Abstract
The detection of Pneumocystis carinii DNA in blood by PCR could be useful for studying the natural history of pneumocystosis and could also be a noninvasive diagnostic method. The results of previous studies are nevertheless conflicting. In our study, we compared three commercially available DNA extraction kits (GeneReleaser, QIAamp Tissue Kit, and ReadyAmp Genomic DNA Purification System) and proteinase K and proteinase K-phenol-chloroform treatments for the extraction of P. carinii DNA from dilutions of a P. carinii f. sp. hominis cyst suspension mixed with human whole blood. A rapid and simple nested PCR protocol which amplifies a portion of the mitochondrial large-subunit rRNA gene was applied to all the extraction products. The QIAmp Tissue Kit was the most effective kit for the isolation of amplification-ready P. carinii DNA and was used with nested PCR for the testing of whole-blood specimens from 35 immunocompetent control patients and 84 human immunodeficiency virus (HIV)-infected patients investigated for pulmonary disease and/or fever. In HIV-infected patients, P. carinii DNA was detected by nested PCR in blood samples from 3 of 14 patients with microscopically proven P. carinii pneumonia, 7 of 22 patients who were considered to be colonized with P. carinii, and 9 of 48 patients who were neither infected nor colonized with P. carinii. P. carinii DNA was not detected in blood specimens from the 35 immunocompetent patients. P. carinii DNA in blood might represent viable P. carinii organisms or DNA complexes released from pulmonary phagocytes. In conclusion, P. carinii DNA may be detected in whole blood from HIV-infected patients, but the nature and the meaning of the circulating form of P. carinii remain to be established.
Pneumocystosis is the most common pulmonary opportunistic infection in AIDS patients. It is almost always limited to the lungs. Cases of extrapulmonary Pneumocystis carinii infection are reported with increasing frequency (21). Their pathogenesis is not well understood. It is not known whether the route of dissemination is lymphatic or hematogenous or both, nor is the circulating form of P. carinii known. Moreover, it is important to know if the dissemination of P. carinii correlates with pulmonary disease, even without clinical signs of extrapulmonary infection. If this is the case, the detection of P. carinii in blood samples may be an interesting noninvasive diagnostic procedure for patients with suspected P. carinii pneumonia.
Using PCR, Kitada et al. (10) were the first to report on the presence of P. carinii DNA in blood specimens from nude mice infected with P. carinii and from an AIDS patient with P. carinii pneumonia (PCP). The results of previous studies (1, 4, 9–11, 13, 17–20, 22) are conflicting since the frequency of detection of P. carinii DNA by PCR in blood showed great variability in the various studies.
The aims of our study were to compare three commercially available DNA extraction kits and proteinase K and proteinase K-phenol-chloroform treatments for the isolation of amplification-ready P. carinii DNA from blood samples and to search for P. carinii DNA in blood specimens from human immunodeficiency virus (HIV)-infected patients and immunocompetent control patients by a nested PCR protocol (15).
MATERIALS AND METHODS
Clinical specimens.
Eighty-four blood samples were obtained from 84 HIV-infected patients from 1 March 1995 to 31 December 1995. These patients were investigated for pulmonary disease and/or fever. They partook in a prospective study comparing Giemsa and methenamine silver stains with PCR for the detection of P. carinii in bronchoalveolar lavage (BAL) specimens (15). BAL and blood specimens were collected before the start of therapy. Thirty-five blood specimens were also obtained from 35 immunocompetent control patients.
Specimen processing.
Blood was collected in tubes containing EDTA as anticoagulant (for retrieval of whole blood and cell fraction) and in clotting tubes (for retrieval of serum). The sample in the EDTA-containing tube was divided: 1 ml of whole blood was used for DNA amplification, and the remaining portion was decanted. The plasma and the cell fraction were frozen separately at −20°C, as was serum from the clotting tube.
DNA extraction. (i) Digestion with proteinase K.
Two hundred microliters of whole blood was incubated overnight in a mixture containing 50 mM KCl, 10 mM Tris HCl (pH 8.3), 2.5 mM MgCl2, 0.05% gelatin, 0.25% Tween 20, and 60 μg of proteinase K per ml. Part of the extracted DNA was directly amplified. The other part was purified with phenol-chloroform, precipitated with ethanol, and dissolved in water.
(ii) Extraction with commercially available kits.
The procedure with the Gene Releaser kit (BioVentures Inc., Murfreesboro, Tenn.), described by the manufacturer as a “cellular enrichment protocol,” was used. Two hundred microliters of whole blood was mixed with 200 μl of Triton X-100 in a microtube and then centrifuged for 1 min at 12,000 × g. The sediment was washed twice in 200 μl of PCR buffer (50 mM KCl, 10 mM Tris HCl [pH 8.3], 2.5 mM MgCl2). Twenty microliters of GeneReleaser was added to the sediment, which was then denatured in a thermocycler (MJ Research Inc., Watertown, Md.).
In the procedure with QIAamp Tissue Kit (Qiagen, Santa Clarita, Calif.), 200 μl of whole blood was incubated at 55°C for 1 h in 200 μl of lysis buffer (provided in the kit) containing 25 μl of proteinase K. The purification procedure was carried out with QIAmp spin columns; the DNA was adsorbed onto the QIAmp silica membrane during a brief centrifugation step, washed twice, and eluted with 200 μl of distilled water.
In the procedure with the ReadyAmp Genomic DNA Purification System (Promega Corp., Madison, Wis.), 200 μl of whole blood was incubated at room temperature for 10 min and then centrifuged for 2 min at 12,000 × g. The sediment was incubated with 200 μl of ReadyAmp resin for 20 min at 56°C, vortexed at high speed for 5 to 10 s, and incubated again for 8 min at 100°C. The sample was centrifuged for 2 min at 12,000 × g. The isolated DNA was recovered in the supernatant.
Nested PCR.
Nested PCR was performed by a rapid amplification protocol described elsewhere (15). Briefly, DNA amplification was carried out in a reaction mixture (100 μl) containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 0.25 mM (each) dATP, dTTP, dCTP, and dGTP, 0.25 μM (each) the oligonucleotide primers, and 2.5 U of Taq polymerase (Promega Corp.). Primers pAZ102-E and pAZ102-H were used for amplification of a part of the mitochondrial gene encoding for the large-subunit (mtLSU) rRNA (23). After an initial 5-min denaturation step, DNA was amplified for 40 cycles with a final extension period of 5 min at 72°C. Each cycle consisted of 20 s of denaturation at 94°C, 20 s of annealing at 56°C, and 20 s of extension at 72°C. The second amplification step was performed with 1 μl of the amplified product from the first step by using pAZ102-E as the external primer and pAZ102-L2 (23) as the internal primer. The reaction mixture and the amplification protocol were the same as those for the first PCR, except that the primers were used at 0.75 μM and the annealing temperature was 50°C. As a precaution to prevent template contamination, preparation of reaction solutions, DNA extraction, and the two amplification procedures were performed in three separate rooms and aerosol-barrier pipette tips were used to handle all reagent transfers. Positive controls (BAL specimens obtained from patients with microscopically proven PCP) and multiple negative controls (water) were included in each experimental run. The two amplified products were subjected to electrophoresis in a 1.5% agarose gel and visualized after ethidium bromide staining. The expected P. carinii-specific bands consisted of 346- and 120-bp fragments.
Comparison of current DNA extraction methods and commercially available DNA extraction kits.
Tenfold dilutions of a P. carinii f. sp. hominis suspension containing 1.8 × 104 cysts/μl was mixed with whole blood from an immunocompetent patient. DNA extraction was performed with four dilutions (10−3, 10−5, 10−7, and 10−9) with proteinase K alone, proteinase K-phenol-chloroform, and the three kits. The nested PCR protocol was applied to the extraction products.
RESULTS
DNA extraction.
By extraction with proteinase K alone and GeneReleaser, the amplified products were visible only at a dilution of 10−3. By extraction with proteinase K-phenol-chloroform, the QIAamp Tissue Kit, and the ReadyAmp Genomic DNA Purification System, the lower detection limit was the 10−5 dilution. The QIAamp Tissue Kit was used for the rest of the present study because purified DNA, which may be stored at −20°C if it is not used directly, may be obtained.
Nested PCR with whole-blood specimens.
Following extraction with the QIAamp Tissue Kit and amplification by nested PCR, P. carinii DNA was detected in blood specimens from (i) 3 of 14 patients with microscopically proven PCP, (ii) 7 of 22 patients whose BAL specimens were negative by staining and positive by nested PCR, and (iii) 9 of 48 patients whose BAL specimens were negative by staining and nested PCR (Fig. 1). P. carinii DNA was not detected in blood samples from the 35 immunocompetent patients.
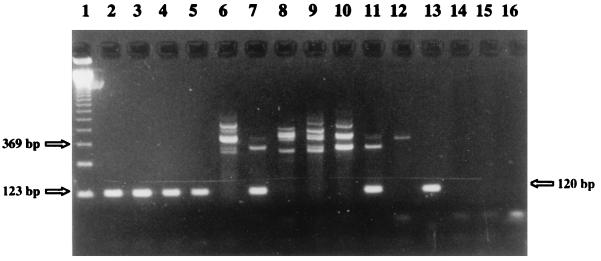
FIG. 1.
DNA extraction with the QIAamp Tissue Kit and nested PCR results. The products of the second amplification steps were subjected to agarose gel electrophoresis, stained with ethidium bromide, and vizualized under UV light. Lane 1, molecular mass marker; lanes 2 to 12, whole-blood specimens from HIV-infected patients neither infected nor colonized with P. carinii (positive specimens are in lanes 2 to 5, 7, and 11 and negative specimens are in lanes 6, 8 to 10, and 12); lane 13, positive control (P. carinii f. sp. hominis cysts); lanes 14 to 16, negative controls (water) for single PCR and nested PCR.
Clinical data were obtained by retrospective medical chart review for the 16 patients without microscopically proven PCP. The diagnosis of PCP for the seven patients whose BAL specimens were positive by nested PCR was discussed in a previous report (15): the diagnosis for one patient was possible PCP, and the six other patients were considered to be colonized with P. carinii or to have a low level of infection. Among the nine remaining patients whose BAL specimens were negative by nested PCR, eight were AIDS patients with a CD4+ lymphocyte counts of 0 to 72 × 106/liter. Among these eight patients, five presented with one or more opportunistic infections (esophageal candidiasis, cryptococcosis, cryptosporidiosis, cytomegalovirus infection, disseminated Mycobacterium avium infection), three presented with Kaposi’s sarcoma, one presented with AIDS dementia complex, and one presented with AIDS nephropathy. The ninth patient did not yet have clinical AIDS, since his CD4+ lymphocyte count was 216 × 106/liter and he had no opportunistic infection, but he presented with bronchial adenocarcinoma. All nine patients were receiving prophylactic therapy against P. carinii infection at the time when the blood sample has been taken: four with aerosolized pentamidine and the others with trimethoprim-sulfamethoxazole (TMP-SMX). The outcomes are known for these nine patients. Six of them died within 3 days (one patient), 1 to 3 months (four patients), and 11 months (one patient) after the blood sample that was positive by nested PCR was taken. Three patients are still alive.
DISCUSSION
Eleven studies about the detection of P. carinii by PCR in blood specimens from human or animals have been published (Table 1). Some investigators have obtained positive results (1, 10, 13, 18, 19), but others have observed negative results (4, 9, 11, 17, 20, 22).
TABLE 1.
Search for P. carinii DNA in blood samples by PCRa
| Host | Blood fraction | DNA extraction method | PCR target | Results | Reference |
|---|---|---|---|---|---|
| Nude mice (5 with PCP) and patients (2 HIV+ with PCP) | Cell pellet, serum | Proteinase K | 5S rRNA | For mice, cell pellet, 3 +; serum, 0 +; for patients, cell pellet, 1 + | 10 |
| Rats (16 IS with PCP, 6 IC housed near IS rats with PCP) | Serum | Proteinase K | DHFR | For IS rats with PCP, 7 of 8 + at 6 weeks and 8 of 8 + at 8 weeks; for IC rats, 6 + at 10 weeks of contact | 19 |
| Rats (17 IS including 14 with PCP) and patients (11 with PCP and 3 with EPP) | Cell pellet, MNC | Proteinase K-phenol-chloroform | 16S rRNA | For rats, cell pellet, 11 of 14 + with PCP; for patients with PCP, cell pellet, 1 +; for patients with EPP, cell pellet, 1 +; MNC, 2 + | 11 |
| Rats (10 IS with PCP) and patients (16 HIV+, including 10 with PCP, and 9 IC) | Serum | Proteinase K | DHFR | For rats, 10 + at 8 weeks; for patients with PCP, 12 +; for HIV+ patients without PCP, 0 +; for IC patients, 0 + | 18 |
| Patients (195 HIV+, including 14 with PCP; 10 IS HIV− without PCP; and 19 IC) | Serum, buffy coat | Proteinase K-phenol-chloroform | mtLSU rRNA | 1 + (patient with EPP) | 17 |
| Patients (27 HIV+ with PCP and 25 IC) | Serum | Proteinase K-phenol-chloroform | ITS | For HIV+ patients, 26 +; for IC patients, 0 + | 1 |
| Rats (6 IS) and patients (2 HIV+ with PCP) | Buffy coat | Proteinase K | mtLSU rRNA | 0 + | 9 |
| Rats (10 IS) | Buffy coat | Proteinase K-phenol-chloroform | MSG | 4 + at 8 weeks | 4 |
| Patients (15 HIV+ with PCP) | Serum, MNC, PMNC | Proteinase K, QIAamp Blood Kit | MtLSU rRNA | For serum, 0 +, for MNC, 1 + with QIAamp Blood Kit; for PMNC, 1 + with proteinase K treatment and 4 + with QIAamp Blood Kit | 20 |
| Patients (65 IS HIV−, including 10 with PCP) | Serum | Proteinase K | 5S rRNA | With PCP, 10 +; Without PCP, 4 + | 13 |
| Patients (19 HIV+, including 9 with PCP; 10 IS HIV−, including 4 with PCP; and 7 IC) | Serum | Proteinase K-phenol-choroform | mtLSU rRNA | HIV+ with PCP, 1 +; HIV− with PCP, 0 +; HIV− without PCP, 4 +; IC, 0 + | 22 |
Abbreviations and symbols: IS, immunosuppressed; IC, immunocompetent; EPP, extrapulmonary pneumocystosis, MNC, monomorphonuclear cells; PMNC, polymorphonuclear cells; +, positive; −, negative
In our study, by using the QIAamp Tissue Kit for DNA extraction and nested PCR, P. carinii DNA was not found in blood samples from 35 immunocompetent control patients, which is consistent with the results of all previous studies (1, 17, 18, 22). We detected P. carinii DNA in 21% of our patients with PCP, as has already been reported in three other studies (4, 11, 20). P. carinii DNA was also detected in blood specimens from 7 of 22 patients who were considered to be colonized or to have a low level of infection on the basis of the fact that their clinical course was inconsistent with that of PCP and their BAL specimens were negative by staining but positive by nested PCR (15). A search for parasitemia in colonized immunosuppressed patients has not been performed in previous studies except in that of Wagner et al. (22), in which the results were negative for the four patients investigated. Sepkowitz et al. (19) detected P. carinii DNA by PCR in the lungs and serum of six nonimmunocompromised sentinel rats housed near corticosteroid-treated P. carinii-infected rats. P. carinii DNA disappeared rapidly from the lungs and sera after the sentinel rats were isolated from the P. carinii-infected rats. The investigators suggested that subclinical infection or a high level of lung colonization and a low level of parasitemia may result from exposure to P. carinii-infected rats.
We detected P. carinii DNA in blood specimens from 9 of 48 patients whose BAL samples were negative by both staining and nested PCR. These HIV-infected patients were severely immunosuppressed, as attested to by their low CD4+ lymphocyte counts, their opportunistic infections, and the rapidly fatal outcomes for five of them. Schluger et al. (18) did not find parasitemia in six AIDS patients not infected or colonized with P. carinii. Roux et al. (17) obtained positive results for only 3 patients among a population of 205 immunosuppressed patients including 10 HIV-seronegative patients. Two teams published results which are consistent with ours and described immunosuppressed patients, infected or not infected with HIV, who presented with pulmonary illness inconsistent with PCP and whose BAL specimens were negative by PCR; Miyawaki et al. (13) amplified P. carinii DNA from the sera from four HIV-seronegative patients with hematological malignancies, and Wagner et al. (22) amplified P. carinii DNA from the sera from two AIDS patients and one patient with lymphoma. According to Wagner et al. (22), such results could be related to transient pulmonary P. carinii carriage that was favored by the immunosuppression and an acute or chronic lung disease and that was followed by organism destruction by pulmonary phagocytes.
Several explanations can be evoked for the conflicting results of all these studies. First, the targets of the PCR methods used for the detection of P. carinii DNA in blood samples included genes encoding mtLSU rRNA (9, 17, 20, 22), 5S rRNA (10, 13), 16S rRNA (11), dihydrofolate reductase (DHFR) (18, 19), internal transcribed spacers (ITS) of the rRNA operon (1), and major surface glycoprotein (MSG) (4). The sensitivities of these PCR methods are different. The mtLSU rRNA PCR and the ITS PCR are the most sensitive PCR assays (7, 12). This reason alone is probably not sufficient to explain the divergent results but is compatible with the fact that a highly sensitive technique is required because of the low P. carinii load in blood (2 to 25 haploid organisms/μl) (18).
Second, the DNA extraction technique is probably more crucial for extraction of DNA from blood than from BAL specimens. Indeed, blood contains particular Taq polymerase inhibitors, whereas P. carinii DNA amplification has easily been achieved in several studies when DNA was extracted from BAL specimens, whatever DNA extraction method was used. In 6 of the 11 studies cited above, the investigators did not purify the DNA after proteinase K digestion. Our comparison of different DNA extraction protocols showed that the purification with phenol-chloroform enables the detection limit of the nested PCR to be lowered and that the same result may be obtained with two commercially available DNA extraction kits. Tamburrini et al. (20) also used a commercially available kit (QIAmp Blood Kit, Qiagen), which had a higher sensitivity than proteinase K digestion.
Third, the P. carinii form and its localization in blood fractions are unknown. Some investigators searched for P. carinii DNA in serum (1, 13, 19, 22) and others searched for it in buffy coats isolated by density gradient centrifugation (such as Ficoll-Hypaque) (4, 9–11). Kitada et al. (10), Lipschick et al. (11), and Chary-Reddy and Graves (4) tested the cell pellet obtained after centrifugation of heparinized blood. The times and speeds of centrifugation were very different and, moreover, were not always specified.
The most important point in the discussion is the meaning of the presence of P. carinii DNA in blood: does it represent viable organisms or the DNA complexes released from phagocytes as evoked by Wagner et al. (22)? According to Schluger et al. (18), the circulating P. carinii form, which can be filtered through a 0.22-μm-pore-size filter, may be free DNA or protein-DNA aggregates originating from organisms damaged by phagocytosis. On the other hand, the possibility of the existence of viable circulating P. carinii organisms was supported before the publication of molecular studies by (i) the existence of human infections in which P. carinii was obviously transmitted in utero (2, 14), (ii) the description of extrapulmonary localizations, like the kidney or the eye, which are reached only by hematogenous spread from the lung (3, 16), and (iii) the observation of P. carinii cysts within the lumen and the wall of blood vessels in histological sections from the lung or other tissues (6, 8). In an experimental model, Chary-Reddy and Graves (4) demonstrated that the P. carinii DHFR gene is actively transcribed in extrapulmonary sites of infection, indicating the presence of viable P. carinii forms. Using PCR analysis with MSG primers, they also detected P. carinii DNA in blood samples from their immunosuppressed rats. All these results suggest that a relation might exist between circulating P. carinii forms and viable P. carinii organisms in extrapulmonary tissues. Moreover, Contini et al. (5) obtained short-term propagation of P. carinii in cell cultures with buffy coats purified from blood specimens taken from HIV-infected patients with PCP or disseminated P. carinii infection. This would also lend support to the hypothesis of the existence of viable circulating P. carinii forms.
The appearance of P. carinii DNA in blood follows its appearance in lung tissue (4, 18, 19). After the initiation of therapy with TMP-SMX (4, 19) or the termination of corticosteroid treatment (19), P. carinii DNA disappears rapidly but reappears when corticosteroids are reinitiated (4, 19). In patients, the blood sample positive by PCR was often taken at the time when PCP was clinically suspected but before the start of therapy. In 12 of 13 patients treated with TMP-SMX, Atzori et al. (1) did not detect P. carinii DNA in serum, but the delay between the initiation of therapy and retrieval of a blood sample was not specified.
The interest in the detection of P. carinii in blood by PCR is obvious. It may help to clarify the pathophysiology of pneumocystosis, on the condition that the nature of the circulating P. carinii form is elucidated. From a clinical point of view, it could be a particularly simple and noninvasive method of diagnosis and evaluation of the effectiveness of therapy. We reported here the results of our search for P. carinii DNA in blood samples, which are preliminary results. We detected P. carinii DNA in whole blood from 19 patients, and we are searching for P. carinii DNA in cell pellet, plasma, and serum samples from the same patients.
ACKNOWLEDGMENTS
This work was supported by research grants from the “Hospices Civils de Lyon” and the association “Ensemble Contre le SIDA.”
We thank Philippe Hauser for helpful suggestions.
REFERENCES
- 1.Atzori C, Lu J J, Jiang B, Bartlett M S, Orlando G, Queener S F, Smith J W, Cargnel A, Lee C H. Diagnosis of Pneumocystis carinii pneumonia in AIDS patients by using polymerase chain reactions on serum specimens. J Infect Dis. 1995;172:1623–1626. doi: 10.1093/infdis/172.6.1623. [DOI] [PubMed] [Google Scholar]
- 2.Bazaz G R, Manfredi O L, Howard R G, Claps A A. Pneumocystis carinii pneumonia in three full-term siblings. J Paediatr. 1970;76:767–769. doi: 10.1016/s0022-3476(70)80301-8. [DOI] [PubMed] [Google Scholar]
- 3.Boldorini R, Guzzetti S, Meroni L, Quirino T, Cristina S, Monga G. Acute hepatic and renal failure caused by Pneumocystis carinii in patients with AIDS. J Clin Pathol. 1995;48:975–978. doi: 10.1136/jcp.48.10.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chary-Reddy S, Graves D C. Identification of extrapulmonary Pneumocystis carinii in immunocompromised rats by PCR. J Clin Microbiol. 1996;34:1660–1665. doi: 10.1128/jcm.34.7.1660-1665.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contini C, Mastrantoni S, Romani R, Cultrera R, Delia S. Evidence of Pneumocystis carinii in cell line cultures infected with peripheral blood mononuclear cells isolated from AIDS patients with P. carinii pneumonia. J Med Microbiol. 1995;42:394–398. doi: 10.1099/00222615-42-6-394. [DOI] [PubMed] [Google Scholar]
- 6.Davey R T, Margolis D, Kleiner D, Deyton L, Travis W. Digital necrosis and disseminated Pneumocystis carinii infection after aerosolized pentamidine prophylaxis. Ann Intern Med. 1989;111:681–682. doi: 10.7326/0003-4819-111-8-681. [DOI] [PubMed] [Google Scholar]
- 7.De Luca A, Tamburrini E, Ortona E, Mancarini P, Margutti P, Antinori A, Visconti E, Siracusano A. Variable efficiency of three primer pairs for the diagnosis of Pneumocystis carinii pneumonia by the polymerase chain reaction. Mol Cell Probes. 1995;9:330–340. doi: 10.1016/s0890-8508(95)91636-9. [DOI] [PubMed] [Google Scholar]
- 8.Dyner S D, Lang W, Bush D F, Gordon P R. Intravascular and pleural involvement by Pneumocystis carinii in a patient with the acquired immunodeficiency syndrome (AIDS) Ann Intern Med. 1989;111:94–95. doi: 10.7326/0003-4819-111-1-94. [DOI] [PubMed] [Google Scholar]
- 9.Evans R, Joss A L W, Pennington T H, Ho-Yen D O. The use of a nested polymerase chain reaction for detecting Pneumocystis carinii from lung and blood in rat and human infection. J Med Microbiol. 1995;42:209–213. doi: 10.1099/00222615-42-3-209. [DOI] [PubMed] [Google Scholar]
- 10.Kitada K, Oka S, Kimura S, Shimada K, Serikawa T, Yamada J, Tsunoo H, Egawa K, Nakamura Y. Detection of Pneumocystis carinii sequences by polymerase chain reaction: animal models and clinical application to non invasive specimens. J Clin Microbiol. 1991;29:1985–1990. doi: 10.1128/jcm.29.9.1985-1990.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipschick G Y, Gill V J, Lungren J D, Andrawis V A, Nelson N A, Nielsen J O, Ognibene F P, Kovacs J A. Improved diagnosis of Pneumocystis carinii infection by polymerase chain reaction on induced sputum and blood. Lancet. 1992;340:203–206. doi: 10.1016/0140-6736(92)90469-j. [DOI] [PubMed] [Google Scholar]
- 12.Lu J J, Chen C H, Bartlett M S, Smith J W, Lee C H. Comparison of six different PCR methods for detection of Pneumocystis carinii. J Clin Microbiol. 1995;33:2785–2788. doi: 10.1128/jcm.33.10.2785-2788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyawaki H, Fujita J, Hojo S, Harada M, Yamaji Y, Suguri S, Takhara J. Detection of Pneumocystis carinii sequences in serum by polymerase chain reaction. Respir Med. 1996;90:153–157. doi: 10.1016/s0954-6111(96)90157-2. [DOI] [PubMed] [Google Scholar]
- 14.Pavlica F. Erste Beobachfung von angeborener Pneumozystenpneumonie bei einem Reifen, ausgetragenen totgeborenen Kind. Zentbl Allg Pathol. 1962;103:236–241. [Google Scholar]
- 15.Rabodonirina M, Raffenot D. D, Cotte L, Boibieux A, Mayençon M, Bayle G, Persat F, Rabatel F, Trepo C, Peyramond D. D, Piens M A. Rapid detection of Pneumocystis carinii in bronchoalveolar lavage specimens from HIV-infected patients: use of a simple DNA extraction procedure and nested polymerase chain reaction. J Clin Microbiol. 1997;35:2748–2751. doi: 10.1128/jcm.35.11.2748-2751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao N A, Zimmermann P L, Boyer D, Biswas J, Causey D, Beniz J, Nichols P W. A clinical, histopathologic, and electron microscopic study of Pneumocystis carinii choroiditis. Am J Ophthalmol. 1989;107:218–228. doi: 10.1016/0002-9394(89)90303-6. [DOI] [PubMed] [Google Scholar]
- 17.Roux P, Lavrard I, Poirot J L, Chouaid C, Denis M, Olivier J L, Nigou M, Miltgen M. Usefulness of PCR for detection of Pneumocystis carinii DNA. J Clin Microbiol. 1994;32:2324–2326. doi: 10.1128/jcm.32.9.2324-2326.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schluger N, Goldwin T, Sepkowitz K, Armstrong D, Bernard E, Rifkin M, Cerami A, Bucala R. Application of DNA amplification to pneumocystosis: presence of serum Pneumocystis carinii DNA during human and experimentally induced Pneumocystis carinii pneumonia. J Exp Med. 1992;176:1327–1333. doi: 10.1084/jem.176.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sepkowitz K, Schluger N, Godwin T, Armstrong D, Cerami A, Bucala R. DNA amplification in experimental pneumocystosis: characterization of serum Pneumocystis carinii DNA and potential P. carinii carrier states. J Infect Dis. 1993;168:421–426. doi: 10.1093/infdis/168.2.421. [DOI] [PubMed] [Google Scholar]
- 20.Tamburrini E, Mencarini P, Visconti E, Zolfo M, De Luca A, Siracusano A, Ortona E, Wakefield A E. Detection of Pneumocystis carinii DNA in blood by PCR is not of value for diagnosis of P. carinii pneumonia. J Clin Microbiol. 1996;34:1586–1588. doi: 10.1128/jcm.34.6.1586-1588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telzak E E, Armstrong D. Extrapulmonary infection and other unusual manifestations of Pneumocystis carinii. In: Walzer P D, editor. Pneumocystis carinii pneumonia. 2nd ed. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 361–378. [Google Scholar]
- 22.Wagner D, Königer J, Kern W V, Kern P. Serum PCR of Pneumocystis carinii DNA in immunocompromised patients. Scand J Infect Dis. 1997;29:159–164. doi: 10.3109/00365549709035877. [DOI] [PubMed] [Google Scholar]
- 23.Wakefield A E, Pixley F J, Banerji S, Sinclair K, Miller R F, Moxon E R, Hopkin J M. Amplification of mitochondrial ribosomal RNA sequences from Pneumocystis carinii DNA of rat and human origin. Mol Biochem Parasitol. 1990;43:69–76. doi: 10.1016/0166-6851(90)90131-5. [DOI] [PubMed] [Google Scholar]