Abstract
Objective
Chaperonin‐containing tailless complex polypeptide 1 subunit 6A (CCT6A) is reported to be an efficient prognostic biomarker in various cancers, but it is rarely reported in astrocytoma. Thus, this study aimed to evaluate the expression of CCT6A and its correlation with disease features and prognosis in astrocytoma patients.
Methods
Totally, 198 astrocytoma patients who received surgery treatment were enrolled. CCT6A protein expression was determined in the tumor tissues fixed in formalin and embedded in paraffin (FFEP) by immunohistochemistry (IHC) assay. In addition, 133 out of 198 astrocytoma patients had fresh tumor tissues frozen in the liquid nitrogen for the determination of CCT6A mRNA expression by reverse transcription‐quantitative polymerase chain reaction.
Results
Sixty‐nine (34.8%), 70 (35.4%), 46 (23.2%), and 13 (6.6%) astrocytoma patients had the CCT6A immunohistochemistry (IHC) score of 0–3, 4–6, 7–9, and 10–12, respectively. CCT6A protein expression was correlated with increased World Health Organization (WHO) grade (P < 0.001) and less isocitrate dehydrogenase (IDH) mutation (P = 0.002); meanwhile, CCT6A mRNA expression was only related to elevated WHO grade (P = 0.001). However, CCT6A protein and mRNA expression were not correlated with other clinical features and subsequent treatment modalities (all P > 0.05). Moreover, CCT6A protein high and CCT6A mRNA high were related to shorter accumulating overall survival (OS; both P < 0.05). CCT6A protein high was an independent factor for predicting the worse OS (hazard ratio: 1.821, P = 0.012).
Conclusion
Chaperonin‐containing tailless complex polypeptide 1 subunit 6A correlates with elevated WHO grade and less IDH mutation; besides, CCT6A high expression is independently associated with unfavorable accumulating OS of astrocytoma patients.
Keywords: astrocytoma, chaperonin‐containing tailless complex polypeptide 1 subunit 6A, isocitrate dehydrogenase mutation, overall survival, World Health Organization grade
Chaperonin‐containing tailless complex polypeptide 1 subunit 6A (CCT6A) is reported to be an efficient prognostic biomarker in various cancers, but it is rarely reported in astrocytoma. Thus, this study aimed to evaluate the expression of CCT6A and its correlation with disease features and prognosis in astrocytoma patients. Totally, 198 astrocytoma patients who received surgery treatment were enrolled. CCT6A protein expression was determined in the tumor tissues fixed in formalin and embedded in paraffin (FFEP) by immunohistochemistry (IHC) assay. In addition, 133 out of 198 astrocytoma patients had fresh tumor tissues frozen in the liquid nitrogen for the determination of CCT6A mRNA expression by reverse transcription‐quantitative polymerase chain reaction. Sixty‐nine (34.8%), 70 (35.4%), 46 (23.2%), and 13 (6.6%) astrocytoma patients had the CCT6A immunohistochemistry (IHC) score of 0–3, 4–6, 7–9, and 10–12, respectively. CCT6A protein expression was correlated with increased World Health Organization (WHO) grade (P < 0.001) and less isocitrate dehydrogenase (IDH) mutation (P = 0.002); meanwhile, CCT6A mRNA expression was only related to elevated WHO grade (P = 0.001). However, CCT6A protein and mRNA expression were not correlated with other clinical features and subsequent treatment modalities (all P > 0.05). Moreover, CCT6A protein high and CCT6A mRNA high were related to shorter accumulating overall survival (OS; both P < 0.05). CCT6A protein high was an independent factor for predicting the worse OS (hazard ratio: 1.821, P = 0.012). CCT6A correlates with elevated WHO grade and less IDH mutation; besides, CCT6A high expression is independently associated with unfavorable accumulating OS of astrocytoma patients.
![]()
1. INTRODUCTION
Astrocytoma, one of the most common and aggressive type of primary brain tumors in adults, is a fatal malignant tumor with natural resistance to many drugs due to the inability of anti‐tumor drugs in crossing the blood‐brain barrier.1, 2 Although resection with or without following radiotherapy and chemotherapy remains the leading treatment modality in general astrocytoma patients, the prognosis of astrocytoma patients is diverse according to classification issued by the World Health Organization (WHO), of which, astrocytoma patients with low grade (WHO grade I‐II) have a better survival outcome with a 5‐year survival of 63.8%–65%,3, 4, 5 while high‐grade (WHO grade III‐IV) astrocytoma patients are accompanied with a worse clinical outcome with a 5‐year survival rate of less than 5%.6 Even though great efforts have been given in exploring the biomarkers for prognosis of astrocytoma patients such as isocitrate dehydrogenase (IDH) mutation and deletion of 1p/19q, etc.,7, 8 the urgent needs of finding additional biomarkers for evaluating disease status, predicting prognosis of astrocytoma, and further realizing its individualized treatment are still not met.
Chaperonin‐containing tailless complex polypeptide 1 subunit 6A (CCT6A) is one out of eight different subunits of chaperonin‐containing tailless complex polypeptide 1 (CCT), which is involved in folding actins and tubulins, then further participating in the regulation of proliferation, migration, invasion, and cell cycle of various cancer cells.9, 10, 11, 12 For instance, in hepatocellular carcinoma, CCT6A overexpression leads to cancer cell growth via affecting the G1 to S phase transition9; besides, another study discloses that CCT6A promotes the metastasis of non‐small‐cell lung carcinoma (NSCLC) cell lines via suppressing SMAD2 and TGF‐β signaling pathway.12 Moreover, recent studies have also explicated the prognosis value of CCT6A in various tumors including hepatocellular carcinoma, breast cancer, Ewing sarcoma, and especially in glioblastoma.13, 14, 15 For instance, in glioblastoma which is the most advanced astrocytoma, CCT6A is highly expressed and shows a negative association with survival.16 However, the value of CCT6A severing as a potent biomarker for prognostication in astrocytoma is still unclear. This study aimed to evaluate the expression of CCT6A and its correlation with disease features and prognosis in astrocytoma patients.
2. MATERIALS AND METHODS
2.1. Patients
This study was a retrospective cohort study. Between January 2016 and December 2020, 198 astrocytoma patients who received surgery treatment in our hospitals were retrospectively reviewed in this study. Patients conforming to the inclusion criteria were included as follows: (1) diagnosis of astrocytoma in accordance with the criteria issued by World Health Organization (WHO)17; (2) age within 18–80 years; (3) received the resection treatment; (4) integrated pre‐surgery clinical information and follow‐up data; and (5) had tumor tissues fixed in formalin and embedded in paraffin (FFEP). Patients were excluded whether they had received the neoadjuvant therapy, or had a history of malignant tumor, radiotherapy, or chemotherapy for other malignant tumors before the resection. The Institutional Review Board approved this study, and the written informed consents were collected.
2.2. Characteristics and specimen's collection
Characteristics such as age, and gender were obtained from the electric database; besides, the follow‐up data, adjuvant radiotherapy, or chemotherapy after surgery and overall survival (OS) were also recorded. The FFPE tumor tissues were collected for further evaluation of CCT6A protein expression by immunohistochemistry (IHC) assay. Furthermore, 133 out of 198 astrocytoma patients had fresh tumor tissues frozen in the liquid nitrogen, which were also collected for the further determination of CCT6A mRNA expression by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR).
2.3. Immunohistochemistry assay
Immunohistochemistry (IHC) assay was used for the detection of the CCT6A protein expression in tumor tissues of 198 astrocytoma patients. In brief, the primary antibody used in IHC was the CCT6A Polyclonal Antibody (Catalog #PA5‐101926, Invitrogen), which was diluted into 1:200, and the secondary antibody was the Goat anti‐Rabbit IgG (H+L; Catalog #31466, Invitrogen), which was diluted into 1:60. After the FFPE tissues been deparaffinized and hydrated, the microwave method was applied to retrieve the heat‐induced antigen epitope; meanwhile, EDTA antigen retrieval solution was added to the samples, and they were incubated for 20 min. Then, hydrogen peroxide was added, followed by being incubated with primary antibody at 4°C overnight. Subsequently, a secondary antibody was added in the samples, and they were incubated at 37°C for 60 min. The diaminobenzidine (DAB; Sigma‐Aldrich) was used for staining, and hematoxylin (Sigma‐Aldrich) was used for counterstain. The intensity and density of IHC staining were recorded under a light microscope using a semi‐quantitative scoring method as previous report.18 Two investigators evaluated the intensity and density of IHC staining independently, and the final value of intensity and density of IHC staining was calculated as the mean of two investigators. Furthermore, the IHC score less than or equal to 3 was defined as the CCT6A protein low expression, while IHC score more than 3 was defined as CCT6A protein high expression.
2.4. Reverse transcription‐quantitative polymerase chain reaction assay
The fresh tumor tissues frozen in the liquid nitrogen were used for the detection of the CCT6A mRNA expression by RT‐qPCR assay. In detail, the total RNA was isolated using TRIzol™ Reagent (Catalog #15596026, Invitrogen); besides, the detailed procedure was as follows: after the frozen tumor tissues being grinded, TRIzol reagent was added to the samples to lyse the cell; then, after standing for 5 min, the chloroform was added to the samples followed by centrifuging for 1 min. Then, adding isopropanol or adding ethanol followed by centrifuging were sequentially used for washing the samples. Finally, RNase‐free water was used for dissolving the RNA. After that, cDNA was synthesized by reverse transcription assay using the iScript™ Reverse Transcription Supermix (Catalog #1708840, Bio‐Rad); in detail, firstly, the total RNA was mixed with water and set at 65°C for 5 min. Then, the reaction system was mixed with 5 × RT buffer, enzyme mix, and RT primer, and the mixed reaction system was set at 42°C for 18 min. Lastly, the reaction system was set at 98°C for 5 min; then, the synthesized cDNA was stored at −20°C. Finally, the qPCR was carried out using QuantiNova SYBR Green PCR Kit (Catalog #208057, Qiagen); in detail, the mixed reaction system was set at the followed reaction conditions: 94°C for 5 min for 1 cycle and then 94°C for 5 s followed by 61°C for 30 s for 40 cycles. The expression of CCT6A mRNA was normalized by GAPDH with a 2−ΔΔCt method. The PCR primers used in this study were referred to the previous study.19
2.5. Statistical analysis
Comparisons of variables were determined by Student's t test and Wilcoxon rank sum test as appropriate. Correlation analysis for ordinal variable was examined by the Spearman test. Kaplan‐Meier curves and log‐rank test were applied to display the correlation of CCT6A expression (protein and mRNA expression) with accumulating OS. Cox's proportional hazard regression model was used to evaluate the predictive factors for OS. For subgroup analyses, patients were categorized as patients with low‐grade astrocytoma (WHO I‐II grade) and patients with high‐grade astrocytoma (WHO III‐IV grade), respectively; patients were classified as IDH mutation and IDH non‐mutation subgroups according to the IDH mutation status; meanwhile, they were categorized as WHO grade I, WHO grade II, WHO grade III, and WHO grade IV subgroups according to the WHO grade; besides, they were classified as age ≥60 years and age <60 years subgroups according to their age. SPSS 21.0 statistical software (IBM Corp) and GraphPad Prism 7.02 (GraphPad Software Inc) were used in this study for the data analysis and presentation. All tests were two‐sided, and P < 0.05 indicated a statistical significance.
3. RESULTS
3.1. Baseline characteristics
In total astrocytoma patients, the mean age was 48.7 ± 14.6 years. There were 77 (38.9%) females and 121 (61.1%) males. The distribution of WHO grade in astrocytoma patients was listed as follows: 10 (5.1%) patients with grade I, 75 (37.9%) patients with grade II, 74 (37.3%) patients with grade III, and 39 (19.7%) patients with grade IV. Other baseline characteristics of astrocytoma patients including IDH mutation, Karnofsky performance status (KPS) score, adjuvant radiotherapy, and adjuvant chemotherapy were listed in Table 1.
TABLE 1.
Clinical characteristics of astrocytoma patients
| Items | Astrocytoma patients (N = 198) |
|---|---|
| Age (years), mean ± SD | 48.7 ± 14.6 |
| Gender, No. (%) | |
| Male | 121 (61.1) |
| Female | 77 (38.9) |
| WHO grade, No. (%) | |
| I | 10 (5.1) |
| II | 75 (37.9) |
| III | 74 (37.3) |
| IV | 39 (19.7) |
| IDH mutation, No. (%) | |
| Yes | 91 (46.0) |
| No | 107 (54.0) |
| KPS score (points), mean ± SD | 62.2 ± 11.7 |
| Adjuvant radiotherapy, No. (%) | |
| Yes | 166 (83.8) |
| No | 32 (16.2) |
| Adjuvant chemotherapy, No. (%) | |
| Yes | 135 (68.2) |
| No | 63 (31.8) |
Abbreviations: IDH, isocitrate dehydrogenase; KPS, Karnofsky performance status; SD, standard deviation; WHO, World Health Organization.
3.2. Correlation of CCT6A expression with clinical features and subsequent therapy
The CCT6A protein expression in FFPE tumor tissues was determined by IHC assay (Figure 1A), which showed that 69 (34.8%), 70 (35.4%), 46 (23.2%), and 13 (6.6%) astrocytoma patients had the IHC score of 0–3, 4–6, 7–9, and 10–12, respectively (Figure 1B). As to the correlation of CCT6A protein expression with clinical features, the elevated CCT6A IHC score was correlated with increased WHO grade (P < 0.001, Figure 2C) and less IDH mutation (P = 0.002, Figure 2D), while CCT6A IHC score was not correlated with other clinical features including age (P = 0.178, Figure 2A), gender (P = 0.960, Figure 2B), or KPS score (P = 0.097, Figure 2E).
FIGURE 1.

Chaperonin‐containing tailless complex polypeptide 1 subunit 6A protein expression in astrocytoma patients. Diagram of different IHC grade of CCT6A expression in astrocytoma (X200; A). Distribution of IHC score of CCT6A in astrocytoma patients (B). CCT6A, chaperonin‐containing tailless complex polypeptide 1 subunit 6A; IHC, immunohistochemistry
FIGURE 2.

Correlation of CCT6A protein expression with clinical features in astrocytoma patients. Correlation of CCT6A protein expression with age (A), gender (B), WHO grade (C), IDH mutation (D), and KPS score (E) in astrocytoma patients. CCT6A, chaperonin‐containing tailless complex polypeptide 1 subunit 6A; WHO, World Health Organization; IDH, isocitrate dehydrogenase; KPS, Karnofsky performance status
As to the association of CCT6A mRNA expression with clinical features, increased CCT6A mRNA expression was only related to elevated WHO grade (P = 0.001, Figure 3C), while CCT6A mRNA expression was not related to other clinical features including age (P = 0.378, Figure 3A), gender (P = 0.884, Figure 3B), IDH mutation (P = 0.087, Figure 3D), or KPS score (P = 0.716, Figure 3E).
FIGURE 3.

Correlation of CCT6A mRNA expression with clinical features in astrocytoma patients. Correlation of CCT6A mRNA expression with age (A), gender (B), WHO grade (C), IDH mutation (D), and KPS score (E) in astrocytoma patients. CCT6A, chaperonin‐containing tailless complex polypeptide 1 subunit 6A; WHO, World Health Organization; IDH, isocitrate dehydrogenase; KPS, Karnofsky performance status
Moreover, CCT6A protein and mRNA expressions were not associated with subsequent treatment modalities, no matter adjuvant radiotherapy, or adjuvant chemotherapy (all P > 0.05, Figure 4A‐D).
FIGURE 4.
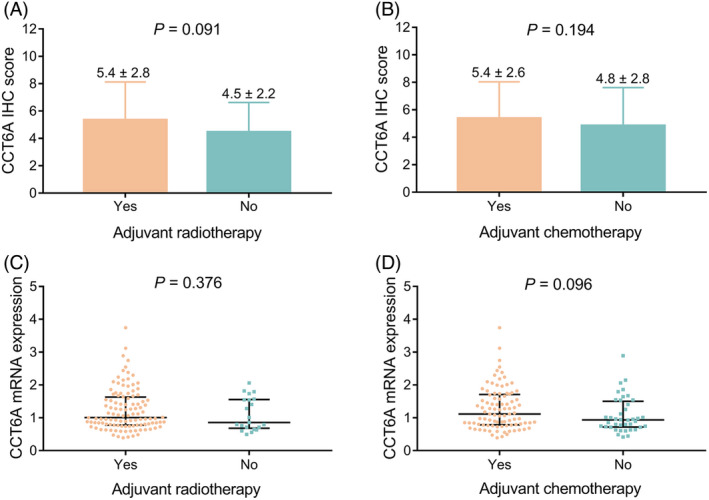
Correlation of CCT6A expression with adjuvant therapy in astrocytoma patients. Correlation of CCT6A protein expression with adjuvant radiotherapy (A) and adjuvant chemotherapy (B) in astrocytoma patients; correlation of CCT6A mRNA expression with adjuvant radiotherapy (C) and adjuvant chemotherapy (D) in astrocytoma patients. CCT6A, chaperonin‐containing tailless complex polypeptide 1 subunit 6A; IHC, immunohistochemistry
3.3. Association of CCT6A expression with survival
Astrocytoma patients were categorized as CCT6A protein high (n = 129) and CCT6A protein low (n = 69) according to the cut‐off value of CCT6A IHC score of 3. Meanwhile, the astrocytoma patients were categorized as CCT6A mRNA high and CCT6A mRNA low according to the median value of CCT6A mRNA expression. Besides, log‐rank test was conducted to evaluate the correlation of CCT6A expression with survival. The findings disclosed that CCT6A protein high was related to shorter accumulating OS (P = 0.007, Figure 5A); meanwhile, the similar tendency was also observed as to the association of CCT6A mRNA expression with accumulating OS (P = 0.012, Figure 5B).
FIGURE 5.

Correlation of CCT6A expression with accumulating OS in astrocytoma patients. Correlation of CCT6A protein (A) and CCT6A mRNA (B) expression with accumulating OS in astrocytoma patients. CCT6A, chaperonin‐containing tailless complex polypeptide 1 subunit 6A; OS, overall survival
For subgroup analyses, astrocytoma patients were classified as low‐grade astrocytoma patients (WHO I‐II grade) and high‐grade astrocytoma patients (WHO III‐IV grade). The analysis indicated that in low‐grade astrocytoma patients, CCT6A expressions (both mRNA and protein, both P > 0.05, Figure 6A,B) were not associated with accumulating OS. In high‐grade astrocytoma patients, CCT6A mRNA high was related to the shorter accumulating OS (P = 0.045, Figure 6D); however, no correlation was found in CCT6A protein expression and accumulating OS (P = 0.793, Figure 6C).
FIGURE 6.

Correlation of CCT6A expression with accumulating OS in low grade and high‐grade astrocytoma patients. Correlation of CCT6A protein (A) and CCT6A mRNA (B) expression with accumulating OS in low‐grade astrocytoma patients; correlation of CCT6A protein (C) and CCT6A mRNA (D) expression with accumulating OS in high‐grade astrocytoma patients CCT6A, chaperonin‐containing tailless complex polypeptide 1 subunit 6A; OS, overall survival
Apart from that, CCT6A was not correlated with accumulating OS no matter in IDH mutation astrocytoma patients or IDH non‐mutation astrocytoma patients' subgroups (All P > 0.05, Figure [Link], [Link]); meanwhile, CCT6A was not associated with accumulating OS in WHO grade I, WHO grade II, WHO grade III, and WHO grade IV subgroups (All P > 0.05, Figure [Link], [Link]); besides, CCT6A only correlated with accumulating OS in patients with age <60 years (All P < 0.05, Figure S3A,B), but not in patients with age ≥60 years (All P > 0.05, Figure S3C,D).
3.4. Independent factors in predicting OS
In order to explicating the factors influencing OS, univariate logistic regression analysis was conducted. As listed in Table 2, CCT6A protein high (hazard ratio [HR]: 1.885, P = 0.008), CCT6A mRNA high (HR: 1.935, P = 0.015), age (≥60 years; HR: 1.641, P = 0.037), and WHO high grade (HR: 2.924, P < 0.001) were associated with shorter OS, while IDH mutation (HR: 0.555, P = 0.007) was correlated with favorable OS.
TABLE 2.
Factors correlated with OS
| Items | Cox's proportional hazard regression model | |
|---|---|---|
| P value | HR (95% CI) | |
| Univariate Cox's regression | ||
| CCT6A protein high | 0.008 | 1.885 (1.179–3.013) |
| CCT6A mRNA high | 0.015 | 1.935 (1.140–3.285) |
| Age (≥60 years) | 0.037 | 1.641 (1.030–2.625) |
| Gender (male) | 0.143 | 0.719 (0.462–1.118) |
| WHO high grade | <0.001 | 2.924 (1.841–4.646) |
| IDH mutation | 0.007 | 0.555 (0.361–0.852) |
| KPS score (≥70) | 0.153 | 0.724 (0.465–1.127) |
| Adjuvant radiotherapy | 0.326 | 1.321 (0.758–2.302) |
| Adjuvant chemotherapy | 0.598 | 1.123 (0.730–1.727) |
| Multivariate Cox's regression (forward stepwise)a | ||
| CCT6A protein high | 0.012 | 1.821 (1.138–2.913) |
| Age (≥60 years) | 0.008 | 1.833 (1.172–2.868) |
CCT6A, chaperonin‐containing tailless complex polypeptide 1 subunit 6A.
Abbreviations: CI, confidence interval; HR, hazard ratio; IDH, isocitrate dehydrogenase; KPS, Karnofsky performance status; OS, overall survival; WHO, World Health Organization.
Considering the multicollinearity, all factors except “CCTA6 mRNA high” and “WHO high grade” were included in the multivariate Cox's regression analysis with forward stepwise method.
Considering the multicollinearity among CCT6A mRNA high, WHO high grade, and CCT6A protein high, only CCT6A protein high was chosen to be included in the multivariate Cox's regression analysis with forward stepwise method along with other factors such as age and gender in univariate logistic regression model. Moreover, it indicated that CCT6A protein high (HR: 1.821, P = 0.012) and age (≥60 years; HR: 1.833, P = 0.008) were independently associated with worse OS (Table 2).
4. DISCUSSION
CCT, a cytosolic chaperonin in eukaryotes, has been best elucidated in its interactions with the cytoskeletal proteins actin and tubulin9; meanwhile, as to the role of CCT in cancers, extensive studies have disclosed that CCT promotes the cancer cell growth and tumorigenicity through affecting the expression of MYC, cyclin D1, and regulating the Wnt/β‐Catenin signaling pathway.20, 21 CCT6A belongs to the subunit of CCT, which is firstly reported in 2006, among which, CCT6A is identified in drug‐resistant melanoma and is reported to play an important role in the regulation of cancers.15, 22 After that, over the past two decades, the regulatory role of CCT6A in malignant tumor has been gradually explicated, which shows that CCT6A leads to the cancer cell growth and metastasis via affecting the cell cycle and TGF‐β signaling.9, 10, 11, 12 To sum up the evidence mentioned above, the CCT6A serves as a cancer‐promoting gene in the genesis and the progression of tumors. As to astrocytoma, there is only one study concerning the clinical value of CCT6A evaluating the prognosis of glioblastoma, which is the most advanced astrocytoma, and the discovery shows that CCT6A is highly expressed, and the elevated expression of CCT6A exhibited a lower survival, while as to the whole cohort of astrocytoma despite the WHO grade, the relative study is scarce. The present study enrolled 198 astrocytoma patients, meanwhile, evaluated the expression of CCT6A in astrocytoma patients and its correlation with clinical features and survival of astrocytoma patients.
Firstly, we found that CCT6A expression was correlated with increased WHO grade and less IDH mutation. The possible explanations might be as follows: (1) CCT6A acts as a cancer‐promoting gene which might promote the proliferation destiny of astrocytoma cells, and thereby, the elevated expression of CCT6A leads to the increasement of WHO grade12; (2) CCT was reported to be downregulated in the differentiated cells, while as a subunit of CCT, the increased expression of CCT6A was also supposed to be associated with worse differentiation of astrocytoma cells which indicated a higher WHO grade, thus further disclosed a positive association between CCT6A expression and WHO grade23; and (3) about 80% IDH mutation is observed in low‐grade glioma patients including astrocytoma patients, while it is rare in high‐grade astrocytoma patients.24, 25 Meanwhile, elevated CCT6A expression was shown to be correlated with increased WHO grade in our study; thus, the elevated CCT6A expression is correlated with less IDH mutation in astrocytoma patients.
Secondly, we found that CCT6A high was associated with worse OS in astrocytoma patients. This finding is similar with previous studies which disclose that CCT6A expression is negatively correlated with OS in non‐small cell lung carcinoma patients and breast cancer.11, 26 This finding might be explained as follows: (1) CCT6A acts as an inducer in the tumorigenesis of astrocytoma, which implies that the high expression of CCT6A may promote the proliferation, invasion, and metastasis, while inhibits the apoptosis of astrocytoma cells, thereby, causes a worse survival of astrocytoma patients; (2) astrocytoma patients with the low expression of CCT6A may be more sensitivity to the chemotherapy agents such as temozolomide, platinum, and lomustine, while this hypothesis needs further verified; (3) as the correlation of CCT6A expression with clinical features of astrocytoma patients, CCT6A expression was positively correlated with WHO grade; therefore, the increased CCT6A expression might cause a worse survival outcome though contributing to an elevated WHO grade.
There were some limitations in this study: (1) the underlying mechanism of CCT6A in astrocytoma was not clear, which needed further assessment; (2) as we mentioned above, the CCT6A expression might be related to the drug sensitivity of the chemotherapy agents such as temozolomide, platinum, and lomustine, while this hypothesis needed further exploration; (3) the detection of CCT6A expression in peripheral blood might be more convenient and feasible than that in tumor tissues; thus, further study was needed; (4) the disease‐free survival or progression‐free survival was not evaluated in this study, which needed to be explored in the further study; (5) because that the 1p/19q was also a vital prognostic biomarker in astrocytoma patients, while this study did not evaluate the correlation between CCT6A and 1p/19q, which needed a further study to explore this article.
To be conclusive, CCT6A correlates with elevated WHO grade and less IDH mutation; besides, CCT6A high expression is independently associated with unfavorable accumulating OS of astrocytoma patients.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Supporting information
Fig S1
Fig S2
Fig S3
Supplementary Material
ACKNOWLEDGEMENTS
None.
Hu Y, Fu P, Zhao H, et al. Chaperonin‐containing tailless complex polypeptide 1 subunit 6A correlates with increased World Health Organization grade, less isocitrate dehydrogenase mutation, and deteriorative survival of astrocytoma patients. J Clin Lab Anal. 2021;35:e23917. 10.1002/jcla.23917
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- 1.Kapoor M, Gupta V. Astrocytoma. Treasure Island, FL: StatPearls; 2021. [Google Scholar]
- 2.Riddick E, Evans S, Rousch J, et al. Identification of death receptors DR4 and DR5 in HTB‐12 astrocytoma cell lines and determination of TRAIL sensitivity. J Solid Tumors. 2013;3(6):20‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwadate Y, Matsutani T, Hirono S, et al. IDH1 mutation is prognostic for diffuse astrocytoma but not low‐grade oligodendrogliomas in patients not treated with early radiotherapy. J Neurooncol. 2015;124(3):493‐500. [DOI] [PubMed] [Google Scholar]
- 4.Okamoto Y, Di Patre PL, Burkhard C, et al. Population‐based study on incidence, survival rates, and genetic alterations of low‐grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108(1):49‐56. [DOI] [PubMed] [Google Scholar]
- 5.Gimenez M, Marie SK, Oba‐Shinjo S, et al. Quantitative proteomic analysis shows differentially expressed HSPB1 in glioblastoma as a discriminating short from long survival factor and NOVA1 as a differentiation factor between low‐grade astrocytoma and oligodendroglioma. BMC Cancer. 2015;15:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Jin Y, Zou Q, et al. Integrated genomic and transcriptomic analysis suggests KRT18 mutation and MTAP are key genetic alterations related to the prognosis between astrocytoma and glioblastoma. Ann Transl Med. 2021;9(8):713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandner S, Jaunmuktane Z. IDH mutant astrocytoma: biomarkers for prognostic stratification and the next frontiers. Neuropathol Appl Neurobiol. 2019;45(2):91‐94. [DOI] [PubMed] [Google Scholar]
- 8.Speirs CK, Simpson JR, Robinson CG, et al. Impact of 1p/19q codeletion and histology on outcomes of anaplastic gliomas treated with radiation therapy and temozolomide. Int J Radiat Oncol Biol Phys. 2015;91(2):268‐276. [DOI] [PubMed] [Google Scholar]
- 9.Zeng G, Wang J, Huang Y, et al. Overexpressing CCT6A contributes to cancer cell growth by affecting the G1‐to‐S phase transition and predicts a negative prognosis in hepatocellular carcinoma. Onco Targets Ther. 2019;12:10427‐10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sternlicht H, Farr GW, Sternlicht ML, et al. The t‐complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc Natl Acad Sci USA. 1993;90(20):9422‐9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T, Shi W, Tian K, et al. Chaperonin containing t‐complex polypeptide 1 subunit 6A correlates with lymph node metastasis, abnormal carcinoembryonic antigen and poor survival profiles in non‐small cell lung carcinoma. World J Surg Oncol. 2020;18(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying Z, Tian H, Li Y, et al. CCT6A suppresses SMAD2 and promotes prometastatic TGF‐beta signaling. J Clin Invest. 2017;127(5):1725‐1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Liu J, Zhao H. Prognostic power of a chaperonin containing TCP‐1 subunit genes panel for hepatocellular carcinoma. Front Genet. 2021;12:668871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu WX, Song W, Jiang MP, et al. Systematic characterization of expression profiles and prognostic values of the eight subunits of the chaperonin TRiC in breast cancer. Front Genet. 2021;12:637887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J, Liu C, Xu G, et al. CCT6A, a novel prognostic biomarker for Ewing sarcoma. Medicine. 2021;100(4):e24484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallal S, Russell BP, Wei H, et al. Extracellular vesicles from neurosurgical aspirates identifies chaperonin containing TCP1 subunit 6A as a potential glioblastoma biomarker with prognostic significance. Proteomics. 2019;19(1–2):e1800157. [DOI] [PubMed] [Google Scholar]
- 17.Louis DN, Perry A, Reifenberger G, et al. The 2016 World health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803‐820. [DOI] [PubMed] [Google Scholar]
- 18.Shen S, Yao Y. A‐kinase interacting protein 1 is sufficiently expressed and positively associates with WHO grade, meanwhile predicts unfavorable overall survival independently in glioma patients. Medicine. 2021;100(4):e20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Lu X, Ma C, et al. Long non‐coding RNA NEAT1 promotes human glioma tumor progression via miR‐152‐3p/CCT6A pathway. Neurosci Lett. 2020;732:135086. [DOI] [PubMed] [Google Scholar]
- 20.Ghozlan H, Showalter A, Lee E, et al. Chaperonin‐containing TCP1 complex (CCT) promotes breast cancer growth through correlations with key cell cycle regulators. Front Oncol. 2021;11:663877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu H, Zhu F, Dong H, et al. Upregulation of CCT‐3 induces breast cancer cell proliferation through miR‐223 competition and wnt/beta‐catenin signaling pathway activation. Front Oncol. 2020;10:533176. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Tanic N, Brkic G, Dimitrijevic B, et al. Identification of differentially expressed mRNA transcripts in drug‐resistant versus parental human melanoma cell lines. Anticancer Res. 2006;26(3A):2137‐2142. [PubMed] [Google Scholar]
- 23.Vonk WIM, Rainbolt TK, Dolan PT, et al. Differentiation drives widespread rewiring of the neural stem cell chaperone network. Mol Cell. 2020;78(2):329‐345 e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yip S, Butterfield YS, Morozova O, et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol. 2012;226(1):7‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13(5):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang K, Zeng Y, Xie Y, et al. Bioinformatics analysis of the prognostic value of CCT6A and associated signalling pathways in breast cancer. Mol Med Rep. 2019;19(5):4344‐4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Supplementary Material
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


