Abstract
The energy allocation for vegetative and reproductive growth is regulated by developmental signals and environmental cues, which subsequently affects seed output. However, the molecular mechanism underlying how plants coordinate yield-related traits to control yield in changing source–sink relationships remains largely unknown. Here, we discovered the lectin receptor-like kinase LecRK-VIII.2 as a specific receptor-like kinase that coordinates silique number, seed size, and seed number to determine seed yield in Arabidopsis (Arabidopsis thaliana). The lecrk-VIII.2 mutants develop smaller seeds, but more siliques and seeds, leading to increased yield. In contrast, the plants overexpressing LecRK-VIII.2 form bigger seeds, but less siliques and seeds, which results in similar yield to that of wild-type plants. Interestingly, LecRK-VIII.2 promotes the growth of the rosette, root, and stem by coordinating the source–sink relationship. Additionally, LecRK-VIII.2 positively regulates cell expansion and proliferation in the seed coat, and maternally controls seed size. The genetic and biochemical analyses demonstrated that LecRK-VIII.2 acts upstream of the mitogen-activated protein kinase (MAPK) gene MPK6 to regulate silique number, seed size, and seed number. Collectively, these findings uncover LecRK-VIII.2 as an upstream component of the MAPK signaling pathway to control yield-related traits and suggest its potential for crop improvement aimed at developing plants with stable yield, a robust root system, and improved lodging resistance.
A lectin receptor-like kinase regulates yield-related traits and coordinates the source–sink relationship in Arabidopsis.
Introduction
The seed size/number trade-off is an important concept in life-history theory, which contributes to evolutionarily comprehending the astonishing diversity of seed sizes in plant communities (Lord et al., 1995; Moles et al., 2005). The seed size/number trade-off has been observed in different environments and studied for decades in a variety of species, but the molecular mechanism underlying how plants perceive and transduce extracellular signals to organize this process remains obscure. In plants, if the resources supplied for reproduction are fixed, the trade-off is inevitable. The increase in seed size must be compensated by a decrease in seed number (Smith and Fretwell, 1974), whereas several previous studies have shown a large variability, with negative, positive, or even uncorrelated relationships between both variables (GlendA and Mike, 1998; Eriksson, 1999; Richards, 2000; Baker et al., 2006; Koenig et al., 2009; Sõber and Ramula, 2013; Brancalion and Rodrigues, 2014; Lazaro and Larrinaga, 2018). These studies suggest that plants may possess both common and distinct mechanisms to determine their trade-off. Additionally, for seed crops, reproductive output is an accumulated outcome of both source and sink strength during plant growth and development. When the net photosynthetic rate and the rate of photoassimilate remobilization from source tissues to sink fluctuate due to abiotic and biotic stresses, the seed size/number trade-off and seed yield are subsequently affected (reviewed in Smith et al. 2018). However, our understanding about the molecular mechanism underlying how the source–sink relationship influences the trade-off remains limited.
Seed size is determined by the coordinated growth of embryo, endosperm, and maternal tissues. Up to now, numerous signaling pathways that act maternally or zygotically to regulate seed size have been identified, including the ubiquitin-proteasome pathway (Li et al., 2008; Xia et al., 2013; Du et al., 2014; Peng et al., 2015; Dong et al., 2017), G-protein signaling (Fujisawa et al., 1999; Fan et al., 2006; Huang et al., 2009; Mao et al., 2010; Chakravorty, 2011; Roy et al., 2014), mitogen-activated protein kinase (MAPK) cascade signaling (Lukowitz et al., 2004; Duan et al., 2014; Liu et al., 2015; Zhang et al., 2017), the HAIKU pathway (Luo et al., 2005; Zhou et al., 2009; Wang et al., 2010; Cheng et al., 2014), phytohormones (auxin, abscisic acid, brassinosteroid (BR), and cytokinin), and some of their downstream transcriptional factors (reviewed in Li et al., 2019). Seed size is the final result that is coordinated by complex networks involving many different developmental and environmental signals. Many regulators and pathways have been characterized, but our understanding of the regulatory networks of seed size is fragmented. Mainly as a result of this, the key factors upstream of these signaling pathways are still missing, which could integrate some of them as a network.
The receptor-like kinases (RLKs) are believed to act as sensors to mediate the cellular response toward various environmental cues, hormonal signals, and stress (De Smet et al., 2009). The Arabidopsis thaliana genome encodes over 600 RLKs (Shiu and Bleecker, 2001a, 2001b), and these include 75 lectin receptor-like kinases (LecRKs) that are divided into three types: C-type, G-type, and L-type (Vaid et al., 2012). LecRKs are designated by the presence of a legume–lectin protein-like extracellular domain that is structurally similar to carbohydrate-binding proteins and exhibits glucose/mannose specificity (Hervé et al., 1999). LecRKs play important and versatile roles in pollen development (Wan et al., 2008; Peng et al., 2020), seed germination (Cheng et al., 2013), fiber development (Zuo et al., 2004), legume-rhizobia symbiosis (Hirsch, 1999; Navarro-Gochicoa et al., 2003; Labbé et al., 2019), hormone signaling (Deng et al., 2009; Xin et al., 2009; Luo et al., 2017; Tripathi et al., 2018; Zhang et al., 2018), defense against pathogens and insect pests (Chen et al., 2006; Bonaventure, 2011; Gilardoni et al., 2011; Singh et al., 2012; Wang et al., 2016; Gouhier-Darimont et al., 2019; Wang et al., 2019; Luo et al., 2020; Pham et al., 2020; Woo et al., 2020; Xu et al., 2020), and responses to abiotic stresses (He et al., 2004; Joshi et al., 2010; Vaid et al., 2015; Liu et al., 2017). More importantly, recent studies revealed that LecRK-I.9 and LecRK-I.5 act as plant receptors for extracellular ATP (eATP), which are required for eATP-induced calcium response, MAPK activation, and gene expression (Choi et al., 2014; Tripathi et al., 2018; Wang et al., 2018; Pham et al., 2020). Equally, LecRK-I.8 and LecRK-VI.2 function as potential extracellular nicotinamide adenine dinucleotide (eNAD+) receptors that are essential for basal resistance against bacterial pathogens (Wang et al., 2017, 2019). As pentose sugar derivatives, ATP and NAD+ are two of the most universal energy currencies and common signaling molecules in all organisms (Choi et al., 2014; Wang et al., 2017, 2018, 2019; Pham et al., 2020). Considering that the trade-off and the energy allocation between vegetative growth and reproductive output could be involved in carbohydrate (or saccharide derivative)-based signal transduction and depend on RLKs to organize cellular activities between the inside and outside of a cell, we therefore hypothesized that LecRKs could be good candidates to dissect the molecular mechanism coordinating the process.
In this study, we report that LecRK-VIII.2 acts upstream of MPK6 to determine yield by coordinating silique number, seed size, and seed number. The genetic results show that LecRK-VIII.2 increases the organ size of the rosette, stem, and root, and affects their source–sink relationship. LecRK-VIII.2 promotes cell expansion and proliferation to regulate the development of the embryo and seed coat. In general, our findings reveal the roles of LecRK-VIII.2 in regulating yield-related traits and plant growth and development.
Results
LecRK-VIII.2 exhibits specific expression in developing seeds
To test our hypothesis about the roles of LecRKs in coordinating the three interactive, yield-related traits, we performed global expression analyses of all 75 Arabidopsis LecRKs during plant development and responses to various environmental cues. LecRK-VIII.2 showed specific high expression during stage 4–stage 7 of developing seeds, while few transcripts were detected in other tissues (Schmid et al., 2005; Nakabayashi et al., 2005; Klaas and Francine, 2009). The analyses of the microarray data on seed development revealed that LecRK-VIII.2 was predominantly expressed in the seed coat, but less in the embryo and endosperm (Le et al., 2010; Supplemental Figure S1, A and B). Interestingly, both independent datasets showed that LecRK-VIII.2 transcripts exhibited accumulation from the globular to torpedo stage and displayed a common transcriptional peak in the torpedo stage of developing seeds. The patterns of LecRK-VIII.2 were confirmed by our reverse transcription quantitative PCR (RT-qPCR) results, showing its high expression in developing seeds and flowers (Supplemental Figure S1, C and D).
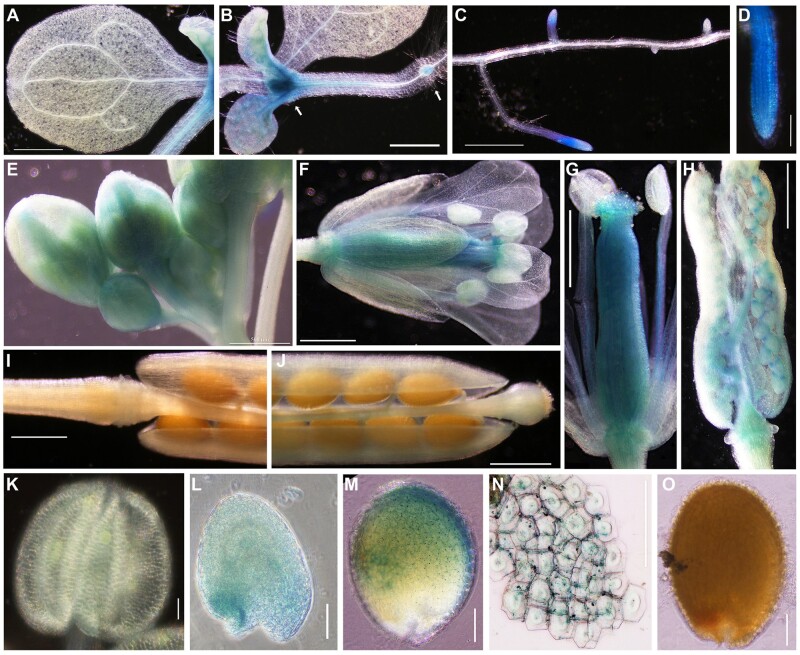
To further define the LecRK-VIII.2 functions during plant development at the protein level, we constructed pLecRK-VIII.2::LecRK-VIII.2-GUS transgenic plants. In 7-d-old seedlings, no β-glucuronidase (GUS) signal was detected in cotyledons, while LecRK-VIII.2-GUS was expressed in the shoot apical meristem, adventitious root primordia, lateral root (elongation zone), and primary root tip (Figure 1, A–D). In flowers, GUS activity was only observed in the pistil, ovule, filament, and receptacle but not in the sepal, carpel, anther, and pollen (Figure 1, E–G and K). After fertilization, LecRK-VIII.2-GUS was expressed in the suspensor, embryo, developing seed, and seed coat, but not in mature seed (Figure 1, H and L–O). In summary, the transcription and GUS staining data suggest that LecRK-VIII.2 could play an important role in seed growth and development.
Figure 1.
Tissue-specific expression of pLecRK-VIII.2::LecRK-VIII.2-GUS. A–O, The expression of pLecRK-VIII.2::LecRK-VIII.2-GUS in cotyledon (7-d-old seedling, A), shoot apical meristem (left arrow, B) and adventitious root primordia (right arrow, B), lateral root (7-d-old seedling, C), primary root tip (7-d-old seedling, D), inflorescence (E), flower (F), pistil and stamen (G), silique (2 DAF, H), mature silique (I, J), pollen (K), embryo (L), developing seed (M), seed coat (N), mature seed (O). Bar = 500 μm (A–C, E, F, G–J), 100 μm (D, K–O).
LecRK-VIII.2 coordinates seed size, silique number, and seed number to determine seed yield
To further investigate the function of LecRK-VIII.2, we identified two T-DNA insertion mutants, lecrk-VIII.2-1 (Salk_051706) and lecrk-VIII.2-2 (Salk_053278), and constructed two LecRK-VIII.2 overexpression lines (OE3 and OE7). After genotyping of the T-DNA events (Figure 2, B, primers listed in Supplemental Table S1), a RT-qPCR assay was performed to check the LecRK-VIII.2 expression level. The transcripts of LecRK-VIII.2 were hardly detected in the T-DNA mutants, while the OE7 plant exhibited a nearly 10-fold transcriptional increase (Figure 2, C and Supplemental Figure S2, B).
Figure 2.
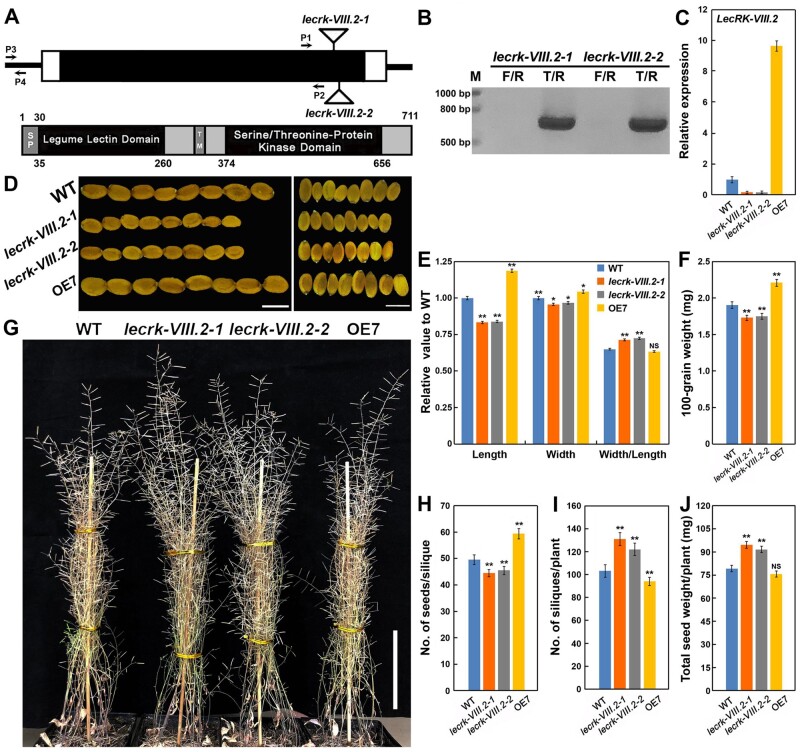
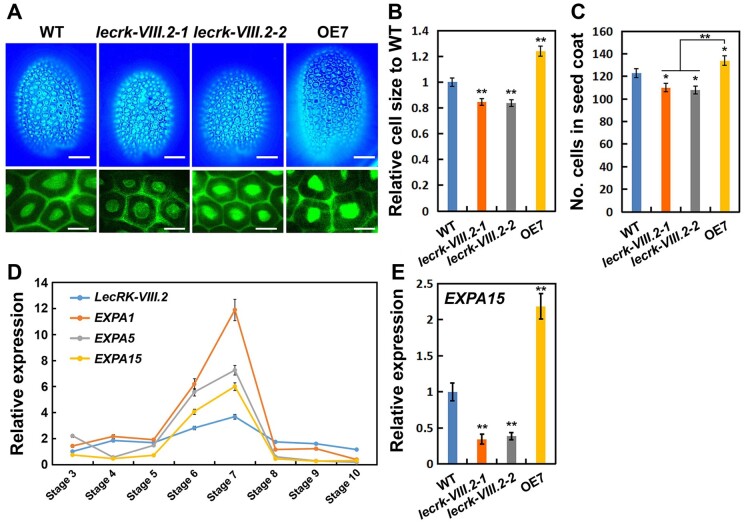
LecRK-VIII.2 coordinates silique number, seed size, and seed number to determine seed yield. A, LecRK-VIII.2 gene structure. Black box represents the exon and white boxes represent the 5′- and 3′-untranslated regions, and lines indicate intergenic regions. The T-DNA insertions sites of the lecrk-VIII.2-1 and lecrk-VIII.2-2 mutants are shown. The arrows of P1/P2 and P3/P4 show the positions of primers. The LecRK-VIII.2 protein contains a signal peptide (SP), legume lectin domain, transmembrane (TM), and serine/threonine protein kinase domain. B, Genotyping T-DNA insertion events in the lecrk-VIII.2-1 and lecrk-VIII.2-2 mutants by PCR. The band from the primers T/R means the T-DNA insertion in LecRK-VIII.2. The primers (lecrk-VIII.2-F/R/T) are listed in Supplemental Table S1. C, Relative expression level of LecRK-VIII.2 in 7-d-old seedlings of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7. The values are based on three biological replicates. The primers (LecRK-VIII.2-qF/R-(P1/2)) are shown in Supplemental Table S1. D, Mature seeds of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7, bar = 500 μm. E, Seed length, width and length/width ratio of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 are shown after being normalized to the mean value of WT. Values are means ± SE (n > 500 seeds) relative to the WT value that is set at 1. F, 100-grain weight of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7. G, 90-d-old plants of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7, bar = 10 cm. H–J, Number of seeds per silique, number of siliques per plant, and total seed weight per plant of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7. Values are means ± SE (E, n > 500; F, n > 500 * 5 seeds; H, n > 30 siliques; I, n = 12 plants; J, n = 12 plants). *P < 0.05, **P < 0.01 compared with the WT, NS means no significance (Student’s t test).
The reproductive output of plants is directly determined by the silique (or spikelet) number, seed size, and seed number (Li et al., 2019). Given the expression patterns of LecRK-VIII.2, we examined the function of LecRK-VIII.2 in controlling seed growth and development. Compared with the dry seeds from wild-type (WT, Columbia-0) plants, the lecrk-VIII.2 mutant seeds exhibited significantly reduced length and width, resulting in seeds with a rounded appearance, while the seeds from the OE7 line were obviously bigger and heavier than the WT seeds (Figure 2, D–F and Supplemental Figure S2, A–D). This demonstrates that LecRK-VIII.2 positively controls seed size and weight. Moreover, the lecrk-VIII.2 mutants produced less seeds per silique, while the OE7 line formed more than WT plants (Figure 2, G and H), indicating that LecRK-VIII.2 increases the number of seeds in each silique. Interestingly, when the number of siliques and total seed yield per plant were measured, we observed that the lecrk-VIII.2 plants obviously formed more siliques and the OE7 lines but produced less than WT plants. The seed output of the lecrk-VIII.2 mutants was significantly higher than that of the WT, whereas OE7 plants exhibited a similar reproductive output to the WT (Figure 2, I and J). In summary, lecrk-VIII.2 mutants develop small seeds, but increase seed yield by forming more siliques and seeds. Plants overexpressing LecRK-VIII.2 produce big seeds, but stabilize their reproductive output by reducing the amount of siliques and seeds. This reveals that LecRK-VIII.2 acts as a key regulator to determine seed yield by coordinating seed size, silique number, and seed number.
LecRK-VIII.2 coordinates the source–sink relationship among shoots, roots, and seeds
Generally, source is defined as the capacity of net carbohydrate export in mature leaves, while sink means seeds, tubers (stems), and fruits that accumulate and consume carbohydrates. Seed yield is the cumulative result of both the strength of source and sink for photoassimilates and nutrients during seed development (reviewed in Smith et al., 2018). Given that the reproductive output was controlled by LecRK-VIII.2, we further asked whether it affected the energy flow between the source and the sink. Thus, we measured the organ size and biomass of the rosette, root, and stem in the different genotypes.
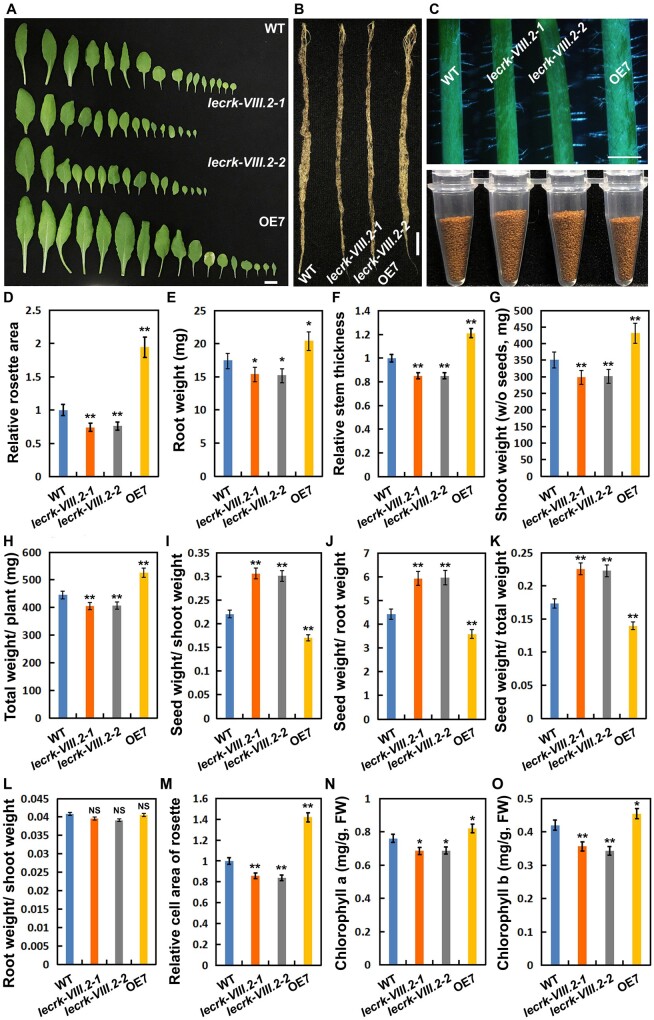
Interestingly, the lecrk-VIII.2 mutants exhibited reduced size of rosettes, roots, and stems, whereas the plants overexpressing LecRK-VIII.2 developed bigger rosettes, more robust roots, and thicker stems (Figure 3, A–F), indicating that LecRK-VIII.2 promoted the growth of these organs. In this study, we focused on its roles in seed output (Figures 2, J and 3, C). Thus, in order to dissect the source–sink relationship, we conducted multiple comparisons of the weight ratio among shoots, roots, and seeds. The lecrk-VIII.2 mutants showed a significantly higher ratio of seed to shoot, seed to root, and seed to total weight, suggesting that loss of function of LecRK-VIII.2 induced energy from photosynthesis to be stored in seeds. On the contrary, the OE lines displayed a prominently decreased ratio of the three pairwise comparisons compared with WT plants, demonstrating that overexpressing LecRK-VIII.2 resulted in more energy to be allocated for the growth of rosettes and roots, but less for seeds (Figure 3, G–K). Moreover, no differences from the ratio of root to shoot were observed between the mutants and the WT (Figure 3, L). In general, these results demonstrate that LecRK-VIII.2 promotes biomass accumulation and regulates the source–sink relationship among shoots, roots, and seeds.
Figure 3.
LecRK-VIII.2 promotes the growth of rosettes, roots, and stems by coordinating the source–sink relationship. A, Rosettes from 35-d-old plants of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7, bar = 1 cm. B, Roots from 35-d-old plants of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7, bar = 1 cm. C, Primary stems from 35-d-old plants of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7, bar = 1 cm (upper panel). Total seeds from each plant of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 (bottom panel). D, Relative rosettes size (35-d-old plants, n = 8) of lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 to WT. Values are means ± SE relative to the WT value that is set at 1. E, Root dry weight per plant of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 (82-d-old plants, n = 12). F, Relative stem thickness of lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 to WT (n = 12). G, Shoot dry weight (without seeds) per plant of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 (82-d-old plants, n = 12). H, Total dry weight per plant of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 (82-d-old plants, n = 12). I and L, Seed weight to shoot weight ratio (I), seed weight to root weight ratio (J), seed weight to total weight ratio (K), root weight to shoot weight ratio (L). M, Relative size of epidermal cells in rosettes from WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 (35-d-old plants, n > 100). N and O, Chlorophyll content in rosettes from 35-d-old plants of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7. FW means fresh weight. Values are means ± SE, n = three biological repeats. The dry weight of shoots and roots was scored after incubation at 80°C for 12 h, while the seed weight was scored after incubation at 37°C for 3 d. Values are means ± SE (E–H, n = 12). *P < 0.05, **P < 0.01 compared with WT (Student’s t test).
Additionally, given the altered rosette area and total biomass of the mutants and OE lines (Figure 3, D and H), we further measured cell size and chlorophyll content of rosettes. The lecrk-VIII.2 mutants developed smaller epidermal cells, and showed lower levels of chlorophyll a and chlorophyll b, while the OE lines exhibited opposite phenotypes (Figure 3, M–O). This suggests that LecRK-VIII.2 promotes cell expansion to increase leaf area, and positively regulates the chlorophyll level in rosettes.
LecRK-VIII.2 positively regulates the size of the integument and embryo
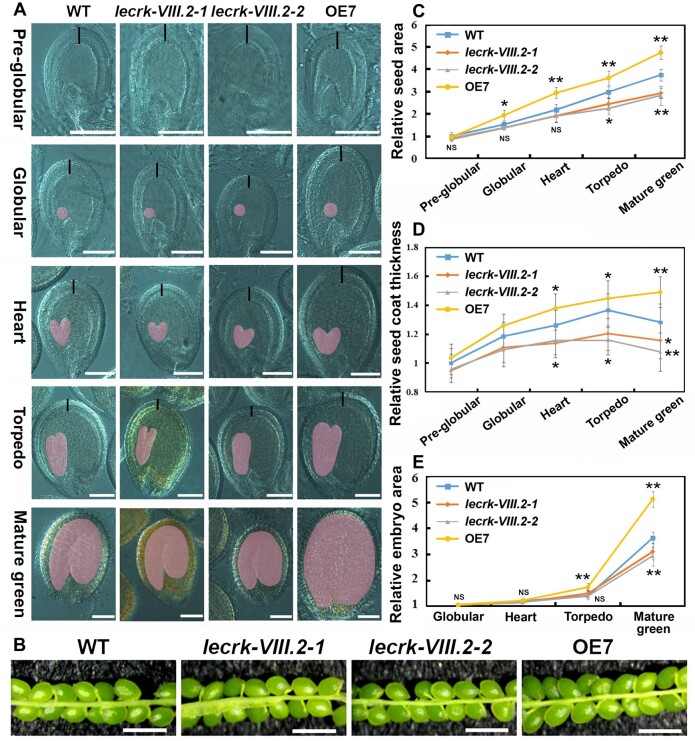
In flowering plants, a seed is comprised of an embryo, endosperm, and seed coat, and seed size is determined by the coordinated growth of maternal sporophytic and/or zygotic tissues (Lopes and Larkins, 1993; Reiser and Fischer, 1993). The integument develops into the seed coat after fertilization, which can restrict seed growth physically to determine the final seed size in Arabidopsis (Schruff et al., 2006; Adamski et al., 2009; Fang et al., 2012). Thus, we observed the development of the seed coat and embryo after clearing treatment. The OE lines displayed faster growth of seeds after the globular stage, while the lecrk-VIII.2 mutants showed a significantly decreased size of seeds compared with WT plants starting from the torpedo stage (Figure 4, A and C). The lecrk-VIII.2 mutants exhibited reduced seed coat thickness at the beginning of the heart stage, while the seed coat of OE7 plants was thicker compared with that of the WT (Figure 4, D). Starting from the torpedo embryo stage, the difference of embryos size between WT and OE7 plants became significant, while the difference between the lecrk-VIII.2 and WT embryos did not emerge until the mature green stage (Figure 4, E). In summary, these results demonstrate that LecRK-VIII.2 promotes the growth of the seed coat and embryo.
Figure 4.
LecRK-VIII.2 positively regulates the size of the integument and embryo. A and B, Developing seeds of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 lines were collected at various stages (Pre-globular, Globular, Heart, Terpedo and Mature green), and observed by a differential interference contrast microscope after fixation and clearing treatment as described in the “Materials and methods” section, bar = 50 μm A and 1 mm B. C–E, Relative size of lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 lines to WT. Values are means ± SE (n > 100 developing seeds) relative to the WT value that is set at 1. *P < 0.05, **P < 0.01 compared with the WT, NS means no significance (Student’s t test).
LecRK-VIII.2 promotes cell expansion and proliferation, and maternally controls seed size
Organ size is determined by cell number and cell size. The GUS profile shows the specific expression of LecRK-VIII.2 in the epidermal cells of the seed coat (Figure 1, N). Therefore, we investigated the cellular basis on altered seed size of the mutants using Fluorescent Brightener 28 staining. The lecrk-VIII.2 mutants produced significantly smaller cells than WT plants, while OE7 lines formed larger cells, indicating that LecRK-VIII.2 positively controls cell expansion in the maternal integument. Further detection of the number of epidermal cells showed that the lecrk-VIII.2 mutant developed slightly less cells, but OE7 lines produced more cells than WT plants (Figure 5, A–C and Supplemental Figure S2, E). These results reveal that LecRK-VIII.2 promotes cell expansion and slightly affects cell proliferation in maternal tissue to determine seed size.
Figure 5.
LecRK-VIII.2 promotes cell expansion and proliferation in the seed coat. A, The size and number of cells in the seed coat were investigated by Fluorescent Brightener 28 staining, bar = 100 μm (upper panel) or 20 μm (lower panel). B, Relative cell size of lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 to WT. Values are means ± SE (n > 120 cells) relative to the WT value that is set at 1. C, The number of cells in the seed coat of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7. Values are means ± SE (n > 120 seeds). D, The expression level of LecRK-VIII.2, EXPA1, EXPA5, and EXPA15 in different stages of developing seeds. Values are means ± SE relative to the LecRK-VIII.2 value that is set at 1. E, The expression level of EXPA15 in the developing seeds of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7. Values are means ± SE (n = three biological replicates). *P < 0.05, **P < 0.01 compared with the WT (Student’s t test).
Furthermore, we attempted to figure out the molecular mechanism of LecRK-VIII.2 in promoting cell expansion. In Arabidopsis, expansins (AtEXPAs) are identified as cell-wall-loosening proteins that mediate pH-dependent extension of the cell wall and induce cell expansion (Lee et al., 2001; Cosgrove, 2015; Nardi et al., 2015). Considering the potential association based on co-localization between EXPAs and LecRK-VIII.2, we compared the expression profiles of all 36 AtEXPAs during seed development. Interestingly, EXPA1, EXPA5, and EXPA15 exhibited similar patterns to LecRK-VIII.2, showing a common transcriptional peak in stage 7 of developing seeds (Figure 5, D). Next, we performed a RT-qPCR assay to investigate their relationship. The significantly reduced level of EXPA15 was detected in the lecrk-VIII.2 mutants, while overexpressing LecRK-VIII.2 obviously induced EXPA15 transcription (Figure 5, E). However, evidence of EXPA1 and EXPA5 regulated by LecRK-VIII.2 was not observed in developing seeds. In general, this suggests that LecRK-VIII.2 probably promotes cell expansion of the seed coat by regulating the expression of EXPA15.
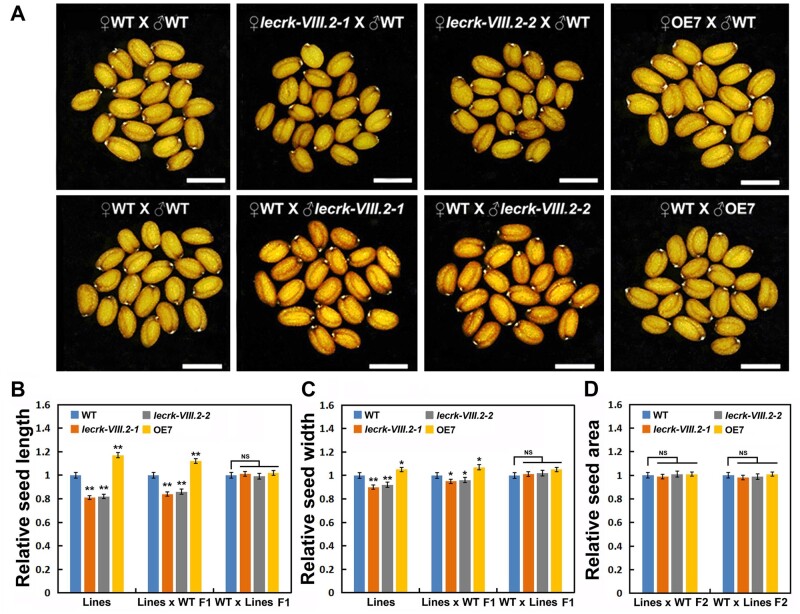
Additionally, we conducted reciprocal cross experiments to test whether LecRK-VIII.2 acted maternally. We pollinated lecrk-VIII.2 (or OE7) plants with WT pollen. The length and width of F1 seeds from such crosses were comparable to that of self-pollinated lecrk-VIII.2 mutants (or OE7). In contrast, the size of seeds from WT plants pollinated by lecrk-VIII.2 mutant (or OE7) pollen was undistinguishable from that of the self-pollinated WT plants (Figure 6, A–C). This indicates that LecRK-VIII.2 functions maternally to determine seed size. We further investigated the size of lecrk-VIII.2-2/Col-0 F2 and Col-0/lecrk-VIII.2-2 F2 seeds. Seeds from lecrk-VIII.2-2/Col-0 F1 and Col-0/lecrk-VIII.2-2 F1 plants contain WT, lecrk-VIII.2, and lecrk-VIII.2/+ embryos within lecrk-VIII.2/+ seed coats. The size of lecrk-VIII.2/Col-0 F2 and Col-0/lecrk-VIII.2 F2 seeds was comparable with that of WT seeds (Figure 6, D). These results indicate that the embryo and endosperm genotypes for LecRK-VIII.2 have no effect on seed size, and LecRK-VIII.2 is essential in sporophytic tissue of the mother plant to determine seed size.
Figure 6.
LecRK-VIII.2 maternally regulates seed size. A, Mature F1 seeds from the reciprocal cross experiments among WT, lecrk-VIII.2-1, lecrk-VIII.2-2, and OE7 lines, bar = 500 μm. B and C, Relative seed length and width of the F1 seeds of reciprocal cross experiments. D, Relative seed area of the F2 seeds of reciprocal cross experiments. Values are means ± SE (n > 500 seeds) relative to the WT value that is set at 1. **P < 0.01 compared with the WT (Student’s t test).
LecRK-VIII.2 regulates yield-related traits in a MPK6-dependent manner
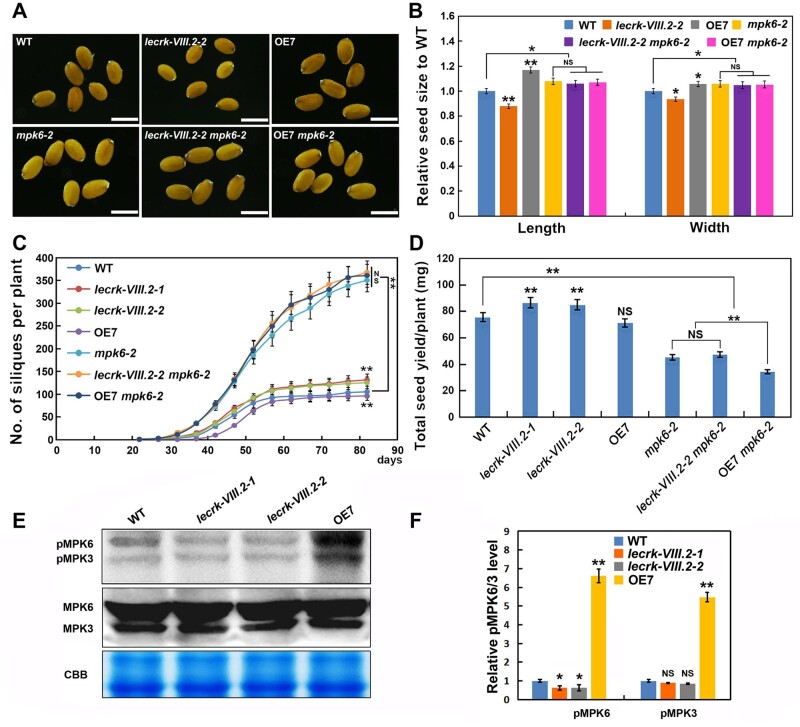
MAPKs cascades play essential roles in plant development and stress responses (Xu and Zhang, 2015). Recent studies indicate that MPK3 and MPK6, as conserved downstream factors of RLKs, regulate inflorescence architecture, ovule integument development, and seed development (Bush and Krysan, 2007; Wang et al., 2008; López-Bucio et al., 2013; Zhang et al., 2017). In light of this, we constructed lecrk-VIII.2 mpk6-2, OE7 mpk6-2, lecrk-VIII.2 mpk3-1, and OE7 mpk3-1 lines by crossing (Supplemental Figure S3). Interestingly, seeds from the mpk6-2 mutant display a range of phenotypes, ∼67% WT-like bigger seeds (mpk6wb seeds), ∼25% raisin-like seeds, and ∼6% seeds with burst-out embryo (burst seeds; Bush and Krysan, 2007; López-Bucio et al., 2013; Zhang et al., 2017; Creff et al., 2019), indicating the complex function of MPK6 in controlling seed size and development. In order to remove the interference of the embryo/seed development defect on seed size, we only selected the WT-like seeds to explore the genetic relationship between LecRK-VIII.2 and MPK6. Surprisingly, the length and width of lecrk-VIII.2-2 mpk6-2 and OE7 mpk6-2 seeds were undistinguishable from those of mpk6wb seeds (Figure 7, A and B), suggesting that mpk6 was epistatic to lecrk-VIII.2. Additionally, the genetic results also reveal that LecRK-VIII.2 acts upstream of MPK6 to control silique number and seed yield (Figure 7, C and D). In general, LecRK-VIII.2 functions in a common pathway with MPK6 to regulate seed size, seed number, and silique number. In addition, the epistatic role of MPK6 to LecRK-VIII.2 in controlling flower size by promoting cell expansion was observed (Supplemental Figure S4, A and B), also confirming that LecRK-VIII.2 acts upstream of MPK6 to determine organ size.
Figure 7.
LecRK-VIII.2 acts upstream of MPK6 to control yield-related traits. A, WT-like mature seeds of WT, lecrk-VIII.2-2, OE7, mpk6-2, lecrk-VIII.2-2 mpk6-2, and OE7 mpk6-2, bar = 500 μm. B, Relative seed length and width of the mutants to WT. Values are means ± SE (n > 500 seeds) relative to the WT value that is set at 1. C, The number of siliques per plant of WT, lecrk-VIII.2-2, OE7, mpk6-2, lecrk-VIII.2-2 mpk6-2, and OE7 mpk6-2. The number of siliques was counted every 5 d, starting from the bolting of the mpk6-2 mutant (the earliest flowering line among analyzed plants) to the 82nd d. Values are means ± SE (n = 12 plants). D, Total seed yield per plant of WT, lecrk-VIII.2-2, OE7, mpk6-2, lecrk-VIII.2-2 mpk6-2, and OE7 mpk6-2. Values are means ± SE (n = 12 plants). E and F, Phosphorylation level of MPK3 and MPK6 in developing seeds (∼10 DAF) of WT, lecrk-VIII.2-1, lecrk-VIII.2-2, OE7, mpk3-1, and mpk6-2. Values are means ± SE (n = three biological replicates) relative to the WT value that is set at 1. *P < 0.05, **P < 0.01 compared with the WT, NS means no significance (Student’s t test).
To further investigate the molecular basis of interplay between LecRK-VIII.2 and MPK6, we detected the phosphorylation status of MPK6 in developing seeds. We found reduced phosphorylated MPK6 (pMPK6) levels in the lecrk-VIII.2 mutant, while overexpressing LecRK-VIII.2 increased pMPK6 levels (Figure 7, E and F). Considering the genetic relationship between them, this demonstrates that LecRK-VIII.2 regulates the three yield-related traits by adjusting the phosphorylation level of MPK6. Interestingly, the OE lines exhibited a significantly increased level of pMPK3, but no alteration of pMPK3 was observed in lecrk-VIII.2 mutants. Previous studies indicate that MPK6 and MPK3 usually work redundantly to regulate plant growth and development (reviewed in Xu and Zhang, 2015). Thus, we compared the size of seeds and flowers from mpk3-1, OE7 mpk3-1, and mpk3-1 plants. The results showed there was no significant change in the size of seed and flower of the mpk3-1 mutant compared with that of the WT, indicating that MPK3 may not play an important role in seed development. However, the seed and flower size of the OE7 mpk3-1 line was significantly bigger than those of the mpk3-1 mutant, suggesting that MPK3 may not be the downstream factor of LecRK-VIII.2 to regulate seed and flower size (Supplemental Figure S4, C and D). As a result, LecRK-VIII.2 controls the size of seeds and flowers upstream of MPK6, but independent of MPK3.
LecRK-VIII.2 regulates the development of the pistil and ovule
Recent studies revealed that the development of the flower organ is closely linked with yield-related traits. For example, inflorescence architecture is implicated in the number of spikelets or siliques (reviewed in Li et al., 2019). Therefore, we dissected the flowers and observed the development of the pistil, stamen, and pollen. The lecrk-VIII.2 mutants developed shorter pistils with less ovules, whereas the OE plants formed longer pistils containing more ovules than the WT. The knockout mutants showed reduced length of siliques with less seeds, while the OE lines exhibited opposite phenotypes (Supplemental Figure S5). Nevertheless, the knockout and OE plants all displayed no developmental defects of the pistil, ovule, stamen, and pollen, and were undistinguishable in pollen size and pollen activity (Supplemental Figures S5, S6). In summary, these results demonstrate that LecRK-VIII.2 positively regulates pistil length and ovule number per pistil, but does not affect anther and pollen development. This is consistent with the GUS pattern that LecRK-VIII.2 is expressed in the pistil and ovule, but not in the anther or pollen (Figure 1, G–K). Importantly, the difference of pistil length and ovule number per pistil could have a direct effect on silique length and seed number per silique.
LecRK-VIII.2 regulates the accumulation of storage protein and carbohydrate in seeds
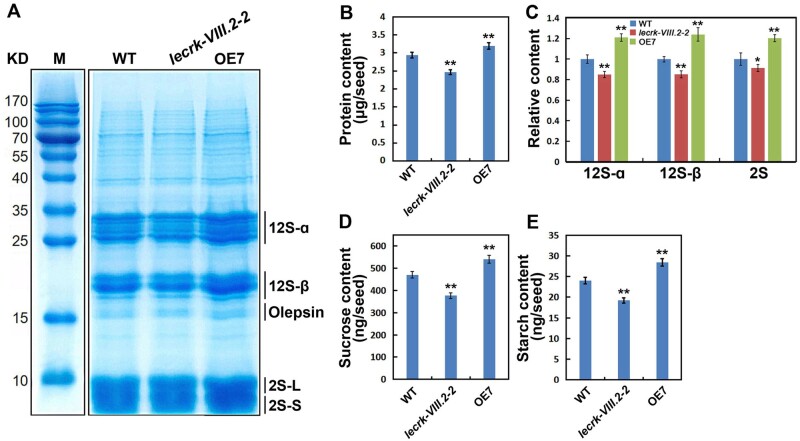
Larger seeds usually show increased germination activity and fitness, mainly as a result of higher storage protein and carbohydrate in seeds (Michelle and Richards 2000; Li et al., 2019). Therefore, we determined the content of storage protein and carbohydrate in dry seeds. LecRK-VIII.2 loss-of-function mutants exhibit reduced content of total protein per seed, whereas OE lines contain much more than WT plants (Figure 8, B), indicating that LecRK-VIII.2 promotes the accumulation of storage protein. Next, in order to further check the profiles of seed storage protein, total protein extracted from dry seeds was separated on 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels. The gels showed that lecrk-VIII.2 mutant developed seeds with significantly less 12S-ɑ and 12S-β (12S globulin subunits) than WT plants, while they contained a comparable content of 2S albumin subunits (2S-L and 2S-S). OE7 seeds accumulated a remarkably higher level of 12S and 2S subunits than WT (Figure 8, A and C). Additionally, the loss of function of LecRK-VIII.2 decreased the content of sucrose and starch per seed, whereas OE lines accumulated more sucrose and starch in seeds (Figure 8, E and F). This indicates that LecRK-VIII.2 positively regulates the carbohydrate content in seeds. In general, LecRK-VIII.2 promotes the accumulation of protein and carbohydrate in seeds, and regulates the relative proportion and content of the subunits of storage protein.
Figure 8.
LecRK-VIII.2 promotes the accumulation of storage protein and carbohydrate in seeds. A, Profiles of storage protein in dry seeds from WT, lecrk-VIII.2-2, and OE7 plants. Each lane contains protein extracted from 10 seeds. Total proteins extracted from dry seeds were separated on 15% SDS-PAGE and stained with Coomassie Blue. 12S-ɑ and 12S-β, 12S globulin subunits; 2S-L and 2S-S, 2S albumin subunits. B, Protein content per seed of WT, lecrk-VIII.2-2, and OE7 plants. C, Relative content of 12S-ɑ, 12S-β, and 2S in the seeds from WT, lecrk-VIII.2-2, and OE7 plants. D, Sucrose content per seed of WT, lecrk-VIII.2-2, and OE7 plants. E, Starch content per seed of WT, lecrk-VIII.2-2, and OE7 plants. Values are means ± SE of three (B–E) biological replicates. *P < 0.05, **P < 0.01 compared with the WT, NS means no significance (Student’s t test).
Discussion
The energy allocation between vegetative and reproductive organs is regulated by developmental signals and environmental cues, which subsequently affect the seed size/number trade-off to determine seed yield. This phenomenon has been observed and studied for decades in a variety of species (reviewed in Smith and Fretwell, 1974; Lazaro and Larrinaga, 2018; Li et al., 2019), but the regulatory mechanism underlying how plants perceive and transduce extracellular or environmental signals to organize this process remains largely obscure. The major reason is that the key factors, especially RLKs upstream of the pathways, are still undiscovered, which could connect the signals and integrate some identified signaling into a network.
In this study, we report that LecRK-VIII.2 is a specific RLK to determine seed yield by coordinating silique number, seed size, and seed number. The lecrk-VIII.2 mutants form smaller seeds, but produce an increased number of siliques and seeds, leading to significantly increased yield. In contrast, the OE lines develop bigger seeds, but less siliques and seeds, which results in a similar yield to that of the WT. Interestingly, the OE lines exhibit an elevated rosette area, more robust roots, and thicker stem compared with WT plants (Figures 2, 3, A–F, and 9). This indicates that LecRK-VIII.2 has potential as a promising target for crop improvement, which aims to cultivate crops with stable grain yield, enhanced use efficiency of water and fertilizer, and improved lodging resistance.
Figure 9.
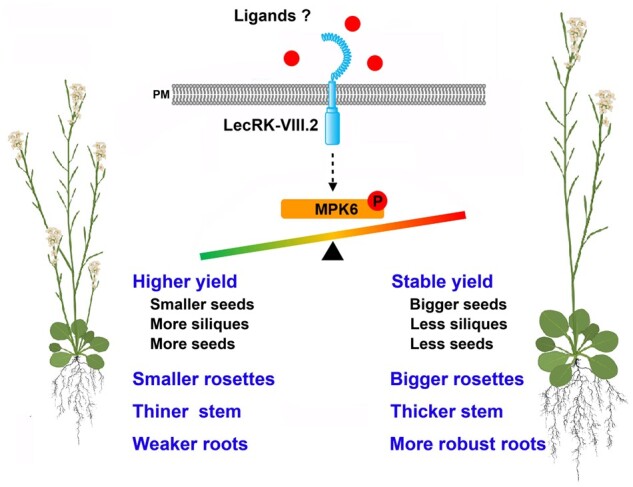
A genetic and molecular framework for LecRK-VIII.2/MPK6-mediated regulation of yield-related traits and organ size. In the presence of unknown ligands as developmental signals, LecRK-VIII.2 phosphorylates MPK6, and coordinates silique number, seed size, and seed number to determine reproductive output. lecrk-VIII.2 mutants with reduced pMPK6 level exhibit increased seed yield, but decreased size of rosettes, stems, and roots. Plants overexpressing LecRK-VII.2 with an elevated pMPK6 level can maintain stable yield, and develop bigger rosettes, thicker stems, and more robust roots, compared with WT plants. Like some RLKs, LecRK-VIII.2 is able to form a homodimer, suggesting that the signaling downstream of LecRK-VIII.2 could start from its autophosphorylation. LecRK-VIII.2 and its homolog LecRK-VIII.1 can compose a heterodimer, suggesting that they could play similar roles during plant development.
Additionally, in OE plants, the elevated total biomass could result from increased leaf area and chlorophyll level, which subsequently lead to increased photosynthesis. The higher amount of photoassimilates is allocated for the growth of rosettes and roots, rather than accumulated in seeds (Figure 3, G–O). It should be noted that the lecrk-VIII.2 mutants show a reduced size of rosette, root, and stem, but increased yield. This indicates that the LecRK-VIII.2 loss-of-function plants store more energy in seeds at the cost of the growth of rosettes and roots. Given that LecRK-VIII.2 function as a potential receptor, we hypothesize the existence of a ligand(s) to control the energy allocation between vegetative and reproductive growth. The energy fluctuation inputted to reproductive growth could lead to the alteration of silique number, seed size, and seed number, which subsequently determine seed output.
A previous study revealed that the legume-lectin protein-like extracellular domain of L-type LecRKs is structurally similar to that of carbohydrate-binding proteins and exhibited glucose/mannose specificity (Hervé et al., 1999). Interestingly, no GUS activity was observed in 7-d-old cotyledons (Figure 1, A), whereas strong signal was detected after mannose and glucose treatment. Additionally, previous studies revealed that in developing seeds, a low sucrose to hexose (fructose and glucose) ratio is closely associated with cell division at the early stage, while a high ratio is required for cell elongation later (Weber et al., 1996, 1997; Ohto et al., 2005). Generally, considering the roles of LecRK-VIII.2 both in cell expansion and proliferation, we therefore infer that the potential ligands of LecRK-VIII.2 are some kind of hexose or sugar derivatives. Furthermore, recent studies indicate that in the LecRK family, LecRK-I.9 functions as a receptor for eATP, and LecRK-I.8 acts as an eNAD+ sensor (Choi et al., 2014; Wang et al., 2017, 2018; Tripathi et al., 2018). Thus, nucleotides, as signal substances and/or energy molecules, also should be considered as the potential ligands of LecRK-VIII.2. Metabolomics study on saccharides and nucleotides during seed development or response to sugar stimulus could be required for fully understanding the molecular mechanism of signal transduction and energy allocation in a LecRK-VIII.2-dependent manner. Moreover, it would be interesting to study the action of LecRK-VIII.2 in photosynthesis.
The investigation of seed development shows that LecRK-VIII.2 promotes the growth of the integument and embryo (Figure 4). Given that the difference of embryo size emerges later than that in the integument, we speculate that the reduced thickness of the integument in lecrk-VIII.2 mutants sets the physical limitation, and this inhibition emerges after fertilization and constantly accumulates, which subsequently leads to the observable suppression of embryo expansion starting from the torpedo stage. Interestingly, the OE lines display faster growth of seeds beginning at the globular stage, while LecRK-VIII.2 loss-of-function mutants show decreased seed size compared with WT plants beginning at the torpedo stage (Figure 4, C). Similarly, the LecRK-VIII.2-GUS expression profiles showed high activity in the pistil (valve), but 2 d after fertilization, the signal emerged from the embryo, along with decreased expression in the valve (Figure 1). These asynchronous phenotypes suggest the roles of LecRK-VIII.2 to regulate the timing of key events during pistil and seed development. Further studies are needed to dissect the regulatory mechanism of LecRK-VIII.2 before and after fertilization.
The genetic and biochemical results demonstrate that LecRK-VIII.2 works upstream of MPK6 to determine yield by coordinating silique number, seed size, and seed number (Figures 7, 9). Previous studies reveal that the YDA(MAPKKK4)-MKK4/MKK5-MPK3/MPK6 cascade plays important roles in embryo development, localized cell division, root development, and seed size (Bergmann, 2004; Lukowitz et al., 2004; Wang et al., 2007; Meng et al., 2013; Musielak and Bayer, 2014; Xu and Zhang, 2015). Given that the mutants of YDA, MKK4/MKK5, MPK3/MPK6 develop seeds with a burst embryo, it is possible that LecRK-VIII.2 is involved in the YDA-MKK4/5-MPK3/6 signaling pathway. Interestingly, LecRK-VIII.2, MKK4/5, and MPK6 all exhibit maternal control of seed development, further suggesting the possibility. However, we were not able to identify YDA as an interaction factor with LecRK-VIII.2-3xFLAG fusion protein by a co-immunoprecipitation (Co-IP)/mass spectrometry (MS) assay. LecRK-VIII.2 could interact with other MAPKKKs to transduce the signals via a phosphorylation cascade. A recent study demonstrated that the G-protein beta subunit, AGB1 acts as a linker between RLKs and the YDA-MKK4/5-MPK3/6 cascade to control zygote and fruit development (Yuan et al., 2017), and AGB1 plays an essential role in regulating seed size (Li et al., 2012). Thus, it would be interesting to investigate whether LecRK-VIII.2 is linked to the MAPK pathway by recruiting the G-protein complex as a scaffold. Interestingly, we identified some BR-responsive leucine-rich repeat receptor like kinases (LRR-RLKs) as interaction proteins with LecRK-VIII.2 by Co-IP/MS and yeast two-hybrid (Y2H) assays. Given that BRASSINOSTEROIDS INSENSITIVE 1 (BRI1) and BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) have been implicated to interact with G proteins and regulate sugar-responsive growth and development (Peng et al., 2018), further studies are needed to identify the association among the LecRK-VIII.2-MAPKs module, G proteins, and BR signaling.
Recent studies revealed that rice (Oryza sativa) ERECTA1 (OsER1) and the dual-specific MAPK PHOSPHATASE1 (OsMKP1) regulate the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice (Guo et al., 2018, 2020; Xu et al., 2018). This greatly extends our understanding of the molecular mechanism of the trade-off. Considering the conserved role of MPK6 in Arabidopsis and rice (Bush and Krysan, 2007; López-Bucio et al., 2013; Yi et al., 2016; Zhang et al., 2017; Guo et al., 2018, 2020; Xu et al., 2018), and the potential of AtLecRK-VIII.2 for crop improvement, we therefore searched against the dataset of Viridiplantae in EggNOG v4.5.1 (Jaime et al., 2015). We found that Os05g0125200 and Os06g0285400 shared high sequence similarity and conserved domains with AtLecRK-VIII.2, and more homologs of AtLecRK-VIII.2 in crops are shown in Supplemental Figures S7, S8. Some of them in rice, soybean (Glycine max), maize (Zea mays), and potato (Solanum tuberosum) exhibit common high expression during grain (or fruit) development (Supplemental Figure S9), suggesting the conserved roles of LecRK-VIII.2 in the crops. It would be interesting to dissect their functions in grain (or fruit) yield and plant development, making it a candidate for crop improvement or design.
In summary, we discovered that LecRK-VIII.2 is a specific RLK that regulates seed yield and the source–sink relationship. Further studies on uncovering the ligands and new component(s) in the LecRK-VIII.2-MPK6 signaling pathway would further contribute to clarifying their regulatory mechanism of energy allocation and promoting crop improvement.
Materials and methods
Plant growth conditions and plant materials
Arabidopsis thaliana ecotype Col-0 was used as the WT. Mutants lecrk-VIII.2-1 (Salk_051706) and lecrk-VIII.2-2 (Salk_053278) were obtained from the Arabidopsis Biological Resource Center. The mpk3-1 (Salk_151594) and mpk6-2 (Salk_073907) mutants were obtained from the Nottingham Arabidopsis Stock Centre (NASC). The double mutants lecrk-VIII.2-2 mpk6-2, OE7 mpk6-2, lecrk-VIII.2-2 mpk3-1, and OE7 mpk3-1 were identified from the F2 generation after crossing. Seeds were plated on half-strength Murashige and Skoog (1/2 MS) medium supplemented with 1% (w/v) sucrose and kept at 4°C for 3 d before transferring to a 22°C chamber under long day conditions (16-h light/8-h dark) for 7 d. Then, seedlings were transplanted into soil and cultured in a greenhouse (22°C, 16-h/8-h light cycle). The LecRK-VIII.2 coding sequence (CDS) was amplified by the primers listed in Supplemental Table S1 and was recombined into pLEELA (Invitrogen), driven by the CaMV 35S promoter. Then, the 35S::LecRK-VIII.2 recombinant vector was transformed into Agrobacterium tumefaciens strain GV3101 and the positive colony was cultured for the subsequent floral dipping transformation into WT plants. Basta-resistant T1 transgenic plants were screened and the homozygous OE lines with a single-copy insertion were obtained after self-cross and Basta screening.
GUS histochemical staining
GUS activity was analyzed by staining various tissues of pLecRK-VIII.2-2::LecRK-VIII.2-2-GUS transgenic plants for 24 h at 37°C in staining solution (1.5 mg/mL X-GlcA, 50 mM sodium phosphate buffer, pH 7, 0.1% (v/v) Triton X-100, 0.5 mM potassium ferricyanide, and 0.5 mM potassium ferrocyanide). Samples were photographed using the NIKON ECLIPSE Ti2 microscope after destaining by ethanol.
Phenotype observation and analyses
For observing seeds and flowers, green seeds were separated from siliques (before turning brown), dry seeds were dehydrated in a 37°C incubator for 7 d, and flowers at stage 15 were photographed using a stereo microscope equipped with an Olympus DP12 digital camera. The pistils and stamens from flowers at stage 15 were dissected and observed by the same lens. The Digimizer software (https://www.digimizer.com/index.php) was used for organ size measurement.
Differential interference contrast analyses
Seeds were collected at various development stages and dipped in fixative solution (methanol/acetic acid 3:1, v/v) at 4°C overnight. Fixed seeds were then incubated in Hoyer’s solution for 40 min and observed by differential interference contrast optics of a Zeiss Scope A1 microscope equipped with an Olympus DP12 digital camera. Embryo area, and integument perimeter and thickness were measured using Digimizer software as described above.
To count the ovule number, flowers of stage 12 that had not been fertilized were cleared in a chloralhydrate/glycerol/water solution (8 g:1 mL:3 mL) overnight, and dissected under the Nikon Inverted Microscope Eclipse Ti-S and the ovule number was counted directly.
Pollen size and germination assay
Pollens from stage 15 flowers were harvested and observed by a Nikon inverted microscope eclipse Ti-S. Pollen size was measured using Digimizer software as described above. For pollen germination in vitro, pollens were collected from the stage 15 flowers and placed onto medium containing 0.36 mg/mL CaCl2, 0.01 mg/mL myo-inositol, 1% (w/v) gelatin, 0.08 mg/mL H3BO3, 20% (w/v) sucrose, and 1% (w/v) agar. The plates were cultured at 22°C under moist conditions in the dark. The same Nikon microscope was used to observe pollen grains. The pollen germination rate was calculated by dividing the number of germinated tubes by the number of grains (Zhu et al., 2014).
Cell size observation
For petal cells observation, petals were separated from flowers at stage 14 and fixed in fixative solution, then photographed by using a Zeiss Scope A1 microscope. For seed coat cell measurement, mature seeds were stained using a 1% (w/v) solution of Fluorescent Brightener 28 (Sigma) according to the method (Ursache et al., 2018). Seeds were then photographed under the excitation of UV light using a Zeiss Scope A1 microscope. Cell size and number were measured via Digimizer software.
RNA isolation and RT-qPCR analyses
Total RNA was extracted from the plant tissues (homogenized by multi-sample tissuelyser, Jingxin Technology), using the plant RNA extraction kit (R401-01, Vazyme Biotech Co., Ltd), and the first chain of cDNA was synthesized by cDNA synthesis superMix (AE301-02, Transgen Biotech). Quantitative PCR was performed with a QuantStudio 5 Real-Time PCR System (Applied Biosystems) using SYBR RT-PCR Master Mix (-21, Transgen Biotech). Primers used in the qPCR are shown in Supplemental Table S1 and the mRNA level of genes in each sample (three technical replicates and three biological replicates) was normalized to that of WT (ACTIN2 was used as internal control).
MAPK phosphorylation analyses
Plant tissues were collected and ground with liquid nitrogen by a multi-sample tissuelyser (Jingxin Technology). Then total protein was extracted with homogenization buffer (50 mM HEPES pH 7.5, 250 mM sucrose, 15 mM EDTA, 5% (w/v) glycerol, 0.5% (w/v) polyvinylpyrrolidone, 3 mM DTT, and 1× protease inhibitor cocktail) and quantified with Brandford reagent (E211-01, Vazyme Biotech Co., Ltd). Equal protein samples were separated using 10% (W/V) SDS-PAGE and transferred to polyvinylidene difluoride membranes. After blocking with block buffer (PBS buffer, 3% (w/v) BSA, 0.01% (v/v) Tween 20), transblots were incubated in Rabbit Anti-Phospho-p44/p42 MAPK antibody (1:2000, Cell Signaling Technology, #9101) diluted with PBS buffer at 4°C overnight, and immunoreactive bands were visualized with FluorChem system (Protein Simple) after incubation in PBS buffer with 1:1000 anti-rabbit-IgG-HRP antibody (Cell Signaling Technology, #7074) at room temperature for 1 h. The protein level was represented by immunoblot analysis using anti-MPK3 (M8318) and anti-MPK6 (A7104) antibodies from Sigma. Coomassie brilliant blue (CBB) staining was used to confirm equal protein loading.
In order to quantify the relative level between pMPK6 or pMPK3 and MPK6 or MPK3, Image J was used to measure the mean gray value of the bands. The value of pMPK6/MPK6 or pMPK3/MPK3 in WT plants was set to 1.
Chlorophyll content determination
Fresh leaves (0.1 g) were homogenized by tissuelyser in liquid nitrogen. Total chlorophyll was extracted using 95% (v/v) ethanol by vortex for 10 min in darkness. After centrifuging at 15,000 rpm for 10 min at 4°C, the supernatant was transferred to a new tube and diluted 10-fold with 95% ethanol. The absorbance of the diluted solution was determined at 665 and 649 nm using a Shimadzu 1800 spectrophotometer. The chlorophyll content was calculated according to the following formula: Chl-a = (13.95A665−6.88A649) * V/1000W and Chl-b = (24.96*A649−7.32*A665)*V/1000W.
Quantification of protein and carbohydrate in seeds
Forty mature dry seeds from WT plants or mutants were homogenized with 80 μL of extraction buffer (100 mM Tris–HCl, pH 8, 0.5% [v/v] SDS, 10% [v/v] glycerol, and 20% [v/v] 2-mercaptoethanol) by tissuelyser, followed by boiling for 5 min and centrifugation for 10 min at 15,000 rpm. Five microliters of the supernatant was used for total protein quantification (E211-01, Vazyme Biotech Co., Ltd) according to the manufacturer’s protocol. Twenty microliters of each extract was used for 15% SDS-PAGE (Carine et al., 2015; Di et al., 2018). Samples of 20 dry seeds were used for carbohydrate analyses according to the previous protocol (Baud et al., 2002). Seeds were ground twice in 1000 µL of 80% (v/v) ethanol at 4°C. After centrifugation (15,000 g for 10 min at 4°C), the supernatant containing the soluble carbohydrates was collected, evaporated at room temperature under vacuum, and then dissolved in 200 µL of water before analysis. The pellet left from the extraction was used for starch determination. The sucrose and starch measurements were performed with kits (#BC2465 and #BC0700 from Solarbio Life Sciences).
Analyses of expression profiles and AtLecRK-VIII.2 homologs
The expression profiles of 75 LecRKs during plant development were retrieved and analyzed by Arabidopsis eFP browser of the Bio-Array Resource (BAR) website (http://bar.utoronto.ca/; Winter et al., 2007) and AtGenExpress Visualization Tool (Redman et al., 2004; Schmid et al., 2005; Kilian et al., 2007). To identify homologs of AtLecRK-VIII.2, its amino acid sequence was searched against the dataset of Viridiplantae in EggNOG v4.5.1. The analysis of alignment, phylogenetic tree, and conserved domain was performed by the online tools curated by EggNOG v4.5.1 (Jaime et al., 2015).
Statistical tests
Three biological repeats were used in all RT-qPCR and western blot assays. To score the embryo/seed phenotypes, we randomly picked at least five siliques, phenotyped all embryos/seeds (>100), and calculated the percentages of each phenotype. Triplicates were performed to obtain mean values with standard deviations. P values were calculated by two-sided Student’s t test. Asterisks above the columns were used to indicate differences that are statistically significant (*P < 0.05) and very significant (**P < 0.01), respectively.
Accession numbers
Sequence data used in this article can be obtained from The Arabidopsis Information Resource (TAIR) or GenBank/EMBL datasets by using the following accession numbers: LecRK-VIII.2 (AT5G03140), LecRK-VIII.1 (AT3G53380), MPK6 (AT2G43790), MPK3 (AT3G45640), EXPA1 (AT1G69530), EXPA5 (AT3G29030), EXPA15 (AT2G03090), ACTIN2 (AT3G18780).
Supplemental data
Supplemental Figure S1. Transcription patterns of LecRK-VIII.2 during plant growth and development.
Supplemental Figure S2. Overexpression of LecRK-VIII.2 affects organ size and seed development.
Supplemental Figure S3. LecRK-VIII.2 acts upstream of MPK6 to control the growth of the rosette and stem.
Supplemental Figure S4. LecRK-VIII.2 controls the size of seeds and flowers in an MPK6-dependent manner, but independent of MPK3.
Supplemental Figure S5. LecRK-VIII.2 regulates the development of flowers and siliques.
Supplemental Figure S6. LecRK-VIII.2 exhibits no effect on the development of stamen and pollen.
Supplemental Figure S7. Phylogenetic analysis of AtLecRK-VIII.2 homologs in crops.
Supplemental Figure S8. Alignment of amino acid sequences of AtLecRK-VIII.2 and its homologs in higher plants.
Supplemental Figure S9. The expression profiles of the homologous genes of AtLecRK-VIII.2.
Supplemental Table S1. Primers used in this work.
Supplementary Material
Acknowledgments
The authors thank Yongliang Li for technical assistance, and ABRC and NASC for seeds used in this work.
Funding
This research was supported by grants from the National Natural Science Foundation of China (31872866 and 31540064), the China Hunan Provincial Department of Science and Technology (2019NK2081, 2019RS2019, and 2020JJ3007), the National Key Research and Development Program (2017YFF0210301), and the Ministry of Education of China (Q2019173).
Conflict of interest statement. We declare no conflict of interest.
Present address: College of Biology, Hunan University, Changsha 410082, China.
W.X. and S.H. performed the expression profiles analyses of the LecRK gene family, genotyped lecrk-VIII.2 T-DNA mutants and generated OE lines, measured the seed size, and carried out the observation of developing seeds. W.X. and R.C. performed the GUS staining and quantitative PCR (qPCR) assays. W.X., S.H., R.C., and R.L. performed the yield-related traits analyses and the observation of seed shape. S.H., W.X., and X.Z. conducted the reciprocal cross experiment and measured the F1 seed size. W.X., S.H., and R.L. performed the FB28 staining assays. W.X., R.C., and R.L. determined the content of storage materials and protein profiles of seeds. W.X., S.H., and R.C. performed the analyses of LecRK-VIII.2-MPK3/6 genetic association. W.X. contributed to the detection of pMPK3/6, performed homologs analyses of LecRK-VIII.2, and drafted the manuscript. W.X., R.L., and X.Z. conducted the subcellular location assay. W.X., S.H., X.G., and R.Y. designed the experiments and analyzed the data. All authors reviewed and approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Ruifeng Yao (ryao@hnu.edu.cn) and Xinhong Guo (gxh@hnu.edu.cn).
References
- Adamski NM, Anastasiou E, Eriksson S, O’Neill CM, Lenhard M (2009) Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc Natl Acad Sci USA 106: 20115–20120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K, Richards AJ, Tremayne M (2006) Fitness constraints on flower number, seed number and seed size in the dimorphic species Primula farinosa L. and Armeria maritima (Miller) Willd. New Phytol 128: 563–570 [DOI] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Bergmann DC (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bonaventure G (2011) The Nicotiana attenuata LECTIN RECEPTOR KINASE 1 is involved in the perception of insect feeding. Plant Signal Behav 6: 2060–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancalion PHS, Rodrigues RR (2014) Seed size-number trade-off in Euterpe edulis in plant. Sci Agric 71: 226–231 [Google Scholar]
- Bush SM, Krysan PJ (2007) Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J Exp Bot 58: 2181–2191 [DOI] [PubMed] [Google Scholar]
- Carine D, Isabelle B, Halima M, Alain G, Martine M, Marine F, Thierry C, Sabine D (2015) Ubiquitin-mediated proteasomal degradation of oleosins is involved in oil body mobilization during Post-germinative seedling growth in Arabidopsis. Plant Cell Physiol 56: 1374–1387 [DOI] [PubMed] [Google Scholar]
- Chakravorty D (2011) An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J 67: 840–851 [DOI] [PubMed] [Google Scholar]
- Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46: 794–804 [DOI] [PubMed] [Google Scholar]
- Cheng X, Wu Y, Guo J, Du B, Chen R, Zhu L, He G (2013) A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J 76: 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZJ, Zhao XY, Shao XX, Wang F, Zhou C, Liu YG, Zhang Y, Zhang XS (2014) Abscisic acid regulates early seed development in Arabidopsis by ABI5-Mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26: 1053–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G (2014) Identification of a plant receptor for extracellular ATP. Science 343: 290–294 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2015) Plant expansins: Diversity and interactions with plant cell walls. Curr Opin Plant Biol 25: 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creff A, Brocard L, Joubès J, Taconnat L, Doll NM, Marsollier AC, Pascal S, Galletti R, Boeuf S, Moussu S, et al. (2019) A stress-response-related inter-compartmental signalling pathway regulates embryonic cuticle integrity in Arabidopsis. PLoS Genet 15: e1007847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Voss U, Jürgens G, Beeckman T (2009) Receptor-like kinases shape the plant. Nat Cell Biol 11: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Deng K, Wang Q, Zeng J, Guo X, Zhao X, Tang D, Liu X (2009) A lectin receptor kinase positively regulates ABA response during seed germination and is involved in salt and osmotic stress response. J Plant Biol 52: 493–500 [Google Scholar]
- Di BJ, Anne M, Adeline B, Kohki Y, Gwendal C, Fabien C, Céline MD, Michèle RC (2018) Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J Exp Bot 69: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Dumenil J, Lu FH, Na L, Vanhaeren H, Naumann C, Klecker M, Prior R, Smith C, McKenzie N, et al. (2017) Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Genes Dev 31: 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Li N, Chen L, Xu Y, Li Y, Zhang Y, Li C, Li Y (2014) The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 26: 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P, Rao Y, Zeng D, Yang Y, Li Y (2014) SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4 (MKK4), influences grain size in rice. Plant J 77: 547–557 [DOI] [PubMed] [Google Scholar]
- Eriksson O (1999) Seed size variation and its effect on germination and seedling performance in the clonal herb Convallaria majalis. Acta Oecol 20: 61–66 [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Fang W, Wang Z, Cui R, Li J, Li Y (2012) Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J 70: 929–939 [DOI] [PubMed] [Google Scholar]
- Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y (1999) Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA 96: 7575–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardoni PA, Hettenhausen C, Baldwin IT, Bonaventure G (2011) Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell 23: 3512–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GlendA V, Mike R (1998) Sources and consequences of seed mass variation in Banksia marginata (Proteaceae). J Ecol 86: 563–573 [Google Scholar]
- Gouhier-Darimont C, Stahl E, Glauser G, Reymond P (2019) The Arabidopsis lectin receptor kinase LecRK-I 8 is involved in insect egg perception. Front Plant Sci 10: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Chen K, Dong NQ, Shi CL, Ye WW, Gao JP, Shan JX, Lin HX (2018) GRAIN SIZE and NUMBER1 negatively regulates the OsMKKK10–OsMKK4–OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30: 871–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Lu ZQ, Shan JX, Ye WW, Dong NQ, Lin HX (2020) ERECTA1 acts upstream of the OsMKKK10–OsMKK4–OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell 32: 2763–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Zhang ZG, Yan DQ, Zhang JS, Chen SY (2004) A salt-responsive receptor-like kinase gene regulated by the ethylene signaling pathway encodes a plasma membrane serine/threonine kinase. Theor Appl Genet 109: 377–383 [DOI] [PubMed] [Google Scholar]
- Hervé C, Serres J, Dabos P, Canut H, Barre A, Rougé P, Lescure B (1999) Characterization of the Arabidopsis lecRK-a genes: Members of a superfamily encoding putative receptors with an extracellular domain homologous to legume lectins. Plant Mol Biol 39: 671–682 [DOI] [PubMed] [Google Scholar]
- Hirsch AM (1999) Role of lectins (and rhizobial exopolysaccharides) in legume nodulation. Curr Opin Plant Biol 2: 320–326 [DOI] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41: 494–497 [DOI] [PubMed] [Google Scholar]
- Jaime HC, Damian S, Kristoffer F, Helen C, Davide H, Walter MC, Thomas R, Mende DR, Shinichi S, Michael K (2015) EggNOG 45: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44: D286–D293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Dang HQ, Vaid N, Tuteja N (2010) Pea lectin receptor-like kinase promotes high salinity stress tolerance in bacteria and expresses in response to stress in planta. Glycoconjugate J 27: 133–150 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, Angelo CD, Bornberg-Bauer E, Kudla JR, Harter K (2007) The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Klaas B, Francine G (2009) Arabidopsis L-type lectin receptor kinases: Phylogeny, classification, and expression profiles. J Exp Bot 60: 4383–4396 [DOI] [PubMed] [Google Scholar]
- Koenig WD, Knops JM, Carmen WJ, Sage RD (2009) No trade‐off between seed size and number in the valley oak (Quercus lobata) Am Nat 173: 682–688 [DOI] [PubMed] [Google Scholar]
- Labbé J, Muchero W, Czarnecki O, Wang J, Wang X, Bryan AC, Wang D (2019) Mediation of plant–mycorrhizal interaction by a lectin receptor-like kinase. Nat Plants 5: 676–680 [DOI] [PubMed] [Google Scholar]
- Lazaro A, Larrinaga AR (2018) A multi-level test of the seed number/size trade-off in two Scandinavian communities. PLoS ONE 13: e201175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Choi D, Kende H (2001) Expansins: Ever-expanding numbers and functions. Curr Opin Plant Biol 4: 527–532 [DOI] [PubMed] [Google Scholar]
- Li N, Xu R, Li Y (2019) Molecular networks of seed size control in plants. Annu Rev Plant Biol 70: 111–1130 [DOI] [PubMed] [Google Scholar]
- Li S, Liu Y, Zheng L, Chen L, Li N, Corke F, Lu Y, Fu X, Zhu Z, Bevan MW (2012) The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol 194: 690–703 [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng L, Corke F, Smith C, Bevan MW (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev 22: 1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Hua L, Dong S, Chen H, Zhu X, Jiang JE, Chen F (2015) OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J 84: 672–681 [DOI] [PubMed] [Google Scholar]
- Liu S, Wang J, Chen K, Zhang Z, Zhang P (2017) The L-type lectin receptor-like kinase (PnLecRLK1) from the Antarctic moss Pohlia nutans enhances chilling-stress tolerance and abscisic acid sensitivity in Arabidopsis. Plant Growth Regul 81: 409–418 [Google Scholar]
- Lopes MA, Larkins BA (1993) Endosperm origin, development, and function. Plant Cell 5: 1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio JS, Dubrovsky JG, Raya-González J, Ugartechea-Chirino Y, López-Bucio J, de Luna-Valdez LA, Ramos-Vega M, León P, Guevara-García AA (2013) Arabidopsis thaliana mitogen-activated protein kinase 6 is involved in seed formation and modulation of primary and lateral root development. J Exp Bot 65: 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J, Westoby M, Leishman M (1995) Seed size and phylogeny in six temperate floras: Constraints, niche conservatism, and adaptation. Am Nat 146: 349–364 [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Wu W, Liang Y, Xu N, Wang Z, Zou H, Liu J (2020) Tyrosine phosphorylation of the lectin receptor‐like kinase LORE regulates plant immunity. EMBO J 39: e102856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Xu N, Huang J, Gao F, Zou H, Boudsocq M, Coaker GL, Liu J (2017) A lectin receptor-like kinase mediates pattern-triggered salicylic acid signaling. Plant Physiol 174: 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci USA 107: 19579–19584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Wang H, He Y, Liu Y, Walker JC, Torii KU, Zhang S (2013) A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moles AT, Ackerly DD, Webb CO, Tweddle JC, Dickie JB, Westoby M (2005) A brief history of seed size. Science 307: 576–580 [DOI] [PubMed] [Google Scholar]
- Musielak TJ, Bayer M (2014) YODA signalling in the early Arabidopsis embryo. Biochem Soc Trans 42: 408–412 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nardi CF, Villarreal NM, Rossi FR, Martínez S, Martínez GA, Civello PM (2015) Overexpression of the carbohydrate binding module of strawberry expansin2 in Arabidopsis thaliana modifies plant growth and cell wall metabolism. Plant Mol Biol 88: 101–117 [DOI] [PubMed] [Google Scholar]
- Navarro-Gochicoa MT, Camut S, Timmers AC, Niebel A, Hervé C, Boutet E, Cullimore JV (2003) Characterization of four lectin-like receptor kinases expressed in roots of Medicago truncatula. Structure, location, regulation of expression, and potential role in the symbiosis with Sinorhizobium meliloti. Plant Physiol 133: 1893–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Wang M, Li Y, Yan W, Chang Z, Chen Z, Xu C, Yang C, Deng XW, Wu J, Tang X (2020) Lectin receptor kinase OsLecRK‐S.7 is required for pollen development and male fertility. J Integr Plant Biol 62: 1227–1245 [DOI] [PubMed] [Google Scholar]
- Peng Y, Chen L, Li S, Zhang Y, Li Y (2018) BRI1 and BAK1 interact with G proteins and regulate sugar-responsive growth and development in Arabidopsis. Nat Commun 9: 1215–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Chen L, Lu Y, Wu Y, Dumenil J, Zhu Z, Bevan MW, Li Y (2015) The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27: 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham AQ, Cho SH, Nguyen CT, Stacey G (2020) Arabidopsis lectin receptor kinase P2K2 is a second plant receptor for extracellular ATP and contributes to innate immunity. Plant Physiol 183: 1364–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD (2004) Dev. and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38: 545–561 [DOI] [PubMed] [Google Scholar]
- Reiser L, Fischer RL (1993) The ovule and the embryo sac. Plant Cell 5: 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelle AT, Richards AJ (2000) Seed weight and seed number affect subsequent fitness in outcrossing and selfing primula species. New Phytol 148: 127–142 [DOI] [PubMed] [Google Scholar]
- Ursache R, Andersen TG, Marhavý P, Geldner N (2018) A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J 93: 399–412 [DOI] [PubMed] [Google Scholar]
- Roy CS, Riesselman AJ, Pey S (2014) Constitutive or seed-specific overexpression of Arabidopsis G-protein gamma subunit 3 (AGG3) results in increased seed and oil production and improved stress tolerance in Camelina sativa. Plant Biotechnol J 12: 49–59 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001a) Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 113: e22. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001b) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Kuo YC, Mishra S, Tsai CH, Chien CC, Chen CW, Desclos-Theveniau M, Chu PW, Schulze B, Chinchilla D (2012) The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 24: 1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CC, Fretwell SD (1974) The optimal balance between size and number of offspring. Am Nat 108: 499–506 [Google Scholar]
- Smith MR, Rao IM, Merchant A (2018) Source–sink relationships in crop plants and their influence on yield development and nutritional quality. Front Plant Sci 9: 1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sõber V, Ramula S (2013) Seed number and environmental conditions do not explain seed size variability for the invasive herb Lupinus polyphyllus. Plant Ecol 214: 883–892 [Google Scholar]
- Tripathi D, Zhang T, Koo AJ, Stacey G, Tanaka K (2018) Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiol 176: 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid N, Pey P, Srivastava VK, Tuteja N (2015) Pea lectin receptor-like kinase functions in salinity adaptation without yield penalty, by alleviating osmotic and ionic stresses and upregulating stress-responsive genes. Plant Mol Biol 88: 193–206 [DOI] [PubMed] [Google Scholar]
- Vaid N, Pey PK, Tuteja N (2012) Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice. Plant Mol Biol 80: 365–388 [DOI] [PubMed] [Google Scholar]
- Wan J, Patel A, Mathieu M, Kim S, Xu D, Stacey G (2008) A lectin receptor-like kinase is required for pollen development in Arabidopsis. Plant Mol Biol 67: 469–482 [DOI] [PubMed] [Google Scholar]
- Wang A, Garcia D, Zhang H, Feng K, Luo M (2010) The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J 63: 670–679 [DOI] [PubMed] [Google Scholar]
- Wang C, Huang X, Li Q, Zhang Y, Li JL, Mou Z (2019) Extracellular pyridine nucleotides trigger plant systemic immunity through a lectin receptor kinase/BAK1 complex. Nat Commun 10: 4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhou M, Zhang X, Yao J, Zhang Y, Mou Z (2017) A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. eLife 6: e25474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu A, Bruffett AK, Lee AJ, Hause BG (2008) Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20: 602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wilkins KA, Davies JM (2018) Arabidopsis DORN1 extracellular ATP receptor; Activation of plasma membrane K- and Ca-permeable conductances. New Phytol 218: 1301–1304 [DOI] [PubMed] [Google Scholar]
- Wang Y, Nsibo DL, Juhar HM, Govers F, Bouwmeester K (2016) Ectopic expression of Arabidopsis L-type lectin receptor kinase genes LecRK-I.9 and LecRK-IX.1 in Nicotiana benthamiana confers phytophthora resistance. Plant Cell Rep 35: 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U (1996) Controlling seed development and seed size in Vicia faba: A role for seed coat-associated invertases and carbohydrate state. Plant J 10: 823–834 [Google Scholar]
- Weber H, Borisjuk L, Wobus U, Borisjuk L, Wobus U (1997) Sugar import and metabolism during seed development. Trends Plant Sci 2: 169–174 [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JY, Kim YJ, Paek KH (2020) CaLecRK-S.5, a pepper L-type lectin receptor kinase gene, accelerates Phytophthora elicitin-mediated defense response. Biochem Biophys Res Commun 524: 951–956 [DOI] [PubMed] [Google Scholar]
- Xia T, Li N, Dumenil J, Li J, Kamenski A, Bevan MW, Gao F, Li Y (2013) The ubiquitin receptor DA1 interacts with the e3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25: 3347–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Wang A, Yang G, Gao P, Zheng Z (2009) The Arabidopsis a4 subfamily of lectin receptor kinases negatively regulates abscisic acid response in seed germination. Plant Physiol 149: 434–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang S (2015) Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci 20: 56–64 [DOI] [PubMed] [Google Scholar]
- Xu R, Duan P, Yu H, Li J, Zhang G, Zhuang S, Tian Z, Yao S, Li Y (2018) Control of grain size and weight by the OsMKKK10–OsMKK4–OsMAPK6 signaling pathway in rice. Mol Plant 11: 860–873 [DOI] [PubMed] [Google Scholar]
- Xu N, Luo X, Wu W, Xing Y, Liang Y, Liu Y, Zou H, Wei HL, Liu J (2020) A plant lectin receptor-like kinase phosphorylates the bacterial effector AvrPtoB to dampen its virulence in Arabidopsis. Mol Plant 13: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Yi J, Lee YS, Lee DY, Cho MH, An G (2016) OsMPK6 plays a critical role in cell differentiation during early embryogenesis in Oryza sativa. J Exp Bot 67: w52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Li H, Yang W (2017) The integration of Gβ and MAPK signaling cascade in zygote development. Sci Rep 7: 8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Guo XH, Xie HL, Li JY, Liu XQ, Zhu BD, Liu SC, Li HL, Li ML, He MQ, et al. (2018) Quantitative phosphoproteomics of lectin receptor-like kinase VI.4 dependent abscisic acid response in Arabidopsis thaliana. Physiol Plantarum 165: 728–745 [DOI] [PubMed] [Google Scholar]
- Zhang C, Li JY, Guo XH, Zhu BD, Xiao WJ, Wang P, Jiang M, Hu S, Lu XT, He Z, et al. (2017) LecRK-VII.1, a lectin receptor-like kinase, mediates the regulation of salt stress and jasmonic acid response in Arabidopsis. J Plant Growth Regul 36: 385–401 [Google Scholar]
- Zhang M, Wu H, Su J, Wang H, Zhu Q, Liu Y, Xu J, Lukowitz W, Zhang S (2017) Maternal control of embryogenesis by MPK6 and its upstream MKK4/MKK5 in Arabidopsis. Plant J 92: 1005–1019 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Kang X, Zhao X, Zhang X, Ni M (2009) SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wu X, Yuan S, Qian D, Nan Q, An LZ, Xiang Y (2014) Annexin5 plays a vital role in Arabidopsis pollen development via Ca2+-dependent membrane trafficking. PLoS ONE 9: e102407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo K, Zhao J, Wang J, Sun X, Tang K (2004) Molecular cloning and characterization of GhlecRK, a novel kinase gene with lectin-like domain from Gossypium hirsutum. DNA Seq 15: 58–65 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.