Abstract
Background
Polygenic risk scores (PRSs) have been demonstrated to identify women of European, Asian, and Latino ancestry at elevated risk of developing breast cancer (BC). We evaluated the performance of existing PRSs trained in European ancestry populations among women of African ancestry.
Methods
We assembled genotype data for women of African ancestry, including 9241 case subjects and 10 193 control subjects. We evaluated associations of 179- and 313-variant PRSs with overall and subtype-specific BC risk. PRS discriminatory accuracy was assessed using area under the receiver operating characteristic curve. We also evaluated a recalibrated PRS, replacing the index variant with variants in each region that better captured risk in women of African ancestry and estimated lifetime absolute risk of BC in African Americans by PRS category.
Results
For overall BC, the odds ratio per SD of the 313-variant PRS (PRS313) was 1.27 (95% confidence interval [CI] = 1.23 to 1.31), with an area under the receiver operating characteristic curve of 0.571 (95% CI = 0.562 to 0.579). Compared with women with average risk (40th-60th PRS percentile), women in the top decile of PRS313 had a 1.54-fold increased risk (95% CI = 1.38-fold to 1.72-fold). By age 85 years, the absolute risk of overall BC was 19.6% for African American women in the top 1% of PRS313 and 6.7% for those in the lowest 1%. The recalibrated PRS did not improve BC risk prediction.
Conclusion
The PRSs stratify BC risk in women of African ancestry, with attenuated performance compared with that reported in European, Asian, and Latina populations. Future work is needed to improve BC risk stratification for women of African ancestry.
Inherited genetic variation contributes to breast cancer (BC) risk, with approximately 30% of the total variability in liability due to genetic factors (1). Genome-wide association studies (GWAS) have discovered approximately 180 common risk variants for BC at genome-wide statistical significance levels (2-4), and polygenic risk scores (PRSs) comprised of multiple common variants have been demonstrated to stratify women at different levels of risk of developing BC (5). In the Breast Cancer Association Consortium (BCAC), a 313-variant PRS constructed using data from European-ancestry populations identified 1% of women with 4.4-fold and 2.8-fold increased risks of estrogen receptor (ER)-positive and ER-negative BC, respectively (6), compared with the population average (40th-60th PRS percentile). The incorporation of PRS into existing risk prediction models with nongenetic factors also improved risk stratification, with predicted lifetime risk over 30% for women in the top 1% of the PRS (7‐9).
The discovery of risk variants for BC and subsequent PRS development has been based on studies primarily conducted among women of European ancestry. Broad clinical application of the PRS will require performance evaluation and enhancement across diverse racial and ethnic populations.
In this study, we assembled the largest genetic data set of BC in women of African ancestry to date, including 9241 cases (4299 with ER-positive and 2636 with ER-negative disease) and 10 193 controls, to examine the performance of the most current PRS panels for BC [179 (3,4) and 313 (6) variants] in stratifying risk in this population. We also recalibrated the PRSs by replacing index variants with markers that better captured BC risk in women of African ancestry and evaluated performance in this population.
Methods
Study Participants
This includes women of African ancestry from 4 BC consortia and 1 additional study, each genotyped with a different GWAS array. Detailed descriptions of each consortium and study are provided in Supplementary Table 1 (available online). The African American Breast Cancer consortium (10,11) includes genetic data from 3007 cases (1518 ER-positive, 987 ER-negative) and 2720 controls in the analysis; the African American Breast Cancer Epidemiology and Risk consortium (12) includes data from 1407 cases (952 ER-positive, 385 ER-negative) and 2408 controls; the BCAC/Genetic Associations and Mechanisms in Oncology (GAME-ON) OncoArray consortium (13) includes 2271 cases (1130 ER-positive, 613 ER-negative) and 1406 controls; the GWAS of Breast Cancer in the African Diaspora consortium (14) includes data for 1657 cases (403 ER-positive, 374 ER-negative) and 2029 controls, and the Ghana Breast Health Study (GBHS) includes 899 cases (296 ER-positive, 277 ER-negative) and 1630 controls. The study was approved by institutional review boards at each of the study sites.
Risk Variant Characteristics
We evaluated 2 PRSs. The first, PRS313, is a 313-variant PRS developed by BCAC in European-ancestry populations (6). Briefly, the 313 variants were determined by P values and linkage disequilibrium threshold-based filtering followed by a stepwise forward selection. The second, PRS179, included 179 known common risk variants that reached genome-wide statistical significance in GWAS analyses (3,4). Comparing these 2 sets of variants, 54 variants overlapped and another 52 were highly correlated (r2 ≥ 0.8 in the 1000 Genomes Project (1KGP) European-ancestry populations). In total, 163 of the 179 (91.1%) variants and 224 of the 313 (71.6%) variants had imputation scores (r2) ≥ 0.8 across all consortia and GBHS (Supplementary Table 2, available online). For missing variants (not genotyped or imputed) in 1 study, we assigned each individual in that study the expected dosage derived from the remainder of studies; details are provided in the Supplementary Methods (available online).
Statistical Analysis
Association Testing of Individual Risk Alleles
Risk Allele Frequencies (RAFs) were derived by averaging the RAF in controls of each consortium and GBHS, weighted by the corresponding control numbers. We excluded variants with minor allele frequency (MAF) less than 0.1% or imputation quality score less than 0.3 in each consortium or study. Power calculations were conducted using odds ratios (ORs) in previous European GWAS (4) and RAFs in women of African ancestry. The odds ratio and P value of each variant were estimated using unconditional logistic regression adjusting for relevant covariates (Supplementary Table 1, available online) in each consortium and in GBHS, and the results were combined using a fixed-effect meta-analysis with inverse variance weights. Consistent directionality of effect was determined as alleles with odds ratios in the same direction as those previously reported (ie, OR > 1). A nominal P value of .05 was used to determine statistical significance.
PRS Analyses
For each individual i, a PRS was constructed as , where is the risk allele dosage at variant m, is the weight for variant m, and C defines a set of risk loci. The weights for PRS313 were those derived by Mavaddat et al. (6) in the development of PRS313, while the weights for the 179 variant PRS (PRS179) were the log odds ratios from GWAS of BC in women of European ancestry (4). For PRSs of BC subtypes, we used the corresponding subtype-specific weights from women of European ancestry (4,6). An ER-negative specific PRS was constructed using 15 risk alleles, which were identified to have stronger associations with triple-negative or nonluminal BC subtypes than ER-positive BC through a cluster analysis (PRS15) (15). We categorized PRSs by percentile (<1%, 1%-5%, 5%-10%, 10%-20%, 20%-40%, 40%-60%, 60%-80%, 80%-90%, 90%-95%, 95%-99%, ≥99%) in controls, and the risk for each category was estimated relative to the 40%-60% reference group using logistic regression adjusting for the first 10 principal components (PCs), age, and study. We estimated odds ratios per unit SD and the area under the receiver operating characteristic curves (AUC) to compare with results from previous studies. We also computed theoretical AUC and the precision-recall curve (AUPRC) (16); details are provided in the Supplementary Methods (available online). In sensitivity analyses, excluding variants based on their imputation quality did not affect the performance of PRSs (Supplementary Table 3, available online).
We examined whether age modified the association between PRS and BC risk in stratified models by age category (<40 years, 40-49 years, 50-59 years, 60-69 years, ≥70 years) and the interaction with age. We compared distributions of PRSs in controls and the performance of PRSs between African countries (Ghana, Nigeria in Breast Cancer in the African Diaspora consortium, and the Women of African Ancestry Breast Cancer Study (WAABCS) study in the OncoArray consortium) and those of admixed US and Barbadian origin. We also assessed the interaction between PRS with ancestry (PC1) and family history on BC risk in African Americans and Barbadians; family history information was not available for samples from African countries.
The lifetime absolute risk of BC by PRS category among women with African and European ancestry were estimated taking into account the competing risk of dying from causes other than BC, which was described elsewhere (5). Inputs included the odds ratios of PRS313 estimated in women of African ancestry and European ancestry (6); age-specific BC incidence rates from the Surveillance, Epidemiology and End Results program (2000-2016); and mortality rates from the National Center for Health Statistics, Centers for Disease Control and Prevention (CDC) (2000-2016). The absolute risk for BC subtypes did not account for the competing risk of other subtypes. We also computed the lifetime risk with the odds ratios of continuous PRS313 (per SD) within the age range of 35 to 85 years using the R package iCARE 1.18.0 (17).
Recalibrated PRS
For the 179-variant PRS, we examined whether the performance could be improved by constructing recalibrated PRSs. To avoid overestimating performance, we implemented 150 repeated fourfold cross-validations in which samples in each consortium or study were randomly split into 4 nonoverlapping parts; in each round, 1 part was left out as the testing set and the other 3 parts were used as training sets. We estimated log odds ratios (weights) and selected markers in training sets and then applied the results to testing sets. First, we examined weights based on the marginal log odds ratios derived from women of African ancestry (PRS179.AFR) and from multi-ethnic populations of African and European ancestry (PRS179.ME). Second, for each risk variant, we considered alternative markers within each region that might be more informative than the index variant (“better markers”) in women of African ancestry (PRS179.AFR-better) or in the multi-ethnic populations (PRS179.ME-better). We assessed the variability of log odds ratios of the 179 risk loci in training sets by calculating the root-mean-squared deviation from effect sizes estimated using all samples. Details are provided in the Supplementary Methods (available online).
All statistical analyses were conducted using R v.4.0.0. All tests for statistical significance used a 2-sided alpha of .05.
Results
Participant Characteristics
The analysis included 9241 BC cases (4299 ER-positive and 2636 ER-negative) and 10 193 controls. The characteristics of the participants by study are described in Supplementary Table 1 (available online). The mean age of cases ranged from 45 years to 71 years across studies.
Association Testing of Individual Risk Alleles
The comparison of RAF and effect size of individual variants between women of European and African ancestry are summarized in Supplementary Figure 1 and Supplementary Tables 4 and 5 (available online). We had 80% power to detect the reported European effect sizes of overall BC for 42 of the 179 variants and for 48 of the 313 variants. Of the 179 previously reported BC risk loci, 177 were polymorphic with MAF of at least 0.1% and imputation quality score of at least 0.3 in at least 2 of the 4 consortia or studies. Of the 177, 133 (75.1%) had consistent direction of effect on overall BC risk, and 29 (16.4%) were nominally statistically significant (P < .05). Of the 313 variants in PRS313, 311 had a MAF greater than 0.1% and imputation score greater than 0.3 in at least 2 of the 4 consortia or studies, among which 215 (69.1%) showed directional consistency and 47 (15.1%) were nominally statistically significantly associated with overall BC risk.
Polygenic Risk Score
Both PRS179 and PRS313 were associated with risk, with slightly greater effect sizes and AUCs observed for PRS313 (Table 1). The actual threshold of percentiles of PRSs was shown in Supplementary Table 6 (available online). Women in the top 10% and 1% of the PRS313 had a 1.54-fold (95% CI = 1.38-fold to 1.72-fold) and a 2.01-fold (95% CI = 1.53-fold to 2.63-fold) elevated risk compared with women at average risk (PRS in 40th-60th percentiles), respectively (Table 1; Figure 1). The odds ratio per 1 SD of PRS313 was 1.27 (95% CI = 1.23 to 1.31), the AUC was 0.571 (95% CI = 0.562 to 0.579), and the AUPRC was 0.539 (baseline = 0.476).
Table 1.
Association between PRS and overall breast cancer risk in women of African ancestry
| PRS and AUCa | Controls, No. | PRS179 |
PRS313 |
||||
|---|---|---|---|---|---|---|---|
| Cases, No. | OR (95% CI)b | P c | Cases, No. | OR (95% CI)b | P c | ||
| PRS category, % | |||||||
| <1 | 106 | 58 | 0.62 (0.44 to 0.89) | .009 | 60 | 0.63 (0.44 to 0.89) | .009 |
| 1-5 | 406 | 248 | 0.70 (0.58 to 0.84) | 1.43 × 10−4 | 235 | 0.70 (0.58 to 0.85) | 2.29 × 10−4 |
| 5-10 | 508 | 306 | 0.67 (0.56 to 0.79) | 2.25 × 10−6 | 309 | 0.72 (0.61 to 0.85) | 9.51 × 10−5 |
| 10-20 | 1020 | 687 | 0.74 (0.65 to 0.84) | 2.84 × 10−6 | 681 | 0.74 (0.65 to 0.84) | 3.52 × 10−6 |
| 20-40 | 2038 | 1598 | 0.89 (0.81 to 0.99) | .03 | 1560 | 0.84 (0.76 to 0.93) | 6.92 × 10−4 |
| 40-60 | 2038 | 1808 | 1.00 (Referent) | — | 1802 | 1.00 (Referent) | — |
| 60-80 | 2038 | 2033 | 1.10 (1.00 to 1.22) | .045 | 1952 | 1.03 (0.94 to 1.14) | .50 |
| 80-90 | 1019 | 1093 | 1.17 (1.04 to 1.32) | .008 | 1197 | 1.29 (1.15 to 1.45) | 1.22 × 10−5 |
| 90-95 | 508 | 645 | 1.42 (1.23 to 1.65) | 1.80 × 10−6 | 643 | 1.38 (1.20 to 1.60) | 1.28 × 10−5 |
| 95-99 | 406 | 593 | 1.60 (1.37 to 1.87) | 2.15 × 10−9 | 602 | 1.61 (1.38 to 1.87) | 1.32 × 10−9 |
| >99 | 106 | 172 | 1.83 (1.39 to 2.40) | 1.56 × 10−5 | 200 | 2.01 (1.53 to 2.63) | 3.69 × 10−7 |
| Continuous PRS per 1 SD | 10 193 | 9241 | 1.26 (1.22 to 1.30) | 2.59 × 10−45 | 9241 | 1.27 (1.23 to 1.31) | 4.23 × 10−49 |
| AUC | 0.568 (0.56 to 0.576) | — | 0.571(0.562 to 0.579) | — | |||
AUCs were adjusted for study and the first 10 principal components. AUC = area under the receiver operating characteristic curve; CI = confidence interval; GBHS = Ghana Breast Health Study; OR = odds ratio; PRS = polygenic risk score.
Odds ratios were estimated using unconditional logistic regression model, adjusting for age, study, and the first 10 principal components in each consortium and in the GBHS and then combined using a fixed-effect meta-analysis with inverse variance weights.
Two-sided P values were Wald P value from fixed-effect meta-analysis.
Figure 1.
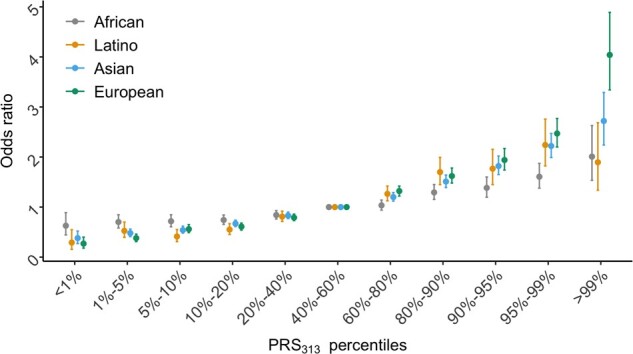
Association between the categorical 313-variant polygenic risk score (PRS313) and overall breast cancer risk by population. The x-axis indicates the PRS313 percentiles. The y-axis represents odds ratio (OR) values for the indicated PRS313 percentiles compared with the 40%-60% category of PRS313 as the reference (population average risk). Dots represent odds ratios and error bar lines represent standard error estimated in each population. The grey line represents results for women of African ancestry, which were estimated in this study. The yellow line represents results for Latinas obtained from a previous Latino PRS study (10). The blue line represents results for Asian women obtained from an Asian PRS study (18). The green line represents results for women of European ancestry obtained from a previous European ancestry PRS study (6). African ancestry results are also provided in Table 1.
For ER-positive BC, compared with the population average, women in the top 10% and 1% of PRS313 had a 1.85-fold (95% CI = 1.61-fold to 2.13-fold) and a 2.16-fold (95% CI = 1.56-fold to 3.00-fold) increased risk, respectively (Table 2). The odds ratio per 1 SD of PRS313 was 1.37 (95% CI = 1.32 to 1.43), the AUC was 0.588 (95% CI = 0.577 to 0.599), and the AUPRC was 0.368 (baseline = 0.297). The theoretical AUC of PRS313 was 0.588 for African-ancestry women and 0.643 for women of European ancestry. For ER-negative BC, compared with women at average risk, those in the top 10% and 1% of PRS313 had a 1.47-fold (95% CI = 1.25-fold to 1.74-fold) and a 2.18-fold (95% CI = 1.50-fold to 3.16-fold) increased risk, respectively (Table 3). The PRS179 performed slightly better than PRS313, with an AUC of 0.578 vs 0.562, an AUPRC of 0.256 vs 0.246 (baseline = 0.205), and an odds ratio per 1 SD of 1.31 vs 1.21, respectively. The PRS15 performed similarly to PRS179 (Table 3). The theoretical AUC of PRS313 was 0.554 for African-ancestry women and 0.604 for European-ancestry women.
Table 2.
Association between PRS and ER-positive breast cancer risk in women of African ancestry
| PRS and AUCa | Controls, No. | PRS179 |
PRS313 |
PRS313 in European womend |
||||
|---|---|---|---|---|---|---|---|---|
| Cases, No. | OR (95% CI)b | P c | Cases, No. | OR (95% CI)b | P c | OR (95% CI) | ||
| PRS category, % | ||||||||
| <1 | 106 | 25 | 0.54 (0.33 to 0.87) | .01 | 28 | 0.60 (0.37 to 0.95) | .03 | 0.16 (0.09 to 0.30) |
| 1-5 | 406 | 93 | 0.51 (0.39 to 0.66) | 4.75 × 10−7 | 89 | 0.56 (0.43 to 0.73) | 1.92 × 10−5 | 0.32 (0.25 to 0.40) |
| 5-10 | 508 | 152 | 0.62 (0.50 to 0.77) | 2.10 × 10−5 | 128 | 0.63 (0.50 to 0.79) | 7.12 × 10−5 | 0.50 (0.42 to 0.60) |
| 10-20 | 1020 | 323 | 0.71 (0.60 to 0.83) | 4.11 × 10−5 | 288 | 0.69 (0.59 to 0.82) | 3.17 × 10−5 | 0.61 (0.53 to 0.69) |
| 20-40 | 2038 | 736 | 0.83 (0.73 to 0.95) | .007 | 723 | 0.86 (0.75 to 0.98) | .02 | 0.77 (0.70 to 0.85) |
| 40-60 | 2038 | 856 | 1.00 (Referent) | — | 807 | 1.00 (Referent) | — | 1.00 (Referent) |
| 60-80 | 2038 | 918 | 1.07 (0.94 to 1.21) | .32 | 934 | 1.13 (1.00 to 1.29) | .05 | 1.40 (1.28 to1.52) |
| 80-90 | 1019 | 496 | 1.13 (0.97 to 1.31) | .11 | 541 | 1.30 (1.12 to 1.51) | 6.18 × 10−4 | 1.59 (1.44 to 1.76) |
| 90-95 | 508 | 302 | 1.43 (1.19 to 1.72) | 1.71 × 10−4 | 350 | 1.75 (1.46 to 2.10) | 1.52 × 10−9 | 2.17 (1.93 to 2.44) |
| 95-99 | 406 | 306 | 1.80 (1.48 to 2.19) | 2.56 × 10−9 | 315 | 1.89 (1.56 to 2.29) | 7.97 × 10−11 | 2.68 (2.37 to 3.03) |
| >99 | 106 | 92 | 2.21 (1.58 to 3.09) | 3.31 × 10−6 | 96 | 2.16 (1.56 to 3.00) | 4.12 × 10−6 | 4.37 (3.59 to 5.33) |
| Continuous PRS per 1 SD | 10 193 | 4299 | 1.33 (1.27 to 1.38) | 4.35 × 10−41 | 4299 | 1.37 (1.32 to 1.43) | 8.63 × 10−51 | 1.74 (1.66 to 1.82) |
| AUC | 0.576 (0.566 to 0.585) | — | 0.588 (0.577 to 0.599) | — | 0.651 | |||
AUCs were adjusted for study and the first 10 principal components. AUC = area under the receiver operating characteristic curve; CI = confidence interval; GBHS = Ghana Breast Health Study; OR = odds ratio; PRS = polygenic risk score.
Odds ratios were estimated using unconditional logistic regression model, adjusting for age, study, the first 10 principal components in each consortium and in the GBHS, and then combined using a fixed-effect meta-analysis with inverse variance weights.
Two-sided P values were Wald P value from fixed-effect meta-analysis.
Results were obtained from a previous PRS study in women of European ancestry (6).
Table 3.
Association between PRS and ER-negative breast cancer risk in women of African ancestry
| PRS and AUCa | Controls, No. | PRS179 |
PRS15 |
PRS313 |
PRS313 in European womend | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases, No. | OR (95% CI)b | P c | Cases, No. | OR (95% CI)b | P c | Cases, No. | OR (95% CI)b | P c | OR (95% CI) | ||
| PRS category , % | |||||||||||
| <1 | 106 | 17 | 0.68 (0.39 to 1.20) | .19 | 15 | 0.63 (0.35 to 1.12) | .12 | 22 | 0.86 (0.52 to 1.42) | .55 | 0.27 (0.09 to 0.86) |
| 1-5 | 406 | 56 | 0.58 (0.42 to 0.80) | 7.78 × 10−4 | 71 | 0.73 (0.55 to 0.97) | .03 | 90 | 0.94 (0.72 to 1.23) | .66 | 0.70 (0.47 to 1.05) |
| 5-10 | 508 | 75 | 0.57 (0.44 to 0.76) | 9.26 × 10−5 | 87 | 0.74 (0.57 to 0.96) | .02 | 86 | 0.71 (0.54 to 0.92) | .01 | 0.74 (0.52 to 1.05) |
| 10-20 | 1020 | 192 | 0.75 (0.61 to 0.91) | .003 | 185 | 0.78 (0.64 to 0.95) | .01 | 191 | 0.78 (0.64 to 0.94) | .01 | 0.83 (0.64 to 1.08) |
| 20-40 | 2038 | 462 | 0.90 (0.77 to 1.04) | .16 | 463 | 0.95 (0.82 to 1.11) | .54 | 454 | 0.87 (0.75 to 1.01) | .08 | 0.85 (0.69 to 1.04) |
| 40-60 | 2038 | 523 | 1.00 (Referent) | — | 492 | 1.00 (Referent) | 517 | 1.00 (Referent) | — | 1.00 (Referent) | |
| 60-80 | 2038 | 537 | 1.06 (0.91 to 1.22) | .46 | 563 | 1.14 (0.98 to 1.32) | .09 | 551 | 1.02 (0.88 to 1.18) | .80 | 1.32 (1.09 to 1.59) |
| 80-90 | 1019 | 339 | 1.30 (1.10 to 1.55) | .002 | 341 | 1.33 (1.12 to 1.58) | .001 | 324 | 1.20 (1.01 to 1.43) | .03 | 1.51 (1.22 to 1.87) |
| 90-95 | 508 | 190 | 1.42 (1.15 to 1.75) | .001 | 201 | 1.59 (1.29 to 1.96) | 1.27 × 10−5 | 165 | 1.20 (0.96 to 1.50) | .11 | 2.24 (1.76 to 2.85) |
| 95-99 | 406 | 187 | 1.80 (1.44 to 2.24) | 1.51 × 10−7 | 172 | 1.84 (1.47 to 2.30) | 1.01 × 10−7 | 175 | 1.66 (1.32 to 2.07) | 9.12 × 10−6 | 2.39 (1.86 to 3.07) |
| >99 | 106 | 58 | 2.05 (1.41 to 2.97) | 1.72 × 10−4 | 46 | 1.93 (1.31 to 2.86) | 9.72 × 10−4 | 61 | 2.18 (1.50 to 3.16) | 4.08 × 10−5 | 2.78 (1.83 to 4.24) |
| Continuous PRS per 1 SD | 10 193 | 2636 | 1.31 (1.24 to 1.37) | 5.53 × 10−27 | 2636 | 1.28 (1.22 to 1.34) | 9.29 × 10−25 | 2636 | 1.21 (1.15 to 1.27) | 3.39 × 10−15 | 1.47 (1.37 to 1.58) |
| AUC | 0.578 (0.564 to 0.591) | — | 0.571 (0.557 to 0.583) | — | 0.562 (0.551 to 0.573) | — | 0.611 | ||||
AUCs were adjusted for study and the first 10 principal components. AUC = area under the receiver operating characteristic curve; CI = confidence interval; GBHS = Ghana Breast Health Study; OR = odds ratio; PRS = polygenic risk score.
Odds ratios were estimated using unconditional logistic regression model, adjusting for age, study, the first 10 principal components in each consortium, and in the GBHS, and then combined using a fixed-effect meta-analysis with inverse variance weights.
Two-sided P values were Wald P value from fixed-effect meta-analysis.
Results were obtained from a previous PRS study in women of European ancestry (6).
We did not observe a statistically significant interaction between either PRS and age at diagnosis for overall or subtype-specific BC risk (Supplementary Table 7; Supplementary Figure 2, available online). The average PRS was greater in controls from studies in Africa compared with those from studies of US and Barbadian origins (P < 2.2 × 10−16 for both PRS313 and PRS179; Supplementary Figure 3, available online). The performance of the PRSs by country of origin, ancestry proportion defined by PC1, or family history are shown in Supplementary Tables 7-9.
The distribution and root-mean-square deviation (RMSD) of log odds ratios of the “better markers” and the 179 known risk variants in the 600 training sets are shown in Supplementary Figure 4 and Supplementary Table 10 (available online). When estimating the 4 recalibrated PRSs for overall BC, we found the PRS179.ME-better, which used “better markers” and variant weights derived from the multi-ethnic training sets, had the largest AUC but did not differ from that of PRS179 (average AUC = 0.569 vs AUC = 0.566; Supplementary Figure 5, available online).
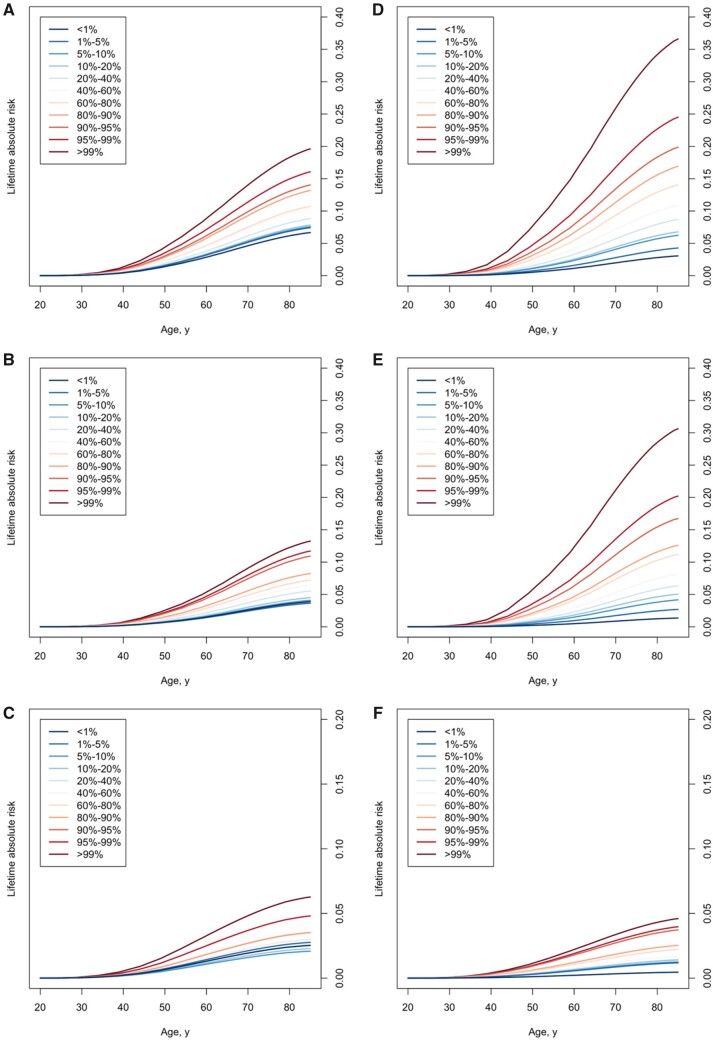
The estimated lifetime absolute risks for African American and European-ancestry women by PRS313 categories for overall and subtype-specific BC are shown in Figure 2. By age 85 years, the absolute risk of overall BC was 19.6% for women in the top 1% of PRS313 and 6.7% for women in the lowest 1%. The density plots of lifetime absolute risk of ER-positive and ER-negative BC in European and African American women are shown in Supplementary Figure 6 (available online). The average lifetime risk of ER-positive BC from 35 years to 85 years is 9.1% (SD = 4.6%) for European-ancestry women and 6.6% (SD = 2.0%) for African American women, and for ER-negative BC the risks are 1.9% (SD = 0.7%) for European-ancestry women and 2.9% (SD = 0.6%) for African American women.
Figure 2.
Lifetime absolute risk of breast cancer by polygenic risk score (PRS) category in African American women. Lifetime absolute risk of developing breast cancer for the 313-variant PRS (PRS313) in African American women for overall breast cancer (A), estrogen receptor (ER)-positive breast cancer (B) and ER-negative breast cancer (C) and in European-ancestry women for overall breast cancer (D), ER-positive breast cancer (E), and ER-negative breast cancer (F). The x-axis represents age and the y-axis is the absolute risk of breast cancer by a given age. The different colored lines represent the corresponding PRS strata. See Methods for details about the calculation of absolute risks.
Discussion
We evaluated the performance of 2 PRSs developed in women of European ancestry in women of African ancestry and found both PRSs to be statistically significantly associated with overall and subtype-specific BC risk, with odds ratios per SD of 1.21 to approximately 1.37 and AUCs ranging between 0.57 and 0.59. Women in the top 10% of either PRS had a 1.54-fold elevated risk of BC. Women in the top 1% of the PRSs had a 1.83-fold to 2.01-fold increase in risk and were estimated to have an 18% to 20% lifetime risk of developing BC. However, these estimates are markedly lower than what have been reported for other racial or ethnic populations.
The authors of the largest PRS study of BC in women of European ancestry examined multiple PRSs using between 77 and 3820 risk variants, reported odds ratios per SD between 1.49 and 1.71, and AUCs between 0.61 and 0.64 for overall BC (6). The authors of the largest PRS study in Latinas examined the performance of a 71- and 180-variant PRS for overall BC and reported odds ratios per SD from 1.51 to 1.58 and AUCs of 0.61 to 0.63 (19) (Figure 1). Researchers examined in East Asians a 67-variant PRS for overall BC and reported an odds ratio per SD of 1.44 and an AUC of 0.61 (20,21). The authors of the largest PRS study in Asians examined a 287-variant PRS (derived from the 313 variants in the European ancestry study) and reported an odds ratio per SD of 1.51 and AUC of 0.62 for overall BC (18). The weaker performance of PRS in women of African ancestry is consistent with observations for other cancers and other chronic diseases (22,23). Factors likely to be underlying the difference include variation in linkage disequilibrium patterns, allele frequencies, and potential effect heterogeneity between populations. The reduction in performance is also in agreement with previous studies that have demonstrated the decline in PRS performance with increasing genetic divergence from the training population (24‐26).
To note, even though the AUC was worse in African-ancestry women than that in European-ancestry women, the risk stratification in the population, which depends also on the underlying rates of disease, may not be worse. This was demonstrated in comparison of ER-negative BC in our study: the AUC was greater in European-ancestry women (theoretical AUC = 0.604) than in African-ancestry women (theoretical AUC = 0.554); however, because of the greater baseline risk of ER-negative disease in African-ancestry women than in European-ancestry women (2.9% vs 1.9% average lifetime risk, respectively), for any given risk threshold, a larger percentage of African-ancestry women than European-ancestry women would be identified as being at elevated risk of this disease.
For BC subtypes, PRS313 performed better in the prediction of ER-positive BC than ER-negative BC, which is consistent with the previous study in women of European ancestry (6). Both PRS179 and PRS15 performed better than PRS313, which suggests PRS313 is not optimal for ER-negative disease in women of African ancestry. The previous investigations of the PRS in women of European ancestry reported a weak nonlinear decline in effect with increasing age for ER-positive BC. However, we did not observe an interaction with age for overall or BC subtypes in this study. The previous BCAC study reported an attenuated odds ratio of PRS313 for ER-positive BC in women with family history (6). We detected a suggestive interaction of the PRS with family history, although larger studies will be needed to improve the power to detect interactions in women of African ancestry.
Based on the current sample size of African-ancestry women in this study, we found European weights provided optimal PRS performance—the performance could not be improved using weights estimated in women of African ancestry or by replacing an index variant with a “better African marker” in each risk region. This could be due to the training set not being large enough to provide accurate estimates of effect or to distinguish causal variants from correlated markers in each risk region. We observed a minor improvement in PRS using multi-ethnic weights, which is corroborated by a simulation study showing that multi-ethnic training populations substantially outperformed PRS using a single training population (27). The current multi-ethnic samples predominantly consisted of women of European ancestry. We expect that PRS performance may be further improved when including more samples of African ancestry.
The current guideline for BC screening classifies those at high risk based on BRCA1 or BRCA2 mutations and family history–based lifetime risk assessment (28‐30). Several studies have reported that incorporating PRS into these existing risk models could increase discrimination accuracy and improve calibration (8,9,31‐34). These studies and models were predominantly conducted in European ancestry populations, and similar assessments are needed in larger African-ancestry studies to assess the value of incorporating common germline variation into the decision-making process of screening recommendations. Of note, incidence rates of BC in West African countries are substantially lower than that in the United States (35). And given the differences in access to health care, the harm-to-benefit ratio and cost-effectiveness of introducing risk-stratified BC screening programs at the population level should be carefully assessed.
In conclusion, although PRSs developed in women of European ancestry can identify women at elevated risk of developing BC, they currently substantially underperform in African-ancestry women compared with women of European, Asian, and Latino ancestry.
Funding
This work was supported by NIH grants R01CA228198, R01CA202981, R01CA058420, U01CA164974, R01CA098663, and R01CA228357. The GBHS has been funded with federal funds from the US National Cancer Institute (NCI), National Institutes of Health, under the Intramural Research Program and NCI Contract No. 75N910D00024. Genotyping of the OncoArray was principally funded from 3 sources: the PERSPECTIVE project, funded by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the “Ministère de l’Économie, de la Science et de l’Innovation du Québec” through Genome Québec, and the Quebec Breast Cancer Foundation; the NCI Genetic Associations and Mechanisms in Oncology (GAME-ON) initiative and Discovery, Biology and Risk of Inherited Variants in Breast Cancer (DRIVE) project (NIH Grants U19 CA148065 and X01HG007492); and Cancer Research UK (C1287/A10118 and C1287/A16563). BCAC was funded by Cancer Research UK (C1287/A16563), by the European Community’s Seventh Framework Programme under grant agreement 223175 (HEALTH-F2-2009–223175) (The Collaborative Oncological Gene-environment Study, COGS), and by the European Union’s Horizon 2020 Research and Innovation Programme under grant agreements 633784 (B-CAST) and 634935 (BRIDGES). The authors acknowledge the research contributions of the Cancer Genomics Research Laboratory in the GBHS for their expertise, execution, and support of this research in the areas of project planning, wet laboratory processing of specimens, and bioinformatics analysis of genotyping data. The authors acknowledge the LAGENO Consortium for providing the Latino PRS results for comparison. The Sister Study was funded by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES044005).
Notes
Role of the funders: The study funders and sponsors did not participate in the collection, analysis, or interpretation of data, or in the writing of the manuscript.
Disclosures: The authors have no conflicts of interest to disclose.
Author contributions: Conceptualization: CAH, DH, MGC, JRP; Supervision: CAH; Data curation or Investigation: TA, KLL, GZ, EARN, SAH, EMJ, LB, WZ, JJH, RGZ, SN, EVB, SAI, MFP, SLD, JLRG, DPS, JAT, QW, CRW, CMK, WB, KLN, AH, BN, SA, LESC, JTB, SJC, AFO, CBA, OIO, MKB, JD, AMD, DFE, KM, PDPP, TOO, OO, JNCL, FD, LE, VV, JY, EAdjei, FA, BAwuah, JO, EOB, NT, MA, BAW, EAbaidoo; Formal analysis: ZD, GG, TA; Funding acquisition: CAH, DH; Methodology: DVC, MGC, LB, TA, JF, SW, DA, KN, RB; Resources: BA, MAT, PPC, JF, EMJ, LB, WZ, JJH, RGZ, SN, EVB, SAI, MFP, SLD, JLRG, DPS, JAT, QW, CRW, CMK, WB, KLN, AH, BN, SA, LESC, JTB, SJC, AFO, CBA, OIO, TOO, JY, BA, BAW, JRP, MGC, DH, JNCL, JY, BAwuah, BAW; Project administration and data curation: ET, NOOO, VO, BArhin, IB, MF, PA, MB, AT; Writing—original draft: ZD, DVC, MGC, CAH; Writing—review & editing: all authors.
Disclaimers: The contents of this article are solely the responsibility of the authors and do not reflect the official views of the National Institutes of Health.
Acknowledgements: The study protocol was approved by the Institutional Review Boards of participating cancer registries, as required. The GBHS Study Team: Florence Dedey (Korle Bu Teaching Hospital, Accra, Ghana), Richard Biritwum (University of Ghana, Accra, Ghana), Lawrence Edusei (Korle Bu Teaching Hospital, Accra, Ghana), Verna Vanderpuye (Korle Bu Teaching Hospital, Accra, Ghana), Ernest Adjei (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Francis Aitpillah (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Joseph Oppong (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Margaret Frempong (Peace and Love Hospital, Kumasi, Ghana), Jonine Figueroa (University of Edinburgh, Edinburgh, Scotland), Louise Brinton (US National Cancer Institute, Bethesda, MD, USA), Thomas Ahearn (U.S. National Cancer Institute, Bethesda, MD, USA), Ernest Osei-Bonsu (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Nicholas Titiloye (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Michelle Brotzman (US National Cancer Institute, Bethesda, MD, USA), Ann Truelove (Westat, Inc, MD, USA), Evelyn Tay (Korle Bu Teaching Hospital, Accra, Ghana), Naomi Oyoe Ohene Oti (Korle Bu Teaching Hospital, Accra, Ghana), Victoria Okyne (Korle Bu Teaching Hospital, Accra, Ghana), Isaac Boakye (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Bernard Arhin (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Marion Alcpaloo (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Emma Abaidoo (Peace and Love Hospital, Kumasi, Ghana), Prince Agyapong (Peace and Love Hospital, Kumasi, Ghana), Joe Nat Clegg-Lamptey (Korle Bu Teaching Hospital, Accra, Ghana), Joel Yarney (Korle Bu Teaching Hospital, Accra, Ghana), Kofi Nyarko (University of Ghana, Accra, Ghana), Daniel Ansong (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Baffour Awuah (Komfo Anoyke Teaching Hospital, Kumasi, Ghana), Seth Wiafe (Peace and Love Hospital, Kumasi, Ghana), Beatrice Wiafe-Addai (Peace and Love Hospital, Kumasi, Ghana), Montserrat Garcia-Closas (US National Cancer Institute, Bethesda, MD, USA).
Prior presentation: Parts of this study have been presented during the poster session at the American Association for Cancer Research Annual Meeting, Virtual, June 22-24. 2020.
Data Availability
The datasets are publicly available via dbGaP for AABC (phs000851.v1.p1), ONCO (phs001265.v1.p1), AMBER (phs000669.v1.p1), GBHS (phs002387.v1.p1) and ROOT (phs000383.v1.p1).
Supplementary Material
References
- 1.Mucci LA, Hjelmborg JB, Harris JR, et al. Familial risk and heritability of cancer among twins in Nordic countries. JAMA. 2016;315(1):68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park SL, Cheng I, Haiman CA.. Genome-wide association studies of cancer in diverse populations. Cancer Epidemiol Biomarkers Prev. 2018;27(4):405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milne RL, Kuchenbaecker KB, Michailidou K, et al. ; ABCTB Investigators. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet. 2017;49(12):1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michailidou K, Lindström S, Dennis J, et al. NBCS Collaborators. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mavaddat N, Pharoah PDP, Michailidou K, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107(5). doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavaddat N, Michailidou K, Dennis J, et al. ; NBCS Collaborators. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104(1):21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Rice M, Tworoger SS, et al. Addition of a polygenic risk score, mammographic density, and endogenous hormones to existing breast cancer risk prediction models: a nested case–control study. PLoS Med. 2018;15(9):e1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med. 2019;21(8):1708–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pal Choudhury P, Wilcox AN, Brook MN, et al. Comparative validation of breast cancer risk prediction models and projections for future risk stratification. J Natl Cancer Inst. 2020;112(3):278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen F, Chen GK, Stram DO, et al. A genome-wide association study of breast cancer in women of African ancestry. Hum Genet. 2013;132(1):39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Rhie SK, Huo D, et al. Characterizing genetic susceptibility to breast cancer in women of African ancestry. Cancer Epidemiol Biomarkers Prev. 2017;26(7):1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer JR, Ambrosone CB, Olshan AF.. A collaborative study of the etiology of breast cancer subtypes in African American women: The AMBER Consortium. Cancer Causes Control. 2014;25(3):309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amos CI, Dennis J, Wang Z, et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol Biomarkers Prev. 2016;26(1):126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huo D, Feng Y, Haddad S, et al. Genome-wide association studies in women of African ancestry identified 3q26.21 as a novel susceptibility locus for oestrogen receptor negative breast cancer. Hum Mol Genet. 2016;25(21):4835–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahearn TU, Zhang H, Michailidou K, et al. Common variants in breast cancer risk loci predispose to distinct tumor subtypes. bioRxiv. 2020:733402. doi: 10.1101/733402. [DOI] [PMC free article] [PubMed]
- 16.Saito T, Rehmsmeier M.. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLOS One. 2015;10(3):e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhury PP, Maas P, Wilcox A, et al. iCARE: an R package to build, validate and apply absolute risk models. PLOS One. 2020;15(2):e0228198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho W-K, Tan M-M, Mavaddat N, et al. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat Commun. 2020;11(1):3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shieh Y, Fejerman L, Lott PC, et al. A polygenic risk score for breast cancer in U.S. Latinas and Latin-American women. bioRxiv. 2019:598730. doi: 10.1101/598730. [DOI] [PMC free article] [PubMed]
- 20.Wen W, Shu X, Guo X, et al. Prediction of breast cancer risk based on common genetic variants in women of East Asian ancestry. Breast Cancer Res. 2016;18(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu X, Long J, Cai Q, et al. Identification of novel breast cancer susceptibility loci in meta-analyses conducted among Asian and European descendants. Nat Commun. 2020;11(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conti DV, Wang K, Sheng X, et al. Two novel susceptibility loci for prostate cancer in men of African ancestry. J Natl Cancer Inst. 2017;109(8):djx084. doi: 10.1093/jnci/djx084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manichaikul A, Peres LC, Wang X-Q, et al. ; The African American Breast Cancer Consortium (AABC). Identification of novel epithelial ovarian cancer loci in women of African ancestry. Int J Cancer. 2020;146(11):2987–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scutari M, Mackay I, Balding D.. Using genetic distance to infer the accuracy of genomic prediction. PLoS Genet. 2016;12(9):e1006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin AR, Gignoux CR, Walters RK, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100(4):635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ.. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Márquez-Luna C, Loh P-R, Price AL; SIGMA Type 2 Diabetes Consortium. Multi-ethnic polygenic risk scores improve risk prediction in diverse populations. Genet Epidemiol. 2017;41(8):811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Practice Bulletin No. 122: Breast cancer screening : obstetrics and gynecology. https://journals.lww.com/greenjournal/Fulltext/2011/08000/Practice_Bulletin_No__122__Breast_Cancer_Screening.40.aspx. Accessed February 27, 2020. [DOI] [PubMed]
- 29.Oeffinger KC, Fontham ETH, Etzioni R, et al. ; American Cancer Society. Breast cancer screening for women at average risk: 2015 guideline update from the American Cancer Society. JAMA. 2015;314(15):1599–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.US Preventive Services Task Force. Screening for breast cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2009;151(10):716–726. [DOI] [PubMed] [Google Scholar]
- 31.Cuzick J, Brentnall AR, Segal C, et al. Impact of a panel of 88 single nucleotide polymorphisms on the risk of breast cancer in high-risk women: results from two randomized tamoxifen prevention trials. J Clin Oncol. 2017;35(7):743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakeman IMM, Hilbers FS, Rodríguez-Girondo M, et al. Addition of a 161-SNP polygenic risk score to family history-based risk prediction: impact on clinical management in non-BRCA1/2 breast cancer families. J Med Genet. 2019;56(9):581–589. [DOI] [PubMed] [Google Scholar]
- 33.Evans DGR, Harkness EF, Brentnall AR, et al. Breast cancer pathology and stage are better predicted by risk stratification models that include mammographic density and common genetic variants. Breast Cancer Res Treat. 2019;176(1):141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilcox AN, Choudhury PP, Gao C, et al. Prospective evaluation of a breast cancer risk model integrating classical risk factors and polygenic risk in 15 cohorts from six countries. medRxiv. 2019:19011171. doi: 10.1101/19011171. [DOI] [PMC free article] [PubMed]
- 35.Adeloye D, Sowunmi OY, Jacobs W, et al. Estimating the incidence of breast cancer in Africa: a systematic review and meta-analysis. J Glob Health. 2018;8(1):010419. doi: 10.7189/jogh.08.010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets are publicly available via dbGaP for AABC (phs000851.v1.p1), ONCO (phs001265.v1.p1), AMBER (phs000669.v1.p1), GBHS (phs002387.v1.p1) and ROOT (phs000383.v1.p1).