Abstract
Background
Increasing studies reported that long non‐coding RNAs are involved in regulating breast cancer (BRCA) progression. However, the specific roles and mechanisms of lncRNAs in BRCA remain largely unknown. Here, we sought to explore the functions and mechanisms of AC016405.3 in BRCA progression.
Methods
Bioinformatic analysis for AC016405.3, miR‐22‐3p, and ERBB3 were performed on starBase. The expressions of AC016405.3, miR‐22‐3p, and ERBB3 were examined by RT‐qPCR. The functions of AC016405.3 on the proliferation, migration, and invasion of cells were evaluated by conducting CCK‐8, colony formation, wound‐healing, and Transwell assays. The subcellular distribution of AC016405.3 in BRCA cells was identified by performing fluorescence in situ hybridization (FISH) and subcellular fractionation techniques. Dual‐luciferase assay was applied to validate the interactions of miR‐22‐3p with AC016405.3 or ERBB3. The interaction between ERBB3 and miR‐22‐3p was also tested by Anti‐Ago2 RNA immunoprecipitation (RIP) assay.
Results
The results showed that AC016405.3 is highly expressed in BRCA tissues as well as cells and positively correlated with poor prognosis in BRCA patients. Silencing AC016405.3 obviously repressed the malignant behaviors of BRCA cells. Mechanistically, AC016405.3 functioned as a competing endogenous RNA (ceRNA) for miR‐22‐3p in the cytoplasm and sponged miR‐22‐3p to release its suppression of ERBB3. Rescue experiments revealed that the suppression role induced by AC016405.3 depletion on malignant behaviors of BRCA cells could be obviously counter by inhibiting miR‐22‐3p or overexpressing ERBB3.
Conclusion
AC016405.3 promotes BRCA progression by the derepression of ERBB3 via sponging miR‐22‐3p, which may represent a potential target for BRCA treatment.
Keywords: AC016405.3, breast cancer, competing endogenous RNA network, ERBB3, miR‐22‐3p
AC016405.3 is involved in BRCA progression by up‐regulating ERBB3 expression via miR‐22‐3p. BRAC cells (MCF7 and ZR7530) were co‐transfected si‐AC016405.3 with empty vector (EV) or ERBB3 overexpression vector (ERBB3). The proliferation of BRAC cells from different groups was examined by (A) CCK‐8 and (B) colony formation assays. (C) Wound‐healing assay assessed the migration ability of BRAC cells from different groups. (D) Transwell assay evaluated both the migration and invasion of BRAC cells from different groups. (E) A schematic diagram exhibited the proposed mechanism that cytoplasmic AC016405.3 sponges miR‐22‐3p to promote ERBB3 expression thus contributes to BRCA progression. Note: ** p < 0.01 and *** p < 0.005. Scale bar = 100μm.
1. INTRODUCTION
As the most common malignancy among women worldwide, breast cancer (BRCA) is a highly heterogeneous disease, which has substantial genetic and epigenetic changes during the progression of tumor.1 Currently, BRCA is mainly classified based on the expression level of several receptors (ER, PR, and HER2) in BRCA cells.2 Even though various therapeutic strategies have been developed based on different BRCA subtypes, for BRCA patients the outcomes remain unsatisfactory since the intratumor heterogeneity.3 Increasing studies indicated that the intrinsic gene expression profile of the primary tumor can effectively predict the response to therapy and prognosis for patients.4, 5 Therefore, understanding the genetic and epigenetic mechanisms behind BRCA progression is of great significance for the development of novel efficient BRCA therapeutic strategies.
With the rapid development of genome‐wide sequencing, huge progress has been achieved in the identification of biomarkers in various diseases. In recent years, numerous long non‐coding RNAs (lncRNAs) have been found of aberrant expression in diverse cancers, including BRCA.6 lncRNAs refer to a type of transcripts with length exceeding 200 nucleotides, which are lacking in the capacity of protein coding. Despite not involved in protein coding, lncRNAs are capable of regulating pathological processes at epigenetic, transcriptional, and post‐transcriptional levels, thereby playing crucial roles in disease progression.7, 8 A well‐studied lncRNA, MALAT1, can induce cell proliferation, migration, and invasion, which has been proven to be closely correlated with tumor progression and metastasis in BRCA.9 It has been reported that lncRNA can regulate gene expression indirectly by acting as a competing endogenous RNA (ceRNA) which is defined as the regulatory network among RNAs. It is widely accepted that microRNA (miRNA) exerts a regulatory role on target genes, usually by binding to the 3' UTR of targets.10 In a ceRNA regulatory network, lncRNA sponges miRNA thus reducing the pool of available miRNA in the cell, which in turn to up‐regulate the target genes of miRNA.
Zhao, Yu11 indicated that lncRNA‐Xist competed with miR‐101 to regulate C/EBPα and KLF6 expression, thereby suppressing the proliferation and migration abilities of breast tumor. Besides, Dong, Hu12 demonstrated that lncRNA TINCR contributed to trastuzumab resistance and epithelial‐mesenchymal transition (EMT) through sponging miR‐125b in BRCA. Based on an online analysis platform, we found that lncRNA AC016405.3 was significantly up‐regulated in BRCA and closely correlated with poor prognosis. However, there is no report on the biological functions of AC016405.3 in BRCA so far. Therefore, this study sought to investigate the effects of AC016405.3 on BRCA cells and further explore the potential regulatory mechanism behind its functions.
2. MATERIALS AND METHODS
2.1. Bioinformatic analysis online tools
StarBase (http://starbase.sysu.edu.cn/panCancer.php)13 was applied to perform: (1) the differential expression analysis of AC016405.3 and miR‐22‐3p; (2) the survival analysis of AC016405.3; (3) the co‐expression analysis for AC016405.3/miR‐22‐3p and miR‐22‐3p/ERBB3; and (4) the prediction of the target miRNAs of AC016405.3 and the target genes of miR‐22‐3p. StarBase is designed for decoding Pan‐Cancer Networks of lncRNAs, miRNAs, pseudogenes, snoRNAs, RNA‐binding proteins (RBPs), and all protein‐coding genes by analyzing their expression profiles across 32 cancer types integrated from TCGA project.
DIANA‐LncBase v3.0 (http://www.microrna.gr/LncBase)14 was utilized to predict the target miRNAs of AC016405.3.
TargetScan (http://www.targetscan.org/vert_72/)15 was applied to perform the prediction of potential binding sites of miR‐22‐3p with AC016405.3 and ERBB3.
Both the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer‐pku.cn)16 and miRDB (http://mirdb.org)17 online databases were applied to predict the target genes of miR‐22‐3p.
2.2. Cell lines and culture
The human BRCA cell lines (T47D, MDAMB231, MCF7, and ZR7530) and non‐tumorigenic breast epithelial cell line (MCF10A) were cultured in RPMI‐1640 medium (Gibco) containing 5% FBS (Gibco) and 1% penicillin‐streptomycin (Gibco) at 37°C in a 5% CO2 incubator. This study has been approved by the Ethics Committee of International Peace Maternity and Child Health Hospital, School of Medicine, Shanghai Jiao Tong University.
2.3. RT‐qPCR analysis
Total RNA was isolated to conduct reverse transcription. Afterward, RT‐qPCR analysis was performed on the 7500 Real‐Time PCR System (Applied Biosystems) to determine the expression levels of AC016405.3, miR‐22‐3p, miR‐576‐5p, AKT3, PTEV, ESR1, P53, and ERBB3. GAPDH and U6 were utilized for normalization; the expressions of studied RNAs were further analyzed based on the 2–ΔΔCt approach.18 The primer sequences were described in Table S1.
2.4. Cell transfection
Small interference RNA (siRNA) against AC016405.3 (si‐AC016405.3), miRNA mimics and inhibitor for miR‐22‐3p, and the overexpression plasmid of ERBB3 were used to transfect into MCF7 and ZR7530 cells (table S2). Briefly, cells were cultured in plates to the confluence of 80%–90%, followed by transfected with different plasmids by Lipofectamine® 2000 reagent (Invitrogen). Transfection efficiencies were determined by RT‐qPCR.
2.5. CCK‐8 assay
Cells were seeded into 96‐well plates and, respectively, cultivated for 24, 48, 72, and 96 h. CCK‐8 (cat.no ab228554; Abcam) was used to determine the cell viability. At designed time points, 10% (v/v) CCK‐8 solution was added to each well. After further 2‐h incubation, the absorbance values at 450 nm were detected with a Microplate reader (Bio‐Rad).
2.6. Colony formation assay
1.8 × 102 cells were seeded into each well of six‐well plates and incubated for two weeks. Cell colonies were fixed with 4% PFA prior to staining with 1% crystal violet for 15 min. Finally, plates were gently rinsed with distilled water. BX51 microscope (Olympus) was applied to count stained colonies.
2.7. Wound‐healing assay
Cells were seeded into six‐well plates and cultured until a monolayer of cells had formed. The cell monolayers were scraped using a 200‐μl pipette tip. Then, scratched cells were removed by gently rinsing PBS thrice; the remaining cells were further cultured for 36 h. The scratched fields were photographed by a BX51 microscope at 0 and 36 h to determine the cell migration.
2.8. Transwell migration/invasion assay
By using uncoated and Matrigel‐coated membrane of Transwell chambers, migration and invasion assays were conducted, respectively. In brief, cells were suspended with serum‐free medium. 200 μl cell suspensions were added to the upper chamber; and 700 μl medium (10% FBS) was added to the lower chamber. Next, cells were cultivated at 37°C in incubator. 24 h later, the cells passed through membrane were fixed with 4% PFA before staining with 1% crystal violet, after removing cells on the other side of the membrane. Stained cells were counted in five random fields under a BX51 microscope.
2.9. Fluorescence in situ hybridization
The subcellular localization of AC016405.3 was investigated by Fluorescence in situ hybridization (FISH). The experiment was performed as Soares, Maglieri19 previously described. In brief, after the fixation and permeabilization of cells, Cy3‐labeled probe designed against AC016405.3 was utilized to incubate with cells for hybridization overnight. Then, the cells were gently rinsed thrice and stained with DAPI for 10 min. Finally, the cells were rinsed thrice, followed by photographed and observed under a BX51 microscope.
2.10. Dual‐luciferase assay
The AC016405.3 and ERBB3 fragments containing binding site for miR‐22‐3p were cloned into the luciferase reporter vector (Promega) (AC016405.3 WT and ERBB3 WT). Mutation (Mut) sites were also designed (AC016405.3 Mut and ERBB3 Mut). By using Lipofectamine 2000, cells were co‐transfected with the luciferase reporter vector and miR‐22‐3p mimics. Mimics NC was served as negative control. 48h later, the cells were collected to detect the activities of firefly and Renilla luciferase. Relative luciferase activity was represented as the ratio of Renilla/firefly.
2.11. Western blot
Total protein extracted from cells was separated by SDS‐PAGE and transferred onto PVDF membrane. Then, the membrane was blocked with skimmed milk and subsequently incubated with the Anti‐ERBB3 (cat.no ab32121) and Anti‐GAPDH antibodies (cat.no ab8245) overnight. After washing, the membrane was incubated with Goat Anti‐Rabbit IgG H&L (HRP) (cat.no ab6721) for 2 h and visualized by ECL Substrate Kit. The reagents and antibodies used in Western blot (WB) analysis were all obtained from Abcam.
2.12. Anti‐Ago2‐RNA immunoprecipitation assay
Anti‐Ago2‐ RNA immunoprecipitation (RIP) assay was conducted by using a Magna RIP Kit (Millipore, MA, USA) according to the guidelines provided by manufacturers. Briefly, cell extract was immunoprecipitated with magnetic beads conjugated with antibody against Ago2 (#ab32381, Abcam). Then, RNAs that immunoprecipitated by Ago2 were analyzed qRT‐PCR. IgG was served as negative control.
2.13. Statistical analysis
All data were presented as mean ± standard deviation (SD) in this study. All statistical analysis was performed on GraphPad Prism 8.0.1. Students' test or one‐way ANOVA with post hoc test (Bonferroni) was performed to analyze the difference among the groups. It is considered to be statistically significant when p < 0.05.
3. RESULTS
3.1. AC016405.3 is highly expressed in BRCA and correlated with poor prognosis
Based on the bioinformatics analysis from starBase, we found that the expression of AC016405.3 in BRCA samples was significantly higher than that in normal samples (Figure 1A). Afterward, we examined the expression level of AC016405.3 using qPCR on a panel of BRCA cell lines (T47D, MDAMB231, MCF7, and ZR7530) and a non‐tumorigenic breast epithelial cell line MCF10A. All these BRCA cell lines showed a significant increase in AC016405.3 expression relative to MCF10A cells (Figure 1B). In addition, survival analysis showed that BRCA patients with lower AC016405.3 expression had longer survival time, hinting that AC016405.3 is related to poor prognosis in BRCA. These data suggested that AC016405.3 may contribute to the progression of BRCA.
FIGURE 1.
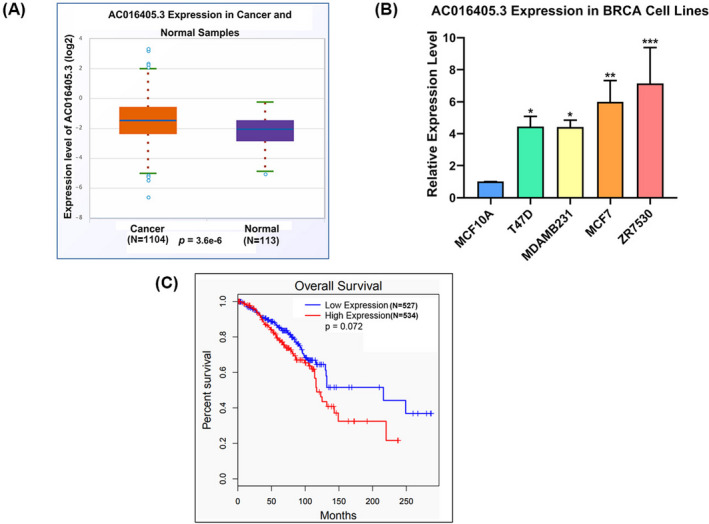
AC016405.3 is highly expressed in BRCA. (A) Differential analysis of AC016405.3 based on starBase database. (B) Basic levels of AC016405.3 in normal breast epithelial cell line (MCF10A) and different BRCA cell lines, as detected with RT‐qPCR. (C) Survival analysis based on starBase database. Note: *p < 0.05, **p < 0.01, and ***p < 0.005
3.2. Knockdown of AC016405.3 could suppress the malignant behaviors of BRAC cells
In order to investigate the roles of AC016405.3 in BRAC, we silenced the expression of AC016405.3 in MCF7 and ZR7530 cells. RT‐qPCR analysis confirmed the successful transfection of si‐AC016405.3 in MCF7 and ZR7530 cells (Figure 2A). CCK‐8 and colony formation assays showed that the knockdown of AC016405.3 obviously decreased the proliferation of MCF7 and ZR7530 cells (Figure 2B and C). Metastasis is the lethal progression of BRCA, so we also assessed the effect of AC016405.3 on the BRCA cell migration and invasion. The results showed that silencing AC016405.3 could significantly reduce not only the motility but also the invasive ability of MCF7 and ZR7530 cells (Figure 2D and E). These results suggested that AC016405.3 could promote the malignant behaviors of BRAC cells.
FIGURE 2.
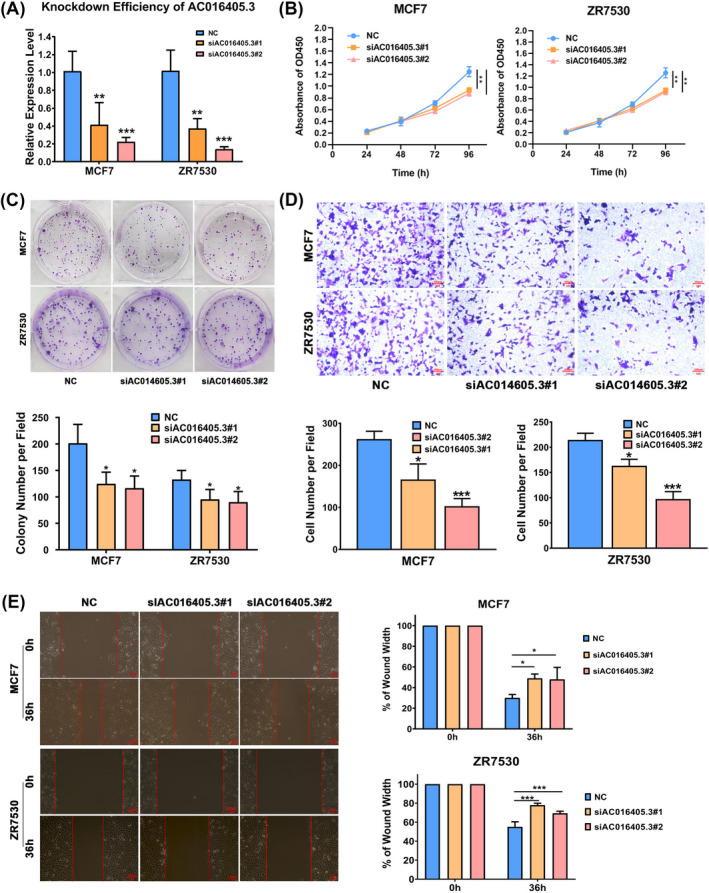
Silencing AC016405.3 could suppress the malignant behaviors of BRAC cells. BRAC cells (MCF7 and ZR7530) were transfected with si‐AC016405.3#1 or si‐AC016405.3#2. (A) Transfection efficiency of si‐AC016405.3 verified by RT‐qPCR. The proliferation of BRAC cells from different groups was examined by (B) CCK‐8 and (C) colony formation assays. The invasion and migration ability of BRAC cells from different groups were detected by (D) Transwell and (E) wound‐healing assays, respectively. Note: *p < 0.05, **p < 0.01 and ***p < 0.005, vs. NC group. Scale bar = 100 μm
3.3. AC016405.3 is mainly distributed in the cytoplasm and directly targets miR‐22‐3p
Since the function of lncRNA is closely related to its subcellular distribution,20 we observed the subcellular localization of AC016405.3 in MCF7 cells by performing FISH. We found that AC016405.3 largely distributed in the cytoplasm (Figure 3A). Consistent with the result of FISH, transcriptome analysis indicated that most AC016405.3 (72.3%) was distributed in the cytoplasm (Figure 3B). As reported, multiple lncRNAs function as ceRNAs by sponging miRNAs in cytoplasm. Hence, we further searched for the interacted miRNAs of AC016405.3 in order to explore the underlying mechanisms by which AC016405.3 regulated the malignant behaviors of BRCA cells. By using starBase and LncBase, nine and thirty‐six target miRNAs were predicted respectively; we subsequently obtained two miRNAs (miR‐576‐5p and miR‐22‐3p) by intersecting the results of two databases (Figure 3C). The expressions of miR‐576‐5p and miR‐22‐3p in MCF7 cells were significantly increased after the silencing AC016405.3, confirmed that a regulatory relationship between AC016405.3 and miR‐576‐5p/miR‐22‐3p (Figure 3D). miR‐22‐3p was selected to perform the further analysis. After predicting the potential binding sites between AC016405.3 and miR‐22‐3p (Figure 3E), their direct interaction was verified by a dual‐luciferase assay (Figure 3F). Contrary to AC016405.3, differential analysis revealed that miR‐22‐3p expression in BRCA samples was statistically lower than that in normal samples (Figure 3G). It is worth noting that miR‐22‐3p was inversely correlated with AC016405.3 in BRCA (Figure 3H). Furthermore, we also investigated miR‐22‐3p expression in MCF10A, T47D, MDAMB231, MCF7, and ZR7530 cell lines. Compared with MCF10A cell line, miR‐22‐3p expression was dramatically decreased in BRCA cell lines (Figure 3I). All these data suggested that AC016405.3 may facilitate the progression of BRCA by reducing miR‐22‐3p expression.
FIGURE 3.
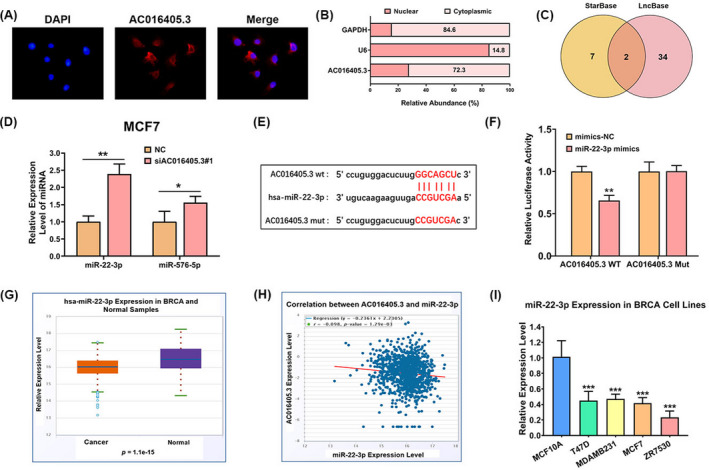
AC016405.3 is mainly distributed in the cytoplasm and directly targets miR‐22‐3p. (A) FISH indicated the subcellular location of AC016405.3 in MCF7 cells (red). (B) Relative expression levels of AC016405.3 in nuclear and cytosolic fractions of MCF7 cells. GAPDH and U6 were served as cytosolic and nuclear controls, respectively. (C) Target miRNAs of AC016405.3 obtained by intersecting the miRNAs predicted by starBase and LncBase. (D) The expression levels of miR‐22‐3p and miR‐576‐5p in MCF7 cells with or without the transfection of si‐AC016405.3. (E) The putative binding sites between AC016405.3 and miR‐22‐3p. (F) The direct interaction between AC016405.3 and miR‐22‐3p verified by dual‐luciferase assay. (G) Differential analysis of miR‐22‐3p and (H) correlation analysis between AC016405.3 and miR‐22‐3p based on starBase database. (I) Basic levels of miR‐22‐3p in normal breast epithelial cell line (MCF10A) and different BRCA cell lines, as detected with RT‐qPCR. Note: *p < 0.05, **p < 0.01, and ***p < 0.005
3.4. AC016405.3 contributes to the malignant phenotypes of BRCA cells via sponging miR‐22‐3p
To verify our hypothesis that AC016405.3 contributes to malignant phenotypes via sponging miR‐22‐3p, we transfected si‐AC016405.3 alone or combination with miR‐22‐3p inhibitor into MCF7 and ZR7530 cells. Initially, RT‐qPCR analysis confirmed the knockdown efficiency of miR‐22‐3p inhibitor (Figure 4A). Consistent with previous experiments, silencing AC016405.3 significantly reduced the proliferation of MCF7 and ZR7530 cells. However, this effect was almost blocked after co‐transfecting with miR‐22‐3p inhibitor (Figure 4B and C). In the meantime, wound‐healing assay and Transwell assay exhibited that miR‐22‐3p inhibitor could effectively restore the decreasing metastatic capacity of BRCA cells induced by AC016405.3 silence (Figure 4D and E). Moreover, miR‐22‐3p inhibitor also remarkably weakened the suppression effect of AC016405.3 silence on the invasion of BRCA cells (Figure 4F). Altogether, the inhibition of miR‐22‐3p contributes to AC016405.3‐mediated malignant phenotypes in BRCA cells.
FIGURE 4.
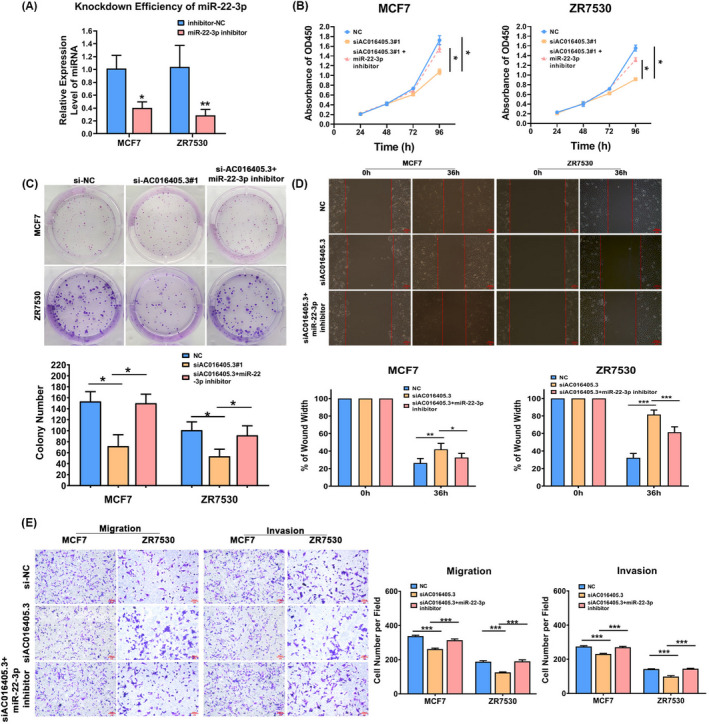
AC016405.3 contributes to the malignant phenotypes of BRCA cells via sponging miR‐22‐3p. BRAC cells (MCF7 and ZR7530) were transfected si‐AC016405.3 alone or combination with miR‐22‐3p inhibitor. (A) Transfection efficiency of miR‐22‐3p inhibitor tested by RT‐qPCR. The proliferation of BRAC cells from different groups was examined by (B) CCK‐8 and (C) colony formation assays. The invasion and migration ability of BRAC cells from different groups were detected by (D) Transwell and (E) wound‐healing assays, respectively. Note: *p < 0.05 and **p < 0.01, vs. si‐AC016405.3 group. Scale bar = 100 μm
3.5. ERRB3 directly targets miR‐22‐3p in BRCA
To reveal the mechanism behind the effects of AC016405.3/miR‐22‐3p on BRCA, we further identified the target genes of miR‐22‐3p. Three bioinformatic tools (miRDB, starBase, and GEPIA) were applied and jointly predicted that 68 genes may be the potential targets of miR‐22‐3p (Figure 5A). Then, AKT3, PTEN, ESR1, P53, and ERBB3 were chosen for verification. Among these five genes, only P53 and ERBB3 expressions were significantly reduced in MCF7 cells after the transfection of miR‐22‐3p mimics (Figure 5B). Then, we selected ERBB3 as the studied objective for further analysis. Correlation analysis based on starBase database exhibited a significantly negative correlation between ERBB3 and miR‐22‐3p in BRCA (Figure 5C). WB analysis indicated that the expression of ERBB3 in MCF7 and ZR7530 cells could be suppressed by miR‐22‐3p mimics, while elevated by miR‐22‐3p inhibitor (Figure 5D). The direct interaction between miR‐22‐3p and ERBB3 was subsequently confirmed (Figure 5F and G). Furthermore, we performed Ago2‐RIP assay, which revealed that miR‐22‐3p and ERBB3 were both enriched in AGO2‐containing immunoprecipitated complexes (Figure 5E). This means, the AGO2 protein bound to ERBB3 and miR‐22‐3p directly in BRCA cells.
FIGURE 5.
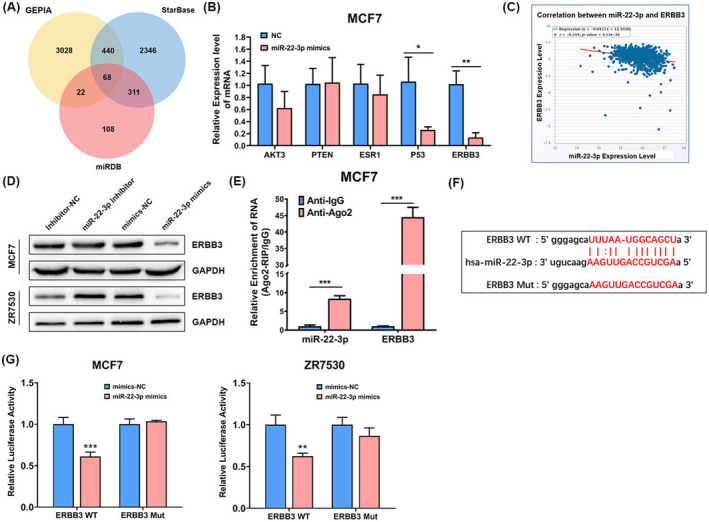
ERRB3 directly targets miR‐22‐3p in BRCA. (A) A Venn diagram showed that 68 genes may be functional targets of miR‐22‐3p. (B) The expression of AKT3, PTEN, ESR1, P53, and ERBB3 in MCF7 cells detected by RT‐qPCR after the transfection of miR‐22‐3p mimics. (C) Correlation analysis between ERBB3 and miR‐22‐3p based on starBase database. (D) The expression of ERBB3 protein in MCF7 cells detected by WB after the transfection of miR‐22‐3p inhibitor or mimics. (E) Ago2‐RIP assay assessed endogenous Ago2 binding to ERBB3 and miR‐22‐3p in MCF7 cells. (F) The binding sites between ERBB3 and miR‐22‐3p predicted by TargetScan. (G) Dual‐luciferase assay confirmed the direct interaction between ERBB3 and miR‐22‐3p. Note: *p < 0.05, **p < 0.01, and ***p < 0.005
3.6. AC016405.3 facilitates BRCA progression by up‐regulating ERBB3 expression
We have validated both AC016405.3 and ERBB3 bond to miR‐22‐3p in BRCA. We wondered whether AC016405.3 contributes to BRCA progression by regulating ERBB3. Then, MCF7 and ZR7530 cells were co‐transfected with si‐AC016405.3 and ERBB3 overexpression vector (ERBB3) or empty vector (EV). CCK‐8 and colony formation assay revealed that ERBB3 obviously blocked the suppression role of si‐AC016405.3 on the proliferation of BRCA cells (Figure 6A and B). Besides, ERBB3 also countered the effects of si‐AC016405.3 on the migration and invasive ability of BRCA cells (Figure 6C‐E). These data collectively suggested that cytoplasmic lncRNA AC016405.3 may sponge miR‐22‐3p to up‐regulating ERBB3, which in turn to promote the progression of BRCA (Figure 6G).
FIGURE 6.
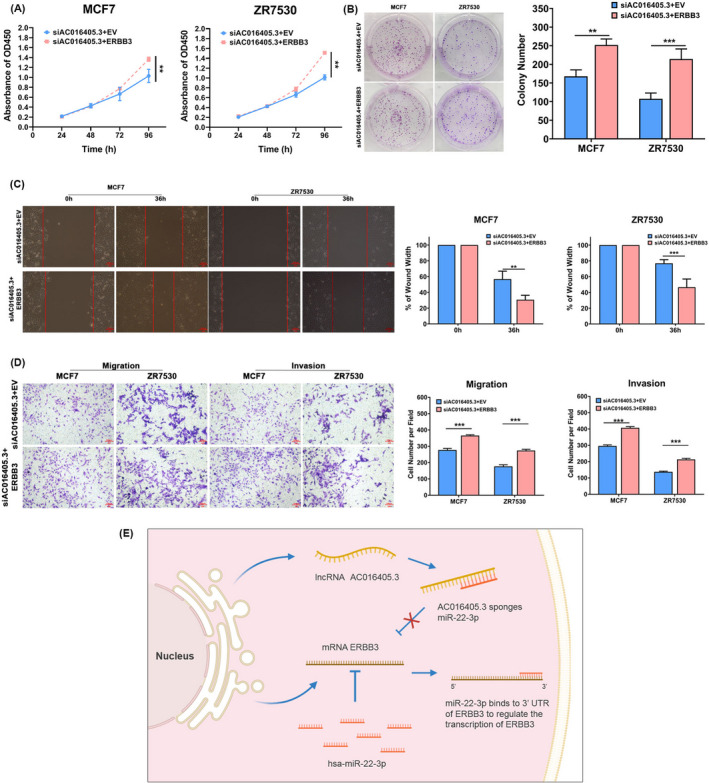
AC016405.3 is involved in BRCA progression by up‐regulating ERBB3 expression via miR‐22‐3p. BRAC cells (MCF7 and ZR7530) were co‐transfected si‐AC016405.3 with empty vector (EV) or ERBB3 overexpression vector (ERBB3). The proliferation of BRAC cells from different groups was examined by (A) CCK‐8 and (B) colony formation assays. (C) Wound‐healing assay assessed the migration ability of BRAC cells from different groups. (D) Transwell assay evaluated both the migration and invasion of BRAC cells from different groups. (E) A schematic diagram exhibited the proposed mechanism that cytoplasmic AC016405.3 sponges miR‐22‐3p to promote ERBB3 expression thus contributes to BRCA progression. Note: **p < 0.01 and ***p < 0.005. Scale bar = 100μm
4. DISCUSSION
In recent years, multiple ceRNA regulatory axes have been reported to be involved in BRCA development and progression, such as SNHG1/miR‐448/IDO,21 SNORD3A/miR‐185‐5p/UMPS,22 and LINC00963/miR‐324‐3p/ACK123 axes. The present study, for the first time, uncovers an AC016405.3/miR‐22‐3p/ERRB3 axis that contributes to the progression of BRCA. Based on an online bioinformatic analysis, the significantly differential expression of AC016405.3 was observed between BRCA and normal samples. AC016405.3 is also named as RP11‐44N11.2, which has been proven to suppress the cell proliferation and metastasis in glioblastoma multiforme (GBM).24 However, there is no study on the roles of AC016405.3 in BRCA progression so far. In contrast to the findings in GBM, survival analysis suggested that AC016405.3 may function as an oncogenic lncRNA in BRCA. Then, a series of functional analyses on BRCA cells with the silence of AC016405.3 were performed and validated the oncogenic potential of AC016405.3, showing that the depletion of AC016405.3 could effectively repress the proliferation, migration, and invasion of BRCA cells.
According to previous studies, lncRNAs with different subcellular localizations exert different functions. In general, lncRNAs distributed in nuclear mainly participate in transcriptional regulation and chromatin modification by recruiting transcription factors or modification enzymes, while lncRNAs distributed in cytoplasm usually act as ceRNAs for miRNAs in turn to modulate the expression of related mRNAs 20. To better understand the molecular mechanisms by which AC016405.3 regulated downstream effectors in BRCA, FISH and subcellular fractionation assays were applied to identify the subcellular localization of AC016405.3 in MCF7 cells. Then, our study first revealed that AC016405.3 was mainly distributed in cytoplasm of MCF7 cells. By using bioinformatic tools, we identified two miRNAs (miR‐22‐3p and miR‐576‐5p) that may target AC016405.3. In our study, silencing AC016405.3 significantly increased the expression of both miR‐576‐5p and miR‐22‐3p in MCF7 cells. Multiple studies demonstrated that miR‐22‐3p serves as an anti‐oncogenic gene in diverse cancers,25 including BRCA.26, 27 Nakamura, Hayashi28 found that the up‐regulation of miR‐22‐3p could increase radiosensitivity in cervical cancer. Recently, Yang, Su29 indicated that overexpressing miR‐22 could suppress breast cancer cell proliferation, metastasis, and invasion, as well as tumorigenesis by targeting eEF‐2K in triple‐negative BRCA. Based on the biological functions of miR‐22‐3p in cancers, we speculated that AC016405.3 may promote BRCA progression by sponging miR‐22‐3p. According to the results obtained from starBase, there is a significantly negative correlation between AC016405.3 and miR‐22‐3p in BRCA specimens. Dual‐luciferase assay also confirmed their direct interaction. Moreover, inhibiting miR‐22‐3p could obviously reverse the suppression effects of AC016405.3 depletion on the proliferation, migration, and invasion of BRCA cells. Therefore, the promotion role of AC016405.3 in BRCA progression, at least in part, is depending on sponging miR‐22‐3p.
In order to better understand how AC016405.3/miR‐22‐3p contributes to the malignant behaviors of BRCA cells, we further searched for the direct targeted genes of miR‐22‐3p. Based on four prediction online tools, a total of 68 target genes were obtained in our study. Among these genes, we selected five (AKT3, PTEN, ESR1, P53, and ERBB3) to investigate the relationship between them and miR‐22‐3p, given that they were widely reported to be associated with BRCA progression.30, 31, 32, 33, 34 Ling, Wang35 indicated that miR‐22‐3p could obviously inhibit cell proliferation and metastasis in lung cancer through directly targeting ERBB3. Our study found that overexpressing miR‐22‐3p could significantly suppress both the mRNA and protein expression levels of ERBB3. Then, by using dual‐luciferase assay, we also confirmed that ERBB3 is a direct target of miR‐22‐3p in BRCA cells. Additionally, RIP assay showed that both ERBB3 and miR‐22‐3p are enriched in the Ago2‐containing immunoprecipitate, which exerts a critical role in gene silencing mediated by miRNA.36 Therefore, ERBB3 may be a crucial downstream effector of AC016405.3/miR‐22‐3p regulating BRCA progression. To verify this point, rescue experiments were further carried out. The results showed that the overexpression of ERBB3 could obviously block the inhibitory effects on proliferation, migration, and invasion of BRCA cells induced by AC016405.3 depletion. These findings collectively imply that AC016405.3 promotes the progression of BRCA via modulating miR‐22‐3p/ERBB3. Further in vivo studies are needed, however, to validate the functions of AC016405.3/miR‐22‐3p/ERRB3 in BRCA models.
In summary, our study identified a novel regulatory axis (AC016405.3/miR‐22‐3p/ERRB3) that modulates the malignant phenotypes of BRCA cells, which may help expand the existing understanding of the molecular mechanisms on BRCA progression, and provide some inspiration in developing novel therapeutic strategies.
CONFLICT OF INTERESTS
The authors declare that they have no competing interests.
Supporting information
Table S1‐2
Wei M, Wang J, He Q, Liu L, Wang Z. AC016405.3 functions as an oncogenic long non‐coding RNA by regulating ERBB3 via sponging miR‐22‐3p in breast cancer. J Clin Lab Anal. 2021;35:e23952. 10.1002/jcla.23952
Funding information
This study was supported in part by grant funding from Natural Science Foundation of China (grant number 81101847); Doctoral Fund of Ministry of Education of China (grant number 20110073120089); Shanghai Committee of Science and Technology, China (grant number 124119a4801); NUST Research Funding (grant number 2010ZYTS068); and Fund of International Peace Maternity and Child Health Hospital (grant number GFY 5502; GFY 9307)
DATA AVAILABILITY STATEMENT
The data used to support the findings of this study are available from the corresponding author upon request.
REFERENCES
- 1.Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev. 2017;26:444‐457. [DOI] [PubMed] [Google Scholar]
- 2.Waks AG, Winer EP. Breast cancer treatment: a review. Jama. 2019;321:288‐300. [DOI] [PubMed] [Google Scholar]
- 3.Tong CWS, Wu M, Cho WCS, To KKW. Recent advances in the treatment of breast cancer. Front Oncol. 2018;8:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrell JC, Prat A, Parker JS, et al. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat. 2012;132:523‐535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey LA, Berry DA, Cirrincione CT, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol. 2016;34:542‐549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Zhuang J, Liu L, et al. Integrative transcriptome data mining for identification of core lncRNAs in breast cancer. PeerJ. 2019;7:e7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dykes IM, Emanueli C. Transcriptional and post‐transcriptional gene regulation by long non‐coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non‐coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arun G, Spector DL. MALAT1 long non‐coding RNA and breast cancer. RNA Biol. 2019;16:860‐863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Z, Rajewsky N. The impact of miRNA target sites in coding sequences and in 3'UTRs. PLoS ONE. 2011;6:e18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Yu Z, Ma R, et al. lncRNA‐Xist/miR‐101‐3p/KLF6/C/EBPα axis promotes TAM polarization to regulate cancer cell proliferation and migration. Mol Ther Nucleic Acids. 2021;23:536‐551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong H, Hu J, Zou K, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial‐mesenchymal transition by targeting MicroRNA‐125b in breast Cancer. Mol Cancer. 2019;18:3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA‐ceRNA, miRNA‐ncRNA and protein‐RNA interaction networks from large‐scale CLIP‐Seq data. Nucleic Acids Res. 2014;42:D92‐D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karagkouni D, Paraskevopoulou MD, Tastsoglou S, et al. DIANA‐LncBase v3: indexing experimentally supported miRNA targets on non‐coding transcripts. Nucleic Acids Res. 2020;48:D101‐D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98‐W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127‐D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25:402‐408. [DOI] [PubMed] [Google Scholar]
- 19.Soares RJ, Maglieri G, Gutschner T, et al. Evaluation of fluorescence in situ hybridization techniques to study long non‐coding RNA expression in cultured cells. Nucleic Acids Res. 2018;46:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR‐448/IDO. Int J Biol Macromol. 2018;118:24‐30. [DOI] [PubMed] [Google Scholar]
- 22.Luo L, Zhang J, Tang H, et al. LncRNA SNORD3A specifically sensitizes breast cancer cells to 5‐FU by sponging miR‐185‐5p to enhance UMPS expression. Cell Death Dis. 2020;11:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N, Zeng X, Sun C, et al. LncRNA LINC00963 promotes tumorigenesis and radioresistance in breast cancer by sponging miR‐324‐3p and inducing ACK1 expression. Mol Ther Nucleic Acids. 2019;18:871‐881. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Ren S, Xu Y. AC016405.3, a novel long noncoding RNA, acts as a tumor suppressor through modulation of TET2 by microRNA‐19a‐5p sponging in glioblastoma. Cancer Sci. 2019;110:1621‐1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Wang YS, Mugiyanto E, Chang WC, Yvonne Wan YJ. MiR‐22 as a metabolic silencer and liver tumor suppressor. Liver Res. 2020;4:74‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorur A, Bayraktar R, Ivan C, et al. ncRNA therapy with miRNA‐22‐3p suppresses the growth of triple‐negative breast cancer. Mol Ther Nucleic Acids. 2021;23:930‐943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong LM, Liao CG, Zhang Y, et al. A regulatory loop involving miR‐22, Sp1, and c‐Myc modulates CD147 expression in breast cancer invasion and metastasis. Can Res. 2014;74:3764‐3778. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura M, Hayashi M, Konishi H, et al. MicroRNA‐22 enhances radiosensitivity in cervical cancer cell lines via direct inhibition of c‐Myc binding protein, and the subsequent reduction in hTERT expression. Oncol Lett. 2020;19:2213‐2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Su W, Li Y, et al. MiR‐22‐3p suppresses cell growth via MET/STAT3 signaling in lung cancer. Am J Transl Res. 2021;13:1221‐1232. [PMC free article] [PubMed] [Google Scholar]
- 30.Chin YR, Yoshida T, Marusyk A, Beck AH, Polyak K, Toker A. Targeting Akt3 signaling in triple‐negative breast cancer. Can Res. 2014;74:964‐973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangel R, Lee SC, Hon‐Kim Ban K, et al. Transposon mutagenesis identifies genes that cooperate with mutant Pten in breast cancer progression. Proc Natl Acad Sci USA. 2016;113:E7749‐E7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol.. 2015;12:573‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertheau P, Lehmann‐Che J, Varna M, et al. p53 in breast cancer subtypes and new insights into response to chemotherapy. Breast (Edinburgh, Scotland). 2013;22(Suppl 2):S27‐S29. [DOI] [PubMed] [Google Scholar]
- 34.Morrison MM, Hutchinson K, Williams MM, et al. ErbB3 downregulation enhances luminal breast tumor response to antiestrogens. J Clin Investig. 2013;123:4329‐4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling B, Wang GX, Long G, Qiu JH, Hu ZL. Tumor suppressor miR‐22 suppresses lung cancer cell progression through post‐transcriptional regulation of ErbB3. J Cancer Res Clin Oncol. 2012;138:1355‐1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golden RJ, Chen B, Li T, et al. An Argonaute phosphorylation cycle promotes microRNA‐mediated silencing. Nature. 2017;542:197‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐2
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


