Abstract
Arabidopsis (Arabidopsis thaliana) CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) and members of the SUPPRESSOR OF PHYTOCHROMEA-105 (SPA) protein family form an E3 ubiquitin ligase that suppresses light signaling in darkness by polyubiquitinating positive regulators of the light response. COP1/SPA is inactivated by light to allow photomorphogenesis to proceed. Mechanisms of inactivation include light-induced degradation of SPA1 and, in particular, SPA2, corresponding to a particularly efficient inactivation of COP1/SPA2 by light. Here, we show that SPA3 and SPA4 proteins are stable in the light, indicating that light-induced destabilization is specific to SPA1 and SPA2, possibly related to the predominant function of SPA1 and SPA2 in dark-grown etiolating seedlings. SPA2 degradation involves cullin and the COP10-DEETIOLATED-DAMAGED-DNA BINDING PROTEIN (DDB1) CDD complex, besides COP1. Consistent with this finding, light-induced SPA2 degradation required the DDB1-interacting Trp-Asp (WD)-repeat domain of SPA2. Deletion of the N-terminus of SPA2 containing the kinase domain led to strong stabilization of SPA2 in darkness and fully abolished light-induced degradation of SPA2. This prevented seedling de-etiolation even in very strong far-red and blue light and reduced de-etiolation in red light, indicating destabilization of SPA2 through its N-terminal domain is essential for light response. SPA2 is exclusively destabilized by phytochrome A in far-red and blue light. However, deletion of the N-terminal domain of SPA2 did not abolish SPA2-phytochrome A interaction in yeast nor in vivo. Our domain mapping suggests there are two SPA2-phytochrome A interacting domains, the N-terminal domain and the WD-repeat domain. Conferring a light-induced SPA2-phyA interaction only via the WD-repeat domain may thus not lead to COP1/SPA2 inactivation.
Light inactivates the COP1/SPA2 repressor of photomorphogenesis through cullin- and CDD-mediated degradation of SPA2, whereas the family members SPA3 and SPA4 are stable in the light.
Introduction
Plants constantly adjust their growth, development, and metabolism to the ambient light environment and seasons. This includes seed germination, seedling de-etiolation, the shade avoidance response, and photoperiodic induction of flowering. To sense the light, plants have evolved several classes of photoreceptors. In particular, phytochromes and cryptochromes are responsible for seedling de-etiolation in red (R), far-red (FR), and blue light (B). In Arabidopsis (Arabidopsis thaliana), phytochromes are encoded by a small gene family comprising five genes. Phytochrome B (phyB) is the most important phytochrome active in continuous R (Rc), while phytochrome A (phyA) is the sole photoreceptor that initiates responses in continuous FR (FRc). Blue light is sensed by phyA as well as by the two cryptochromes cry1 and cry2 (Kami et al., 2010).
In darkness, light signaling is actively suppressed by three repressor systems, the PHYTOCHROME-INTERACTING FACTORS (PIFs), the CONSTITUTIVE PHOTOMORPHOGENESIS 1/SUPPRESSOR OF PHYTOCHROME A-105 (COP1/SPA) complex, and the COP10-DE-ETIOLATED1 (DET1)-DAMAGED DNA-BINDING PROTEIN (DDB1) (CDD) complex. Mutations in any of the three components cause constitutive photomorphogenesis in darkness, with seedlings exhibiting features of light-grown seedlings in complete darkness (Huang et al., 2014; Hoecker, 2017; Favero, 2020). The COP1/SPA complex acts as a CULLIN4 (CUL4)-dependent E3 ubiquitin ligase that polyubiquitinates positive regulators of the light response, mainly transcription factors, in darkness, thereby causing their degradation in the 26S proteasome. In light-grown plants, the light-activated phytochromes and cryptochromes directly bind to the COP1/SPA complex, leading to the inactivation of the COP1/SPA E3 ubiquitin ligase activity and subsequent stabilization of a number of transcription factors, such as ELONGATED HYPOCOTYL5 (HY5), which promotes seedling de-etiolation, CONSTANS (CO), which is involved in photoperiodic flowering, or PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1) and PAP2, which activate anthocyanin biosynthesis (Osterlund et al., 2000; Laubinger et al., 2006; Jang et al., 2008; Liu et al., 2008; Maier et al., 2013). The extent of COP1/SPA inactivation positively corresponds to the fluence rate of light and also involves polyubiquitination of photoreceptors, in particular phyA and cry2, as a negative feedback mechanism (Seo et al., 2004; Weidler et al., 2012; Debrieux et al., 2013).
The COP1/SPA complex is a tetramer consisting of two COP1 and two SPA proteins, with any combination of the four SPA proteins (SPA1–SPA4) possible (Zhu et al., 2008). Both, COP1 and SPA proteins are necessary for the activity of the COP1/SPA complex. This is evidenced by the finding that both cop1 and spa quadruple mutants undergo constitutive photomorphogenesis in darkness (Deng et al., 1991; Laubinger et al., 2004; Ordonez-Herrera et al., 2015). The four SPA genes have overlapping but also partially distinct functions during growth and development (Menon et al., 2016). SPA1 and SPA2 are the primary SPA genes responsible for suppression of photomorphogenesis in dark-grown seedlings. In light-grown seedlings, SPA1, SPA3, and SPA4, but not SPA2, prevent overstimulation by light. SPA3 and SPA4 mainly regulate leaf expansion, while SPA1 and SPA4 are the primary regulators of photoperiodic flowering (Laubinger et al., 2004, 2006; Fittinghoff et al., 2006).
The COP1 protein carries three functional domains, an N-terminal RING-finger domain, which likely is responsible for recruiting ubiquitin-conjugated E2 for in vitro ubiquitination assays, a central coiled-coil domain responsible for COP1 homodimerization and COP1/SPA heterodimerization, and a C-terminal WD-repeat domain which binds substrates as well as DDB1 of the CUL4-DDB1-RBX1 scaffold (Deng et al., 1992; Huang et al., 2014; Uljon et al., 2016). SPA proteins also contain a WD-repeat domain closely related to the WD-repeat of COP1 and with apparent similar functions. They carry a coiled-coil domain responsible for the COP1/SPA complex formation and an N-terminal kinase domain with weak sequence similarity to Ser/Thr protein kinases (Hoecker et al., 1999; Hoecker, 2017). Recently, it was shown that SPA1 exhibits kinase activity towards PIF1 (Paik et al., 2019).
Light inactivates the COP1/SPA complex via at least four distinct mechanisms (Podolec and Ulm, 2018; Ponnu, 2020). It causes nuclear exclusion of COP1, disruption of the COP1–SPA1 interaction by the photoreceptors phyA, phyB, and cry1, cry-mediated competitive displacement of substrates from COP1, and by destabilization of SPA1 and SPA2 proteins. SPA2 stands out from the other SPAs because it is most efficiently inactivated by light when compared to the other three SPAs. A spa1 spa3 spa4 triple mutant retaining only SPA2 function etiolates like wild-type in darkness but resembles a spa quadruple mutant once exposed to even extremely low fluence rates of light (Laubinger et al., 2004; Balcerowicz et al., 2011). This effective light-induced inactivation of SPA2 corresponds to a very rapid degradation of SPA2 in light-exposed seedlings (Balcerowicz et al., 2011). A short pulse of light with a very low fluence rate is sufficient to cause degradation of SPA2. Rapid SPA2 degradation is dependent on phytochromes but independent of cryptochromes, even in B (Chen et al., 2015). This indicates that light-induced SPA2 degradation is specifically mediated by phytochrome photoreceptors. SPA2 destabilization requires COP1 and the COP1-interacting coiled-coil domain of SPA2, suggesting that COP1 is the ubiquitin ligase responsible for ubiquitinating SPA2 in light-grown plants (Chen et al., 2015). SPA1 is destabilized by light as well but much less effectively than SPA2 (Balcerowicz et al., 2011). Moreover, the light-induced degradation of SPA1 is counteracted by a light-induced increase in SPA1 expression, while the transcript levels of SPA2 are not affected by light (Hoecker et al., 1999; Fittinghoff et al., 2006). These observations are consistent with the lower rate of SPA1 inactivation by light when compared to SPA2, as evidenced from the phenotype of spa2 spa3 spa4 and spa1 spa3 spa4 mutants (Balcerowicz et al., 2011). Domain swap approaches have shown that the distinct protein stabilities of SPA1 and SPA2 as well as the retained repressor activity in light-grown seedlings maps to the N-terminal domains of SPA1 and SPA2 (Chen et al., 2016). Indeed, the sequence divergence between SPA1 and SPA2 is highest in their respective N-terminal domains (Laubinger and Hoecker, 2003).
Here, we have addressed the mechanism of light-induced SPA2 degradation by examining the roles of E3 ligase components and specific domains in the SPA2 protein as well as their consequence on SPA2 activity in darkness and in the light. Moreover, we analyzed the protein stability of SPA3 and SPA4 to test whether there is further functional specificity among SPAs with respect to their stability in the light.
Results
SPA2 instability depends on CUL4, CSN, and the CDD complex
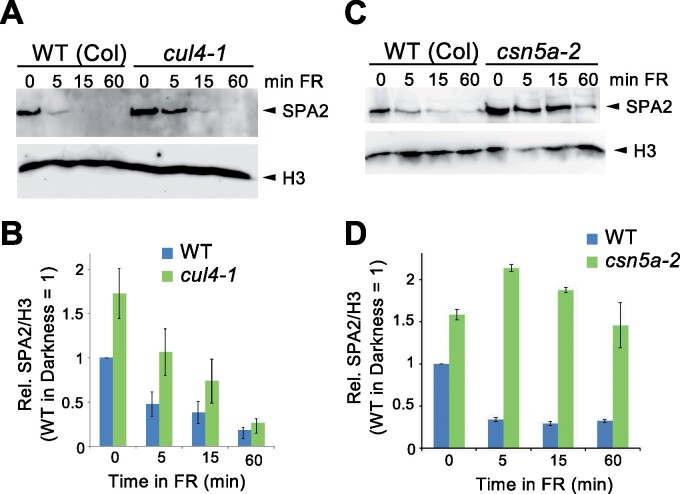
We have shown previously that light-induced degradation of SPA2 requires COP1 and the COP1-interacting coiled-coil domain of SPA2 (Chen et al., 2015), suggesting that COP1 is the E3 ubiquitin ligase responsible for SPA2 degradation in the light. In vivo, COP1 acts as a CUL4-based E3 ubiquitin ligase towards its transcription factor targets (Chen et al., 2010). However, in vitro, COP1 is capable of ubiquitinating targets without the addition of CUL4 complex components; its RING finger domain is sufficient to recruit E2 (Saijo et al., 2003). Hence, it cannot be excluded that there are substrates of COP1 that do not require the CUL4 scaffold. We therefore investigated whether SPA2 degradation requires CUL4 and the COP9 SIGNALOSOME (CSN), a complex that regulates the activity of cullins (Schwechheimer and Isono, 2010). SPA2 protein levels were higher in the hypomorphic cul4-1 mutant than in the wild-type. This was most evident in dark-grown seedlings, while light-induced degradation of SPA2 was still observed in cul4-1 (Figure 1, A and B). In the csn5a mutant which is defective in a core component of the CSN (Jin et al., 2014), SPA2 protein levels were also higher in dark-grown seedlings when compared to the wild-type. In addition, SPA2 protein levels remained high upon exposure to FR, indicating that light-induced degradation was strongly impaired in the absence of CSN activity (Figure 1, C and D). Taken together, these results indicate that normal SPA2 destabilization requires cullin activity in the light as well as in darkness. The observation that light-induced degradation of SPA2 was relatively normal in cul4-1 may be due to the hypomorphic nature of the cul4-1 allele (Bernhardt et al., 2006). In agreement with this interpretation, cul4-1 mutants exhibit only very weak constitutive photomorphogenesis in darkness (Bernhardt et al., 2006).
Figure 1.
Normal SPA2 destabilization depends on CUL4 and CSN5. A and C, SPA2 protein levels in WT and cul4-1 (A) and csn5a-2 (C) mutant seedlings that were grown in darkness for 4 d and then transferred to far-red light (FR) (0.35 µmol m−2 s−1) for the indicated time. SPA2 was detected in nuclear-enriched extracts using α-SPA2 antibodies. Histone H3 (H3) detected by specific antibodies was used as loading control. B and D, Quantification of relative SPA2/H3 protein levels. SPA2/H3 levels in the dark-grown WT was set to 1. Error bars indicate the SEM of three (B) or two (D) independent experiments.
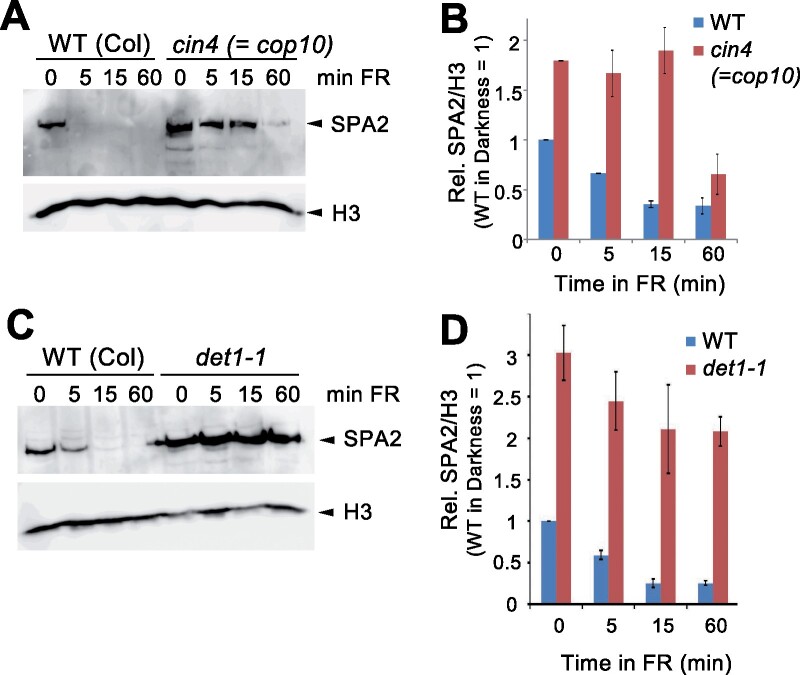
In addition to COP1, the CDD complex is required for the suppression of photomorphogenesis in darkness (Lau and Deng, 2012). We therefore determined SPA2 protein levels in cop10 and det1-1 mutants, which are defective in two components of the CDD complex. SPA2 levels were higher in dark-grown and FR-exposed cop10 mutants when compared to the wild-type. In particular, light-induced degradation of SPA2 occurred much more slowly in cop10 mutants than in the wild-type (Figure 2, A and B). To an even larger extent, this observation was also made in the det1-1 mutant: SPA2 protein levels were much higher in dark-grown det1-1 mutant seedlings when compared to wild-type seedlings and degradation of SPA2 was almost absent upon exposure to FR (Figure 2, C and D). These results indicate that the CDD complex destabilizes SPA2 in darkness and is required for normal light-induced SPA2 degradation.
Figure 2.
Normal SPA2 destabilization depends on COP10 and DET1. A and C, SPA2 protein levels in WT and cop10 (A) and det1-1 (C) mutant seedlings that were grown in darkness for 4 d and then transferred to far-red light (FR) (0.35 µmol m−2 s−1) for the indicated time. SPA2 was detected in nuclear-enriched extracts using α-SPA2 antibodies. Histone H3 (H3) detected by specific antibodies was used as loading control. B and D, Quantification of relative SPA2/H3 protein levels. SPA2/H3 levels in the dark-grown WT was set to 1. Error bars indicate the SEM of two independent experiments.
The WD-repeat domain and the N-terminal domain of SPA2 are required for light-induced degradation of SPA2
Since the interaction of SPA proteins with the CUL4–DDB1–RBX1 complex depends on an intact WD-repeat domain of SPAs (Chen et al., 2010), we asked whether the WD-repeat of SPA2 is required for light-induced SPA2 degradation. To this end, we expressed SPA2ΔWD lacking the WD-repeat under the control of the native SPA2 promoter and 3′-untranslated region (UTR) in transgenic spa1 spa2 spa3 mutant plants. The SPA2 promoter is constitutive and not affected by light (Fittinghoff et al., 2006; Balcerowicz et al., 2011). For protein detection, an HA tag was added to the SPA2 coding sequence (Figure 3A). As expected, SPA2ΔWD-HA did not complement the spa1 spa2 spa3 mutant phenotype, while the expression of full-length SPA2 restored seedling etiolation in darkness (Figure 3B; Balcerowicz et al., 2011). The SPA2ΔWD-HA protein was stable after exposure to light, while full-length SPA2-HA was undetectable upon short exposure to light (Figure 3C). Thus, light-induced degradation of SPA2 requires the WD-repeat of SPA2.
Figure 3.
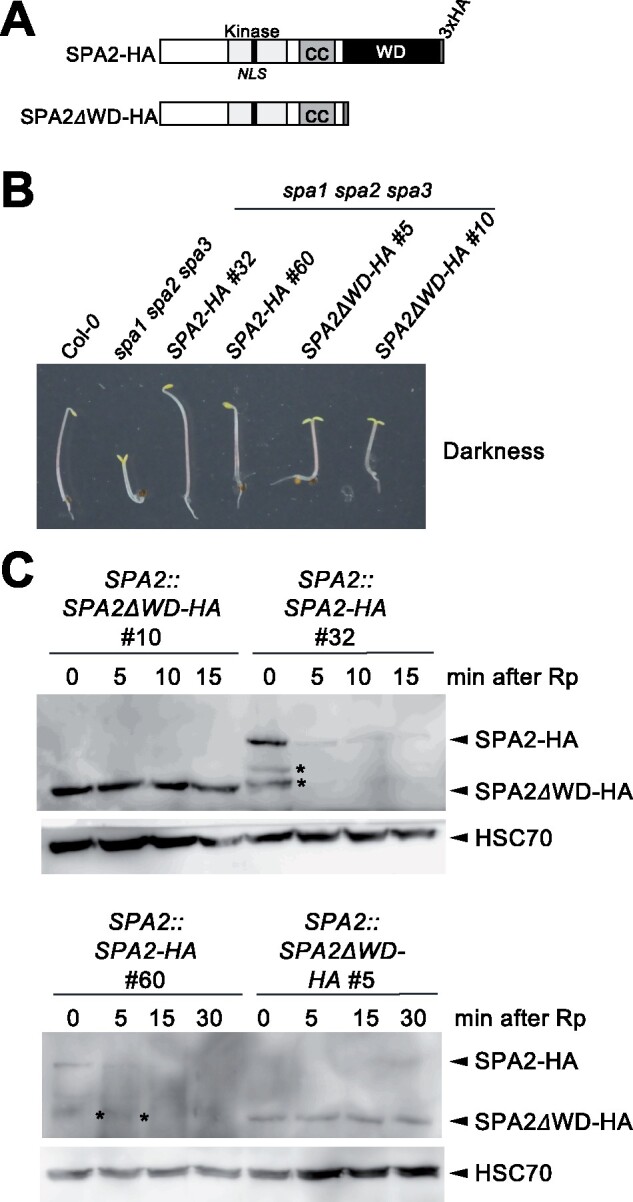
The WD-repeat domain of SPA2 is necessary for the light-induced degradation of SPA2. A, Schematic representation of SPA2-HA and SPA2ΔWD-HA. Both were expressed under the control of the SPA2 promoter in the spa1 spa2 spa3 mutant. B, Representative spa1 spa2 spa3 mutant seedlings expressing SPA2-HA or SPA2ΔWD-HA. Seedlings were grown in darkness for 5 d. C, SPA2-HA or SPA2ΔWD-HA protein levels of transgenic seedlings grown in darkness for 4 d and subsequently exposed to a pulse of red light (Rp) of 30 µmol m−2 s−1 for 200 s followed by darkness for the indicated period of time. SPA2-HA proteins were detected using α-HA antibodies. HSC70 detected by specific antibodies was used as loading control. Asterisks indicate SPA2-HA degradation products.
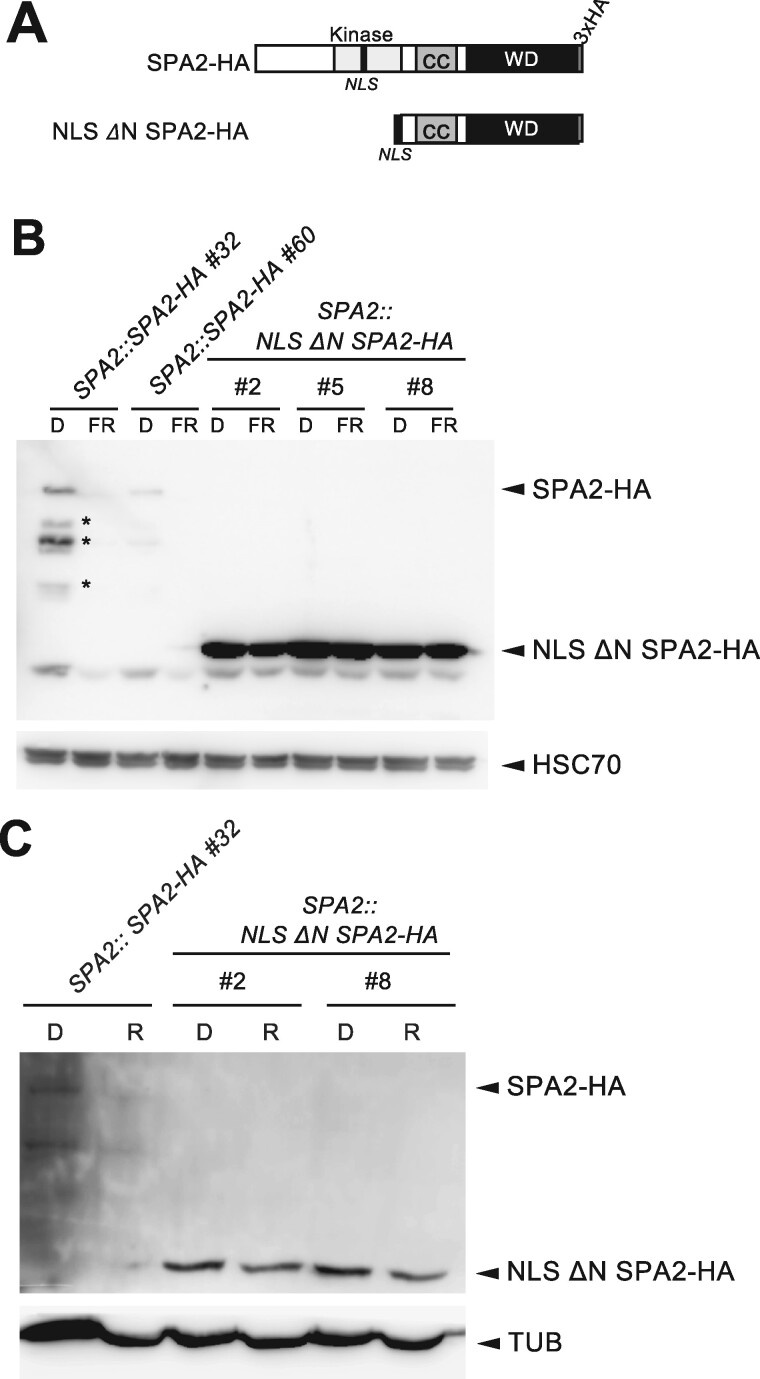
We also examined whether the N-terminal kinase domain of SPA2 is involved in SPA2 stability. We therefore expressed ΔN SPA2-HA lacking the complete N-terminal domain—but retaining the coiled-coil domain in transgenic seedlings. An artificial nuclear localization sequence (NLS) was fused to ΔN SPA2 to replace the missing NLS located in the N-terminal domain of full-length SPA2 (Figure 4A). NLS ΔN SPA2-HA accumulated to much higher levels than full-length SPA2-HA in dark-grown as well as in seedlings grown in FRc or Rc (Figure 4, B and C). In particular, no light-induced degradation of NLS ΔN SPA2-HA was observed. Even very high fluence rates of 5 µmol m−2 s−1 FRc or 30 µmol m−2 s−1 Rc over a period of 4 d did not destabilize NLS ΔN SPA2-HA (Figure 4, B and C). These results demonstrate that the N-terminal domain of SPA2 is required for light-induced degradation of SPA2 as well as for destabilization of SPA2 in darkness.
Figure 4.
The N-terminal kinase domain of SPA2 is necessary for the light-induced degradation of SPA2. A, Schematic representation of SPA2-HA and NLS ΔN SPA2-HA. Both were expressed under the control of the SPA2 promoter in transgenic spa1 spa2 spa3 mutants. An artificial NLS was fused to ΔN SPA2 to replace the missing NLS located in the N-terminal domain of full-length SPA2. B and C, SPA2-HA and NLS ΔN SPA2-HA protein levels in transgenic seedlings grown in darkness (D) or continuous far-red light (FRc) (5 µmol m−2 s−1) (B) or R (30 µmol m−2 s−1) (C) for 4 d. SPA2-HA was detected using α-HA antibodies. HSC70 (B) or tubulin (TUB) (C) detected by specific antibodies were used as loading controls. Asterisks indicate SPA2-HA degradation products.
Seedlings expressing NLS ΔN SPA2 are insensitive to FR and B and exhibit a reduced sensitivity to R
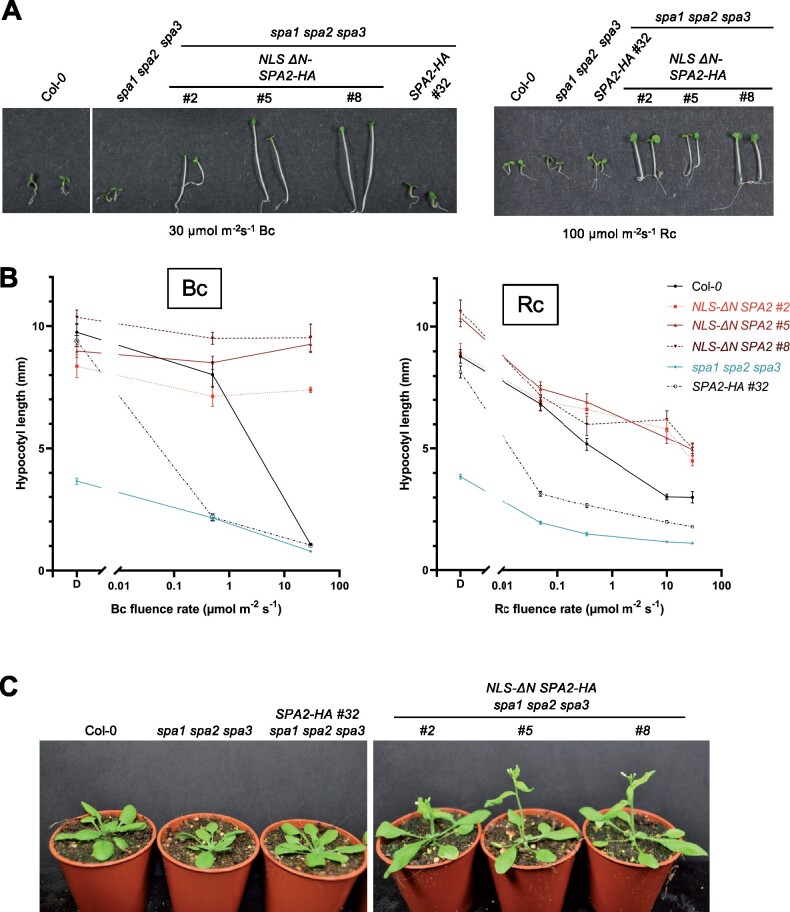
To assess the activity of the NLS ΔN SPA2-HA protein as a repressor of photomorphogenesis, we analyzed seedling growth. NLS ΔN SPA2-HA is expressed in the spa1 spa2 spa3 mutant, which shows constitutive photomorphogenesis in darkness (Laubinger et al., 2004). Transgenic spa1 spa2 spa3 mutant seedlings expressing NLS ΔN SPA2-HA under the control of the native SPA2 promoter showed full complementation of the spa1 spa2 spa3 mutant phenotype in the dark: transgenic seedlings fully etiolated and exhibited the same phenotype as the wild-type and spa1 spa2 spa3 seedlings expressing full-length SPA2-HA (Figure 5, A and B). When grown in FRc, NLS ΔN SPA2-HA seedlings did not de-etiolate but retained a hypocotyl length similar to dark-grown seedlings. Even at very high fluence rates of FRc (30 µmol m−2 s−1), these seedlings remained in a mostly etiolated state with only slightly opened cotyledons (Figure 5, A and B). In contrast, spa1 spa2 spa3 seedlings expressing full-length SPA2-HA were very responsive to FRc, fully de-etiolating already at very low FRc fluence rates, as also reported previously (Balcerowicz et al., 2011; Chen et al., 2016).
Figure 5.
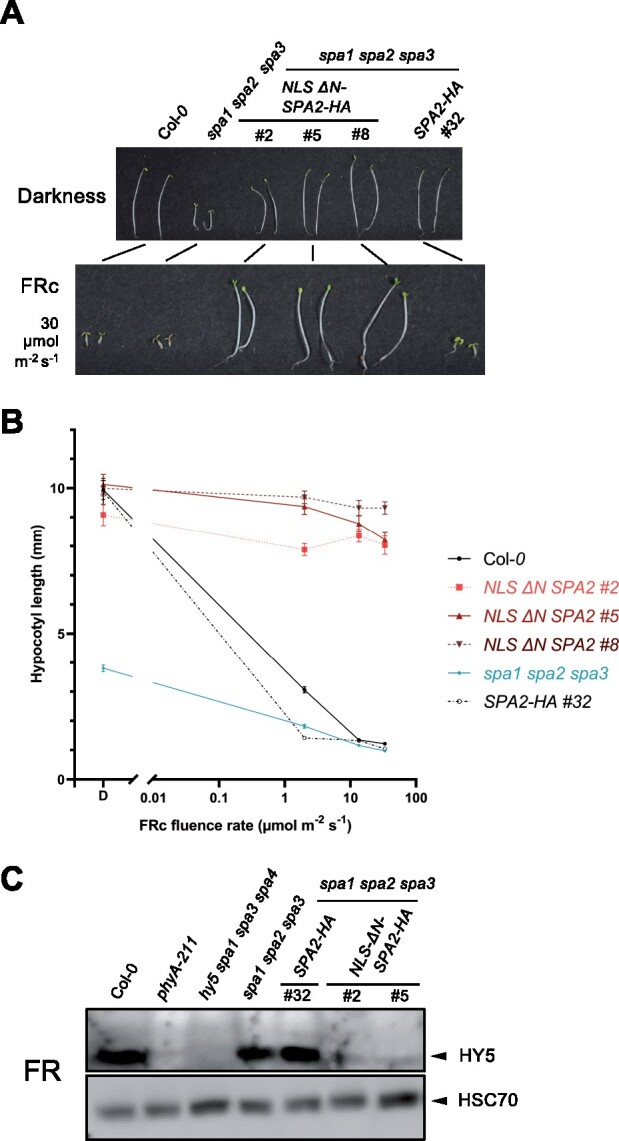
Seedlings expressing NLS-ΔN SPA2 are insensitive to far-red light. A, Visual phenotype of the indicated genotypes grown in darkness or in 30 μmol m−2 s−1 continuous far-red light (FRc) for 4 d. B, Hypocotyl length of seedlings of the indicated genotypes grown under various fluence rates of FRc for 4 d. SPA2-HA and NLS ΔN SPA2-HA seedlings are in the spa1 spa2 spa3 mutant background. Error bars indicate the sem. n= 9–25 seedlings. C, Immunodetection of HY5 protein levels. Seedlings were grown in darkness for 4 d and then shifted to FRc (1 µmol m−2 s−1) for 6 h. HYS was detected using α-HY5 antibody. HSC70 levels, detected by α-HSC70, are shown as a loading control. All transgenes were expressed under the control of the constitutive SPA2 promoter. ΔN SPA2 carried an artificial NLS to replace the missing NLS located in the N-terminal domain of full-length SPA2.
To assess a molecular phenotype, we determined HY5 protein levels in FR-exposed seedlings. HY5 did not accumulate in spa1 spa2 spa3 mutant seedlings expressing NLS ΔN SPA2-HA (Figure 5C) which agrees with the etiolated phenotype of these seedlings when grown in FRc. spa1 spa2 spa3 seedlings expressing full-length SPA2-HA, in contrast, accumulated HY5 protein. Similarly, other de-etiolating genotypes, such as the wild-type and the spa1 spa2 spa3 mutant, exhibited high HY5 protein levels. The FR-insensitive phyA mutant and the hy5 mutant did not accumulate any detectable HY5 protein, as expected (Figure 5C). These results indicate that NLS ΔN SPA2 prevents a light-induced accumulation of HY5 by causing constitutive degradation of HY5, even in the light. Taken together, these results demonstrate that ΔN SPA2 is an active repressor of photomorphogenesis, which cannot be inactivated by FRc. Thus, the N-terminal domain of SPA2 is required for FR-induced inactivation of SPA2.
When grown in Bc, NLS ΔN SPA2-expressing seedlings failed to de-etiolate as well (Figure 6, A and B). The hypocotyl length at high Bc fluence rates was similar to that of dark-grown seedlings. Only hook and cotyledons opened in response to very high Bc (Figure 6A). In Rc, NLS-ΔN SPA2 seedlings de-etiolated, but with a much reduced sensitivity when compared to full-length SPA2-HA-expressing seedlings (Figure 6, A and B). At very high Rc fluence rates of 100 µmol m−2 s−1, expression of ΔN SPA2 allowed only partial de-etiolation while spa1 spa2 spa3, wild-type and spa1 spa2 spa3 mutants expressing full-length SPA2-HA exhibited maximum de-etiolation with an extremely short hypocotyl. In summary, deletion of the N-terminal domain of SPA2 caused strong insensitivity to FRc as well as Bc and a reduction in the sensitivity to Rc.
Figure 6.
Transgenic lines expressing NLS-ΔN SPA2 exhibit reduced sensitivity to red and blue light and flower early. A, Visual phenotype of the indicated genotypes grown in continuous blue (Bc) or red light (Rc) for 4 d. B, Hypocotyl length of seedlings of the indicated genotypes grown under various fluence rates of Bc or Rc for 4 d. SPA2-HA and NLS-ΔN SPA2-HA were expressed under the native SPA2 promoter in the spa1 spa2 spa3 mutant background. Error bars indicate the sem. n = 9–24 seedlings. C, Visual phenotype of the indicated genotypes grown in long day.
Adult plants expressing ΔN SPA2 mimic a phyB mutant
Adult plants expressing NLS ΔN SPA2 exhibited elongated petioles and slightly earlier flowering when compared to the wild-type, the progenitor spa1 spa2 spa3 and spa1 spa2 spa3 mutants expressing full-length SPA2 (Figure 6C;Supplemental Figure S1, A–C). Adult plants of the progenitor spa1 spa2 spa3 behave almost like the wild-type due to the activity of SPA4 (Laubinger et al., 2004; Ordonez-Herrera et al., 2015). Therefore, expression of full-length SPA2 in spa1 spa2 spa3 is not expected to alter this phenotype. In contrast, NLS ΔN SPA2 caused a phenotype that is very similar to that observed in phyB mutant plants (Reed et al., 1993; Supplemental Figure S1, A–C). Hence, it might reflect negative regulation of phyB signaling.
Domain mapping suggests two interacting domains between SPA2 and phyA
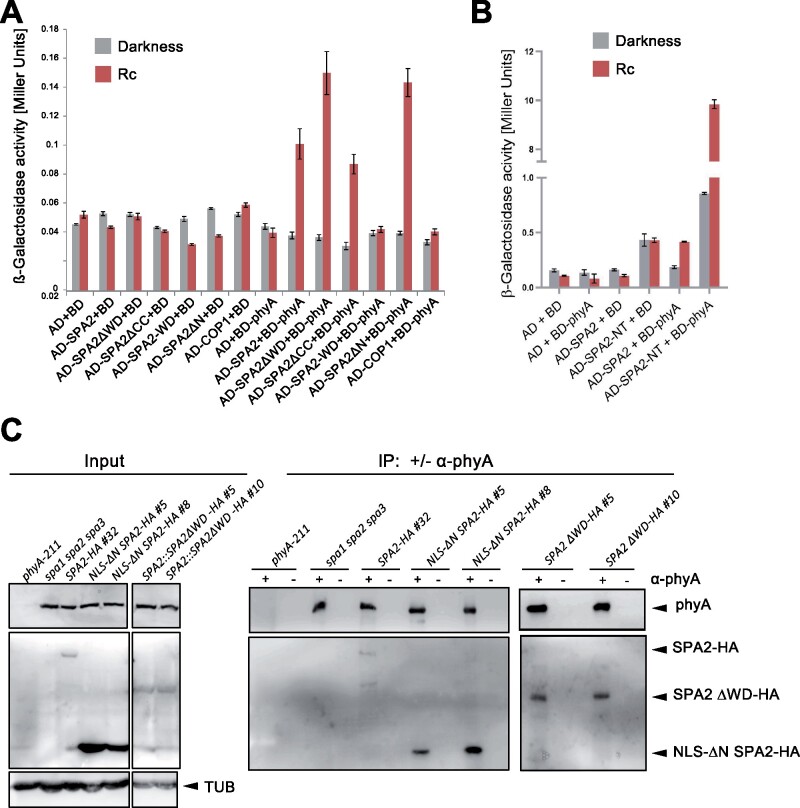
We have shown previously that phyA is the primary photoreceptor initiating SPA2 degradation in FR, R, and B (Chen et al., 2015). Since phyA is also a substrate of COP1/SPA (Seo et al., 2004; Debrieux et al., 2013), we first tested whether phyA accumulates in NLS ΔN SPA2-expressing seedlings. Immunoblot analysis showed that phyA levels were not dramatically altered in NLS ΔN SPA2-expressing seedlings when compared to seedlings expressing full-length SPA2 (Supplemental Figure S2). Hence, the FR-insensitivity of NLS ΔN SPA2-expressing seedlings is not due to a failure to accumulate phyA. We therefore asked whether NLS ΔN SPA2 may be incapable of interacting with phyA. To this end, we conducted a yeast two-hybrid assay using various deletion-derivatives of SPA2. Figure 7A shows that phyA interacted with full-length SPA2, but also with NLS ΔN SPA2 and SPA2ΔWD, in an R-dependent fashion. SPA2 lacking a coiled-coil domain also interacted with phyA. These results indicate that none of the three SPA2 domains is essential for an interaction with phyA. In conclusion, there are likely two phyA-interacting domains in SPA2, the N-terminal domain as well as the WD repeat domain. Indeed, the N-terminal domain of SPA2 comprising the kinase-like domain but lacking the coiled-coil domain was sufficient to interact with phyA. It interacted much more strongly in yeast than full-length SPA2 (Figure 7, A and B). However, the WD-repeat domain was not sufficient to interact with phyA since WD-SPA2 showed no interaction with phyA (Figure 7A). Hence, intra- or intermolecular cooperativity may be needed for WD-SPA2 to interact with phyA in the yeast two-hybrid assay.
Figure 7.
The N-terminal and WD-repeat domains of SPA2 are not necessary for the SPA2-phyA interaction in yeast two-hybrid and in vivo. A and B, Yeast two-hybrid assays expressing the indicated proteins. Yeast cells were grown in darkness or in 1 μmol m−2 s−1 continuous red light (Rc). Error bars indicate the sem. C, Co-immunoprecipitation of SPA2-HA and SPA2-HA deletion proteins by phyA. Seedlings of the indicated genotypes were grown in darkness for 4 d, incubated in MG132 for 3 h to prevent SPA2-HA degradation and then transferred to 5 μmol m−2s−1 Rc for 10 min to allow nuclear accumulation of phyA. After further 10 min in darkness, phyA was immunoprecipitated using protein-A beads with (+) or without (−) a coupled phyA antibody. Co-immunoprecipitated SPA2-HA or SPA2-HA deletion proteins were detected using an α-HA antibody. phyA was detected using an α-phyA antibody. Tubulin (TUB) served as a loading control for the input samples. All input samples were run on the same gel with one lane removed. The IP samples were run on two separate gels.
We subsequently performed in vivo co-immunoprecipitation experiments using the SPA2-HA lines described above. phyA was used as the bait by immunoprecipitating it from extracts of R-exposed seedlings using α-phyA-coupled beads (Figure 7C). Successful and specific immunoprecipitation was demonstrated by immunodetection of phyA in all samples except for the phyA-211 mutant. phyA co-immunoprecipitated full-length SPA2-HA, NLS ΔN SPA2-HA, as well as SPA2-ΔWD-HA. No co-immunoprecipitation of these proteins was observed in samples lacking the α-phyA antibody. These results confirm the observations from the yeast two-hybrid experiment, indicating that neither the N-terminal domain nor the WD-repeat domain of SPA2 is essential for an association of SPA2 with phyA. Taken together, these results indicate that the FR-insensitivity of NLS ΔN SPA2-expressing seedlings is not due to a failure to bind phyA. However, it is possible that the phyA-ΔN SPA2 interaction is nonproductive with respect to light-induced degradation and inactivation of SPA2.
SPA3 and SPA4 are light-stable proteins
We had observed previously that SPA1 and SPA2 differ in their light-induced destabilization, with SPA2 being much less stable than SPA1 (Balcerowicz et al., 2011; Chen et al., 2016). To extend the analysis to SPA3 and SPA4 we expressed HA-tagged SPA3 and SPA4 under the control of the light-independent SPA2 promoter in transgenic plants, as we had done before for SPA1-HA and SPA2-HA (Balcerowicz et al., 2011). SPA3-HA and SPA4-HA fusion proteins were functional as indicated by the increased hypocotyl elongation observed in the transgenic seedlings (Figure 8A). Figure 8B shows that SPA3-HA and SPA4-HA protein levels did not change when dark-grown seedlings were exposed to FR. SPA1-HA and SPA2-HA levels, in contrast, dramatically decreased upon FR irradiation (Balcerowicz et al., 2011; Figure 4). These results indicate that SPA3 and SPA4 are light-stable proteins and, therefore, differ from SPA1 and SPA2.
Figure 8.
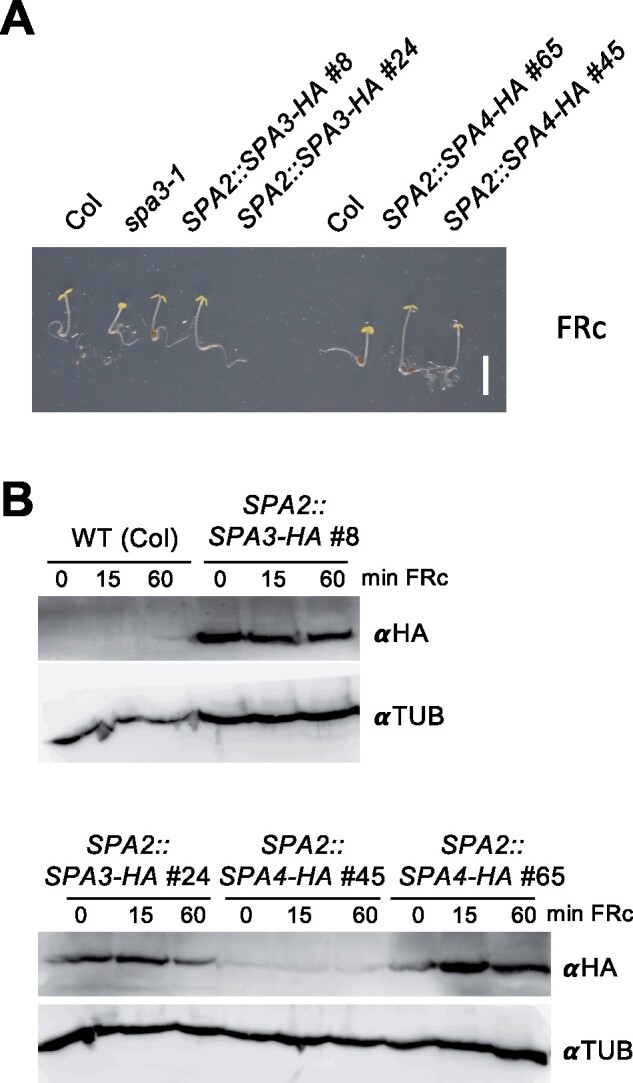
SPA3 and SPA4 are light-stable proteins. A, Phenotype of seedlings expressing SPA3-HA or SPA4-HA under the control of the constitutive SPA2 promoter. SPA3-HA lines were in the spa3-1 background, SPA4-HA lines were in the Col background. Seedlings were grown in 0.5 µmol m−2 s−1 continuous far-red (FRc) light for 4 d. Scale bar indicates 5 mm. B, SPA3-HA or SPA4-HA levels in seedlings of the indicated genotypes. Seedlings were grown in darkness for three days and subsequently transferred to 1 µmol m−2 s−1 FRc for 15 or 60 min. SPA3-HA and SPA4-HA were detected using an αHA antibody. Tubulin (αTUB) detected by a specific antibody was used as a loading control.
Discussion
The four SPA proteins act in complexes with COP1 to suppress photomorphogenesis in dark-exposed plants. While COP1 also exists in animals, SPA genes are limited to the green lineage (Han et al., 2019), suggesting that SPAs may be key to the light-induced inhibition of COP1/SPA activity found in plants. Indeed, SPA proteins are intricate to the light regulation of COP1/SPA activity: they dissociate from COP1 in response to light (Lian et al., 2011; Liu et al., 2011; Lu et al., 2015; Sheerin et al., 2015), they are required for light-induced nuclear exclusion of COP1 (Balcerowicz et al., 2017) and for an in vivo association of COP1 with CRY1 in blue light (Holtkotte et al., 2017). Moreover, SPA1 and SPA2 are rapidly destabilized in response to light. SPA2, in particular, is a highly instable protein in light-grown seedlings (Balcerowicz et al., 2011).
We have shown previously that COP1 is the likely E3 ubiquitin ligase responsible for the SPA2 degradation in the light (Chen et al., 2015). COP1 might act on SPA2 via two possible ways: via its own RING finger domain which can recruit ubiquitin-charged E2 or as part of the CUL4–DDB1–RBX1 complex in which the RING finger protein RBX1 recruits E2-ubiquitin. We found that cul4 and csn5 mutations impair SPA2 degradation, suggesting that SPA2, like transcription factor targets of COP1, is destabilized by a CUL4-DDB1-RBX1COP1/SPA2 E3 ubiquitin ligase. Mutations in the CDD complex also strongly increased SPA2 levels. This indicates that the CDD complex is also involved in SPA2 degradation. A similar result was recently observed for COP1 (Cañibano et al., 2020). Hence, a CUL4-dependent CDD E3 ubiquitin ligase may directly polyubiquitinate COP1 and SPA2, in concert with COP1. Alternatively, the CDD complex might indirectly enhance the activity of COP1 via a thus far unknown mechanism. spa1 and det1 mutations were also shown to act synergistically in regulating photomorphogenesis and HY5 degradation (Nixdorf and Hoecker, 2010). Mutations in CSN5, DET1, and COP10 clearly impaired the light-induced degradation of SPA2, but also stabilized SPA2 in dark-grown seedling, though to a much lesser extent. A similar observation was made in the cop1-4 mutant (Chen et al., 2015). Hence, light-exposure enhanced the activity of these E3 ligases towards SPA2 while at the same time reducing the E3 ligase activity toward the transcription factor targets of the COP1/SPA E3 ligase.
All three domains of SPA2 were required for light-induced degradation of SPA2, i.e. the levels of SPA2 deletion-derivatives lacking either the N-terminal domain, the coiled-coil domain, or the WD-repeat domain were not responsive to light. The WD-repeat domain confers binding to DDB1 (Chen et al., 2010); hence it may be required due to its interaction with the CUL4–DDB1–RBX1 complex. The coiled-coil domain is required for COP1 interaction and is, therefore, likely required (Chen et al., 2015). A lack of the N-terminal domain very strongly stabilized SPA2 in dark-grown and light-grown seedlings when compared to full-length SPA2. ΔN SPA2 was present at very high levels in light-grown seedlings, corresponding to the failure to accumulate HY5. Similarly, ΔN SPA2-expressing seedlings did not de-etiolate in response to FR and B, even at very high fluence rates, and showed a reduced de-etiolation in R. These results indicate that a tight downregulation of SPA2 protein stability is essential for light responsiveness. ΔN SPA1 also accumulated to higher levels than full-length SPA1 in dark- as well as light-grown seedlings, though a direct comparison between light- and dark-grown seedlings has not been conducted so far (Fittinghoff et al., 2006; Yang and Wang, 2006; Holtkotte et al., 2016). However, in contrast to the mostly light-unresponsive ΔN SPA2-expresssing seedlings, ΔN SPA1-expressing seedlings de-etiolated in the light. Hence, it would be interesting to analyze whether ΔN SPA1 levels are reduced upon light-exposure. The exact role of the N-terminal domain in SPA proteins is thus far unknown, but it was recently shown to exhibit kinase activity (Paik et al., 2019). Whether the kinase activity of the N-terminal domain regulates SPA1 and SPA2 protein stability remains to be tested. However, missense mutations in the predicted kinase domain did not change SPA1 protein stability, suggesting that the kinase activity per se is not involved (Holtkotte et al., 2016).
We hypothesized that ΔN SPA2 is not degraded in response to light because it may fail to bind photoreceptors. We therefore investigated the interaction with phyA, the main photoreceptor controlling SPA2 degradation under all light conditions (Chen et al., 2015). ΔN SPA2 interacted with phyA in the yeast two-hybrid system as well as in vivo. SPA2ΔWD also retained an interaction with phyA, indicating that two domains of SPA2 interact with phyA, the N-terminal domain and the WD-repeat domain. However, NT-SPA1, but not WD-SPA1, was sufficient to interact with phyA in yeast and, moreover, NT-SPA1 exhibited a very strong interaction with phyA in yeast. Hence, although deletion of the N-terminal domain of SPA2 did not abolish an interaction with phyA, it might play a primary role in binding phyA in the light. Hence, the deletion of the N-terminus of SPA2 might cause a non-productive interaction that does not lead to SPA2 degradation and subsequent light response. Interestingly, two previous studies obtained seemingly contrasting results, defining the N-terminal domain or the WD-repeat domain of SPA1, respectively, as the phyA-interacting domain using yeast two-hybrid assays (Lu et al., 2015; Sheerin et al., 2015). This supports the idea that there are two interaction hubs between SPAs and phyA.
We showed here that SPA3 and SPA4 are light-stable proteins and thus behave very differently from SPA1 and SPA2. Hence, following gene duplication during evolution the four SPAs diverged with respect to protein stability. This may have facilitated fine-tuning of the diverse light responses to environmental conditions, in particular with respect to the rapidity of response to sudden light exposure. SPA1 and SPA2 have the most important activities during seedling etiolation in darkness, which needs to be terminated quickly once a seedling grows out of the soil to allow rapid opening of cotyledons and the onset of photosynthesis. SPA3 and SPA4, in contrast, mainly function in the expansion of true leaves, which likely does not require light responses within minutes. SPA3 and SPA4 proteins are very similar in sequence (74% sequence identity) and form a subclass distinct from SPA1 and SPA2 (Laubinger and Hoecker, 2003). To determine the ancestral regulation of SPA protein stability, i.e. whether light-induced instability was gained or lost following gene duplications, evolutionarily more ancient SPA proteins would need to be analyzed. Domain-swap analyses between SPA1 and SPA2 mapped the distinct protein stability of SPA1 and SPA2 mainly to the respective N-terminal domain (Chen et al., 2016). Indeed, the N-termini of SPA3 and SPA4 share only very weak sequence similarity with those of SPA1 and SPA2, while the four WD-repeat domains are very similar. In addition, the N-terminal domains of SPA3 and SPA4 are approximately 200 amino acids smaller than those of SPA1 and SPA2 (Laubinger and Hoecker, 2003). Hence, the divergence in the N-terminal sequence of the SPA3/SPA4 pair may be responsible for the light-stable behavior of SPA3 and SPA4. Domain swap experiments between SPA2 and SPA3 or SPA4 can test this hypothesis in the future.
Materials and methods
Plant material, growth conditions, and phenotypic analysis
The Arabidopsis (Arabidopsis thaliana) mutants spa1-7 spa2-1 spa3-1 (Fittinghoff et al., 2006), cul4-1 (Bernhardt et al., 2006), csn5a-2 (Dohmann et al., 2005), det1-1 (Chory et al., 1989), cin4 (Vogel et al., 1998) were described previously. All mutants are in the Col accession. The transgenic lines expressing full-length SPA2-HA under the control of SPA2 promoter and 3′-UTR (SPA2::SPA2-HA) were described in Balcerowicz et al. (2011). SPA2::NLSΔN SPA2-HA expresses an artificial NLS, amino acids 462 until the C-terminus of SPA2 followed by a triple HA tag. SPA2::SPA2ΔWD-HA expresses amino acids 1–720 followed by a triple HA tag. Both SPA2 deletion-proteins are expressed in the spa1-7 spa2-1 spa3-1 mutant background and under the control of the same SPA2 promoter and 3′-UTR as full-length SPA2-HA. SPA3-HA and SPA4-HA fusion proteins were expressed in the spa3-1 and Col wild-type background, respectively, under the control of the constitutive SPA2 promoter and 3′-UTR. Details on the construction of the vectors used for Arabidopsis transformation are described in the Supplemental Materials and Methods. Primer sequences are provided in Supplemental Table S1.
Seedlings and plants were grown and their phenotypes were analyzed as described in Laubinger et al. (2004,, 2006). LED light sources (CLF Plant Climatics, Wertingen, Germany) were used with a λmax of 670 nm (R), 745 nm (FR), and 470 nm (B).
Immunoblot analyses
Nuclear-enriched fractions for SPA2 detection using α-SPA2 antibodies were obtained as described in Cheng et al. (2009). α-SPA2 antibodies were described previously (Maier et al., 2013). α-histone H3 antibodies (Abcam, Cambridge, MA, USA) were used as loading controls. For detection of HA-tagged proteins, total protein was extracted from seedlings and quantified as described in Chen et al. (2015), except that no MG132 was added to the extraction buffer. For the detection of HY5 and phyA protein levels, total protein was extracted from seedlings using 2× sodium dodecyl sulphate (SDS) buffer (0.125 M Tris–HCl [pH 6.8], 4% (w/v) SDS, 20% glycerol, 1× cocktail of protease inhibitors, and 1-mM phenylmethylsulfonyl fluoride (PMSF)) and quantified via bicinchoninic acid (BCA) assay (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific) as described in Ponnu et al. (2019). HY5 was detected using α-HY5 antibodies. phyA was detected using α-phyA antibodies (Hirschfeld et al., 1998). α-HA-HRP antibodies (Roche, Mannheim, Germany) were used for the detection of HA-tagged proteins. α-HSC70 (Stressgen Biotechnologies, San Diego, USA) or α–α-tubulin (Sigma-Aldrich) antibodies were used as loading controls. All immunoblots were performed according to standard procedures.
Co-immunoprecipitation
Four-day-old seedlings were pretreated with 50-μM MG132 for 5 h and treated with 5-μmol m−2 s−1 R for 10 min followed by 10-min dark incubation. Total protein was isolated using extraction buffer (50-mM Tris–HCl pH 8.0, 150-mM NaCl, 10% glycerol, 1-mM DTT, 25-mM β-glycerophosphate, 0.1% Nonidet P-40, 1-mM PMSF, 50-μM MG132, 1× protease inhibitor cocktail (Sigma-Aldrich)). Total protein lysate (0.5–1 mg) was incubated with 50-μL protein A magnetic μMACSTM MicroBeads (Miltenyi Biotec, Germany) and 1-μL α-phyA antibody (Agrisera) in a microtube on a rotator at 4°C for 2 h. After incubation, it was transferred onto μ columns that had been preincubated with extraction buffer. The columns were washed four times with 1-mL ice cold extraction buffer containing 0.2% Nonidet P-40. Bound proteins were eluted according to manufacturer’s protocol using 50-μL pre-warmed 1× Laemmli buffer. Input and co-immunoprecipitated fractions were separated by SDS–polyacrylamide gel electrophoresis; proteins were detected using α-phyA and α-HA antibodies.
Yeast two-hybrid assays
BD-PHYA (D153AH-phyA; Hiltbrunner et al., 2006) and respective AD plasmids were co-transformed into yeast (Saccharomyces cerevisiae) strain Y187. Transformed colonies were selected on drop-out medium lacking Leu and Trp. Four to five transformed single colonies were picked and suspended in 1-mL sterile water. Three biological replicates for each transformation were prepared. The yeast suspensions were diluted to an optical density OD600 of 0.1 and plated in spots of 10 μL on drop-out plates lacking leucine and tryptophan, supplemented with 20-μM phycocyanobilin (Livchem Logistics, Frankfurt a.M., Germany). Plates were incubated at 26°C in constant R (1 μmol m−2s−1) or in darkness for 2 d. After incubation, the colonies were collected and resuspended in ice cold sterile water. For each biological replicate two technical replicates were generated. LacZ activity was measured using o-Nitrophenyl-β-D-galactopyarnoside (ONPG) as a substrate as described previously (Laubinger and Hoecker, 2003).
Accession numbers
SPA2 (At4g11110), SPA3 (At3g15354), SPA4 (At1g53090), SPA1 (At2g46340), COP1 (At2g32950), phyA (At1g09570), HY5 (At5g11260), CUL4 (At5g46210), CSN5a (At5g46210), COP10 (At5g46210), DET1 (At5g46210).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transgenic lines expressing NLS-ΔN SPA2 exhibit longer petioles and early flowering.
Supplemental Figure S2. Immunodetection of phyA protein levels.
Supplemental Materials and Methods. Plasmid constructions.
Supplemental Table 1. Oligonucleotides.
Supplementary Material
Acknowledgments
We are grateful to the Arabidopsis Biological Resource Center for providing seed, to Hanjo Hellmann for providing cul4-1 seed, to Peter Quail and Xing-Wang Deng for the gifts of a phyA and HY5 antibody, respectively. We thank Andreas Hiltbrunner and David Sheerin for the gift of the D153AH-phyA plasmid and help with the yeast two-hybrid protocol. We thank Klaus Menrath, his green house staff and many undergraduate students for expert care of our plants.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG) HO2793/3-3 and under Germanýs Excellence Strategy EXC 2048/1, project ID: 390686111 to U.H.
Conflict of interest statement. The authors declare no conflict of interest.
T.S. and U.H. planned the original research, T.S., S.C., L.T., and C.S. performed the experiments. All authors discussed the results. U.H. and T.S. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Ute Hoecker (hoeckeru@uni-koeln.de).
References
- Balcerowicz M, Fittinghoff K, Wirthmueller L, Maier A, Fackendahl P, Fiene G, Koncz C, Hoecker U (2011) Light exposure of Arabidopsis seedlings causes rapid de-stabilization as well as selective post-translational inactivation of the repressor of photomorphogenesis SPA2. Plant J 65: 712–723 [DOI] [PubMed] [Google Scholar]
- Balcerowicz M, Kerner K, Schenkel C, Hoecker U (2017) SPA proteins affect the subcellular localization of COP1 in the COP1/SPA ubiquitin ligase complex during photomorphogenesis. Plant Physiol 174: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt A, Lechner E, Hano P, Schade V, Dieterle M, Anders M, Dubin MJ, Benvenuto G, Bowler C, Genschik P, et al. (2006) CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J 47: 591–603 [DOI] [PubMed] [Google Scholar]
- Cañibano E, Bourbousse C, Garcia-Leon M, Wolff L, Garcia-Baudino C, Barneche F, Rubio V, Fonseca S (2020) DET1-mediated COP1 regulation avoids HY5 activity over second-site targets to tune plant photomorphogenesis. bioRxiv: 2020.2009.2030.318253 [DOI] [PubMed]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al. (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lory N, Stauber J, Hoecker U (2015) Photoreceptor specificity in the light-induced and COP1-mediated rapid degradation of the repressor of photomorphogenesis SPA2 in Arabidopsis. PLoS Genet 11: e1005516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wirthmueller L, Stauber J, Lory N, Holtkotte X, Leson L, Schenkel C, Ahmad M, Hoecker U (2016) The functional divergence between SPA1 and SPA2 in Arabidopsis photomorphogenesis maps primarily to the respective N-terminal kinase-like domain. BMC Plant Biol 16: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YT, Germain H, Wiermer M, Bi D, Xu F, García AV, Wirthmueller L, Després C, Parker JE, Zhang Y, et al. (2009) Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F (1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991–999 [DOI] [PubMed] [Google Scholar]
- Debrieux D, Trevisan M, Fankhauser C (2013) Conditional involvement of CONSTITUTIVE PHOTOMORPHOGENIC1 in the degradation of phytochrome A. Plant Physiol 161: 2136–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X-W, Caspar T, Quail PH (1991) cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev 5: 1172–1182 [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Dohmann EM, Kuhnle C, Schwechheimer C (2005) Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17: 1967–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero DS (2020) Mechanisms regulating PIF transcription factor activity at the protein level. Physiol Plant 169: 325–335 [DOI] [PubMed] [Google Scholar]
- Fittinghoff K, Laubinger S, Nixdorf M, Fackendahl P, Baumgardt RL, Batschauer A, Hoecker U (2006) Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J 47: 577–590 [DOI] [PubMed] [Google Scholar]
- Han X, Chang X, Zhang Z, Chen H, He H, Zhong B, Deng XW (2019) Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol Plant 12: 847–862 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Tscheuschler A, Viczián A, Kunkel T, Kircher S, Schäfer E (2006) FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA (1998) Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U (2017) The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr Opin Plant Biol 37: 63–69 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH (1999) SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496–499 [DOI] [PubMed] [Google Scholar]
- Holtkotte X, Dieterle S, Kokkelink L, Artz O, Leson L, Fittinghoff K, Hayama R, Ahmad M, Hoecker U (2016) Mutations in the N-terminal kinase-like domain of the repressor of photomorphogenesis SPA1 severely impair SPA1 function but not light responsiveness in Arabidopsis. Plant J 88: 205–218 [DOI] [PubMed] [Google Scholar]
- Holtkotte X, Ponnu J, Ahmad M, Hoecker U (2017) The blue light-induced interaction of cryptochrome 1 with COP1 requires SPA proteins during Arabidopsis light signaling. PLoS Genet 13: e1007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Deng XW (2014) Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr Opin Plant Biol 21C: 96–103 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, Li B, Deng XW, Wei N (2014) Plant COP9 signalosome subunit 5, CSN5. Plant Sci 224: 54–61 [DOI] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Hoecker U (2003) The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J 35: 373–385 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222 [DOI] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ (2015) Red light-dependent interaction of phyB with SPA1 promotes COP1–SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, Rubio V, Uhrig JF, Hulskamp M, Hoecker U (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74: 638–651 [DOI] [PubMed] [Google Scholar]
- Menon C, Sheerin DJ, Hiltbrunner A (2016) SPA proteins: SPAnning the gap between visible light and gene expression. Planta 244: 297–312 [DOI] [PubMed] [Google Scholar]
- Nixdorf M, Hoecker U (2010) SPA1 and DET1 act together to control photomorphogenesis throughout plant development. Planta 231: 825–833 [DOI] [PubMed] [Google Scholar]
- Ordonez-Herrera N, Fackendahl P, Yu X, Schaefer S, Koncz C, Hoecker U (2015) A cop1 spa mutant deficient in COP1 and SPA proteins reveals partial co-action of COP1 and SPA during Arabidopsis post-embryonic development and photomorphogenesis. Mol Plant 8: 479–481 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Paik I, Chen F, Ngoc Pham V, Zhu L, Kim JI, Huq E (2019) A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat Commun 10: 4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec R, Ulm R (2018) Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr Opin Plant Biol 45: 18–25 [DOI] [PubMed] [Google Scholar]
- Ponnu J, Riedel T, Penner E, Schrader A, Hoecker U (2019) Cryptochrome 2 competes with COP1-substrates to repress COP1 ubiqultin ligase activity during Arabidopsis photomorphogenesis. Proc Natl Acaid Sci USA 116: 27133–27141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnu J (2020) Molecular mechanisms suppressing COP1/SPA E3 ubiquitin ligase activity in blue light. Physiol Plant 169: 418–429 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Isono E (2010) The COP9 signalosome and its role in plant development. Eur J Cell Biol 89: 157–162 [DOI] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uljon S, Xu X, Durzynska I, Stein S, Adelmant G, Marto JA, Pear WS, Blacklow SC (2016) Structural basis for substrate selectivity of the E3 ligase COP1. Structure 24: 687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Schuerman P, Woeste K, Brandstatter I, Kieber JJ (1998) Isolation and characterization of Arabidopsis mutants defective in the induction of ethylene biosynthesis by cytokinin. Genetics 149: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidler G, Zur Oven-Krockhaus S, Heunemann M, Orth C, Schleifenbaum F, Harter K, Hoecker U, Batschauer A (2012) Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A. Plant Cell 24: 2610–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang H (2006) The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J 47: 564–576 [DOI] [PubMed] [Google Scholar]
- Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW (2008) Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.