Abstract
To investigate single nucleotide polymorphism (SNP) loci associated with yearling wool traits of fine-wool sheep for optimizing marker-assisted selection and dissection of the genetic architecture of wool traits, we conducted a genome-wide association study (GWAS) based on the fixed and random model circulating probability unification (FarmCPU) for yearling staple length (YSL), yearling mean fiber diameter (YFD), yearling greasy fleece weight (YGFW), and yearling clean fleece rate (YCFR) by using the whole-genome re-sequenced data (totaling 577 sheep) from the following four fine-wool sheep breeds in China: Alpine Merino sheep (AMS), Chinese Merino sheep (CMS), Qinghai fine-wool sheep (QHS), and Aohan fine-wool sheep (AHS). A total of 16 SNPs were detected above the genome-wise significant threshold (P = 5.45E-09), and 79 SNPs were located above the suggestive significance threshold (P = 5.00E-07) from the GWAS results. For YFD and YGFW traits, 7 and 9 SNPs reached the genome-wise significance thresholds, whereas 10 and 12 SNPs reached the suggestive significance threshold, respectively. For YSL and YCFR traits, none of the SNPs reached the genome-wise significance thresholds, whereas 57 SNPs exceeded the suggestive significance threshold. We recorded 14 genes located at the region of ±50-kb near the genome-wise significant SNPs and 59 genes located at the region of ±50-kb near the suggestive significant SNPs. Meanwhile, we used the Average Information Restricted Maximum likelihood algorithm (AI-REML) in the “HIBLUP” package to estimate the heritability and variance components of the four desired yearling wool traits. The estimated heritability values (h2) of YSL, YFD, YGFW, and YCFR were 0.6208, 0.7460, 0.6758, and 0.5559, respectively. We noted that the genetic parameters in this study can be used for fine-wool sheep breeding. The newly detected significant SNPs and the newly identified candidate genes in this study would enhance our understanding of yearling wool formation, and significant SNPs can be applied to genome selection in fine-wool sheep breeding.
Keywords: Chinese fine-wool sheep, FarmCPU, GWAS, re-sequencing, yearling wool traits
Introduction
The wool obtained from fine-wool sheep is a natural performance material that possesses several properties (such as stain resistance, softness, sun-safe, warm, and cool). Hence, wool occupies an important position in textile processing. In studies on fine-wool sheep breeding, the evaluation of wool traits and wool quality has mainly focused on the research of adult wool traits and yearling wool traits. Genetic correlations between measurements at the yearling and adult ages for the same trait range from moderate to high (Brown et al., 2013). Therefore, research on yearling wool traits is essential for fine-wool sheep breeders. For instance, fiber diameter, which is the key characteristic applied in wool sheep breeding, typically accounts for 75%–80% of the per-unit value of fleece (Shahinfar and Kahn, 2018). The diameter of wool fiber is closely associated with the morphogenesis and development of wool follicles in the sheep’s skin (Li et al., 2020). Therefore, to improve wool properties, studying and understanding the molecular genetic mechanisms involved in the formation of wool traits are essential and expected to hasten the optimization of the fine-wool sheep breeding efficiency. In addition, sheep is considered as an extra animal model to further understand human health under normal and diseased conditions, especially in terms of hair growth (Li et al., 2018).
With the rapid development in sequencing technologies, researchers can now accurately identify the quantitative trait loci through genome-wide association studies (GWASs), which has been widely used to map causal variants in humans and livestock (Zhang et al., 2019; Freebern et al., 2020; Matoba et al., 2020). Through the use of sheep medium density DNA chips, the GWAS conducted in Chinese Merino sheep (CMS) detected some single nucleotide polymorphism (SNP) markers that were associated with adult wool traits (Wang et al., 2014). The imputed SNPs from the Ovine InfiniumHD SNP BeadChip and 50K Ovine SNP Chip were used to perform the GWAS on the production and quality wool traits in yearlings and adults of Merino and Merino crossbred sheep, which identified that genes near SNPs with pleiotropic effects on the wool traits were identified (Bolormaa et al., 2017). Whole-genome sequence (WGS) data offer the advantage of theoretically covering all causal mutations that can lead to genetic variation among polygenic traits (Moghaddar et al., 2019). The linkage disequilibrium (LD) observed in sheep was lower than in other domestic species (Kijas et al., 2014). Low LD suggests that denser marker genotypes can improve genomic research accuracy (Daetwyler et al., 2012). Therefore, the GWAS with high-density SNPs is required to understand the genetic architecture of important complex traits in fine-wool sheep. Previous studies have shown that the application of re-sequencing data can improve the accuracy of genome research when compared with the use of 50K Chip on GWAS (Moghaddar et al., 2019). GWAS on growth traits using WGS data has revealed that some SNPs and candidate genes are related to body weight in Chinese fine-wool sheep (Lu et al., 2020). Currently, there are no GWAS available on yearling wool traits by using WGS.
The application of a mixed linear model (MLM) by fitting the Q (population structure) + K (kinship) is popular in GWAS (Yu et al., 2006). The MLM-based GWAS algorithm involves one-dimensional genome scanning by testing one marker at a time (Wang et al., 2016). However, this model is not conducive to the estimation of the marker effect, because most of the quantitative traits are controlled by multiple loci, and confounding problems inevitably occur between loci (Liu et al., 2016). In addition, previous researches have indicated that these confounding problems reduce the power of MLM in the detection of candidate genes among populations with a certain population structure (Tang et al., 2019). In fixed and random model circulating probability unification (FarmCPU), which is a multiple loci model, the MLM is split into a separated fixed effect (population structure) model and a random effect (multiple sets of pseudo-Quantitative Trait Nucleotides [QTNs]) model and both are used iteratively (Liu et al., 2016; Kusmec and Schnable, 2018); this modified version has achieved good results among livestock by solving the confounding problems (Meng et al., 2017; Tang et al., 2019).
Since previous GWAS analyses for wool traits were based on a single breed, the genes that contributed to the variation among the breeds could have been overlooked. To investigate the common genetic architecture of yearling wool traits across the fine-wool sheep breeds, we conducted GWAS by using the FarmCPU model on four yearling wool traits using the whole-genome re-sequenced data from four fine-wool sheep breeds in China. Our objective was to identify SNPs associated with yearling wool traits of fine-wool sheep to optimize marker-assisted selection and dissection of the genetic architecture of wool traits.
Materials and Methods
Ethical note
The experimental protocol was in accordance with the guidelines on the care and use of experimental animals issued by the State Council of the People’s Republic of China (Approval number:2006-398). In addition, the current study was approved by the Animal Management and Ethics Committee of the Lanzhou Institute of Husbandry and Pharmaceutical Sciences of Chinese Academy of Agricultural Sciences (Permit No.SYXK-2016-0039).
Populations and phenotype
In this study, wool traits were sampled from 577 yearling fine-wool sheep (age: 14±1 months) belonging to four fine-wool sheep breeds from different regions of China. Briefly, 337 Alpine Merino sheep (AMS) were sourced from Huangcheng, Gansu province; 60 Aohan fine-wool sheep (AHS) from Chifeng, Inner Mongolia Autonomous Region; 60 Qinghai fine-wool sheep (QHS) from Sanjiaocheng, Qinghai province; and 120 CMS from Gongnaisi, Xinjiang Uygur Autonomous Region. The sheep were farmed on pasture, with appropriate feed supplemented in the winter season. The sheep were selected randomly without considering any pedigree information.
For each animal, the mid-side wool sample (weight: 95–105 g) was collected from the shoulder blades of sheep using shears and then dispatched to the National Animal and Rural Ministry of Animal and Fur Quality Supervision and Inspection Center (Lanzhou, China). The inspection process standards were implemented in accordance with the relevant national standards (GB/T 10685-2007 and GB/T 6978-2007) for yearling mean fiber diameter (YFD) and yearling clean fleece rate (YCFR), respectively. The yearling staple length (YSL) and yearling greasy fleece weight (YGFW) were measured on the local ranch. The staples in the middle on the right were selected, and the furthest natural staple length close to the skin was measured with a ruler as an individual’s YSL. The method of measuring YGFW included laying the sheep on the left side of the shearing table, with the back of the sheep against the shearer, and the abdomen facing outward. The shearer was used to cut the right wool of the fine-wool sheep from the back to the front, followed by turning the fine-wool sheep, so that it lay down on the right side, after which the left wool was cut from the sheep longitudinally over a long distance. The wool obtained was then weighed and recorded as an individual’s YGFW.
Genotype and quality control
Genomic DNA was extracted by the phenol-chloroform method using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) from sheep whole blood (5 mL) collected through the jugular vein into vacutainers containing anticoagulant K2EDTA. All samples used for genome sequencing were processed on the Illumina HiSeq Xten platform. The re-sequencing data of 460 out of 577 samples were obtained from previously published genotype data (Lu et al., 2020). Additional 117 samples were obtained following the protocol described by Lu et al (2020), totaling 577 samples for analysis in this study.
The raw data were filtered in accordance with the following conditions:
1) Reads containing the linker sequence were filtered;
2) The N content in single-ended access reads exceeding 10% of the read length was set as the standard for deleting paired reads; and
3) When the number of low-quality (≤5) bases contained in the single-ended sequencing read exceeded 50% of the length of the read length, the paired reads were removed.
High-quality sequencing data were mapped to the Oar_v4.0 reference sequence using the BWA software (Parameter: mem-t 4-K 32-M) (Li and Durbin, 2009). The duplicates were removed by using the SAMtools (parameter: rmdup) (Li et al., 2009). We performed SNP calling on a population scale using a Bayesian approach as implemented in the package SAMtools. We then calculated the genotype likelihoods from reads for each individual at each genomic location and the allele frequencies in the sample with a Bayesian approach. The “mpileup” command was used to identify SNPs with the parameters as “-q 1 -C 50 -S -D -m 2 -F 0.002 -u.” The nucleotide variants were filtered based on the quality requirement with the read depth (dp > 2), missing rate (Miss < 0.1), and MAF > 0.05) using the SAMtools.
Population stratification
The experimental yearling fine-wool sheep population belonged to four different breeds. In order to minimize the impact of the population structure, principal component analysis (PCA) was performed with the variants identified across the autosomal genome, and then the principal components (PCs) were added as covariates when conducting the GWAS. We used the Prcomp function in R to perform the PCA. A total of 9,181,115 SNPs and 577 individuals were used to perform the PCA.
Estimation of genetic parameters
We used the Average Information Restricted Maximum likelihood algorithm (AI-REML) in the “HIBLUP” (https://github.com/xiaolei-lab/hiblup) package of the R software to estimate the heritability and variance components (Gilmour et al., 1995). The BLUP model is depicted as follows:
where y is the vector of phenotypic values; b and u represent the fixed effects (sex, breed, and farm) and breeding values, respectively; X and Z are design matrices for b and u, respectively; e is the residual error vector with a normal distribution of e~N(0,Iσe2), I is an identity matrix, and σe2 is a residual variance. u~N(0,Gσu2) in which σu2 is additive genetic variance and G is derived from genomic information and constructed by the Van Raden method (VanRaden, 2008).
Genome-wide association studies
Association tests were performed by the FarmCPU method. The top three columns of PCs, including sex, farm effects, and pseudo QTNs, were added as covariates in the fixed-effect model for the association tests. The analysis model was written as follows:
where y is a phenotypic observation vector; P is a matrix of fixed effects, including the top three PCs, sex, and farm effects; Mt is the genotype matrix of t pseudo QTNs that were used as fixed effects; bp and bt are the relevant design matrices for P and Mt, respectively; Si is the ith marker to be tested and dj is the corresponding effect; and e is the residual effect vector and e~N(0, Iσ2e). The random effect model is used for selecting the most appropriate pseudo QTNs. The model can be written as follows:
where y and e stay the same as in a fixed-effect model; u is the genetic effect and u~N (0, Kσ2u), in which K is the relationship matrix defined by pseudo QTNs.
The Bonferroni correction threshold for multiple tests, which was employed for detecting the genome-wise significant SNPs, was defined as P = α/N (α = 0.05 and N is the number of SNPs). The suggestive significant threshold was set at P = 5.00E-07 in this study (Consortium, 2007; Panagiotou et al., 2012).
A previous research has reported that the LD level of sheep is low, usually when the r2 = 0.1, and the genetic distance is approximately 50 kb (Liu et al.,2017). Therefore, the genes located at the region, ±50 kb around the significant SNPs, were considered as candidate genes and were identified by using the gene annotation information of sheep reference genome Ovis aries v4.0 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000298735.2).
Results
Summary information of phenotypic data and genotypic data
The phenotypic data from 577 sheep belonging to four Chinese fine-wool sheep breeds (namely, AMS, CMS, QHS, and AHS, respectively) were studied in total. Among them, YFD with the largest sample size was 575 individuals, whereas the YCFR with the smallest sample size was 507. The descriptive statistical outcomes of YSL, YFD, YGFW, and YCFR traits for all breeds are shown in Table 1. The mean YSL values of all breeds were 10.46 cm, indicating that the mean YSL value was within the normal range when compared with that for Chinese superfine Merino sheep (Di et al., 2014). The mean YFD value of all breeds was 19.07 μm, which is similar to that of the Trangie QPLUS$ Flock established at the Agricultural Research Centre, Trangie NSW, but higher than that of the CSIRO Fine-Wool Flock established at “Longford” Field Station, Armidale NSW (Mortimer et al., 2008). All breeds’ mean YGFW and YCFR values were 5.21 kg and 56.86%, respectively. The correlations and distributions of phenotypes for all breeds are presented in Supplementary Figure S1. We noted that YGFW and YFD had a weak correlation (correlation coefficient = 0.33), which conforms to other past reports (Swan et al., 2008). On the other hand, YGFW and YCFR showed a negative correlation, with a correlation coefficient of −0.43.
Table 1.
The descriptive statistical of four yearling wool traits for all breeds
| Trait1 | Breed | Mean ± SD | Median | CV | Population size |
|---|---|---|---|---|---|
| YSL | AMS | 10.55 ± 0.98 | 10.5 | 0.09 | 335 |
| AHS | 10.03 ± 1.33 | 10 | 0.13 | 60 | |
| QHS | 10.63 ± 1.14 | 10.5 | 0.11 | 60 | |
| CMS | 10.33 ± 1.08 | 10 | 0.10 | 119 | |
| All | 10.46 ± 1.07 | 10.5 | 0.10 | 574 | |
| YFD | AMS | 18.57 ± 1.55 | 18.6 | 0.08 | 337 |
| AHS | 19.72 ± 1.98 | 19.5 | 0.10 | 60 | |
| QHS | 20.91 ± 1.63 | 20.8 | 0.08 | 59 | |
| CMS | 19.25 ± 1.80 | 19.5 | 0.09 | 119 | |
| All | 19.07 ± 1.81 | 19.1 | 0.09 | 575 | |
| YGFW | AMS | 5.10 ± 1.42 | 4.67 | 0.28 | 329 |
| AHS | 6.23 ± 0.81 | 6.3 | 0.13 | 60 | |
| QHS | 5.83 ± 0.99 | 6 | 0.17 | 30 | |
| CMS | 4.85 ± 1.21 | 4.65 | 0.25 | 118 | |
| All | 5.21 ± 1.36 | 4.9 | 0.26 | 537 | |
| YCFR | AMS | 56.79 ± 6.08 | 57.35 | 0.11 | 329 |
| AHS | 47.49 ± 6.68 | 45.66 | 0.14 | 30 | |
| QHS | 49.62 ± 5.41 | 48.7 | 0.11 | 30 | |
| CMS | 61.38 ± 5.22 | 59.16 | 0.09 | 118 | |
| All | 56.86 ± 6.90 | 57.85 | 0.12 | 507 |
1YSL, yearling staple length, cm; YFD, yearling mean fiber diameter, μm; YGFW, yearling clean fleece weight, kg; YCFR, yearling clean fleece rate, %. All: four breeds in total.
After resequencing, 10.727 Tb of raw data was generated (average: 18.591 Gb for each sample), and 10.638 Tb of filtered clean data was obtained (average: 18.438 Gb for each sample) (Supplementary Table S1). The sequencing quality was high with an average Q20 of 96.84% and an average Q30 of 92.47%. The distribution of GC content in the 460 samples ranged from 41.58% to 47.31%, indicating successful library construction and sequencing. Based on our mapping results (Supplementary Table S2), the average mapping rate reached 99.04% (range: 97.44%–99.41%). The average coverage depth was 8.37X. A total of 12,725,769 SNPs were detected after calling the variant. Following filtration and screening, 9,181,115 SNP sites met the requirements of genome-wide resequencing. The SNP density plot of each chromosome is illustrated in Supplementary Figure S2.
Estimation of genetic parameters
Genetic variance, residual variance, and the heritability of YSL, YFD, YGFW, and YCFR were estimated by the AI-REML algorithm for data of the four tested breeds. The variance component and heritability estimations are shown in Table 2. The estimates of heritabilities (h2) were high for YSL (0.6208), YFD (0.7460), YGFW (0.6758), and YCFR (0.5559). The heritabilities of yearling wool traits in our study were similar to and slightly greater than the genetic parameters for wool traits in fine-wool Australian Merino sheep (Mortimer et al., 2008; Swan et al., 2008; Mortimer et al., 2017), which may be related to the difference in the algorithms used.
Table 2.
Estimation of genetic parameters of the yearling wool traits
| Trait1 | Additive genetic variance ± SE | Residual variance ± SE | h2 ± SE |
|---|---|---|---|
| YSL | 0.6937 ± 0.1700 | 0.4237 ± 0.1490 | 0.6208 ± 0.1376 |
| YFD | 1.7280 ± 0.3841 | 0.5868 ± 0.2974 | 0.7460 ± 0.1308 |
| YGFW | 0.5823 ± 0.1359 | 0.2794 ± 0.1155 | 0.6758 ± 0.1389 |
| YCFR | 15.9128 ± 4.2869 | 12.7107 ± 3.758 | 0.5559 ± 0.1360 |
1YSL, yearling staple length; YFD, yearling mean fiber diameter; YGFW, yearling clean fleece weight; YCFR, yearling clean fleece rate.
Population stratification
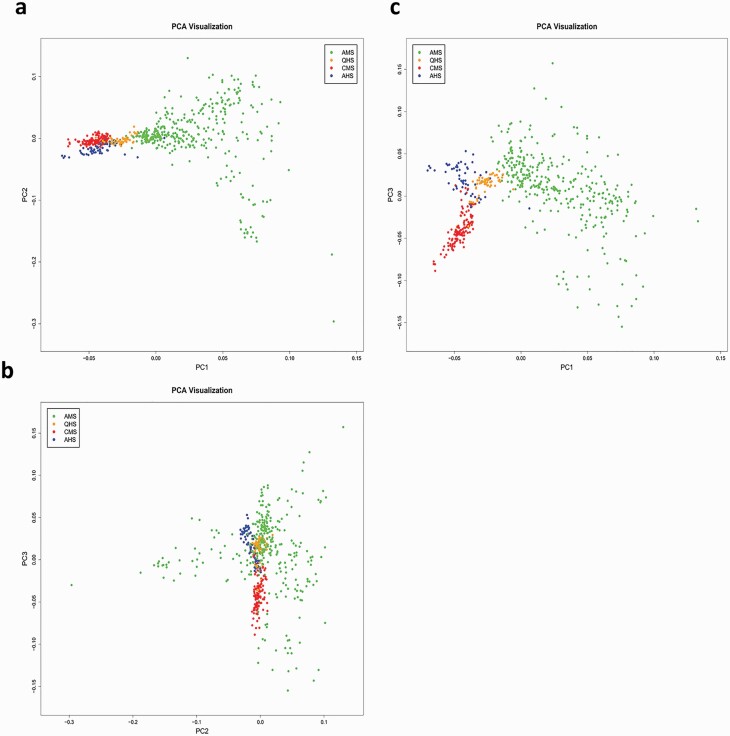
The principal component analysis (PCA) scatterplots illustrate a clear population structure for the 577 individuals from four Chinese fine-wool sheep breeds studied in the present study (Figure 1). In the scatterplot of PC1 and PC2 (Figure 1a), the majority of individuals in AMS were completely separated from the other three breeds. Similarly, most individuals from CMS and AHS were split into two sub-groups, which was more obvious in the scatterplots of PC1 and PC3 (Figure 1c). In contrast, we noticed that, in the three separate clusters, a small number of individuals gathered with QHS. This mixed cluster indicated that, although individuals may come from different breeds, they still retain close genetic relationships. As the breeding of fine-wool sheep in China is mainly based on the introduction of Merino blood fine-wool sheep, breeds in the neighboring areas are relatively closely related.
Figure 1.
Population structure inferred from the principal component analysis.
GWAS on four wool traits for the aggregated dataset
The summary of GWAS outcomes is provided in Tables 3 and 4. The nature of Bonferroni is conservative due to the ignorance of linkages between SNPs, especially for the sequencing data where several adjacent SNPs are highly linked. To overcome this issue, some authors have used a modification of correction that considers the LD between the markers (Gao et al., 2008). However, in order to reduce the amount of calculation, we used 2-threshold lines: the first one was the genome-wise significant threshold (0.05/N), which corresponded to P = 5.45E-09, and the suggestive significant threshold which was set at P = 5.00E-07 in this study to screen out SNPs (Wellcome Trust Case Control Consortium, 2007; Panagiotou et al., 2012). In the GWAS results, a total of 16 SNPs were detected above the genome-wise significance threshold, and 79 SNPs were located above the suggestive significance threshold for the four yearling wool traits. Eventually, 14 genes were located at the region ±50-kb near the genome-wise significant SNPs and 59 genes located at the region of ±50-kb near the suggestive significance SNPs.
Table 3.
List of SNPs exceeding the genome-wise significance threshold (P = 5.45E-09) and the suggestive significance threshold (P = 5.00E-07) for the YSL and YFD traits
| Trait1 | Chr | Position | Ref | Alt | Candidate gene | Distance | P value | Effect |
|---|---|---|---|---|---|---|---|---|
| YSL | 8 | 8641258 | T | G | FAM46A | intergenic (dist=17641) | 1.77E-08 | 0.36 |
| 8 | 8641623 | C | A | FAM46A | intergenic (dist=17276) | 2.80E-08 | 0.36 | |
| 8 | 8641539 | T | A | FAM46A | intergenic (dist=17360) | 9.37E-08 | 0.34 | |
| 10 | 11152096 | T | A | — | — | 1.10E-07 | −0.67 | |
| 3 | 28662360 | A | G | LOC105607652 | ncRNA_intronic | 1.35E-07 | 0.43 | |
| 5 | 64605486 | G | A | SGCD | intronic | 1.43E-07 | 0.51 | |
| 22 | 14975520 | G | A | PLCE1 | intronic | 1.46E-07 | 0.40 | |
| 16 | 20699162 | T | C | RAB3C | intronic | 1.54E-07 | 0.56 | |
| 8 | 8639864 | G | C | — | — | 1.57E-07 | 0.34 | |
| 21 | 1902899 | G | T | — | — | 1.78E-07 | −0.66 | |
| 8 | 30591134 | C | T | AIM1 | Intronic | 2.05E-07 | 0.54 | |
| 21 | 11812855 | T | C | FAT3 | intronic | 2.49E-07 | 0.53 | |
| 6 | 94762098 | G | A | C6H4orf22 | intronic | 2.62E-07 | −0.32 | |
| 2 | 148667359 | A | G | PLA2R1 | UTR3 | 2.73E-07 | −0.60 | |
| 21 | 15148252 | T | A | ANKRD42 | intronic | 3.03E-07 | −0.33 | |
| 3 | 137153518 | A | G | KANSL2 | intronic | 3.17E-07 | 0.70 | |
| 2 | 123635120 | C | T | ZNF804A | intronic | 3.26E-07 | 0.69 | |
| 1 | 74292684 | C | A | DPYD | intergenic (dist=17235) | 3.33E-07 | 0.52 | |
| 1 | 108593576 | G | C | LOC101114228, LOC101122569 | intergenic (dist=4913,dist=4510) | 3.54E-07 | 0.36 | |
| 25 | 34231711 | C | A | — | — | 3.54E-07 | 0.54 | |
| 2 | 149652380 | C | T | LOC105607993 | downstream (dist=344) | 4.05E-07 | −0.45 | |
| 2 | 148667427 | C | G | PLA2R1 | UTR3 | 4.26E-07 | −0.56 | |
| 13 | 24682512 | C | A | LOC105611292, PRTFDC1 | intergenic (dist=26319,dist=8511) | 4.35E-07 | −0.77 | |
| 4 | 32636538 | G | T | LOC101122517 | Intronic | 4.44E-07 | 0.58 | |
| 21 | 42401897 | T | A | C21H11orf85, BATF2 | intergenic (dist=7695,dist=14155) | 4.46E-07 | 0.63 | |
| 7 | 60170543 | A | G | — | — | 4.52E-07 | −0.49 | |
| 2 | 122794998 | C | T | — | — | 4.76E-07 | 0.77 | |
| YFD | 25 | 1201090 | C | T | — | — | 3.74E-12 | −0.68 |
| 1 | 35791750 | G | A | LOC106990409 | ncRNA_intronic | 1.75E-11 | −0.42 | |
| 2 | 239362082 | A | G | ARID1A | intergenic (dist=36217) | 2.01E-11 | 0.41 | |
| 22 | 47736601 | A | G | MGMT | intergenic (dist=43353) | 3.75E-11 | −0.78 | |
| 1 | 261193346 | C | G | U2AF1, CRYAA | intergenic (dist=7077,dist=30262) | 5.35E-11 | −0.76 | |
| 16 | 945395 | C | G | SLIT3 | Intronic | 1.22E-10 | 0.53 | |
| 2 | 165483199 | A | G | ARHGAP15 | Intronic | 2.54E-09 | −0.47 | |
| 6 | 58999238 | G | A | N4BP2 | Intronic | 5.99E-09 | 0.32 | |
| 15 | 77422591 | G | A | TRNAG-UCC | intergenic (dist=24659) | 1.04E-08 | 0.34 | |
| 13 | 81753224 | G | A | LOC105611432 | intergenic (dist=13119) | 1.13E-08 | 0.58 | |
| 19 | 15779888 | G | T | LOC105603404 | intergenic (dist=13359) | 1.18E-08 | −0.63 | |
| 13 | 51218682 | A | G | SLC4A11 | UTR3 | 2.22E-08 | −0.30 | |
| 5 | 98901530 | G | C | — | — | 7.58E-08 | −0.43 | |
| 18 | 22871804 | A | C | LOC105603112 | intergenic (dist=25116) | 2.19E-07 | 0.35 | |
| 7 | 84539935 | G | A | — | — | 2.40E-07 | −0.30 | |
| 13 | 65685700 | T | G | LOC105606895 | ncRNA_intronic | 2.66E-07 | 0.37 | |
| 10 | 34025023 | C | T | LOC101115632 | intergenic (dist=40244) | 3.74E-07 | −0.34 |
1YSL, yearling staple length; YFD, yearling mean fiber diameter; Distance, The base distance between the significant site and the candidate gene.
Table 4.
List of SNPs exceeding the genome-wise significance threshold (P = 5.45E-09) and the suggestive significance threshold (P = 5.00E-07) for the YGFW and YCWR traits
| Trait1 | Chr | Position | Ref | Alt | Candidate gene | Distance | P value | Effect |
|---|---|---|---|---|---|---|---|---|
| YGFW | 1 | 188612124 | C | A | MUC4 | intronic | 2.69E-19 | 0.31 |
| 1 | 269150914 | C | T | — | — | 3.51E-13 | 0.34 | |
| 5 | 23426107 | A | G | — | — | 1.18E-11 | -0.31 | |
| 13 | 27054774 | C | T | SEPHS1 | intergenic (dist=44369) | 2.94E-10 | 0.34 | |
| 1 | 196961811 | A | G | LPP | intronic | 7.91E-10 | 0.22 | |
| 17 | 68414934 | G | T | ZMAT5 | intronic | 1.36E-09 | -0.19 | |
| 4 | 87989420 | C | G | WASL | intronic | 1.67E-09 | 0.21 | |
| 1 | 67506930 | A | G | ZNF644 | intronic | 2.58E-09 | -0.28 | |
| 1 | 85049097 | A | G | FAM102B | intronic | 3.97E-09 | -0.41 | |
| 8 | 25217533 | G | A | LOC105609598 | ncRNA_intronic | 6.98E-09 | 0.24 | |
| 13 | 38899880 | G | A | CFAP61 | intronic | 1.47E-08 | -0.25 | |
| 4 | 77762584 | G | C | HECW1 | intronic | 1.61E-08 | 0.18 | |
| 14 | 11297683 | T | C | IRF8 | intronic | 6.74E-08 | 0.22 | |
| 13 | 7921804 | T | C | MACROD2 | intronic | 9.04E-08 | -0.23 | |
| 22 | 38670661 | G | A | BAG3 | upstream (dist=597) | 1.26E-07 | 0.27 | |
| 5 | 1100634 | T | C | C5H5orf45 | intronic | 1.30E-07 | 0.27 | |
| 9 | 94074802 | C | T | COL14A1, MRPL13 | intergenic (dist=5227, dist=10886) | 1.76E-07 | 0.23 | |
| 5 | 83206038 | C | A | — | — | 2.28E-07 | -0.36 | |
| 21 | 18809253 | C | T | — | — | 2.61E-07 | 0.39 | |
| 2 | 220658191 | G | A | — | — | 3.21E-07 | -0.38 | |
| 11 | 5170923 | G | T | LOC105606925, HLF | intergenic (dist=45751, dist=19483) | 4.71E-07 | -0.23 | |
| YCFR | 1 | 142555360 | G | T | ROBO2 | Intronic | 1.22E-08 | -2.27 |
| 19 | 28423809 | C | T | PDZRN3 | intergenic (dist=4107) | 2.87E-08 | -2.08 | |
| 6 | 113475592 | T | C | AFAP1 | Intronic | 3.01E-08 | 1.84 | |
| 3 | 67874237 | A | G | — | — | 3.76E-08 | 3.74 | |
| 9 | 59105226 | T | A | EXT1 | Intronic | 1.24E-07 | 4.29 | |
| 20 | 2784078 | A | G | PRIM2 | Intronic | 1.25E-07 | 3.32 | |
| 11 | 25670079 | T | G | DERL2, DHX33 | intergenic (dist=5783, dist=2829) | 1.38E-07 | -3.04 | |
| 3 | 59617099 | C | T | IL1RN, IL1F10 | intergenic (dist=28452, dist=21619) | 1.39E-07 | 2.71 | |
| 20 | 2784141 | A | C | PRIM2 | Intronic | 1.46E-07 | 3.62 | |
| 17 | 62088000 | A | G | CCDC64 | Intronic | 1.61E-07 | -1.89 | |
| 3 | 59652444 | A | G | IL1F10 | Intronic | 1.68E-07 | 2.76 | |
| 1 | 183909699 | G | A | STXBP5L | Intronic | 2.02E-07 | 2.32 | |
| 1 | 183909695 | A | G | STXBP5L | Intronic | 2.49E-07 | 2.32 | |
| 23 | 40010684 | G | A | — | — | 2.55E-07 | -1.77 | |
| 17 | 68219473 | C | T | NEFH | UTR3 | 2.63E-07 | 2.05 | |
| 20 | 3348869 | C | G | BEND6 | intergenic (dist=26031) | 2.85E-07 | -2.86 | |
| 20 | 3348874 | G | C | BEND6 | intergenic (dist=26036) | 2.85E-07 | -2.86 | |
| 2 | 234131875 | C | T | ZBTB8A | Intronic | 2.90E-07 | -3.56 | |
| 6 | 69923844 | C | T | — | — | 3.16E-07 | -2.44 | |
| 23 | 9368217 | G | A | — | — | 3.25E-07 | -3.29 | |
| 18 | 27749841 | C | G | TARSL2 | intergenic (dist=48020) | 3.31E-07 | -2.66 | |
| 18 | 17758342 | T | G | — | — | 3.66E-07 | 1.82 | |
| 20 | 2784118 | C | T | PRIM2 | intronic | 3.70E-07 | 2.94 | |
| 18 | 27894106 | A | G | TM2D3 | intergenic (dist=36225) | 3.92E-07 | -2.17 | |
| 3 | 67897772 | G | A | — | — | 4.61E-07 | 3.69 | |
| 9 | 38397717 | C | A | — | — | 4.68E-07 | -2.60 | |
| 3 | 28449359 | A | G | LDAH | intergenic (dist=35449) | 4.82E-07 | -3.35 | |
| 3 | 77756128 | C | T | EPAS1 | intronic | 4.83E-07 | -2.35 | |
| 1 | 123177590 | G | A | KRTAP6-1 | intergenic (dist=29165) | 4.87E-07 | -3.21 | |
| 1 | 126462294 | G | A | — | — | 5.00E-07 | -3.64 |
1YGFW, yearling clean fleece weight; YCFR, yearling clean fleece rate; Distance, The base distance between the significant site and the candidate gene.
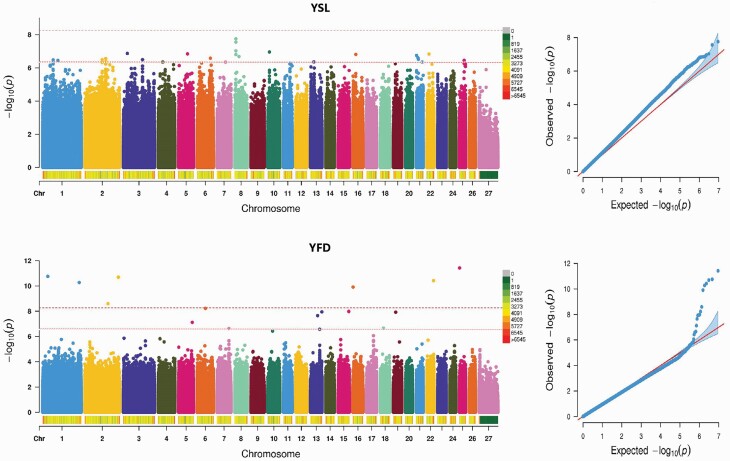
For the YSL trait, none of the SNPs reached the genome-wise significance threshold, whereas 27 SNPs exceeded the suggestive significance threshold. Among the suggestive significance SNPs associated with YSL trait, the most significant SNP was located on Ovis aries chromosome (OAR) 8, and interestingly, three loci were located closely on the OAR 8 and were extracted from the same gene FAM46A. Two adjacent SNPs located on OAR2 were co-distributed in the 3′UTR region of PLA2R1 and one locus at the 344-bp downstream region of LOC105607993. Unfortunately, consultation of the QTL database revealed no QTL near the suggestive significant sites. For the YFD trait, 7 genome-wise significance SNPs and 10 suggestive significance SNPs exceeded the threshold lines. Among the genome-wise significance SNPs associated with YFD trait, the most significant SNP was located on OAR 25 extracted none candidate gene, and one site was located in the ncRNA intronic region of LOC106990409 on OAR 1. As discovered from the distribution of interesting sits above the suggestive significance level, the most significant SNP associated with YFD trait was located in the intronic region of N4BP2 on OAR 6, and one site was located in the ncRNA intronic region of LOC105606895 on OAR13. One site on OAR 13 was located in the 3′UTR region of SLC4A11. The detailed information about significant SNPs for the YSL and YFD traits is provided in Table 3 and Figure 2, respectively.
Figure 2.
Manhattan plots and QQ plots of YSL and YFD traits. The threshold of genome-wise and suggestive significance was set at P = 5.45E-09 and P = 5.00E-07, respectively.
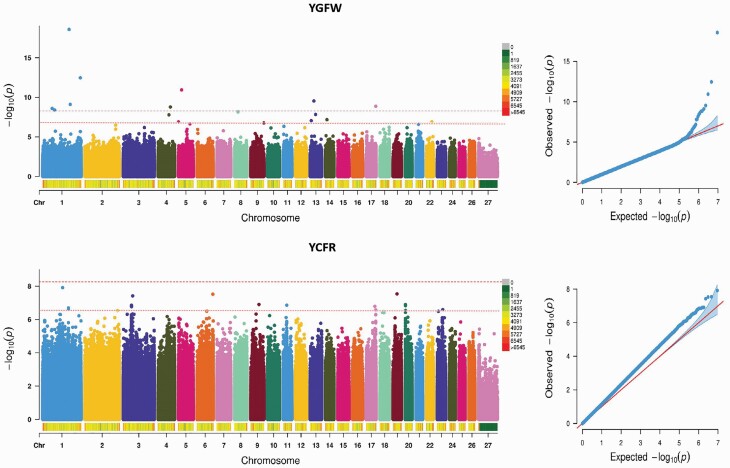
For the YGFW trait, 9 genome-wise significance SNPs and 12 suggestive significance SNPs associated with YGFW traits exceeded the threshold line. Among the genome-wise significance SNPs associated with YGFW trait, the most significant SNP was located in the intronic region of MUC4 on OAR 1. Six sites were located in the intronic region of different genes on different chromosomes. From the point of view of the distribution of the suggestive significance SNPs, the most significant SNP associated with YGFW trait was located in the ncRNA intronic region of LOC105609598 on OAR 8, whereas another one site was located 597-bp upstream of the BAG3 on OAR 22. For the YCFR trait, none of the SNPs reached the genome-wise significance threshold, whereas 30 exceeded the suggestive significance threshold. The most suggestive significant SNP associated with YCFR trait was located in the intronic region of ROBO2 on OAR 1. Interestingly three sites were located in the same gene PRIM2 on OAR20. Moreover, one site on OAR17 was located in the 3′UTR region of NEFH. The detailed information about the significant SNPs for YGFW and YCFR traits is provided in Table 4 and Figure 3, respectively.
Figure 3.
Manhattan plots and QQ plots of YGFW and YCFR traits. The threshold of genome-wise and suggestive significance was set at P = 5.45E-09 and P = 5.00E-07, respectively.
Discussion
The majority of wool traits in fine-wool sheep are quantitative traits that are regulated via complex genetic mechanisms (Bidinost et al., 2008). For fine-wool sheep breeding, revealing the candidate genes underlying the wool traits of fine-wool sheep is an important cornerstone. Since GWAS was first applied to the study of macular degeneration of the retina, it has become the mainstream method for detecting candidate genes underlying target traits (Klein et al., 2005). Therefore, we employed GWAS to detect genes that affect wool traits in fine-wool sheep. Whole-genome re-sequencing involves detection at the whole-genome level, while simultaneously obtaining comprehensive information with high accuracy. The amount of information of a standard Ovine chip, such as Illumia OvineSNP50K or OvineSNP600K chip, is relatively small when compared with whole-genome re-sequencing data. The chip’s locus information and quantity are relatively fixed, which make it difficult to obtain further information about other locations on the sample genome. Considering that most of the previous studies on fine-wool sheep used SNP chip data and considering that the sheep’s LD level is generally particularly low, in order to accurately detect the genes that affect the phenotype, we used re-sequencing data to analyze the phenotype of the yearling wool traits in GWAS analysis. However, as the number of samples is limited in this study, the use of WGSeq for GWAS was challenged by reduced statistical power. Previous studies have shown that, in a large population sample, the statistical power of GWAS is higher if the WGSeq method is used (Wang et al., 2017). In order to reduce the impact of this problem, we adopted the FarmCPU model, which is a multi-loci GWAS model: this model uses the bin method selection in the SUPER (Settlement of MLM Under Progressively Exclusive Relationship) model (Liu et al., 2016). The size of bins and a certain number of bins are used for optimization, which reduces the duplication of information between possible associated sites to a certain extent, thereby improving the statistical effectiveness of the GWAS model.
Moreover, in order to reduce the impact of confounding problems on GWAS, the FarmCPU model uses the feedback information of the detected genetic markers to re-select the markers employed for the calculation of the kinship matrix or to screen the possible association sites employed to enter or exit the model in order to solve the confounding problems of the variables and the detected markers in the model as well as to reduce the chances of false- negative outcomes. In this study, we used 9,181,115 SNPs obtained from the re-sequencing data after quality filtering and subjected them to GWAS for four yearling wool traits of 577 fine-wool sheep.
The heritabilities of the four yearling wool traits were estimated by HIBLUP using the pooling data of the four breeds in this study. The estimated heritabilities of YSL, YFD, YGFW, and YCFR were 0.6208, 0.7460, 0.6758, and 0.5559, respectively. The heritabilities of YSL and YFD were generally consistent with those of reported previously researches on Australian Merino sheep (Swan et al., 2016; Mortimer et al., 2017). When compared with the corresponding previous research reports, the accuracy of the heritability estimation of YSL and YFD in the present study was relatively high. However, the comparative accuracy of YGFW and YCFR was relatively low. In general, the heritability of yearling wool traits in this study was at a medium-high level. The heritabilities of YGFW and YCFR were slightly higher than those reported in previous studies, possibly because we used the genomic data instead of pedigree information or because of the difference in the breeds.
Population structure is normally represented by proportions of individuals belonging to sub-populations by PCs derived from genetic markers covering the whole genome. The habits and lifestyle from each subgroup could present a correlation with several traits of interest. Population stratification is an important issue that can lead to false-positive findings in population-based GWAS (Wu et al., 2011). Therefore, population stratification must be corrected in GWAS. In this research, the experimental samples were sourced from four different fine-wool sheep breeds in China. Based on the PCA results, we obtained a clear population structure, as shown in Figure 1. AMS is separated from the other three groups and presents a dispersed state, which is consistent with the breeding situation learned from the breeding history of the four fine-wool sheep breeds. CMS was bred in the Xinjiang Uygur Autonomous Region in 1985, and AHS was bred by introducing the Merino blood in Inner Mongolia Autonomous Region in 1983. In addition, QHS was bred by imported foreign blood in the Qinghai Province in 1965, whereas AMS was bred by introducing the Merino blood in the Gansu Province in 2005. The genomic inflation factor (λ) of YSL, YFD, YGFW, and YCFR was 1.1679, 0.9445, 0.9756, and 1.2144, respectively. Both the Q–Q plot and the inflation factor of each trait (Supplementary Table S3) exhibited the same trend.
The results of each GWAS are shown in Tables 2 and 3, and the number of genome-wise significant SNPs detected for YFD and YGFW was 7 and 9, respectively. The number of suggestive significant SNPs detected in the four yearling wool traits was 27 (YSL), 10 (YFD), 12 (YGFW), and 30 (YCFW), respectively. On the right side of the Manhattan plot, the quantile–quantile (Q–Q) plots of the test statistics were drawn (Figures 2 and 3), and no overall systemic bias was indicated in the analysis based on a single-marker analysis. We provide below a discussion of the most promising results reported here for the different analyzed traits according to the genome-wise significance and suggestive significance thresholds below.
Yearling staple length
The staple length generally determines the end-use (weaving or knitting) of wool (Cottle, 1991). Above the suggestive significance threshold line, we recorded a significant SNP (14975520, P = 1.46E-07) located on OAR 22 in the intron region of 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 (PLCE1) associated with the YSL trait. PLCE1 is a bifunctional enzyme that also regulates small GTPases of the Ras superfamily through its Ras guanine-exchange factor (RasGEF) activity (Ada-Nguema et al., 2006). Previous research reported that RasGEF plays an important role in the epidermal growth factor receptor signaling pathway (Hamilton and Wolfman, 1998). RasGEF1a has been reported to be related to wool traits in the CMS wool (Liu et al., 2017). Therefore, PLCE1 may be indirectly related to wool traits. PLCE1 plays an essential role in the activation of HRASI (HRas proto-oncogene, GTPase). Mutations in the proto-oncogene HRASI form the basis of the development of Costello syndrome. The main presentations of this syndrome are rough facial skin, premature skin aging, slow hair growth, and easy hair loss (Aoki et al., 2005; Lorenz et al., 2013). During the process of activation, HRASI binds to effector proteins such as serine/threonine RAF kinases, the catalytic subunits of phosphoinositide 3-kinase (PI3K), and phospholipase C1 (PLCE1), and enters the activated state. In addition, a significant SNP located at the 3′UTR of PLA2R1 (secretory phospholipase A2 receptor) on OAR 2 exceeded the suggestive significance threshold line. The differentiation of the mesenchymal stem cells into keratinocytes occurs through the JAK2 signaling pathway (Jiang et al., 2019). The past research displays that PLA2R1 regulates the replicative senescence, a telomerase-dependent proliferation arrest, and mediates tumor suppression via the activation of the JAK2 pathway (Vindrieux et al., 2013; Bernard and Vindrieux, 2014). We thus speculated that PLA2R1 may play a key role in the process of differentiation of mesenchymal stem cells into keratinocytes, thereby affecting the length of wool. This observation suggests that PLA2R1 may play an important role in the process of wool growth. It should be indicated that the most suggestive significant SNPs were on OAR8 and within FAM46A. Although there is no directly related literature report supporting that FAM46A is related to hair growth, previous research has indicated FAM46A as a trans-regulator for leptin (Carayol et al., 2017). Leptin stimulates keratinocytes to proliferate and participates in hair follicle morphogenesis and cycle (Mercati et al., 2019). So we speculate that FAM46A may be indirectly related to hair growth.
Yearling fiber diameter
Fiber diameter is one of the most economically significant attributes of sheep wool. For the YFD trait, we noted a significant site (945395, P = 1.22E-10) located in the intron region of slit homolog 3 protein (SLIT3) on OAR 16, exceeding the genome-wise significance threshold line. SLIT3 was present predominantly in fibrillar collagen-producing cells and negatively regulated cell growth (Marlow et al., 2008; Gong et al., 2020). Previous studies on knockout SLIT3 mice have reported a significant reduction in the hair follicle density in comparison with that in normal mice (Gong et al., 2020). In addition, a high negative correlation between wool fiber diameter and wool follicle density has been demonstrated (Adelson et al., 2002). We found a SNP (51218682, P = 2.22E-08) located at the 3′UTR of sodium bicarbonate transporter-like protein 11 (SLC4A11) on OAR 13 suggestive of significant relation to YFD. SLC4A11 is also referred to as sodium borate cotransporter 1 (NaBC1). A previous study asserted that borate can activate the MAPK pathway to stimulate cell growth and proliferation. Moreover, the knockdown of NaBC1 was found to halted cell growth and proliferation (Park et al., 2004).
Yearling greasy fleece weight
The hair follicle morphology and hair follicle growth cycle has been shown to have a direct impact on the rate of wool growth and greasy fleece weight gain. Marker (87989420, P = 1.67E-09) on OAR 4 that reached the significance threshold line has been associated with the YGFW trait. This marker is located in WASP-like actin nucleation promoting factor (WASL); this gene is also known as N-WASP. According to past reports, the N-WASP protein encoded by this gene is related to keratinization, and it can delay the morphogenesis of hair follicles and abnormal hair follicle circulation. In addition, it is related to circulatory alopecia and long-term growth and activation periods. In addition, this gene was identified as a new element of hair cycle control in the related past studies on the function of N-WASP for regulating the anti-proliferation and pro-apoptotic pathways (TGFβ pathways) in keratinocytes both in vivo and in vitro (Lefever et al., 2010). Moreover, the role of N-WASP in epidermal homeostasis and skin biology was demonstrated in mice with N-WASP knockout, which presented symptoms of stunted growth, abnormal hair loss, and dry skin. Thickened epidermis and abnormal stratum corneum are known to seriously affect the growth of hair clumps, which is undoubtedly extremely important in greasy fleece weight gain (Kalailingam et al., 2017). We also detected a SNP located in the intron region of Mucin-4 (MUC4) on OAR 1, which was significantly related to YGFW. MUC4, a member of the transmembrane mucin family, is expressed in epithelial cells (Kohli et al., 2019). Previous research has confirmed that MUC4 plays an important role in cell proliferation and differentiation of epithelial cells (Kargı et al., 2006; Zhang et al., 2006). The cancer cells use mucin for cell proliferation (Narashiman et al., 2014). Therefore, MUC4 may affect the differentiation and proliferation of hair follicle stem cells, resulting in different wool yields.
Yearling clean fleece rate
Clean fleece rate reflects the cleanliness of wool, which is closely related to the fecal material, plant material, skin homeostasis, and secretion of sweat and oil. Among these influencing factors, skin homeostasis is controlled by complex interactions between tightly regulated transcription factors and signal transduction pathways; moreover, it depends on whether the differentiation and keratinization of keratinocytes were normal, which occurs through programmed cell death and generally does not cause inflammation (Lachner et al., 2017). However, when homeostasis is disrupted, skin inflammation can occur. In the present study, we detected a suggestive significance marker from OAR 3 (59617099, P = 1.39 E-07), which was located within the IL1F10 and was associated with the YCFR trait. A previous study on epidermal homeostasis revealed that, during the terminal differentiation of keratinocytes, the expression levels of IL1A and IL1B were downregulated, whereas that of anti-inflammatory IL1F members interleukin-37 (IL1F7) and interleukin-1 family member 10 (IL1F10) was strongly induced in differentiated keratinocytes. It is thus suggested that anti-inflammatory IL1F members such as IL1F10 play a crucial role in the regulation of skin homeostasis (Lachner et al., 2017). The KRTAP family not only encodes the main structural protein of epithelial tissues and hair but is also particularly involved in the differentiation of sweat glands and sebaceous glands (Donet et al., 2008; Hu et al., 2020). In this study, we discovered a suggestive significance marker (123177590, P = 4.87E-07) located in KRTAP6-1 on OAR1, which was associated with the YCFR trait. Factors such as the abnormal function of the sweat glands and sebaceous glands could affect normal sweating and oil secretion of the skin, resulting in wool pollution. There is a suggestive significant locus in the intron region of Exostosin-1 (EXT1) on OAR 9 related to YCFR. EXT1 is a glycosyltransferase required for the biosynthesis of heparan-sulfate (HS) (Duncan et al., 2001). In the past study, the knockout of HS led to the morphogenesis and hyperplasia of the sebaceous glands in mature mice, which in turn led to exacerbated sebum production on the skin surface (Coulson-Thomas et al., 2014). Therefore, we speculated that the secretion of lanolin may affect the YCFR trait.
Conclusions
In this study, we employed re-sequencing data to perform GWAS by using the FarmCPU model for four yearling wool traits of four Chinese fine-wool sheep breeds. A total of 16 SNPs were detected above the significant threshold line, and 79 SNPs were above the suggestive threshold line, whereas 14 genes were located at the region ±50-kb near the significant SNPs and 59 genes were located at the region ±50-kb near the suggestive significant SNPs. Among them, PLCE1, through its RasGEF activity, plays an important role in the epidermal growth factor receptor. SLIT3 negatively regulates cell growth, and previous studies on SLIT3 knockout have revealed a significant reduction in the mice’s hair follicle density. The newly detected significant SNPs and the newly identified candidate genes in this study may enhance our understanding of yearling wool traits in fine-wool sheep that can now be applied for marker-assisted selection in fine-wool sheep breeding. Furthermore, we believe that our study provides a reference value for the study of hair growth and hair follicle stem cells.
Supplementary Material
Acknowledgments
Bohui Yang and Yaojing Yue played an equally important role in the financial support, experimental design and guidance of this research. This work was supported by the Major Output Research Topic of Chinese Academy of Agricultural Sciences (CAAS-ZDXT2018006), the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2015-LIHPS), and the Modern China Wool Cashmere Technology Research System (CARS-39-02).
Glossary
Abbreviations
- GWAS
genome-wide association studies
- AI-REML
Average Information Restricted Maximum likelihood algorithm
- WGS
Whole-genome sequence
- LD
linkage disequilibrium
- MLM
mixed linear model
- QTNs
quantitative trait nucleotides
- PCA
principal component analysis
- OAR
Ovis aries chromosome
Authors’ Contributions
B.Y. obtained the funding; B.Y. and Y.Y. designed this project; H.Z., S.Z., and T.G. analyzed the content of the data with the help of M.H., B.C., G.Q., and C.Y.; J.L., Z.L., W.S., T.W., F.L., Y.Z., and F.H. provided assistance with sample and data collection. H.Z. drafted the manuscript with the help of B.Y. and Y.Y. All authors came to an agreement for publication.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this research article.
Literature Cited
- Ada-Nguema, A. S., Xenias H., Hofman J. M., Wiggins C. H., Sheetz M. P., and Keely P. J.. . 2006. The small GTPase R-Ras regulates organization of actin and drives membrane protrusions through the activity of PLCepsilon. J. Cell Sci. 119(Pt 7):1307–1319. doi: 10.1242/jcs.02835. [DOI] [PubMed] [Google Scholar]
- Adelson, D., Hollis D., and Brown G.. . 2002. Wool fibre diameter and follicle density are not specified simultaneously during wool follicle initiation. Aust. J. Agric. Res. 53(9):1003–1009. doi: 10.1071/AR01200. [DOI] [Google Scholar]
- Aoki, Y., Niihori T., Kawame H., Kurosawa K., Ohashi H., Tanaka Y., Filocamo M., Kato K., Suzuki Y., Kure S., . et al. 2005. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- Bernard, D., and Vindrieux D.. . 2014. PLA2R1: expression and function in cancer. Biochim. Biophys. Acta 1846:40–44. doi: 10.1016/j.bbcan.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Bidinost, F., Roldan D., Dodero A., Cano E., Taddeo H., Mueller J., and Poli M.. . 2008. Wool quantitative trait loci in Merino sheep. Small Rum. Res. 74(1–3):113–118. doi: 10.1016/j.smallrumres.2007.04.005 [DOI] [Google Scholar]
- Bolormaa, S., Swan A. A., Brown D. J., Hatcher S., Moghaddar N., van der Werf J. H., Goddard M. E., and Daetwyler H. D.. . 2017. Multiple-trait QTL mapping and genomic prediction for wool traits in sheep. Genet. Sel. Evol. 49:62. doi: 10.1186/s12711-017-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D., Swan A. A., and Gill J. S.. . 2013. Genetic correlations across ages for greasy fleece weight and fibre diameter in Merino sheep. Assoc. Adv. Anim. Breed. Genet. 20:110–103. [Google Scholar]
- Carayol, J., Chabert C., Di Cara A., Armenise C., Lefebvre G., Langin D., Viguerie N., Metairon S., Saris W. H. M., Astrup A., . et al. 2017. Protein quantitative trait locus study in obesity during weight-loss identifies a leptin regulator. Nat. Commun. 8:2084. doi: 10.1038/s41467-017-02182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, W. T. C. C. 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447(7145):661. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottle, D. J. 1991. Australian sheep and wool handbook. Melbourne: Inkata Press. [Google Scholar]
- Coulson-Thomas, V. J., Gesteira T. F., Esko J., and Kao W.. . 2014. Heparan sulfate regulates hair follicle and sebaceous gland morphogenesis and homeostasis. J. Biol. Chem. 289:25211–25226. doi: 10.1074/jbc.M114.572511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler, H. D., Kemper K. E., van der Werf J. H., and Hayes B. J.. . 2012. Components of the accuracy of genomic prediction in a multi-breed sheep population. J. Anim. Sci. 90:3375–3384. doi: 10.2527/jas.2011-4557. [DOI] [PubMed] [Google Scholar]
- Di, J., Ainiwaer L., Xu X.-M., Zhang Y.-H., Yu L.-J., and Li W.-C.. . 2014. Genetic trends for growth and wool traits of Chinese superfine Merino sheep using a multi-trait animal model. Small Rum. Res. 117(1):47–51. doi: 10.1016/j.smallrumres.2013.12.001 [DOI] [Google Scholar]
- Donet, E., Bayo P., Calvo E., Labrie F., and Pérez P.. . 2008. Identification of novel glucocorticoid receptor-regulated genes involved in epidermal homeostasis and hair follicle differentiation. J. Steroid Biochem. Mol. Biol. 108:8–16. doi: 10.1016/j.jsbmb.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Duncan, G., McCormick C., and Tufaro F.. . 2001. The link between heparan sulfate and hereditary bone disease: finding a function for the EXT family of putative tumor suppressor proteins. J. Clin. Invest. 108:511–516. doi: 10.1172/JCI13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freebern, E., Santos D. J. A., Fang L., Jiang J., Parker Gaddis K. L., Liu G. E., VanRaden P. M., Maltecca C., Cole J. B., and Ma L.. . 2020. GWAS and fine-mapping of livability and six disease traits in Holstein cattle. BMC Genomics 21:41. doi: 10.1186/s12864-020-6461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X., Starmer J., and Martin E. R.. . 2008. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- Gilmour, A. R., Thompson R., and Cullis B. R.. . 1995. Average information REML: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 51:1440–1450. doi: 10.2307/2533274. [DOI] [Google Scholar]
- Gong, L., Wang S., Shen L., Liu C., Shenouda M., Li B., Liu X., Shaw J. A., Wineman A. L., and Yang Y.. . 2020. SLIT3 deficiency attenuates pressure overload–induced cardiac fibrosis and remodeling. JCI insight 5(12):136852. doi: 10.1172/jci.insight.136852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, M., and Wolfman A.. . 1998. Oncogenic Ha-Ras-dependent mitogen-activated protein kinase activity requires signaling through the epidermal growth factor receptor. J. Biol. Chem. 273:28155–28162. doi: 10.1074/jbc.273.43.28155. [DOI] [PubMed] [Google Scholar]
- Hu, Y., Song Z., Chen J., and Caulin C.. . 2020. Overexpression of MYB in the Skin Induces Alopecia and Epidermal Hyperplasia. J. Invest. Dermatol. 140:1204–1213.e5. doi: 10.1016/j.jid.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., Wu F., Xu Y., Yan J. X., Wu Y. D., Li S. H., Liao X., Liang J. X., Li Z. H., and Liu H. W.. . 2019. A novel role of angiotensin II in epidermal cell lineage determination: Angiotensin II promotes the differentiation of mesenchymal stem cells into keratinocytes through the p38 MAPK, JNK and JAK2 signalling pathways. Exp. Dermatol. 28:59–65. doi: 10.1111/exd.13837. [DOI] [PubMed] [Google Scholar]
- Kalailingam, P., Tan H. B., Jain N., Sng M. K., Chan J. S. K., Tan N. S., and Thanabalu T.. . 2017. Conditional knock out of N-WASP in keratinocytes causes skin barrier defects and atopic dermatitis-like inflammation. Sci. Rep. 7:7311. doi: 10.1038/s41598-017-07125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargı, A., Dinç Z. A., Başok O., and Üçvet A.. . 2006. MUC4 expression and its relation to ErbB2 expression, apoptosis, proliferation, differentiation, and tumor stage in non-small cell lung cancer (NSCLC). Pathol. Res. Pract. 202(8):577–583. doi: 10.1016/j.prp.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Kijas, J. W., Porto-Neto L., Dominik S., Reverter A., Bunch R., McCulloch R., Hayes B. J., Brauning R., and McEwan J.; International Sheep Genomics Consortium . 2014. Linkage disequilibrium over short physical distances measured in sheep using a high-density SNP chip. Anim. Genet. 45:754–757. doi: 10.1111/age.12197. [DOI] [PubMed] [Google Scholar]
- Klein, R. J., Zeiss C., Chew E. Y., Tsai J. Y., Sackler R. S., Haynes C., Henning A. K., SanGiovanni J. P., Mane S. M., Mayne S. T., . et al. 2005. Complement factor H polymorphism in age-related macular degeneration. Science 308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli, M., Shivam A. K., Ahuja P., and Dutta J.. . 2019. Mucin-4: a novel marker for oral cancer. J. Oral Maxillofac. Pathol. 23:49–53. doi: 10.4103/jomfp.JOMFP_175_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmec, A., and Schnable P. S.. . 2018. FarmCPUpp: efficient large-scale genomewide association studies. Plant Direct. 2:e00053. doi: 10.1002/pld3.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner, J., Mlitz V., Tschachler E., and Eckhart L.. . 2017. Epidermal cornification is preceded by the expression of a keratinocyte-specific set of pyroptosis-related genes. Sci. Rep. 7:17446. doi: 10.1038/s41598-017-17782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefever, T., Pedersen E., Basse A., Paus R., Quondamatteo F., Stanley A. C., Langbein L., Wu X., Wehland J., Lommel S., . et al. 2010. N-WASP is a novel regulator of hair-follicle cycling that controls antiproliferative TGF{beta} pathways. J. Cell Sci. 123(Pt 1):128–140. doi: 10.1242/jcs.053835. [DOI] [PubMed] [Google Scholar]
- Li, S., Chen W., Zheng X., Liu Z., Yang G., Hu X., and Mou C.. . 2020. Comparative investigation of coarse and fine wool sheep skin indicates the early regulators for skin and wool diversity. Gene 758:144968. doi: 10.1016/j.gene.2020.144968. [DOI] [PubMed] [Google Scholar]
- Li, H., and Durbin R.. . 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Zheng X., Nie Y., Chen W., Liu Z., Tao Y., Hu X., Hu Y., Qiao H., Qi Q., . et al. 2018. Defining key genes regulating morphogenesis of apocrine sweat gland in sheepskin. Front. Genet. 9:739. doi: 10.3389/fgene.2018.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., He S., Chen L., Li W., Di J., and Liu M.. . 2017. Estimates of linkage disequilibrium and effective population sizes in Chinese Merino (Xinjiang type) sheep by genome-wide SNPs. Genes Genom. 39:733–745. doi: 10.1007/s13258-017-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Huang M., Fan B., Buckler E. S., and Zhang Z.. . 2016. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 12:e1005767. doi: 10.1371/journal.pgen.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, S., Lissewski C., Simsek-Kiper P. O., Alanay Y., Boduroglu K., Zenker M., and Rosenberger G.. . 2013. Functional analysis of a duplication (p.E63_D69dup) in the switch II region of HRAS: new aspects of the molecular pathogenesis underlying Costello syndrome. Hum. Mol. Genet. 22:1643–1653. doi: 10.1093/hmg/ddt014. [DOI] [PubMed] [Google Scholar]
- Lu, Z., Yue Y., Yuan C., Liu J., Chen Z., Niu C., Sun X., Zhu S., Zhao H., and Guo T.. . 2020. Genome-wide association study of body weight traits in chinese fine-wool sheep. Animals 10(1):170. doi: 10.3390/ani10010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow, R., Strickland P., Lee J. S., Wu X., Pebenito M., Binnewies M., Le E. K., Moran A., Macias H., Cardiff R. D., . et al. 2008. SLITs suppress tumor growth in vivo by silencing Sdf1/Cxcr4 within breast epithelium. Cancer Res. 68:7819–7827. doi: 10.1158/0008-5472.CAN-08-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba, N., Akiyama M., Ishigaki K., Kanai M., Takahashi A., Momozawa Y., Ikegawa S., Ikeda M., Iwata N., Hirata M., . et al. 2020. GWAS of 165,084 Japanese individuals identified nine loci associated with dietary habits. Nat. Hum. Behav. 4:308–316. doi: 10.1038/s41562-019-0805-1. [DOI] [PubMed] [Google Scholar]
- Meng, Q., Wang K., Liu X., Zhou H., Xu L., Wang Z., and Fang M.. . 2017. Identification of growth trait related genes in a Yorkshire purebred pig population by genome-wide association studies. Asian-Australas. J. Anim. Sci. 30:462–469. doi: 10.5713/ajas.16.0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercati, F., Dall’Aglio C., Timperi L., Scocco P., De Felice E., and Maranesi M.. . 2019. Epithelial expression of the hormone leptin by bovine skin. Eur J Histochem 63(1):2993. doi: 10.4081/ejh.2019.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddar, N., Khansefid M., van der Werf J. H. J., Bolormaa S., Duijvesteijn N., Clark S. A., Swan A. A., Daetwyler H. D., and MacLeod I. M.. . 2019. Genomic prediction based on selected variants from imputed whole-genome sequence data in Australian sheep populations. Genet. Sel. Evol. 51:72. doi: 10.1186/s12711-019-0514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer, S. I., Hatcher S., Fogarty N. M., van der Werf J. H. J., Brown D. J., Swan A. A., Greeff J. C., Refshauge G., Edwards J. E. H., and Gaunt G. M.. . 2017. Genetic parameters for wool traits, live weight, and ultrasound carcass traits in Merino sheep. J. Anim. Sci. 95:1879–1891. doi: 10.2527/jas.2016.1234. [DOI] [PubMed] [Google Scholar]
- Mortimer, S. I., Robinson D. L., Atkins K., Brien F., Swan A., Taylor P., and Fogarty N.. . 2008. Genetic parameters for visually assessed traits and their relationships to wool production and liveweight in Australian Merino sheep. Anim. Prod. Sci. 49(1):32–42. [Google Scholar]
- Narashiman, S., Narasimhan M., and Venkatraman G.. . 2014. Expression of Mucin 4 in leukoplakia and oral squamous cell carcinoma: an immunohistochemical study. J. Oral Maxillofac. Pathol. 18:25–31. doi: 10.4103/0973-029X.131887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotou, O. A., and Ioannidis J. P.; Genome-Wide Significance Project . 2012. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int. J. Epidemiol. 41:273–286. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- Park, M., Li Q., Shcheynikov N., Zeng W., and Muallem S.. . 2004. NaBC1 is a ubiquitous electrogenic Na+ -coupled borate transporter essential for cellular boron homeostasis and cell growth and proliferation. Mol. Cell 16:331–341. doi: 10.1016/j.molcel.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Shahinfar, S., and Kahn L.. . 2018. Machine learning approaches for early prediction of adult wool growth and quality in Australian Merino sheep. Comput. Electron Agric. 148:72–81. doi: 10.1016/j.compag.2018.03.001 [DOI] [Google Scholar]
- Swan, A. A., Brown D. J., and van der Werf J. H.. . 2016. Genetic variation within and between subpopulations of the Australian Merino breed. Anim. Prod. Sci. 56(1):87–94. [Google Scholar]
- Swan, A., Purvis I. W., and Piper L.. . 2008. Genetic parameters for yearling wool production, wool quality and bodyweight traits in fine wool Merino sheep. Aust. J Exp. Agric. 48(9):1168–1176. [Google Scholar]
- Tang, Z., Xu J., Yin L., Yin D., Zhu M., Yu M., Li X., Zhao S., and Liu X.. . 2019. Genome-wide association study reveals candidate genes for growth relevant traits in pigs. Front. Genet. 10:302. doi: 10.3389/fgene.2019.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden, P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Vindrieux, D., Augert A., Girard C. A., Gitenay D., Lallet-Daher H., Wiel C., Le Calvé B., Gras B., Ferrand M., Verbeke S., . et al. 2013. PLA2R1 mediates tumor suppression by activating JAK2. Cancer Res. 73:6334–6345. doi: 10.1158/0008-5472.CAN-13-0318. [DOI] [PubMed] [Google Scholar]
- Wang, Z., and Chatterjee N.. . 2017. Increasing mapping precision of genome-wide association studies: to genotype and impute, sequence, or both? Genome Biol. 18:118. doi: 10.1186/s13059-017-1255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. B., Feng J. Y., Ren W. L., Huang B., Zhou L., Wen Y. J., Zhang J., Dunwell J. M., Xu S., and Zhang Y. M.. . 2016. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 6:19444. doi: 10.1038/srep19444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Zhang H., Yang H., Wang S., Rong E., Pei W., Li H., and Wang N.. . 2014. Genome-wide association study for wool production traits in a Chinese Merino sheep population. PLoS One 9:e107101. doi: 10.1371/journal.pone.0107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., DeWan A., Hoh J., and Wang Z.. . 2011. A comparison of association methods correcting for population stratification in case-control studies. Ann. Hum. Genet. 75:418–427. doi: 10.1111/j.1469-1809.2010.00639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Pressoir G., Briggs W. H., Vroh Bi I., Yamasaki M., Doebley J. F., McMullen M. D., Gaut B. S., Nielsen D. M., Holland J. B., . et al. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Chen Z., Ye S., He Y., Huang S., Yuan X., Chen Z., Zhang H., and Li J.. . 2019. Genome-wide association study for reproductive traits in a duroc pig population. Animals 9(10):732. doi: 10.3390/ani9100732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Yasin M., Carraway C. A., and Carraway K. L.. . 2006. MUC4 expression and localization in gastrointestinal tract and skin of human embryos. Tissue Cell 38:271–275. doi: 10.1016/j.tice.2006.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.