Abstract
Background
Blood stream infections (BSI) caused by Extended Spectrum Beta-Lactamases (ESBLs) producing Enterobacteriaceae is a clinical challenge leading to high mortality, especially in developing countries. In this study, we sought to describe the epidemiology of ESBL-producing Escherichia coli strains isolated from Vietnamese individuals with BSI, to investigate the concordance of genotypic-phenotypic resistance, and clinical outcome of ESBL E. coli BSI.
Methods
A total of 459 hospitalized patients with BSI were screened between October 2014 and May 2016. 115 E. coli strains from 115 BSI patients were isolated and tested for antibiotic resistance using the VITEK®2 system. The ESBL phenotype was determined by double disk diffusion method following the guideline of Clinical and Laboratory Standards Institute. Screening for beta-lactamase (ESBL and carbapenemase) genes was performed using a multiplex-PCR assay.
Results
58% (67/115) of the E. coli strains were ESBL-producers and all were susceptible to both imipenem and meropenem. Resistance to third-generation cephalosporin was common, 70% (81/115) were cefotaxime-resistant and 45% (52/115) were ceftazidime-resistant. blaCTX-M was the most common ESBL gene detected (70%; 80/115) The sensitivity and specificity of blaCTX-M-detection to predict the ESBL phenotype was 87% (76–93% 95% CI) and 54% (39–48% 95% CI), respectively. 28%% (22/80) of blaCTX-M were classified as non-ESBL producers by phenotypic testing for ESBL production. The detection of blaCTX-M in ESBL-negative E. coli BSI was associated with fatal clinical outcome (27%; 6/22 versus 8%; 2/26, p = 0.07).
Conclusion
A high prevalence of ESBL-producing E. coli isolates harbouring blaCTX-M was observed in BSI patients in Vietnam. The genotypic detection of blaCTX-M may have added benefit in optimizing and guiding empirical antibiotic therapy of E. coli BSI to improve clinical outcome.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12941-021-00466-3.
Keywords: Antimicrobial resistances, Blood stream infections, Sepsis, ESBL, AMR, CTX-M, Drug-resistant Escherichia coli
Introduction
The emergence and spread of antimicrobial resistance (AMR) are of growing importance worldwide. With a population of more than 96 million and a high burden of infectious diseases, Vietnam faces since a decade a significant increase of AMR [1, 2]. Bloodstream infections (BSI) caused by multidrug-resistant bacteria frequently leads to fatal treatment failure [3, 4]. In Vietnam, BSIs are mostly caused by multidrug-resistant pathogens listed in the Global Antimicrobial Resistance Surveillance System (GLASS) report, including those classified as critical and high priority levels by the World Health Organization [5], such as extended spectrum beta-lactamase- (ESBL) and carbapenemase-resistant Enterobacteriaceae (CREs) [6, 7]. The most common cause of both community- and hospital-acquired BSI in South-East Asia is Escherichia coli, a member of the Enterobacteriaceae family [8, 9].
Wide dissemination of beta-lactamase producing E. coli, and increased incidences of hospital-acquired ESBL-producing E. coli infections have been documented [10–14]. In Italy, the incidences of community- and hospital-acquired infections by ESBL-producing E. coli increased between 2007 and 2015 from 29 to 42% and 25 to 35%, respectively [10]. Reports for the years 2002 until 2011 suggest an exponential increase in ESBL resistances from 20 to 45% in the Asia–Pacific region and from 39 to 55% in South-East Asia [15]. Most resistant bacterial strains were identified in patients treated at intensive care units (ICUs). In South-East Asia, Philippines and Vietnam have reported a high burden of ESBL-producing E. coli infections of 59% and 81%, respectively [16]. The most common resistance factors in clinical settings worldwide are the CTX-M-type beta-lactamases [17]. Furthermore, CRE carrying the transmissible carbapenemase genes blaKPC, blaNDM and blaOXA-48 are well characterized across various geographical regions [18]. Local epidemiological data on AMR is essential in guiding empirical antibiotic therapies to improve clinical management and outcome of BSIs, while supporting antibiotic stewardship measures to combat the spread of AMR [9]. However, epidemiological data on AMR from low- and middle-income countries (LMIC) are scarce compared to industrialized nations with a well-functioning surveillance system, so that the AMR burden in LMIC may be underestimated.
The present study aims to describe the molecular epidemiology and resistance patterns of E. coli strains isolated from Vietnamese individuals with BSI using a retrospective study cohort. We also evaluated the concordance of genotypic and phenotypic resistance in E. coli causing BSI towards molecular detection of ESBL-genes to predict cephalosporin resistance in E. coli and to optimize the management of E. coli BSI in a high ESBL-prevalent setting.
Materials and methods
Patient recruitment
The study group included patients with BSI admitted to the 108 Military Central Hospital in Hanoi, Vietnam, between October 2014 and May 2016. The 108 Military Central Hospital is a hospital of maximum care for Northern Vietnam, providing medical care for patients in Hanoi and for those being referred from various regional hospitals. In the latter case patients´ previous antibiotic treatment scheme was provided by the referring hospital. 459 BSI patients with bacterial pathogens diagnosed by blood cultures were hospitalized in the 108 Military Central Hospital during the study period. Of those, 115 patients with blood cultures positive for E. coli were included in this study. Diagnoses of BSI followed the criteria of the Surviving Sepsis Campaign guidelines [19] and the sepsis-related organ failure assessment (SOFA) score.
Blood culture
Two independent venous blood samples of approximately 8 mL per blood culture bottle were collected from both arms of the patients for blood culture with the BACTEC™ Plus Aerobic/F system (Becton–Dickinson, Franklin Lakes, NJ, USA). Blood culture bottles were incubated in the BD BACTEC™ 9120 device (Becton–Dickinson, Franklin Lakes, NJ, USA) at 36 °C for 18–72 h. Positive cultures were subjected to bacterial species identification and antimicrobial susceptibility testing using the VITEK® 2 compact automated system (BioMérieux, Lyon, France). Bacterial strains were stored with 20% glycerol at − 80 °C for further molecular testing.
Antimicrobial susceptibility testing
The minimum inhibitory concentration (MIC) for a wide spectrum of antibiotics were tested using VITEK AST-N204 cards and the VITEK® 2 compact automated system (bioMérieux, Lyon, France). The interpretation of susceptibility patterns was done according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [20]. The following antimicrobial substances were included: Ampicillin (AM), Amoxicillin/Clavulanic acid (AMC), Piperacillin/Tazobactam (TZP), Cefotaxime (CTX), Ceftazidime (CAZ), Cefepime (FEP), Ertapenem (ETP), Imipenem (IPM), Meropenem (MEM), Amikacin (AN) and Ciprofloxacin (CIP).
Phenotypic confirmation of ESBL production was achieved by double disk diffusion methods following the guideline of CLSI [20]. The CTX (30 µg) and CAZ (30 µg) disks alone and in combination with clavulanate (10 µg) were used for testing of ESBL phenotype E. coli on Mueller–Hinton agar. The tests were considered positive if the difference in the zone of clearance between CTX and CTX with clavulanate or CAZ and CAZ with clavulanate was ≥ 5 mm.
Detection of beta-lactamases
Genes encoding ESBLs and carbapenemases were screened using multiplex PCR assays described previously [21]. Six genes encoding ESBL (blaSHV, blaTEM, blaCTX-M [ESBL-1]; blaVEB, blaGES, blaPER [ESBL-2]) and nine genes encoding carbapenamases (blaNDM-1, blaSPM, blaVIM [CARBA-1]; blaIMP, blaAIM, blaKPC [CARBA-2]; blaOXA-23, blaOXA-48-like, blaOXA-58 [CARBA-3]) can be detected using the multiplex PCR panels (Additional file 1: Table S1). The primers were designed to detect the most common variants of the beta-lactamase genes irrespective of the potency of the beta-lactamase activity. Bacterial isolates were incubated at 35 ± 1 °C and colonies were picked and diluted in 300 μl TE buffer, followed by DNA extraction. In detail, the suspension was immersed in 300 μl universal lysis solution containing 200 mM NaOH and 1% SDS and incubated at 95 °C for 5 min. An equal volume of 1 M Tris–HCl was added to achieve a pH of 7.5. Subsequently, the solution was transferred to a 1.5 mL Eppendorf tube and an equal volume of phenol–chloroform-isoamyl alcohol mixture (25/24/1) (ThermoFisher Scientific, Invitrogen, Waltham, MA, USA) was added. The mixture was vortexed for 5 min and centrifuged at 13,200 rpm. The upper aqueous phase was transferred to an Eppendorf tube and an equal volume of isopropanol was added. After mixing, the solution was centrifuged at 13,200 rpm for 30 min to pellet the DNA. Precipitated DNA was washed twice with 70% ethanol and reconstituted in 150 μl TE buffer (25 mM Tris with 0.1 mM EDTA). Multiplex PCR assays were performed in 25 μl reaction volumes containing hot start master mix (2×) (Promega, San Luis Obispo, CA, USA) and individual primer pairs with varying concentrations (Additional file 1: Table S1). Thermal cycling conditions were denaturation at 95 °C for 2 min followed by 40 cycles of 94 °C for 30 s denaturation, 61 °C for 30 s annealing, followed by an extension at 72 °C for 40 s and a final extension step of 72 °C for 5 min. Amplicons were visualized on 1.2% agarose gels (Additional file 1: Figure S1).
Statistical analysis
Statistical analyses were performed using the SPSS software v.23.0 (IBM Corporation, Chicago, IL, USA). Continuous variables are presented as mean ± standard deviation. Categorical variables are given as frequencies with percentages and comparisons of categorical variables between groups were performed using chi-square and Fisher’s exact tests. The level of significance was set at p-values < 0.05.
Results
Clinical characteristics of study subjects
Of the 115 patients, 70 (61%) were males. The mean age of the patients was 62 years; 62/115 patients were > 60 years old. In 73 patients (64%) pre-existing conditions (solid cancer, hypertension, diabetes, liver cirrhosis) were recorded and eight of them were receiving immunosuppressive therapy (Table 1). A primary source of infection was identified in 96 (84%) patients, with urine tract infection (40/96; 42%) and infection of the bile duct system (36/96; 38%) as the main sources, followed by respiratory and post interventional infections. The mean SOFA score was 3.36 ± 3.0 and the median procalcitonin level was 10.19 (0.24–100) ng/L. Shock and mortality rates were 17% and 16%, respectively (Table 1).
Table 1.
Baseline characteristics of BSI patients with Escherichia coli infections
| Baseline characteristics | Values as n (%) |
|---|---|
| Sex (male) | 70/115 (61%) |
| Mean age in years | 62.3 ± 16.2 |
| ≥ 60 years old | 62(54%) |
| Pre-existing conditions | 73/115 (64%) |
| Solid cancer | 35/73 (48%) |
| Hypertension | 33/73 (45%) |
| Diabetes | 20/73 (27%) |
| Liver cirrhosis | 14/73 (19%) |
| Immunosuppressive therapy | 8/73 (11%) |
| Primary source infection n (%) | 96/115 (84%) |
| Urine tract infection | 40/96 (42%) |
| Bile duct infection | 36/96 (38%) |
| Respiratory infection | 8/96 (8%) |
| After intervention | 5/96 (5%) |
| Others | 7/96 (7%) |
| Mean SOFA (points) | 3.36 ± 3.05 |
| While Blood Cells (G/L) | 16.9 ± 13.7 |
| Neutrophile (G/L) | 13.7 ± 9.9 |
| Platelets (G/L) | 190 ± 118 |
| Prothrombin (%) | 76.5 ± 22.8 |
| Pro-Calcitonin median (range)(ng/L) | 10.19 (0.24–100) |
| pH | 7.33 ± 0.13 |
| Lactate (mmol/L) | 5.34 ± 3.53 |
| Length of hospital stay (days) | 19.9 ± 14.7 |
| Shock n (%) | 19/115 (17%) |
| Mortality n (%) | 18/115 (16%) |
Phenotypic antimicrobial susceptibility
Of the 115 E. coli isolates, 67 (58%) produced ESBLs. The phenotypic resistances of all study isolates are summarized in Table 1. The most prevalent resistance to beta-lactam antibiotics was observed for AM (109/115; 95%), followed by CTX (81/115; 70%), CAZ (52/115; 45%), AMC (32/115; 28%), FEP (28/109; 26%) and TZP (8/115; 7%) (Table 2). Three E. coli strains exhibited reduced susceptibility to ETP; one of them resistant with a MIC value of 2 mg/L, and two with intermediate susceptibility with MIC values of 1 mg/L. All isolates were phenotypically susceptible to IPM and MEM. With regard to other antibiotics, 67% (77/115) of strains were resistant to CIP, but 99% (114/115) of strains were still susceptible to AN (Table 2).
Table 2.
Phenotypic resistance of Escherichia coli causing BSI in Vietnam
| Overall, n = 115 | ESBL+, n = 67 | ESBL−, n = 48 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | S | I | R | ||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | |
| AM | 5 | 4.3 | 1 | 0.9 | 109 | 94.8 | 0 | – | 0 | – | 67 | 100.0 | 5 | 10.4 | 1 | 2.1 | 42 | 87.5 |
| AMC | 67 | 58.3 | 16 | 13.9 | 32 | 27.8 | 43 | 64.2 | 12 | 17.9 | 12 | 17.9 | 24 | 50.0 | 4 | 8.3 | 20 | 41.7 |
| TZP | 93 | 80.9 | 14 | 12.2 | 8 | 7.0 | 59 | 88.1 | 7 | 10.4 | 1 | 1.5 | 34 | 70.8 | 7 | 14.6 | 7 | 14.6 |
| CAZ | 62 | 53.9 | 1 | 0.9 | 52 | 45.2 | 31 | 46.3 | 1 | 1.5 | 35 | 52.2 | 31 | 64.6 | 0 | – | 17 | 35.4 |
| CTX | 34 | 29.6 | 0 | 0.0 | 81 | 70.4 | 0 | 0.0 | 0 | 0.0 | 67 | 100.0 | 34 | 70.8 | 0 | – | 14 | 29.2 |
| FEPa | 68 | 62.4 | 13 | 11.9 | 28 | 25.7 | 34 | 55.7 | 11 | 18.0 | 16 | 26.2 | 34 | 70.8 | 2 | 4.2 | 12 | 25.0 |
| ETP | 112 | 97.4 | 2 | 1.7 | 1 | 0.9 | 66 | 98.5 | 1 | 1.5 | 0 | – | 46 | 95.8 | 1 | 2.1 | 1 | 2.1 |
| IPM | 115 | 100.0 | 0 | – | 0 | – | 67 | 100.0 | 0 | – | 0 | – | 48 | 100.0 | 0 | – | 0 | – |
| MEM | 115 | 100.0 | 0 | – | 0 | – | 67 | 100.0 | 0 | – | 0 | – | 48 | 100.0 | 0 | – | 0 | – |
| AN | 113 | 98.3 | 1 | 0.9 | 1 | 0.9 | 67 | 100.0 | 0 | – | 0 | – | 46 | 95.8 | 1 | 2.1 | 1 | 2.1 |
| GM | 73 | 63.5 | 3 | 2.6 | 39 | 33.9 | 43 | 64.2 | 1 | 1.5 | 23 | 34.3 | 30 | 62.5 | 2 | 4.2 | 16 | 33.3 |
| CIP | 37 | 32.2 | 1 | 0.9 | 77 | 67.0 | 17 | 25.4 | 1 | 1.5 | 49 | 73.1 | 20 | 41.7 | 0 | – | 28 | 58.3 |
AMC: Amoxicillin/Clavulanic acid; TZP: Piperacillin/Tazobactam; ESBL: extended spectrum beta lactamase; CTX: Cefotaxime; CAZ: Ceftazidime; FEP: Cefepime; AN: Amikacin; CIP: Ciprofloxacin; S: susceptible; I: intermediate; R: resistant
amissing values for FEP susceptibility, n = 6 in ESBL-producers (ESBL+)
Detection of beta-lactamases
The proportion of strains carrying blaCTX-M, blaTEM, blaSHV was 70% (80/115), 58% (67/115) and 1% (1/115), respectively. Of the 115 strains, 105 (91%) carried at least one of the three resistance genes (blaCTX-M, blaCTX-M, blaSHV); none of the strains carried all three genes. The combination of blaCTX-M and blaTEM was detected in 42 (37%) of the 115 strains (Table 3). 9/115 (8%) strains were positive for blaNDM and 5/115 (4%) carried the blaVIM gene.
Table 3.
Concordance of genotypic and phenotypic resistance in blaCTX-M-harbouring Escherichia coli
| Antibiotic substanceb | blaCTX-M, n = 80a | blaCTX-M-positive ESBL-producers, n = 58a | blaCTX-M-positive non ESBL producers, n = 22a | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | S | I | R | ||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | |
| AM | 0 | – | 0 | – | 80 | 100 | 0 | – | 0 | – | 58 | 100 | 0 | – | 0 | – | 22 | 100 |
| AMC | 48 | 60 | 9 | 11.3 | 23 | 28.8 | 40 | 69 | 9 | 15.5 | 9 | 15.5 | 8 | 36.4 | 0 | – | 14 | 63.6 |
| TZP | 64 | 80 | 9 | 11.3 | 7 | 8.8 | 51 | 87.9 | 6 | 10.3 | 1 | 1.7 | 13 | 59.1 | 3 | 13.6 | 6 | 27.3 |
| CAZ | 35 | 43.8 | 1 | 1.3 | 44 | 55 | 27 | 46.6 | 1 | 1.7 | 30 | 51.7 | 8 | 36.4 | 0 | – | 14 | 63.6 |
| CTX | 11 | 13.8 | 0 | – | 69 | 86.3 | 0 | – | 0 | – | 58 | 100 | 11 | 50 | 0 | – | 11 | 50 |
| FEPc | 37 | 50 | 10 | 13.5 | 27 | 36.5 | 29 | 55.8 | 8 | 15.4 | 15 | 28.8 | 8 | 36.4 | 2 | 9.1 | 12 | 54.5 |
| ETP | 77 | 96.3 | 2 | 2.5 | 1 | 1.3 | 57 | 98.3 | 1 | 1.7 | 0 | – | 20 | 90.9 | 1 | 4.5 | 1 | 4.5 |
| IPM | 80 | 100 | 0 | – | 0 | – | 58 | 100 | 0 | – | 0 | – | 22 | 100 | 0 | – | 0 | – |
| MEM | 80 | 100 | 0 | – | 0 | – | 58 | 100 | 0 | – | 0 | – | 22 | 100 | 0 | – | 0 | – |
| AN | 78 | 97.5 | 1 | 1.3 | 1 | 1.3 | 58 | 100 | 0 | – | 0 | – | 20 | 90.9 | 1 | 4.5 | 1 | 4.5 |
| GM | 52 | 65 | 3 | 3.8 | 25 | 31.3 | 39 | 67.2 | 1 | 1.7 | 18 | 31 | 13 | 59.1 | 2 | 9.1 | 7 | 31.8 |
| CIP | 18 | 22.5 | 1 | 1.3 | 61 | 76.3 | 14 | 24.1 | 1 | 1.7 | 43 | 74.1 | 4 | 18.2 | 0 | – | 18 | 81.8 |
AMC: Amoxicillin/Clavulanic acid; TZP: Piperacillin/Tazobactam; ESBL: extended spectrum beta lactamase; CTX: Cefotaxime; CAZ: Ceftazidime; FEP: Cefepime; AN: Amikacin; CIP: Ciprofloxacin; S: susceptible; I: intermediate; R: resistant
aMay not add up to 100% due to rounding errors to 1 decimal place
bInterpretation of resistance according to the clinical breakpoints of CLSI
c6 missing values for cefepime
The blaNDM gene was detected in 2 isolates carrying the blaCTX-M gene, in 2 isolates carrying the blaTEM, and in 5 isolates harbouring both blaCTX-M and blaTEM genes. The blaVIM gene was detected in 4 isolates harbouring the blaCTX-M, and in one isolate harboring both blaCTX-M and blaTEM genes.We did not detect strains carrying blaKPC, blaVEB, blaPER, blaGES, blaIMP, blaSMP, blaAIM, blaOXA-23, blaOXA-48-like and blaOXA-58.
Concordance of resistance genotypes and phenotypic susceptibility
The concordance of genotypic and phenotypic resistance is summarized in Table 3. 14 out of 81 phenotypically CTX-resistant isolates were non-ESBL-producers according to the double disk method. However, 5 out of the 14 harboured blaCTX-M, 2 harboured blaTEM, 6 harboured both blaCTX-M and blaTEM, and one isolate did not harbour any beta-lactamase genes in our test-panel. There was a discrepancy between the detection of blaCTX-M and the phenotypic testing for ESBL, 22 out of 80 (27.5%) of isolates with blaCTX-M were classified as non-ESBL-producers by the double disk method. In our study setting, the sensitivity and specificity of blaCTX-M detection to predict the ESBL phenotype was 87% (95% CI 76–93%) and 54% (95% CI 39–68), respectively. A summary of the sensitivities and specificity of blaCTX-M detection to predict phenotypic resistance to CAZ, CTX and FEP are summarized in Table 4.
Table 4.
Performance of blaCTX-M detection to predict phenotypic resistance of Escherichia coli
| Antibiotic | Sensitivity % (95% CI) |
Specificity % (95% CI) |
PPV % (95% CI) |
NPV % (95% CI) |
|---|---|---|---|---|
| ESBL | 87 (76–93) | 54 (39–68) | 73 (61–82) | 74 (56–87) |
| CTX | 85 (75–92) | 68 (49–82) | 86 (76–93) | 66 (48–80) |
| CAZ | 85 (72–93) | 44 (31–57) | 57 (45–67) | 77 (59–89) |
| FEP | 90 (76–97) | 46 (34–58) | 50 (38–62) | 89 (72–96) |
ESBL: Extended spectrum beta lactamases; CTX: Cefotaxime; CAZ: Ceftazidime; FEP: Cefepime; and CIP: Ciprofloxacin; PPV: positive predictive value; NPV: negative predictive value; 95% CI: 95% confidence interval
Strikingly, all isolates carrying blaNDM and blaVIM were phenotypically susceptible to all carbapenems (ETP, IPM and MEM), which was unusual. The prevalence of quinolone resistance was overall very high in our isolate collection. However, there was an over-representation of quinolone resistance in blaCTX-M-carrying isolates.
blaCTX-M in cephalosporin-susceptible E. coli associated with unfavorable clinical outcome
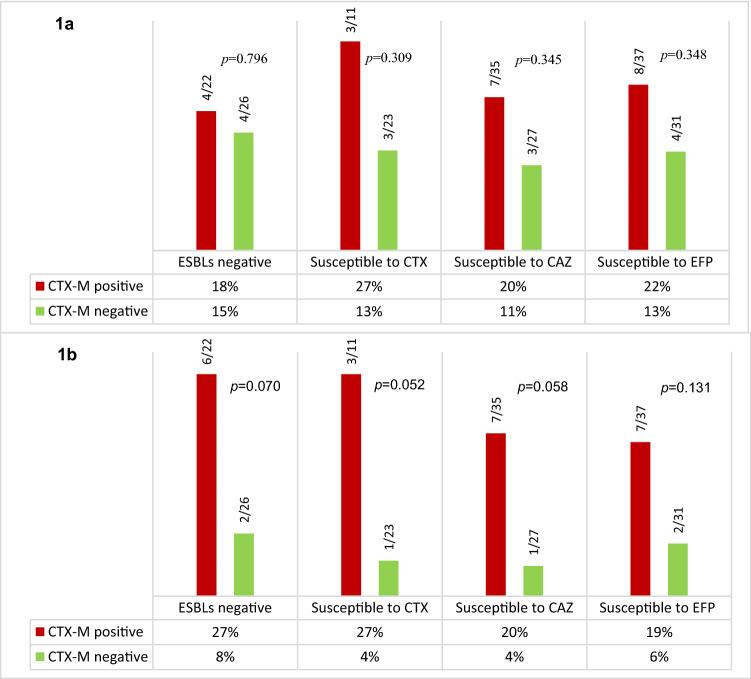
Since not all blaCTX-M-habouring isolates exhibited phenotypic resistance to third- and fourth-generation cephalosporins tested, we performed an analysis comparing the clinical outcome (shock and fatality) of BSI with cephalosporin-susceptible E. coli with blaCTX-M as an exposure variable. Our analysis suggested that patients with blaCTX-M-negative E. coli BSI had better clinical outcomes than patients with blaCTX-M-positive E. coli BSI. In particular, the presence of blaCTX-M in phenotypically cephalosporin-susceptible E. coli BSI is associated with fatal outcome of the infection for CTX-susceptible phenotype (4% versus 27%, p = 0.05), and with a statistical trend for ESBL-negative phenotype (8% versus 27%, p = 0.07), for CAZ-susceptible phenotype (4% versus 20%, p = 0.06) for ESBL-negative phenotype (4% versus 27%, p = 0.05), for FEP-susceptible phenotype (6 versus 19%, p = 0.1) (Fig. 1, Additional file 1: Table S2). In addition, the duration of hospital stay was significantly longer in patients with CTX-M-positive E. coli infections than in patients with CTX-M-negative E. coli BSI (Additional file 1: Table S2). For discrepant genotypic-phenotypic carbapenemase susceptibility, there was no association between the discrepancy and the clinical outcome (neither shock nor death). This analysis was performed with very small number of cases so that further investigations are needed before drawing any definitive conclusions.
Fig. 1.
The contribution of blaCTX-M to the outcome of BSI patients caused by cephalosporin susceptible Escherichia coli. a For ratio of shock and b for fatality. Red column is ratio of shock (a) or death (b) of BSI patients caused by blaCTX-M positive E. coli and green column is ratio of shock (a) or death (b) of BSI patients caused by blaCTX-M negative E. coli
Discussion
In this study, we reported a high prevalence (58%) of ESBL-producers in E. coli causing BSI in Vietnamese patients. Our findings are in accordance with previously published data on the epidemiology of ESBL-producing E. coli from Vietnam, which suggested a comparable (48–55%) prevalence [9, 16, 22]. Similar findings have been reported from studies conducted in in China (56–67%) [22, 23] and other regions in South-East Asia, such as Thailand (50%) [22]. In comparison, studies from industrialized (middle to high income) countries reported lower rates of ESBL-producers in clinical E. coli strains. A study from Canada has reported a far lower rate of 4% ESBL-producing E. coli strains [24]. Another study conducted in Sweden reported similar low rates of ESBL-producing E. coli strains with 2%, 3% and 4% of cases in 2007, 2009 and 2011, respectively [25]. Taken together, these discrepancies illustrate and highlight the increasing AMR problematic in the Asian region and in LMIC, in general. Surveillance, antibiotic use and higher compliance with antibiotic stewardship might account for these differences [26]. In many South-East Asian countries including Vietnam, antibiotics, especially of broad-spectrum cephalosporins can be purchased over the counter without prescription. Thus, inappropriate indications for antibiotic therapy may have an influence on the high rates of resistance [27, 28].
Almost all (97%) ESBL-producing E. coli in our study harboured at least one beta-lactamase gene, with blaCTX-M and blaTEM as the most common, consistent with published studies from Thailand and Vietnam [29, 30]. Indeed, among the 300 ESBL subtypes [31], the TEM-, SHV- and CTX-M-types are predominant and of particular relevance [32]. The CTX-M-type beta-lactamases play a major role as emerging and the most widely spread resistance factors in Enterobacteriaceae on a global scale [17]. Strikingly, we found a significant proportion (46%) of non ESBL-producers, as determined by phenotypic testing for ESBL, carrying the blaCTX-M gene. Since our multiplex PCR was designed to detect the most common CTX-M types, we cannot rule out that this discrepancy is attributed to CTX-M subtypes with weaker beta-lactamase activity. Overall, the concordance between phenotypic and genotypic susceptibility was good for blaCTX-M-carrying isolates. However, the presence of blaCTX-M alone cannot predict phenotypic resistance to cephalosporins perfectly. Not all CTX-M subtypes are equally potent; depending on the expression, the beta-lactamase activity levels may be variable [17, 33]. Furthermore, the presence of insertion sequences, such as ISE cp1B and IS903D, can modify the expression of the ESBL genes [34]. This highlights the necessity of implementing high-resolution molecular typing methods, such as whole-genome sequencing, to study the molecular epidemiology of AMR and multidrug-resistant pathogens in high-prevalent settings.
Carbapenem-resistance is low in E. coli causing BSI in our study population. Our study shows 98% ETP and 100% IPM and MEM susceptibility of E. coli isolates. However, we detected the presence of the Ambler class B metallo-beta-lactamase in 12 isolates, 8 with blaNDM and 4 with blaVIM. Oddly, the presence of these genes did not correlate with the phenotypic resistance to any carbapenems tested, which we could not explain. Indeed, discrepancies between genotypic und phenotypic resistances have been described previously [35]. The primers for both blaNDM and blaVIM were not specific for a particular variant; therefore, it is possible that nucleotide variations leading to a non-functional protein cannot be ruled out. In addition, the expression of beta-lactamase genes may be affected by promoter regions or other genetic element of the plasmid carrying these AMR genes [36]. Nonetheless, the high susceptibility rates for carbapenems and TZP indicate that these substances may be appropriate for the empirical antibiotic therapy of E. coli BSI in Vietnam. Of note, CIP resistance rate of 67% can be considered high in comparison to other studies [37], which may have been the result of frequent use of CIP.
The molecular detection of blaCTX-M in non-ESBL and cephalosporin-susceptible E. coli was significantly associated with a worse clinical outcome (septic shock and fatality) of E. coli BSI. This finding was unexpected since phenotypic susceptibility is considered as the gold standard for guiding antimicrobial therapies. The increased expression of antimicrobial resistance genes under antibiotic pressure may be an explanation for this observation [38]. Exposure to an antimicrobial substance may induce stress response (SOS-response) in bacteria, which causes several genes including AMR genes, such as blaCTX-M to be up-regulated thus increasing tolerance to various substance classes (cross-resistance) [39, 40]. In such cases, antibiotic therapy guided by phenotypic resistance only may not be suitable (i.e., false susceptible) and, thus, may lead to a poor outcome as a consequence of undertreatment. In this study cohort, we did not observe any significant differences in the clinical outcome for cases with discrepant phenotypic and genotypic carbapenem susceptibility. Due to the small number of cases (n = 14), we would refrain from drawing any conclusions. Nonetheless, this finding is unexpected and warrants further scrutiny. The time to antibiotic therapy initiation may influence the outcome of an infection. Therefore, the acceleration of time to report is of particular interest in the routine microbiological diagnostics [41]. Conventional blood culture diagnostics often take more than 24–48 h until antibiotic susceptibility profiles are available for the attending physicians, the introduction of genotypic PCR-based AMR detection may accelerate this process and thus potentially increasing the accuracy of calculated empirical antimicrobial therapy [42]. However, further validation in a larger cohort is needed prior to implementation in the routine setting.
Our study has limitations, the small sample size, especially for the clinical outcome analysis, may limit the generalizability of our findings. Nonetheless, with our small cohort, we could show that the presence of blaCTX-M in cephalosporin-susceptible E. coli correlated with a poor outcome and warrant further investigations. Furthermore, our PCR panel encompasses multiple variants of the beta-lactamase genes. The primers were designed to detect the most common variants of the respective beta-lactamase genes, which may have resulted in discordance in the genotypic-phenotypic susceptibility for low-activity beta-lactamases, such as blaTEM-1, blaTEM-2 and blaSHV-1 [43]. Due to this limitation, the study focused on blaCTX-M.
Taken together, our data contributes to epidemiological data on the AMR burden in South-East Asia, which needs attention and should be closely monitored. The detection of AMR gene does not necessarily correlate with the phenotypic resistance in E. coli and warrant further scrutiny. Easy to implement PCR-based AMR detection method may have added benefit, especially in high-prevalent settings, to accelerate, optimize and guide antimicrobial therapy. The surveillance of the spread of multidrug resistance in LMIC is still suboptimal and access to high-resolution molecular typing methods may help combat the spread of AMR genes on a global scale.
Supplementary Information
Additional file 1: Table S1. Primer sequences used for screening of beta-lactamase encoding genes. Table S2. Duration of hospital stay of patients with BSIs caused by cephalosporin susceptible E. coli. Figure S1: Agarose gel electrophoresis of blaCTX-M (739 bp), blaCTX-M (590 bp), blaTEM (422 bp) genes; M50 Marker (50–1000 bp); (–) negative control; (+) positive control. Samples 1,2,3,5,6,9,13 are positive for blaTEM and blaCTX-M; samples 4,11,12 are positive for blaCTX-M; samples 7 and 10 are negative for all (blaSHV, blaCTX-M and blaTEM)
Acknowledgements
The authors thank the Vietnam National Foundation for Science and Technology Development (NAFOSTED) for funding this research.
Authors' contributions
TVS, LHS, NTT, TPV designed and supervised the studies, TVS, DTQ and NTT conducted the experiments. TVS, NDM, VVS evaluated the clinical data and provided the clinical samples. TVS, LHS, TPV, CGM, NTT, NDM, NTKP, MHB, DN analyzed the data, wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under the Grant number 108.06-2017.21. The funding agency had no role in the study design, data collection and analysis, decision to publish, and/or preparation of the manuscripts.
Availability of data and materials
All data are available within this manuscript.
Declarations
Ethics approval and consent to participate
Ethical approval was obtained from the Ethics Committee and the Review Board of the 108 Military Central Hospital (Approval No: 108 MCH 2018/036-ICID-Ver 2). The Ethics Committee also approved a waiver of informed consent to participate in this study because of its retrospective design. All patient data was anonymized prior to the analysis.
Consent for publication
All authors have read the manuscript and have consented for publication.
Competing interests
All authors disclose no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Trinh Van Son and Nguyen Dang Manh shared equal contributions
Mai Hong Bang and Le Huu Song shared senior authors
References
- 1.Paoli CJ, Reynolds MA, Sinha M, Gitlin M, Crouser E. Epidemiology and costs of sepsis in the United States-an analysis based on timing of diagnosis and severity level. Crit Care Med. 2018;46(12):1889–1897. doi: 10.1097/CCM.0000000000003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McPherson D, Griffiths C, Williams M, Baker A, Klodawski E, Jacobson B, Donaldson L. Sepsis-associated mortality in England: an analysis of multiple cause of death data from 2001 to 2010. BMJ open. 2013;3(8):e002586. doi: 10.1136/bmjopen-2013-002586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiratisin P, Chongthaleong A, Tan TY, Lagamayo E, Roberts S, Garcia J, Davies T. Comparative in vitro activity of carbapenems against major Gram-negative pathogens: results of Asia-Pacific surveillance from the COMPACT II study. Int J Antimicrob Agents. 2012;39(4):311–316. doi: 10.1016/j.ijantimicag.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Song JH, Jung SI, Ko KS, Kim NY, Son JS, Chang HH, Ki HK, Oh WS, Suh JY, Peck KR, et al. High prevalence of antimicrobial resistance among clinical Streptococcus pneumoniae isolates in Asia (an ANSORP study) Antimicrob Agents Chemother. 2004;48(6):2101–2107. doi: 10.1128/AAC.48.6.2101-2107.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO: Global Antimicrobial Resistance Surveillance System: Manual for Early Implementation. In.: World Health Organisation; 2015: 1–44. https://apps.who.int/iris/bitstream/handle/10665/188783/9789241549400_eng.pdf?sequence=9789241549401.
- 6.Dagher GA, Saadeldine M, Bachir R, Zebian D, Chebl RB. Descriptive analysis of sepsis in a developing country. Int J Emerg Med. 2015;8:19–25. doi: 10.1186/s12245-015-0068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanberger H, Antonelli M, Holmbom M, Lipman J, Pickkers P, Leone M, Rello J, Sakr Y, Walther SM, Vanhems P, et al. Infections, antibiotic treatment and mortality in patients admitted to ICUs in countries considered to have high levels of antibiotic resistance compared to those with low levels. BMC Infect Dis. 2014;14:513–513. doi: 10.1186/1471-2334-14-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Southeast Asia Infectious Disease Clinical Research N Causes and outcomes of sepsis in southeast Asia: a multinational multicentre cross-sectional study. Lancet Glob Health. 2017;5(2):e157–67. doi: 10.1016/S2214-109X(17)30007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dat VQ, Vu HN, Nguyen HT, Hoang LB, Vu Tien Viet D, Bui CL, Van Nguyen K, Nguyen TV, Trinh DT, et al. Bacterial bloodstream infections in a tertiary infectious diseases hospital in Northern Vietnam: aetiology, drug resistance, and treatment outcome. BMC Infect Dis. 2017;17(1):493–493. doi: 10.1186/s12879-017-2582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Angelis G, Fiori B, Menchinelli G, D'Inzeo T, Liotti FM, Morandotti GA, Sanguinetti M, Posteraro B, Spanu T. Incidence and antimicrobial resistance trends in bloodstream infections caused by ESKAPE and Escherichia coli at a large teaching hospital in Rome, a 9-year analysis (2007–2015) Eur J Clin Microbiol Infect Dis. 2018;37(9):1627–1636. doi: 10.1007/s10096-018-3292-9. [DOI] [PubMed] [Google Scholar]
- 11.Tian L, Sun Z, Zhang Z. Antimicrobial resistance of pathogens causing nosocomial bloodstream infection in Hubei Province, China, from 2014 to 2016: a multicenter retrospective study. BMC Public Health. 2018;18(1):1121. doi: 10.1186/s12889-018-6013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDanel J, Schweizer M, Crabb V, Nelson R, Samore M, Khader K, Blevins AE, Diekema D, Chiang HY, Nair R, et al. Incidence of extended-spectrum beta-Lactamase (ESBL)-producing Escherichia coli and Klebsiella infections in the United States: a systematic literature review. Infect Control Hosp Epidemiol. 2017;38(10):1209–1215. doi: 10.1017/ice.2017.156. [DOI] [PubMed] [Google Scholar]
- 13.Hongsuwan M, Srisamang P, Kanoksil M, Luangasanatip N, Jatapai A, Day NP, Peacock SJ, Cooper BS, Limmathurotsakul D. Increasing incidence of hospital-acquired and healthcare-associated bacteremia in northeast Thailand: a multicenter surveillance study. PloS ONE. 2014;9(10):e109324. doi: 10.1371/journal.pone.0109324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama T, Ueda S, Huong BT, le Tuyen D, Komalamisra C, Kusolsuk T, Hirai I, Yamamoto Y. Wide dissemination of extended-spectrum beta-lactamase-producing Escherichia coli in community residents in the Indochinese peninsula. Infect Drug Resist. 2015;8:1–5. doi: 10.2147/IDR.S74934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrissey I, Hackel M, Badal R, Bouchillon S, Hawser S, Biedenbach D. A review of ten years of the study for monitoring antimicrobial resistance trends (SMART) from 2002 to 2011. Pharmaceuticals. 2013;6(11):1335–1346. doi: 10.3390/ph6111335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suwantarat N. Carroll KCJAR, Control I: Epidemiology and molecular characterization of multidrug-resistant Gram-negative bacteria in Southeast Asia. Antimicrob Resist Infect Control. 2016;5(1):15. doi: 10.1186/s13756-016-0115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type beta-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol. 2013;303(6–7):305–317. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Codjoe FS, Donkor ES. Carbapenem resistance: a review. Med Sci. 2018;6(1):1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, Sevransky JE, Sprung CL, Douglas IS, Jaeschke R, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41(2):580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 20.CLSI: Performance standards for antimicrobial susceptibility testing; twenty fourth informational supplement, M100S24. Clinical and Laboratory Standards Institute. 2014.
- 21.Trung NT, Hien TT, Huyen TT, Quyen DT, Binh MT, Hoan PQ, Meyer CG, Velavan TP, le Song H. Simple multiplex PCR assays to detect common pathogens and associated genes encoding for acquired extended spectrum betalactamases (ESBL) or carbapenemases from surgical site specimens in Vietnam. Ann Clin Microbiol Antimicrob. 2015;14:23. doi: 10.1186/s12941-015-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang YT, Coombs G, Ling T, Balaji V, Rodrigues C, Mikamo H, Kim MJ, Rajasekaram DG, Mendoza M, Tan TY, et al. Epidemiology and trends in the antibiotic susceptibilities of Gram-negative bacilli isolated from patients with intra-abdominal infections in the Asia-Pacific region, 2010–2013. Int J Antimicrob Agents. 2017;49(6):734–739. doi: 10.1016/j.ijantimicag.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Quan J, Zhao D, Liu L, Chen Y, Zhou J, Jiang Y, Du X, Zhou Z, Akova M, Yu Y. High prevalence of ESBL-producing Escherichia coli and Klebsiella pneumoniae in community-onset bloodstream infections in China. J Antimicrob Chemother. 2016;72(1):273–280. doi: 10.1093/jac/dkw372. [DOI] [PubMed] [Google Scholar]
- 24.Denisuik AJ, Lagace-Wiens PR, Pitout JD, Mulvey MR, Simner PJ, Tailor F, Karlowsky JA, Hoban DJ, Adam HJ, Zhanel GG. Molecular epidemiology of extended-spectrum beta-lactamase-, AmpC beta-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007–11. J Antimicrob Chemother. 2013;68(Suppl 1):i57–65. doi: 10.1093/jac/dkt027. [DOI] [PubMed] [Google Scholar]
- 25.Brolund A, Edquist PJ, Mäkitalo B, Olsson-Liljequist B, Söderblom T, Tegmark Wisell K, Giske CG. Epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in Sweden 2007–2011. Clin Microbiol Infect. 2014;20(6):O344–352. doi: 10.1111/1469-0691.12413. [DOI] [PubMed] [Google Scholar]
- 26.Cars O, Molstad S, Melander A. Variation in antibiotic use in the European Union. Lancet. 2001;357(9271):1851–1853. doi: 10.1016/S0140-6736(00)04972-2. [DOI] [PubMed] [Google Scholar]
- 27.Thu TA, Rahman M, Coffin S, Harun-Or-Rashid M, Sakamoto J, Hung NV. Antibiotic use in Vietnamese hospitals: a multicenter point-prevalence study. Am J Infect Control. 2012;40(9):840–844. doi: 10.1016/j.ajic.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen KV, Thi Do NT, Chandna A, Nguyen TV, Pham CV, Doan PM, Nguyen AQ, Thi Nguyen CK, Larsson M, Escalante S, et al. Antibiotic use and resistance in emerging economies: a situation analysis for Viet Nam. BMC Public Health. 2013;13:1158–1167. doi: 10.1186/1471-2458-13-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiratisin P, Apisarnthanarak A, Laesripa C, Saifon P. Molecular characterization and epidemiology of extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob Agents Chemother. 2008;52(8):2818–2824. doi: 10.1128/AAC.00171-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan NPH, Hien NH, Phuong TLT, Thanh DP, Thieu NTV, Ngoc DTT, Tuyen HT, Vinh PV, Ellington MJ, Thwaites GE, et al. Phenotypic and genotypic characteristics of ESBL and AmpC producing organisms associated with bacteraemia in Ho Chi Minh City, Vietnam. Antimicrob Resist Infect Control. 2017;6:105. doi: 10.1186/s13756-017-0265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghafourian S, Sadeghifard N, Soheili S, Sekawi Z. Extended spectrum beta-lactamases: definition, classification and epidemiology. Curr Issues Mol Biol. 2015;17:11–21. [PubMed] [Google Scholar]
- 33.Liu L, Zhang X, Yang S, Zhai Y, Liu W, Wang X, Zhang Z, Gao Z. Differential CTX-M expression from a conserved promoter: role of promoter-associated spacer sequences downstream of the blaCTX-M regulon. J Mol Microbiol Biotechnol. 2016;26(4):284–290. doi: 10.1159/000445950. [DOI] [PubMed] [Google Scholar]
- 34.Poirel L, Decousser JW, Nordmann P. Insertion sequence ISEcp1B is involved in expression and mobilization of a bla(CTX-M) beta-lactamase gene. Antimicrob Agents Chemother. 2003;47(9):2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urmi UL, Nahar S, Rana M, Sultana F, Jahan N, Hossain B, Alam MS, Mosaddek ASM, McKimm J, Rahman NAA, et al. Genotypic to phenotypic resistance discrepancies identified involving beta-lactamase genes, blaKPC, blaIMP, blaNDM-1, and blaVIM in uropathogenic Klebsiella pneumoniae. Infect Brug Resist. 2020;13:2863–2875. doi: 10.2147/IDR.S262493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kocer K, Klein S, Hildebrand D, Krall J, Heeg K, Boutin S, Nurjadi D. Pitfalls in genotypic antimicrobial susceptibility testing caused by low expression of blaKPC in Escherichia coli. J Antimicrob Chemother. 2021 doi: 10.1093/jac/dkab267. [DOI] [PubMed] [Google Scholar]
- 37.Biedenbach DJ, Bouchillon SK, Hoban DJ, Hackel M, Phuong DM, Nga TT, Phuong NT, Phuong TT, Badal RE. Antimicrobial susceptibility and extended-spectrum beta-lactamase rates in aerobic gram-negative bacteria causing intra-abdominal infections in Vietnam: report from the Study for Monitoring Antimicrobial Resistance Trends (SMART 2009–2011) Diagn Microbiol Infect Dis. 2014;79(4):463–467. doi: 10.1016/j.diagmicrobio.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Singh T, Singh PK, Das S, Wani S, Jawed A, Dar SA. Transcriptome analysis of beta-lactamase genes in diarrheagenic Escherichia coli. Sci Rep. 2019;9(1):3626. doi: 10.1038/s41598-019-40279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu G, Bogaj K, Bortolaia V, Olsen JE, Thomsen LE. Antibiotic-induced, increased conjugative transfer is common to diverse naturally occurring ESBL plasmids in Escherichia coli. Front Microbiol. 2019;10:2119. doi: 10.3389/fmicb.2019.02119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazar V, Nagy I, Spohn R, Csorgo B, Gyorkei A, Nyerges A, Horvath B, Voros A, Busa-Fekete R, Hrtyan M, et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat Commun. 2014;5:4352. doi: 10.1038/ncomms5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolk DM, Johnson JK. Rapid diagnostics for blood cultures: supporting decisions for antimicrobial therapy and value-based care. J Appl Lab Med. 2019;3(4):686–697. doi: 10.1373/jalm.2018.028159. [DOI] [PubMed] [Google Scholar]
- 42.Anjum MF, Zankari E, Hasman H. Molecular methods for detection of antimicrobial resistance. Microbiol Spectr. 2017;5(6):1–17. doi: 10.1128/microbiolspec.ARBA-0011-2017. [DOI] [PubMed] [Google Scholar]
- 43.Livermore DM. beta-Lactamases: quantity and resistance. Clin Microbiol Infect. 1997;3(Suppl 4):S10–S19. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primer sequences used for screening of beta-lactamase encoding genes. Table S2. Duration of hospital stay of patients with BSIs caused by cephalosporin susceptible E. coli. Figure S1: Agarose gel electrophoresis of blaCTX-M (739 bp), blaCTX-M (590 bp), blaTEM (422 bp) genes; M50 Marker (50–1000 bp); (–) negative control; (+) positive control. Samples 1,2,3,5,6,9,13 are positive for blaTEM and blaCTX-M; samples 4,11,12 are positive for blaCTX-M; samples 7 and 10 are negative for all (blaSHV, blaCTX-M and blaTEM)
Data Availability Statement
All data are available within this manuscript.