Abstract
The transcription factor E2F-1 directs the expression of genes that induce or regulate cell division, and a role for E2F-1 in driving cells into apoptosis is the subject of intense discussion. Recently it has been shown that E2F-1 binds and coprecipitates with the mouse double-minute chromosome 2 protein (Mdm2). A domain of E2F-1 (amino acids 390 to 406) shows striking similarity to the Mdm2 binding domain of the tumor suppressor protein p53. It is known that interaction of Mdm2 with p53 through this domain is required for Mdm2-dependent degradation of p53. We show here that E2F-1 protein is upregulated in response to DNA damage. The kinetics of induction are dependent upon the source of DNA damage, i.e., fast and transient after irradiation with X rays and delayed and stable after irradiation with UVC, and thus match the kinetics of p53 induction in response to DNA damage. We show further that E2F-1 is also upregulated by treatment with the transcription inhibitor actinomycin D and with the kinase inhibitor DRB, as well as by high concentrations of the kinase inhibitor H7, all conditions which also upregulate p53. In our experiments we were not able to see an increase in E2F-1 RNA production but did find an increase in protein stability in UVC-irradiated cells. Upregulation of E2F-1 in response to DNA damage seems to require the presence of wild-type p53, since we did not observe an increase in the level of E2F-1 protein in several cell lines which possess mutated p53. Previous experiments showed that p53 is upregulated after microinjection of an antibody which binds to a domain of Mdm2 that is required for the interaction of Mdm2 with p53. Microinjection of the same antibody also increases the expression of E2F-1 protein, while microinjection of a control antibody does not. Furthermore, microinjection of Mdm2 antisense oligonucleotides upregulates E2F-1 protein, while microinjection of an unrelated oligonucleotide does not. These data suggest that E2F-1 is upregulated in a similar way to p53 in response to DNA damage and that Mdm2 appears to play a major role in this pathway.
E2F-1 is a member of a family of transcription factors whose target genes control entry into and progression through the S phase of the cell cycle (11). For high-affinity binding to the E2F-1 consensus sites at promoters of target genes, E2F-1 needs to heterodimerize with a member of the DP family of transcription factors. Binding to pRb, on the other hand, regulates the activity of E2F-1 (reviewed in reference 24): hypophosphorylated pRb binds to the activation domain of E2F-1, rendering the protein inactive (14, 20, 25). Before entry into S phase, pRb becomes phosphorylated by cyclin-dependent kinases, disrupting the interaction between pRb and E2F-1 and allowing the transcription of E2F-1 target genes (54). Deregulation of the activity of E2F family members appears to be a hallmark of all human cancers (2, 52). Transcription of E2F-1 is induced in late G1, and Johnson et al. have shown that overexpression of E2F-1 is sufficient for entry into S phase and DNA synthesis (32). However, unscheduled E2F-1 activity during S phase leads to cell cycle arrest and apoptosis (28, 31, 35, 51), and studies of mice nullizygous for E2F-1 suggest that E2F-1 can exert tumor-suppressing activity (13, 57, 58).
Recent studies have shown that E2F-1 is regulated by the ubiquitin proteasome pathway (8, 22, 27), and the carboxyl terminus is thought to control protein stability. Interestingly, E2F-1 physically interacts with Mdm2 (41), a protein which is known to target the tumor suppressor protein p53 for rapid degradation by the ubiquitin proteasome pathway (23, 36).
When normal cells are exposed to DNA-damaging agents, p53 accumulates and transcription of p53-responsive target genes is activated. This leads to upregulation of WAF/p21, GADD45, cyclin G, Bax, and Mdm2 (1, 12, 21, 33, 44, 47, 56); induction of cell cycle arrest (37); or apoptosis. The mechanism of p53 accumulation in response to DNA damage is still not understood, but it requires, at least in part, protection against proteolysis, since the half-life of the protein is prolonged (40). p53 degradation is regulated by the ubiquitin-proteolysis system (39), which requires a ubiquitin-target protein adduct formation. This complex is built up by the activity of three enzymes: a ubiquitin-activating enzyme, a ubiquitin-conjugating enzyme, and a ubiquitin ligase. In cells infected with human papillomavirus type 16 (HPV-16) or HPV-18, papillomavirus E6 protein and cellular E6-AP protein form a complex and function as a ubiquitin ligase for p53 (50), abolishing the normal p53-dependent stress response in HPV-infected cells.
Since tight regulation of p53 is critical not only for various stress responses but also for normal cell growth and genetic stability, the ubiquitin ligase for p53 was of intense interest until it was recently discovered that the oncoprotein Mdm2 was the missing piece of the puzzle (29). Thus, p53 abundance seems to be regulated by an autoregulatory feedback loop, involving Mdm2: the mdm2 gene is transcriptionally activated by binding of p53 to an internal promoter within the gene (1, 56), and Mdm2 protein binds to p53 and targets it for degradation. Support for the importance of this autoregulatory loop in regulating p53 expression levels comes from experiments in which the p53-Mdm2 interaction has been interrupted, with the result that p53 protein accumulated and p53-responsive reporter genes were activated (6, 43). Recently it has been shown that p53 has to be exported to the cytoplasm in order to be degraded and that Mdm2 acts as the scavenger for this process by providing the nuclear export signal (17, 49). The presence of Mdm2 alone, however, seems to be insufficient for degradation of p53. In cells, most of the endogenous Mdm2 protein is complexed with the histone acetylase p300, and Grossman et al. have shown that the specific interaction between p300 and Mdm2 is required for the degradation of p53 (18). Interestingly, p300 also binds to and acetylates p53 (19). It is, however, still unclear if the formation of a ternary complex is sufficient for p53 degradation or if Mdm2 needs to be acetylated by p300.
Since E2F-1 is, like p53, targeted by the ubiquitin-dependent degradation system and binds to Mdm2 (22, 41), we asked if it was also regulated like p53, in response to DNA damage. We found induction of the E2F-1 protein following DNA damage. There were striking differences in the kinetics and magnitude in response to X rays and UVC that were equivalent to the different p53 responses to these signals (38). Induction of E2F-1 was not the result of increased transcription of the E2F-1 gene; instead, E2F-1 protein was stabilized in UVC-irradiated cells. In our experiments, not only was E2F-1 expression induced by DNA damage, but also inhibition of protein kinase activity could upregulate E2F-1. This upregulation of E2F-1 was accompanied by downregulation of Mdm2 and supported the idea that interruption of Mdm2–E2F-1 complexes could upregulate E2F-1 expression, just as interruption of Mdm2-p53 complex formation upregulates p53 expression (6, 43). The induction of E2F-1 protein after microinjection of mdm2 antisense oligonucleotides and anti-Mdm2 antibody 3G5 (which binds to an epitope at the amino terminus of Mdm2 and inhibits binding of Mdm2 to p53) supported this concept. These observations suggest that E2F-1 may be regulated similarly or even identically to p53. Both proteins are stabilized in response to DNA damage, both proteins are degraded by the ubiquitin-dependent degradation system, and both proteins bind to Mdm2, suggesting that Mdm2 is a key regulator of p53 and E2F-1 expression in response to DNA damage.
MATERIALS AND METHODS
Cell lines and their treatment.
U2-OS cells (6), A431 cells (43), T47D cells (45), HAKAT cells (7), MCF-7 cells (43), and OSA cells (6) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS), penicillin (100 U/ml), and streptomycin (100 μg/ml). GM02184D cells (NIGMS, Camden, N.J.), were grown in RPMI medium supplemented with 15% heat-inactivated FCS and antibiotics (100 U of penicillin per ml, 100 μg of streptomycin per ml). All cells were maintained at 37°C and 6% CO2 in a humidified atmosphere. They were treated 3 days after plating, at 80% confluence.
X-ray irradiation was performed in culture medium with 5 Gy in a TORREX 150D X-ray source (Astrophysics Research Corp., Long Beach, Calif.) at a dose rate of 5 Gy/min with settings at 5 mA and 145 kV. Prior to UVC irradiation, the culture medium was removed, and the cell layer was then irradiated with 30 J/m2 with a Stratalinker (Stratagene) and further cultured in the original conditioned medium. Actinomycin D, solubilized in ethanol, was added to the culture medium at a final concentration of 2 μg/ml or 60 ng/ml. 1-(5-Isoquinolinesulfonyl)-2-methylpiperazine (H7), solubilized in H2O, was added at a final concentration of 10 or 100 μM. 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazol (DRB), solubilized in dimethyl sulfoxide, was added at a final concentration of 250 μM, and cycloheximide, solubilized in water, was added at a final concentration of 20 μg/ml.
Western blotting.
Cells were washed twice in ice-cold phosphate-buffered saline (PBS), scraped into PBS, and centrifuged at 1,000 × g for 5 min. The cells were lysed in 250 mM Tris (pH 7.8)–60 mM KCl–1 mM EDTA–1 mM dithiothreitol–1 mM phenylmethylsulfonyl fluoride by three cycles of freezing and thawing. The cell extracts were centrifuged at 13,000 × g for 10 min at 4°C, and the protein concentration of the supernatant (protein extract) was determined by the Bradford method (Bio-Rad). A 45-μg portion of total protein (unless otherwise indicated) was boiled for 5 min in sample buffer (2% sodium dodecyl sulfate [SDS], 80 mM Tris [pH 6.8], 10% glycerol, 5% 2-mercapthoethanol, 0.001% bromophenol blue), separated on an SDS–10% polyacrylamide gel, and transferred onto a polyvinylidene difluoride membrane (Millipore). The membrane was blocked for 30 min in 5% dry milk–0.2% Tween 20 in PBS. Primary antibodies C-20 (anti-E2F; Santa Cruz), diluted 1:500, CM-1 (anti-p53) (42) diluted 1:1,000, PC-10 (anti-proliferating-cell nuclear antigen [anti-PCNA] ascites) (53) diluted 1:3,000, and 4B2 (anti-Mdm2) (9) at 2.9 μg/ml were used. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit immunoglobulin G (IgG) (DAKO) diluted 1:1,000 were used as secondary antibodies. All antibodies were diluted in 5% dry milk–0.2% Tween 20 in PBS at the indicated concentrations, incubated for 90 min, and given three 5-min washes with PBS–0.2% Tween 20. The Western blots were developed by the enhanced chemiluminescence method.
Northern blotting.
Cells were washed twice in ice-cold PBS, scraped in PBS, and centrifuged at 1,000 × g for 5 min. Poly(A)+ mRNA was prepared with the QuickPrepR Micro mRNA purification kit (Promega) as specified by the manufacturer. A 4.5-μg portion of poly(A)+ mRNA was resolved on a 1.4% agarose–formaldehyde gel, transferred onto a Hybond N+ nylon membrane, and hybridized with a SalI-XhoI fragment of the human E2F-1 gene (provided by Ed Harlow, Boston, Mass.) or with a HindIII fragment of human mdm2. The filters were reprobed with a PstI fragment of the open reading frame of mouse glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) cDNA (16).
Microinjection into cells.
Cells grown on 22-mm glass coverslips were injected with protein A-purified monoclonal antibodies 3G5 and SMP 14 (1 mg/ml) or mdm2 antisense and control oligonucleotides (1 mg/ml) (10) together with an unrelated monoclonal antibody (1 mg/ml) by using the Eppendorf 5242 microinjector and 5170 micromanipulator mounted to an Axiovert 35 M with heated stage. After a 20-h incubation, the cells were fixed for 10 min in 1% paraformaldehyde, permeabilized in 1% Nonidet P-40 (NP-40) in PBS, and blocked in 10% FCS in PBS for 30 min. They were incubated for 1 h with C-20 diluted 1:300 in 10% FCS in PBS, washed with PBS, and incubated for 1 h with fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG and Texas red-conjugated goat anti-mouse IgG (Jackson Immunochemicals) diluted 1:500 in 10% FCS in PBS. They were then washed four times in PBS, stained with 4′,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml; Sigma) for 2 min, and washed again with PBS; the coverslips were mounted on microscope slides with Hydromount (National Diagnostics)–2.5% 1,4-diazabicyclo[2.2.2]octane (Sigma).
RESULTS AND DISCUSSION
Principle of the study.
Since the 1984 study by Maltzman and Czyzyk (40), it has been known that the tumor suppressor protein p53 is upregulated in response to irradiation by prolongation of the half-life of the protein. Today the mechanism leading to this stabilization is still not completely understood. Recently, it has been shown that overexpression of Mdm2 reduces the amount of endogenous p53 (36) and that cotransfection of mdm2 and p53 into human cell lines reduces p53 expression compared to transfection of p53 alone (23). These findings suggest that Mdm2 can downregulate p53 expression. Honda et al. showed that Mdm2 is a ubiquitin ligase for p53, thus marking p53 for rapid degradation by proteasomes (29), and we found that expression of a synthetic miniprotein that competes with p53 for Mdm2 binding induces p53 expression and activates the p53 response (6). Furthermore, increased stability of exogenous mutated p53 stably expressed in tumor cells turned out not to be dependent on individual mutations but to depend strictly on the binding of p53 to Mdm2 (43). The mdm2 gene is transcriptionally activated by binding of p53 to an internal promoter within the gene (5, 56). However, p53 is frequently mutated in tumor cells, and many mutations target the function of p53 as a transcription factor. As a consequence, Mdm2 expression is downregulated in many tumors and it can no longer target p53 for rapid degradation. This evidence led to the idea that Mdm2 might be a key regulator of p53 stability and that inhibition of the interaction of p53 and Mdm2, e.g., by posttranslational modifications, could result in the stabilization of p53, such as that observed after DNA damage. It should be noted that all the above observations were obtained with genetically engineered cells and that one should be careful interpreting the data in relation to the processes occurring in normal cells.
The recent analysis of p53 constructs in which all known phosphorylation sites have been point mutated or deleted and which have been expressed in eucaryotic cells showed that phosphorylation of p53 is not likely to be required for the stabilization of p53 in response to DNA damage (4, 26). However, if p53 itself is not modified by DNA damage in a way that abolishes its degradation by the ubiquitin-dependent degradation process, another protein of this system may potentially be modified in such a way as to impair the degradation of p53. Mdm2 is a very attractive candidate as a target for such modifications. If, however, modifications of Mdm2 interfere with the degradation of p53, other proteins which bind to Mdm2 could be regulated similarly. We were therefore particularly interested in identifying a protein which binds to Mdm2 and which is regulated in a way similar to p53, because this would further support our idea that binding to Mdm2 is crucial for p53 instability and that inhibition of this interaction stabilizes p53, e.g., after DNA damage. A recent report showed that Mdm2 functionally cooperates with the transcription factor E2F-1; moreover, it was reported that the p53 binding domain of Mdm2 also binds directly and specifically to E2F-1 (41). The E2F-1 protein is therefore a good candidate for Mdm2-dependent regulation of stability.
E2F-1 is differentially upregulated in response to X-ray and UVC irradiation.
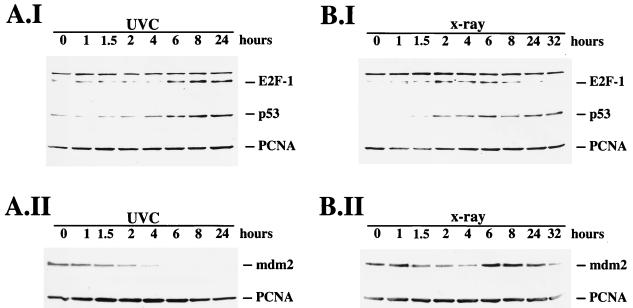
We wished to investigate whether E2F-1 might be upregulated following DNA damage in a similar way to the upregulation of p53. We therefore irradiated U2-OS cells either with 30 J of UVC light per m2 or with 5 Gy of X rays, harvested the cells at different time points after irradiation, and analyzed the cell extracts for E2F-1 expression by Western blotting. We found that both ionizing radiation and UVC irradiation increased the expression level of E2F-1 (Fig. 1), but we noted a striking difference in the kinetics of E2F-1 expression following the two forms of radiation. The increase in the level of E2F-1 protein was detectable 4 to 6 h after exposure to UVC, increased with time, and remained high for at least 24 h (Fig. 1A.I). In contrast, induction after X-ray application was more rapid, showing a response as early as 1.5 to 2 h and reaching a plateau after 2 h. The most striking difference, however, was the decrease in E2F-1 levels 6 to 8 h after irradiation with X rays (Fig. 1B.I). The overall increase in E2F-1 protein expression after UVC irradiation was much higher than the response to X rays. E2F-1 expression in response to irradiation thus showed a striking similarity to p53 expression in response to the same stimuli, which is also differentially induced after ionizing and UVC irradiation (38). To confirm that p53 and E2F-1 show the same kinetics in response to irradiation, we rehybridized the Western blot membranes with an antibody recognizing p53. As expected, the kinetics of p53 induction were very similar to the kinetics of induction of E2F-1, showing a rapid and transient increase after irradiation with X rays and a delayed and persistent induction after irradiation with UVC, which exceeded the induction of p53 at the peak of the X-ray response. Overall, the p53 induction seemed to be slightly faster than the E2F-1 induction, since it was already clearly detectable 2 to 4 h after irradiation with UVC. Interestingly, p53 displayed a second wave of induction at 24 to 32 h after irradiation with X rays in this cell line, which is clearly distinct from E2F-1 expression (Fig. 1B.I). While p53 and E2F-1 accumulated during the time course after UVC irradiation, Mdm2 expression decreased continually in U2-OS cells, becoming undetectable 6 h after UVC irradiation. Thus, the decrease in Mdm2 expression exactly preceded the increase in E2F-1 and p53 expression (Fig. 1A.II). Although the mdm2 gene is a p53 target gene, we were not able to detect induction of Mdm2 by p53 in response to UVC irradiation during the time course in these cells. This result is consistent with the observation of Wu and Levine (55) that the induction of Mdm2 in response to high-dose UV irradiation is quite late and delayed compared, e.g., to the induction of p21, another p53 target gene, which is already accumulating 2 to 5 h after UVC irradiation in some cell lines. In response to irradiation with X rays, there was also a slight initial decrease in Mdm2 expression in U2-OS cells. This decrease in Mdm2 expression preceded the induction of E2F-1 and p53. At 4 to 6 h after X-ray irradiation, Mdm2 expression increased, probably due to activated p53, whose expression reached maximal levels just before the increase in Mdm2 protein became visible. This increased Mdm2 expression then correlated with a decrease in the expression of E2F-1 and p53 8 h after X-ray irradiation. These data suggest that high expression of p53 and E2F-1 and high expression of Mdm2 are mutually exclusive, pointing to a potential function of Mdm2 not only in the regulation of p53 but also in the regulation of E2F-1. However, the question whether the initial decrease in Mdm2 expression in response to X rays is sufficient for the upregulation of E2F-1 and p53 or whether further modifications of the proteins involved are required remains to be elucidated in further experiments.
FIG. 1.
Time course of E2F-1 induction in response to DNA damage. U2-OS cells were irradiated either with UVC (A) or X rays (B). At the indicated time points, the cells were harvested and 45 μg of protein was separated on a 10% polyacrylamide minigel. After Western transfer, the membranes were consecutively hybridized with antibodies recognizing E2F-1 (C-20), p53 (CM-1), and PCNA (PC10) (A.I and B.I) or with antibodies recognizing Mdm2 (4B2) and PCNA (A.II and B.II). The Western blots were developed by the enhanced chemiluminescence method. The time course of E2F-1 induction and the time course of p53 induction in response to irradiation are almost identical.
Rehybridization of the membranes with an antibody recognizing PCNA showed no major differences in PCNA signals during the time courses, confirming that approximately equal amounts of proteins had been loaded onto the gel (Fig. 1). In our experiment, we were not able to confirm the results of Huang et al. (30), who detected upregulation of E2F-1 from 6 to 24 h after X-ray treatment. The reason for this might be the different cell types used.
We extended our study to another cell line to confirm that the upregulation of E2F-1 in response to irradiation is not restricted to U2-OS cells. We used a human lymphoblastoid cell line (GM02184D) originating from a healthy donor, irradiated the cells, and analyzed them at different times after irradiation for E2F-1, p53, and Mdm2 expression. From previous experiments, we knew that p53 was strongly induced in this cell line both at 1.5 and 4 h after irradiation with X rays and at 4 and 9 h after irradiation with UVC. As expected, E2F-1 was upregulated in this cell line at both time points after UVC and X-ray irradiation (Fig. 2). We probed also for p53 expression and found that p53 was induced simultaneously (Fig. 2). The analysis of Mdm2 expression in these cells showed that Mdm2 expression was also increased at both 1.5 and 4 h after X-ray irradiation and at 9 h after UVC irradiation (Fig. 2) and thus that induction of Mdm2 is much faster in these cells than in U2-OS cells.
FIG. 2.
E2F-1 upregulation in response to DNA damage is not restricted to a particular cell line. GM02184D cells (human lymphoblast cell line from a healthy donor) were irradiated with X rays (5 Gy) or UVC (30 J/m2) or treated with actinomycin D (Act. D; 60 ng/ml or 2 μg/ml, as indicated). The cells were harvested 0, 1.5, and 4 h after irradiation with X rays or 0, 5, and 9 h after irradiation with UVC or after treatment with actinomycin D. Proteins (75 μg) were separated on a 10% polyacrylamide gel and analyzed by the Western blot method for expression of Mdm2, E2F-1, and p53 protein by using the 4B2 anti-Mdm2, C-20 anti-E2F-1, and CM-1 anti-p53 antibodies.
p53 is very strongly induced not only by DNA damage caused by irradiation but also by actinomycin D, an agent which intercalates into DNA, resulting in DNA strand breaks. The kinetics of p53 induction by actinomycin D is very similar, if not identical, to the kinetics of p53 induction in response to UVC (4). If p53 and E2F-1 are induced after DNA damage by the same mechanism, E2F-1 should also be upregulated by actinomycin D. To test this prediction, we treated the cells with two different concentrations of actinomycin D, harvested the cells at two different time points after treatment, and analyzed the cell extracts for both E2F-1 and p53 expression. Figure 2 shows that both p53 and E2F-1 are strongly induced by both concentrations of actinomycin D at both time points and that Mdm2 protein levels are decreased after treatment with actinomycin D and are undetectable 9 h after the cells were treated with 60 ng of actinomycin D per ml and 5 and 9 h after the cells were treated with 2 μg of actinomycin D per ml.
Immunofluorescence staining of cells showed that p53 upregulation in response to irradiation is very heterogeneous among individual cells (38). While most cells show very little enhanced nuclear p53 staining, in a small percentage of the cells there is an intense accumulation of p53. We analyzed E2F-1 expression in individual cells in response to UVC irradiation by immunofluorescence staining and found that E2F-1 expression in response to DNA damage is as heterogeneous as is p53 expression. Moreover, intense nuclear E2F-1 and p53 staining colocalized in the very same cell (data not shown). This observation strongly indicates that the two proteins are coregulated: whatever factor causes variation between cells in the p53 response causes exactly the same variation in the E2F-1 response.
E2F-1 upregulation is due to prolongation of the protein half-life.
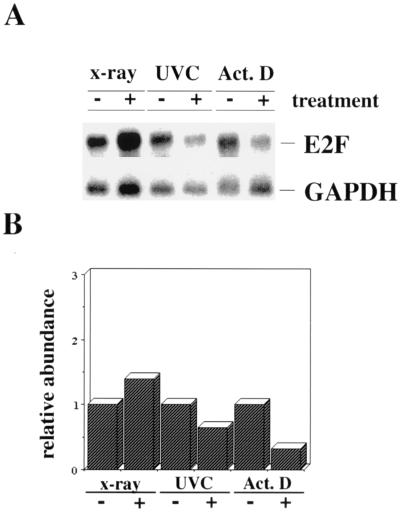
We investigated whether the increase in E2F-1 expression is preceded by an upregulation of E2F-1 RNA. Accumulation of p53 protein after DNA damage is due to protein stabilization and not to enhanced synthesis of the protein or to increased transcription of p53 RNA (4, 40). If the two proteins are indeed coregulated, we would not expect an upregulation of E2F-1 RNA after DNA damage. To test this prediction, we irradiated U2-OS cells with X rays or UVC or treated the cells with actinomycin D and analyzed poly(A)+ mRNA for E2F-1 expression. Simultaneously, we analyzed the proteins for upregulation of E2F-1 in Western blots (data not shown). In our experiments, the E2F-1 RNA level remained constant after irradiation with X rays (Fig. 3). The slight induction of E2F-1 RNA in Fig. 3B was not consistently seen in all experiments. Also, after irradiation with UVC or treatment with actinomycin D, we did not observe any detectable accumulation of E2F-1 RNA. Instead, we repeatedly found a slight reduction in E2F-1 RNA expression after these treatments. The signal produced by the GAPDH RNA displays the amount of poly(A)+ mRNA that was transferred onto the membrane, confirming that there is no enhanced expression of E2F-1 RNA as a result of the various treatments.
FIG. 3.
No induction of E2F-1 RNA in response to DNA damage. (A) U2-OS cells were irradiated with X rays (5 Gy) and harvested after 2.5 h, irradiated with UVC (30 J/m2) and harvested after 8 h, or treated with actinomycin D (Act. D; 60 ng/ml) and harvested after 8 h or mock treated for the same times for control experiments. Poly(A)+ mRNAs were prepared, and 4.5 μg was resolved on a 1.4% agarose–formaldehyde gel. After transfer to a Hybond N+ blotting membrane, the membrane was probed consecutively with 32P-labelled E2F-1 and GAPDH cDNAs and exposed to X-ray film to approximately the same band intensity. (B) The relative levels of E2F-1 RNA were normalized by using the GAPDH RNA signal and plotted in arbitrary units.
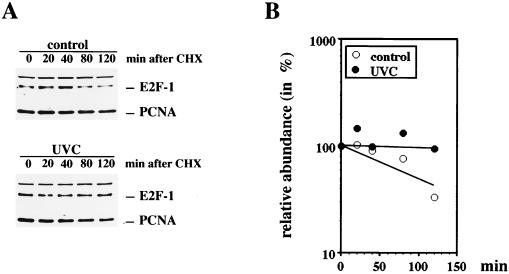
Since p53 is upregulated after DNA damage by prolongation of the half-life of the protein, we attempted to test if E2F-1 is also upregulated by stabilization of the protein in response to DNA damage. Due to the very low abundance of E2F-1 protein in the cell lines we have analyzed, we were not able to label E2F-1 metabolically and monitor the decay of the protein in the normal, untreated cellular environment. Instead, we blocked the overall protein synthesis of the cells by addition of cycloheximide and monitored the degradation of E2F-1 protein by Western blot analysis in UVC-irradiated and nonirradiated cells (Fig. 4). While E2F-1 protein disappeared with a half-life of 100 to 120 min in nonirradiated cells, it was stable throughout the experiment when the cells had been irradiated with UVC 6 to 16 h before the addition of cycloheximide. p53 behaved accordingly under these experimental conditions, with the difference that in nonirradiated cells it disappeared with a half-life of 20 min (data not shown). Although the difference in the half-life of E2F-1 in nonirradiated cells compared to UVC-irradiated cells is striking, one should be aware that determination of the half-life of E2F-1 under these conditions can be only a rough estimation. By treating the cells with cycloheximide, not only is the de novo synthesis of E2F-1 blocked but also the synthesis of proteins required for proteolysis, which influences the half-life of all cellular proteins, is blocked.
FIG. 4.
Induction of E2F-1 in response to UVC irradiation is mediated by prolongation of the half-life of E2F. (A) U2-OS cells irradiated with UVC (30 J/m2) or an unirradiated control was further incubated for 16 h before addition of 20 μg of cycloheximide (CHX) per ml. After a further incubation for the indicated periods, the cells were harvested and 45 μg (control) or 20 μg (UVC irradiated) of cellular protein was resolved on a 10% polyacrylamide minigel. The blots were probed for E2F-1 expression by using the rabbit polyclonal anti-E2F-1 antibody C-20. The blots were reprobed with an anti-PCNA antibody (PC10) to show loading of the samples. (B) E2F-1 expression was quantified, and the mean value of E2F-1 expression of two independent experiments was plotted.
Downregulation of Mdm2 correlates negatively with p53 and E2F-1 expression.
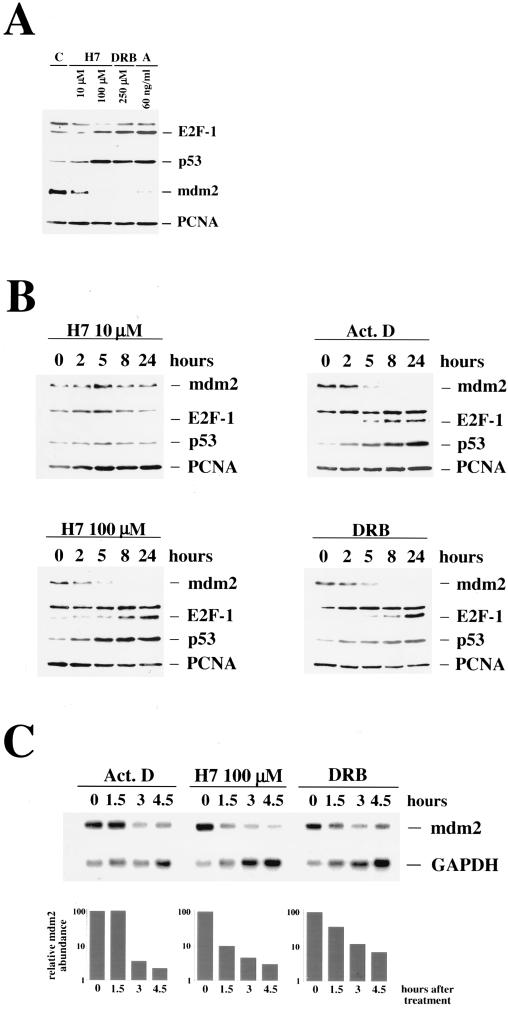
Recent reports have shown that p53 is not induced only by DNA damage but also by the protein kinase inhibitor H7 (48). Interestingly, concentrations which inhibit protein kinase C specifically are not able to upregulate p53, whereas concentrations at least 1 log unit above the concentration required to inhibit protein kinase C can upregulate p53. These observations point to the involvement of a protein kinase distinct from protein kinase C which maintains low expression of p53. In previous experiments, we found that the protein kinase inhibitor DRB also upregulates p53 expression, supporting the idea that inhibition of a particular protein kinase activity can increase p53 stability without damaging the DNA. We wanted to know if inhibition of kinase activity by H7 or DRB is also able to upregulate E2F-1. We treated GM02184D cells with 10 μM H7, a concentration that specifically inhibits protein kinase C and should not induce p53, with 100 μM H7, a concentration that has been shown to induce p53 expression (48), and with 250 μM DRB. Actinomycin D was used as an internal control. The differentially treated cells were harvested 18 h after stimulation. While treatment with 10 μM H7 had no effect on E2F-1 expression, we found a strong induction of E2F-1 after using 100 μM H7 or 250 μM DRB. Rehybridization of the membranes with an anti-p53 antibody showed the same dose dependency of H7 for p53 as well as upregulation of p53 by DRB and actinomycin D (Fig. 5A). We analyzed Mdm2 abundance under these conditions, which upregulated p53 and E2F-1, by rehybridizing the membranes from our experiments with the kinase inhibitors with an anti-Mdm2 antibody. We found that the basal Mdm2 expression was only marginally reduced after treating the cells with concentrations of H7 sufficient to block protein kinase C activity. However, when we treated the cells with concentrations of H7 which upregulated p53 and E2F-1, Mdm2 disappeared completely (Fig. 5A). Mdm2 expression was also not detectable in cells which had been treated with DRB. In cells which had been treated with 60 ng of actinomycin D per ml, Mdm2 was detectable in some experiments only as a faint band (Fig. 5A) while it was undetectable in others, and this band was lost in every experiment when we used higher concentrations of actinomycin D (Fig. 2). As in irradiated U2-OS cells (Fig. 1), accumulation of E2F-1 and p53 correlated with the disappearance of Mdm2.
FIG. 5.
Inhibitors of transcription and protein kinase inhibitors upregulate E2F-1 expression. GM02184D cells were treated with H7 (10 and 100 μM, as indicated), DRB (250 μM), and actinomycin D (60 ng/ml). (A) At 18 h after treatment, the cells were harvested and 45 μg of protein extract was separated on an SDS–10% polyacrylamide gel. The proteins were transferred to a polyvinylidene difluoride membrane and analyzed for Mdm2, E2F-1, and p53 expression and for expression of PCNA for the loading control. (B) Cells were harvested after the indicated time and analyzed for the expression of Mdm2, E2F-1, p53, and PCNA for the loading control. (C) Cells were harvested at the indicated time points, and poly(A)+ mRNAs were prepared. mRNAs (4.5-μg portions) were resolved on a 1.4% agarose–formaldehyde gel and transferred to a Hybond N+ blotting membrane. The membrane was probed consecutively with 32P-labelled mdm2 and GAPDH cDNAs and exposed to X-ray film. The relative levels of mdm2 RNAs were normalized by using the GAPDH RNA signal and plotted in arbitrary units. Upregulation of E2F-1 and p53 correlates with the disappearance of Mdm2.
We further investigated whether the virtual mutual exclusion of Mdm2 expression and E2F-1/p53 accumulation was also reflected in a time course experiment. We treated GM02184D cells with 100 μM H7, 60 ng of actinomycin D per ml, or 250 μM DRB and harvested them after increasing time intervals. We found that all three agents decreased Mdm2 expression to barely detectable levels in less than 5 h. As soon as Mdm2 protein was decreased to an almost undetectable level, we saw an increase in E2F-1 expression (Fig. 5B). The increase in p53 expression occurred slightly earlier, exactly as it did after UVC irradiation (Fig. 1A.I). We measured the half-life of Mdm2 protein in cells which had been treated with these inhibitory agents; however, we found no increased degradation of Mdm2 protein under these conditions (data not shown), which could be responsible for the rapid disappearance of the protein. Northern analysis, however, revealed that the decrease in Mdm2 protein expression was strictly correlated with a rapid decrease in mdm2 RNA levels (Fig. 5C). DRB inhibits mRNA elongation by inhibiting the transcription factor IIH (TFIIH)-associated protein kinase (59), and the decrease in mdm2 RNA levels is probably due to this activity. H7 at 100 μM inhibits various protein kinases quite nonspecifically, and it is conceivable that it can inhibit the TFIIH-associated kinase as well. H7 could therefore easily cause the same phenotype as DRB, as we see it in our experiments. It should be noted that the signal for GAPDH RNA increases during the time course when cells have been treated with DRB or high concentrations of H7 but also with 60 ng of actinomycin D per ml. This is probably due to inhibition of transcription or inhibition of RNA elongation. De novo synthesis is blocked under these conditions, while the degradation, at least at early time points after treatment, is not affected. Thus, transcripts with a short half-life are depleted from the RNA pool while transcripts with a long half-life, like the GAPDH RNA, are enriched.
In all our experiments, we found an absolute conformity between p53 and E2F-1 induction and Mdm2 disappearance when using different concentrations and different inhibitory agents. Mdm2 is supposed to be the ubiquitin ligase for p53 (29), and thus the disappearance of the p53 ubiquitin ligase could well be the reason for the increased stability of p53.
Our results which show that not only p53 but also E2F-1 is stabilized when Mdm2 disappears, together with the evidence from Martin et al. (41) for a physical interaction between Mdm2 and E2F-1, speak persuasively in favor of both p53 and E2F-1 being targeted by Mdm2 for degradation.
E2F-1 upregulation in response to DNA is impaired in cells with mutated p53.
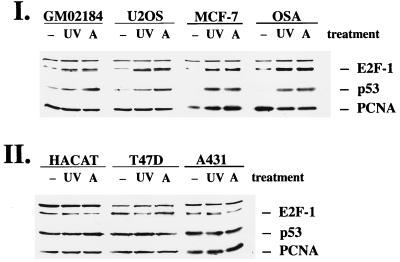
To further support our theory that Mdm2 plays a regulatory role in the accumulation not only of p53 but also of E2F-1, we sought to analyze more cell lines with different levels of Mdm2 expression. We used GM02184D and U2-OS cells as cell lines with wild-type p53 and normal expression levels of Mdm2. We also used MCF-7 cells, which are derived from a breast tumor and which overexpress Mdm2 at the protein level (5), and OSA cells, a human osteosarcoma cell line with highly elevated Mdm2 levels due to gene amplification (15), as cell lines which possess wild-type p53 but which express Mdm2 at unusually high levels. A431 cells, HAKAT cells, and T47D cells were used as cell lines which possess mutated p53 and virtually no detectable Mdm2. We confirmed the relative Mdm2 levels in these different cell lines by immunoprecipitation and Western blotting (data not shown).
The cells were irradiated with 30 J of UVC per m2 or treated with actinomycin D at a final concentration of 60 ng/ml and harvested 18 h posttreatment, and the protein extracts were analyzed in Western blots for E2F-1 and p53 expression (Fig. 6). As expected, U2-OS cells and GM02184D cells accumulated both p53 and E2F-1 in response to UVC irradiation and in response to treatment with actinomycin D. Although MCF-7 cells and OSA cells overexpress Mdm2, they still accumulated p53 in response to DNA damage, confirming earlier experiments (6a, 43). Both cell lines also induced E2F-1 protein in response to UVC irradiation or treatment with actinomycin D. HAKAT cells, A431 cells, or T47D cells, in which the p53 gene is mutated, failed repeatedly to upregulate E2F-1 or p53 in response to these agents. We used three different tumor cell lines because cell lines with mutated p53 are genomically unstable and quite frequently acquire secondary mutations. By analyzing three different cell lines with mutant p53, we wanted to rule out the possibility that other mutations, apart from the mutation of p53, are responsible for the failure to induce E2F-1. It should be noted that there was no obvious increase in basal expression of E2F-1 in these cell lines whereas p53 expression was markedly increased in the cell lines with mutated p53. We do not completely understand the difference in basal expression of E2F-1 and p53 in cell lines in which p53 is mutated, but since several factors contribute to the basal expression of a protein, we assume that Mdm2 is only one of the regulators of E2F-1 expression which become important when the DNA of the cell is damaged. Previous experiments have shown that Mdm2 levels are profoundly decreased in MCF-7 and OSA cells after UVC irradiation (data not shown) and probably also after treatment with actinomycin D. This reduction in Mdm2 expression is presumably responsible for decreased degradation of p53 and E2F-1 in these cells after UVC irradiation or addition of actinomycin D. Therefore, the enhanced Mdm2 expression under normal conditions does not interfere with upregulation of p53 and E2F-1 in response to irradiation with UVC or treatment with actinomycin D. A431, HAKAT, and T47D cells, on the other hand, possess mutated p53 with a markedly decreased turnover of p53. Interestingly, transfection of wild-type p53 into one of these tumor cell lines (A431) has been reported to result in stable endogenous and exogenous p53, although the same product was obviously degraded in cell lines harboring wild-type p53. Other experiments showed that mutant p53 accumulated in response to DNA damage in MCF-7 cells which possess endogenous wild-type p53 (43). This strongly indicates that point mutations of p53 do not result in intrinsically stable p53, which cannot accumulate further. Moreover, loss of induction of mutant p53 in response to DNA damage in tumor cell lines such as A431 is the result of the cell environment. Our data suggest that E2F-1 accumulation is dependent on the same cell environment as p53 and that it accumulates only in response to DNA damage in an environment possessing wild-type p53. Experiments reported by Midgley and Lane (43) have shown that expression of Mdm2 after microinjection reduces p53 stability in A431 cells. We tried a similar approach by using A431 cells stably transfected with an inducible mdm2 expression vector. Unfortunately, the basal E2F-1 expression turned out to be too low to monitor a further reduction by ectopic Mdm2 expression.
FIG. 6.
E2F-1 is induced in response to UVC or actinomycin D in cells with wild-type p53 but not in cells with mutated p53. (I) Cell lines with wild-type p53 (GM02184D, U2-OS, MCF-7, and OSA cells). (II) Cell lines with mutated p53 (HAKAT, T47D, and A431 cells). The cells were irradiated with UVC (UV; 30 J/m2), treated with actinomycin D (A; 60 ng/ml), or left untreated for control experiments (−). At 18 h after treatment, the cells were harvested and analyzed for the expression of E2F-1, p53, and PCNA as described in the legend to Fig. 1.
Microinjection of 3G5 antibody or mdm2 antisense oligonucleotides induce E2F-1 protein expression.
Although we observed a striking correlation between downregulation of Mdm2 and accumulation of both E2F-1 and p53 under various conditions, we were not able to rule out the contribution of other cellular pathways to the upregulation of E2F-1. If, however, Mdm2 is the molecule that targets E2F-1 for degradation, as it does for p53, specific disruption of this pathway would be predicted to cause accumulation of E2F-1. We used two similar approaches to disrupt a predicted interaction of Mdm2 and E2F-1: (i) microinjection of mdm2 antisense oligonucleotides and (ii) microinjection of anti-Mdm2 antibody 3G5 as previously described (6, 43).
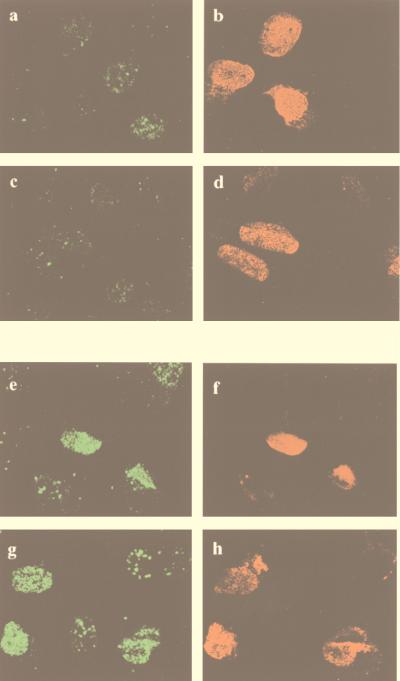
Antisense oligonucleotides often act by inducing RNase H cleavage at the heteroduplex region, resulting in increased degradation of the target mRNA and reduction in the expression of the target protein. Antisense phosphothiorate oligodeoxynucleotides which specifically bind to human mdm2 RNA inhibit Mdm2 protein expression and Mdm2-p53 complex formation even in tumor cells containing mdm2 gene amplifications (10). We microinjected antisense oligonucleotides at 1 mg/ml in the presence of a mouse monoclonal antibody, to facilitate the detection of microinjected cells, and analyzed E2F-1 expression by immunofluorescence. Figure 7 shows that E2F-1 accumulated repeatedly in cells which were injected with mdm2 antisense oligonucleotides (Fig. 7a and c) but not in cells which were injected with unrelated oligonucleotides (Fig. 7e and g).
FIG. 7.
E2F-1 protein is upregulated after microinjection of mdm2 antisense oligonucleotides. U2-OS cells were microinjected with mdm2 antisense oligonucleotides (a to d) or unrelated oligonucleotides for control experiments (e to h), together with an unrelated mouse monoclonal antibody to identify microinjected cells. At 20 h after microinjection, the cells were fixed with 1% paraformaldehyde, permeabilized with 1% NP-40, and stained with rabbit anti-E2F-1 antiserum (C-20) followed by FITC-conjugated donkey anti-rabbit IgG and Texas red-conjugated goat anti-mouse IgG. The right-hand panels (in red) show successful injection of oligonucleotides by staining of coinjected mouse monoclonal antibodies with Texas red-conjugated anti-mouse IgG (b, d, f, and h). The left-hand panels (in green) show expression of E2F-1 (a, c, e, and g). Arrowheads indicate microinjected cells. Cells microinjected with mdm2 antisense oligonucleotides upregulate E2F-1 protein, while microinjection with control oligonucleotides has no effect on E2F-1 expression.
To further support our experimental evidence that E2F-1 is regulated via its interaction with Mdm2, we microinjected two different monoclonal antibodies recognizing two different domains of Mdm2 protein. The 3G5 anti-Mdm2 antibody binds to an epitope at the amino terminus of Mdm2, involving the p53 binding pocket, and the SMP 14 anti-Mdm2 antibody is directed against an epitope in the central part of the Mdm2 protein. It has been shown that the 3G5 antibody is able to block the interaction of p53 with Mdm2 (5), and after microinjection, 3G5 upregulates p53 expression (6, 43). p53 and E2F-1 were coregulated throughout our experiments, and thus we expected that Mdm2 would interact with E2F-1 via the same domain as it interacts with p53. To determine whether anti-Mdm2 antibodies are able to disrupt the interaction of Mdm2 and E2F-1, leading to the accumulation of E2F-1, we microinjected U2-OS cells with 1 mg of 3G5 per ml or with 1 mg of SMP 14 per ml and costained them with anti-mouse IgG, to facilitate detection of microinjected cells, and with the C-20 anti-E2F-1 antibody. Only cells microinjected with 3G5 showed increased nuclear staining for E2F-1 (Fig. 8e and g), while cells microinjected with SMP 14 showed no difference in E2F-1 expression in comparison with noninjected cells (Fig. 8a and c). Microinjection of antibodies directed against pRb were used as a further control, but these antibodies were also unable to induce E2F-1 expression (data not shown). These experiments demonstrate that it is specifically the interruption of Mdm2–E2F-1 complexes which induces E2F-1 expression, since it has been shown that interruption of p53-Mdm2 complexes, and not the microinjection process or the presence of antibodies in the nucleus, induces p53 expression (43).
FIG. 8.
Microinjection of anti-Mdm2 3G5 antibody induces the expression of E2F-1. U2-OS cells were microinjected with mouse monoclonal antibodies SMP 14 (a to d) or 3G5 (e to h), recognizing different epitopes on Mdm2. At 20 h after microinjection, the cells were fixed with 1% paraformaldehyde, permeabilized with 1% NP-40, and incubated with a rabbit anti-E2F-1 antibody (C-20, diluted 1:300) followed by FITC-conjugated donkey anti-rabbit IgG (a, c, e, and g) and Texas red-conjugated goat anti-mouse IgG (b, d, f, and h), recognizing the microinjected anti-Mdm2 antibody. Cells microinjected with 3G5 upregulate E2F-1 protein, while cells microinjected with SMP 14 do not affect E2F-1 expression.
Our data obtained by various microinjection experiments provide direct evidence that E2F-1 and p53 display the same kinetics after irradiation and every inhibitory agent used and, moreover, that the expression and stability of both proteins are mediated through their interaction with cellular Mdm2.
Conclusions.
The experiments presented here demonstrate that p53 and E2F-1 are coregulated under various conditions. Previous studies have shown that both proteins are degraded by the ubiquitin-dependent degradation system (22, 39), and Mdm2 was identified as a ubiquitin ligase for p53 (29). Additionally, Mdm2 is required for the export of p53 into the cytoplasm, where it is degraded by proteasomes (17, 49). Future studies must investigate whether Mdm2 is also a ubiquitin ligase for E2F-1 and whether E2F-1, like p53, must be exported into the cytoplasm in order to be degraded.
Despite repeated efforts, we were unable to detect any decrease in E2F-1 expression in cells cotransfected with E2F-1 and mdm2 or in cells cotransfected with DP-1, E2F-1, and mdm2, compared to cells transfected with E2F-1 alone. This is in contrast to the clear decrease seen in p53 expression when cotransfected with mdm2 (4a, 23). Microinjection of the 3G5 anti-Mdm2 antibody, however, shows that the p53 interaction domain of Mdm2 is required for degradation of E2F-1. We know from previous studies that only p53 tetramers are targeted for degradation by Mdm2 (4), and so we assume that E2F-1 has to oligomerize with other proteins distinct from DP-1 to be targeted for degradation by Mdm2. Alternatively, Mdm2 could act more indirectly by regulating a protein required for E2F-1 degradation.
Since E2F-1 and p53 are both induced by DNA damage and since both proteins interact physically with Mdm2, it becomes likely that upregulation of p53 and E2F-1 in response to DNA damage is caused by disruption of Mdm2-p53 and Mdm2–E2F-1 complexes. As a consequence, p53 and E2F-1 are no longer subjected to rapid degradation and accumulate in the cell. Recently, Nip et al. have shown that DNA damage caused by topoisomerase II inhibition induces apoptosis in a cell line overexpressing E2F-1 (46). Although the authors did not confirm the E2F-1 expression levels following etoposide treatment, which are presumably increased, their data suggest that upregulation of E2F-1 in response to DNA damage has functional consequences. Interestingly, E2F-1-specific induction of apoptosis is blocked by coexpression of Mdm2 (34). Although the authors did not determine whether inhibition of E2F-1-specific apoptosis by Mdm2 occurs at the level of E2F-1 expression, E2F-1 activity, or both, they confirmed the role of Mdm2 in the regulation of E2F-1.
ACKNOWLEDGMENTS
This work was funded by the Cancer Research Campaign. D.P.L. is a Gibb Fellow of the Cancer Research Campaign.
We thank Ed Harlow for providing the E2F-1 cDNA, Arnold Levine for providing the 3G5 and 4B2 antibodies, and Sudhir Agrawal for providing control and mdm2 antisense oligonucleotides. We are grateful to our colleagues Carol Midgley for providing the A431 cell line with an inducible mdm2 expression plasmid, Ralf Dahm for helping with the confocal images, and Dimitris Xirodimas for being always ready to help.
REFERENCES
- 1.Barak Y, Juven T, Haffner R, Oren M. Mdm2 expression is induced by wild type-p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartek J, Bartkova J, Lukas J. The retinoblastoma protein pathway and the restriction point. Curr Opin Cell Biol. 1996;8:805–814. doi: 10.1016/s0955-0674(96)80081-0. [DOI] [PubMed] [Google Scholar]
- 3.Bartek J, Vojtesek B, Grand R J A, Gallimore P H, Lane D P. Cellular-localization and T-antigen binding of the retinoblastoma protein. Oncogene. 1992;7:101–108. [PubMed] [Google Scholar]
- 4.Blattner C, Tobiasch E, Litfin M, Rahmsdorf H J, Herrlich P. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene. 1999;18:1723–1733. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 4a.Blattner, C. Unpublished data.
- 5.Blaydes J P, Gire V, Rowson J M, Wynford-Thomas D. Tolerance of high levels of wild-type p53 in transformed epithelial cells dependent on auto-regulation by mdm-2. Oncogene. 1997;14:1859–1868. doi: 10.1038/sj.onc.1201018. [DOI] [PubMed] [Google Scholar]
- 6.Bottger A, Bottger V, Sparks A, Liu W L, Howard S F, Lane D P. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 6a.Bottger, A. Personal communication.
- 7.Boukamp P, Petrussevska R T, Breitkreutz D, Hornung J, Markham A, Fusenig N E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell-line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campanero M R, Flemington E K. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRb tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J D, Marechal V, Levine A J. Mapping of the p53 and Mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L H, Agrawal S, Zhou W Q, Zhang R W, Chen J D. Synergistic activation of p53 by inhibition of Mdm2 expression and DNA damage. Proc Natl Acad Sci USA. 1998;95:195–200. doi: 10.1073/pnas.95.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F-1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eldeiry W S, Harper J W, Oconnor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y S, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kinzler K W, Vogelstein B. Waf1/Cip1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 13.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W G, Livingston D M, Orkin S H, Greenberg M E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 14.Flemington E K, Speck S H, Kaelin W G. E2F-1-mediated transactivation is inhibited by complex-formation with the retinoblastoma susceptibility gene-product. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florenes V A, Maelandsmo G M, Forus A, Andreassen A, Myklebost O, Fodstad O. Mdm2 gene amplification and transcript levels in human sarcomas—relationship to the p53 gene status. J Natl Cancer Inst. 1994;86:1297–1302. doi: 10.1093/jnci/86.17.1297. [DOI] [PubMed] [Google Scholar]
- 16.Fort P, Marty L, Piechaczyk M, Elsabrouty S, Dani C, Jeanteur P, Blanchard J M. Various rat adult tissues express only one major messenger-RNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by Mdm2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/Mdm2 complexes participate in Mdm2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 19.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 20.Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in-vivo and TBP binding in-vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 22.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Degradation of E2F by the ubiquitin-proteasome pathway: regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 23.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 24.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 25.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hengstermann A, Whitaker N J, Zimmer D, Zentgraf H, Scheffner M. Characterization of sequence elements involved in p53 stability regulation reveals cell type dependence for p53 degradation. Oncogene. 1998;17:2933–2941. doi: 10.1038/sj.onc.1202282. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann F, Martelli F, Livingston D M, Wang Z Y. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 28.Holmberg C, Helin K, Sehested M, Karlstrom O. E2F-1-induced p53-independent apoptosis in transgenic mice. Oncogene. 1998;17:143–155. doi: 10.1038/sj.onc.1201915. [DOI] [PubMed] [Google Scholar]
- 29.Honda R, Tanaka H, Yasuda H. Oncoprotein Mdm2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y Y, Ishiko T, Nakada S, Utsugisawa T, Kato T, Yuan Z M. Role for E2F is DNA damage-induced entry of cells into S phase. Cancer Res. 1997;57:3640–3643. [PubMed] [Google Scholar]
- 31.Hunt K K, Deng J, Liu T J, WilsonHeiner M, Swisher S G, Clayman G, Hung M C. Adenovirus-mediated overexpression of the transcription factor E2F-1 induces apoptosis in human breast and ovarian carcinoma cell lines and does not require p53. Cancer Res. 1997;57:4722–4726. [PubMed] [Google Scholar]
- 32.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F-1 induces quiescent cells to enter S-phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 33.Kastan M B, Zhan Q M, Eldeiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J. A mammalian-cell cycle checkpoint pathway utilizing p53 and Gadd45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 34.Kowalik T F, DeGregori J, Leone G, Jakoi L, Nevins J R. E2F-1-specific induction of apoptosis and p53 accumulation, which is blocked by Mdm2. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 35.Kowalik T F, Degregori J, Schwarz J K, Nevins J R. E2F-1 overexpression in quiescent fibroblasts leads to induction of cellular DNA-synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 37.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Wild-type p53 is a cell-cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X, Lane D P. Differential induction of transcriptionally active p53 following UV or ionizing-radiation—defects in chromosome instability syndromes. Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 39.Maki C G, Huibregtse J M, Howley P M. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 40.Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor-antigen in nontransformed mouse cells. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin K, Trouche D, Hagemeier C, Sorensen T S, LaThangue N B, Kouzarides T. Stimulation of E2F-11/DP-1 transcriptional activity by Mdm2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 42.Midgley C A, Fisher C J, Bartek J, Vojtesek B, Lane D, Barnes D M. Analysis of p53 expression in human tumors—an antibody raised against human p53 expressed in Escherichia coli. J Cell Sci. 1992;101:183–189. doi: 10.1242/jcs.101.1.183. [DOI] [PubMed] [Google Scholar]
- 43.Midgley C A, Lane D P. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 44.Miyashita T, Reed J C. Tumor-suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 45.Nigro J M, Baker S J, Preisinger A C, Jessup J M, Hostetter R, Cleary K, Bigner S H, Davidson N, Baylin S, Devilee P, Glover T, Collins F S, Weston A, Modali R, Harris C C, Vogelstein B. Mutations in the p53 gene occur in diverse human tumor types. Nature. 1989;342:705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 46.Nip J, Strom D K, Zambetti G, Cleveland J L, Hiebert S W. E2F-1-mediated induction of p53 in myeloid progenitor cells suppresses topoisomerase II alpha. Blood. 1997;90:205. [Google Scholar]
- 47.Okamoto K, Beach D. Cyclin-G is a transcriptional target of the p53 tumor-suppressor protein. EMBO J. 1994;13:4816–4822. doi: 10.1002/j.1460-2075.1994.tb06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ronca F, Chan S L, Yu V C. 1-(5-Isoquinolinesulfonyl)-2-methylpiperazine induces apoptosis in human neuroblastoma cells, SH-SY5Y, through a p53-dependent pathway. J Biol Chem. 1997;272:4252–4260. doi: 10.1074/jbc.272.7.4252. [DOI] [PubMed] [Google Scholar]
- 49.Roth J, Dobbelstein M, Freedmann D, Shenk T, Levine A. Nucleo-cytoplasmic shuttling of the Hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus Rev protein. EMBO J. 1998;17:554–556. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 51.Shan B, Lee W H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 53.Waseem N H, Lane D P. Monoclonal-antibody analysis of the proliferating cell nuclear antigen (PCNA)—structural conservation and the detection of a nucleolar form. J Cell Sci. 1990;96:121–129. doi: 10.1242/jcs.96.1.121. [DOI] [PubMed] [Google Scholar]
- 54.Weinberg R A. The retinoblastoma protein and cell-cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 55.Wu L, Levine A J. Differential regulation of the p21/WAF-1 and mdm2 genes after high-dose UV irradiation: p53-dependent and p53-independent regulation of the mdm2 gene. Mol Med. 1997;3:441–451. [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X W, Bayle J H, Olson D, Levine A J. The p53 Mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 57.Yamasaki L, Bronson R, Williams B O, Dyson N J, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−) mice. Nat Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 58.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 59.Yankulov K, Yamashita K, Roy R, Egly J M, Bentley D L. The transcriptional elongation inhibitor 5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein-kinase. J Biol Chem. 1995;270:23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]