Abstract
Coronaviruses have emerged as alarming pathogens owing to their inherent ability of genetic variation and cross-species transmission. Coronavirus infection burdens the endoplasmic reticulum (ER.), causes reactive oxygen species production and induces host stress responses, including unfolded protein response (UPR) and antioxidant system. In this study, we have employed a neurotropic murine β-coronavirus (M-CoV) infection in the Central Nervous System (CNS) of experimental mice model to study the role of host stress responses mediated by interplay of DJ-1 and XBP1. DJ-1 is an antioxidant molecule with established functions in neurodegeneration. However, its regulation in virus-induced cellular stress response is less explored. Our study showed that M-CoV infection activated the glial cells and induced antioxidant and UPR genes during the acute stage when the viral titer peaks. As the virus particles decreased and acute neuroinflammation diminished at day ten p.i., a significant up-regulation in UPR responsive XBP1, antioxidant DJ-1, and downstream signaling molecules, including Nrf2, was recorded in the brain tissues. Additionally, preliminary in silico analysis of the binding between the DJ-1 promoter and a positively charged groove of XBP1 is also investigated, thus hinting at a mechanism behind the upregulation of DJ-1 during MHV-infection. The current study thus attempts to elucidate a novel interplay between the antioxidant system and UPR in the outcome of coronavirus infection.
Keywords: Mouse Hepatitis Virus, Neuro-COVID, Microglia, Astrocyte, Oxidative stress, XBP1-DJ-1-Nrf2 axis
1. Introduction
Successful virus infection and completion of the viral life cycle, including replicating genome and assembly of functional virus particles, are satisfied at the expense of host cell metabolism [1]. A number of viruses have evolved the ability to alter host cellular environments by inducing cellular stress responses like endoplasmic reticulum (E.R.) stress and oxidative stress responses, and unfolded protein response (UPR) [2]. Coronaviruses (CoVs) are a family of enveloped, positive-strand RNA viruses that have been associated with several animal and human diseases [3,4]. The Coronavirus Disease 2019 (COVID19), originated in Wuhan, China, is an ongoing pandemic that has aggressively spread worldwide [5,6]. Interestingly, this is the third example of zoonotic transmission and the coronavirus outbreak after SARS-CoV in 2003 and MERS-CoV in 2012 [7]. Thus, the highly contagious nature of CoVs and their ability to spill over different hosts makes them an important subject of investigation even in terms of host stress responses [8]. Coronavirus infection is known to up-regulate the expression of E.R. stress markers like GRP78, GRP94 in the host cells [9], [10], [11]. It is hypothesized that CoVs might cause E.R. stress in several ways [12], [13], [14], [15], [16] and to reinstate E.R. homeostasis, the host cells have evolved UPR.
Another host stress response is the generation of reactive oxygen species (ROS). ROS are produced at all times, even during homeostatic conditions, as oxygen serves as the final electron acceptor in the mitochondrial energy metabolism. In fact, a substantial amount of ROS is generated as a by-product of protein synthesis and folding in the E.R., which can have detrimental effects on the host cell [17]. Therefore, the body has developed several defense mechanisms to keep the ROS in check. However, increased viral load and cellular stress can shift the redox balance between the pro-oxidants and antioxidants towards oxidative stress. On the one hand, this oxidative stress induces UPR to promote cell survival and preserve host cell functions [18]. On the other hand, an imbalance in the redox potential can directly or indirectly affect the E.R. protein folding process. Studies have shown that overexpression of SARS-CoV 3CL-Proprotein induces ROS and NFκB expression [19]. SARS-CoV accumulation in the E.R. can trigger all three major UPR signaling pathways, promoting an inflammatory response [20]. Recent findings on SARS-CoV-2 also report the induction of an inflammatory cytokine storm in response to rapid viral replication [21]. Nuclear factor erythroid 2-related factor 2 (Nrf2), which is also called the master regulator of cellular redox homeostasis, has shown implications as a therapeutic target in SARS-CoV-2 infection owing to its reported antioxidant, anti-inflammatory effects, and transcriptional repressor activity [22,23].
Our study is focused on identifying the putative role of DJ-1, a ROS sensing molecule, upon M-CoV (murine-CoV) infection of the glial cells in the outcome of E.R. and oxidative stress responses. Neurotropic M-CoV MHV-infection in mice is widely used as an experimental animal model to study viral-induced direct neuroglia dystrophy, innate neuro-inflammation and chronic progressive CNS myelin pathology and axonal loss. DJ-1 is a 189 amino acid long dimeric protein ubiquitously expressed in all cells [24]. It is encoded by the Park7 gene and is involved in a variety of signaling cascades [25]. DJ-1 has been found to play essential roles in neuro-protection under several disease conditions like Parkinson's disease, Alzheimer's disease, and Multiple Sclerosis [26], [27], [28]. Upon activation, DJ-1 stimulates Nrf2 [29], which regulates the expression of several antioxidant genes like Heme oxygenase 1 (HMOX-1), Catalase, Thioredoxin reductase 1 (txnrd1), NAD(P)H dehydrogenase (quinone) 1 (Nqo1) [30], as well as heat shock response related heat shock factor 1 (HSF1) [31]. HSF1 serves as a major transcription factor for several heat shock proteins. DJ-1 activation is also known to play essential roles in E.R. stress and UPR by modulating activating transcription factor 4 (ATF4) and decreasing the expression of its downstream signaling molecules CHOP and BiP [32]. While both DJ-1 and Nrf2 are relatively unexplored in CoV infections, X-box protein 1 (XBP1), a UPR transcription factor, has profound and established functions in modulating high E.R. stress upon CoV infection both in vitro and in vivo [16]. XBP1 works in the IRE1 branch of the UPR, the most conserved UPR signaling pathway that is also known to regulate oxidative stress [33]. Upon activation, XBP1 translocate to the nucleus, where it enhances the expression of various UPR genes, including molecular chaperons like HSF1 and Hsp70, that help to restore E.R. stress [33].
In the current study, we have investigated the Nrf2-DJ-1-XBP1 axis during Mouse Hepatitis Virus (MHV- M-CoV) infection. RSA59, an isogenic spike gene recombinant strain of MHV-A59, is a well-established prototypic group 2- murine β coronavirus (M-CoV). It infects the liver and CNS and is a CoV model used to investigate the mechanisms of neuroinflammation, viral pathogenesis, host stress responses, as well as anti-viral immune responses [34,35]. Intracranial infection of M-CoV causes a biphasic disease in C57Bl/6 mice, an acute stage characterized by meningoencephalomyelitis (days 5/6 post-infection) and chronic stage demyelination and axonal loss [34]. Our previous studies have also shown that M-CoV infection induces optic neuritis by promoting infiltration of peripheral inflammatory cells of mixed populations during the acute stage [36]. Optic nerves from M-CoV infected mice develop demyelination and axonal loss during the chronic stage, accompanied by a significant loss of retinal ganglionic cells in the optic nerve [37]. Further, M-CoV leads to mitochondrial ROS accumulation in the infected optic nerves during peak inflammation and 30 days post-infection. ROS accumulation in infected mice correlates with decreased levels of proteins associated with mitochondrial function and biogenesis [37].
The results show that M-CoV infection in the mouse brain results in profuse viral replication and increased viral transcript denoted by nucleocapsid gene, activation of astrocytes and microglia during acute infection, up-regulation of inflammatory cytokines, and chemokines, and the up-regulation of antioxidant and UPR genes. As the virus titer starts to decline and inflammation is resolved at day 10 p.i., a significant progressive increase was observed in the ROS sensing molecule DJ-1 along with the up-regulation of its downstream effectors, selected heat shock responsive genes, and UPR marker XBP1. We have also attempted an in silico approach to show the interaction of DJ-1 promoter with XBP1. The result of such analysis will throw some light on the mechanism of DJ-1 up-regulation and the dynamic communication between ROS, cellular stress, and UPR during M-CoV infection.
2. Materials and methods
2.1. Viruses
RSA59 viruses were used in the study. MHV-A59 is a dual tropic murine β Coronavirus that infects the liver and the CNS. Intracranial inoculation of MHV-A59 in C57BL/6 mice is used as an experimental animal model to understand the mechanisms of neuro degeneration and demyelination as observed in the human neurological disorder Multiple Sclerosis (MS). RSA59 is an isogenic recombinant, demyelinating strain of MHV-A59 from our previous studies where the spike gene is introduced by targeted RNA recombination. Spike gene encodes an envelope glycoprotein that mediates virus-host attachment and various biological properties of MHV, including virus-cell and cell-cell fusion, viral spread, anti-viral host responses, and pathogenicity. Additionally, in RSA59, gene 4 is replaced by enhanced green fluorescence protein (EGFP) described elsewhere [38], [39], [40]. Throughout the paper, RSA59 is referred as M-CoV.
2.2. Inoculation of mice
The animals and all experimental procedures were reviewed and approved by the Indian Institute of Science Education and Research-Kolkata (IISERK). Animal protocols were followed according to the Committee's guidelines for the purpose of control and supervision of experiments on animals (CPCSEA), India.
Four-week-old, M-CoV-seronegative C57BL/6 male mice were intra-cranially inoculated with 50% of the LD50 dose of M-CoV (20000 PFU) [38,39]. Mice were monitored daily for clinical disease symptoms. Parallel controls were inoculated with PBS-BSA (Mock).
2.3. Tissue harvesting
Mice were sacrificed on day 5 post-infection (peak of inflammation) for histo pathological analyses. For RNA and protein studies, mice were sacrificed on days 3, 5 10 post-infection. Mice were transcardially perfused with DEPC (diethyl polycarbonate) treated PBS, and liver, brain, and spinal cord tissues were harvested for experimentation. For immunofluorescence analysis, tissues were post fixed in 4% PFA for 48- 72 hours, processed, and embedded in paraffin [38,39].
2.4. Immunofluorescence on paraffin sections
5 µm thick serial sections from mock and RSA59 infected brains were subjected to immunofluorescence analysis. Briefly, tissue sections were deparaffinized and dehydrated and subjected to antigen unmasking (Vector Laboratories, Burlingame, California, USA). After blocking in blocking solution (1XPBS+ 2.5% Goat Serum + 0.5% TritonX-100) for 1 h at 37ᴼC, the sections were incubated overnight at 4ᴼC with primary antibody. The primary antibody was washed in 1X PBS, and subsequently, the sections were labeled with a secondary antibody for 90 min at 37ᴼC. All incubations were carried out in a humidified chamber. After PBS washing, the sections were mounted with mounting medium containing 4’6-diamidino-2-phenylindole (DAPI; Vectashield, Vector Laboratories, Burlingame, California, USA) and visualized under Nikon Eclipse TS2 Epifluorescence microscope and analyzed in Nikon NIS-Elements software (Nikon Corporation, Tokyo, Japan).
The dilutions of primary antibodies used were- 1:500 dilution of anti-Iba1 (FUJIFILM WAKO Chemicals, Richmond, Virginia, USA), 1:500 dilution of anti-GFAP (Sigma Aldrich, Saint Louis, Missouri, USA), and 1:40 dilution of a monoclonal antibody directed against the nucleocapsid protein (N) of MHV-JHM (monoclonal antibody clone 1-16-1 provided by Julian Leibowitz, Texas A&M University). GFAP is astrocyte specific marker and Iba1 is microglia specific marker. The dilutions of secondary antibodies (fluorescent) used are 1:250 dilution of Alexa Fluor 488 conjugated goat anti-mouse antibody (Thermo Scientific, Waltham, Massachusetts, USA) (for detecting Viral N protein) and 1:250 dilution of Alexa Fluor 568 conjugated donkey anti-rabbit antibody (Thermo Scientific, Waltham, Massachusetts, USA) (for detecting GFAP/Iba1 protein).
2.5. RNA extraction, cDNA preparation, and qRT- PCR analysis
RNA isolation was performed from mock and M-CoV infected brain tissues using standard TRIzol protocol (Ambion ®, Austin, Texas, USA) as per the manufacturer's instructions. The concentration of RNA was measured using Nanodrop-2000 (Thermo Scientific, Waltham, Massachusetts, USA). 1 µg of RNA was used for cDNA preparation using the High-Capacity Reverse Transcription Kit protocol (Applied Bio systems, Inc. Foster City, California, USA). qRT- PCR was performed using DyNAmo ColourFlash Probe PCR Kit (Thermo Scientific, Waltham, USA) in Applied Biosystem 7900HT Fast Real-Time PCR System (Applied Biosystem, Foster City, USA) under the following conditions: initial denaturation at 950C for 7 min, 40 cycles of 950C for 10 s, 60°C for 30 s, melting curve analysis at 60°C for 30 s. Reactions were performed in quadruplets. The sequence of primers used is given in Table 1 . Del Ct values were used to analyze changes in the expression pattern of respective genes.
Table 1.
Primer sequence
| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| Real-Time PCR Primers | |
| DJ-1 Forward | AACACACCCACTGGCTAAGG |
| DJ-1 Reverse | CTCCACAATGGCTAGTGCAA |
| Nrf2 Forward | GATCCGAGATATACGCAGGAGAGGTAAGA |
| Nrf2 Reverse | GCTCGACAATGTTCTCCAGCTTCC |
| HMOX-1 Forward | GCCCCACCAAGTTCAAACAGCTCTA |
| HMOX-1 Reverse | CTCTGTCAGCATCACCTGCAGC |
| Xbp1(s) Forward | GAGTCCGCAGCAGGTG |
| Xbp1(s) Reverse | GTGTCAGAGTCCATGGGA |
| Nqo1 Forward | GCGTTTCTGTGGCTTCCAGGTCTT |
| Nqo1 Reverse | ATAGAGTGGGGTCTCCTCCCAGAC |
| Txnrd1 Forward | AGTCACATCGGCTCGCTGAACT |
| Txnrd1 Reverse | CGATGAGGAACCGCTCTGCTGAA |
| HSF1 Forward | GACTCCAAGCTCCTGGCCATGAA |
| HSF1 Reverse | CAGAGGGATCTTTCTCTTCACCCCC |
| Hsp70 Forward | AGGTGAACTACAAGGGCGAGAGC |
| Hsp70 Reverse | TGCCGCTGAGAGTCGTTGAAGTAG |
| IL6 Forward | AGTTGCCTTCTTGGGACTGA |
| IL6 Reverse | TCCACGATTTCCCAGAGAAC |
| IFNα Forward | CTTCCACAGGATCACTGTGTACCT |
| IFNα Reverse | TTCTGCTCTGACCACCTCCC |
| IFNβ Forward | CTGGCTTCCATCATGAACAA |
| IFNβ Reverse | AGAGGGCTGTGGTGGAGAA |
| IFNγ Forward | ATCTGGAGGAACTGGCAAAA |
| IFNγ Reverse | TTCAAGACTTCAAAGAGTCTGAGG |
| CSF2 Forward | GCATGTAGAGGCCATCAAAGA |
| CSF2 Reverse | CGGGTCTGCACACATGTTA |
| CCL2 Forward | CATCCACGTGTTGGCTCA |
| CCL2 Reverse | GATCATCTTGCTGGTGAATGAGT |
| CCL5 Forward | CCA ATC TTG CAG TCG TGT TTG T |
| CCL5 Reverse | CCA ATC TTG CAG TCG TGT TTG T |
| CXCL9 Forward | CTTTTCCTCTTGGGCATCAT |
| CXCL9 Reverse | GCATCGTGCATTCCTTATCA |
| CXCL10 Forward | GACGGTCCGCTGCAACTG |
| CXCL10 Reverse | CTTCCCTATGGCCCTCATTCT |
| GAPDH Forward | GCCCCTTCTGCCGATGC |
| GAPDH Reverse | CTTTCCAGAGGGGCCATCC |
| N gene Forward | AGGATAGAAGTCTGTTGGCTCA |
| N gene Reverse | GAGAGAAGTTAGCAAGGTCCTACG |
| 18s RNA Forward | ATTGACGGAAGGGCACCACCAG |
| 18s RNA Reverse | CAAATCGCTCCACCAACTAAGAACG |
2.6. 3D Model Generation, Docking and Molecular Dynamic Simulation
To look into the role of XBP1 in the regulation of DJ-1 expression, in silico analysis of interaction between XBP1 protein and the DJ-1 promoter is attempted (supplementary section). XBP1 protein structure was generated by homology modeling using I-TASSER software [41]. XBP1 sequence with sequence accession number 035426 (protein database from NCBI website (https://www.ncbi.nlm.nih.gov/protein/O35426.2)) was used for the modeling with no additional restraints or assigned templates. Along with that, no particular template was excluded from the I-TASSER template library, and no secondary structure was assigned for specific residues. Following this, molecular dynamics simulation was performed using GROMACS 5.1 software [42]. In summary, the protein model was solvated in the SPC water model inside a cube with a 1 nm distance from the edges. Charge neutralization was performed using eight Chloride ions and energy minimization was performed using the Steepest Descent Method. The obtained structure was subjected to 2 ns of NVT (number, volume, temperature) equilibration using leapfrog integrator and modified Berendsen thermostat, followed by 2 ns of NPT (number, pressure, temperature) equilibration with the addition of Parrinello-Rahman barostat for pressure coupling. After this, Molecular dynamics simulation was performed using the OPLS-AA force field under periodic boundary conditions for 50 ns extracting frames every 2.5 ns, and the surface charge distribution of XBP1 protein was calculated using APBS plugin for PyMOL [43].
The DJ-1 promoter region was chosen from -456 to -427 bp (DJ-1 promoter sequence was taken from gene id: 57320 - nucleotide database from NCBI website (https://www.ncbi.nlm.nih.gov/gene/?term=57320)), and the B-DNA structure was generated by using 3D DART software [44], with no specific nucleic acid modeling parameters. Initial docking of the two obtained structures was done using NP Dock Server according to previously published methodology [45], and using default values (Number of decoys generated with GRAMM= 20000; Number residues of interfaces in contact= 1; Number of the best-scored model used for clustering= 100; RMSD cut-off for clustering= 5 Å). The docked structures were subjected to refinement with Monte Carlo by considering the default parameters (Number of steps of simulation= 1000; Temperature of the first step of simulation= 15000 K; Temperature of the last step of simulation= 295 K). Every accepted step was saved. The best-refined structure was selected, and the interacting structures were analyzed, and images were obtained using PyMOL (http://www.pymol.org).
2.7. Statistical Analysis
A scattered plot was chosen for graphical representation of mean ± S.D. (Standard Deviation). One-way ANOVA and uncorrected Fisher's LSD Multiple Comparison Test was performed on the normalized value to evaluate the data's statistical significance containing three or more variables. A student's unpaired t-test was performed to analyze the significance of the differences observed in the comparisons between the two groups. Outliers were identified and excluded by the ROUT test. Fold changes were calculated for all M-CoV infected set with respect to the mock infected set. All statistical analyses were done using GraphPad Prism 6 (La Jolla, CA) for all performed experiments; the statistical significance parameter was p-value < 0.05.
3. Declarations
3.1. Ethics Statement
All experimental procedures and animal care and use were strictly regulated and reviewed following good animal ethics approved by the Institutional Animal Care and Use Committee at the Indian Institute of Science Education, and Research Kolkata (AUP No. IISERK/IAEC/AP/2017/15) Experiments were performed following the guidelines of the CPCSEA, India.
4. Results
Upon M-CoV infection, 4-week-old C57BL/6 mice developed acute stage hepatitis illustrated by necrotic and non-necrotic lesions, inflammation in the meninges (meningitis) and encephalitis characterized by perivascular cuffing and microglial nodule as described previously [38,39]. Neuroinflammation induced by M-CoV, RSA59, provides a stage for studying the nexus between oxidative stress and the UPR pathway at different days post-infection.
4.1. M-CoV infection activates microglia and astrocytes at day 5 post-infection
On day 5 p.i., M-CoV infected brain tissue sections were subjected to immuno labeling by using anti-viral nucleocapsid, anti-Iba1, and anti-GFAP antibodies for examining in situ presence of viral particles, microglia and astrocyte activation, respectively. Astrocyte activation (marked by GFAP staining) was observed upon M-CoV infection (marked by Viral N staining) in the basal forebrain (Supplementary figure 1B) and hypothalamus (Supplementary figure 1F) regions of the brain. The mock infected counterparts did not show any astrocyte activation in basal forebrain (Supplementary figure 1A) and hypothalamus (Supplementary figure 1E). Microglial activation (marked by Iba1 staining) was observed upon M-CoV infection (marked by Viral N staining) in the basal forebrain (Supplementary figure 1D) and hypothalamus (Supplementary figure 1H) regions of the brain. The mock infected counterparts did not show any astrocyte activation in basal forebrain (Supplementary figure 1C) and hypothalamus (Supplementary figure 1G). Similar glial cell activation was also observed upon M-CoV infection in cortex, caudate putamen, and midbrain region of the brain (data not shown).
This observation suggests that the viral spread across the brain parenchyma leads to the activation of microglia and astrocytes. RT-qPCR examination revealed the up-regulation of inflammatory cytokines IL6, IL1β, IFNα, IFNβ, IFNγ, and CSF2 (Supplementary figure 1I) as well as chemokines CCl2, CCl5, CXCL9, and CXCL10 (Supplementary figure 1J) upon M-CoV infection. Viral infection and CNS resident glial cell activation are the two critical checkpoints for neuro-inflammation. These glial cells upon inflammation produce ROS and have a vast repertoire of innate immune markers, including pattern recognition receptors (PRRs), which can signal via UPR (e.g., IRE1α and XBP1) [46]. Given the activation of glial cells, we sought to investigate the role of glial cell activation in XBP1 mediated cellular stress responses.
4.2. M-CoV infection induces cellular stress responses in vivo
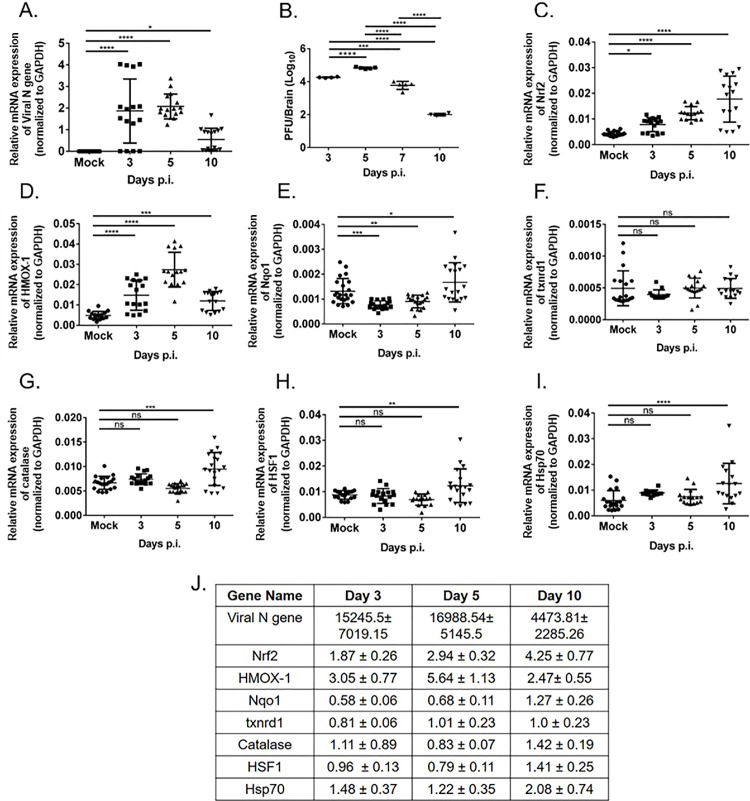
Viral N gene expression is observed on days 3, 5, and 10 p.i. (Fig. 1 A). Viral titer, as assessed by routine plaque assay, increased at the onset of inflammation (day 3 p.i) and reached its peak at day 5 p.i. The virus titre starts to decline by day 7 p.i. as reported earlier (Fig. 1B) [38,47]. During the acute phase, i.e., days 3, 5, and 10 p.i. until the viral titer drops down below detection limit, RT-qPCR analysis was performed for Nrf2, HMOX-1, Catalase, Nqo1, txnrd1, HSF1, Hsp70. Genes showed differential regulation at different times post-infection. Results revealed that at early time points during the peak of inflammation (days 3 and 5 p.i.), antioxidant genes Nrf2, HMOX-1, Nqo1, and catalase showed most significant up-regulation when infectious viral particles were below the detection limit and acute neuro-inflammation is resolved (Fig. 1C, D, E, G). However, antioxidant gene txnrd1 did not show any change upon M-CoV infection (Fig. 1F). The heat shock responsive genes (HSF1 and Hsp70) showed significant upregulation, (Fig. 1H, I) [1]. Thus, the most striking increase in the cellular stress pathway genes is detected when infection and acute inflammation are resolved. This also marks the bridging between innate and adaptive immunity. The fold change in mRNA expressions is shown in Fig. 1J.
Fig. 1.
M-CoV infection resulted in the differential regulation of host stress response genes in vivo. Quantitative real-time PCR analysis of the brain tissues of M-CoV infected mice at day 3, 5, and 10 p.i. was used to examine the mRNA expression level viral N-gene (A), Nrf2 (C), HMOX-1 (D), Nqo1 (E), txnrd1 (F), Catalase(G), HSF1 (H), Hsp70 (I) compared to mock-infected. Viral titre estimation was performed on brain tissues of days 3, 5, 7, and 10 post infected mice (B). The fold change of all the genes expression at different day points p.i. with respect to the mock-infected set were tabulated in J. All the experiments were done with N = 4. Solid lines represented the differences in mean values. Asterix (*) indicated values that were statistically significant by One-Way ANOVA analysis and uncorrected Fisher's LSD Multiple Comparison Test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
4.3. M-CoV infection induces cellular stress responses in primary microglia and astrocytes
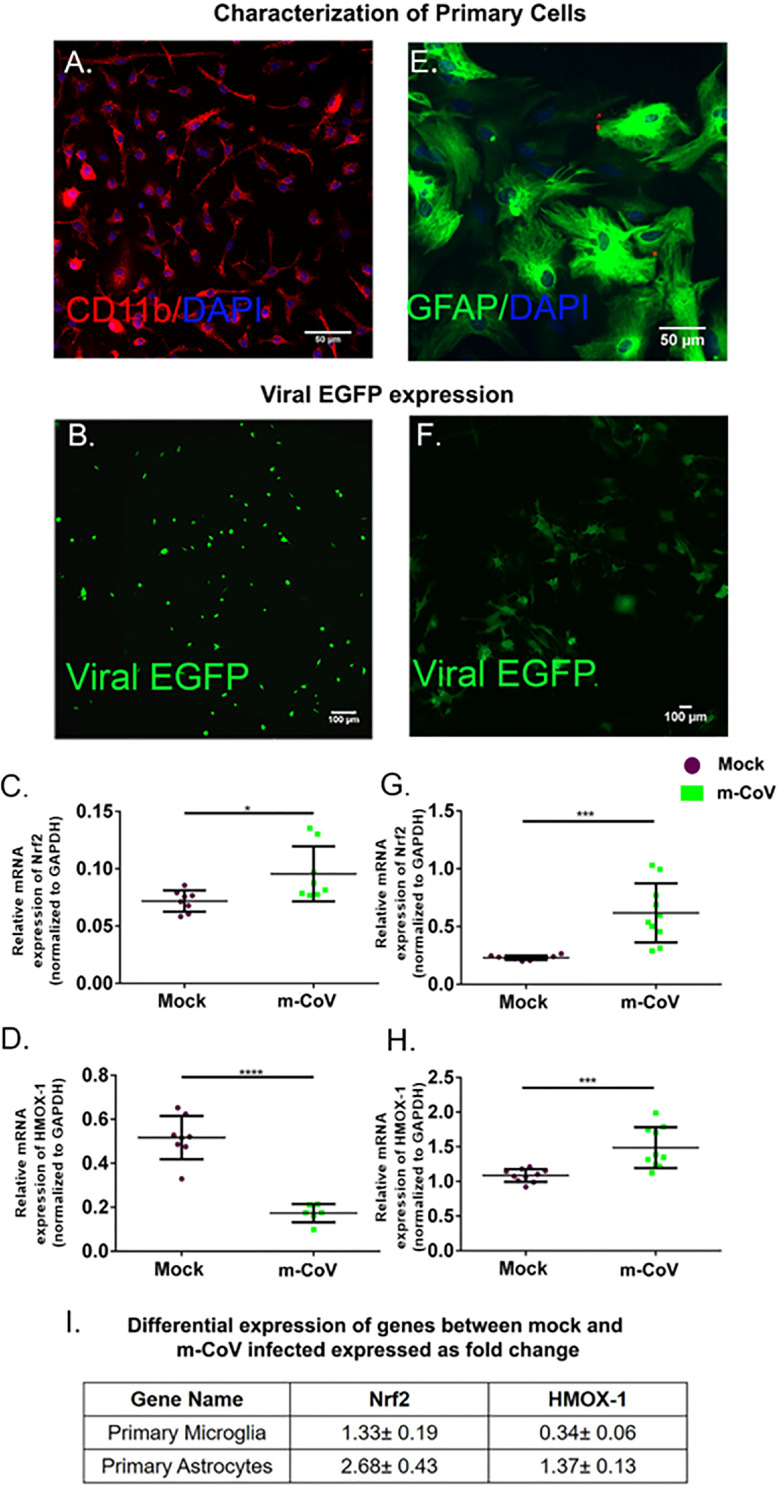
To understand the glial cell-specific cellular response in a reductionist approach, we investigated antioxidant response genes' expression in mixed glial cultures enriched in microglia or astrocytes. Both primary microglia and astrocytes were characterized by the expression of CD11b or GFAP respectively (Fig. 2 A, E) and infected with M-CoV at 2 MOI (Fig. 2B, F) as discussed in the materials and methods section. At 24 h p.i., RT-qPCR analysis was performed for antioxidant response pathway markers Nrf2 and HMOX-1, which showed significant up-regulation in infected mouse brain. Results revealed a significant up-regulation of Nrf2 in both primary microglia and astrocytes (Fig. 2C, G). However, HMOX-1 was up-regulated in primary astrocytes but down-regulated in microglia (Fig. 2D, H). The fold change in mRNA expression is shown in Fig. 2I.
Fig. 2.
M-CoV infection resulted in differential regulation of oxidative stress-related genes in both primary astrocytes and microglia culture. Primary astrocytes and primary microglia were characterized by CD11b (for primary microglia) (A) and GFAP (for primary astrocytes) (E) from neonatal mice brain. M-CoV infection was confirmed in primary microglia (B) and primary astrocytes (F) by EGFP expression. Differential expression was plotted for Nrf2 (C), HMOX-1 (D) by Quantitative real-time PCR analysis upon M-CoV infection in primary microglia. Differential expression was plotted for Nrf2 (G), HMOX-1 (H) by Quantitative real-time PCR analysis upon M-CoV infection in primary astrocytes. The fold change in expression of both the genes with respect to the mock-infected set was tabulated in I. All the experiments were conducted with N = 4-5. Solid lines represented the means of mRNA expression. Asterix (*) indicated values that were statistically significant by student's unpaired T-test analysis for all experiments. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
4.4. M-CoV infection alters the expression of DJ-1 and XBP1 in vivo and in primary cells
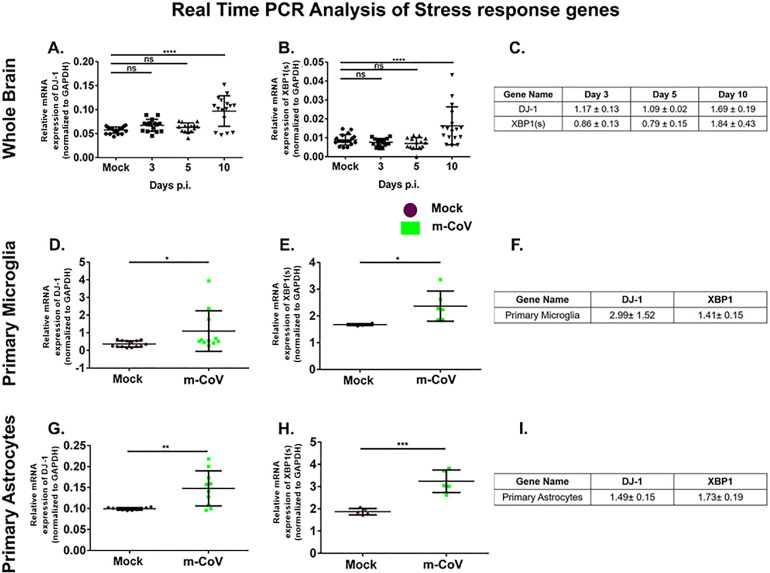
The regulatory role of ROS sensing molecule DJ-1 and UPR pathway marker, XBP1 was assessed both in the brain and individually in microglia and astrocytes by examining the mRNA expression. Such roles will be important in understanding the possible crosstalk between the oxidative stress and UPR pathways in the process of microglial activation. Results showed significant up-regulation of DJ-1 and XBP1 mRNA with days of post infection until day 10 as observed in mice brains (Fig. 3 A, B) and fold change in mRNA expression is shown in Fig. 3C. Likewise, both primary microglia (Fig. 3D, E) and primary astrocytes (Fig. 3G, H) displayed a significant induction of DJ-1 and XBP1 mRNA upon M-CoV infection at 24 h post-infection. The fold change in expressions of mRNA is shown in Fig. 3F, I.
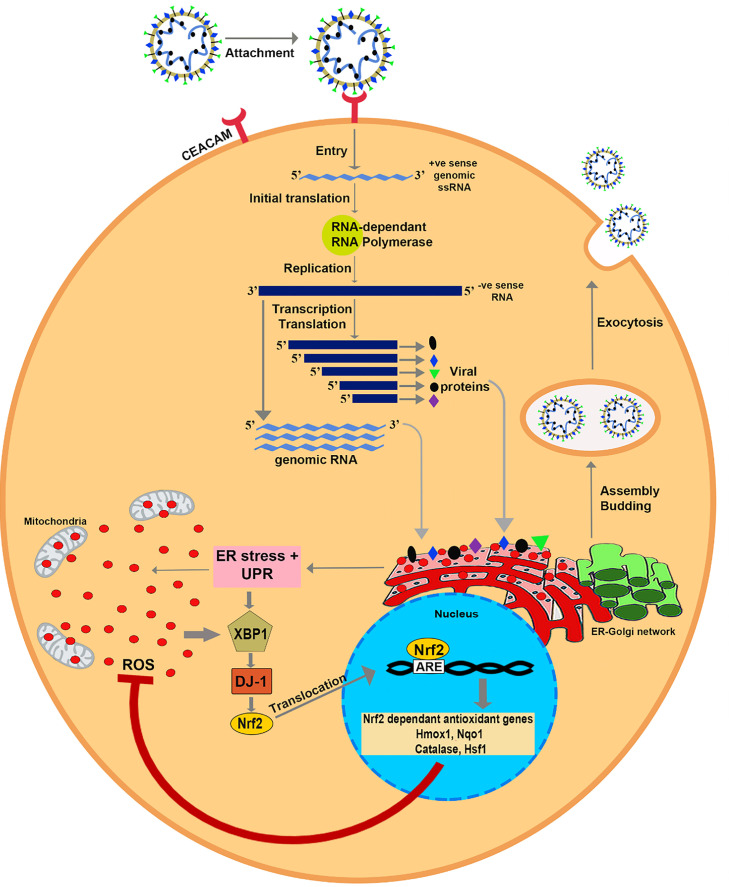
Fig. 4.
Schematic showing the interaction between E.R. stress, UPR, and antioxidant system in response to M-CoV infection. Graphical abstract summarizes our new findings that DJ-1 under the influence of XBP-1 can regulate the reactive oxygen species (ROS) imbalance in virus-infected CNS cells. The assembly of the individual structural proteins into a virion induces E.R. stress and UPR. E.R. stress, UPR, and mitochondria in virus-infected cells tend to generate ROS in large quantities. On the contrary, the host cell system generates XBP-1 in response to E.R. stress and UPR. XBP-1 binds to the DJ-1 promoter and increases the expression of DJ-1. DJ-1, in turn, up regulates Nrf2 and facilitates its translocation to the nucleus. The transcription factor Nrf2 binds to the antioxidant responsive element (ARE) in the promoter region and induces the expression of Nrf2 dependent antioxidant genes HMOX-1, Nqo1, Catalase, and HSF1. These antioxidant genes play a significant role in quenching the ROS and protect the cell from ROS mediated cellular damage and death.
Fig. 3.
M-CoV infection resulted in up-regulation of DJ-1 and XBP1 both in vivo and in vitro. Quantitative real-time PCR analysis was used to examine the mRNA expression level of DJ-1, and XBP1 in the brain tissues of M-CoV infected mice at days 3, 5, and 10 p.i. A and B respectively; in primary microglia D and E respectively and primary astrocytes G and H respectively. The fold change of all the genes expression at different day points p.i. with respect to the mock-infected set were tabulated in C, F, and I. All the experiments were done with N=4. Solid lines represented the differences in mean values. Asterix (*) indicated values that were statistically significant by One-Way ANOVA analysis and uncorrected Fisher's LSD Multiple Comparison Test (*p < 0.05, ***p < 0.001, ****p < 0.0001).
Our study from the virus-host-response implies that DJ-1 and XBP1 are significantly up-regulated both in vivo in brain tissues and in vitro in mixed glial cultures enriched in either microglia or astrocytes. Such changes can indicate a possible crosstalk between the UPR and oxidative stress pathways taking place during M-CoV infection in CNS.
4.5. Murine coronavirus infection-induced DJ-1- XBP1 interactome complex as a regulator of oxidative stress
Our results have shown that M-CoV infection causes an up-regulation of the UPR pathway associated XBP1 and antioxidant pathway DJ-1. A previous report [48] showed that XBP1 activation was involved in the up-regulation of DJ-1 mRNA by direct interaction with the DJ-1 promoter to prevent cell death. Therefore, we used the in silico approach to identify DJ-1 and XBP1 as interaction partners in modulating host stress response to M-CoV.
For in silico analysis, we performed molecular dynamics simulation on the structure of XBP1 protein generated by iterative threading-based modeling using I-TASSER software. Molecular dynamics simulation was performed using GROMACS 5.1 for 50 ns, and frames were extracted every 2.5 ns. The protein structure was stable, with an average backbone root-mean-square-deviation (RMSD) value of 0.4097 ± 0.02982 Å, and showed a highly positively charged groove (denoted by blue region). Encouraged by the highly positive charged site in the XBP1 protein capable of DNA binding, we generated a B-DNA model of the DJ-1 promoter region. Following this, docking was performed using NP Dock Server (http://genesilico.pl/NPDock). Since NP Dock performs rigid docking, we needed to consider the DNA binding grove's intrinsic flexibility. This was achieved by using XBP1 protein structures extracted every 2.5 ns during the 50 ns M.D. simulation. Docking using the NP Dock server gave several docked models. Still, the best-refined structure, as provided by the software, showed stable binding between the DJ-1 promoter and the positively charged groove of XBP1 protein at the same site. The fit of the structure of the DJ-1 promoter in the XBP1 protein groove was visually analyzed using PyMOL (Supplementary Figure 2).
Our recent findings on the up-regulation of DJ-1 and XBP1 both in vivo and in vitro in combination with in silico study predicts that XBP1 may promote DJ-1 expression and provide cyto-protective function.
5. Discussion
M-CoV induced glial cell activation heightens the secretion of a complex array of chemokines, cytokines, and production of oxygen radicals directing the host anti-viral response [49], [50], [51], [52], [53]. In turn, activated glial cells fine-tune E.R. stress and oxidative stress responses to preserve the host tissues' integrity [18,54,55]. While activated glial cells work towards promoting virus clearance, prolonged neuronal damage by the increased virus assembly can result in the loss of redox balance, which further induces a shift towards glial cells mediated pro-inflammatory host responses via NF-kB signaling [56,57]. While there is ample evidence indicating the involvement of microglia and astrocytes in the induction of host immune responses, oxidative stress, E.R. stress stimulation and UPR upon M-CoV infection, and their interdependence is underexplored.
This study summarizes the nexus between oxidative stress, E.R. stress, and the UPR pathway mediated by DJ-1 and XBP1 upon M-CoV induced activation of CNS resident glial cells. We observed: i) M-CoV infection activates astrocytes and microglia during the acute stage of infection and significantly increases the level of pro-inflammatory cytokines and chemokines; ii) antioxidant genes Nrf2, HMOX-1, and Nqo1 show a significant increase in the brains of M-CoV infected mice; iii). M-CoV infection-induced up-regulation of DJ-1 possibly under its transcriptional regulator XBP1 along with a marked increase in heat shock response genes HSF1 and Hsp70.
Our observation demonstrating the alteration of DJ-1, an oxidant sensing molecule, is the first instance in M-CoV pathogenesis. This finding is corroborated by a considerable amount of research conducted in familial Parkinson's [27,29,58], where DJ-1 is shown to play a prominent protective role by balancing the cellular redox potential [59]. Likewise, as expected, DJ-1 up-regulation is associated with the increase in its downstream signaling molecule Nrf2, a master regulator of oxidative stress and detoxification processes [29]. Moreover, our findings reveal the increased expression of cyto-protective and detoxifying genes, including Nqo1, Catalase, and HMOX-1, upon M-CoV infection, which is induced by Nrf2 and is in line with previous findings [30].
Many studies in M-CoV infection have acknowledged the functional importance of XBP1, which is a transcription regulator of UPR [60] known to be induced in response to the accumulation of unfolded proteins and other stress factors by IRE1 mediated splicing [33]. IRE1-XBP1 is the most conserved UPR signaling pathway known to regulate oxidative stress[33]. In our studies, we observed an up-regulation of XBP1 along with DJ-1 on day 10 p.i. However, the demonstration of possible crosstalk between the oxidative stress response and UPR in M-CoV infection in mounting host protection is the highlight of our study. Previous research had identified consensus sites on XBP1 for DJ-1 binding in humans and mice [48]. However, we have taken an in silico approach to gain insight into the charge distribution of the interaction site of the XBP1 protein with the DJ-1 promoter (Supplementary Figure. 2). Correlation between UPR and oxidative stress pathways in the context of M-CoV MHV infection is an important field of research to understand the changes taking place in neuroglial pathology from the virus-host interaction standpoint. Previously, we have shown that MHV-A59 induced oxidative stress in the optic nerve of infected mice [37]. Treatment of mice with SRTAW04, a compound that activates SIRT1, an NAD-dependent deacetylase that sense cellular stress, significantly reduced neuronal loss during optic nerve inflammation by attenuating accumulation of mitochondrial ROS while promoting the expression of mitochondrial enzymes associated with ROS sensing. SIRT1 activating compound significantly up-regulated the levels of oxygen sensing enzymes such as succinate dehydrogenase (SDH) and superoxide dismutase 2 (SOD2) in MHV-A59 infected mice. The peroxisome proliferator-activated receptor (PPAR) co-activator 1-α (PGC1-α), a regulator of mitochondrial biogenesis, gets decreased during MHV-A59 infection, and treatment with SRTAW04 significantly increased the protein levels. Results suggest that one of the mechanisms of viral-induced neuronal loss involves the generation of oxidative stress, which can be attenuated by promoting mitochondrial function. Figure 6 illustrates the putative interaction between E.R. stress and UPR and host stress responses where DJ-1, Nrf2, and XBP-1 axis can be instrumental in understanding the communication between the various stress responses and serve as a platform for further investigation.
A line of studies in CoV infections has demonstrated the detrimental effect of redox imbalance on membrane-bound organelles such as mitochondria and E.R. through lipid peroxidation [61]. Thus, it is apparent that both E.R. and oxidative stress pathways collaborate towards promoting protective host responses. The current study enlightens that the DJ-1-XBP1 axis is crucial in governing M-CoV induced cellular stress responses both in vivo in brain and in vitro in primary astrocytes and microglia cultures. DJ-1 via Nrf2, the master transcription regulator of antioxidant genes, can maintain the redox balance.
CoVs are a group of medically and economically important viruses that have raised considerable interest from the entire scientific community. M-CoV is the best prototypic viral model employed vastly for studying the deeper aspects of dys-regulated host stress responses in CoV infections. It is of utmost importance that we take insights from the current findings to validate and further investigate the antioxidant system's scope, especially DJ-1, as a potential target in CoV infections.
6. Conclusion
In our work, we have hypothesized that M-CoV infection in the CNS of mice results in glial cell activation in different brain regions and cellular stress response factors are regulated in the process. we had also hypothesized that the stress response factors are interlinked in response to viral induced cellular stress. We have observed that M-CoV infection results in activation of microglia and astrocytes. Glial cell activation is linked with oxidative stress response, and we have observed an up-regulation of cellular antioxidant DJ-1-Nrf2 pathway in astrocytes and microglia, which in turn regulates the heat shock response pathway. This up-regulation of DJ-1 upon M-CoV infection is predicted to be under the effect of its transcriptional regulator XBP1, which is found to be activated upon viral infection. Therefore, the outcome of our study can be explored to identify newer interplays between different stress response pathways underlying the mechanism of glial cell activation upon mouse coronavirus infection.
Author Contribution
SK and JDS designed and planned all the experiments. S.K., GRA, VAR, R.K., S.S., and L.S. performed the experiments. S.K., VAR, and D.P. performed the in-silico experiment. S.K., F.S., S.S., and JDS performed data analysis and data interpretation. F.S., S.S., and M.K. blindly analyzed the data and helped S.K. and JDS write the manuscript. JDS critically reviewed the manuscript and supervised this work. All authors reviewed the manuscript.
Declaration of Competing Interests
The authors declare that they have no competing interests.
Declaration of Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the Department of Biotechnology (DBT Grant No.: BT/PR20922/MED/122/37/2016, BT/PR4530/MED/30/715/2012), and Indian Institute of Science Education and Research Kolkata (IISER Kolkata). The authors would like to thank the IISER Kolkata Animal facility for providing all the necessary support. The authors also thank the members of the LRI Imaging Core, Cleveland Clinic, for the histo-pathological and cytokine and chemokine data. S.K. and M.K. were supported by University Grant Commission (UGC). F.S. was supported by Council for Scientific and Industrial Research (CSIR). The DST-INSPIRE fellowship supported GRA, VAR, and R.K. under the banner of IISER Kolkata. S.S. and L.S. were supported by the Ministry of Human Resource Development (MHRD) fellowship under the banner of IISER Kolkata.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dscb.2021.100021.
Appendix. Supplementary materials
References
- 1.Bechill J., et al. Coronavirus Infection Modulates the Unfolded Protein Response and Mediates Sustained Translational Repression. J. Virol. 2008;82(9):4492. doi: 10.1128/JVI.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L., Wang A. Virus-induced ER stress and the unfolded protein response. Front. Plant Sci. 2012;3 doi: 10.3389/fpls.2012.00293. 293-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denison M.R., et al. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the pre-fusion conformation. Science. 2020;367(6483):1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walls A.C., et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292.e6.. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehr A.R., Perlman S. In: Coronaviruses: Methods and Protocols. Maier H.J., Bickerton E., Britton P., editors. Springer New York; New York, NY: 2015. Coronaviruses: An Overview of Their Replication and Pathogenesis; pp. 1–23. Editors. [Google Scholar]
- 9.Liao Y., et al. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 2013;87(14):8124–8134. doi: 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang B.S., et al. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J. Virol. 2005;79(10):6180–6193. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Versteeg G.A., et al. The coronavirus spike protein induces endoplasmic reticulum stress and up-regulation of intracellular chemokine mRNA concentrations. J. Virol. 2007;81(20):10981–10990. doi: 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fung T.S., Huang M., Liu D.X. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus-host interactions. Virus Res. 2014;194:110–123. doi: 10.1016/j.virusres.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldsmith C.S., et al. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10(2):320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snijder E.J., et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fung T.S., Liao Y., Liu D.X. Regulation of stress responses and translational control by coronavirus. Viruses. 2016;8(7) doi: 10.3390/v8070184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao S.S., Kaufman R.J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox. Signal. 2014;21(3):396–413. doi: 10.1089/ars.2014.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozgur R., et al. Interplay between the unfolded protein response and reactive oxygen species: a dynamic duo. J. Exp. Bot. 2018;69(14):3333–3345. doi: 10.1093/jxb/ery040. [DOI] [PubMed] [Google Scholar]
- 19.Wen C.-L., et al. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol. Med. Microbiol. 2006;46:375–380. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan C.P., et al. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2006;80(18):9279–9287. doi: 10.1128/JVI.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay M.Z., et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olagnier D., et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020;11(1):4938. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zinovkin R.A., Grebenchikov O.A. Transcription Factor Nrf2 as a potential therapeutic target for prevention of cytokine storm in COVID-19 patients. Biochemistry (Moscow) 2020;85(7):833–837. doi: 10.1134/S0006297920070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagakubo D., et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 1997;231(2):509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 25.van der Brug M.P., et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc. Natl. Acad. Sci. U S A, 2008;105(29):10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baulac S., et al. Increased DJ-1 expression under oxidative stress and in Alzheimer's disease brains. Mol. Neurodegener. 2009;4:12. doi: 10.1186/1750-1326-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lev N., et al. Role of DJ-1 in Parkinson's disease. J. Mol. Neurosci. 2006;29(3):215–225. doi: 10.1385/jmn:29:3:215. [DOI] [PubMed] [Google Scholar]
- 28.van Horssen J., et al. Nrf2 and DJ1 are consistently up-regulated in inflammatory multiple sclerosis lesions. Free Radic. Biol. Med. 2010;49(8):1283–1289. doi: 10.1016/j.freeradbiomed.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 29.Clements C.M., et al. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. U S A, 2006;103(41):15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho H.-Y., et al. Gene expression profiling of NRF2-mediated protection against oxidative injury. Free Radical Biol. Med. 2005;38(3):325–343. doi: 10.1016/j.freeradbiomed.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Paul S., et al. NRF2 transcriptionally activates the heat shock factor 1 promoter under oxidative stress and affects survival and migration potential of MCF7 cells. J. Biol. Chem. 2018;293(50):19303–19316. doi: 10.1074/jbc.RA118.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J., et al. DJ-1 modulates the unfolded protein response and cell death via upregulation of ATF4 following ER stress. Cell Death Dis. 2019;10(2):135. doi: 10.1038/s41419-019-1354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Zhang C., Wang A. Divergence and Conservation of the Major UPR Branch IRE1-bZIP Signaling Pathway across. Eukaryotes. Sci. Rep. 2016;6:27362. doi: 10.1038/srep27362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das Sarma J. A mechanism of virus-induced demyelination. Interdiscip. Perspect. Infect. Dis. 2010;2010 doi: 10.1155/2010/109239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das Sarma J. Microglia-mediated neuroinflammation is an amplifier of virus-induced neuropathology. J. Neurovirol. 2014;20(2):122–136. doi: 10.1007/s13365-013-0188-4. [DOI] [PubMed] [Google Scholar]
- 36.Shindler K.S., et al. Experimental Optic Neuritis Induced by a Demyelinating Strain of Mouse Hepatitis Virus. J. Virol. 2008;82(17):8882. doi: 10.1128/JVI.00920-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan R.S., et al. SIRT1 Activating compounds reduce oxidative stress mediated neuronal loss in viral induced CNS demyelinating disease. Acta Neuropathol.Commun. 2014;2(1):3. doi: 10.1186/2051-5960-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakravarty D., et al. CD4 deficiency causes poliomyelitis and axonal blebbing in murine coronavirus induced neuro-inflammation. J. Virol. 2020 doi: 10.1128/JVI.00548-20. JVI.00548-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das Sarma J., et al. Mechanisms of primary axonal damage in a viral model of multiple sclerosis. The J. Neurosci. 2009;29(33):10272. doi: 10.1523/JNEUROSCI.1975-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das Sarma J., et al. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 2002;8(5):381–391. doi: 10.1080/13550280260422686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abraham M.J., et al. GROMACS: High performance molecular stimulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1(2):19–25. [Google Scholar]
- 43.Delano, W.L., PyMOL: An Open-Source Molecular Graphics Tool. 2002.
- 44.van Dijk M., Bonvin A.M. 3D-DART: a DNA structure modeling server. Nucleic Acids Res. 2009;37(Web Server issue):W235–W239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuszynska I., et al. NPDock: a web server for protein-nucleic acid docking. Nucleic Acids Res. 2015;43(W1):W425–W430. doi: 10.1093/nar/gkv493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Gonzalez P., et al. Interplay between the unfolded protein response and immune function in the development of neurodegenerative diseases. Front Immunol. 2018;9:2541. doi: 10.3389/fimmu.2018.02541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das Sarma J., et al. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 2002;8(5):381–391. doi: 10.1080/13550280260422686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duplan E., et al. ER-stress-associated functional link between Parkin and DJ-1 via a transcriptional cascade involving the tumor suppressor p53 and the spliced X-box binding protein XBP-1. J. Cell Sci. 2013;126(Pt 9):2124–2133. doi: 10.1242/jcs.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller K.D., Schnell M.J., Rall G.F. Keeping it in check: chronic viral infection and antiviral immunity in the brain. Nat. Rev. Neurosci. 2016;17(12):766–776. doi: 10.1038/nrn.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Streit W.J., Mrak R.E., Griffin W.S.T. Microglia and neuroinflammation: a pathological perspective. J. Neuroinflamm. 2004;1(1) doi: 10.1186/1742-2094-1-14. 14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rempel J.D., et al. Differential regulation of innate and adaptive immune responses in viral encephalitis. Virology. 2004;318(1):381–392. doi: 10.1016/j.virol.2003.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth-Cross J.K., Bender S.J., Weiss S.R. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008;82(20):9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., et al. Coronavirus neuro-virulence correlates with the ability of the virus to induce pro-inflammatory cytokine signals from astrocytes and microglia. J. Virol. 2004;78(7):3398–3406. doi: 10.1128/JVI.78.7.3398-3406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang J., et al. Microglia polarization and endoplasmic reticulum stress in chronic social defeat stress induced depression mouse. Neurochem. Res. 2018;43(5):985–994. doi: 10.1007/s11064-018-2504-0. [DOI] [PubMed] [Google Scholar]
- 55.Sanchez C.L., et al. Endoplasmic reticulum stress differentially modulates the IL-6 family of cytokines in murine astrocytes and macrophages. Sci. Rep. 2019;9(1):14931. doi: 10.1038/s41598-019-51481-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rojo A.I., et al. Redox control of microglial function: molecular mechanisms and functional significance. Antioxid. Redox Sig. 2014;21(12):1766–1801. doi: 10.1089/ars.2013.5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhatia T.N., et al. Astrocytes do not forfeit their neuro-protective roles after surviving intense oxidative stress. Front. Mol. Neurosci. 2019;12(87) doi: 10.3389/fnmol.2019.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kahle P.J., Waak J., Gasser T. DJ-1 and prevention of oxidative stress in Parkinson's disease and other age-related disorders. Free Radic. Biol. Med. 2009;47(10):1354–1361. doi: 10.1016/j.freeradbiomed.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Wilson M.A. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid. Redox. Signal. 2011;15(1):111–122. doi: 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida H., et al. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 61.Fung, T.S. and D.X. Liu, Coronavirus infection, ER stress, apoptosis and innate immunity. 2014. 5(296). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.