-D (Figure 1). In the control cells, cell density and cell weight showed typical exponential growth characteristics with maximum values on the fifth day. Auxin-starved cells showed no cell division during the 7 d of growth cycle (Figure 1, A and C). The weight of auxin-free-cultured cells increased slightly, with a maximum on the sixth day (an approximately 11-fold increase in fresh weight compared to day zero; Figure 1, C). High cell viability (approximately 95%) was observed in the 2,4-D-supplemented culture during the whole growth cycle (Figure 1, B). Auxin-deprived cells maintained a stable level of living cells during the first 3 d of cultivation and then dropped to approximately 80% and 60% ratios of viable cells in the 4th and 7th d, respectively.
Figure 1.
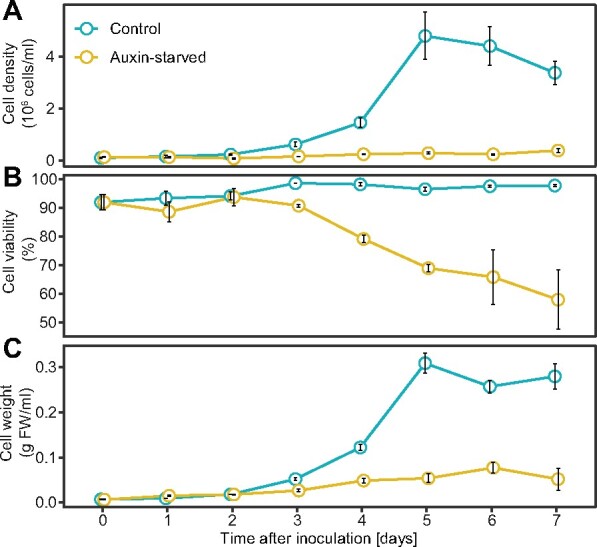
Growth parameters of BY-2 cells in auxin-free medium. Cell density (A), viability (ratio of living cells) (B), and weight (C) were monitored during the cultivating period of N. tabacum L. cv Bright Yellow 2 cells cultured in the presence (control) and absence (Auxin-starved) of synthetic auxin 2,4-D. Means ± SE are shown (n = 3).
These results show that 2,4-D-supplemented and auxin-starved BY-2 cells show comparable growth parameters during the first 2 d of cultivation; therefore, we decided to perform the analysis of auxin metabolites and auxin metabolism 2 d after inoculation.
oxIAA-Asp is the main auxin metabolite in BY-2 cells
Levels of endogenous IAA and its metabolites were determined by liquid chromatography mass spectrometry (LCMS) analysis in 2-d-old BY-2 cells cultured in the presence (control) or absence (auxin-starved, AS) of synthetic auxin 2,4-D. In both conditions, the major IAA metabolite was oxIAA-Asp, which exhibited levels almost 100-fold higher compared with other metabolites oxIAA, oxIAA-GE, and IAA itself (Figure 2, A). While the level of IAA was similar in both conditions, the level of oxIAA was three-fold lower in auxin-starved cells. However, auxin-starved cells showed slightly higher abundances of oxidized forms of IAA conjugates oxIAA-Asp and oxIAA-GE (1.6- and 1.1-fold, respectively). Levels of IAA-GE, IAA-Asp, and IAA-Glu were below the detection limit of the instrument. To exclude the possibility that metabolites are exported out from the cells, we harvested and analyzed the culture media, where no measurable levels of either IAA or its metabolites were found.
Figure 2.
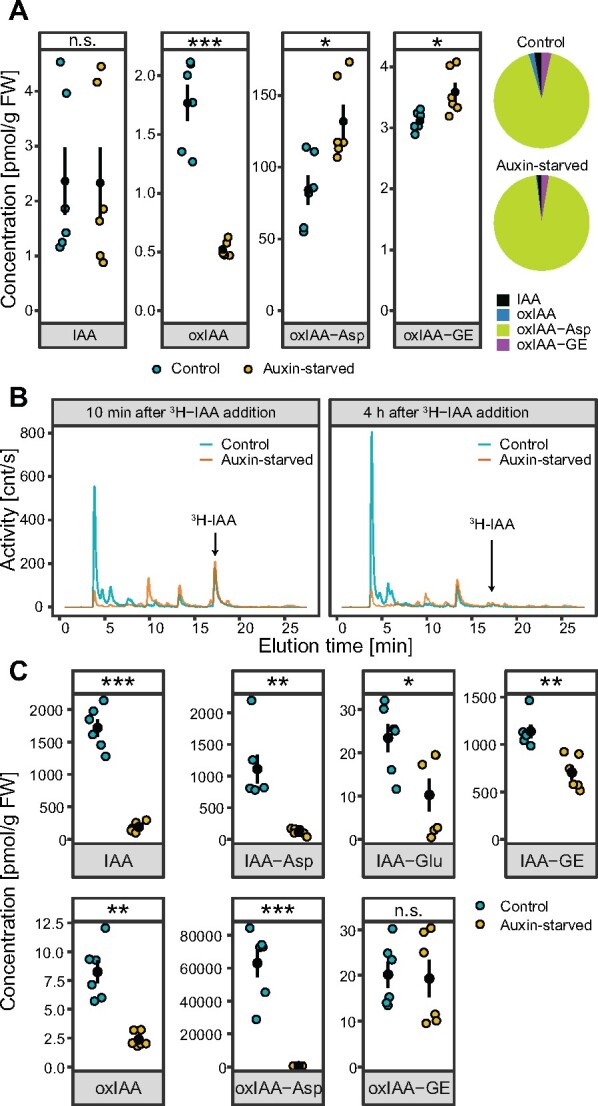
Distinct auxin metabolism in 2,4-D-supplemented and auxin-starved BY-2 tobacco cell cultures. A, The endogenous contents of free IAA and its metabolites were measured in 2-d-old control (2,4-D-supplemented) and auxin-free-cultivated BY-2 cell culture. Data points and error bars indicating mean ± SE are shown. Pie charts represent relative contributions of detected metabolites to the overall content of auxin in the cells. B, Auxin metabolite profiling of control and auxin-starved cells performed by HPLC chromatography 10 min and 4 h after application of 3H-IAA into the media. C, Mass-spectrometry-based quantification of free IAA and its metabolites in control and auxin-starved cells 2 h after application of 1 µM IAA into the media. Data points and error bars indicating mean ± SE are shown. A and C, Student’s t test P-values: *P < 0.05, **P < 0.01, ***P < 0.001.
2,4-D controls the capability of BY-2 cells to metabolize exogenously applied IAA
Two-day-old control and auxin-starved BY-2 cells were further compared for their ability to metabolize exogenously applied IAA. Qualitative and quantitative analyses of IAA and its metabolites were performed by high-performance liquid chromatography (HPLC) with online radioactivity detection (HPLC-RAD), and LCMS, respectively. Qualitative analysis was done upon cell incubation with radioactively labeled IAA (3H-IAA) for 10 min and 4 h, followed by HPLC-RAD. The dominant metabolite in the control cells, represented by a peak eluted at 4 min, was present with a very low abundance in the auxin-starved sample. 3H-IAA, represented by a peak eluted at 17 min (chromatogram data for standard 3H-IAA are shown in Supplemental Data S1), was completely metabolized in both control and auxin-starved conditions 4 h after its application. Based on the chromatograms, all 3H-IAA metabolites were generally present at much lower levels in auxin-starved cells (Figure 2, B).
LCMS was used to validate the qualitative results of HPLC analysis (Figure 2, C). Levels of IAA and its metabolites were measured in BY-2 cells 2 h after adding IAA into the media. Higher abundance of all metabolites except for oxIAA-GE was detected in control cells compared with auxin-starved cells. The most striking difference was observed in the case of oxIAA-Asp, which was the dominant metabolite in the control, showing 150-fold higher abundance compared with auxin-starved cells. In conclusion, we found that cells cultured in auxin-free conditions displayed substantially quantitative and qualitative changes in their spectra of metabolic intermediates, whether 3H-IAA or IAA was applied.
High-throughput analysis of RNA and protein levels in control and auxin-starved BY-2 cells
To understand the molecular background responsible for the auxin-dependent differences in IAA metabolic capacity of BY-2 cells, transcriptomic and proteomic analyses were carried out. RNA-seq was performed for seven control and three auxin-free-cultured BY-2 cell culture samples harvested 2 d after inoculation. Quality of RNA and total number of reads per biological replicate are summarized in Supplemental Table S1.
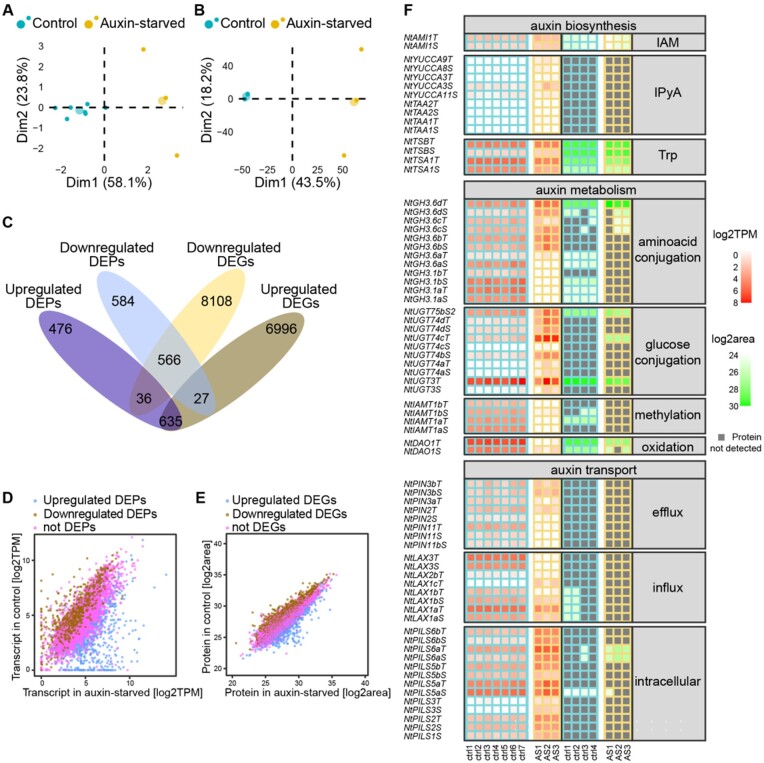
Reads were quality-filtered and mapped against N. tabacum v1.0 cDNA reference dataset (Edwards et al., 2017) using the software salmon (Patro et al., 2017). Quality of the analysis was assessed by principal component analysis and hierarchical clustering (Figure 3, A and Supplemental Figure S1, A). Transcripts with TPM value higher than 0.5 in either control or auxin-starved were considered as detected (totally 40,285 transcripts out of 69,500 in the reference dataset). DESeq2 and Sleuth R packages were used for identification of differentially expressed genes (DEGs). 23,348 transcripts were agreed upon by both algorithms as being significantly differently expressed (q-value ≤ 0.05). Of these transcripts, 8770 and 7598 were considered as up- and down-regulated, respectively (fold-change ≥ 2, summarized in Supplemental Table S2). The full dataset including transcripts with TPM values and results of statistical evaluation is shown in Supplemental Data S2. In brief, in auxin-free-cultured BY-2 cells, members of the extensin gene family and stress-related heme peroxidases were among the most up-regulated genes, while members of the AUX/IAA and GH3 families belonged to the most down-regulated genes.
Figure 3.
Transcriptomic and proteomic characterization of 2,4-D-supplemented and auxin-starved tobacco BY-2 cell cultures. Principal component analysis of RNA-seq (A) and mass-spectrometry-defined proteome (B) of 2 d old BY-2 cell culture. Small dots represent individual samples, large dots show mean coordinates of the individuals in the group. Axes labels show percentage of variances explained by each principal component. RNA-seq was performed for seven control (2,4-D-supplemented) and three auxin-starved samples. Proteomic analysis was done using four control and three auxin-starved samples. Venn diagram (C) and scatter plots (D and E) summarize DEGs and proteins (DEGs and DEPs) and their overlap in both methods. Plot (D) relates transcription abundance (TPM) in control and auxin-starved (AS) samples, and color of dots represents results of statistics for a particular protein in proteome analysis. Plot (E) relates protein abundance (log2 transformed area of peaks), and color of dots represents result of statistics in RNA-seq experiment. In (F), a heat map representing transcription and protein levels of selected genes involved in the regulation of auxin activity (biosynthesis, metabolism, and transport) is shown. Transcription of several genes representing auxin transport and auxin conjugation was validated by RT-qPCR (Supplemental Figure S2).
To address the biological processes supported by the DEGs, we used the GO statistical overrepresentation test of PantherDB (Mi et al., 2019). In summary, processes associated with succinate transmembrane transport, calcium-dependent signaling, beta-glucan biosynthesis, plant-type cell wall biogenesis, and cytoskeleton-dependent cytokinesis were enriched in genes up-regulated in auxin-starved cells. Processes associated with mitochondrial translation and membrane organization, protein synthesis (ribosome biogenesis, translation, protein folding, and methylation), and cell division (DNA replication, chromosome segregation and organization, and DNA repair and recombination) were enriched in down-regulated set of transcripts. Complete results of PANTHER Overrepresentation tests are shown in Supplemental Tables S3, S4).
Proteomic characterization of BY-2 cells in the auxin-supplemented and auxin-starved conditions was performed via label-free quantitative LC–MS/MS analysis. Quality control of the analysis was done by principal component analysis and hierarchical clustering (Figure 3, B and Supplemental Figure S1, B). In total, 6547 proteins were identified in 2-d-old BY-2 cells (q-value ≤ 0.05). 2324 proteins were determined to be differentially expressed (DEPs, q-value ≤ 0.05, fold-change ≥ 2), with 1147 and 1177 showing up- and down-regulated patterns, respectively (Supplemental Data S3). Peroxidases, extensins, annexin, methyltransferase, and proteinase inhibitor represent the most up-regulated proteins, whereas histone H2A, phospholipase D, helicase, several members of ribosomal proteins, or glyceraldehyde-3-dehydrogenase are among those most down-regulated. Combining the transcriptomic and proteomic data indicated that at both the transcript and protein levels, 566 genes were downregulated and 635 genes were upregulated. Thirty-six genes were upregulated at the protein level and downregulated at transcript level, and 27 genes showed the opposite pattern (summarized in Venn diagram in Figure 3, C). Visualization of gene transcript and protein levels and their corresponding result of statistical determination is shown as scatter plots in Figure 3, D and E. Altogether, our data provide a high-throughput analysis of the BY-2 cell line with very robust characterization of transcript and protein levels.
Distinct regulation of genes involved in auxin metabolism
To better understand the molecular background of the different auxin metabolic capacities of control and auxin-starved BY-2 cells, RNA-seq and proteomic data were filtered to select genes related to auxin metabolism (biosynthesis, conjugation, and degradation) and auxin transport. Transcription of several genes representing auxin transport and auxin conjugation was validated by RT-qPCR (Supplemental Figure S2).
Auxin biosynthesis-related genes were represented by homologs of tryptophan aminotransferases (TAAs), flavin-binding monooxygenases (YUCCAs), amidase1 (AMI1), aldehyde oxidase 1 (AAO1), and tryptophan synthases (TSA and TSB). Genes coding for enzymes involved in indole-3-pyruvic acid pathway (TAAs and YUCCAs) showed very little transcription in both control and auxin-starved conditions. None of the proteins coded by these genes was detected at all. The putative indole-3-acetamide pathway was represented by AMI1, which was expressed at similar levels in both conditions. Genes involved in tryptophan synthesis showed higher expression in control cells (Figure 3, F).
Next, we focused on genes related to auxin conjugation and degradation, in particular members of the GH3 gene family involved in IAA amino acid conjugation, DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1) which is involved in the oxidation of IAA, IAA carboxymethyltransferase (IAMT), and selected UDP-glucosyl transferases (UGTs). Since plant genomes harbor many genes coding for UGTs, we selected homologs of AtUGT84B1 (AT2G23260), which is responsible for IAA glycosylation in Arabidopsis (Jackson et al., 2001), and NtUGT3, a homolog of AtUGT74D1 (AT2G31750), which participates in glycosylation of oxIAA (Tanaka et al., 2014). Expressions of NtUGT3, several GH3 members, as well as tobacco homologs of IAMT and DAO1, were significantly lower in auxin-starved BY-2 cells compared with control. Surprisingly, GH3.6b, c, and d isoforms of the GH3 family showed no change in auxin-starved conditions, or even increased in the expression level. Furthermore, NtGH3.6dT showed the highest protein expression among GH3 members in both control and auxin-starved BY-2 cells (Figure 3, F).
Genes coding for auxin transporters were represented by genes coding for auxin influx carriers (AUX/LAXes, Peret et al., 2012), auxin efflux carriers (PINs), and putative endomembrane auxin carriers (PILSes; Sauer and Kleine-Vehn, 2019). The highest expression at both transcript and protein levels was found in the case of PILS5 and PILS6 homologs, and their abundances were significantly higher in auxin-starved cells. In contrast, in auxin-starved cells, genes of the PIN family showed generally lower transcript levels, and all expressed members except for PIN3b were downregulated. LAX genes were represented by the dominant transcripts of LAX1a and LAX3. Auxin starvation caused downregulation of most tobacco AUX/LAX genes and induction of LAX1c expression, confirming the results published in Müller et al. (2019).
To conclude, our analysis of transcriptome and proteome changes induced by the absence of auxin in the culture media clearly showed that, in contrast to largely insignificant changes in the expression of genes coding for enzymes involved in auxin biosynthesis, multiple genes coding for auxin-metabolizing enzymes exhibited different expression at transcript and protein levels. The amino acid conjugating pathway, represented by the GH3 family, showed both up- and down-regulation. Genes from the family of UDP-glucosyltransferases, coding for putative IAA glycosylating enzymes, were generally more expressed in the auxin-starved cells. However, NtUGT3, the most abundant UGT at both the transcript and protein levels, was less expressed in the auxin-free-cultured cells, suggesting its role in auxin metabolism. In auxin-starved conditions, both homologs of DAO1 showed significant downregulation, at both transcript and protein levels. We propose that the different expression of DAO1 is a crucial factor in the different metabolic profiles, particularly the different levels of oxIAA-Asp between the control and auxin-free-cultured BY-2 cells.
Localization studies confirmed auxin-dependent transcriptional regulation of NtDAO1
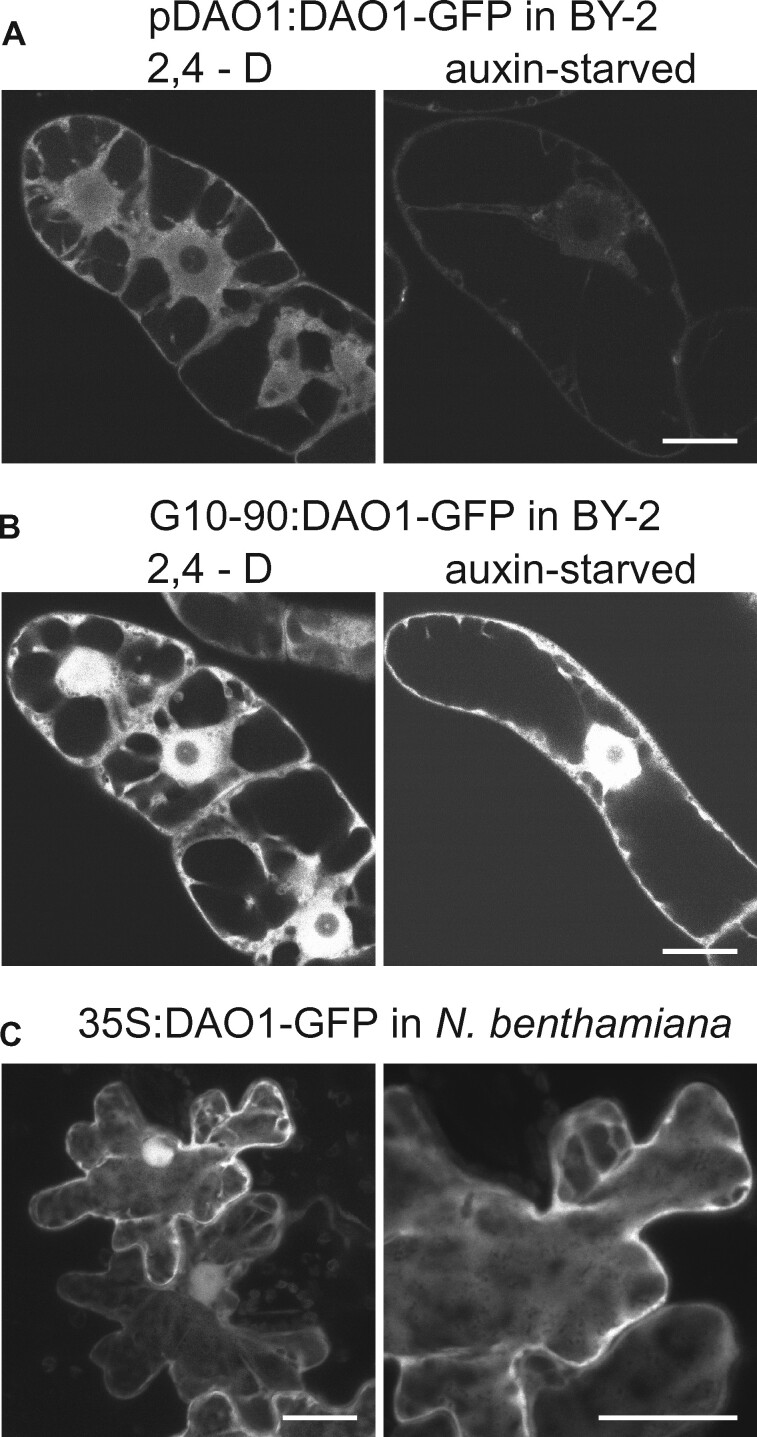
To address potential auxin-dependent posttranslational regulation of tobacco DAO1, transgenic BY-2 lines carrying a GFP-translational fusion to the coding sequence of NtDAO1T (GenBank ID XM_016591349.1) under native promoter (pDAO1T:DAO1-GFP) or under the strong constitutive promoter G10-90 (G10-90:DAO1-GFP) were prepared. Images of 2-d-old BY-2 cells cultured in control or auxin-starvation conditions were obtained by confocal microscopy. In auxin-supplemented conditions, the GFP signal was strong in the cytosol and nucleus in G10-90:DAO1-GFP as well as in pDAO1T:DAO1-GFP line (Figure 4, A and B and Supplemental Figure S3, A and B). In auxin-free conditions, the signal was weak in the pDAO1:DAO1-GFP line, whereas the G10-90:DAO1-GFP line showed a GFP signal of an intensity similar to auxin-supplemented conditions (Figure 4, A and B and Supplemental Figure S3, A and B). Next, we screened our proteomic data for possible ubiquitination or phosphorylation sites. While the signal of DAO1 protein was high in LC–MS/MS spectrum, samples from control and auxin-starved cells showed no DAO1 signal among the list of phosphorylated or ubiquitinylated proteins. We concluded that the auxin-dependent pattern is largely controlled at the transcription level; however, post-transcriptional or post-translational regulation cannot be ruled out. No signal was observed in control WT BY-2 line (Supplemental Figure S3, C). Cytosolic and nuclear localization of NtDAO1T was further confirmed in leaves of Nicotiana benthamiana infiltrated with Agrobacterium tumefaciens carrying a 35S:DAO1-GFP gene construct (Figure 4, C). To exclude a presence of cleaved GFP in nucleus, Western Blot using anti-GFP antibody was performed on extracts from N. benthamiana leaves. A faint band corresponding to free GFP (approximately 27 kDa) in comparison to strong band of DAO1-GFP (61 kDa) indicates, that the DAO1-GFP must be the major source of fluorescence in nucleus of transformed cells (Supplemental Figure S4).
Figure 4.
Subcellular localization of DAO1-GFP in BY-2 cells and in tobacco leaves. A and B, Confocal images of 2-d-old N. tabacum L. cv Bright Yellow 2 transgenic pDAO1T:DAO1-GFP (A) and G10-90:DAO1-GFP (B) lines in 2,4-D-supplemented or auxin free conditions are shown. DAO1-GFP, regardless of its promoter, is localized in the cytosol and nucleus (but not the nucleolus) in auxin-supplemented conditions (A and B—2,4-D). In auxin-starved cells, DAO1-promoter-driven DAO1-GFP signal is very weak (A—auxin-starved). Individual optical sections are presented. Identical scanning parameters were used during acquisition so that fluorescence intensities should reflect expression levels in cell lines presented in A–C. C, Agrobacterium-infiltrated N. benthamiana leaves expressing 35S:DAO1-GFP. Maximum intensity projection of confocal z-stacks. A–C, Bars 20 µm. See also Supplemental Figures S3, S4.
Auxin metabolite profiling in tobacco cells and Arabidopsis seedlings carrying mutations in DAO1 reveals DAO1 activity on IAA amino acid conjugates
To test the role of DAO1 in the metabolization of IAA, we tested IAA metabolism in a set of BY-2 mutants carrying either siRNA-silenced or CRISPR-Cas9-mutated NtDAO1, as well as in dao1-1 Arabidopsis plants. The BY-2 line with knocked-down DAO1 activity (siDAO1) was prepared by β-estradiol-inducible expression of DAO1-specific siRNA. Reduced expression of DAO1 in β-estradiol-treated cells (approximately two- and three-fold lower compared with WT and DMSO-treated control, respectively) was verified by RT-qPCR (Supplemental Figure S5).
siDAO1 and crisprDAO1 transgenic lines were tested for their capacity to metabolize exogenously applied IAA. Concentrations of intracellular IAA and its metabolites were measured by LCMS 2 h after application of 1 µM IAA. Figure 5, A shows that the crisprDAO1 line metabolized IAA primarily into amino acid conjugates IAA-Asp and IAA-Glu (76- and 22-fold increases compared with WT control), whereas the abundance of oxidized forms (represented by oxIAA, oxIAA-Asp, and oxIAA-GE) was 2.5-, 120-, and 3-fold lower, respectively, in comparison to WT BY-2 cells. Substantiating the effects of modulated DAO1 activity on auxin metabolism, β-estradiol-induced siDAO1 cells exhibited concentrations of IAA-Asp and IAA-Glu that were, respectively, 10- and 6-fold higher in comparison to DMSO-treated siDAO1 control, while all other metabolites were present at similar levels. These measurements support the role of DAO1 in the oxidation of IAA to form oxIAA, but also present evidence for a role in the oxidation of IAA-Asp to form oxIAA-Asp.
Figure 5.
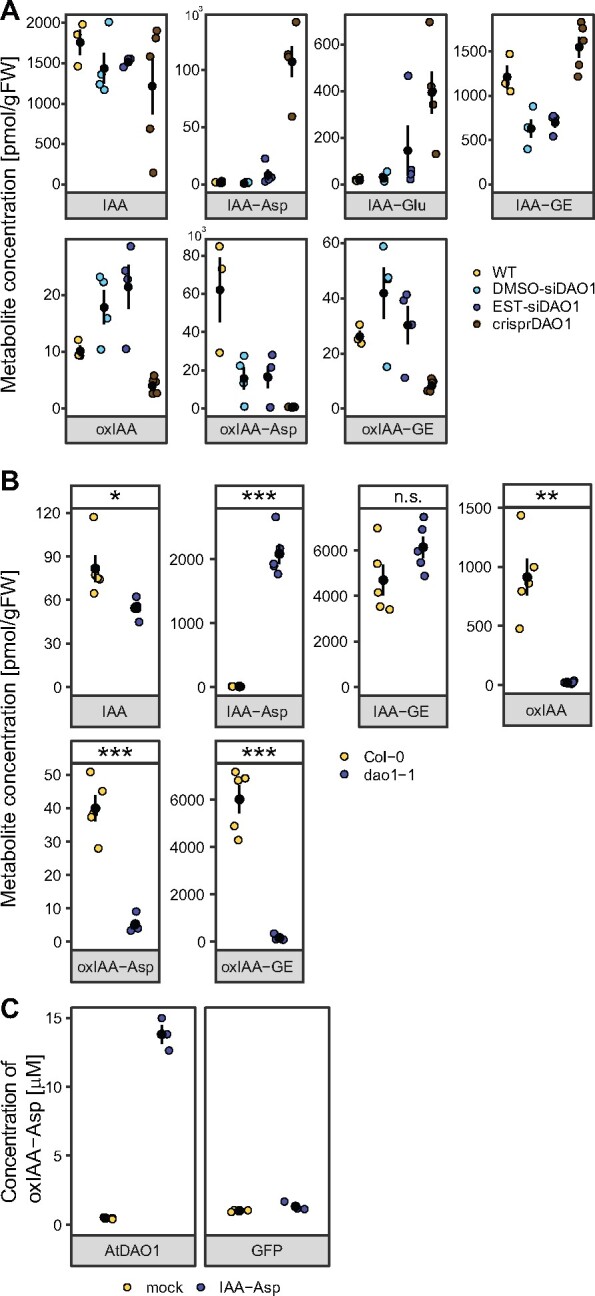
Activity of DAO1 corresponds with the levels of oxIAA as well as of oxidized conjugated forms of IAA, that is oxIAA-Asp and oxIAA-GE. Mass-spectrometry-based quantification of IAA and its metabolites in BY-2 cell lines (A) and seedlings of A. thaliana (B). In (A), metabolites were measured in control BY-2 cell culture (WT), DMSO-treated β-estradiol-inducible DAO1-silencing cell line (DMSO-siDAO1), β-estradiol-induced DAO1-silencing cell line (EST-siDAO1), and CRISPR-Cas9 mutated DAO1 cell line (crisprDAO1) 2 h after adding 1 µM IAA to the media. Valid statistics are not shown due to low number of measurements (n = 3–4) and high variance of measured metabolite levels. See also Supplemental Figure S5 for RT-qPCR analysis of NtDAO1 transcription in the siDAO1 cell line. In (B), IAA and its metabolites were measured in Arabidopsis Col-0 and dao1-1 KO mutants. Data points and error bars indicating mean ± SE are shown. In (C), oxIAA-Asp was measured in supernatant fractions from AtDAO1- or GFP-producing bacterial cultures supplemented with 0.1 mM IAA-Asp and DAO cofactor mixture for 6 h at 20°C. Bacterial cultures without IAA-Asp and the DAO cofactor mixture were used as mock samples. Mean ± SD (n = 3). For B, Student’s t test P-values: *P < 0.05, **P < 0.01, ***P < 0.001.
The role of DAO1 was further examined in Arabidopsis plants by profiling of endogenous IAA metabolites. In the dao1-1 mutant, significantly lower levels of oxIAA and oxIAA-GE were accompanied by a significantly lower level of oxIAA-Asp (approximately 10-fold lower in dao1-1 compared with Columbia-0 plants; Figure 5, B). To validate AtDAO1-mediated IAA-Asp oxidation, we expressed recombinant AtDAO1 protein in Escherichia coli. AtDAO1-producing bacterial culture was tested in a reaction with or without 0.1 mM IAA-Asp in combination with a DAO cofactor mixture, as these conditions were previously optimized for AtDAO1-mediated oxIAA production (Brunoni et al., 2019). A high level of oxIAA-Asp was detected in the AtDAO1 samples, in stark contrast to the GFP samples (Figure 5, C), confirming that AtDAO1 is able to oxidize IAA-Asp. Based on these results, we conclude that the role of DAO1 in auxin metabolism is broader than previously believed and includes oxidative activity on amino acid conjugates identified here.
Discussion
The concentration and activity of auxins are key factors determining many processes in plant growth and development. Several mechanisms regulating auxin activity were described, including transport (cellular influx, efflux, and intracellular translocation across endomembranes), reversible and irreversible conjugation, or degradation via oxidative pathways. Tobacco BY-2 cell lines are frequently used for providing a proof of function and for unraveling the molecular background of various proposed regulatory mechanisms, for example during the discovery of the PIN family of auxin efflux carriers (Petrášek et al., 2006) or the effect of altered expression of putative auxin endomembrane transporters on auxin metabolism (Mravec et al., 2009; Barbez et al., 2012). Despite these studies, information concerning the ability of BY-2 cells to regulate auxin activity was very limited. Recently, we revealed a homeostatic mechanism whereby BY-2 cells maintain auxin concentration through transcriptional regulation of auxin efflux and influx carriers (Müller et al., 2019). However, insights into the natural capacity of BY-2 cells to metabolize IAA are still largely limited. Hošek et al. (2012) reported rapid metabolism of the synthetic auxin NAA, while 2,4-D, which is a mandatory component of the cultivation media for BY-2 cell cultures, is metabolized only very weakly.
Here, we show that BY-2 cells maintain very low levels of IAA and its metabolites, and that auxin deprivation does not lead to increased IAA biosynthesis. While the levels of endogenous IAA in Arabidopsis seedlings varied between 20 and 40 ng/g FW, BY-2 cells maintain about 0.5 ng/g FW. A major IAA metabolite in BY-2 cells, oxIAA-Asp, was present at similar levels in BY-2 cells and Arabidopsis seedlings (approximately 30–60 ng/g FW), while IAA-GE, the dominant metabolite in Arabidopsis, was not detected in the BY-2 cells. This suggests an independence of BY-2 cells from IAA achieved through the presence of 2,4-D in the medium, securing the necessary auxin signal. In 2,4-D-free conditions however, BY-2 cells cannot compensate for the absence of auxin by synthesis of IAA, as indicated by the unchanged levels of IAA in control and auxin-deprived cells (Figure 2, A), and also by the absence of changes in the transcription of the auxin biosynthetic apparatus (Figure 3, F).
In contrast to IAA biosynthesis, proteins involved in the auxin metabolic machinery are present at very high levels in control BY-2 cells (Figure 3, F). Exogenously applied IAA is rapidly metabolized mainly to oxIAA-Asp (Figure 2, B and C). In auxin-starved cells, the IAA metabolic profile, as well as the expression of genes involved in auxin metabolism, differs significantly. In general, 2 h after application of 1 µM IAA, levels of all metabolites except oxIAA-GE were significantly lower in auxin-starved cells compared with control (Figure 2, C). The metabolic profile of radioactively labeled IAA revealed several peaks corresponding to unknown metabolites in auxin-deprived cells. Overall lower levels of IAA metabolites in the auxin-starved cells are probably caused by lower influx of auxin from the media. Lower transcript and protein levels of several members of AUX/LAX auxin influx carriers (LAX1a, LAX1b, and LAX3) in auxin-deprived cells (Figure 3, F) confirm this hypothesis, but involvement of other mechanisms (e.g. rapid efflux of metabolites into the media or the presence of here undetected metabolites) cannot be excluded.
Next, we aimed to explain the most striking difference in IAA metabolites between the control and auxin-deprived BY-2 cells, namely, the abundance of oxIAA-Asp. Although formation of oxIAA-Asp in plants has already been reported (Östin et al., 1998), here we present a determination of the oxIAA-Asp level by LC-MS/MS in IAA-treated plant cells, as well as the level of endogenous oxIAA-Asp in plants. This metabolite is a product of amino acid conjugation and oxidation. GH3 genes, coding for the enzymes catalyzing amino acid conjugation, were identified throughout the plant kingdom and they are involved in regulation of many developmental processes (reviewed for example in Staswick, 2005; Casanova-Sáez and Voß, 2019). Strong expression induction of many GH3 genes after auxin treatment classified GH3 among early auxin responsive genes, but an opposite regulation of some members was reported, for example during fruit development (Matsuo et al., 2018). Our transcriptomic and proteomic analysis identified the most abundant GH3 member (NtGH3.6dT, homolog of Arabidopsis GH3.6) to be upregulated in auxin-free conditions. The same regulation pattern was observed for other homologs of AtGH3.6, while homologs of GH3.1 showed significant downregulation in auxin-starved cells (Figure 3, F).
Complex regulation of auxin amino acid conjugation cannot explain the differences in auxin-dependent IAA metabolism; therefore, we focused on the oxidation pathway represented by tobacco homologs of recently identified DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1) (Porco et al., 2016; Zhang et al., 2016). Auxin metabolic profiling in DAO1 knock-out and knock-down tobacco and Arabidopsis mutants confirmed increased levels of IAA-Asp (as already reported in Mellor et al., 2016). Moreover, significantly lower abundance of oxIAA-Asp in mutants compared with WT controls and the enzyme assay of AtDAO1-expressing bacteria provide direct evidence of DAO1-mediated oxidation of IAA-Asp. Our results thus provide insight into the biological aspects of DAO1 function while helping to elucidate the coordination of oxidative and conjugating pathways of auxin metabolism.
The sequence of amino acid conjugation and oxidation has not yet been explored, but the data indicate that the oxIAA-Asp-forming pathway starts with amino acid conjugation followed by DAO1-mediated oxidation. A recent study describes allosteric regulation of OsDAO, during which the enzyme multimerizes in the presence of IAA (Takehara et al., 2020). The dimeric form showed a higher reaction rate and affinity toward IAA compared with the OsDAO monomer; however, the in vitro-measured IAA oxidation rate of the “active” form was still about four-fold lower compared with the reaction rate of GH3-catalyzed IAA amino acid conjugation.
While oxIAA-Asp seems to be the most abundant IAA metabolite in tobacco BY-2 cells, Arabidopsis plants mainly convert IAA into the oxIAA-GE form by glycosylation of oxIAA (Tanaka et al., 2014; Porco et al., 2016). To conclude, to determine whether this difference is caused by species-specific capacity of the auxin metabolic machinery, additional metabolic profiles (including levels of oxidated amino acid conjugates) of diverse plant species, as well as tissues, will be required. The large content of oxIAA-Asp in IAA-treated tobacco cells, as well as that of oxIAA-GE in Arabidopsis plants, both formed during DAO1-dependent reactions, indicating a more important role of DAO1 during auxin homeostasis than previously thought.
Furthermore, it is obvious that a final resolution on the balance of oxidation and conjugation rates and ratios can be declared only after all involved pathways and metabolites are known. It has been, for instance, generally accepted that IAA-Asp and IAA-Glu conjugates cannot be hydrolyzed, and thus do not represent storage forms of auxin (LeClere et al., 2002). Our results raise an interesting hypothesis that rapid DAO1-mediated oxidation, by depleting the pool of amino acid conjugates, might prevent their possible cleavage by hydrolases. Further research into the specificity of the DAO1 and IAA metabolic conversion network in dao1 mutants is needed to validate our hypothesis and bring further insights in this field of auxin homeostasis.
Conclusions
Here, we show a multi-omical characterization of tobacco BY-2 cell culture cultivated in the presence of exogenously supplemented synthetic auxin 2,4-D in comparison with cell culture cultivated for 48 h in auxin-free conditions. Both conditions were characterized by very low levels of auxin metabolites as well as insignificant transcription of auxin biosynthetic genes, suggesting no or minimal auxin biosynthesis under both conditions. By contrast, we demonstrated significant differences in expression of auxin-metabolizing enzymes at transcript and proteins levels. Knockout and knockdown mutants of tobacco DAO1 verified its importance in IAA metabolism and revealed its role in conversion of IAA-Asp into oxIAA-Asp.
Materials and methods
Plant material and chemicals
Tobacco cell line BY-2 (N. tabacum L. cv Bright Yellow 2) was maintained in liquid Murashige and Skoog (MS) medium (3% sucrose (w/v), 4.3 g/L MS salts, 100 mg/L myo-inositol, 1 mg/L thiamin, 0.2 mg/l 2,4-D, and 200 mg/l KH2PO4; pH 5.8), in the dark, at 27°C, under continuous shaking (150 rpm; orbital diameter 30 mm), and was sub-cultured every 7 d. Auxin-free-cultured cells were inoculated to medium lacking 2,4-D. Transgenic cells and calli were maintained on the same medium supplemented with 100 µg/mL cefotaxime and 20 µg/mL hygromycin (BY-2 siDAO1) or 50 µg/mL kanamycin (BY-2 crisprDAO1). Aliquots (2 mL) of 2-d-old cell suspensions were filtered, immediately frozen in liquid nitrogen, and stored at −80°C until extraction or use for further treatments.
Arabidopsis thaliana ecotype Columbia 0 and dao1 T-DNA insertion mutant (obtained from Nottingham Arabidopsis Stock Centre, SALK_093162) were grown on square agar plates (4.4 g/L MS, 10 g/L sucrose, and 10 g/L plant agar, pH 5.7). Plates were stratified for 4 d and placed vertically in long-day conditions (16-h light/8-h dark) at 22°C. Seven-day-old whole seedlings were collected in five replicates and weighed (approximately 5 mg per replicate), immediately frozen in liquid nitrogen, and stored at −80°C until extraction.
Unless stated otherwise, all chemicals and kits were obtained from Sigma–Aldrich Inc.
RNA isolation and sequencing analysis
Total RNA was isolated from 50 to 100 mg of BY-2 cells using RNeasy Plant Mini kit (Qiagen) and treated with DNA-Free kit (Thermo Fischer Scientific). RNA purity, concentration, and integrity were evaluated on 0.8% agarose gels (v/w) and by the RNA Nano 6000 Assay Kit using Bioanalyzer instrument (Agilent Technologies). For RNA-seq analysis, approximately 5 µg of RNA isolated from seven control and three auxin-starved replicates were submitted for the service procedure provided by GATC-Biotech Company. The analysis resulted in at least 20 million 50 bps long reads. Rough reads were quality-filtered using FastQC and Trimmomatic (parameters: leading: 3, trailing: 3, slidingwindow: 4:20, minlen: 40). Transcript abundances (transcripts per million—TPM) were determined using Salmon (Patro et al., 2017) with parameters –seqBias, –gcBias, –numBootstraps 30. Index was built from N. tabacum v1.0 cDNA dataset (Edwards et al., 2017). Visualization and quality control of data analysis were done using R packages Factoextra and FactoMineR (Lê et al., 2008). DEGs were selected using sleuth (version 0.29.0) and DeSeq2 (version 1.18.0) packages in R (Love et al., 2014; Pimentel et al., 2017). Transcripts with q-value ≤ 0.05 determined by both packages and log2 fold change ≥1 (upregulated) or ≤ −1 (downregulated) were considered to be significantly differentially expressed. Gene ontology analysis (statistical overrepresentation test) was done using Panther Classification System (Mi et al., 2019).
Proteomic analysis
Plant cell callus was lysed with a combination of sonication and high temperature detergent-assisted lysis. Proteins were trypsin-digested, and nanoflow liquid chromatography was used for separation of the resulting peptides. Next, analysis of the samples by tandem mass spectrometry (nLC-MS2) on a Thermo Orbitrap Fusion (q-OT-qIT, Thermo Scientific) instrument was performed as described elsewhere (Erban et al., 2019). The mass spectrometry data were analyzed and quantified using MaxQuant software (version 1.5.3.8; Cox et al., 2014) with false discovery rate set to 1% for both proteins and peptides, with a minimum length specified at seven amino acids. The Andromeda search engine was used for the MS/MS spectra search against the tobacco protein dataset (Edwards et al., 2017), and normalized intensity values were then further processed using the Perseus software (Tyanova et al., 2016).
Genes and protein annotation
Annotation of genes and corresponding proteins followed the state of the art information on the genome of N. tabacum available at Sol Genomics Network under N. tabacum TN90 and N. tabacum v1.0 datasets (Sierro et al., 2014; Edwards et al., 2017). For enhanced clarity, selected genes related to auxin biosynthesis and auxin metabolism were named according to the closest homolog in A. thaliana. Endings of the name (S or T) correspond to the closest homolog in either N. sylvestris or N. tomentosiformis.
Reverse transcription quantitative PCR
Approximately 1 µg of DNAse-treated RNA was reverse-transcribed using M-MLV reverse transcriptase, RNase H-, point mutant (Promega). Quantitative real-time PCR was performed using GoTaq qPCR Master Mix (Promega) at 58°C annealing temperature on a LightCycler480 instrument (Roche). PCR efficiency was estimated using serial dilution of template cDNA. Tobacco aminopeptidase-like, fructokinase2, and ribokinase-like genes (GenBank accession numbers XM_016640300, KJ683760, and XR_001650512, respectively) were used as references in validation of control versus auxin-starved RNA-seq data. These were chosen for their stable transcriptions in the BY-2 line in our experimental setup. Relative transcript abundances were then calculated using the equation:
where effref and efftarget stand for the qPCR efficiencies of reference and target genes, respectively, and CPref and CPtarget stand for the respective crossing points of reference and targets genes. Positive transcript levels and the quality of PCR products were verified by melting curve analysis. Verification of transgenic lines by RT-qPCR was performed after induction of gene expression with 1 µM β-estradiol. DMSO-treated cells were used as a non-induced control. As genes are in allotetraploid N. tabacum usually present in highly homologous pairs reflecting N. tomentosiformis and N. sylvestris parental genomes, primers for RT-qPCR were designed to be specific to both copies. Their sequences are shown in Supplemental Table S5.
IAA metabolic profiling
Two-day-old control BY-2 cells and BY-2 cells cultivated in auxin-free medium were used for IAA metabolite profiling in experiments aimed at endogenous hormone determination. To exclude the possibility of secretion of auxin metabolites, the cultivation media were also analyzed (in both conditions). In auxin feeding experiments, control and auxin-free-cultured BY-2 cells were incubated with 20 nM 3H-IAA (specific activity 25 Ci/mmol, American Radiolabeled Chemicals) for 10 min and 4 h, or with 1 µM non-radiolabeled IAA for 2 h. Transgenic siDAO1 and crisprDAO1 BY-2 cell lines were treated with 1 µM non-labeled IAA for 2 h.
Auxin metabolites were extracted and purified according to Ivanov Dobrev and Kamínek (2002) and Dobrev et al. (2017), followed by a quantitative analysis on LCMS (endogenous hormone analysis, IAA-feeding experiments) or by a separation on HPLC and detection on Ramona 2000 flow-through radioactivity detector (3H-IAA feeding experiments). The metabolites were separated on Kinetex EVO C18 column (2.6 µm, 150 × 2.1 mm, Phenomenex). Mobile phases consisted of A) 5 mM ammonium acetate in water and B) 95/5 acetonitrile/water (v/v). The following gradient program was applied: 5% B (v/v) in 0 min, 7% B (v/v) in 0.1–5 min, 10 to 35% (v/v) in 5.1–12 min, 100% B (v/v) at 13–14 min, and 5% B (v/v) at 14.1 min. The LCMS system consisted of UHPLC 1290 Infinity II (Agilent Technologies) coupled to 6495 Triple Quadrupole mass spectrometer (Agilent Technologies). MS analysis was done in the MRM mode, using isotope dilution method. Data acquisition and processing was done with Mass Hunter software version B.08 (Agilent Technologies).
Measurement of endogenous auxin metabolites in A. thaliana Col-0 and dao1-1 plants was performed following the methods described by Pěnčík et al. (2018). Samples were extracted with 1 mL of 50 mM phosphate buffer (pH 7.0) containing 0.1% sodium diethyldithiocarbamate (v/w) and a mixture of stable-isotope-labeled internal standards. Two hundred microliter aliquots of the extract was acidified with 1 M HCl to pH 2.7 and purified by in-tip micro solid phase extraction (in-tip µSPE). After evaporation under reduced pressure, the samples were analyzed using the HPLC system 1260 Infinity II (Agilent Technologies) equipped with a Kinetex C18 column (50 × 2.1 mm, 1.7 µm; Phenomenex) and linked to 6495 Triple Quad detector (Agilent Technologies). Auxin levels were quantified using stable-isotope-labeled internal standards as a reference.
Preparation of constructs and transformation of BY-2 cells
All vectors for transformation were prepared using GoldenBraid 2.0 cloning system (Sarrion-Perdigones et al., 2013) and their sequences are listed in Supplemental Data S4. Basic DNA parts were selected from GoldenBraid 2.0 kit from Diego Orzaez (Addgene kit # 1000000076) or MoClo Toolkit, from Sylvestre Marillonnet (Addgene kit # 1000000044). Specific parts (target gRNA or siRNA sequence) were prepared in the Laboratory of Hormonal Regulations in Plants IEB or Laboratory of Virology IEB, Prague. The cloning was performed using Type IIS restriction enzymes BsmBI, BsaI (TermoFischer Scientific) and T4 ligase (Promega). Transformation of BY-2 cells was done by co-cultivation with A. tumefaciens strain GV2260 (An et al., 1985). Transgenic lines were harvested after 4 weeks, cultured on solid media with kanamycin, and tested for the presence of CRISPR-Cas9 activity. Suspension cultures were established from selected lines validated for NtDAO1 mutation rates.
NtDAO1 enzymes in the tobacco genome are encoded by two very similar genes (95.8% pairwise identity) reflecting N. sylvestris and N. tomentosiformis parental genomes. We labeled these copies according to a rule described above as NtDAO1T (GenBank accession number XM_016591349.1) and NtDAO1S (GenBank accession number XM_016618186.1). For CRISPR-Cas9 editing of NtDAO1 enzymes, a vector containing 35S:hCas9:NosT transcriptional unit (TU) and two guide RNAs (guide1: 5′-AGACCCAAAGCCTCATAAAGTGG-3′, guide2: 5′-CTACAGCTTCTGGAGTGAAGTGG-3′) TUs was used. (Both guide RNA sequences were specific to both DAO1 copies.) Verification of the crisprDAO1 line was done by PCR amplification of the CRISPR-targeted DAO1 genomic sequence followed by cloning of the product into a pJET1.2 vector and Sanger sequencing. Since CRISPR-Cas9 BY-2 mutants do not enable a simple way to cross out Cas9 machinery, stability of the crisprDAO1 BY-2 mutants was monitored in two independent lines in several subsequent inoculation intervals.
For inducible silencing of NtDAO1 transcription, a β-estradiol-inducible expression (Zuo et al., 2000) of NtDAO1-specific (GenBank ID XM_016591349, region 549-785) hairpin RNA gene construct was prepared. BY-2 cell transformation was performed as described above. Ten independent lines were tested by RT-qPCR for the expression of NtDAO1. A line with stable downregulation of the target was used for auxin metabolic profiling.
GFP-tagged NtDAO1 constructs were generated by fusion of a GFP-coding sequence to the C-terminus of the coding sequence of NtDAO1T (GenBank accession number XM_016591349.1). Regulation of DAO1-GFP expression was controlled by the native promoter region of NtDAO1T (pDAO1, 2000 bps upstream from the start codon) in pDAO1:DAO1-GFP line, by synthetic promoter G10-90 in G10-90:DAO1-GFP line or by CaMV-35S promotor from GoldenBraid 2.0 kit in 35S:DAO1-GFP line.
Light microscopy
For in vivo microscopy, a Zeiss LSM 880 inverted confocal laser scanning microscope with a 40× C-Apochromat objective (NA = 1.2 W) was used (http://www.zeiss.com/). GFP fluorescence was detected either in single images or in z-stacks with acquisition parameters: excitation 488 nm (2.6% laser), emission 490–570 nm, pinhole 0.92 AU, pixel dwell 7.91 µm, and gain 750. Maximum intensity projection of z-stacks was created using ZEN black software (Zeiss).
Western blot analysis
For protein extraction, leaf disc samples were homogenized and resuspended in extraction buffer (50mM Tris pH 6.8; 5% (v/v) glycerol; 1.5% (w/v) insoluble poly-vinylpolypyrrolidone; 150 mM KCl; 5 mM Na EDTA; 5 mM Na EGTA; 1 mM DTE; 50 mM NaF; 20 mM beta-glycero phosphate; 1 mM benzamidine; 1 mM PMSF; and 1 Roche Complete Mini Protease Inhibitor tablet per 10 mL). Samples were cleared by centrifugation (2100g for 2 min). The supernatant was mixed 1:1 with 2× Laemmli sample loading buffer (Bio-Rad), separated by SDS PAGE with Mini-PROTEAN precast gels TGX Stain-Free (Bio-Rad), transferred to nitrocellulose membranes, blocked with 3% BSA solution (v/w), and probed with monoclonal anti-GFP (1:2000; Roche), followed by HRP-conjugated goat-anti-mouse IgG (1:30,000; Promega). For determination of protein amounts, we used the picture of the TGX Stain-Free showing total protein amount loaded.
Enzyme assay
Recombinant protein expression was carried out according to Brunoni et al. (2019). Briefly, AtDAO1-producing E. coli BL21 (DE3) bacteria were grown at 37°C in Luria Broth medium containing 50 μg/mL kanamycin to the mid-log phase, followed by IPTG induction at 20°C over-night. To test the enzymatic activity of recombinant AtDAO1 protein, overnight induced bacterial culture was supplemented with or without 0.1 mM IAA-Asp and with DAO cofactor mixture [5 mM 2-oxoglutarate, 5 mM ascorbate, 0.5 mM (NH4)2Fe(SO4)2] for 6 h at 20°C. One milliliter of bacterial culture was harvested in three technical and three biological replicates 6 h after incubation with and without the substrate, centrifuged to pellet (4000 rpm for 5 min), and supernatant fractions were analyzed for oxIAA-Asp production by UHPLC-ESI-MS/MS as described in Brunoni et al. (2019). Samples from GFP-producing bacterial culture were used as negative control.
Accession numbers
Rough sequencing data are available through GEO Series accession number GSE160438 at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). Accession numbers of genes important for the study are presented in format protein acronym/GenBank accession/Arabidopsis homolog as follows: DAO1/XM_016591349.1/AT1G14130.1; GH3.1a/XM_016616759.1/AT2G14960.1; GH3.6a/XM_016643055.1/AT5G54510.1; GH3.6b/XM_016643194.1/AT5G54510.1; GH3.6c/XM_016606102.1/AT5G54510.1; GH3.6d/XM_016606101.1/AT5G54510.1; LAX1a/XM_016646001.1/AT2G38120.1; LAX1c/XM_016619322.1/AT2G38120.1; PILS5a/XM_016657384.1/AT2G17500.4; PILS6a/XM_016596938.1/AT5G01990.1; PIN11/XM_016622595.1/AT1G73590.1; PIN2/XM_016643000.1/AT5G57090.1; UGT3/XM_016625843.1/AT3G21790.1. Additional meta information regarding the genes are available in corresponding datasets (Supplemental Data S2, S3).
Supplemental data
The following supplemental materials are available.
Supplemental Figure S1. Hierarchical clustering analysis, supplemental quality control to transcriptomic and proteomic profiling.
Supplemental Figure S2. Validation of RNA-seq data by RT-qPCR.
Supplemental Figure S3. Subcellular localization of DAO1-GFP in BY-2 cells.
Supplemental Figure S4. Western blot analysis of DAO1-GFP.
Supplemental Figure S5. Quantification of NtDAO1 transcription in the siDAO1 line.
Supplemental Table S1. List of RNA samples submitted for RNA-seq including quality (RIN value) and obtained read counts (50 bps single read sequencing).
Supplemental Table S2. Summary of RNA-seq analysis.
Supplemental Table S3 . Complete results of GO-slim biological process terms from PANTHER overrepresentation test of transcripts upregulated in auxin-starved cells.
Supplemental Table S4 . Complete results of GO-slim biological process terms from PANTHER overrepresentation test of transcripts downregulated in auxin-starved cells.
Supplemental Table S5. List of RT-qPCR primers.
Supplemental Data S1. Elution time and corresponding radioactivity of standard 3H-IAA after separation on HPLC and detection on Ramona 2000 flow-through radioactivity detector.
Supplemental Data S2 . Complete dataset of transcript abundances (TPM) and results of statistical evaluation (DeSeq and Sleuth algorithms).
Supplemental Data S3. Complete dataset of proteins identified by LC-MS/MS analysis.
Supplemental Data S4. Sequences of plasmids used for plants and cell culture transformations.
Funding
The work was supported by The European Regional Development Fund-Project “Centre for Experimental Plant Biology” (No. CZ.02.1.01/0.0/0.0/16_019/0000738) and by the Ministry of Education, Youth and Sports of the Czech Republic (Grant No. LTC18047). Computational resources were supplied by the Ministry of Education, Youth and Sports of the Czech Republic under the Projects CES-NET (Project No. LM2015042) and CERIT-Scientific Cloud (Project No. LM2015085) provided within the program Projects of Large Research, Development and Innovations Infrastructures. Microscopy was done at Imaging Facility of the Institute of Experimental Botany AS CR supported by the MEYS CR (LM2018129).
Conflict of interest statement. None declared.
Supplementary Material
K.Mü., J.P., and P.I.D. conceived the original screening and research plans. P.I.D., K.Ho., A.P., O.N., and R.F. performed the metabolic analyses. Z.V., L.H., and K.Mü. prepared the material and measured transcript levels. K.Ma. and T.M. performed the localization experiments. K.Mü. and P.H. performed the bioinformatics analyses. F.B. performed the enzyme assay. K.Ha. and K.R. participated in the protein analysis. K.Mü. wrote the manuscripts with contributions from P.H., K.Ho., and J.P. K.Mü. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Karel Müller (muller@ueb.cas.cz).
References
- Adamowski M, Friml J (2015) PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27: 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Watson BD, Stachel S, Gordon MP, Nester EW (1985) New cloning vehicles for transformation of higher plants. EMBO J 4: 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi Y, Tanaka K, Cook SD, Hayashi KI, Kasahara H (2020) GH3 auxin-amido synthetases alter the ratio of indole-3-acetic acid and phenylacetic acid in Arabidopsis. Plant Cell Physiol 61: 596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, et al. (2012) A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485: 119–122 [DOI] [PubMed] [Google Scholar]
- Brunoni F, Collani S, Šimura J, Schmid M, Bellini C, Ljung K (2019) A bacterial assay for rapid screening of IAA catabolic enzymes. Plant Methods 15: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova-Sáez R, Voß U (2019) Auxin metabolism controls developmental decisions in land plants. Trends Plant Sci 24: 741–754 [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrev PI, Hoyerová K, Petrášek J (2017) Analytical determination of auxins and cytokinins. Methods Mol Biol 1569: 31–39 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Fernandez-Pozo N, Drake-Stowe K, Humphry M, Evans AD, Bombarely A, Allen F, Hurst R, White B, Kernodle SP, et al. (2017) A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erban T, Sopko B, Talacko P, Harant K, Kadlikova K, Halesova T, Riddellova K, Pekas A (2019) Chronic exposure of bumblebees to neonicotinoid imidacloprid suppresses the entire mevalonate pathway and fatty acid synthesis. J Proteomics 196: 69–80 [DOI] [PubMed] [Google Scholar]
- Hošek P, Kubeš M, Laňková M, Dobrev PI, Klíma P, Kohoutová M, Petrášek J, Hoyerová K, Jiřina M, Zažímalová E (2012) Auxin transport at cellular level: New insights supported by mathematical modelling. J Exp Bot 63: 3815–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov Dobrev P, Kamínek M (2002) Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J Chromatogr A 950: 21–29 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Lim EK, Li Y, Kowalczyk M, Sandberg G, Hogget J, Ashford DA, Bowles DJ (2001) Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F (2008) FactoMineR: An R package for multivariate analysis. J Stat Softw 25: 1–18 [Google Scholar]
- LeClere S, Tellez R, Rampey RA, Matsuda SPT, Bartel B (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem 277: 20446–20452 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J (2014) Auxin and the Interaction Between Plants and Microorganisms. In Zažímalová E., Petrášek J., Benková E., eds, Auxin Its Role Plant Dev. Springer; Vienna, Vienna, pp 413–434 [Google Scholar]
- Ludwig-Müller J (2011) Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot 62: 1757–1773 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo S, Kikuchi K, Nagasuga K, Ueno H, Imanishi S (2018) Transcriptional regulation of auxin metabolic-enzyme genes during tomato fruit development. Sci Hortic (Amsterdam) 241: 329–338 [Google Scholar]
- Mellor N, Band LR, Pěnčík A, Novák O, Rashed A, Holman T, Wilson MH, Voß U, Bishopp A, King JR, et al. (2016) Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc Natl Acad Sci USA 113: 11022–11027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2019) PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47: D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, Petrášek J, Zhang J, Gaykova V, Stierhof YD, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Müller K, Hošek P, Laňková M, Vosolsobě S, Malínská K, Čarná M, Fílová M, Dobrev PI, Helusová M, Hoyerová K, et al. (2019) Transcription of specific auxin efflux and influx carriers drives auxin homeostasis in tobacco cells. Plant J 100: 627–640 [DOI] [PubMed] [Google Scholar]
- Östin A, Kowalyczk M, Bhalerao RP, Sandberg G (1998) Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol 118: 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S, Weijers D (2016) Q&A: Auxin: The plant molecule that influences almost anything. BMC Biol 14: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pěnčík A, Casanova-Sáez R, Pilařová V, Žukauskaitė A, Pinto R, Luis Micol J, Ljung K, Novák O (2018) Ultra-rapid auxin metabolite profiling for high-throughput mutant screening in Arabidopsis. J Exp Bot 69: 2569–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peret B, Swarup K, Ferguson A, Seth M, Yang Y, Dhondt S, James N, Casimiro I, Perry P, Syed A, et al. (2012) AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wiśniewska J, Tadele Z, Kubeš M, Čovanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science (80-) 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L (2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690 [DOI] [PubMed] [Google Scholar]
- Porco S, Pěnčík A, Rasheda A, Vo U, Casanova-Sáez R, Bishopp A, Golebiowska A, Bhosale R, Swarupa R, Swarup K, et al. (2016) Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc Natl Acad Sci USA 113: 11016–11021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reemmer J, Murphy A (2014) Intercellular transport of auxin. In Zažímalová E., Petrášek J., Benková E., eds, Auxin Its Role Plant Dev. Springer Vienna, Vienna, pp 75–100 [Google Scholar]
- Sakai A, Miyazawa Y, Saito C, Nagata N, Takano H, Hirano H-Y, Kuroiwa T (1999) Amyloplast formation in cultured tobacco cells. III Determination of the timing of gene expression necessary for starch accumulation. Plant Cell Rep 18: 589–594 [Google Scholar]
- Sanchez Carranza AP, Singh A, Steinberger K, Panigrahi K, Palme K, Dovzhenko A, Dal Bosco C (2016) Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci Rep 6: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Vazquez-Vilar M, Palací J, Castelijns B, Forment J, Ziarsolo P, Blanca J, Granell A, Orzaez D (2013) Goldenbraid 2.0: A comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol 162: 1618–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Kleine-Vehn J (2019) PIN-FORMED and PIN-LIKES auxin transport facilitators. Development 146: dev168088. [DOI] [PubMed]
- Sierro N, Battey JND, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov N V. (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Skůpa P, Viaene T, Zwiewka M, Tejos R, Klíma P, Čarná M, Rolčík J, De Rycke R, Moreno I, et al. (2016) PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol 211: 65–74 [DOI] [PubMed] [Google Scholar]
- Staswick PE (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell Online 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerszen JB, Szczyglowski K, Bandurski RS (1994) iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265: 1699–1701 [DOI] [PubMed] [Google Scholar]
- Takehara S, Sakuraba S, Mikami B, Yoshida H, Yoshimura H, Itoh A, Endo M, Watanabe N, Nagae T, Matsuoka M, et al. (2020) A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin. Nat Commun 11: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Hayashi KI, Natsume M, Kamiya Y, Sakakibara H, Kawaide H, Kasahara H (2014) UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in arabidopsis. Plant Cell Physiol 55: 218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740 [DOI] [PubMed] [Google Scholar]
- Westfall CS, Sherp AM, Zubieta C, Alvarez S, Schraft E, Marcellin R, Ramirez L, Jez JM (2016) Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc Natl Acad Sci USA 113: 13917–13922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winicur ZM, Zhang GF, Staehelin LA (1998) Auxin deprivation induces synchronous Golgi differentiation in suspension-cultured tobacco BY-2 cells. Plant Physiol 117: 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lin JE, Harris C, Campos Mastrotti Pereira F, Wu F, Blakeslee JJ, Peer WA (2016) DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc Natl Acad Sci USA 113: 11010–11015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peer WA (2017) Auxin homeostasis: the DAO of catabolism. J Exp Bot 68: 3145–3154 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Chua N-H (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.