A bryophyte-specific gene regulates apical dominance, leaf length, plasmodesma frequency, auxin distribution pattern and governs overall gametophore development in moss.
Abstract
Convergent evolution of shoot development across plant lineages has prompted numerous comparative genetic studies. Though functional conservation of gene networks governing flowering plant shoot development has been explored in bryophyte gametophore development, the role of bryophyte-specific genes remains unknown. Previously, we have reported Tnt1 insertional mutants of moss defective in gametophore development. Here, we report a mutant (short-leaf; shlf) having two-fold shorter leaves, reduced apical dominance, and low plasmodesmata frequency. UHPLC-MS/MS-based auxin quantification and analysis of soybean (Glycine max) auxin-responsive promoter (GH3:GUS) lines exhibited a striking differential auxin distribution pattern in the mutant gametophore. Whole-genome sequencing and functional characterization of candidate genes revealed that a novel bryophyte-specific gene (SHORT-LEAF; SHLF) is responsible for the shlf phenotype. SHLF represents a unique family of near-perfect tandem direct repeat (TDR)-containing proteins conserved only among mosses and liverworts, as evident from our phylogenetic analysis. Cross-complementation with a Marchantia homolog partially recovered the shlf phenotype, indicating possible functional specialization. The distinctive structure (longest known TDRs), absence of any known conserved domain, localization in the endoplasmic reticulum, and proteolytic cleavage pattern of SHLF imply its function in bryophyte-specific cellular mechanisms. This makes SHLF a potential candidate to study gametophore development and evolutionary adaptations of early land plants.
Introduction
Physcomitrium patens (previously known as Physcomitrella patens), a model moss, is extensively studied to understand the evolution and development of the gametophytic generation (Kammerer et al\., 2014). In bryophytes, this generation is photosynthetic, long-lived, and has diverse tissue types. This dominant generation of moss consists of filamentous tissues such as chloronema, caulonema, buds, and shoot-like gametophores (Kofuji and Hasebe, 2014). The leaf-like organs, known as phyllids (hereafter leaves), are spirally arranged around the gametophores. Though the morphological features of moss and certain liverwort gametophores closely resemble the flowering plant shoot, they are not analogous structures. Since gametophores and shoots of flowering plant have evolved independently, the underlying genetic network governing their development is distinct (Fujita et al., 2008; Sakakibara et al., 2008). However, they exhibit similar developmental responses to hormones such as auxin (Coudert et al., 2015).
A striking similarity between gametophore and flowering plant shoots is the apical dominance phenomenon (Fujita et al., 2008); auxin synthesized at the shoot apex controls the dormancy of axillary buds (Cline, 1997). Decapitation of the shoot apex removes the auxin source and promotes axillary bud outgrowth; a phenotype that can be suppressed upon application of exogenous auxin to the apex (Thimann and Skoog, 1933). Similar decapitation experiments in moss showed that the gametophore apex inhibits branching, and exogenous auxin can maintain the inhibitory effect (Von Maltzahn, 1959; Coudert et al., 2015). In flowering plants, polar auxin transport (PAT) governs apical dominance. However, PAT has not been detected in P. patens gametophores (Fujita et al., 2008). Previously, Han et al. (2014) showed that auxin transport through plasmodesmata (PD) influences a phototropic response in the Arabidopsis hypocotyl (Han et al\., 2014). Later, Coudert et al. (2015) proposed a model that considered the gametophore apex as an auxin source and its diffusion through PD instead of PAT in moss. Upon application of the callose synthesis inhibitor, 2-deoxy-glucose (DDG), the authors observed reduced gametophore branching, presumably because of higher auxin transport owing to less callose deposition at the PD. Moreover, analysis of moss PIN mutants revealed their lack of significant involvement in apical dominance. Though the major mode of auxin transport is not conserved between the moss gametophore and the flowering plant shoot, other pathways such as auxin metabolism (TRYPTOPHAN AMINOTRANSFERASE-RELATED (TARs), YUCCA (YUC), SHORT INTERNODE/STYLISH (SHI/STY)), signaling (TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX (TIR1/AFB), Auxin/INDOLE-3-ACETIC ACID (Aux/IAA)), and the response (AUXIN RESPONSE FACTOR (ARF)) are conserved (Thelander et al., 2018). For example, the soybean auxin response promoter (GH3:GUS) has been shown to faithfully report the cellular auxin concentrations (Bierfreund et al., 2003; Fujita et al., 2008). Auxin also exerts profound effects on gametophore development. The role of auxin in gametophore initiation and leaf and rhizoid development is well established (Thelander et al., 2018).
As a consequence of convergent evolution in shoot development, the function of several crucial gene regulatory networks of flowering plants is not conserved in moss gametophores. For example, class-I KNOX genes are known as master regulators of flowering plant shoot apical meristem (Vollbrecht et al., 1991) but have no role in gametophore development (Sakakibara et al., 2008). However, AP2-type transcription factors, which are known to regulate flowering plant root meristem and embryogenesis, are indispensable for gametophore apical cell formation (Boutilier et al., 2002; Aida et al., 2004). Lack of functional conservation suggests that bryophytes may have acquired novel mechanisms through clade-specific genes. For example, the PDLP5 gene regulates PD permeability in flowering plants (Lee et al., 2011), but the moss genome lacks the PDLP gene family (Brunkard and Zambryski, 2017). Stevenson et al. (2016) identified an ABA NON-RESPONSIVE (ANR) gene specific to non-vascular land plants. The ANR gene codes for a protein with a unique domain architecture and functions as an ABA response regulator. However, the mode of action of its PAS domain remains unknown. Our knowledge of bryophyte-specific genes and their contribution to gametophore development is extremely limited. Forward genetic screens can enable the identification and functional characterization of clade-specific genes; however, understanding their molecular mechanism remains a challenge due to their unique features.
Using a Tnt1-mediated forward genetics approach, we previously reported insertional mutants of P. patens defective in moss gametophore development (Mohanasundaram et al., 2019). Here, we describe a Tnt1 insertional mutant (short-leaf; shlf) in moss that demonstrates the involvement of a novel bryophyte-specific gene in apical dominance, leaf development, PD frequency, and auxin distribution pattern in the moss gametophore. Based on a comprehensive phenotypic analysis, differential auxin accumulation and response at the gametophore apex, and TEM-based PD frequency determination, we show that the reduced apical dominance in shlf gametophores is associated with impaired auxin distribution. Whole-genome sequencing (WGS) and functional characterization of candidate genes revealed that a unique bryophyte-specific gene, SHLF, is responsible for the shlf (short-leaf) phenotype. Phylogenetic analysis established that SHLF represents a novel family of tandem direct repeat (TDR)-containing proteins that are conserved only among mosses and liverworts. However, a distantly related Marchantia SHLF homolog could only partially rescue the shlf phenotype, possibly due to functional specialization. In summary, we demonstrate that the novel bryophyte-specific gene SHLF regulates apical dominance, leaf length, PD frequency, auxin distribution pattern, and governs overall gametophore development in moss.
Results
The Tnt1 insertional mutant (short-leaf) has short leaves and exhibits reduced apical dominance
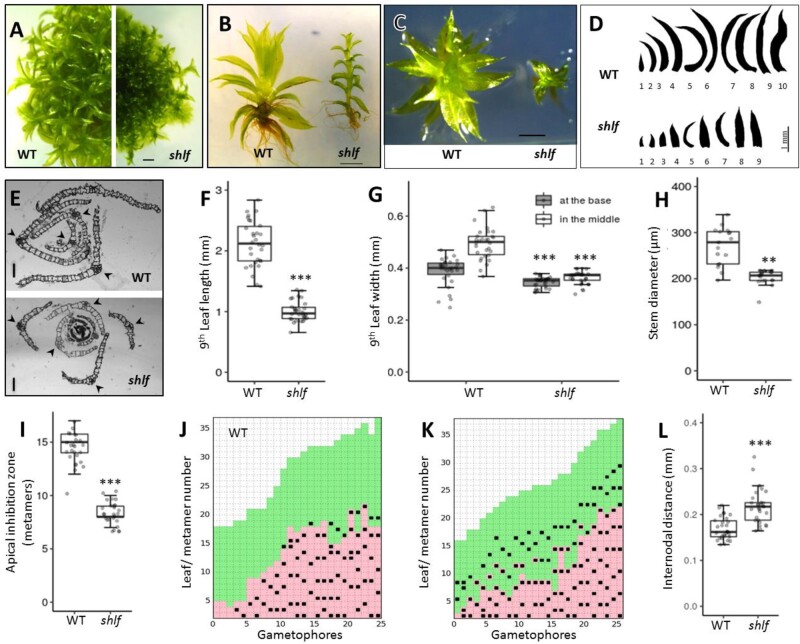
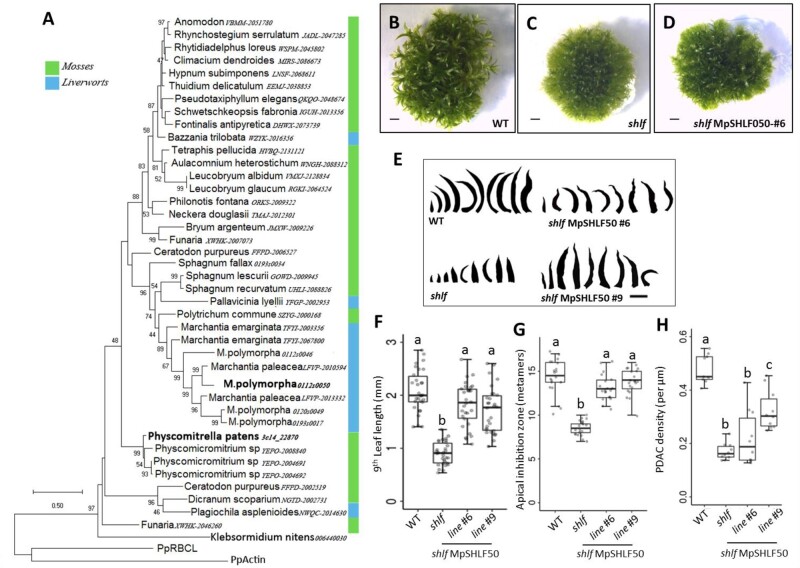
The shlf mutant was isolated from a morphological screen on the Tnt1 retrotransposon insertional mutant population of the moss P. patens (Mohanasundaram et al., 2019). As the name suggests, the mutant gametophores produced two-fold shorter leaves than wild-type (WT) (Figure 1, A–F). When we compared the first nine leaves of WT and shlf gametophores, it was observed that the leaf length deviation starts as early as the second leaf. The leaf length of both gametophores saturates around the 9th leaf stage (Figure 1, D and Supplemental Figure S1, A). Hence, we chose the ninth leaf to represent mature gametophore leaves in further experiments. shlf leaf length and width at the center of the proximo-distal axis were reduced by a factor of 2, but the width at the base of the leaf remained unaffected (Figure 1, E–G). Hence, shlf leaves are lanceolate-shaped (Supplemental Figure S1, C). Reduction in leaf cell number along the proximo-distal, medio-lateral axes (Supplemental Figure S1, B), and the whole leaf area (Supplemental Figure S1, D–H) together contributed to the shorter length and width of shlf leaves. Similarly, the shlf gametophore diameter was also reduced due to low cell expansion, whereas the number of layers and the number of cells in the layers remained unchanged (Figure 1, H and Supplemental Figure S1, J). Apart from the striking short-leaf phenotype, shlf gametophores exhibited reduced apical dominance, where branch initials began as early as eighth metamer (on an average) as opposed to 13th metamer in WT (Figure 1, I–K). In WT gametophores, new branch initials are observed mostly in the metamers with mature leaves and rhizoids (Figure 1, J). However, in shlf, branch initials are observed even in metamers with young leaves (Figure 1, K). We also noticed that the internodal distance of shlf gametophores has increased (Figure 1, L). These results indicate that several aspects of gametophore development, such as leaf size and shape, gametophore diameter, and branching pattern, are affected in the shlf mutant.
Figure 1.
Short-leaf (shlf) mutant exhibits pleiotropic phenotypes and suppressed growth of gametophores. A–D, Comparison of 3 weeks old WT and shlf colonies (A), gametophores (B and C), and leaf arrays (D). Scale bars, 1 mm. E, Transverse sections of WT and shlf gametophore apices showing the difference in the number of leaf blade cells. Arrow head marks the midrib flanked by single cell-layered leaf blades. Scale bars, 50 µm. F–H, WT and shlf 9th leaf length (n = 30) (F), width at the base as well as at the middle of medio-lateral axis (n = 30) (G) and stem diameter (n = 3) (H) were analyzed. I, The number of metamers from the apex till the first branch is known as apical inhibition zone (n = 26) was analyzed. J and K, Hinton diagram showing differences in the branching pattern between WT (J) and shlf (K) gametophores. The lowest and highest metamer numbers represent the base and the apex of the gametophore, respectively. Each branch/branch initial is marked as a black square. The young leaves are marked as green box and mature leaves with rhizoids are marked as pink box. L, The internodal distance (n = 30) of WT and shlf gametophores was also analyzed. All statistical analysis were performed using Mann–Whitney Wilcox test and P-value <0.001 and <0.0001 were marked as ** and ***, respectively.
The shlf gametophore exhibits a differential auxin distribution pattern and low PD frequency
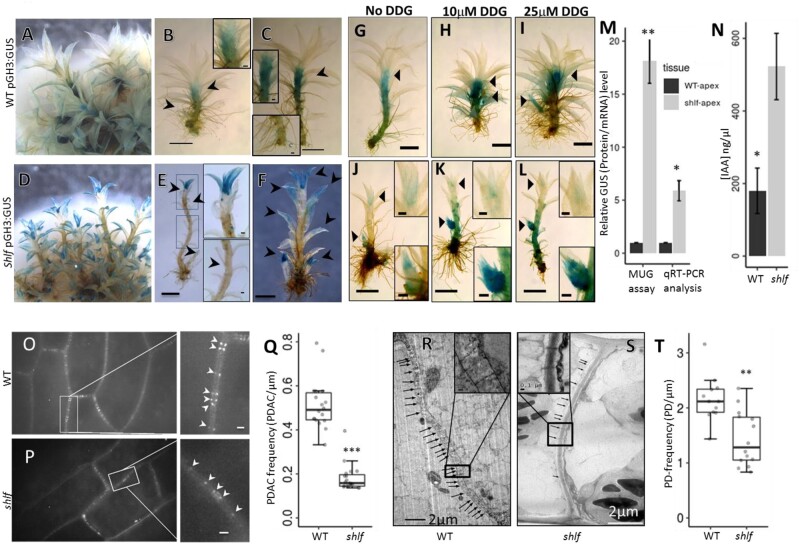
The reduced apical dominance phenotype of the shlf mutant prompted us to study the auxin accumulation pattern. A widely used auxin-responsive soybean GH3 promoter–reporter (GH3:GUS) construct (Fujita et al., 2008) was transformed in WT and shlf backgrounds (Supplemental Figure S2). The transgenic lines had similar phenotypes to their respective parental lines. WT gametophores showed high GUS activity at the sub-apical region (Figure 2, A–C). By contrast, shlf gametophores had high GUS activity at the apex, side branch initials, young leaves, and leaf tips; however, the sub-apical region had no GUS activity (Figure 2, D–F). To study the impact of symplastic auxin transport on the GH3:GUS expression pattern in shlf gametophores, we treated the gametophores with 10 and 25 µM DDG, a callose biosynthesis inhibitor. The DDG treatment did not produce any drastic change in the GUS activity of WT GH3:GUS lines (Figure 2, G–I). However, in the case of the shlf GH3:GUS lines, DDG treatments resulted in an increased GUS activity in the sub-apical region of the gametophore (Figure 2, J–L). This effect was more pronounced in the side branches. 4-methyl-umbelliferyl-β-d-glucuronide (MUG) assay and RT-qPCR analysis of GUS affirmed that the shlf apex had greater soybean GH3:GUS activity than WT (Figure 2, M). Our UHPLC-MS/MS-based quantification of auxin further confirmed that the auxin response at the shlf apex is due to increased auxin accumulation (more than two-fold; Figure 2, N). The expression pattern of auxin metabolism genes (biosynthesis regulators (SHI1/2), catabolism genes (GH3-1), efflux carriers (PINs), and response regulators (IAAs)) in the shlf apex was comparable with WT (Supplemental Figure S1, I). These results suggest that increased auxin accumulation at the apex could explain the reduced apical dominance observed in shlf gametophores.
Figure 2.
Differential auxin distribution and low PD frequency in shlf mutant. A–F, Auxin responsive promoter (soybean GH3:GUS) expression showing differential auxin response in WT (A–C) compared with mutant gametophores (D–F). Arrow heads mark GUS activity in meristem and leaf (scale bars, 1 mm). Arrow heads indicate GUS-stained gametophores region. G–L, Effect of callose synthesis inhibitor (DDG) on auxin distribution pattern in WT and shlf. WT GH3:GUS (G–I) and shlf GH3:GUS lines (J–L) grown on BCDAT media containing 0, 10, and 25 μM of DDG, respectively. Inset (upper) represents GUS-stained gametophore apex whereas inset (lower) represents stained auxiliary branch initial. Scale bars, 1 mm. All scale bars in the inset represent 0.1 mm. M, Relative GUS protein and mRNA expression pattern in apical tissue of GH3:GUS lines of WT and the shlf background. N, The differential auxin accumulation between WT and shlf gametophore apices (n = 3) was quantified using UHPLC-MS/MS. O–Q, Aniline blue-based PDAC staining and quantification showed reduced PD frequency in shlf leaves. Figure in inset shows pairs of PDAC punctate (arrow head) corresponding to pairs of PD-neck regions in the cell wall. Scale bars, 0.5 µm. R–T, Transmission electron microscopy images of protonemal cell wall sections were used to quantify the PD frequency in WT and shlf. Scale bars, 2 µm. Figure in inset shows PD channels (arrow head) crossing the cell wall. Scale bars, 1 µm. All statistical analyses were performed using either Student’s t test (M and N) or Mann–Whitney Wilcox test (Q and T). P-value < 0.05, <0.001, and <0.0001 were marked as *, **, and ***, respectively. The error bars represent standard deviation.
One plausible explanation for increased auxin accumulation in the shlf apex is impaired PD-dependent auxin distribution. Aniline blue staining of PD-associated callose (PDAC; Zavaliev and Epel, 2015) and TEM-based PD quantification revealed significantly reduced PDAC (three-fold) and PD frequencies in shlf leaves and protonemal filaments, respectively (Figure 2, O–T and Supplemental Figure S3). These results highlight a possible link between increased auxin accumulation and low PD frequency phenotypes of shlf.
The SHLF gene (Pp3c14_22870) is the genetic cause for short-leaf phenotype
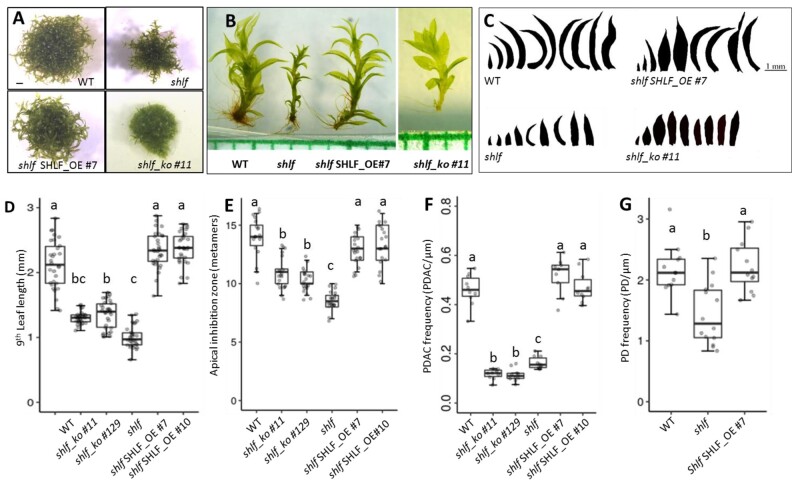
WGS of the shlf mutant and an in-house pipeline identified three Tnt1 and one T-DNA insertion in the shlf genome (Supplemental Table S1 and Supplemental Figure S4). Two of the three Tnt1 insertions were in genic regions (Pp3c14_22870; PpSHLF and Pp3c1_9390; PpEXTENSIN) and confirmed by genomic DNA PCR. Using RT-PCR, the insertion inside the coding region of Pp3c14_22870 (PpSHLF) was confirmed. Further, RT-qPCR expression analysis using N-terminal-specific primers of SHLF revealed that the SHLF expression in the mutant is reduced by 28-fold due to the Tnt1 insertion (Supplemental Figure S5, D). Moreover, C-terminal-specific primers of SHLF flanking the Tnt1 insertion failed to amplify SHLF transcript in the mutant (Supplemental Figure S5, D). Additionally, Pp3c1_9390 (PpEXTENSIN) transcripts were not detected either in mutant or WT (Supplemental Figures S5, S6). Moreover, PpEXTENSIN overexpression in the shlf mutant background did not revert the phenotype (Supplemental Figure S7). Notably, transgenic lines over-expressing PpSHLF in the shlf mutant (shlf SHLF_OE #7 and shlf SHLF_OE #10) recovered gametophore-related phenotypes (leaf length, width, apical dominance, internodal distance, and PDAC and PD frequency; Figure 3 and Supplemental Figures S3, S8, S10–S11). The knockout lines of PpSHLF in WT background (shlf_ko #11 and shlf_ko #129) phenocopied many of the mutant phenotypes (leaf length, apical dominance, and PDAC frequency; Figure 3 and Supplemental Figures S3, S9–S11). Overexpression and knockout studies showed that the Pp3c14_22870 gene (Supplemental Figure S12) is responsible for the shlf phenotype and was named SHORT-LEAF (SHLF).
Figure 3.
SHLF (Pp3c14_22870) OE and knockout (KO) rescues the mutant phenotype. A–C, Comparison of 3-week-old WT, shlf, SHLF OE line (shlf SHLF_OE #7), and knockout (ko) line (shlf_ko #11) colonies (A), gametophores (B), and leaf arrays (C) showing recovery and phenocopy of the mutant phenotype in OE and KO lines, respectively (scale bars, 1 mm). Leaf silhouettes were digitally extracted for comparison. D–G, Leaf (ninth) length (n = 30) (D), apical inhibition zone (n = 20) (E), aniline blue-based PDAC frequency of leaves (n = 10) (F), and transmission electron microscopy-based PD frequency of protonema (n = 12) (G) of WT, shlf, OE, and KO lines were analyzed and shows recovery and phenocopy of the mutant phenotype in OE and KO lines, respectively. Statistical analyses were performed using either ANOVA (E–G) or Kruskal–Wallis test followed by post hoc Dunn test (D). Pairwise comparisons were performed with appropriate P-value adjustments as suggested in R statistical software. Where applicable, data were converted to log scale to meet assumptions of the statistical test (F and G).
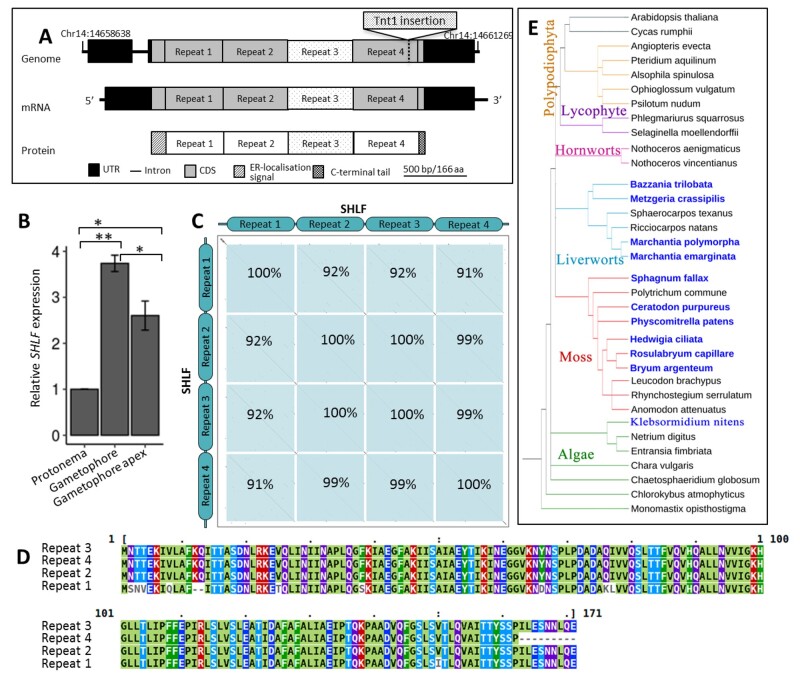
SHLF represents a unique bryophyte-specific TDR-containing multi-gene family
Surprisingly, we noticed that the SHLF gene does not have any previously characterized conserved domain. SHLF is a ∼2.1-kbp TDR-containing protein-coding gene (Figure 4, A). Though SHLF does not have any other paralogs in the P. patens genome, it is transcriptionally very active. For example, SHLF has 16-fold higher expression than the α-ACTIN gene in the gametophore tissue (Figure 4, B). Moss genome assembly (v3.3) showed only three TDRs, each 513 bp long inside the SHLF coding sequence. However, we detected a fourth repeat following the sequencing of the full-length SHLF PCR product (Supplemental Figure S13). We also noted that the tobacco (Nicotiana tabacum) Tnt1 retro-transposon had an insertion at the end of the last repeat (Figure 4, A). Our analysis suggested that the four TDRs of SHLF are near-perfect (99%) at both the nucleotide and protein level, whereas the first repeat is the most divergent among the group (Figure 4, C and D and Supplemental Figure S14). Using available plant genome sequences in Phytozome (v12.1.6) and transcriptome data from the oneKP plant project, SHLF homologs were identified through bioinformatics analysis and marked on a representative cladogram (Figure 4, E). The P. patens genome has only a single SHLF family gene, but we identified three and five SHLF family genes from sphagnum moss (Sphagnum fallax) and Marchantia polymorpha genomes, respectively. The number of TDRs per gene also varied from 3 to 5. Altogether, we found that 30 (out of 40) mosses and 8 (out of 26) liverwort species transcriptomes have SHLF homologs (score > 100), but it is absent in other plant and algal lineages except for two Klebsormidium species (score < 100; Supplemental Figure S15). However, this does not rule out the possibility of having homologs in remaining mosses (10 species) and liverworts (18 species), as the data source for these species were transcriptomes. These results establish that SHLF represents a unique bryophyte-specific, TDR-containing multi-gene family.
Figure 4.
SHLF represents a repeat-containing novel and conserved bryophyte-specific gene family. A, The SHLF (Pp3c14_22870) gene structure and the Tnt1 (∼5.6 kb) insertion site are depicted. In both the genome and mRNA, the SHLF gene contains four TDRs each 513 bp long that translates into a protein with four TDRs, each 171 aa long. B, Tissue-specific expression of SHLF in WT protonema, gametophore, and gametophore apex (Ct value for α-ACTIN and SHLF is 21 and 16, respectively). The error bars represent standard deviation. C and D, The percentage of sequence similarity among the four TDRs at nucleotide level (C) and amino acid level (D) is shown as domain-dot plot and multiple sequence alignment, respectively. E, The presence and absence of SHLF orthologs among the sequenced genomes of all green lineages (blue colored species name indicates presence of SHLF ortholog/s). The near-perfect TDRs in both gene and protein sequences are a unique feature of this bryophyte-specific SHLF gene family.
Phylogenetic analysis of SHLF protein family and partial recovery of shlf phenotypes by a distant Marchantia homolog
To understand the phylogeny and functional conservation among the SHLF family members, we constructed a SHLF phylogenetic tree and expressed a SHLF M. polymorpha homolog in a shlf background (Figure 5, A). A multiple sequence alignment of SHLF homologs revealed positional conservation of amino acids, including the N-terminal and repeat sequences (Supplemental Figure S16). SignalP (5.0) analysis predicted that SHLF and most of its homologs have the N-terminal signal peptide characteristic of proteins entering the conventional secretory pathway. In the alignment, 21% and 25% positions from the repeat region were identical and similar amino acids, respectively (Supplemental Figure S16). These results highlight that SHLF family proteins share a conserved domain. Compared with bryophyte SHLF homologs, sequences from Klebsormidium spp. showed less conservation with P. patens SHLF and lacked TDRs (Supplemental Figure S15, A). For example, the kfl00644_0030 protein (220 aa) from the Klebsormidium nitens genome shared 50% (48 aa) identity and 11% (11 aa) similarity with SHLF over a stretch of 96 amino acids toward the C-terminal (Supplemental Figure S15, B). Also, these proteins from Klebsormidium have the N-terminal signal peptide. We did not detect any SHLF homologs in other algal genomes, including the recently sequenced Chara braunii (Supplemental File S1). The phylogenetic tree constructed through the maximum-likelihood method showed that the moss and liverwort homologs do not form separate clades. When we expressed one out of five M. poylmorpha SHLF homologs (Mapoly0112s0050/MpSHLF50) in moss shlf background, it resulted in only a partial recovery of shlf phenotypes (leaf length and PDAC frequency; Figure 5, B–H and Supplemental Figures S10, S17). However, the internodal distance of shlf reverted completely, suggesting partial functional conservation (Supplemental Figure S11).
Figure 5.
A distant Marchantia SHLF ortholog (MpSHLF50) partially rescues shlf mutant phenotypes in moss. A, A maximum-likelihood-based phylogenetic tree of bryophyte-specific SHLF gene family was constructed based on the amino acid sequences available from 1000 plant genomes project. Each leaf of the tree is labeled with the species name and 1000 plant genome code followed by transcript number. Mosses and liverworts are marked by green and blue bars, respectively. Bootstrap values above 40 are given at respective nodes. B–E, Comparison of 3-weeks-old WT, shlf, and MpSHLF50 OE line (shlf MpSHLF50_OE #6) colonies (B–D) and leaf arrays (E) are presented. All scale bars represent 1 mm (F–H) analysis of leaf (ninth) length (n = 30) (F), apical dominance (G) and aniline blue-based PDAC frequency (n = 10) (H) showed partial recovery of the mutant phenotype. Lines #6 and #9 indicate MpSHLF50 OE in shlf background. F–H, Statistical analysis was performed using ANOVA. Pairwise comparisons were performed with appropriate P-value adjustments as suggested in R statistical software. G and H, Where applicable, data were converted to log scale to meet assumptions of the statistical test.
SHLF protein trafficks to the endoplasmic reticulum
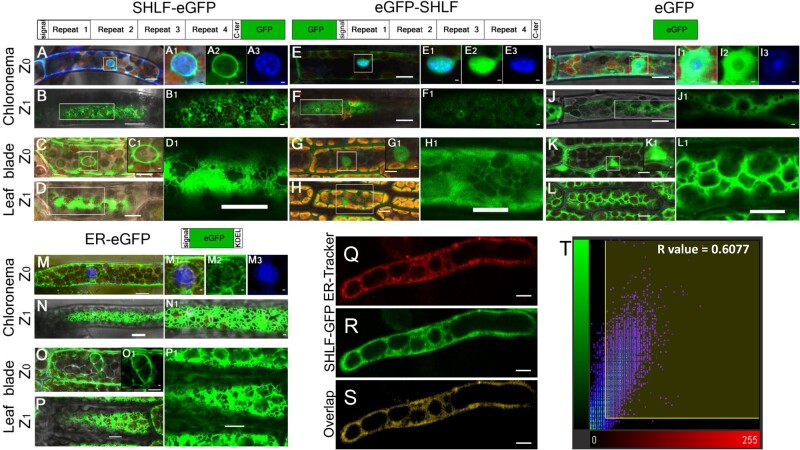
To validate the conserved N-terminal signal peptide of SHLF family proteins and to gain insights into their molecular function, we investigated its subcellular localization. For this purpose, SHLF protein fused with C-terminal (SHLF-eGFP) or N-terminal (eGFP-SHLF) eGFP was expressed under the maize ubiquitin promoter in the shlf background. SHLF-eGFP showed a strong GFP signal in the nuclear membrane and ER of moss protonemal and leaf blade cells (Figure 6, A–D). This signal correlated with localization signals from the ER marker line (Figure 6, M–P). To assess the signal peptide’s role in SHLF trafficking, we used the eGFP-SHLF line, with a eGFP tag that masks the N-terminal signal peptide. We observed that the SHLF protein could not enter the ER and remained in the cytosol and inside the nucleus (Figure 6, E–H), which matched the cytoplasmic eGFP localization pattern observed in the 35S:eGFP line (Figure 6, I–L). Additionally, we treated SHLF-eGFP protonemal filaments with ER-Tracker dye, which co-localized with the SHLF-eGFP in the ER (Figure 6, Q–T). These results suggest that the SHLF protein trafficks to the ER through the conventional secretory pathway.
Figure 6.
SHLF protein has a conserved signal peptide and traffics to ER in moss. A, C, M, and O, In tissue types, such as chloronema and leaf blade cells, at Z0 (optical plane bisecting the nucleus), the C-terminal GFP fusion line (SHLF-eGFP) (A and C) and ER marker line show GFP signal along with the nuclear membrane (M and O). E, G, I, and K, However, both N-terminal (eGFP-SHLF) eGFP fusion line (E and G) and eGFP line (I and K) show GFP signal in cytoplasm and inside the nucleus. The magnified view of the nucleus is a merged (A1, C1, E1, G1, M1, I1, and K1) image of eGFP (A2, E2, I2, and M2) and DAPI (A3, E3, I3, and M3) signals shown in green and blue colors, respectively. B, D, N, and P, Similarly at Z1 (optical section underneath the plasma membrane), C-terminal GFP fusion line (SHLF-eGFP) (B and D) and ER marker line (N and P) showed GFP signal in the cortical ER. F, H, J, and L, Both N-terminal eGFP fusion line (eGFP-SHLF) (F and H) and eGFP line (J and L) show GFP signal in cytoplasm and inside the nucleus. Insets are magnified images for better visualization. Q–S, Co-localization of ER-Tracker dye (Q) and SHLF-eGFP (R) signals depicted in the overlap (S) in protonemal cells (scale bar = 10 μm). T, Scatter plot showing the co-localization of the ER-Tracker and SHLF-eGFP signals with a Pearson’s coefficient of 0.6077. All scale bars represent 10 µm. The scale bars in the insets represent 1 µm except D1, H1, and L1 which are of 10 µm.
SHLF protein undergoes proteolytic cleavage
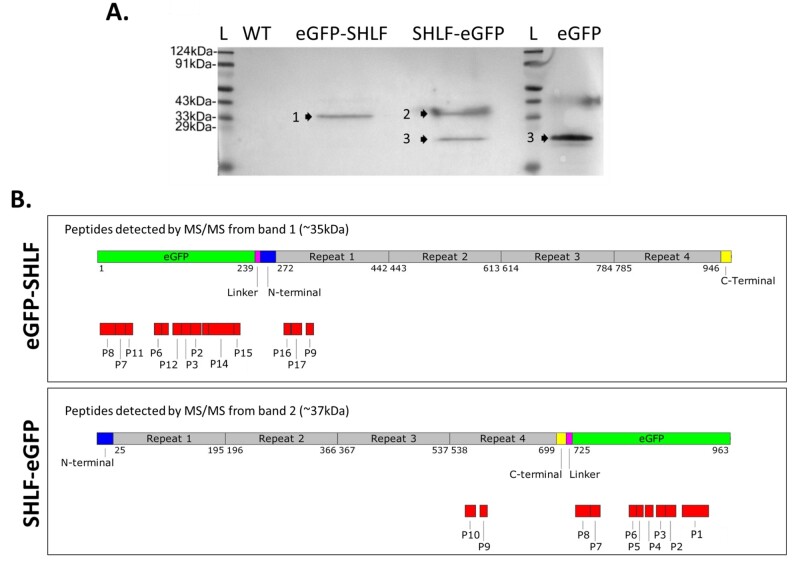
Our Western blot analysis with anti-GFP antibody did not reveal the expected 105-kDa band for full-length SHLF fused to GFP (Figure 7, A). Instead, we observed a single band (∼35 kDa) in eGFP-SHLF but two bands (∼27 and ∼37 kDa) in SHLF-eGFP lines. The ∼27-kDa band corresponds to the GFP positive control band. These banding patterns remained consistent with and without the addition of protease inhibitors (Supplemental Figure S18). All three bands were subjected to MS/MS-based peptide analysis to predict the probable cleavage sites on SHLF peptides (Figure 7, B). The peptides from the ∼27-kDa band matched specifically to eGFP (data not shown), whereas peptides from the ∼35- and ∼37-kDa bands matched with the N-terminal or C-terminal of SHLF, respectively. Hence, these two bands resulted from proteolytic cleavage of SHLF at a minimum of two different sites. Since the GFP tag was cleaved off in the ER, we could not trace SHLF’s trafficking beyond the ER. Our findings suggest that the SHLF protein trafficks to the ER where it undergoes proteolytic cleavage.
Figure 7.
SHLF undergoes cleavage and produces several peptides. A, Western blot analysis of total protein from eGFP-SHLF and SHLF-eGFP lines to detect SHLF cleavage pattern using anti-GFP antibodies. Total protein isolated from 35S:eGFP line was used as a control. B, Schematics showing mapped peptides of corresponding bands (bands 1 and 2) identified through MS/MS analysis. Band 3 marks eGFP band. All corresponding peptide sequences are listed in Supplemental File S3. Schematics is drawn to scale.
Discussion
SHLF regulates gametophore development in moss
The shlf gametophore exhibited phenotypes such as short-leaf, reduced apical dominance, increased internodal distance, and low PDAC frequency (Figures 1, 2). WGS of shlf identified the causal gene of these phenotypes. Two out of three Tnt1 insertions were in genic regions (Pp3c14_22870 and Pp3c1_9390; Supplemental Table S1), but only the Pp3c14_22870 (SHLF) locus has detectable mRNA expression (Figure 4, B). Notably, overexpression of the Pp3c14_22870 gene recovered all shlf phenotypes and its knockout lines phenocopied leaf length, apical dominance, and low PDAC frequency (Figure 3). However, the overexpression of the Pp3c1_9390 (PpEXTENSIN) gene could not recover any of the shlf phenotypes (Supplemental Figure S7). These results establish that Pp3c14_22870 (SHLF) is the causal gene for the mutant gametophore phenotypes. This led us to further investigate the cellular and molecular mechanisms underlying the shlf phenotypes.
Our results suggest that shlf phenotypes related to leaf size, leaf shape, and gametophore diameter are due to a suppressed cell division and expansion process (Figure 1, F–H). Previous studies showed that exogenous auxin treatments to a developing moss leaf resulted in similar phenotypes (Coudert et al., 2015). The leaves of PppinaPppinb double mutants were narrower due to reduced cell division in the medio-lateral axis (Bennett et al\., 2014; Viaene et al\., 2014). Also, the highly orchestrated spatio-temporal cell division and expansion pattern of a growing moss leaf coincided with PIN (auxin efflux carrier) expression. These studies indicate that in shlf, perturbation of cellular auxin concentration during leaf development could have caused the reduced cell division and expansion.
The shlf gametophores also exhibited reduced apical dominance (Figure 1, I–K): a phenomenon known to be controlled by auxin. Previously, Coudert et al. (2015) used auxin biosynthesis and transport mutants of moss to study the influence of auxin on gametophore branching. The auxin-deficient mutant (shi 2-1) exhibited reduced apical dominance, whereas the mutant with elevated auxin levels (SHI ox-5) showed increased apical dominance (Coudert et al., 2015). Apical dominance was not affected in the auxin transport mutant (PppinaPppinb). Hence, reduced apical dominance observed in shlf (Figure 1, I–K) implies that either suboptimal net auxin synthesis or inefficient auxin distribution might have caused the phenotype. The comparable expression pattern of genes involved in auxin metabolism (biosynthesis regulators (SHI1/2), catabolism (GH3-1), efflux carriers (PINs), and response regulators (IAAs)) in shlf and WT rules out the possibility of the former (Supplemental Figure S1, I). Moreover, greater than two-fold auxin accumulation and high auxin response (GH3:GUS activity) in shlf apex points toward the latter. This notion was further validated by the application of callose synthesis inhibitor DDG (10 and 25 µM) to shlf GH3:GUS lines, which resulted in increased sub-apical GUS activity (Figure 2, J–L). From these observations, we infer that the permeability of the limited number of PD in shlf might have increased (reduced callose deposition) upon DDG treatment. Together these results indicate that high auxin accumulation at the shlf apex could explain the short-leaf and reduced apical-dominance phenotypes. Also, low PDAC and PD frequency observed in leaf and protonema tissue types can potentially affect the auxin distribution pattern. Only a detailed investigation on the molecular function of the SHLF gene can validate the causal link between these mutant phenotypes. Since the SHLF protein coded by the Pp3c14_22870 locus has only conserved domains of unknown function, the molecular function of SHLF remains to be determined experimentally.
SHLF, a member of a unique bryophyte-specific TDRs containing multi-gene family, trafficks to ER
Unique sequence features and phylogeny of SHLF holds crucial information about its evolution and molecular function. The presence of multiple SHLF homologs in sphagnum moss and M. polymorpha genomes suggests that SHLF represents a bryophyte-specific multi-gene family. Variation in the number of TDRs across SHLF homologs indicates that the number of repeats may affect the SHLF function quantitatively. The conserved N-terminal signal peptide of the SHLF family hints at the trafficking of its members to the ER and secretion through the conventional secretory pathway (Figure 4, A). The four unique (513 bp/171 aa) near-perfect TDRs of SHLF are particularly intriguing. To the best of our knowledge, near-perfect TDRs with repeat unit size as long as 513 bp/171 aa have not been reported in any organism. A well-known example of near-perfect TDRs is the poly-Ubiquitin gene with a repeat unit size of 228 bp/76 aa. Each repeat of the poly-ubiquitin protein is a functional ubiquitin moiety that gets conjugated onto a target protein (Özkaynak et al., 1984). The P. patens genome codes for more than 21% tandem repeat-containing proteins, including armadillo (Coates, 2007), pentatricopeptide (Lurin et al., 2004), ankyrin (Becerra et al., 2004), and WD40 (Van Nocker and Ludwig, 2003), known to function in various cellular pathways (Schaper and Anisimova, 2015). However, tandem repeats of a protein are rarely identical to each other at the amino acid level (Jorda et al., 2010). Phylogenetic analyses provided crucial insights regarding the evolution of the SHLF family. Klebsormidium nitens is closely related to streptophycean algal ancestors of land plants, and its genome has a SHLF-like gene (kfl00644_0030; Figure 4, E). The presence of amino acid sequence similarity, a conserved N-terminal signal peptide, and the lack of TDRs suggests that kfl00644_0030 could be an ortholog of bryophyte SHLF genes and may closely resemble the ancestral SHLF gene. The availability of more algal genomic resources may help to validate this hypothesis. Among the bryophytes, mosses and liverworts have SHLF homologs; however, many liverworts do not develop a shoot-like gametophore. Our attempt to recover the moss shlf mutant with the liverwort SHLF homolog (MpSHLF50) resulted in only a partial recovery of mutant gametophore phenotypes. The possible functional specialization and divergence of MpSHLF genes is perhaps the reason for this (Figure 5).
Sub-cellular localization and post-translational processing provided additional insights about SHLF function. Detection of SHLF-eGFP signal in the nuclear membrane and the cortical ER network validated that the conserved N-terminal signal peptide is functional and shows that SHLF enters the conventional secretory pathway. Lack of a 105-kDa band in the Western blot suggests that the C-terminal GFP tag is cleaved off in the ER, and SHLF may not be an ER-resident protein (Figures 6, 7). The absence of any cell organelle-specific transit signal/s prompted us to consider that SHLF might follow the conventional secretory pathway. Our observation corroborates the report that detected SHLF as an abundant protein in the P. patens secretome (Lehtonen et al., 2013). All the evidence suggests that SHLF trafficks to the ER and maybe ultimately secreted into the extracellular matrix.
In summary, we show that the novel bryophyte-specific gene SHLF influences gametophore development by regulating auxin distribution in moss gametophores. There could be two possible explanations for other phenotypes. First, the mutated copy of SHLF in the mutant affects PD development and potentially influences symplastic auxin transport in moss gametophore. Second, the mutated SHLF may have caused the differential auxin distribution pattern in the shlf gametophores and resulted in the secondary pleiotropic phenotypes, including lower PD frequency. This investigation provides crucial insights and has opened up many interesting avenues concerning SHLF gene family evolution and functions. Only future research can unravel the molecular role of this protein family. We hypothesize that the bryophyte-specific SHLF family of proteins have been involved in adaptations unique to non-vascular land plants and are potential candidates to study plant terrestrialization.
Materials and methods
Plant culture and maintenance
Physcomitrium patens (moss) ecotype “Gransden” was procured from the International Moss Stock Center (IMSC), University of Freiburg, Germany, and maintained under standard growth conditions (Cove et al., 2009). Moss tissues were incubated in growth incubators at 16:8 h (light:dark) cycle at 24°C for all experiments. Takaragaike-1 (Tak-1; Male) and Takaragaike-2 (Tak-2; Female) strains of M. polymorpha were a kind gift from Prof. Takayuki Kohchi (Kyoto University, Japan). Marchantia lines were grown aseptically at 24°C in half-strength B5 solid media (Tanaka et al., 2016).
Phenotypic characterization of shlf mutant
Seven-day-old protonemal filaments were cultured on BCDAT medium and incubated for 3 weeks (Cove et al., 2009). The ninth leaf from the apex of an actively growing gametophore was imaged using Leica S8 APO Stereo-microscope (Leica Microsystems, Germany). Using ImageJ software (Schneider et al., 2012), various phenotypic measurements of the leaf were recorded. Leaf array silhouettes were prepared using Adobe Photoshop CS. For toluidine blue staining of the cell wall, leaves were cleared using 2 M NaOH and then stained in 0.1% (w/v) toluidine blue solution. For gametophore branching pattern analysis, we have carefully considered all branches, including small primary outgrowths at the auxiliary point (the earliest stage of a branch initial). The metamer number of all side branches was noted from gametophores of 3-week-old colonies and plotted as a Hinton diagram using the Matplotlib package (Hunter, 2007).
Cloning and plant transformation
To develop the GH3:GUS construct in the pTFH15.3 backbone, the soybean auxin-responsive promoter–reporter (GH3:GUS) fragment was amplified from pUC19-GH3:GUS (obtained from Prof. Thomas J. Guilfoyle, University of Missouri, USA) plasmid using GH3_KpnI_F and GH3_NosT_XmaI_R primer pair (Supplemental Table S2). Using KpnI and XmaI, the GH3:GUS DNA fragment was cloned into the pTFH15.3 vector (obtained from Prof. Mitsuyasu Hasebe, NIBB Japan) by replacing the rice Actin promoter. The construct was used for PEG-mediated protoplast transformation in moss to generate 10 auxin-responsive marker lines each in WT and shlf background (WT GH3:GUS and shlf GH3:GUS). For validation of the candidate genes disrupted by Tnt1 insertion, a full-length coding region of SHLF (Pp3c14_22870.1; ∼2.1 Kb) and PpEXTENSIN (Pp3c1_9390; ∼1.4 Kb) was amplified from the P. patens genome using the primer pairs Tnt1_sl_ins1_F, Tnt1_sl_ins1_R and Tnt1_sl_ins2_F, Tnt1_sl_ins2_R cloned into the pTFH15.3 vector using ApaI and AscI restriction enzymes (Supplemental Table S2). To construct the N-terminal and C-terminal eGFP fusion cassettes of SHLF, the full-length gene was amplified using attB1_SHLF_F, attB5r_SHLF_R and attB5_SHLF_F, attB2_SHLF_R primer pairs, respectively (Supplemental Table S2). The former was cloned into pDONR P1P5r, and the latter was cloned into pDONR P5P2 through a BP reaction. To construct the N-terminal and C-terminal eGFP fusion cassettes of SHLF, two sets of vector pairs (pENTR L1R5 SHLF, pENTR L5L2 eGFP) and (pENTR L1R5 eGFP, pENTR L5L2 SHLF) were recombined with the destination vector pTK-UBI-gate through an LR reaction as per the manufacturer’s instructions. All the above constructs were used individually for PEG-mediated P. patens protoplast transformation as per the protocol of Nishiyama et al. (2000) Regenerated protoplasts were selected on G418 (20 mg/L) or hygromycin (20 mg/L) antibiotics, and colonies were subjected to molecular analysis for further verification.
GUS and MUG assay
One-month-old P. patens colonies grown on BCDAT medium were subjected to a β-glucuronidase (GUS) staining assay. Tissues were fixed in 0.3% (v/v) formaldehyde solution for 30 min, washed with liquid BCDAT medium, followed by transfer to the GUS-staining buffer, and incubation at 37°C for 12 h in the dark (Jefferson et al., 1987). GUS-stained tissues were fixed with 5% (v/v) formalin for 10 min, incubated in 5% (v/v) acetic acid for 10 min, and dehydrated using a series of ethanol washes (30%, 50%, 70%, and 100% v/v). Images were obtained using a Leica S8 APO stereomicroscope (Leica, Germany). To quantify GUS expression in the apex of WT and shlf gametophores, MUG assays (Gallagher, 1992) were performed with 3 µg of total protein and measured using a multimode plate reader (Varioskan flash, Thermo Scientific).
RT-qPCR analysis
Total RNA was extracted from protonema and gametophore tissues using RNAiso Plus (Takara Bio USA Inc.). Two micrograms (2 µg) of RNA samples was reverse-transcribed using oligo dT primers and SS-IV reverse transcriptase (Invitrogen). Relative quantification of transcripts was performed using SYBR Premix Ex Taq II (Tli RNaseH Plus) in a Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Relative target gene expression levels were carried out using β-actin as a reference gene. The fold-change (sample value/reference value) was calculated based on the 2−ΔΔCt method (Schmittgen and Livak, 2008). All the primers used for RT-qPCR analysis are mentioned in Supplemental Table S2.
PDAC staining using aniline blue
In order to quantify the PDAC frequency, P. patens gametophores were vacuum infiltrated with 0.1% (w/v) aniline blue solution for 4 h on ice (Ishikawa et al., 2011). Excess stain was washed off in BCDAT liquid media. Gametophores were mounted on slides and observed under a Leica TCS SP8 microscope (Leica, Germany) using a Plan Apochromat 40× oil immersion lens. An excitation laser of 405 nm and an emission filter of 421–575 nm were used to capture the PDAC fluorescence. The mono-layered leaf blade cell walls were covered by Z-stacking, de-convoluted using AutoQuant X3 (Media Cybernetics, USA) and subjected to Imaris based (Oxford Instruments) spot analysis to determine the number of discrete PDAC. Using ImageJ (Schneider et al., 2012), the total cell wall length was manually traced and measured. Total PDAC spots were divided by total cell wall length to calculate the final PDAC frequency.
Sample preparation and LC–MS/MS for auxin analysis
Auxin was extracted from the gametophore apex and quantified as per the protocol with minor modifications (Novák et al., 2016). UHPLC–MS/MS fitted with a C18 column (2.1 mm × 150 mm) with Bruker Ultrahigh-Resolution Q-TOF were used for auxin detection. The acquisition parameter was run in negative ionization mode. To detect IAA, the dried extracts were re-dissolved in 40 μL of 10% (v/v) cold aqueous methanol and transferred to 100 µL insert-equipped vials. For quantification, the IAA standards (10, 50, 100, 500, 1000 ng/mL, and up to 0.1 mg/mL) were analyzed in LC–MS/MS. A linear gradient of the mobile phase was used at a flow rate of 0.2 mL/min with a mix of solvent A (water) and solvent B (100% (v/v) methanol with 0.1% (v/v) formic acid). The column temperature was set to 30°C. Data analysis was performed using Hystar software v. 3.2 (Novák et al., 2016; Shree et al., 2019).
TEM analysis
Moss protonemal tissues were fixed for 3 h at room temperature in 2% (v/v) glutaraldehyde (Sigma–Aldrich) and 1% (v/v) formaldehyde (Hi-media) prepared in 0.5 M sodium cacodylate buffer, pH 7.4, and were rinsed with 0.5 M sodium cacodylate buffer. Post fixation, tissues were incubated with 1% (w/v) osmium tetraoxide (Sigma–Aldrich) for 4 h at room temperature. Fixed tissues were dehydrated through a series of ethanol gradients (30–100% v/v), gradually replaced with 100% (v/v) propylene oxide (Sigma–Aldrich), and further embedded in Dr Spurr’s resin (Sigma–Aldrich). Thin sections (70–80 nm) were made using the Leica EM UC7 ultramicrotome (Leica, Germany) and examined under a JEM-2100 transmission electron microscope (JEOL, Japan). PD frequency was counted using ImageJ (Schneider et al., 2012).
Whole-genome sequencing
Genomic DNA (1 µg) was isolated from the shlf mutant using the protocol (PHYSCObase; http://moss.nibb.ac.jp/protocol.html) and submitted for WGS at Genotypic Technology (Bangalore, India). A total of 150 bp paired-end sequencing was performed using an Illumina platform (Illumina, USA) and approximately 30× depth raw data were generated. A customized bioinformatics in-house pipeline was used to identify the Tnt1 and T-DNA insertion loci in the mutant genome (Supplemental File S3). Tnt1 and T-DNA insertions detected by WGS were confirmed by the genomic DNA PCR and RT-PCR using the primers pairs Tnt1_sl_ins1_F, LTR_qF (1633 bp) and LTR7, Tnt1_sl_ins1_R (179 bp) for insertion in the genomic locus Pp3c14_22870 (Supplemental Figure S5). Similarly, for PpEXTENSIN, primer pairs Tnt1_sl_ins2_F, LTR_qF (1147 bp) and LTR7, Tnt1_sl_ins2_R (491 bp) for insertion in the genomic locus Pp3c1_9390 were used to confirm the nature of integration (Supplemental Figure S6 and Supplemental Table S1).
Bioinformatics analysis
To conduct a comprehensive analysis on SHLF homologs in other plant species, all available non-seed plant transcriptome datasets (Algae: 105 species; Moss: 40 species; Liverwort: 26 species; Hornwort: 7 species) were downloaded from oneKP project (Matasci et al\., 2014). Genomes of S. fallax, M. polymorpha, P. patens, K. nitens and 53 other vascular plants from Phytozome v12.1.6 were used in the analysis. All the transcriptome assemblies were translated and converted into a protein BLAST database using BLASTP 2.2.31+ toolkit (Camacho et al., 2009). SHLF amino acid sequence was blasted against the individual local protein database to detect the presence of homologs. Any hit that has a minimum score (bits) of 100 and an E-value less than one was considered as a homolog. All the organisms were arranged on a cladogram using PhyloT online tool (http://phylot.biobyte.de/) and visualized in iTOL (www.itol.embl.de). PhyloT tool generates trees based on the NCBI taxonomy database.
Phylogenetic analysis
SHLF homologs identified from oneKP database with intact N-terminal and first repeat sequence were considered for phylogenetic analysis. The evolutionary history was inferred using the maximum-likelihood method and Whelan And Goldman + Freq. model (Whelan and Goldman, 2001). The tree with the highest log likelihood (−10097.09) is shown (Figure 5, A). The bootstrap values associated with each taxon is included next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with a superior log-likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 48-aa sequences and there were 421 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018). Since the SHLF family does not bear resemblance to any known protein families, ACTIN and RBCL sequences were chosen as outgroups. We have also constructed another tree without outgroups for reference (Supplemental Figure S19).
Confocal microscopy
Transgenic moss lines of SHLF-eGFP, eGFP-SHLF, ER-eGFP, and eGFP were subjected to microscopic analysis to determine the subcellular localization pattern of SHLF. Protonemal filaments were treated with 1 µg/mL DAPI solution and analyzed under the emission range 493–527 (for eGFP) and 630–761 nm (for chlorophyll) using Leica TCS SP8 confocal laser scanning microscope with 488-nm excitation from an argon laser. Similarly, DAPI fluorescence was captured under the emission range of 440–473 nm and excitation at 405 nm from UV laser.
Co-localization study
ER-Tracker Blue-White DPX dye was acquired from Invitrogen (USA). The moss protonemal cells were stained with 10 μM ER-Tracker dye for 30 min in the dark at 24°C. The cells were washed with BCDAT liquid medium before imaging on Leica TCS SP8 microscope. Argon laser of 488 nm wavelength was used to excite GFP and the subsequent emission was captured at 490–532 nm. The 405-nm UV laser was used to excite the ER-Tracker dye and the subsequent emission was captured at 420–479 nm. Image analysis for the co-localization pattern was carried out using Imaris software. Pearson’s co-localization test was conducted to detect the percentage of co-localizing pixels in the different channels. A threshold was set to rule out the bias due to background noise.
Western blot
Three-week-old colonies of WT, eGFP-SHLF, SHLF-eGFP, and eGFP moss lines were crushed in liquid nitrogen and the total protein was extracted with or without protease inhibitor cocktail (Sigma, USA) and 2× SDS in PBS extraction buffer (20 mM, pH 7.4). The resuspended solution was vortexed vigorously for 5 min. The supernatant collected after centrifugation was concentrated through a 3-kDa cut-off column (Millipore, USA) and an equal quantity of protein from each tissue except for the eGFP line was loaded onto a 12.5% SDS-PAGE gel. All the protein extraction procedures were performed at 4°C. After running the gel, the protein was transferred to the PVDF membrane (GE Healthcare, USA). A standard western blot protocol was followed using primary rabbit anti-GFP polyclonal antibodies (Cloud-Clone Corp., USA) at 1:500 dilution and secondary anti-rabbit goat polyclonal antibodies at 1:7000 dilution. Total protein from eGFP and WT lines was used as positive and negative controls, respectively. An identical SDS-PAGE gel was used for MS/MS-based peptide identification. Protein bands were trypsinized, peptides were extracted and subjected to MS/MS analysis using ABSciex Triple TOF 6600 system (Kelkar et al., 2019)
Statistical analysis
Boxplots were plotted using the ggplot2 package from the R programming language (Wickham, 2016). The whiskers of the box plot mark the closest data point within the 1.5× interquartile range above the first and below third quartile (Tukey’s style). Data points outside the whiskers are outliers and were also included in the statistical analysis. In the case of bar plots, error bars denote the standard error. Student’s t test or ANOVA analysis were performed for the data points following normal distribution and equal variance. In other cases, the Mann–Whitney Wilcox test or Kruskal–Wallis test with post hoc Dunn test were performed (Mann and Whitney, 1947; Dunn, 1964). P-value < 0.05, <0.001, and <0.0001 were marked as *, **, and ***, respectively.
Accession numbers
Genes listed in this article can be found in Phytozome 13 with the following gene IDs: PpACTIN-Pp3c10_17070; PpSHLF-Pp3c14_22870; PpEXTENSIN-Pp3c1_9390; PpGH3-1-Pp3c24_16260; PpIAA1-Pp3c8_14720; PpIAA2-Pp3c24_6610; PpIAA1B-Pp3c8_14720; PpSHI1-Pp3c21_16440; PpSHI2-Pp3c18_8920; PpPINA-Pp3c23_10200; and PpPINB-Pp3c24_2970.
Supplemental data
Supplemental Figure S1. Cell division and elongation processes are suppressed in leaves and stem of shlf mutant.
Supplemental Figure S2. Generation of GH3:GUS lines in WT and shlf background.
Supplemental Figure S3. Callose staining indicates differential PD frequency in moss leaves.
Supplemental Figure S4. In-house pipeline employed to analyze shlf WGS data.
Supplemental Figure S5. PCR confirmation of Tnt1 insertion inside the locus Pp3c14_22870 (PpSHLF) in shlf mutant.
Supplemental Figure S6. PCR confirmation of Tnt1 insertion inside the locus Pp3c1_9390 (PpEXTENSIN).
Supplemental Figure S7. Generation and confirmation of Pp3c1_9390 (PpEXTENSIN) over-expression (OE) lines in the shlf background.
Supplemental Figure S8. Generation and confirmation of Pp3c14_22870 (PpSHLF) OE lines in the shlf background.
Supplemental Figure S9. Generation of shlf_ko lines in WT background.
Supplemental Figure S10. Width comparison of the ninth leaf in various lines.
Supplemental Figure S11. Comparison of internodal distance in various lines.
Supplemental Figure S12. Primary structure of SHLF gene and protein.
Supplemental Figure S13. Full-length SHLF CDS contains four repeats as ascertained by PCR amplification and sequencing.
Supplemental Figure S14. Tandem direct DNA repeats of SHLF.
Supplemental Figure S15. The amino acid level conservation of probable Klebsormidium homolog with other SHLF family members.
Supplemental Figure S16. Multiple sequence alignment of SHLF homologs with intact N-terminal and first repeat.
Supplemental Figure S17. Generation of MpSHLF50 overexpression lines in shlf background.
Supplemental Figure S18. Western blot analysis of C-terminal and N-terminal eGFP-tagged SHLF proteins showing in vivo proteolytic cleavage.
Supplemental Figure S19. Maximum-likelihood-based phylogenetic tree of bryophyte-specific SHLF family genes.
Supplemental Table S1. Tnt1 and T-DNA insertions in shlf mutant genome identified by WGS.
Supplemental Table S2. List of primers used in this study.
Supplemental File S1. Homology search for SHLF among bryophytes transcriptomes.
Supplemental File S2. List of peptides detected in MS/MS analysis.
Supplemental File S3. List of codes used to identify Tnt1 insertions in shlf genome.
Supplementary Material
Acknowledgments
The authors thank Prof. Mitsuyasu Hasebe, NIBB, Japan for sharing the OE and knock-out vectors (pTFH15.3 and pTN186) and acknowledge the late Prof. Tom Guilfoyle, University of Missouri, USA, for providing the soybean auxin-responsive GH3:GUS cassette. Gateway compatible vectors and ER marker lines were a kind gift from Prof. Magdalena Bezanilla, Dartmouth College, USA. We also thank the microscopy facility at IISER Pune for all help related to confocal microscopy. We sincerely thank Director, CCMB, Hyderabad, India, for giving us access to the TEM facility and Mr. A. Harikrishna for his help in TEM imaging. Kind support was received from Dr. Sona Pandey for moss line maintenance at Donald Danforth Plant Science Center, Missouri, USA, and is gratefully acknowledged. Technical help received from Mr Manish Kumar and Ms. Sheeba John is acknowledged. We thank Prof. David Hannapel, Iowa State University, USA for kindly reading our manuscript and providing necessary input.
Funding
This work was supported by a grant (Grant No. EMR/2016/004852) from the Science and Engineering Research Board (SERB), Government of India to A.K.B. Core funding and infrastructure were provided by the Indian Institute of Science Education and Research (IISER) Pune, India.
Conflict of interest statement. None declared.
B.M. and A.K.B. conceived the idea, designed the research, and edited the manuscript. A.J.B. and S.P. have contributed in vector constructions, transgenic line generation, microscopy, and western blot analysis. B.M. and S.P. have carried out all bioinformatics studies. G.C. and B.M. have constructed gateway vectors and analyzed phenotypic data. G.C. has maintained Marchantia culture and generated transgenic lines of overexpressing Marchantia ortholog in the moss mutant. S.P. has established the aniline blue protocol, generated knock-out lines, and also gathered phenotypic data from the mutant analysis. TEM-based PD frequency analysis was carried out by A.J.B. M.L. and S.K.M. have helped in auxin quantification experiments. All authors analyzed the data and wrote their respective experimental parts.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Anjan K. Banerjee (akb@iiserpune.ac.in).
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh Y-S, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Becerra C, Jahrmann T, Puigdomènech P, Vicient CM (2004) Ankyrin repeat-containing proteins in Arabidopsis: characterization of a novel and abundant group of genes coding ankyrin–transmembrane proteins. Gene 340: 111–121 [DOI] [PubMed] [Google Scholar]
- Bennett TA, Liu MM, Aoyama T, Bierfreund NM, Braun M, Coudert Y, Dennis RJ, O’Connor D, Wang XY, White CD, et al. (2014) Plasma membrane-targeted PIN proteins drive shoot development in a moss. Curr Biol 24: 2776–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierfreund NM, Reski R, Decker EL (2003) Use of an inducible reporter gene system for the analysis of auxin distribution in the moss Physcomitrella patens. Plant Cell Rep 21: 1143–1152 [DOI] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu C-M, van Lammeren AAM, Miki BLA, et al. (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkard JO, Zambryski PC (2017) Plasmodesmata enable multicellularity: new insights into their evolution, biogenesis, and functions in development and immunity. Curr Opin Plant Biol 35: 76–83 [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MG (1997) Concepts and terminology of apical dominance. Am J Bot 84: 1064–1069 [PubMed] [Google Scholar]
- Coates JC (2007) Armadillo repeat proteins: versatile regulators of plant development and signalling. In: Bögre L., Beemster G. (eds) Plant Growth Signaling. Plant Cell Monographs, vol 10. Springer, Berlin, Heidelberg. pp 299–314 [Google Scholar]
- Coudert Y, Palubicki W, Ljung K, Novak O, Leyser O, Harrison CJ (2015) Three ancient hormonal cues co-ordinate shoot branching in a moss. eLife 4: e06808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove DJ, Perroud P-F, Charron AJ, McDaniel SF, Khandelwal A, Quatrano RS (2009) The moss Physcomitrella patens: a novel model system for plant development and genomic studies. Cold Spring Harb Protoc 2009: pdb-emo115 [DOI] [PubMed]
- Dunn OJ (1964) Multiple comparisons using rank sums. Technometrics 6: 241–252 [Google Scholar]
- Fujita T, Sakaguchi H, Hiwatashi Y, Wagstaff SJ, Ito M, Deguchi H, Sato T, Hasebe M (2008) Convergent evolution of shoots in land plants: lack of auxin polar transport in moss shoots. Evol Dev 10: 176–186 [DOI] [PubMed] [Google Scholar]
- Gallagher SR (1992) GUS Protocols: using the GUS Gene as a Reporter of Gene Expression. Academic Press, pp 1–4 [Google Scholar]
- Han X, Hyun TK, Zhang M, Kumar R, Koh E, Kang B-H, Lucas WJ, Kim J-Y (2014) Auxin–callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev Cell 28: 132–146 [DOI] [PubMed] [Google Scholar]
- Hunter JD (2007) Matplotlib: A 2D graphics environment. Comput Sci Eng 9: 90–95 [Google Scholar]
- Ishikawa M, Murata T, Sato Y, Nishiyama T, Hiwatashi Y, Imai A, Kimura M, Sugimoto N, Akita A, Oguri Y, et al. (2011) Physcomitrella cyclin-dependent kinase A links cell cycle reactivation to other cellular changes during reprogramming of leaf cells. Plant Cell 23: 2924–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Bevan M, Kavanagh T (1987) The use of the Escherichia coli β-glucuronidase gene as a gene fusion marker for studies of gene expression in higher plants. Biochem Soc Trans 15: 17–18. [DOI] [PubMed] [Google Scholar]
- Jorda J, Xue B, Uversky VN, Kajava AV (2010) Protein tandem repeats—the more perfect, the less structured. FEBS J 277: 2673–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer W, Cove DJ, Schaefer DG, Zrÿd J-P, Prigge MJ, Bezanilla M, Decker EL, Parsons J, Reski R, Parsons J, et al. (2014) Evolutionary crossroads in developmental biology: Physcomitrella patens. Plant Biotechnol J 5: 12225. [Google Scholar]
- Kelkar DS, Ravikumar G, Mehendale N, Singh S, Joshi A, Sharma AK, Mhetre A, Rajendran A, Chakrapani H, Kamat SS (2019) A chemical–genetic screen identifies ABHD12 as an oxidized-phosphatidylserine lipase. Nat Chem Biol 15: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji R, Hasebe M (2014) Eight types of stem cells in the life cycle of the moss Physcomitrella patens. Curr Opin Plant Biol 17: 13–21 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al. (2011) A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen MT, Takikawa Y, Ronnholm G, Akita M, Kalkkinen N, Ahola-Iivarinen E, Somervuo P, Varjosalo M, Valkonen JPT (2013) Protein secretome of moss plants (Physcomitrella patens) with emphasis on changes induced by a fungal elicitor. J Proteome Res 13: 447–459 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR (1947) On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat 18: 50–60 [Google Scholar]
- Matasci N, Hung L-H, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, et al. (2014) Data access for the 1,000 Plants (1KP) project. Gigascience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanasundaram B, Rajmane VB, Jogdand SV, Bhide AJ, Banerjee AK (2019) Agrobacterium-mediated Tnt1 mutagenesis of moss protonemal filaments and generation of stable mutants with impaired gametophyte. Mol Genet Genomics 294: 583–596 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara K, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Novák O, Pěnčik A, Blahoušek O, Ljung K (2016) Quantitative auxin metabolite profiling using stable isotope dilution UHPLC-MS/MS. Curr Protoc Plant Biol 1: 419–430 [DOI] [PubMed] [Google Scholar]
- Özkaynak E, Finley D, Varshavsky A (1984) The yeast ubiquitin gene: head-to-tail repeats encoding a polyubiquitin precursor protein. Nature 312: 663. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Deguchi H, Hasebe M (2008) Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol Dev 10: 555–566 [DOI] [PubMed] [Google Scholar]
- Schaper E, Anisimova M (2015) The evolution and function of protein tandem repeats in plants. New Phytol 206: 397–410 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C T method. Nat Protoc 3: 1101. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shree M, Lingwan M, Masakapalli SK (2019) Metabolite profiling and metabolomics of plant systems using 1H NMR and GC–MS. OMICS Approach Plant Biotechnol 129: 129 [Google Scholar]
- Stevenson SR, Kamisugi Y, Trinh CH, Schmutz J, Jenkins JW, Grimwood J, Muchero W, Tuskan GA, Rensing SA, Lang D, et al. (2016) Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE, a regulator of ABA responses unique to basal land plants and required for desiccation tolerance. Plant Cell 28: 1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka D, Ishizaki K, Kohchi T, Yamato KT (2016) Cryopreservation of Gemmae from the Liverwort Marchantia polymorpha L. Plant Cell Physiol 57: 300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander M, Landberg K, Sundberg E (2018) Auxin-mediated developmental control in the moss Physcomitrella patens. J Exp Bot 69: 277–290 [DOI] [PubMed] [Google Scholar]
- Thimann KV, Skoog F (1933) Studies on the growth hormone of plants: III. The inhibiting action of the growth substance on bud development. Proc Natl Acad Sci USA 19: 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nocker S, Ludwig P (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaene T, Landberg K, Thelander M, Medvecka E, Pederson E, Feraru E, Cooper ED, Karimi M, Delwiche CF, Ljung K, et al. (2014) Directional auxin transport mechanisms in early diverging land plants. Curr Biol 24: 2786–2791 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S (1991) The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243 [DOI] [PubMed] [Google Scholar]
- Von Maltzahn KE (1959) Interaction between kinetin and indoleacetic acid in the control of bud reactivation in Splachnum ampullaceum (L.) Hedw. Nature 183: 60–61 [Google Scholar]
- Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699 [DOI] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York [Google Scholar]
- Zavaliev R, Epel BL (2015) Imaging callose at plasmodesmata using aniline blue: quantitative confocal microscopy. In Plasmodesmata. Springer, pp 105–119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.