A nucleotide exchange factor regulates the activity of a major endoplasmic reticulum luminal chaperone and is involved in the salt stress response in rice.
Abstract
The endoplasmic reticulum (ER) quality control system monitors protein homeostasis and relies on the activity of many molecular chaperones. Binding immunoglobulin protein (BiP) is a major ER luminal chaperone that is involved in most functions of the organelle. BiP activity is tightly regulated by nucleotide exchange factors (NEFs). However, information about NEFs in plants is limited. We obtained a Fes1-like protein (OsFes1C) through isobaric tags for relative and absolute quantitation-based proteomics analysis of ER-stressed rice (Oryza sativa) seeds. Unlike its homologs in yeast and mammals, which are located in the cytosol and respond to heat stress, OsFes1C is an ER membrane protein and responds to ER and salt stresses. OsFes1C interacts directly with OsBiP1 and the interaction is inhibited by ATP but promoted by ADP, suggesting that OsFes1C acts as a potential NEF of OsBiP1 in vivo. Overexpression or suppression of OsFes1C led to hypersensitivity to ER stress and affected the growth of rice. Furthermore, we established that OsFes1C directly interacts with a putative salt response protein and is involved in the salt response. Taken together, our study marks an important step toward elucidating the functional mechanisms of an identified ER stress response factor in rice.
Introduction
Secretory and membrane proteins are folded and modified in the endoplasmic reticulum (ER) under the control of the ER-mediated protein quality control (ERQC) system in eukaryotes. Adverse environmental conditions or defects in physiological processes usually cause accumulation of unfolded, misfolded, unassembled, or damaged proteins, which disturb ER homeostasis inducing ER stress. In response to ER stress, cells invoke unfolded protein response (UPR) signaling pathway to increase the expression of ER chaperones to facilitate protein folding, and ER-associated protein degradation (ERAD) pathway to stimulate the degradation of misfolded or unassembled proteins (Strasser, 2018). ERQC is a complicated regulatory pathway requiring cooperation of multiple factors. Until now, many factors involved in ERQC have been identified, such as binding immunoglobulin protein (BiP), protein disulfide-isomerase (PDI), 3-hydroxy-3-methyl glutaryl coenzyme A reductase degradation protein 1 (Hrd1), and degradation in ER protein 1 (Derlin1; Oikonomou and Hendershot, 2020).
The heat shock protein 70 (Hsp70) chaperone system participates in protein folding and quality control of unfolded proteins. The Hsp70 molecular chaperone system consists of chaperone Hsp70 (DnaK), cochaperone Hsp40 (DnaJ-type), and a nucleotide exchange factor (NEF). Hsp70 chaperones bind to nonnative proteins as substrates to prevent their aggregation (Schilke et al., 2017; Rosenzweig et al., 2019). The binding and release of the substrates are regulated by a cycle of ATP/ADP exchange. Hsp40 and substrate binding accelerate ATP hydrolysis on Hsp70, resulting in tight binding of substrate. Bound substrate is released after dissociation of ADP and rebinding of ATP. NEFs promote the dissociation of ADP from Hsp70, which is the rate-limiting step in the ATPase cycle (Bukau and Horwich, 1998).
To date, most of the studies on NEF of Hsp70 were in yeast (Saccharomyces cerevisiae) and mammals. The first nuclear Hsp70 NEF Fes1 was identified in yeast. In the fes1 mutant, misfolded proteins fail to undergo polyubiquitylation, aggregate, and induce a strong heat shock response (Gowda et al., 2016). Fes1 has two functional isoforms, Fes1L and Fes1S, via alternative splicing of the transcript. Fes1L is actively targeted to the nucleus, whereas Fes1S localizes to the cytosol and is required for the efficient proteasomal degradation of cytosolic misfolded proteins (Gowda et al., 2016). Three classes of Hsp70 NEFs, including HSPBP1 (Fes1p), Hsp110 (Sse1, 2p), and BAG domain protein families (Snl1p), are found in the cytosol involved in ERQC (Bracher and Verghese, 2015). In mitochondria, two putative NEF orthologs, GrpE-like 1 (GrpEL1) and GrpEL2 are reported to form a heterooligomeric subcomplex with mtHSP70 regulating the functions of mtHsp70. The formation of this subcomplex is critical for conferring stability to the NEFs, helping fine-tune mitochondrial protein quality control (Srivastava et al., 2017).
BiP, an ER ortholog of the Hsp70 family, is one of the major chaperones. BiP plays an important role in assisting protein folding and in the processes of ERAD (McCracken and Brodsky, 2003; Denic et al., 2006; Maattanen et al., 2010). BiP also acts as a sensor protein, recognizing the strength of ER stress in cells. Like all Hsp70 proteins, the binding and release of the substrate of BiP are regulated by ATP/ADP exchange (Kleizen and Braakman, 2004; Winter and Jakob, 2004). To date, two NEFs for BiP have been identified, suppressor of inositol-requiring enzyme 1 and lumenal Hsp 70 1 deletion 1 (SIL1) and glucose-regulated protein 170 (Grp170). Yeast SIL1 interacts with secretory 61 (Sec61) and binds preferentially to the ADP-bound karyogamy 2 (Kar2p, yeast BiP; Kabani et al., 2000). SIL1 plays a role in the translocation of proteins into the ER lumen (Behnke et al., 2015). Lumenal Hsp70 1 (Lhs1p, yeast GRP170) serves as a NEF of Kar2p, binding of the nucleotide to Lhs1p can stimulate the interaction with Kar2p and is essential for NEF activity (de Keyzer et al., 2009). Lhs1p binds directly to a variety of incompletely folded protein substrates in the ER, but does not interact with folded secretory proteins (Behnke and Hendershot, 2014).
In contrast to the diverse biological functions of NEF in yeast or mammalian systems, relatively little is known about NEF in plants. To date, only one Fes1 ortholog AtFes1 has been reported in Arabidopsis thaliana. AtFes1A is cytosolic and interacts with cytosolic Hsp70, preventing Hsp70 degradation. The loss of AtFes1A increased heat-sensitivity of Arabidopsis plants (Zhang et al., 2010). Although AtFes1A interacts with Hsp70, it showed no NEF activity in vitro. The rice (Oryza sativa) genome contains at least five OsBiP genes, and OsBiP1 plays major roles. Overexpression (OE) or suppression (Ri) of OsBiP1 in rice seeds induces a severe ER stress response, resulting in a deterioration of grain properties (Yasuda et al., 2009; Wakasa et al., 2011). Until recently, several ER-resident J proteins, such as OsP58A, OsP58B, and OsERdj3B, have been identified as cochaperons interacting with OsBiP1 (Ohta et al., 2013). However, the NEF for OsBiP1 remains unknown.
In this study, we identified an ER-stress response protein OsFes1C via proteomic strategies in rice (Qian et al., 2015). Unlike yeast Fes1p and AtFes1A that located in cytosol, OsFes1C is an ER membrane protein. The expression of OsFes1C is induced by ER and salt stress. Either increase or decrease of the expression level of OsFes1C has exactly the same effect on the growth and development of rice, and both can lead to hypersensitivity to ER stress, even causing the same changes of transcript profiles between OsFes1C transgenic plants and the wild plants. OsFes1C directly interacts with OsBiP1, and the interaction is strengthened by ADP but weakened by ATP. We suggest that OsFes1C acts as a potential NEF of OsBiP1 in vivo.
Results
Identification of OsFes1C protein
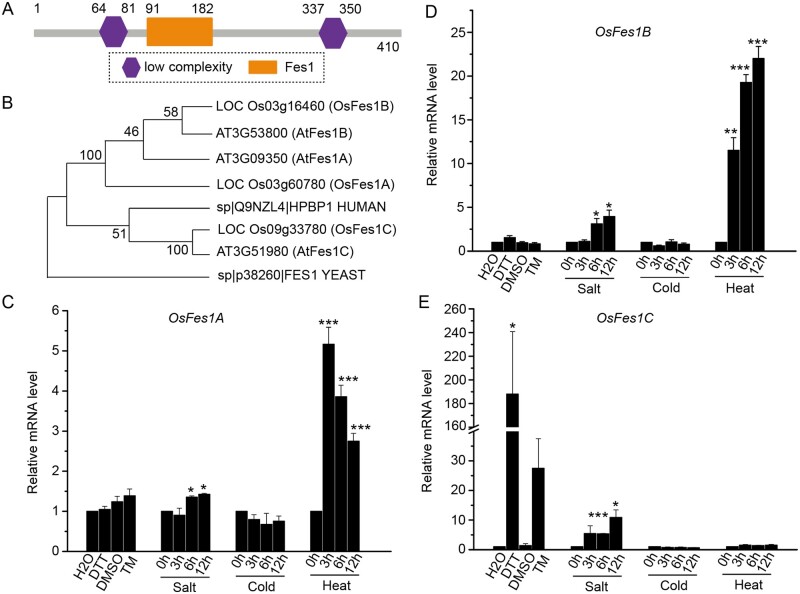
Previously, we analyzed the low salt-soluble proteins from developing rice seeds by the isobaric tags for relative and absolute quantitation method (Qian et al., 2015). The wild-type (WT) and transgenic rice plants with seed-specific ER-stress samples were used to investigate the ER-stress regulation of the proteome. We identified a Fes1-like protein whose expression level in the ER-stressed seed was more than 25-fold higher than that in WT. The Fes1-like protein, containing 410 amino acids, is deduced as a putative NEF. SMART (http://smart.embl-heidelberg.de/) analysis showed that the Fes1-like protein contains a Fes1 domain, and two low complexity domains (Figure 1A). Phylogenetic analysis showed that the Fes1-like protein was closely related to Arabidopsis AtFes1c (Figure 1B); therefore, the Fes1-like was designated OsFes1C (LOC_Os09g33780). A survey of the rice genome revealed two other Fes1 homologs. According to the amino acid similarity to Fes1 in Arabidopsis (Zhang et al., 2010), these two proteins were named as OsFes1A (LOC_Os03g60780) and OsFes1B (LOC_Os03g16460). OsFes1C shares a relatively low degree of sequence identity with OsFes1A and OsFes1B.
Figure 1.
Treatment of OsFes1 by different stress conditions. A, A schematic illustration of domain organization of OsFes1C. It contains a Fes1 domain. B, Phylogenetic tree of OsFes1C with homologs in other species. HPBP1 is the homolog of Fes1 in human. The neighbor-joining method was used for constructing the phylogenetic tree. The bootstrap probability values were shown on the nodes of the phylogenic tree. C, Expression of OsFes1A under different stress conditions. OsFes1A is significantly increased under heat stress. D, Expression of OsFes1B under different stress conditions. OsFes1B is significantly increased under heat stress. E, Expression of OsFes1C under different stress conditions. OsFes1C is significantly increased under ER and salt stresses. The expression of the OsFes1 isoforms in (C–E) has been normalized to ACTIN, then the mRNA level under different stress conditions was relative to the control as 1.0. All data are averages of three independent experiments, and error bars represent sem. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
Expression of OsFes1C is upregulated by ER stress and salt stress
It has been reported that the expression of AtFes1A is strongly induced by heat stress (Zhang et al., 2010). To test whether Fes1 proteins in rice respond to the heat stress, we analyzed the expression of three OsFes1s under the heat stress. The results showed that the expression levels of OsFes1A and OsFes1B increased significantly under heat stress conditions, but that of the OsFes1C showed no obvious difference (Figure 1, C–E). In addition, the expression levels of three OsFes1s were not changed under cold stress condition (Figure 1, C–E).
Our proteomic data showed that OsFes1C was upregulated by ER stress (Qian et al., 2015). To confirm this result, the expression level of OsFes1C in plants treated with dithiothreitol (DTT) or tunicamycin (Tm) was examined by reverse-transcription quantitative PCR (RT-qPCR). The results showed that expression level of OsFes1C was increased by 188- and ∼27.5-fold in plants treated with DTT and Tm for 4 h, respectively (Figure 1E). In contrast, the expression levels of OsFes1A and OsFes1B remained constant under DTT and Tm treatments (Figure 1, C and D). In addition, we found that the expression of OsFes1A, OsFes1B, and OsFes1C can be induced by salt stress, particularly at 6 and 12 h after treatment (Figure 1, C–E).
OsFes1C is an ER membrane protein
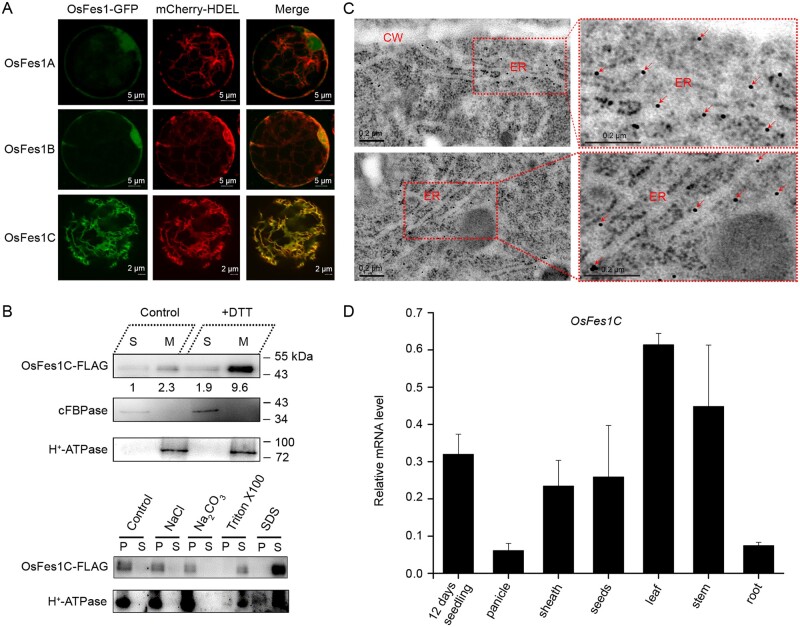
To determine their subcellular localization, we fused green fluorescent protein (GFP) to the C-terminus of OsFes1 proteins and transiently expressed the constructs in rice protoplasts. Confocal laser scanning microscopy revealed that OsFes1A-GFP and OsFes1B-GFP are located in both cytoplasm and nucleus, whereas OsFes1C-GFP showed typical ER localization (Figure 2A). Coexpression of OsFes1C-GFP with the ER marker mCherry-HDEL showed that the green fluorescence of OsFes1C-GFP merged well with the red fluorescence of mCherry-HDEL (Figure 2A), confirming that OsFes1C was located in the ER.
Figure 2.
OsFes1C is an ER membrane protein and tissue-specific expression of OsFes1C. A, Subcellular localization of the three OsFes1 isoforms analyzed by confocal microscopy. OsFes1A-GFP and OsFes1B-GFP are located in both cytoplasm and nucleus, whereas OsFes1C is located in the ER. B, Upper part: OsFes1C-FLAG protein was detected predominantly in the membrane fraction. S, soluble fraction; M, membrane fractions. H+-ATPase is a membrane protein and cFBPase is a soluble protein, which acts as a control. Lower part: The solubilization assays of OsFes1C protein. The OsFes1C protein can be solubilized by ionic and nonionic detergents like SDS and Triton X100, respectively, but not high salt (1 M NaCl) and high pH (100 mM Na2CO3, pH 11.5) conditions. The membrane protein H+-ATPase acts as a control. S, supernatants; P, pellets. C, Immunolocalization of OsFes1C in the ER of root cell. The OsFes1C antibody was labeled with 10-nm immunogold particles. Arrow indicates OsFes1C accumulated in the ER. CW, cell wall. D, Tissue-specific expression of OsFes1C. OsFes1C is mainly expressed in leaf. Seedling, 12 d seedling; other individual tissues came from the filling stage rice. The expression of OsFes1C was normalized to that of ACTIN. Data are averages of three independent experiments, and error bars represent sem.
OsFes1C was predicted to be a membrane protein with a single transmembrane helix (Supplemental Figure S1). To determine the intracellular partition of OsFes1C, we generated transgenic rice expressing OsFes1C-3×FLAG under the control of ubiquitin promoter, and extracted membrane proteins from the transgenic seedling for immunoblot analysis. Western blot with antibody against FLAG showed that OsFes1C was detected predominantly in the membrane fraction (Figure 2B, upper part). Western blot analysis with the membrane protein extracted from the transgenic seedlings under DTT treatment also showed a similar result (Figure 2B, upper part). The membrane fraction was further suspended in control buffer, high-salt buffer (1 M NaCl), alkaline buffer (100 mM Na2CO3, pH 11.5), Sodium dodecyl sulfate (SDS) buffer, and Triton X-100 buffer, ultracentrifuged and subjected to immunoblot analysis with antibody against FLAG. The results showed that OsFes1C could be solubilized by SDS or Triton X-100 buffer, but could not be extracted with high-salt or alkaline buffer (Figure 2B, lower part). Furthermore, the intracellular localizations of OsFes1C in the root cell of OsFes1C-3×FLAG transgenic plants were examined by immunoelectron microscopy. As shown in Figure 2C, the OsFes1C labeled with 10-nm immunogold particles were mainly distributed in ER. These results indicate that OsFes1C is an integral ER membrane protein.
To investigate the expression patterns of OsFes1C, we performed RT-qPCR analysis. Although OsFes1C was universally expressed in all tissues including root, leaf, leaf sheath, stem, panicle, and seed, it was predominantly expressed in leaf and leaf sheath with the lowest expression level in panicle and root (Figure 2D).
To date, all Fes1 proteins reported were located at cytosol or nucleus responding to heat stress (Zhang et al., 2010; Gowda et al., 2016). The features of being an ER membrane protein and its response to ER and salt stresses make OsFes1C a unique protein. Thus, we focused on the study of OsFes1C.
OE or Ri of OsFes1C leads to hypersensitivity to ER stress
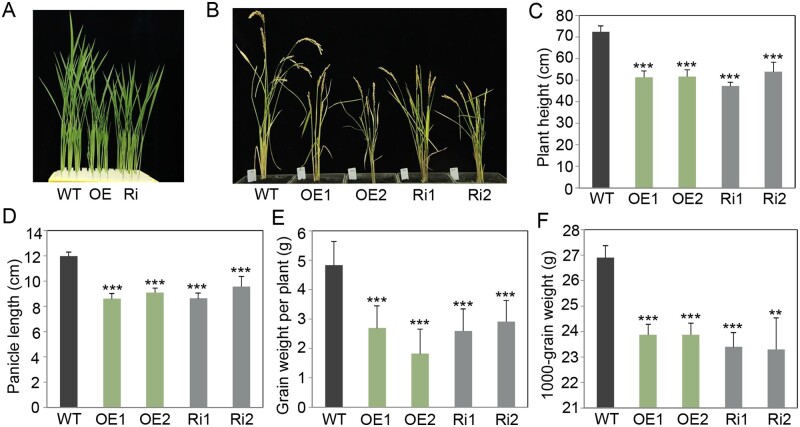
To further elucidate the function of OsFes1C, we generated OsFes1C OE and Ri transgenic plants under the control of the maize (Zea mays) ubiquitin promoter. Positive transgenic plants were identified by PCR, and the expression level of OsFes1C was measured by RT-qPCR. The transcription levels of OsFes1C were lower in Ri leaves and higher in OE leaves than in the leaves of the WT (Supplemental Figure S2), indicating that the OsFes1C was successfully overexpressed or suppressed, respectively. The plant height, panicle length, grain weight per plant, and 1,000-grain weight of the transgenic plants, both OE and Ri, were reduced significantly compared to the WT (Figure 3). However, no obvious differences were found on these traits between OE and Ri plants.
Figure 3.
Effects of OE and repression of OsFes1C on rice growth. Compared to the WT rice, the plants of the OsFes1C transgenic lines had obvious dwarf phenotype at the seedling stage (A) and at the ripening stage (B). OE indicates OsFes1C-overexpressing rice; Ri represents OsFes1C-repressed rice. C–F, Significant genotypic differences in plant height (C), panicle length (D), grain weight per plant (E), and 1,000-grain weight (F) between the OsFes1C transgenic rice and WT. All data are averages of three independent experiments, and error bars represent sem. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
To verify whether OE or Ri of OsFes1C induced UPR, the levels of ER-resident chaperones in leaves were investigated by western blotting analysis. The results showed that the levels of BiP1, PDI2-3, and CNX in the OsFes1C OE and OsFes1C Ri lines were comparable with those in the WT (Supplemental Figure S3). These results suggested that OE or Ri of OsFes1C did not trigger the UPR.
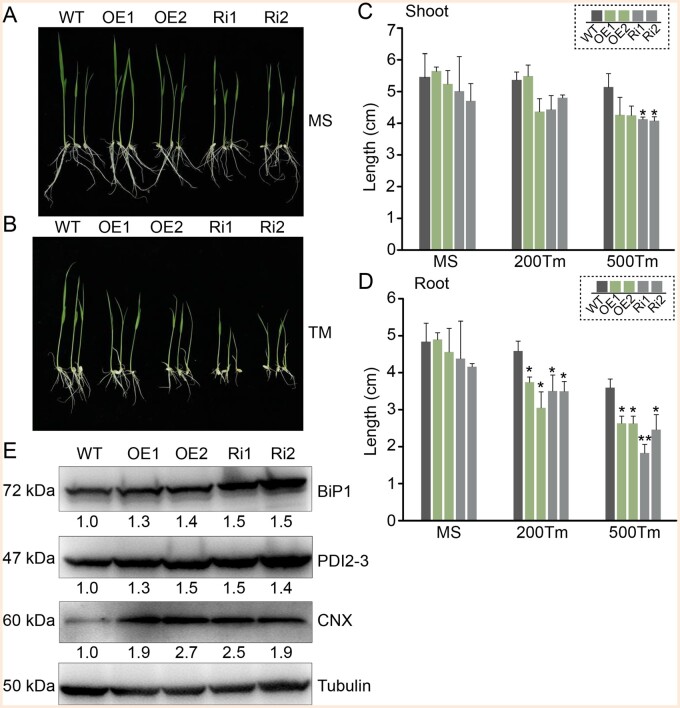
To further investigate the effect of overexpressed or suppressed OsFes1C on the ER-stress responses, the seedlings of OsFes1C OE, OsFes1C Ri, and WT were treated with Tm for 5 d. Under normal condition, the lengths of the shoot and root were slightly different between the seedlings of the WT and OsFes1C transgenic seedlings (Figure 4, A, C, and D). Under ER stress condition, the lengths of the shoot and root of OE and Ri seedlings were significantly shorter than those of the WT (Figure 4, B, C, and D). The OE and Ri plants showed similar phenotype in terms of root and shoot length. Under ER stress condition, the expression levels of BiP1, PDI2-3, and CNX in OE and Ri plants increased substantially compared to the WT (Figure 4E), indicating that alteration of the expression of OsFes1C enhanced the UPR signaling in response to ER stress. These results indicated that alteration of the expression level of OsFes1C led to hypersensitivity to ER stress and affected the growth of rice.
Figure 4.
Phenotype of the WT and transgenic rice under the Tm treatment conditions. The growth of OE and Ri in liquid MS medium without (A) and with (B) Tm was observed over a period of 5 d. WT served as a control. A, Under normal condition, the lengths of the shoot and root were slightly different among the WT and OsFes1C transgenic seedlings. B, After treatment with Tm, the lengths of the shoot and root of OsFes1C transgenic seedlings were significantly shorter than those of the WT. C and D, Quantitative measurements of the lengths of shoots and roots of WT, OE, and Ri seedlings that treated with Tm in three trials. Error bars represent sd. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test). E, Under ER stress, the UPR-activation indicators, such as BiP1, PDI2-3, and CNX, strongly accumulated in OsFes1C transgenic plants compared to the WT. Transgenic calli treated with Tm were used for the immunoblot. Anti-BiP1, anti-CNX, anti-PDI2-3, and anti-tubulin antibodies were used to detect the expression. The numbers below the strip represent the proteins expression level relative to WT as 1.0.
OsFes1C interacts with OsBiP1 in vitro and in vivo
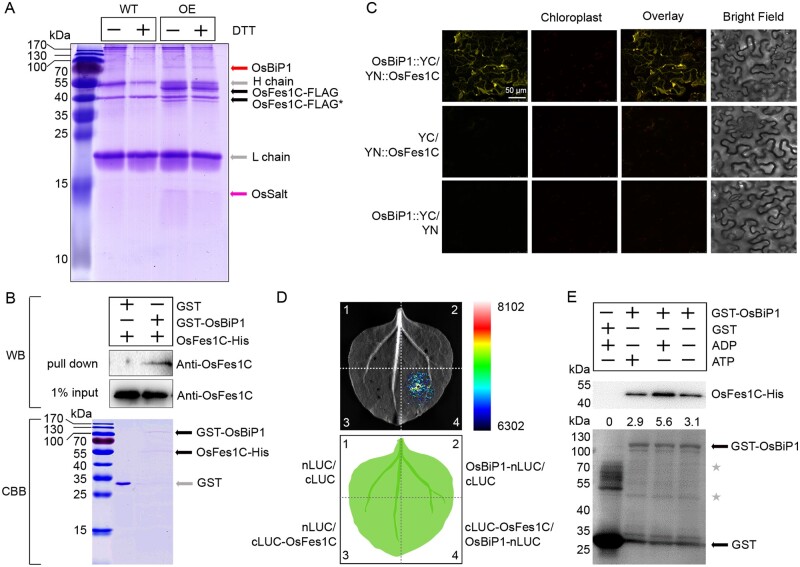
To search for factors interacting with OsFes1C in rice, we performed coimmunoprecipitation (CoIP) experiments, using OsFes1C-3×FLAG-overexpressing rice calli treated with or without DTT. The callus cell extract was incubated with anti-FLAG antibody linked beads, and the WT extract was used as a control. After SDS-Polyacrylamide gel electrophoresis (PAGE) and staining, an intense band and a faint band were detected, with no band observed at the equivalent positions in the control lane (Figure 5A). The different bands were identified by mass spectrometry (Supplemental Table S1). The results showed that the tryptic peptides of the intense band aligned well with the primary sequences deduced from OsBiP1 (LOC_Os02g02410), while that from the faint band aligned with a putative salt response protein (OsSalt, LOC_Os01g24710; Figure 5A). CoIP assays using OsFes1C-3×FLAG seedlings treated with or without DTT also identified the two proteins (Supplemental Figure S4).
Figure 5.
OsFes1C directly interacts with OsBiP1. A, CoIP assay of transgenic OsFes1C-FLAG and endogenous interacting proteins using transgenic calli. All the bands were identified by mass spectrometric analyses, respectively. Two proteins, OsBiP1 and OsSalt, were immunoprecipitated with OsFes1C. The asterisk indicates the degraded bands of OsFes1C-FLAG. B, The GST pull-down assay shows that the OsFes1C-His was pulled down only with GST-OsBiP1, but not with the GST control. C, The interaction between OsFes1C and OsBiP1 in plant cells was confirmed by BiFC. The scale bar (50 μm) for the top left image apply to all images. D, Firefly LCI assay for interaction between cLUC-OsFes1C and OsBiP1-nLUC in N. benthamiana. E, ADP strengthens the interaction between OsFes1C and OsBiP1, and ATP weakens the interaction. The ratios of OsFes1C-His to GST-OsBiP1 in each condition were shown. The asterisks indicate the degraded bands of GST-OsBiP1.
To confirm OsFes1C directly binds to OsBiP1, we performed pull-down experiment. His-tagged OsFes1C and glutathione-S-transferase (GST)-OsBiP1 were expressed in Escherichia coli. The purified His-tagged OsFes1C protein was incubated with GST alone or purified GST-OsBiP1, and the coprecipitated complex was analyzed by western blot using anti-OsFes1C antibody. As shown in Figure 5B, the OsFes1C-His was pulled down only with the GST-OsBiP1, but not with the GST alone. These results indicated that OsFes1C directly interacted with OsBiP1. The interaction between OsFes1C and OsBiP1 was further confirmed by bimolecular fluorescence complementation (BiFC) in plant cells. Full-length OsFes1C cDNA was fused to the N-terminal half of Yellow fluorescent protein (YFP), OsBiP1 was fused to the C-terminal half of YFP, and the two constructs were infiltrated into Nicotiana benthamiana leaf epidermal cells. As shown in Figure 5C, YFP fluorescence was only detected in the cells cotransformed with nYFP-OsFes1C and OsBiP1-cYFP. YFP fluorescence was not detected in the nYFP-OsFes1C and cYFP or OsBiP1-cYFP and nYFP coexpression cells of pairwise expression (Figure 5C). The interaction between OsFes1C and OsBiP1 was further confirmed by firefly luciferase complementation imaging (LCI) assay (Figure 5D). These results indicated that OsFes1C binds to OsBiP1 both in vitro and in vivo.
It has been reported that the AtFes1A was associated with HSP70, and the interaction was inhibited in the presence of ATP (Zhang et al., 2010). Similarly, the interaction between the yeast ortholog Fes1p and Ssa1p was also inhibited by ATP (Kabani et al., 2002). To investigate whether the adenosine nucleotides affect the binding of OsBiP1 to OsFes1C, we performed pull-down experiments as described above incubated with ATP or ADP. The results showed that the interaction between OsFes1C and OsBiP1 was strengthened by ADP but weakened by ATP (Figure 5E). These results indicated that ATP affects the interaction between OsBiP1 and OsFes1C in rice.
OsFes1C interacts with OsSalt and is involved in the salt stress response
To investigate if OsSalt responds to salt stress, we treated 10-d-old rice seedlings with 200 mM NaCl, and measured the expression level of OsSalt. The result showed that the expression level of OsSalt increased dramatically 12 h after treatment. This result suggested that OsSalt was involved in salt stress response (Supplemental Figure S5).
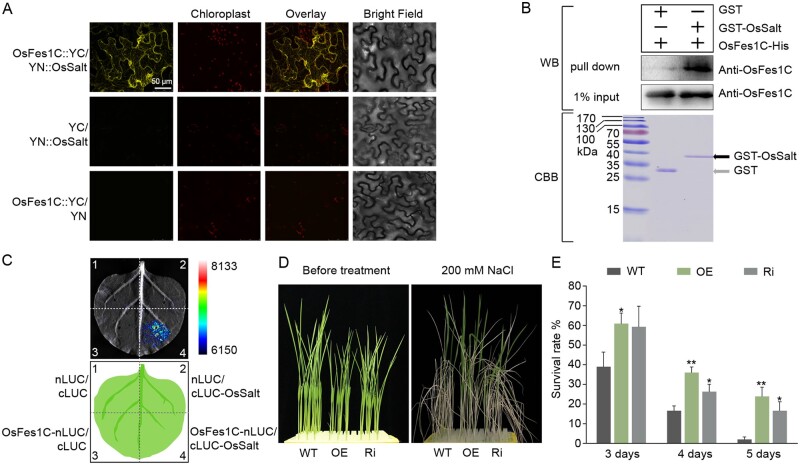
To confirm the interaction between OsFes1C and OsSalt, we performed BiFC experiments. Full-length OsSalt cDNA was fused to the N-terminal half of YFP, while OsFes1C was fused to the C-terminal half of YFP, and the two constructs were infiltrated into N. benthamiana leaf epidermal cells. As a control, the empty vector in combination with each fusion construct was also coinfiltrated into N. benthamiana leaf epidermal cells. YFP fluorescence was only observed in N. benthamiana cells coexpressing OsFes1C-cYFP and nYFP-OsSalt, but not in those coexpressing OsFes1C-cYFP and nYFP or cYFP and nYFP-OsSalt. These results indicated that OsFes1C interacts with OsSalt (Figure 6A). The interaction between OsFes1C and OsSalt was further confirmed by pull-down assay in E. coli, using GST-OsSalt as the bait and OsFes1C-His as the prey. OsFes1C-His was only pulled down with GST-OsSalt, but not with the GST alone (Figure 6B). The interaction between OsFes1C and OsSalt was also confirmed by LCI assay (Figure 6C). In addition, OsSalt was localized in cytoplasm, nucleus, and ER (Supplemental Figure S6). These results confirmed that OsFes1C interacts directly with OsSalt.
Figure 6.
OsFes1C is involved in salt response. A, The interaction between OsFes1C and OsSalt in plant cells was confirmed by BiFC. The scale bar (50 μm) for the top left image apply to all images. B, The GST pull-down assay shows that the OsFes1C-His was pulled down only with GST-OsSalt, but not with the GST control. C, LCI assay for interaction between OsFes1C-nLUC and cLUC-OsSalt in N. benthamiana. D, More transgenic seedlings survived than WT under the NaCl treatment condition. E, Quantitative measurements of the survived WT and transgenic seedlings after treatment with NaCl for 3–5 d. All data are averages of three independent experiments, and error bars represent sem. *P < 0.05, **P < 0.01 (Student’s t test).
We tested whole-plant salt sensitivity of OsFes1C OE and Ri plants by treating 10-d-old seedlings with 200 mM NaCl for 3–5 d. For the salt treatment for 5 d, we observed that almost all leaves of WT rolled, whereas the leaves of OsFes1C OE and Ri seedlings just started rolling (Figure 6D). After recovery for 7 d, almost all the WT wilted, whereas 20% of the OsFes1C OE and Ri seedlings survived (Figure 6E). These results showed that OE or Ri of OsFes1C increased the salt stress tolerance of the plants.
Transcriptome sequencing and proteomics analysis of the OsFes1C OE and Ri transgenic plants
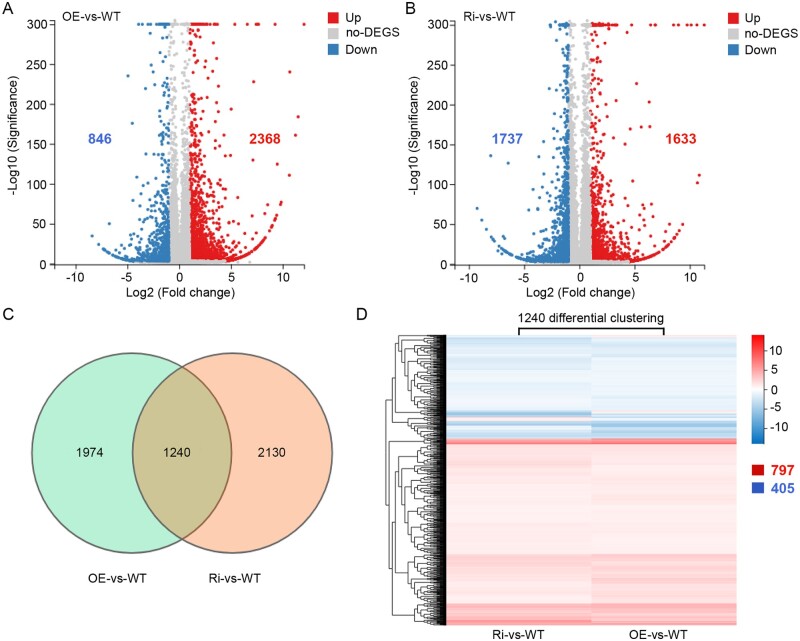
Compared to the WT, transcriptomes of both OsFes1C OE and Ri transgenic lines displayed significant changes (P < 0.05). In total, 2,368 and 846 genes were up- and downregulated with a threshold of two-fold change in the OE plants, whereas in the Ri plants, 1,633 and 1,737 genes were up and downregulated, respectively (Figure 7, A and B; Supplemental Data Sets S1, S2). Venn diagram shows that 1,240 genes were changed both in OE and Ri lines (Figure 7C; Supplemental Data Set S3). Among the 1,240 genes, 1,202 genes showed a similar expression pattern in both OE and Ri lines, with 797 genes upregulated and 405 genes downregulated. Only 38 genes showed opposite expression patterns between the OE and Ri lines (Figure 7D). These results indicated that either increase or decrease of the expression level of OsFes1C leads to a similar response in transcript profiles in rice. In addition, 42 genes were identified as transcription factors among the 1,240 differentially expressed genes (DEGs), which belong to different transcriptional regulator families, including bHLH, WRKY, NAC, and MYB transcription factors (Supplemental Figure S7; Supplemental Data Set S4). To validate these transcriptome sequencing data, 12 genes were randomly selected and analyzed by RT-qPCR. The expression patterns of these genes reflected by RT-qPCR were consistent with the observation by RNA-seq (Supplemental Figure S7).
Figure 7.
Transcriptome sequencing in the OsFes1C OE and OsFes1C Ri transgenic plant. A, Compared to the WT, a total of 2,368 and 846 genes were upregulated and downregulated with a threshold of two-fold change in the OsFes1C OE plants, respectively. B, Compared to the WT, a total of 1,633 and 1,737 genes were upregulated and downregulated with a threshold of two-fold change in the OsFes1C Ri plants, respectively. C, Venn diagram shows that in total of 1,240 genes were changed in both OsFes1C OE and OsFes1C Ri transgenic lines. D, Among these 1,240 differential genes, 797 genes were upregulated and 405 genes were downregulated in both OsFes1C OE and OsFes1C Ri transgenic lines. Only the remaining 38 genes showed opposite expression change patterns between the OsFes1C OE and OsFes1C Ri transgenic lines.
Western blot analysis clearly displayed that OsFes1C was highly expressed in the OE plants and substantially suppressed in the Ri plants compared to the WT (Supplemental Figure S8A). We then performed SWATH-based quantitative proteomic analysis to investigate the changes in protein level of the OE and Ri transgenic plants. Compared to the WT, 83 proteins were changed with a threshold of two-fold both in OE and Ri plants (Supplemental Figure S8B; Supplemental Data Set S5), with 52 proteins upregulated and 31 proteins downregulated both in OE and Ri plants (Supplemental Figure S8C). The similar changes in both the transcriptome and proteome of OE and Ri plants may underlie their similar phenotypes.
Discussion
NEFs are cofactors of HSP70 chaperones, which regulate the functions of chaperones by stimulating release of ADP and allowing ATP to rebind, thereby determining how fast substrates are released from the chaperones. In contrast to the diverse biological functions of NEFs in yeast or mammalian systems, relatively little is known about NEFs in plants. Currently, the only report about plant NEFs is Arabidopsis AtFES1. AtFes1A is located in cytosol, and is involved in heat stress responses (Zhang et al., 2010). Its homolog in yeast, Fes1, is also a cytosolic NEF that is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins (Gowda et al., 2013). OsBiP1 is a major ER luminal chaperon intricately involved in most functions of the organelle through its interactions with a variety of substrates and regulatory proteins. To date, multiple DnaJ-like cofactors, such as OsP58A, OsP58B, OsERdj2, and OsERdj3B, have been identified as cochaperons interacting with OsBiP1 (Ohta et al., 2013). Only SIL1 and GRP170 have been reported to be ER luminal NEFs of BiP (Behnke et al., 2015). The expression of SIL1 is induced dramatically by ER stress in yeast, slightly in Yarrowia lipolytica, but decreased in human cells (Behnke et al., 2015). In this study, we obtained a Fes1-like protein (OsFes1C) from proteomic analysis of ER-stress response proteins (Qian et al., 2015). Our data showed that OsFes1C is an ER membrane protein (Figures 2, A–C). The expression of OsFes1C is induced by ER and salt stresses (Figure 1E). In addition, OsFes1C directly interacts with OsBiP1, and the interaction is strengthened by ADP but weakened by ATP (Figure 5), like that of previously reported NEFs. The interaction of OsFes1C with OsBiP1 is much similar to those of SIL1 to Kar2p/BiP and GRP170 to BiP (Kabani et al., 2000). These features of OsFes1C make it a potential NEF of OsBiP1 in vivo.
We tried to produce an OsFes1C knockout mutant via genomic editing strategy; however, we failed to regenerate plants. These results suggested that OsFes1C is essential for plant growth and development, lacking the protein might be lethal like that of Grp170 in mice (Mus musculus; Kitao et al., 2001). It is noteworthy that OsFes1C OE and OsFes1C Ri lines exhibited similar phenotypes with stunted growth (Figures 4, 6). The profiles of gene expression in OsFes1C OE and OsFes1C Ri lines were also similar, with only 38 among the 1,240 DEGs, compared to WT, showing opposite expression patterns between the transgenic lines (Figure 7). Nevertheless, the functions of the 38 genes were almost unclear. These results indicated that the fine tuning of OsFes1C is crucial for its function. Similar phenomena have been reported for OsBiP1 and OsDER1 in rice, in which either underexpression or OE of OsBiP1 or OsDER1 activates an ER stress response (Wakasa et al., 2011; Qian et al., 2018). It seems likely that the expression level of these ER stress response genes should be precisely controlled in rice, as ER adopts an elaborate surveillance system, known as ERQC, to maintain the protein homeostasis in ER.
It has been reported that Fes1 plays important Hsp70-independent roles in the cell by binding proteins other than Hsp70 (Kumar and Masison, 2019). The growth retardation of OsFes1C OE and OsFes1C Ri plants may be caused by loss of these functions rather than loss of OsBiP1 regulation. In addition, we also identified an OsFes1C-interacting protein other than OsBiP1. Although the protein was annotated as OsSalt, its functions were unknown. The expression of OsSalt is induced by salt stress (Supplemental Figure S5). The OsFes1C OE and Ri transgenic plants are more resistant to salt stress, suggesting that OsFec1C is a functionally diverse gene. However, no known salt response gene could be detected in the 1,240 DEGs (Figure 7). The mechanisms of how OsFes1C contributes to the various biological processes remain to be investigated.
Materials and methods
The binary vector construction and rice transformation
To construct the gene OE vector, the OsFes1C-3×FLAG fragment was inserted in the binary vector pCAMBIA1301 containing maize ubiquitin-1 promoter. To construct the RNAi vector, the specific sequence of OsFes1C was amplified and ligated to the Ubi-pTCK303 vector (Wang et al., 2004). The binary vectors were introduced into rice (O. sativa cv Kitaake) by Agrobacterium tumefaciens-mediated transformation as described (Qu et al., 2005).
All primer sequences are listed in Supplemental Table S2.
Stress treatment
To check the expression level of OsFes1C under various abiotic stresses, the seedlings of Kitaake were grown in liquid Murashige and Skoog (MS) medium for 3 weeks under normal conditions. For salt stress assays, the seedlings were treated with 200-mM NaCl solution for 3, 6, and 12 h. For heat stress assay, the plants were exposed to 42°C for 3, 6, and 12 h. For cold stress assay, the plants were transferred to a growth chamber at 4°C for 3, 6, and 12 h. For ER stress assay, the 8-d-old plants were treated with 2 mM DTT for 4 h or 5 μg mL−1 Tm for 4 h. The treated materials were harvested immediately for total RNA extraction using TRIpure Reagent (Bioteke). RT-qPCR was performed on a LightCycler system (Roche Diagnostics) as described previously (Qian et al., 2015). The primers were designed by Beacon Designer 8.0 (Supplemental Table S2).
For testing the growth of transgenic plants under Tm treatment, positive transgenic and WT plants were grown in one-half-strength MS solid medium with or without Tm for 5 d, and the concentration of Tm was 200 or 500 ng mL−1. The phenotype was photographed, the root length and shoot length were investigated for growth measurements.
To test the salt stress tolerance of transgenic plants at the seedling stage, 10-d-old positive transgenic lines and the WT were treated with 200-mM NaCl solution for 3, 4, or 5 d and allowed to recover for 1 week. Seedlings that could not grow were considered as dead (Xiong and Yang, 2003). All of these salt tolerance experiments were repeated at least three times. The phenotype was photographed, the root length and shoot length were investigated. The values are presented as the mean ± sd. Statistical analysis of the data was performed using Student’s t test to evaluate the statistical significance of differences.
Cellular fractionation and immunoblotting analysis
To isolate the soluble cytoplasm and insoluble membrane, the transgenic seedlings containing the OsFes1C-3×FLAG tag under the control of the ubiquitin promoter were generated. The fusion proteins were extracted as previously described (Duan et al., 2017). Protein concentration was measured using the Coomassie (Bradford) protein assay kit. The soluble fractions and insoluble membrane fraction were brought up to equal concentrations, and the fractions were analyzed by SDS–PAGE with 10% (w/v) polyacrylamide gels and then immunoblotted using FLAG monoclonal antibody (Sigma). Next, a 1:5,000 dilution of H+-ATPase (Agrisera) and a 1:5,000 dilution of cFBPase (Agrisera) were used as plasma membrane and cytosolic markers, respectively.
The insoluble membrane fraction was further resuspended in control buffer, high-salt buffer (1 M NaCl), alkaline buffer (100 mM Na2CO3, pH 11.5), 1% (w/v) SDS, and 1% (v/v) Triton X-100 as reported (Ren et al., 2020). After incubation for 20 min on ice, these resuspension solutions were centrifuged at 100,000g for 1 h at 4°C to obtain the pellet (P) and supernatant (S) fractions for immunoblot analysis. Antibodies against α-tubulin, His, GFP, and FLAG were generated by Sigma. Antibodies against BiP1, PDI2-3, and CNX were kindly provided by Dr Fumio Takaiwa (Wakasa et al., 2011).
Subcellular localization
To examine subcellular localization, GFP was fused to the C-terminus of OsFes1A, OsFes1B, and OsFes1C and inserted into the vector pBI221. mCherry-HDEL served as a marker for ER localization. All the fusion constructs were placed under the control of the 35S promoter and NOS terminator. Then, the chimeric plasmid was cotransformed into rice protoplasts as described (Chen et al., 2006). Fluorescence signals were observed on a confocal microscope (Olympus FV1000 MPE). The GFP fluorophore was excitation with a 488-nm laser, and emitted fluorescence was recorded at 505–525 nm. The mCherry fluorophore was excitation with a 543-nm laser, and emitted fluorescence was recorded at 560–660 nm.
Immunoelectron microscopy
Transverse sections of rice root tip 4 d after germination were fixed in PBS buffer (pH 7.2). Samples were embedded in Spurr’s low-viscosity resin and cut into ultrathin sections. For immunogold localization, ultrathin sections were blocked with 1% (w/v) bovine serum albumin in Tris-buffered saline, and incubated with diluted antibodies. Following removal of nonspecifically bound antibodies, sections were incubated with gold-conjugated secondary antibodies, stained with uranyl acetate, and observed by transmission electron microscope (JEM-1230, Hitachi, Tokyo, Japan). Polyclonal antibody against the OsFes1C was produced in mouse.
Bimolecular fluorescence complementation
The vectors used to produce constructs for the BiFC assays were pXY104 and pXY106, which carry fragments encoding the C- and N-terminal halves of YFP (cYFP and nYFP), respectively (Luo et al., 2014; Lu et al., 2015; Lian et al., 2018). PCR-amplified coding regions of OsFes1C, OsBiP1, and Salt gene were introduced into the vector. The resulting plasmid nYFP-OsFes1C and nYFP-Salt contained the N-terminal part of yellow fluorescent protein, while OsFes1C-cYFP and OsBiP1-cYFP contained the C-terminal region of yellow fluorescent protein. The vectors were transformed into A. tumefaciens strain GV3101. Equal volumes of A. tumefaciens cultures harboring each of the nYFP-OsFes1C and OsBiP1-cYFP constructs, or nYFP-Salt and OsFes1C-cYFP constructs were mixed to a final optical density (OD) at 600 nm of OD600 = 0.3 in infiltration buffer (10 mM MES, pH 5.6, 10 mM MgCl2, and 200 mM acetosyringone). The cultures were infiltrated into fully expanded N. benthamiana leaves using a 1-mL needleless syringe. The agroinfiltrated N. benthamiana plants were grown at 16-h light/8-h dark cycle for 48–72 h at 23°C. The fluorescence was detected by confocal microscopy (Leica). The GFP fluorophore was excitation with a 488-nm laser, and emitted fluorescence was recorded at 500–575 nm. The mCherry fluorophore was excitation with a 543-nm laser, and emitted fluorescence was recorded at 575–630 nm.
CoIP assays
Transgenic calli containing the OsFes1C-3×FLAG tag under the control of the ubiquitin promoter were generated. Successful transgenic calli were screened by anti-FLAG antibodies. In the CoIP assay, the WT served as a negative control; the transgenic calli and WT calli were ground and extracted by lysis buffer (25 mM Tris–HCl, pH 8, 150 mM NaCl, 2.0% [w/v] n-dodecyl-β-d-maltopyranoside, and a protease inhibitor cocktail [Roche]). Briefly, after centrifugation, the supernatant was incubated with anti-FLAG resin for 2 h at 4°C, the immunoprecipitation complexes were washed three times using wash buffer (25 mM Tris–HCl, pH 8, 150 mM NaCl, and 0.02% [w/v] n-dodecyl-β-d-maltopyranoside), and the protein complexes were eluted by 2× SDS–PAGE loading buffer and subjected to immunoblot analysis. The procedure of CoIP assay with the transgenic seedlings was the same as that mentioned above.
GST pull down
The recombinant proteins (GST-OsBiP1, GST-Salt, and OsFes1C-His) were expressed in E. coli. Briefly, all the plasmid constructs were transformed into E. coli BL21 (DE3), respectively. The transformed cells were cultured in LB broth at 37°C till OD600 = 0.8 and then induced with 0.5 mM Isopropyl β-D-Thiogalactoside (IPTG). The expression of protein was carried out at 16°C for 20 h. The induced cells were harvested by centrifugation at 4,000 g and resuspended in phosphate buffer saline (PBS), then lysed by sonication. The lysate was centrifuged at 14,000g and 4 °C for 20 min to remove precipitate.
The supernatant of OsFes1C-His recombinant protein was loaded on a Ni-IDA column (TransGen Biotech). The column was balanced with PBS containing 10 mM imidazole, washed with PBS containing 40, 60, 80, and 100 mM imidazole, eluted with PBS containing 300 mM imidazole. The fractions containing the recombinant protein were collected.
The supernatant of GST-OsBiP1 and GST-Salt was incubated with prepared glutathione Sepharose beads (TransGen Biotech), respectively, on the rotating incubator at 4°C for 2 h, and then washed four times. After removing the supernatant, 1-mL purified OsFes1C-His recombinant protein was incubated with the beads for 2 h. The beads were washed four times with PBS buffer. The target proteins were eluted with reduced glutathione. These elutes were then analyzed and detected by SDS–PAGE and western blotting.
For in vitro protein binding assay, OsFes1C-His and GST-OsBiP1 fusion proteins were purified according to the method mentioned above. A 5 mM ATP or 5 mM ADP was subjected to the process of binding.
RNA-Seq analysis
Sample preparation: the 8-d-old OsFes1C transgenic (OsFes1C OE and OsFes1C Ri) and WT plants were treated with or without 2 mM DTT for 4 h.
Total RNA was extracted from those samples using TRIzol reagent (Invitrogen), and each group was prepared with three parallel replicates. Later, all the samples were sent to BGI Corporation (Shenzhen, China) for further RNA-Seq detection and analysis via Illumina HiSeq sequencer. The Venn analysis and Cluster analysis for DEGs was performed on the Dr Tom network platform of BGI (http://report.bgi.com). The results were then confirmed by RT-qPCR.
LCI assay
For the interaction assay between OsFes1C and OsBiP1, the coding sequence of OsBiP1 was fused upstream of the N-terminus of LUC (nLUC) in the pCAMBIA-nLUC vector, while the coding sequences of OsFes1C were fused downstream of the C-terminus of LUC (cLUC) in the pCAMBIA-cLUC vector. For the interaction assay between OsFes1C and OsSalt, the coding sequence of OsSalt was fused downstream of cLUC in the pCAMBIA-cLUC vector, while the coding sequences of OsFes1C were fused upstream of the nLUC in the pCAMBIA-nLUC vector. Different construct combinations were infiltrated into different positions in the same N. benthamiana leaf. After 2 d at 23°C, the relative LUC activity was measured as previously described (Zhou et al., 2018).
Proteomics analysis
Proteomics analysis was performed as described previously (Qian et al., 2015). The plant total protein extraction kit (SIGMA) was used according to the manufacturer’s instructions, protein samples were extracted from the 12-d-old OsFes1C transgenic (OsFes1C OE and OsFes1C Ri) and WT plants. All operations were performed on ice, and the total protein was quantified using the 2D Quant Kit (GE Healthcare). SWATH analysis was performed on a TripleTOF 5600 system (AB SCIEX; Gillet et al., 2012). A total of three technical replicates and two biological replicates were performed.
Accession numbers
Sequence data from this article can be found in the Rice Genome Annotation Project Database under the following accession numbers: OsFes1A, LOC_Os03g60780; OsFes1B, LOC_Os03g16460; OsFes1C, LOC_Os09g33780; OsBiP1, LOC_Os02g02410; OsSalt, LOC_Os01g24710; PDI2-3, LOC_Os09g27830; and CNX, LOC_Os04g32950.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Prediction of the transmembrane helix of OsFes1C.
Supplemental Figure S2 . Analysis of the OsFes1C transcription levels in transgenic plants.
Supplemental Figure S3 . Analysis of the expression levels of ER-resident chaperones in callus and leaf and by western blotting.
Supplemental Figure S4 . CoIP assay of transgenic OsFes1C-FLAG and endogenous interacting proteins using transgenic OsFes1C-3×FLAG seedlings.
Supplemental Figure S5 . Analysis of the OsSalt mRNA levels in rice seedlings after treatment with 200 mM NaCl.
Supplemental Figure S6 . Subcellular localization of the OsSalt analyzed by confocal microscopy.
Supplemental Figure S7 . Analysis of the 42 differential transcription factors among the 1,240 DEGs.
Supplemental Figure S8 . Proteomics analysis of the OsFes1C OE and OsFes1C Ri transgenic plant.
Supplemental Table S1 . Mass spectrometry analysis of the proteins coprecipitated with OsFes1C-FLAG.
Supplemental Table S2 . The primers for vector construction.
Supplemental Data Set S1 . List of the 3,214 differential genes between OE and WT.
Supplemental Data Set S2 . List of the 3,370 differential genes between Ri and WT.
Supplemental Data Set S3 . List of the 1,240 genes that changed both in OE and Ri lines.
Supplemental Data Set S4 . List of the 42 transcription factors among the 1,240 DEGs.
Supplemental Data Set S5 . List of the 83 differentially expressed proteins both in OsFes1C OE and Ri plants.
Supplementary Material
Acknowledgments
We thank Ms Fengqin Dong and Dr Zhuang Lu from Plant Science Facility of the Institute of Botany, Chinese Academy of Sciences for their technical assistances in IEM and MS/MS.
Funding
This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDA24010400), the National Program of Transgenic Variety Development of China (grant no. 2016ZX08001-006 to L.Q.Q.), and the Natural Science Foundation of China (grant no. 31801267 to D.Q.).
Conflict of interest statement. The authors declare that there is no conflict of interest.
These authors contributed equally to the article (L.T. and L.Q.Q.).
D.Q., L.T., and L.Q.Q. designed the research and analyzed the data; D.Q. performed most the experiments; L.T. performed cellular fractionation and immunoelectron microscopy experiments; S.X., M.L., and L.T. provided technical assistance to D.Q.; D.Q., L.T., and L.Q.Q. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Author (https://academic.oup.com/plphys/pages/general-instructions) is: Le Qing Qu (lqqu@ibcas.ac.cn).
References
- Behnke J, Feige MJ, Hendershot LM (2015) BiP and its nucleotide exchange factors Grp170 and Sil1: mechanisms of action and biological functions. J Mol Biol 427: 1589–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Hendershot LM (2014) The large Hsp70 Grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional Hsp70s. J Biol Chem 289: 2899–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher A, Verghese J (2015) GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones. Subcell Biochem 78: 1–33 [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366 [DOI] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- de Keyzer J, Steel GJ, Hale SJ, Humphries D, Stirling CJ (2009) Nucleotide binding by Lhs1p is essential for its nucleotide exchange activity and for function in vivo. J Biol Chem 284: 31564–31571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS (2006) A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126: 349–359 [DOI] [PubMed] [Google Scholar]
- Duan M, Zhang R, Zhu F, Zhang Z, Gou L, Wen J, Dong J, Wang T (2017) A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 29: 1748–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 11: O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda NK, Kaimal JM, Masser AE, Kang W, Friedlander MR, Andreasson C (2016) Cytosolic splice isoform of Hsp70 nucleotide exchange factor Fes1 is required for the degradation of misfolded proteins in yeast. Mol Biol Cell 27: 1210–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda NK, Kandasamy G, Froehlich MS, Dohmen RJ, Andreasson C (2013) Hsp70 nucleotide exchange factor Fes1 is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins. Proc Natl Acad Sci USA 110: 5975–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Beckerich JM, Gaillardin C (2000) Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol Cell Biol 20: 6923–6934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL (2002) HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett 531: 339–342 [DOI] [PubMed] [Google Scholar]
- Kitao Y, Ozawa K, Miyazaki M, Tamatani M, Kobayashi T, Yanagi H, Okabe M, Ikawa M, Yamashima T, Stern DM, et al. (2001) Expression of the endoplasmic reticulum molecular chaperone (ORP150) rescues hippocampal neurons from glutamate toxicity. J Clin Invest 108: 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B, Braakman I (2004) Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol 16: 343–349 [DOI] [PubMed] [Google Scholar]
- Kumar S, Masison DC (2019) Hsp70-nucleotide exchange factor (NEF) Fes1 has non-NEF roles in degradation of gluconeogenic enzymes and cell wall integrity. PLoS Genet 15: e1008219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H, Xu P, He S, Wu J, Pan J, Wang W, Xu F, Wang S, Huang J, Yang HQ (2018) Photoexcited CRYPTOCHROME 1 interacts directly with G-protein beta subunit AGB1 to regulate the DNA-binding activity of HY5 and photomorphogenesis in Arabidopsis. Mol Plant 11: 1248–1263 [DOI] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Luo Q, Lian HL, He SB, Li L, Jia KP, Yang HQ (2014) COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26: 2441–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maattanen P, Gehring K, Bergeron JJ, Thomas DY (2010) Protein quality control in the ER: the recognition of misfolded proteins. Semin Cell Dev Biol 21: 500–511 [DOI] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL (2003) Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD). Bioessays 25: 868–877 [DOI] [PubMed] [Google Scholar]
- Ohta M, Wakasa Y, Takahashi H, Hayashi S, Kudo K, Takaiwa F (2013) Analysis of rice ER-resident J-proteins reveals diversity and functional differentiation of the ER-resident Hsp70 system in plants. J Exp Bot 64: 5429–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou C, Hendershot LM (2020) Disposing of misfolded ER proteins: a troubled substrate's way out of the ER. Mol Cell Endocrinol 500: 110630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D, Tian L, Qu LQ (2015) Proteomic analysis of endoplasmic reticulum stress responses in rice seeds. Sci Rep 5: 14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D, Chen G, Tian L, Qu LQ (2018) OsDER1 is an ER-associated protein degradation factor that responds to ER stress. Plant Physiol 178: 402–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LQ, Yoshihara T, Ooyama A, Goto F, Takaiwa F (2005) Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222: 225–233 [DOI] [PubMed] [Google Scholar]
- Ren Y, Wang Y, Pan T, Gan L, Wei Z, Wang F, Wu M, Jing R, Wang J, Wan G, et al. (2020) GPA5 encodes a Rab5a effector required for post-Golgi trafficking of rice storage proteins. Plant Cell 32: 758–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B (2019) The Hsp70 chaperone network. Nat Rev Mol Cell Biol 20: 665–680 [DOI] [PubMed] [Google Scholar]
- Schilke BA, Ciesielski SJ, Ziegelhoffer T, Kamiya E, Tonelli M, Lee W, Cornilescu G, Hines JK, Markley JL, Craig EA (2017) Broadening the functionality of a J-protein/Hsp70 molecular chaperone system. PLoS Genet 13: e1007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Savanur MA, Sinha D, Birje A, Vigneshwaran R, Saha PP, D'Silva P (2017) Regulation of mitochondrial protein import by the nucleotide exchange factors GrpEL1 and GrpEL2 in human cells. J Biol Chem 292: 18075–18090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R (2018) Protein quality control in the endoplasmic reticulum of plants. Annu Rev Plant Biol 69: 147–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasa Y, Yasuda H, Oono Y, Kawakatsu T, Hirose S, Takahashi H, Hayashi S, Yang L, Takaiwa F (2011) Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J 65: 675–689 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L). Plant Mol Biol Rep 22: 409–417 [Google Scholar]
- Winter J, Jakob U (2004) Beyond transcription—new mechanisms for the regulation of molecular chaperones. Crit Rev Biochem Mol Biol 39: 297–317 [DOI] [PubMed] [Google Scholar]
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H, Hirose S, Kawakatsu T, Wakasa Y, Takaiwa F (2009) Overexpression of BiP has inhibitory effects on the accumulation of seed storage proteins in endosperm cells of rice. Plant Cell Physiol 50: 1532–1543 [DOI] [PubMed] [Google Scholar]
- Zhang JX, Wang C, Yang CY, Wang JY, Chen L, Bao XM, Zhao YX, Zhang H, Liu J (2010) The role of Arabidopsis AtFes1A in cytosolic Hsp70 stability and abiotic stress tolerance. Plant J 62: 539–548 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Bi G, Zhou JM (2018) Luciferase complementation assay for protein-protein interactions in plants. Curr Protoc Plant Biol 3: 42–50 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.