Abstract
Background
Circular RNAs (circRNAs) have been identified to be involved in onset and progression of multiple malignant tumors. The present study aimed to systematically evaluate the diagnostic values of circRNAs in breast cancer.
Methods
The PubMed, Web of Science, Embase, CNKI, and Wanfang online databases were searched for the relevant studies before December 31, 2020. Statistical analysis of the diagnostic tests was performed based on STATA 16.0, Meta‐DiSc 1.4, and RevMan 5.3 software. The threshold effect and publication bias were measured by the Spearman correlation and Deeks’ funnel plot asymmetry test, respectively.
Results
Twenty‐one studies from 13 articles were included in this meta‐analysis. The pooled sensitivity and specificity were 0.77 and 0.71, respectively. The pooled positive likelihood ratio (PLR), negative likelihood ratio (NLR), and overall diagnostic odds ratio (DOR) were 2.6, 0.33, and 8, respectively. Furthermore, the area under the summary receiver operator characteristic curve was 0.80. In addition, down‐regulated circRNAs achieved a diagnostic performance higher than up‐regulated circRNAs, with area under curve (AUC) values of 0.81 and 0.74, respectively. Studies based on tissue samples presented better diagnostic accuracy than those based on plasma samples, with AUC values of 0.80 and 0.67. In addition, two circRNAs, including circ_0001073 and circTADA2A‐E5/E6, showed higher diagnostic values, with AUC value of 0.990 and 0.937, respectively. According to the results of meta‐regression, the case size (p<0.05) might be the source of the heterogeneity.
Conclusion
CircRNAs exhibited a high diagnostic value for breast cancer and may function as potential diagnostic biomarkers for breast cancer.
Keywords: biomarker, breast cancer, circular RNA, diagnosis, meta‐analysis
Thirteen studies comprised of 21 circRNAs were included in this meta‐analysis. Down‐regulated circRNAs achieved a diagnostic performance higher than up‐regulated circRNAs. Tissue‐based circRNA analysis presented better diagnostic accuracy than plasma‐based analysis.

1. INTRODUCTION
Breast cancer has surpassed lung cancer as the most commonly diagnosed cancer and is the leading cause of cancer death among women worldwide in 2020, followed by colorectal and lung cancer for incidence and vice versa for mortality.1 Despite significant advances in diagnosis, surgical intervention, and local and systemic adjuvant therapies, the overall 5‐year survival rate of breast cancer remains unsatisfied. Previous studies have suggested that accurate detection for early breast cancer significantly reduced breast cancer death rates in the long term.2 Patients with tumors≤2.0 cm have a 5‐year survival of 95% compared with 70% for those with tumors >5 cm.3 Mammography is used as a gold standard for early breast cancer screening; however, the high false‐positive and false‐negative rates, as well as overdiagnosis, remain a major concern in breast cancer screening.4, 5 Apart from screening techniques, a breast biopsy, an invasive method, is generally performed to distinguish between cancerous and benign tissues, but it is time‐consuming and requires skilled labor.6 Some biomarkers have also been clinically used for the diagnosis of breast cancer, such as carbohydrate antigen 153 (CA153) and the serum carcinoembryonic antigen (CEA); however, the sensitivity and specificity of these biomarkers are still unsatisfied.7, 8 Thus, the discovery of effective, noninvasive, and reliable biomarkers is pressing for the diagnosis, prognosis, and treatment of breast cancer.
Circular RNA (circRNA) was first discovered in RNA viruses, which assumes a covalent closed‐loop structure generated by backsplicing of precursor mRNA.9 In the past several decades, circRNAs were defined as the by‐products of splicing errors without biological functions.10 With the development of high‐throughput RNA sequencing technologies and bioinformatics, a larger number of circRNAs have been identified.11 CircRNAs are identified as noncoding circRNAs; however, some circRNAs may be translated into protein, if there is an internal ribosome entry site (IRES) present.12, 13 According to the source of sequence, circRNAs are classified into four categories: exonic circRNAs, composed of exons only and found mainly in the cytoplasm14; intron‐derived circRNAs, composed of introns and mostly expressed in the nucleus15; retained‐intron circRNAs, composed of exons and introns and mainly expressed in the nucleus16; and virus circRNAs, generated by circularization of viral RNA genomes, tRNAs, rRNAs, and snRNAs, among others.17, 18 CircRNAs regulate gene expression by serving as microRNA sponges and interacting with RNA‐binding proteins.19, 20 Emerging evidence shows that circRNAs are essential for the onset and development of malignant tumors.21, 22, 23 Most circRNAs are often specifically expressed in different tissues and at different developmental stages.24 Furthermore, circRNAs are resistant to exonuclease or ribonuclease‐mediated degradation and are more stable than linear mRNAs.25, 26 All these properties suggested that circRNAs may be an extra potential biomarker for early‐stage cancer. In this study, we conducted a systematic review and meta‐analysis to evaluate the value of circRNAs in the diagnosis of breast cancer.
2. MATERIALS AND METHODS
2.1. Literature search strategy
An electronic search of PubMed, Web of Science, Embase, CNKI, and Wanfang was performed for eligible articles that were published before December 31, 2020. No language restrictions were imposed. The following keywords were used to retrieve literature: (“circular RNA” OR “circRNA”) AND (“breast cancer” OR “breast carcinoma” OR “breast tumor” OR “breast neoplasm” OR “mammary cancer”). Then, the title, abstract, and full text are reviewed manually by two researchers (CMY and FYQ) to identify the appropriate studies. We also manually searched the reference lists of all included articles to obtain additional data.
2.2. Inclusion and exclusion criteria
Studies that met the following inclusion criteria were included in the meta‐analysis: (1) patients with a pathological diagnosis of breast cancer; (2) studies that detected the circRNA expression levels in serum, plasma, or tissue; and (3) true positive (TP), false positive (FP), true negative (TN), and false negative (FN) were available or could be calculated indirectly. Studies were excluded if (1) irrelevant to breast cancer, circRNA or diagnosis; or (2) duplicate data as previous studies; or (3) reviews, animal experiments, letters, conference abstracts and meta‐analyses; or (4) with insufficient data.
2.3. Data extraction
The full texts of all eligible studies were reviewed by two researchers (CMY and FYQ) independently. Any disagreements were discussed with a third investigator (JYC) until a consensus was reached. The following data were extracted from each study: (1) first author, publication year, country, type of circRNAs, sample size, control source, and specimen type; (2) altered expression and detection method; and (3) diagnostic sensitivity and specificity of circRNAs. RevMan software was applied to extract the sensitivity, specificity, and area under curve (AUC) indirectly from TP, FP, TN, and FN values if the studies did not present complete diagnostic data.
2.4. Quality assessment
Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS‐2) was adopted to judge the quality and bias of the eligible studies.27 The QUADAS‐2 checklist was composed of two parts, “risk of bias” and “applicability concerns.” The risk of bias is assessed in four key areas: patient selection, index test, reference standard, and flow and timing. Concern for applicability is assessed in three key areas: patient selection, index test, and reference standard. Each item was evaluated as low risk, high risk, or unclear risk.28 If a study is judged “high” or “unclear” in 1 or more domains, then it may be judged “at risk of bias” or as having “concerns regarding applicability.”
2.5. Statistical analysis
Statistical analysis of the diagnostic tests was conducted using STATA 16.0, Meta‐DiSc 1.4, and RevMan 5.3 software. The heterogeneity among each selected study was estimated by the Higgins I 2 statistics and Cochran's Q‐test. I 2 > 50% and Phet <0.05 suggest that there was significant heterogeneity among the included studies, and a random‐effect model was applied to estimate the pooled results; otherwise, a fixed‐effect model was applied.29 To estimate the ability of circRNAs to distinguish between breast cancer cases and controls, we extracted TP, FP, TN, and FN values from each study and used a random‐effects model to quantify the pooled sensitivity [TP/(FN + TP)], specificity [TN/(FP + TN)], positive likelihood ratio (PLR) [(SEN/ (1‐SPE)], negative likelihood ratio (NLR) [(1‐SEN)/SPE)], overall diagnostic odds ratio (DOR) [PLR/NLR], and AUC with their corresponding 95% confidence intervals (CIs) for the diagnostic meta‐analysis. Fagan plot analysis was performed to assess the clinical value of circRNAs. Spearman correlation analysis was conducted to verify the threshold effects. Subgroup analysis and meta‐regression were applied to explore the potential sources of heterogeneity based on specimen (tissue or plasma), sample size (≥100 or <100), and circRNA expression status (up‐regulation or down‐regulation). Sensitivity analysis was performed to assess the stability and reliability of the meta‐analysis results. Additionally, publication bias was evaluated using Deeks’ funnel plot asymmetry test.30 P‐value <0.05 was considered statistically significant.
3. RESULTS
3.1. Search results
The study selection procedure is shown in a flow diagram (Figure 1). A total of 1134 articles were initially retrieved from the online databases according to the search strategy. A total of 536 records remained after removal of duplicates. Upon a careful reading of the titles and abstracts, 178 articles were ruled out due to irrelevant topics. The remaining 358 articles were further examined by review of the full text; as a result, 345 articles were excluded according to exclusion criteria. The remaining 13 studies31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43 were finally included for the subsequent meta‐analysis.
FIGURE 1.
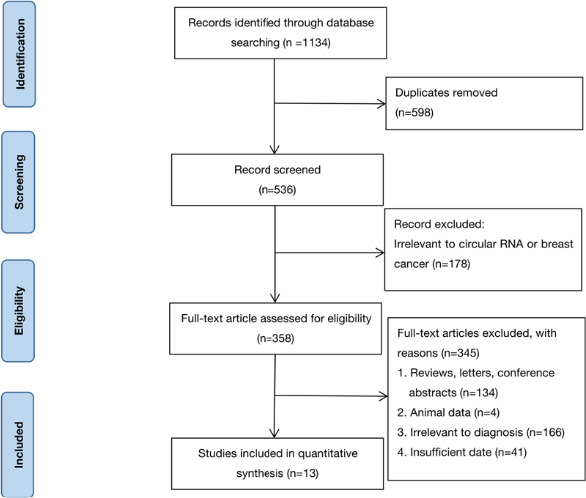
Steps for screening eligible articles
3.2. Study characteristics and quality assessment
The meta‐analysis of 13 articles, involving 1,755 cases and 1,085 controls, has been the largest sample study for predicting the effect of circRNAs on the diagnosis of breast cancer to date. All studies were published between 2017 and 2020 in China as shown in Table 1. In total, thirteen studies comprised of 21 circRNAs were included in the meta‐analysis, among which nine circRNAs were identified as tumor promoters and twelve circRNAs were tumor suppressors. The expression levels of circRNA in all included studies were detected by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) in tissue (n = 17) and plasma (n = 4). Six articles31, 32, 36, 38, 42, 43 directly provided sensitivity, specificity, and the area under the ROC curve; moreover, we calculated the sensitivity and specificity for the other seven studies33, 34, 35, 37, 39, 40, 41 by using RevMan software. Each of the eligible studies was scrutinized via the QUADAS‐2, in the areas of risk of bias and concern for applicability (Table 2 and Figure 2). The greatest risk of bias was most often associated with the items flow and timing and index test. The greatest concern in the category of applicability was the patient selection. The concern for bias and applicability was most often due to failure to provide sufficient data to permit a judgment.
TABLE 1.
Characteristics of studies for diagnosis analysis
| No. | Study | Year | Country | CircRNA signature | Sample size | Control type | Specimen type | Altered expression | Method | AUC | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||||
| 1 | Hu et al.31 | 2020 | China | circ_0008673 | 378 | 102 | Normal breast tissues | Plasma | Up‐regulated | qRT‐PCR | 0.833 |
| 2 | Li et al.32 | 2020 | China | circVRK1 | 350 | 163 | Adjacent non‐tumor tissues | Tissue | Down‐regulated | qRT‐PCR | 0.720 |
| 3 | Li et al.33 | 2020 | China | circ_0104824 | 37 | 37 | Adjacent non‐tumor tissues | Tissue | Down‐regulated | qRT‐PCR | 0.878 |
| 4 | Liu et al.34 | 2020 | China | circ_0043278 | 50 | 50 | Normal breast tissues | Tissue | Down‐regulated | qRT‐PCR | 0.690 |
| 5 | Xing et al.35 | 2020 | China | circIFI30 | 38 | 38 | Adjacent non‐tumor tissues | Tissue | Up‐regulated | qRT‐PCR | 0.733 |
| 6 | Yi et al.36 | 2020 | China | circ_0001073 | 132 | 132 | Adjacent non‐tumor tissues | Tissue | Down‐regulated | qRT‐PCR | 0.990 |
| 7 | Yuan et al.37 | 2020 | China | circ_0068033 | 36 | 36 | Adjacent non‐tumor tissues | Tissue | Down‐regulated | qRT‐PCR | 0.848 |
| 8 | Zheng et al.38 | 2020 | China | circSEPT9 | 60 | 60 | Adjacent non‐tumor tissues | Tissue | Up‐regulated | qRT‐PCR | 0.711 |
| 9 | Xiao et al.39 | 2019 | China | circAHNAK1 | 38 | 38 | Normal breast tissues | Tissue | Down‐regulated | qRT‐PCR | 0.720 |
| 10 | Yang et al.40 | 2019 | China | circAGFG1 | 40 | 40 | Adjacent non‐tumor tissues | Tissue | Up‐regulated | qRT‐PCR | 0.767 |
| 11 | Xu et al.41 | 2019 | China | circTADA2A‐E6 | 115 | 16 | Normal breast tissues | Tissue | Down‐regulated | qRT‐PCR | 0.855 |
| 12 | Xu et al.41 | 2019 | China | circTADA2A‐E5/E6 | 115 | 16 | Normal breast tissues | Tissue | Down‐regulated | qRT‐PCR | 0.937 |
| 13 | Yin et al.42 | 2018 | China | circ_0001785 | 20 | 17 | Healthy individual's blood specimen | Plasma | Up‐regulated | qRT‐PCR | 0.771 |
| 14 | Yin et al.42 | 2018 | China | circ_0108942 | 20 | 17 | Healthy individual's blood specimen | Plasma | Up‐regulated | qRT‐PCR | 0.701 |
| 15 | Yin et al.42 | 2018 | China | circ_0068033 | 20 | 17 | Healthy individual's blood specimen | Plasma | Down‐regulated | qRT‐PCR | 0.619 |
| 16 | Lv et al.43 | 2017 | China | circ_103110 | 51 | 51 | Adjacent non‐tumor tissues | Tissue | Down‐regulated | qRT‐PCR | 0.630 |
| 17 | Lv et al.43 | 2017 | China | circ_104689 | 51 | 51 | Adjacent non‐tumor tissues | Tissue | Down‐regulated | qRT‐PCR | 0.610 |
| 18 | Lv et al.43 | 2017 | China | circ_104821 | 51 | 51 | Adjacent non‐tumor tissues | Tissue | Down‐regulated | qRT‐PCR | 0.600 |
| 19 | Lv et al.43 | 2017 | China | circ_006054 | 51 | 51 | Adjacent non‐tumor tissues | Tissue | Up‐regulated | qRT‐PCR | 0.710 |
| 20 | Lv et al.43 | 2017 | China | circ_100219 | 51 | 51 | Adjacent non‐tumor tissues | Tissue | Up‐regulated | qRT‐PCR | 0.780 |
| 21 | Lv et al.43 | 2017 | China | circ_406697 | 51 | 51 | Adjacent non‐tumor tissues | Tissue | Up‐regulated | qRT‐PCR | 0.640 |
TABLE 2.
Study quality of the diagnostic studies, as judged by the QUADAS‐2 checklist
| Study | Risk of bias | Applicability concerns | |||||
|---|---|---|---|---|---|---|---|
| Patient selection | Index Test | Reference standard | Flow and timing | Patient Selection | Index Test | Reference Standard | |
| Hu et al.31 |

|
? |

|

|
? |

|

|
| Li et al.32 |

|

|

|
? | ? |

|

|
| Li et al.33 |

|
? |

|

|
? |

|

|
| Liu et al.34 |

|
? |

|
? | ? |

|

|
| Xing et al.35 | ? |

|

|
? | ? |

|

|
| Yi et al.36 | ? | ? |

|

|
? |

|

|
| Yuan et al.37 | ? |

|

|

|
? |

|

|
| Zheng et al.38 | ? |

|

|
? |

|

|

|
| Xiao et al.39 |

|
? |

|

|

|

|

|
| Yang et al.40 |

|

|

|
? | ? |

|

|
| Xu et al.41 |

|

|

|

|
? |

|

|
| Yin et al.42 | ? |

|

|

|
? |

|

|
| Lv et al.43 |

|
? |

|
? |

|

|

|
 = low;
= low;  = high; ? = unclear.
= high; ? = unclear.
“Low” means “at low risk of bias” or having “low concern regarding applicability”; “High” means “at risk of bias” or having “concerns regarding applicability”; and “unclear” means insufficient data for judgment.
FIGURE 2.
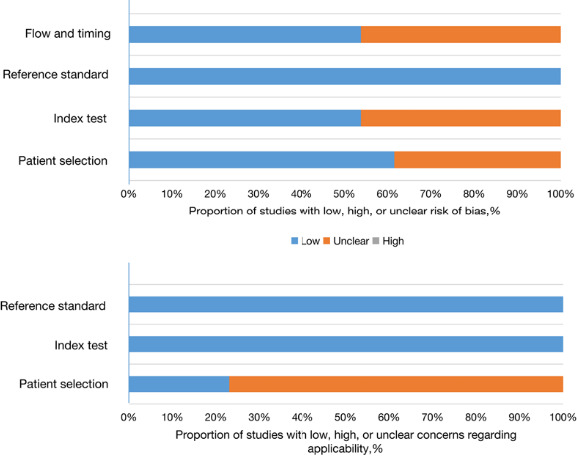
Quality assessment of eligible studies. “Low” means “at low risk of bias” or having “low concern regarding applicability”; “High” means “at risk of bias” or having “concerns regarding applicability”; and “unclear” means insufficient data for judgment
3.3. Diagnostic analysis
As shown in Figure 3, there was significant heterogeneity in the pooled sensitivity (I 2 = 90.56%, Phet < 0.001) and specificity (I 2 = 83.99%, Phet < 0.001); thus, the random‐effects model was applied to analyze the diagnostic parameters. The pooled sensitivity and specificity were 0.77 (95% CI: 0.70–0.83) and 0.71 (95% CI: 0.63–0.78), respectively. In addition, the pooled PLR, NLR, and DOR were 2.6 (95% CI: 2.0–3.5), 0.33 (95% CI: 0.24–0.45), and 8 (95% CI: 5–14), respectively. Furthermore, we drew a summary receiver operator characteristic (SROC) curve and calculated the value of AUC (0.80, 95% CI: 0.77–0.84, Figure 4). Then, the Fagan plot was analyzed to present the clinical value of circRNAs. The pre‐test probability of the left column is 62%, the PLR of the middle column is 3, and the post‐test probability is 81%. The NLR of the middle column is 0.33, and the post‐test probability is 35% (Figure S1). Moreover, we found two circRNAs, including circ_0001073 and circTADA2A‐E5/E6 exhibited high diagnostic potentials for breast cancer, with the AUC values of 0.990 and 0.937, respectively (Table 1). These results indicated that circRNAs have moderate‐high diagnostic accuracy for breast cancer. Additionally, Spearman's correlation coefficient value was −0.197 and the P‐value was 0.392 (>0.05), suggesting that there was no threshold effect as well. It can be equated to the fact that the threshold effect is not a source of heterogeneity.
FIGURE 3.
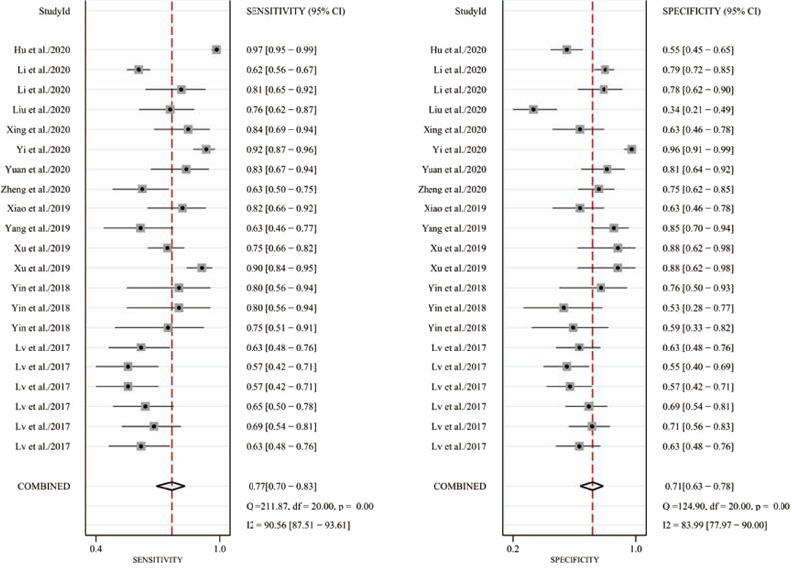
Forest plots of sensitivity and specificity of circRNAs in the diagnosis of breast cancer. (A) Pooled sensitivity for circRNAs. (B) Pooled specificity for circRNAs
FIGURE 4.
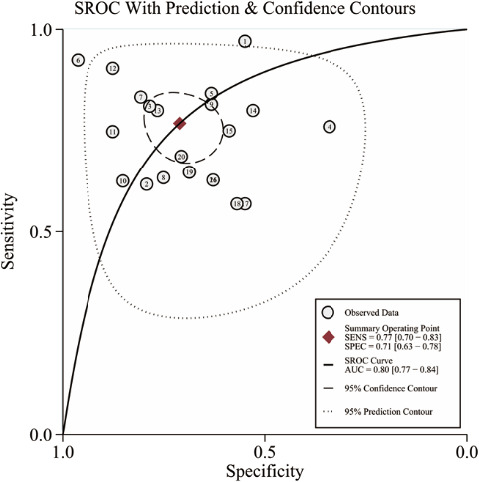
The summary receiver operator characteristic (SROC) curve of circRNAs in the diagnosis of breast cancer. The study case numbers inside of the graphics represent the corresponding articles we used for the meta‐analysis, which can refer to Table 1
3.4. Subgroup analysis and meta‐regression analysis
Subgroup analyses were performed according to specimen (tissue or plasma), sample size (≥100 or <100), and circRNA expression status (up‐regulation or down‐regulation) to explore the potential sources of heterogeneity. As shown in Table 3, down‐regulated circRNAs achieved a diagnostic performance higher than up‐regulated circRNAs, with AUC values of 0.81 [95%CI: 0.78–0.85] and 0.74 [95%CI: 0.70–0.78]. Studies based on tissue samples presented better diagnostic accuracy than those based on plasma samples, with AUC values of 0.80 [95%CI: 0.76–0.83] and 0.67 [95%CI: 0.63–0.71], respectively. The heterogeneity of studies by tissue was higher than the studies by plasma samples (I 2 = 84.9% and I 2 = 76.5%). When subgrouped by sample size, there was no heterogeneity for the studies with a sample size of less than 100. But the significant heterogeneity existed for the studies with a sample size of more than 100 (I 2 = 92.90, Phet <0.01). According to the results of meta‐regression, the case size (P < 0.05) might be the source of the heterogeneity.
TABLE 3.
Subgroup analysis of diagnostic accuracy of circRNAs in breast cancer
| Subgroups | CircRNAs | Sensitivity (95%CI) | Specificity (95%CI) | PLR (95%CI) | NLR (95%CI) | DOR (95%CI) | AUC (95%CI) | Heterogeneity I 2 test (%)/P het | Meta‐regression P‐value |
|---|---|---|---|---|---|---|---|---|---|
| Specimen | |||||||||
| Tissue | 17 | 0.73 (0.67–0.79) | 0.73 (0.65–0.81) | 2.8 (1.9–4.0) | 0.36 (0.27–0.49) | 8 (4–14) | 0.80 (0.76–0.83) | 84.9%/<0.01 | 0.620 |
| Plasma | 4 | 0.88 (0.72–0.95) | 0.60 (0.49–0.71) | 2.2 (1.7–2.9) | 0.20 (0.08–0.49) | 11 (4–31) | 0.67 (0.63–0.71) | 76.5%/<0.01 | |
| Case size | |||||||||
| ≥100 | 5 | 0.88 (0.73–0.95) | 0.85 (0.67–0.94) | 5.7 (2.5–13.0) | 0.15 (0.06–0.33) | 39 (11–136) | 0.93 (0.90–0.95) | 92.9%/<0.01 | 0.001 |
| <100 | 16 | 0.71 (0.65–0.75) | 0.66 (0.59–0.72) | 2.1 (1.7–2.5) | 0.45 (0.37–0.55) | 5 (3–7) | 0.74 (0.70–0.78) | 59.2%/<0.01 | |
| Altered expression | |||||||||
| Up‐regulated | 9 | 0.77 (0.64–0.87) | 0.68 (0.62–0.74) | 2.4 (2.1–2.8) | 0.33 (0.21–0.52) | 7 (4–12) | 0.74 (0.70–0.78) | 75.0%/<0.01 | 0.817 |
| Down‐regulated down‐regulated | 12 | 0.76 (0.67–0.83) | 0.74 (0.61–0.83) | 2.9 (1.8–4.8) | 0.33 (0.21–0.50) | 9 (4–22) | 0.81 (0.78–0.85) | 89.1%/<0.01 | |
| Overall | 21 | 0.77 (0.70–0.83) | 0.71 (0.63–0.78) | 2.6 (2, 0–3.5) | 0.33 (0.24–0.45) | 8 (5–14) | 0.80 (0.77–0.84) | 85.1%/<0.01 | |
3.5. Sensitivity analysis and publication bias
Sensitivity analysis was performed by omitting studies one by one, and results showed that removal of any single study did not alter the combined diagnostic effect (Figure 5), suggesting that the results of this meta‐analysis were relatively stable and reliable. In order to evaluate potential publication bias, the Deeks’ funnel plot asymmetry test was performed. The P‐value of Deeks’ test was 0.18 (Figure 6), illustrating no significant publication bias existed.
FIGURE 5.
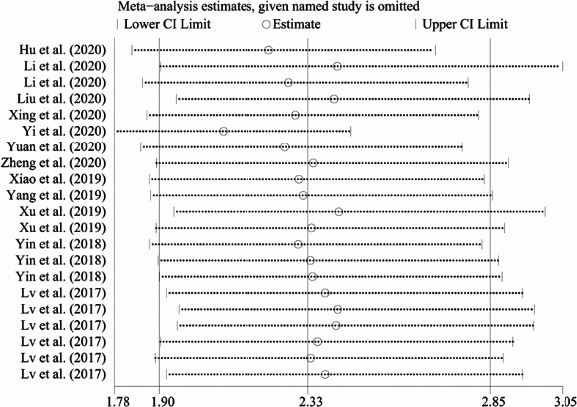
Sensitivity analysis to assess the stability results. Sensitivity analysis was performed by omitting each study one by one, and the omitted studies were shown on the left side of the graphics
FIGURE 6.
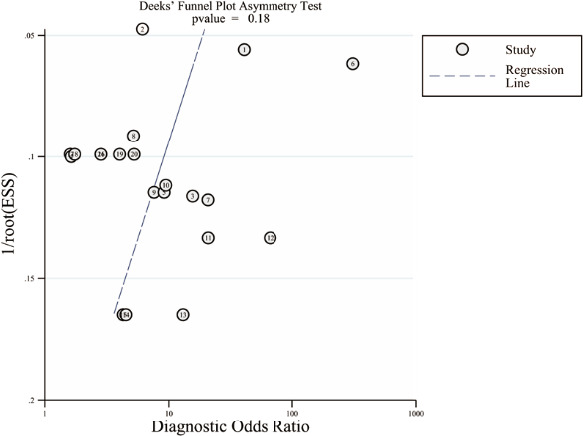
The Deeks’ funnel plot asymmetry test for publication bias. Each point represents a separate study for the indicated association. The study case numbers inside of the graphics represent the corresponding articles we used for meta‐analysis, which can refer to Table 1
4. DISCUSSION
Breast cancer is a major cause of cancer‐related deaths among women worldwide, and the early diagnosis is imperative for improving disease prognosis. However, breast cancer is not easily diagnosed at the outset since no obvious symptoms typically occur at early stage. Thus, developing novel accurate and efficient diagnostic biomarkers is crucial for early intervention. Due to the conserved sequences, high stability, and tissue specificity of circRNAs in tissue or plasma, circRNAs have been considered as excellent candidate biomarkers in many kinds of tumors, such as gastric cancer,44 colorectal cancer,45 hepatocellular carcinoma,46 and lung cancer.47 Previous studies have shown that circRNAs are involved in breast cancer development and may serve as potential biomarkers for breast cancer diagnosis.48 However, there only has been one meta‐analysis focused on the relationship between the circRNAs and breast cancer until now. Because there was insufficient literature at the time of the search, the study of Ma et al. included only 4 diagnostic studies involving the 498 cases and 271 controls.49
Our current study provided an updated systematic review and meta‐analysis of the diagnostic value of circRNAs in breast cancer, which included 13 qualified studies enrolling 1,755 cases and 1,085 controls. In our study, the pooled sensitivity and specificity of circRNAs were 0.77 and 0.71, respectively, suggesting that circRNAs presented well diagnostic accuracy. The AUC of the SROC curve was 0.80, which further reflects high potential diagnostic value for breast cancer. The pooled PLR was 2.6, meaning that patients with breast cancer have 2.6‐fold possibility of altered expression of circRNAs comparing to normal people. In addition, the pooled DOR of circRNAs was 8, suggesting a powerful discriminating capacity of circRNAs to discriminate breast cancer patients from noncancerous controls. In terms of clinical value, the PLR‐post‐test probability was 81% and the NLR‐post‐test probability was 35%. These mean that if a patient is diagnosed with a positive result through circRNAs, the probability of being breast cancer is 81% and if a negative result, the probability of being healthy is 35%. Together, these findings suggest that circRNAs might be effective biomarkers for breast cancer diagnosis. Some circRNAs, such as circ_0001073 and circTADA2A‐E5/E6, exhibited higher diagnostic values in the diagnosis of breast cancer, with AUC values of 0.990 and 0.937, respectively.36, 41 Circ_0008673 possessed higher accuracy than traditional cancer biomarkers such as CA153 and CEA in the diagnosis of breast cancer. In addition, the combined detection of plasma circ_0008673, CA153, and CEA showed greater predictability than circ_0008673 alone, with the AUC value of 0.896.31 Recently, Wang et al. reported that three circRNA panels (circ_0000745, circ_0001531, and circ_0001640) showed better diagnostic value than each individual circRNA.50 All these data strongly supported our conclusion that circRNAs exhibited a high diagnostic value for breast cancer.
As circRNAs with different expression status may exert different functions in breast cancer, we conducted subgroup analyses. Stratified analysis based on circRNAs expression status showed that circRNA, which function as tumor suppressors, achieved a diagnostic performance higher than tumor promoters, and tissue‐based circRNA analysis presented better diagnostic accuracy than plasma‐based analysis. Heterogeneity is inevitable in a meta‐analysis51, 52 and was therefore also evident in our meta‐analysis. According to SROC curve and Spearman's correlation coefficient of −0.197 (P = 0.392), we found that there was no threshold effect. We also explored the possible factors responsible for heterogeneity using the sensitivity analysis and the meta‐regression test. The sensitivity analysis revealed that no individual studies were outliers, suggesting that the heterogeneity of our data is acceptable and the combined effects are reliable. The meta‐regression test traced the factors, such as specimen type, sample size, and circRNA expression status, and revealed that sample size may be a major cause for heterogeneity.
Nevertheless, limitations still exist in our current meta‐analysis. Firstly, cases of studies were all from China and this onefold ethnic group will influence the final results and lead to population selection bias. Secondly, there was significant heterogeneity among included studies. Although we performed subgroup analysis and meta‐regression analysis to explore the potential sources, the results did not fully explain the potential heterogeneity. In summary, the current meta‐analysis highlighted the diagnostic value of circRNAs in breast cancer. However, higher quality studies with more cases and a wider range of populations are required to confirm the clinical value of circRNAs in breast cancer in future.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
Mingyu Chu designed the study, analyzed the data, and drafted the study. Yaqun Fang analyzed the data and drafted the study. Yucui Jin designed the study and approved the final study. All authors read and approved the final study.
Supporting information
Fig S1
Chu M, Fang Y, Jin Y. CircRNAs as promising biomarker in diagnosis of breast cancer: An updated meta‐analysis. J Clin Lab Anal. 2021;35:e23934. 10.1002/jcla.23934
Mingyu Chu and Yaqun Fang authors contributed equally to this work.
DATA AVAILABILITY STATEMENT
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 2.Migowski A. Early detection of breast cancer and the interpretation of results of survival studies. Ciencia & Saude Coletiva. 2015;20(4):1309. [DOI] [PubMed] [Google Scholar]
- 3.Coleman C. Early detection and screening for breast cancer. Semin Oncol Nurs. 2017;33(2):141‐155. [DOI] [PubMed] [Google Scholar]
- 4.Bleyer A, Welch HG. Effect of three decades of screening mammography on breast‐cancer incidence. The New England Journal of Medicine. 2012;367(21):1998‐2005. [DOI] [PubMed] [Google Scholar]
- 5.Drukteinis JS, Mooney BP, Flowers CI, et al. Beyond mammography: new frontiers in breast cancer screening. The American Journal of Medicine. 2013;126(6):472‐479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yen TWF, Li J, Sparapani RA, et al. The interplay between hospital and surgeon factors and the use of sentinel lymph node biopsy for breast cancer. Medicine. 2016;95(31):e4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffy MJ, Evoy D, McDermott EW. CA 15–3: uses and limitation as a biomarker for breast cancer. Clin Chim Acta. 2010;411(23–24):1869‐1874. [DOI] [PubMed] [Google Scholar]
- 8.Patani N, Martin LA, Dowsett M. Biomarkers for the clinical management of breast cancer: international perspective. Int J Cancer. 2013;133(1):1‐13. [DOI] [PubMed] [Google Scholar]
- 9.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerelle C, Mascrez B, Hetuin D, et al. Mis‐splicing yields circular RNA molecules. FASEB Journal. 1993;7(1):155‐160. [DOI] [PubMed] [Google Scholar]
- 11.Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Ruan Y, Zhang H, et al. Tumor‐suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9‐21 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly S, Greenman C, Cook PR, et al. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427(15):2414‐2417. [DOI] [PubMed] [Google Scholar]
- 15.Aucamp J, Bronkhorst AJ, Pretorius PJ. A historical and evolutionary perspective on circulating nucleic acids and extracellular vesicles: circulating nucleic acids as homeostatic genetic entities. Adv Exp Med Biol. 2016;924:91‐95. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256‐264. [DOI] [PubMed] [Google Scholar]
- 17.Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2017;14(8):978‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanford TJ, Mears HV, Fajardo T, et al. Circularization of flavivirus genomic RNA inhibits de novo translation initiation. Nucleic Acids Res. 2019;47(18):9789‐9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lv Q, Wang G, Zhang Y, et al. CircAGAP1 promotes tumor progression by sponging miR‐15‐5p in clear cell renal cell carcinoma. Journal of Experimental & Clinical Cancer Research : CR. 2021;40(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui W, Dang Q, Chen C, et al. Roles of circRNAs on tumor autophagy. Molecular Therapy Nucleic Acids. 2021;23:918‐929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan Y, Wang J, Jin W, et al. CircNR3C2 promotes HRD1‐mediated tumor‐suppressive effect via sponging miR‐513a‐3p in triple‐negative breast cancer. Mol Cancer. 2021;20(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365(2):141‐148. [DOI] [PubMed] [Google Scholar]
- 26.Hsiao KY, Sun HS, Tsai SJ. Circular RNA ‐ New member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136‐1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529‐536. [DOI] [PubMed] [Google Scholar]
- 28.Hegedus EJ, Goode AP, Cook CE, et al. Which physical examination tests provide clinicians with the most value when examining the shoulder? Update of a systematic review with meta‐analysis of individual tests. Br J Sports Med. 2012;46(14):964‐978. [DOI] [PubMed] [Google Scholar]
- 29.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta‐analysis. Stat Med. 2002;21(11):1539‐1558. [DOI] [PubMed] [Google Scholar]
- 30.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58(9):882‐893. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Song Q, Zhao J, et al. Identification of plasma hsa_circ_0008673 expression as a potential biomarker and tumor regulator of breast cancer. J Clin Lab Anal. 2020;34(9):e23393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Li H. Circular RNA VRK1 correlates with favourable prognosis, inhibits cell proliferation but promotes apoptosis in breast cancer. J Clin Lab Anal. 2020;34(1):e22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Ma F, Wu L, et al. Identification of Hsa_circ_0104824 as a potential biomarkers for breast cancer. Technology in CANCER Research & Treatment. 2020;19:1533033820960745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C, Han T, Shi Y. The decreased expression of hsa_circ_0043278 and its relationship with clinicopathological features of breast cancer. Gland Surgery. 2020;9(6):2044‐2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing L, Yang R, Wang X, et al. The circRNA circIFI30 promotes progression of triple‐negative breast cancer and correlates with prognosis. Aging. 2020;12(11):10983‐11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi Z, Li Y, Wu Y, et al. Circular RNA 0001073 attenuates malignant biological behaviours in breast cancer cell and is delivered by nanoparticles to inhibit mice tumour growth. OncoTargets and Therapy. 2020;13:6157‐6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan P, Lei L, Dong S, et al. Circular RNA hsa_circ_0068033 acts as a diagnostic biomarker and suppresses the progression of breast cancer through sponging miR‐659. OncoTargets and Therapy. 2020;13:1921‐1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng X, Huang M, Xing L, et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple‐negative breast cancer. Mol Cancer. 2020;19(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao W, Zheng S, Zou Y, et al. CircAHNAK1 inhibits proliferation and metastasis of triple‐negative breast cancer by modulating miR‐421 and RASA1. Aging. 2019;11(24):12043‐12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang R, Xing L, Zheng X, et al. The circRNA circAGFG1 acts as a sponge of miR‐195‐5p to promote triple‐negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR‐203a‐3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin WB, Yan MG, Fang X, et al. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363‐368. [DOI] [PubMed] [Google Scholar]
- 43.Lv L, Sun J, Shi P, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8(27):44096‐44107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li R, Jiang J, Shi H, et al. CircRNA: a rising star in gastric cancer. Cellular and Molecular Life Sciences : CMLS. 2020;77(9):1661‐1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang X, Sun G, He Q, et al. Circular noncoding RNA circMBOAT2 is a novel tumor marker and regulates proliferation/migration by sponging miR‐519d‐3p in colorectal cancer. Cell Death Dis. 2020;11(8):625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sukowati CHC, Cabral LKD, Tiribelli C, et al. Circulating long and circular noncoding RNA as non‐invasive diagnostic tools of hepatocellular carcinoma. Biomedicines. 2021;9(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Tian W, Wang S, et al. CircRNAs as promising biomarker in diagnostic and prognostic of lung cancer: An updated meta‐analysis. Genomics. 2021;113(1 Pt 1):387‐397. [DOI] [PubMed] [Google Scholar]
- 48.Fu B, Zhang A, Li M, et al. Circular RNA profile of breast cancer brain metastasis: identification of potential biomarkers and therapeutic targets. Epigenomics. 2018;10(12):1619‐1630. [DOI] [PubMed] [Google Scholar]
- 49.Ma Y, Niu X, Yan S, et al. Circular RNA profiling facilitates the diagnosis and prognostic monitoring of breast cancer: A pair‐wise meta‐analysis. J Clin Lab Anal. 2021;35(1):e23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang YW, Xu Y, Wang YY, et al. Elevated circRNAs circ_0000745, circ_0001531 and circ_0001640 in human whole blood: Potential novel diagnostic biomarkers for breast cancer. Exp Mol Pathol. 2021;121: 104661. [DOI] [PubMed] [Google Scholar]
- 51.Higgins JP. Commentary: Heterogeneity in meta‐analysis should be expected and appropriately quantified. Int J Epidemiol. 2008;37(5):1158‐1160. [DOI] [PubMed] [Google Scholar]
- 52.Groenwold RH, Rovers MM, Lubsen J, et al. Subgroup effects despite homogeneous heterogeneity test results. BMC Med Res Methodol. 2010;10:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Data Availability Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.


