Abstract
Cell-free mitochondrial DNA (cf-mtDNA) is a marker of inflammatory disease and a predictor of mortality, but little is known about cf-mtDNA in relation to psychobiology. A systematic review of the literature reveals that blood cf-mtDNA varies in response to common real-world stressors including psychopathology, acute psychological stress, and exercise. Moreover, cf-mtDNA is inducible within minutes and exhibits high intra-individual day-to-day variation, highlighting the dynamic regulation of cf-mtDNA levels. We discuss current knowledge on the mechanisms of cf-mtDNA release, its forms of transport (“cell-free” does not mean “membrane-free”), potential physiological functions, putative cellular and neuroendocrine triggers, and factors that may contribute to cf-mtDNA removal from the circulation. A review of in vitro, pre-clinical, and clinical studies shows conflicting results around the dogma that physiological forms of cf-mtDNA are pro-inflammatory, opening the possibility of other physiological functions, including the cell-to-cell transfer of whole mitochondria. Finally, to enhance the reproducibility and biological interpretation of human cf-mtDNA research, we propose guidelines for blood collection, cf-mtDNA isolation, quantification, and reporting standards, which can promote concerted advances by the community. Defining the mechanistic basis for cf-mtDNA signaling is an opportunity to elucidate the role of mitochondria in brain-body interactions and psychopathology.
Keywords: Psychosocial stress, Mitochondria, mtDNA, cell-free DNA, Non-inflammatory effects, Standard protocol
1. The rise of mitochondria in psychopathology
While mitochondria have historically been known as “the power-house of the cell”, recent research has revealed that mitochondria communicate and exchange information (via proteins, DNA, ions, and metabolites) with each other (Ono et al., 2001; Eisner et al., 2014; Picard et al., 2015a); and with the cell nucleus where they induce epigenetic remodeling (Tian et al., 2016; Lozoya et al., 2018; Minocherhomji et al., 2012; Santos, 2020) and impact gene expression (Picard et al., 2014). Intracellularly, mitochondrial signaling occurs via the release of small molecules including metabolites (Chouchani et al., 2014), proteins (Fiorese et al., 2016; Cardamone et al., 2018; Kim et al., 2018, 2017), RNA (Dhir et al., 2018), and the mitochondrial genome itself (Dhir et al., 2018; Nakahira et al., 2011; Wu et al., 2019; West et al., 2015). But in addition to intracellular signaling, mitochondria also communicate information beyond the cell membrane – systemically – by producing and releasing various small molecules. Mitochondria are the source of all steroid hormones, including the potent sex-defining hormones testosterone, progesterone and estrogens, as well as the stress-induced glucocorticoids that function as endocrine signals to promote stress adaptation (Selvaraj et al., 2018). Beyond steroidogenesis in the mitochondrial matrix, mitochondria contain their own genome from which small mtDNA-encoded peptides are synthesized (e.g., MOTS-c, humanin) (Yen et al., 2020; Lee et al., 2015; Reynolds et al., 2020a), and released in the systemic circulation in response to challenges to optimize metabolic regulation. Thus, mitochondria-derived signaling molecules – or mitokines – likely play a significant role in human physiology and stress adaptation (Reynolds et al., 2020b).
In relation to brain-body processes and psychopathology, mitochondria have been implicated in two main ways: as a target of stress, and as a source of signals that shape resilience and adaptation (Picard et al., 2019; Daniels et al., 2020). Evidence that mitochondria are a downstream target of stress is derived from studies showing that chronic stress influences mitochondrial structure and function (e.g., (Cai et al., 2015), see (Picard and McEwen, 2018) for a systematic review). In humans, positive mood has also been found to predict higher leukocyte mitochondrial energy production capacity within days (Picard et al., 2018), and early life adversity is associated with alterations in mitochondrial respiratory capacity (Boeck et al., 2018, 2016; Gumpp et al., 2020), indicating that mitochondria are likely targets of psychological states.
Mitochondria also regulate stress physiology and behavior. Genetic or pharmacological perturbations of mitochondrial functions influence physiological, metabolic, and transcriptional responses to psychological stress (Picard et al., 2015b). Perturbation of mitochondrial functions also influences behavioral traits such as social dominance behavior (Hollis et al., 2015; van der Kooij et al., 2018), and cause mood disorder-like vegetative and psychomotor symptoms in mice (Kasahara et al., 2006). Moreover, individuals suffering from psychiatric symptoms show genetic and biochemical evidence of mitochondrial dysfunction in blood leukocytes or brain tissue (Rossignol and Frye, 2012; Holper et al., 2019; Wang et al., 2016; Cherix et al., 2020), implicating mitochondrial defects as a potential cause or consequence of psychopathology. In support of this hypothesis, patients with inherited primary mtDNA defects, which directly impair respiratory chain function, show high rates of psychiatric symptoms (Fattal et al., 2007; Morava et al., 2010). Although not free of confounds, these collective data show that mitochondria may contribute to human psychopathology (Pei and Wallace, 2018).
Collectively, the recent evidence implicating mitochondrial signaling in cellular and physiological stress regulation and mental health provides a basis for the rapidly growing interest to map psychobiological mechanisms linking stress, mitochondria, and psychopathology. A major challenge for the field is to develop robust, interpretable, and non-invasive biomarkers of mitochondrial dysfunction and signaling. One such emerging candidate biomarker is the circulating mitochondrial genome.
2. Collecting, measuring, and interpreting blood cell-free mtDNA: An overview
Mitochondria are the only mammalian organelle besides the nucleus to contain their own genome. Each mitochondrion contains multiple copies of the 16.5 kb circular mitochondrial DNA (mtDNA) bound in protein-DNA complexes called nucleoids (Farge and Falkenberg, 2019; Lee and Han, 2017). Surprisingly, mtDNA is detectable in most body fluids, including the liquid fraction of blood – plasma or serum – where it exists as circulating cell-free mtDNA (ccf-mtDNA, or cf-mtDNA when referring to any biofluid). Unlike tissue mtDNA copy number, which represents the number of copies of mtDNA per cell and loosely approximates respiratory capacity in tissues (e.g., leukocytes, brain, muscle) (Filograna et al., 2020), as an extracellular entity, cf-mtDNA does not reflect energy production capacity. Moreover, cf-mtDNA is not correlated with mtDNA copy number in circulating leukocytes (Rosa et al., 2020; Lindqvist et al., 2018). As discussed below, cf-mtDNA concentration may reflect different types of stress responses, but the exact functions of cf-mtDNA in its different forms remain unclear.
In some pathologies, blood cf-mtDNA is consistently elevated. Plasma cf-mtDNA levels are particularly elevated in trauma (Lam et al., 2004; Zhang et al., 2010a), inflammatory diseases (Boyapati et al., 2017; Hu et al., 2019; Kung et al., 2012; Yamanouchi et al., 2013), as well as cancers, myocardial infarction, and sepsis (Li et al., 2016; Schwarzenbach et al., 2011; Bhagirath et al., 2015; Dwivedi et al., 2012; Nakahira et al., 2013). In hospitalized critically ill patients, elevated plasma cf-mtDNA levels are prospectively associated with a 4–8-fold increased risk of mortality (Nakahira et al., 2013), suggesting that cf-mtDNA is a marker of physiological stress.
Because of mitochondria’s bacterial origin (Sagan, 1967), cf-mtDNA is typically considered as a pro-inflammatory antigen capable of triggering inflammatory responses in target immune and non-immune cells (Boyapati et al., 2017; Zhong et al., 2019; West and Shadel, 2017). Indeed, purified mtDNA injected into mice or exposed to cells in culture can induce proinflammatory cytokines (Collins et al., 2004). Consistent with this notion, numerous groups have defined receptor-mediated pathways whereby mtDNA triggers inflammatory signaling (reviewed in (Riley and Tait, 2020) and below). However, unlike the isolated laboratory forms of cf-mtDNA used in these studies, physiological cf-mtDNA is not necessarily membrane free. In fact, recent evidence suggests that most of the cf-mtDNA may be encapsulated within extracellular vesicles, such as microvesicles (Sansone et al., 2017; Lazo et al., 2020), exosomes (Sansone et al., 2017), or whole mitochondria (Al Amir Dache et al., 2020; Song et al., 2020; Stephens et al., 2020) that make mtDNA inaccessible to receptors. Our critical appraisal of the human cf-mtDNA literature reveals that evidence on the inflammatory effect of cf-mtDNA is not uniform, and that the pro-inflammatory nature of physiologically occurring cf-mtDNA in humans should be re-evaluated (see Section 5.2).
Recent evidence also suggests that release mechanisms are not limited to cell death as expected in trauma (reviewed in (Miliotis et al., 2019)). As described in the section below, triggers of cf-mtDNA release occur in several non-life-threatening situations. Both psychological factors and psychopathology can elevate cf-mtDNA. But differences in methodology used to measure cf-mtDNA may have significant impact on their comparability and interpretability. Therefore, standardization of approaches across laboratories is needed.
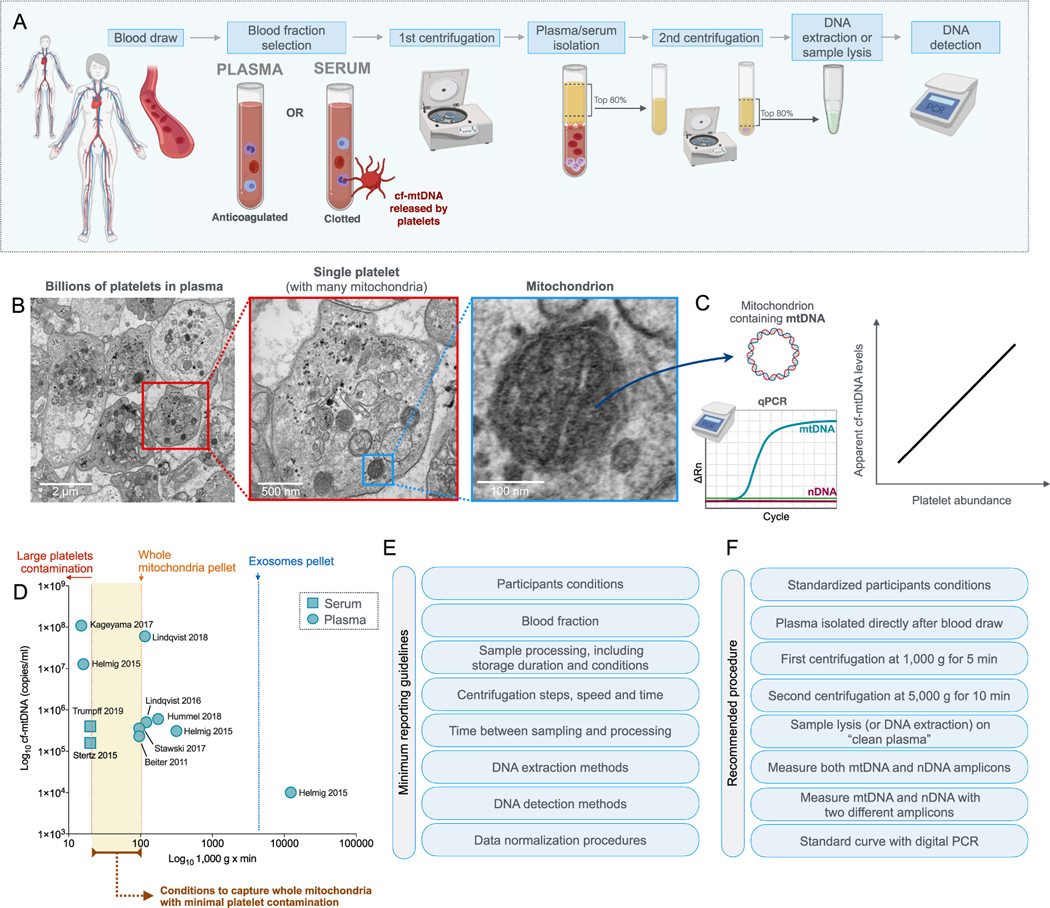
As described in detail in Section 5, there are different forms of transport for blood cf-mtDNA that can be separated to increase biological specificity during sample preparation. Cf-mtDNA is compartmentalized in distinct entities such as platelets, mitochondria or extracellular vesicles, and the duration and speed of centrifugation for plasma or serum isolates different forms of cf-mtDNA (Fig. 1). Slow centrifugation forces that are insufficient to sediment platelets cause contamination with large platelets (which contain mtDNA but no nDNA) in the final plasma, whereas excessively rapid centrifugation and the use of filters deplete potentially meaningful forms of cf-mtDNA, such as whole mitochondria and microvesicles containing mtDNA. Because existing studies have not used standardized methods, their results are not directly comparable. Another discrepancy occurs through the choice of the blood fraction: some studies have used serum (post-coagulation), which is not directly comparable to plasma (coagulation inhibited) because the clotting process may increase serum cf-mtDNA (Xia et al., 2009). Importantly, most studies in the psychopathology literature did not measure cell-free nuclear DNA (cf-nDNA) along with cf-mtDNA, preventing assessment of specific release of cf-mtDNA, where release of both cf-mtDNA and cf-nDNA would be interpreted as arising from tissue damage (Fig. 1C). Adopting standard cf-mtDNA isolation and quantification techniques (see Section 6, Technical Considerations) will optimize the biological specificity of our measurements and ensure reproducibility across laboratories (Meddeb et al., 2019; El Messaoudi et al., 2013). Such methodological standardization will enable the field to make concerted progress towards meaningful biological and clinical insights.
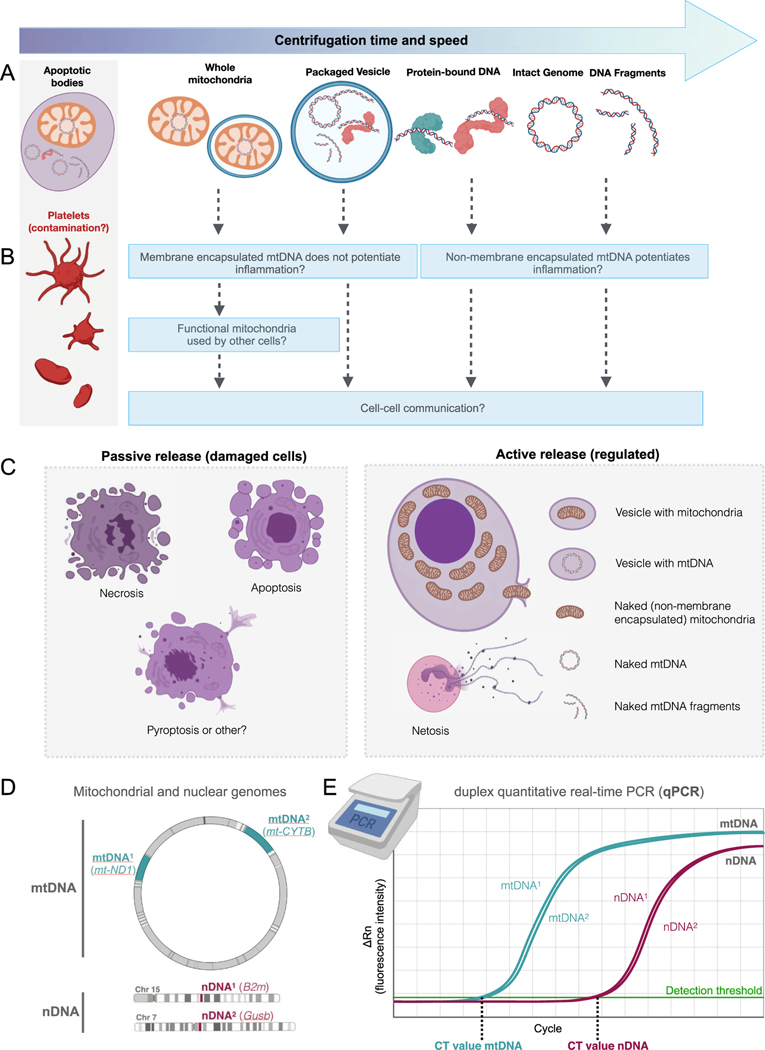
Fig. 1. Potential forms of blood circulating cell-free mtDNA (cf-mtDNA).
(A) Several forms of cf-mtDNA exist in the circulation. During cf-mtDNA isolation, the time and speed (i.e., g force) of the initial and secondary centrifugations determine the specific forms of cf-mtDNA that are isolated and detected. Platelets, which contain abundant mitochondria and mtDNA, can easily contaminate cf-mtDNA preparations and therefore inflate apparent cf-mtDNA levels when centrifugation forces are low. Note that structures in A are not drawn to scale. (B) The different forms of cf-mtDNA may have different physiological consequences. See Section 5 for discussion. (C) Cf-mtDNA release may occur “passively” with different forms of cell death, or “actively” through regulated processes. (D) Quantifying both cf-nDNA and cf-mtDNA determines whether increases in cf-DNA are specific to mtDNA. The selection of two different mtDNA/nDNA amplicons (mtDNA1 and mtDNA2) helps ensure that results are robust to potential inter-individual mtDNA sequence variations. The example mtDNA and nDNA genes/amplicons are from (Trumpff et al., 2019a). (E) The mtDNA/nDNA amplicons can be quantified in duplex quantitative real-time polymerase chain reaction (qPCR) or using other methods. qPCR measures the exponential amplification of target sequences over several cycles of PCR via fluorescence. The crossing of the resulting function in log linear phase of amplification above the detection threshold is called the cycle threshold (Ct) or crossing point, which depends on the number of the target amplicons (mtDNA or nDNA) in the original sample. Ct values are inversely proportional to the abundance of the amplicons in the original sample. By using multiple dilutions of a known amount of standard DNA, a standard curve can be generated of log concentration against the Ct, and the amount of DNA in a sample can be calculated, yielding the number of copies of mtDNA per mL of plasma.
To achieve a mechanistic understanding of the role of cf-mtDNA in mental health, five main questions must be resolved: 1) What are the triggers of cf-mtDNA release? 2) From which tissue(s) or cell type(s) does cf-mtDNA originate? 3) What are the kinetics of cf-mtDNA release and removal? 4) What are the molecular forms of transport of cf-mtDNA in the circulation? 5) What are the functional effects of cf-mtDNA on target cells?
Aiming for an overview of the current state of knowledge, before addressing these questions and discussing current methods available to selectively isolate and quantify specific forms of cf-mtDNA, we first present a systematic review of human studies on the association of blood cf-mtDNA and various real-life stressors. While cf-mtDNA exists in other biofluids, such as cerebrospinal fluid and urine (Varhaug et al., 2017; Kim et al., 2019a), the present systematic review and discussion focuses on blood cf-mtDNA. We then highlight outstanding questions for the field, particularly the popular notion that cf-mtDNA is pro-inflammatory, and discuss current knowledge about the nature and regulation of blood cf-mtDNA to help translational scientists in the design and interpretation of cf-mtDNA dynamics. Finally, we propose standardized procedures that will enhance biological specificity and reproducibility of human cf-mtDNA research.
It is an exciting time for mitochondrial psychobiology: over two decades of investigation into circulating cell-free DNA in various diseases now converge with emerging work on mitochondrial function and cf-mtDNA in psychiatric disorders. This convergence makes the study of cf-mtDNA in mental health a unique interdisciplinary opportunity to gain insight into the brain-mitochondria relationship and other aspect of human health.
3. Systematic review: psychopathology, stress, exercise and cf-mtDNA
We first performed a systematic review of human studies on the association of blood cf-mtDNA and various real-life stressors, namely acute psychological stress, psychopathology, exercise training, and acute physical activity. Here we compared the psychopathology and stress literature with the exercise literature, as it is another form of non-pathological stressor that relies heavily on mitochondrial respiration.
To examine cf-mtDNA levels in psychiatric disorders, acute psychological stress, and exercise, we performed a systematic review using keywords including: cell-free DNA, mtDNA, psychological stress, mental disorders, or exercise (complete search strategy described in Figs. S1 and S2). The search was independently performed by two authors (CT and VT) in Pubmed and Scopus, and was limited to studies published in English between 1960 and January 9, 2020. Study selection was performed according to PRISMA guidelines (Moher et al., 2009). Reasons for study rejection included: literature reviews, preclinical studies, did not measure cf-mtDNA, and other pre-specified criteria outlined in Supplemental Figs. S1 and S2. The studies included are listed and their methods and findings are summarized in Tables 1 and 2. The summary of effect sizes (hedges’g (g) and fold changes) is presented in Figs. 2 and 3.
Table 1.
Results from the systematic review of studies evaluating the effects of psychopathology and acute psychological stress on cf-mtdna levels.
| Study | Blood fraction | Sample processing | Likely source of mtDNA | Participants characteristics | Cf-mtDNA findings | Other findings |
|---|---|---|---|---|---|---|
| PSYCHOPATHOLOGY | ||||||
| ASD (Zhang et al., 2010b)b | Serum | Seruma was stored at − 80 °C until assayed. DNA was extracted (Qiagen DNA Micro extraction kit) and cf-mtDNA and cf-nDNA levels were measured by qPCR. | mtDNA released during clotting from platelets. Cf-mtDNA in whole mitochondria, large EVs, small EVs, and exosomes. | ASD patients (n = 20; 11 males and 3 females; mean age: 3.0 ± 0.4 years old) and HC (n = 12; 11 males and 1 female; mean age: 3 ± 1.2 years old). | ↑ cf-mtDNA (7.5-fold mt-CytB, 4-fold mt-7S) in ASD children vs HC. | cf-nDNA not detected. |
| BD (Stertz et al., 2015) | Serum | Blood allowed to clot, centrifuged at 2,000g for 10 min, serum extracted and kept frozen at − 80 °C. DNA was extracted using QIAmp DNA Mini Kit. Cf-mtDNA and cf-nDNA levels were measured by qPCR. | mtDNA released during clotting from platelets. Cf-mtDNA in whole mitochondria, large EVs, small EVs, and exosomes. | Drug-free BD patients during an acute mood episode (n = 20, 35 ± 13 years old, 60% women), healthy controls (n = 20, 35 ± 13 years old, 65% women), patients with sepsis (n = 20, 59 ± 13 years old, 20% women). | No significant difference between BD and HC (fold change = 1.01, hedges’g = 0.05 (95%CI; − 0.82, 0.93)), ↑ cf-mtDNA in BDvs sepsis (fold change = 1.19, hedges’g = 1.98 (95%CI; 0.91, 3.04)), ↑ cf-mtDNA in HC vs sepsis (fold change = 1.18, hedges’g = 2.01 (95%CI; 0.94, 3.09)). |
↑ cf-nDNA in BD vs HC (fold change = 1.08, hedges’g = 0.98). ↑ cf-nDNA in sepsis vs HC (fold change = 1.15, hedges’g = 2.14). HSP60: ↓ HSP60 in HC vs BD (fold change = − 1.51, hedges’g = − 0.21); ↓ HSP60 in HC vs sepsis (fold change = −3.48, hedges’g = − 0.46) HSP70: ↑ HSP70 in BD vs HC (fold change = 2.35, hedges’g = 0.48); ↓ HSP70 in HC than in sepsis (fold change = − 7.17, hedges’g = − 0.74). HSP90α: ↓ HSP90α in HC vs BD (fold change = − 2.35, hedges’g = − 1.73); ↓ HSP90α in HC vs sepsis (fold change = − 3.8, hedges’g = − 1.65). |
| Nonviolent suicide attempters (Lindqvist et al., 2016) | Plasma | EDTA tubes, blood centrifuged at 2,000g for 10 min, stored at − 70 °C. Plasma samples were thawed, centrifuged at 10,000g for 10 min, DNA extracted (QIAmp 96 DNA Blood Kit), qPCR used to measure cf-mtDNA levels. | Cf-mtDNA in small EVs, and exosomes. | Suicide attempters (n = 37, 39 ± 14 years old, 70% women) and HC (n = 37, 65% women, mean age: 38 ± 17 years old). | ↑ Elevated baseline cf-mtDNA levels in suicide attempters vs HC (fold change = 46.8, cohen’s d = 2.64). Post-DST treatment, ↑ elevated cf-mtDNA levels in suicide attempters vs controls: 8 h post-DST (fold change = 39.4, cohen’s d = 4.01) 15 h post-DST (fold change = 45.5, cohen’s d = 2.55). |
|
| MDD (Lindqvist et al., 2018) | Plasma | Same as (Lindqvist et al., 2016) | Cf-mtDNA in small EVs, and exosomes. | 50 unmedicated MDD (n = 50, 29.6 ± 14.7 years old, 54% women) & HC (n = 55, 37.6 ± 13.9 years old, 60% women). Repeated measures in 19 MDD after 8 weeks of SSRI treatment. | ↑ Elevated levels cf-mtDNA levels in MDD vs HC (fold change = 1.1, cohen’s d = 0.93 (95%CI; 0.47, 1.27)) After 8 weeks, SSRI responders had lower (4.8-fold lower) cf-mtDNA compared to SSRI non-responders (cohen’s d = −1.42 (95%CI; − 2.44, − 0.40) | |
| MDD, BD and SZ (Kageyama et al.,2017) | Plasma | Blood was immediately centrifuged after collection at 1,000g for 15 min to extract plasma and stored at − 80 °C. DNA was extracted using QIAamp DNA Blood Mini Kit. Cf-mtDNA was measured by qPCR. | Cf-mtDNA in whole mitochondria, large EVs, small EVs, and exosomes. | Patients with MDD (n = 109, 46.0 ± 15.9 years old, 53% women), BD (n = 28, 47.3 ± 14.9 years old, 48% women), SZ (n = 17, 33.6 ± 15.7 years old, 47% women) and HC (n = 29, 40.8 ± 14.4 years old). | ↓ cf-mtDNA levels in BD vs HC (fold change = 2.26, cohen’s d = −0.65 (95%CI; − 1.19, − 13)) ↓ cf-mtDNA levels in MDD patients than HC (fold change = 9.40, cohen’s d = −1.18 (95%CI; − 1.75, 0.63)). SZ did not differ from controls (fold change = 1.42, cohen’s d = −0.32 (95%CI; − 0.84, 0.19)). |
cf-mtDNA levels showed a positive correlation with GM-CSF, IL-2 and IL-4 in patients with MDD. No correlation was found with IL-6. |
| ASD (Tsilioni and Theoharides, 2018) | Serum | Blood was collected using serum separator vacutainer tubes, allowed to clot 15–30 min, centrifuged at 1,000–2,000g for 10 min at 4 °C, stored at − 80 °C. Samples were filtered with syringe filters to exclude particles>0.8 μm and isolate extracellular vesicules (EVs). Cf-mtDNA levels were measured by qPCR. | Extracellular Vesicles (some possibly released as a result of clotting). | Children with ASD (n = 20, 20% females, aged 4–12 years old) and HC (n = 8, 25% females, aged 4–12 years old). | ↑ cf-mtDNA levels in ASD vs HC (fold change = 1.26, cohen’s d = 0.92 (95% CI; 0.01, 1.84)). | Serum EVs (5 μg/mL) from ASD patients stimulated cultured human microglia to secrete higher proinflammatory cytokine interleukin IL-1β compared to EVs from HC. |
| ACUTE STRESS | ||||||
| Induced psychological stress (Hummel et al., 2018) A venous catheter was placed 45 min before the stress test, and blood was collected 2 min before and 2, 15, 30, and 40 min after the stress. | Plasma | Blood immediately acentrifuged at 1600g for 10 min, then at 16,000g for 10 min, then plasma was passed into a 0.8 μm filter. DNA was extracted with a QIAamp Circulating Nucleic Acid Kit. cf-mtDNA and cf-nDNA levels were measured by qPCR. | Small extracellular vesicles, exosomes. | Healthy men (n = 20, aged 18–36, mean = 23.3 ± 3.8). | ↑ cf-mtDNA after 2 min (1.7-fold, cohen’s d = 0.5 (95%CI;–0.32,1.46)). | ↑ cf-nDNA after 2 min (1.8-fold, hedges’g = 1.27 (95%CI; 0.31,2.23)). |
| Induced psychological stress (Trumpff et al., 2019a) Blood was collected before, just after the stress, and after 30 min | Serum | Blood allowed to clot, centrifuged at 1,000g for 10 min, frozen at − 80 °C. Sample were thawed, centrifuged at 2000 × g for 5 min. DNA was extracted by proteinase K and ethanol precipitation. cf-mtDNA and cf-nDNA levels were measured by qPCR. | Large EVs, Whole mitochondria, small EVs, exosomes. | Healthy participants (n = 50, 40% women, aged 41–58 years old). | ↑ cf-mtDNA 30 min after stress (2–3 fold, cohen’s d range = 0.85–1.23) at the two sessions separated by 1 month. Effect sizes were stronger in men than women. | No stress induced increase in cf-nDNA. |
Abbreviations: ASD, autism spectrum disorder; BD, bipolar disorder; CI, confidence interval, EVs, extracellular vesicles; GM-CSF, Granulocyte-macrophage colony-stimulating factor; HC, Healthy controls; MDD, Major Depressive Disorders, SZ, schizophrenia.
Centrifugation protocol not described.
Raw data not available for this study to compute effect sizes.
Table 2.
Results From the Systematic Review of Studies Evaluating the Effects of Exercise training and Acute Physical Activity on cf-mtDNA levels.
| Blood fraction | Sample processing | Likely source of cf- mthNA | Participants characteristics | cf-mthNA findings | Other findings | |
|---|---|---|---|---|---|---|
| EXERCISE TRAINING | ||||||
| Regular physical activity (Nasi et al., 2016) | Plasma | Fasting blood draw with EDTA tubes, blood was then centrifuged at 2,850g for 15 min. Cf-mtDNA levels were measured by qPCR. | Exercise sample: male volleyball players over two seasons (n = 12, aged 20–35, mean: 27.5). Control sample: age-matched non-athlete males. (n = 20, aged 19.– 34, mean age: 28). Blood samples were collected from the athlete group during the preseason (T1) and in-season at three timepoints (T2, T3, and T4). Blood samples were collected at baseline in HC. | Baseline cf-mtDNA levels were lower in the control sample than the exercise sample. During season 1, cf-mtDNA decreased from baseline to the end of the first season (fold change = −10.9, cohen’s d = −1.20(95% CI; − 2.43, − 0.03)). During season 2, cf-mtDNA stayed lower than baseline but did not decreased further from the beginning (T1) to the end (T4) (fold change = 1.02, cohen’s d = −0.02 (95% CI; − 0.8, 0.8)). |
||
| ACUTE PHYSICAL ACTIVITY | ||||||
| 30 min exhaustive treadmill test (Beiter et al., 2011) | Plasma | Blood collected with EDTA tubes was centrifuged at 1,600g for 10 min, then at 16,000g for 5 min. DNA was extracted using QIAamp DNA Blood Mini Kit, cf-mtDNA and cf-nDNA measured by qPCR. | Small extracellular vesicles, exosomes, free mtDNA. Potentially some mitochondria given the short centrifugation at 16,000g. Note: blood was drawn immediately after exercise, which may not have captured full change in cf-mtDNA. | Athletes (n = 9 men, mean age: 29.3 ± 8.5). | Immediately after: fold change = 1.2, cohen’s d = 0.29 (95% CI; − 1.03, 1.6) 30 min after: fold change = −1.3, cohen’s d = −0.39 (95% CI; − 1.32, 0.55). | ↑cf-nDNA by 14-fold immediately after and 4.2-fold after 30 min. |
| Incremental treadmill exercise test until exhaustion (Helmig et al., 2015) Blood samples were collected before, immediately after and 3+, 5+, 10+, 30+, and 90+ min after the test as well as after recovery. | Plasma | Blood was centrifuged at 1,600g for 10 min. Plasma was centrifuged at 10,000g for 30 min. Supernatant was filtered through 0.2 μm syringe filters. The supernatant fluid was centrifuged at 100,000g for 2 h. The samples were stored at − 20 °C and DNA was extracted using a QIAamp DNA Blood Mini Kit, cf-mtDNA and cf-nDNA levels were measured by qPCR. | Whole mitochondria, large and small extracellular vesicles, exosomes, free DNA. | Physically active men. (n = 5, mean age: 26.8 ± 2.2). | No difference in cf-mtDNA over time. Post: fold change = −1.1, cohen’s d = −0.13 (95% CI; − 1.4, 1.1) 10+: fold change = 1.2, cohen’s d = 0.28 (95% CI; − 1.0, 1.5) 30+: fold change = −1.1, cohen’s d = −0.12 (95% CI; − 1.4, 1.1) 90+: fold change = 1.2, cohen’s d = 0.24 (95% CI; − 1.0, 1.5). | ↑ in cf-nDNA: Post: fold change = 7.9, cohen’s d = 4.09 (95% CI; 1.9, 6.3) 10+: fold change = 6.9, cohen’s d = 4.51 (95% CI; 2.2, 6.8) 30+: fold change = 2.5, cohen’s d = 2.52 (95% CI; 0.9, 4.2) 90+: fold change = −1.42, cohen’s d = −0.88 (95% CI; − 2.2, 0.4). |
| Treadmill exercise for 90 min at 60% VO2 max (Shockett, Khanal et al. 2016)a Blood samples were collected before, during (at + 18 and + 54 min), immediately after (+90 min), and after recovery from the test. | Plasma | Blood collected with EDTA tubes, centrifugation at 800g for 5 min to extract plasma, second centrifugation at 2,600g for 5–7 min. DNA was extracted using a QIAamp DNA Blood Mini Kit, cf-mtDNA and cf-nDNA levels were measured by qPCR. | Large extracellular vesicles, whole mitochondria, small extracellular vesicles, exosomes, free mtDNA. Note: Decrease in cf-mtDNA suggests two possibilities – either cf-mtDNA is absorbed into cells or is degraded in the blood stream. | Healthy moderately trained young men (n = 7, mean age: 22.4 ± 1.2). | ↓ cf-mtDNA levels reduced during exercise at + 54 and + 90 vs control condition. At + 54 in exercise sample, cf-mDNA levels were 47.5% of baseline. At + 90 in exercise sample, cf-mDNA levels were 61.02% of baseline. | ↓ cf-mDNA were accompanied by increased lactate and followed by an increase in IL-6. |
| Repeated bouts of exhaustive treadmill exercise (Stawski, Walczak et al. 2017) Participants were asked to run on a treadmill to 70% of their VO2 max volume on three separate occasions. Blood samples were taken before and after each bout of exercise. | Plasma | Blood extracted in EDTA tubes was centrifuged at 1,600g for 10 min to extract plasma, second centrifugation at 16,000g for 5 min. Samples were stored at − 80 °C. DNA was extracted using QIAamp DNA Blood Mini Kit, cf-mtDNA and cf-nDNA measured by qPCR. | Small extracellular vesicles, exosomes, free mtDNA. Potentially some mitochondria given the short centrifugation at 16,000g. Note: blood was drawn immediately after exercise, which may not have captured full change in cf-mtDNA. |
Healthy men (n = 11, aged 25–45, mean 34.0 ± 5.2). | ↑ Elevated cf-mtDNA after each bout of exercise, but only a significant increase after 2nd and 3rd bout. 1st bout: fold change = 1.29, cohen’s d = 0.39 (95% CI; 0.46,1.22). 2nd bout: fold change = 1.96, cohen’s d = 0.99 (95% CI; 0.11, 1.88). 3rd bout: fold change = 2.62, cohen’s d = 1.20 (95% CI; 0.3, 2.11). Pre-exercise mt-DNA baseline decreased with each bout of exercise: From 1st to 2nd bout: fold change = −1.5, cohen’s d = −0.79 (95% CI; −0.93, 0.50). From 2nd bout to 3rd bout: fold change = −1.3, cohen’s d = −0.44 (95% CI; − 1.64, 0.75). from 1st to 3rd bout: fold change = −2.0, cohen’s d = −1.05 (95% CI; − 0.24, 2.27). |
↑ cf-nDNA with each bout of exercise. 1st bout: fold change = 11.3, cohen’s d = 2.43 (95% CI; 0.81, 2.79). 2nd bout: fold change = 11.8, cohen’s d = 3.01 (95% CI; 1.29, 3.47). 3rd bout: fold change = 17.3, cohen’s d = 2.44 (95% CI; 0.81, 2.79). |
| Treadmill Exercise (Hummel et al., 2018) A venous catheter was placed 45 min before the test, and blood was collected 2 min before and 2, 15, 30, and 40 min after the test. | Plasma | Blood immediately centrifuged at 1,600g for 10 min, then at 16,000 g for 10 min, then plasma was passed into a 0.8 μm filter. DNA was extracted with a QIAamp Circulating Nucleic Acid Kit. Cf-mtDNA and cf-nDNA measured by qPCR. | Healthy men (n = 20, aged 18–36, mean = 23.3 ± 3.8). | ↑ cf-mtDNA after 2 (1.6- fold, cohen’s d = 0.66 (95% CI; − 0.24,1.56)) and 15 min (1.2-fold, cohen’s d = 0.30 (95% CI;−0.58,1.18)). | ↑ cf-nDNA at 2 min (4- fold, cohen’s d = 2.36 (95% CI;1.21,3.50)), peaking at 15 min (5- fold cohen’s d = 2.62 (95% CI;1.43, 3.82)). | |
Abbreviations: CI, confidence interval, HC, healthy controls, EVs, extracellular vesicles.
Raw data not available for this study to compute effect sizes.
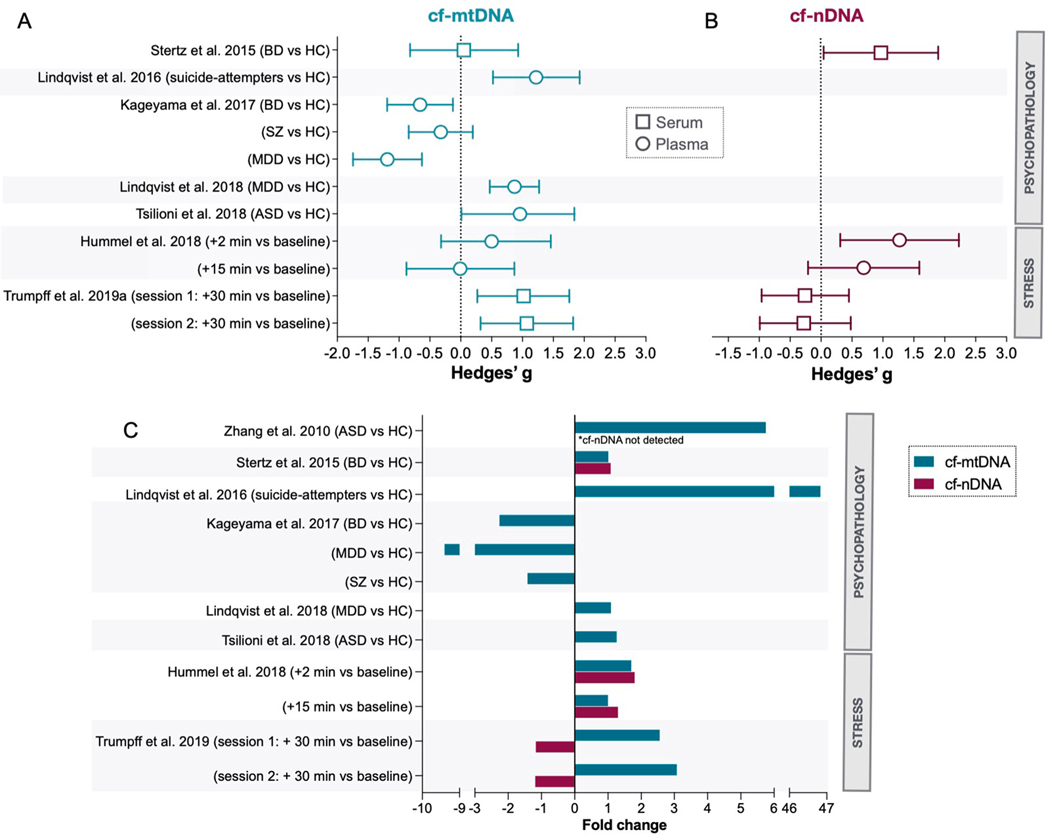
Fig. 2. Systematic review of human studies evaluating the effects of psychopathology and acute psychological stress on cfmtDNA levels.
(A) Summary of effects sizes (Hedges’g, 95% confidence intervals) for cf-mtDNA and (B) cf-nDNA. Circles are used for plasma and squares for serum. Results from Zhang et al. (Zhang et al., 2010b) are not displayed in this figure. (C) Magnitude of group differences or acute responses in cf-mtDNA and cf-nDNA represented as fold changes for psychological stress. Note that not all studies measured cf-nDNA. Abbreviations: ASD, autism spectrum disorder; BD, bipolar disorder; HC, healthy controls; MDD, major depressive disorders; SZ, schizophrenia.
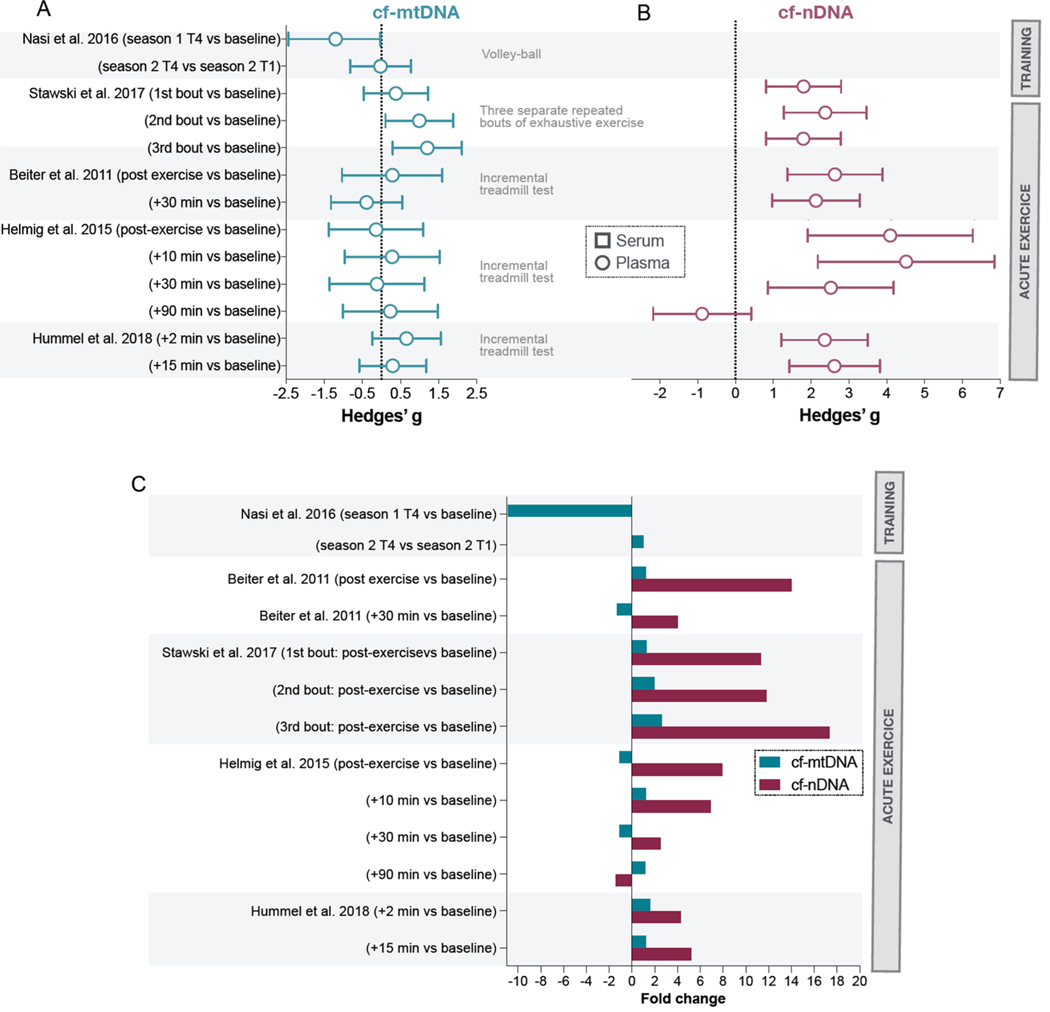
Fig. 3. Systematic review of studies evaluating the effects of exercise training and acute physical activity on cf-mtDNA levels.
(A) Summary of effects sizes (Hedges’g, 95% confidence intervals) in response to exercise for cf-mtDNA levels and (B) cf-nDNA levels. (C) Magnitude of group differences or acute responses in cf-mtDNA and cf-nDNA to exercise stress represented as fold changes. Results from Schockett et al. (2016) are not displayed in these figures.
3.1. Psychopathology and psychological stress
3.1.1. Psychopathology
Eight studies measured cf-mtDNA levels in individuals with psychiatric disorders. Of these, two were conducted in pediatric populations and six in adults. In children, Zhang et al. found that compared to neurotypical children (n = 12), children with autism spectrum disorder (ASD) (n = 20) had 5.7-fold higher serum cf-mtDNA concentrations (Zhang et al., 2010b). Unfortunately, the lack of information on the centrifugation protocol used (speed and duration) in this study limits its interpretability (see Table 1). In this study, cf-nDNA was not detected. Another study reported 1.3-fold higher cf-mtDNA in ASD, specifically from serum-derived extracellular vesicles (EVs) (Tsilioni and Theoharides, 2018). While both studies suggest that blood cf-mtDNA might be elevated in ASD, one study measured cf-mtDNA in whole serum while the other study focused on EVs extracted from serum, which make their findings not directly comparable. Because serum cf-mtDNA level is confounded by the presence of cf-mtDNA derived from coagulation and platelet activation (Boudreau et al., 2014) (see Section 6.1. below), additional confirmatory studies are warranted.
In adults, Lindqvist et al. found that suicide attempters with various psychiatric diagnoses – mood disorders being the most common – (n = 37) had significantly higher plasma cf-mtDNA concentrations (d = 2.64, fold change = 46.8) when compared to healthy controls (n = 37) (Lindqvist et al., 2016). Higher cf-mtDNA levels in suicide attempters correlated with a higher cortisol response to the dexamethasone suppression test (DST), suggesting a link between hypothalamic pituitary adrenal (HPA) axis hyperactivity and cf-mtDNA levels. In 2018, the same group found that patients with major depressive disorders (MDD) (n = 50) had higher plasma cf-mtDNA (d = 0.93, fold change = 1.1) compared to healthy controls (n = 55) (Lindqvist et al., 2018). Additionally, in a small subset of MDD participants who were treated with a selective serotonin reuptake inhibitor (SSRI) (n = 19), SSRI responders had lower cf-mtDNA levels compared to non-responders after 8 weeks of treatment (d = 1.59, fold change = 0.21), though this only reached trend-level significance. Of note, this study reported no significant differences between MDD subjects and healthy controls for PBMC mtDNA copy number, underscoring that cf-mtDNA may represent a more sensitive marker of MDD pathophysiology and of the mechanisms underlying treatment responsiveness. The conclusions around cf-mtDNA from these two studies are strengthened by the use of plasma (see Section 6.1 below) and by an additional centrifugation step ensuring complete removal of cellular debris (see Table 1).
While studies by Lindqvist et al. are in line with the hypothesis of elevated amounts of blood cf-mtDNA in psychiatric disorders, other research groups have found no associations between cf-mtDNA and psychopathology. Stertz et al., found that compared to healthy controls (n = 20), patients with bipolar disorder (BD) (n = 20) did not differ in serum cf-mtDNA concentration (g = 0.05, fold change = 1.01, not statistically significant (ns)) (Stertz et al., 2015). Surprisingly, BD patients (g = 1.98, fold change = 1.19) and healthy controls (g = 2.01, fold change = 1.18) showed higher serum cf-mtDNA concentrations compared to a group of patients with sepsis (n = 20). This result raises questions because prior studies have consistently found higher plasma cf-mtDNA concentrations in patients with sepsis compared to controls (e.g., (Kung et al., 2012; Yamanouchi et al., 2013; Nakahira et al., 2013)). The discrepancy may come from specific characteristics of the control population, the use of serum, or from the cf-mtDNA isolation protocol, which lacked a secondary centrifugation step usually required to remove cellular contaminants that may artificially inflate mtDNA content (see Section 6 below for more details). Another study by Kageyama et al. compared plasma cf-mtDNA concentrations in individuals with MDD (n = 109), BD (n = 28), schizophrenia (SZ) (n = 17), and healthy controls (n = 29) (Kageyama et al., 2017). Compared to healthy controls, MDD (d = −1.35, fold change = −9.40) and BD patients (d = −1.22, fold change = − 2.26) presented lower cf-mtDNA concentration (Kageyama et al., 2017). Patients with SZ did not differ significantly from healthy controls (d = −0.33, fold change = −1.42, ns) (Kageyama et al., 2017). Here, cf-mtDNA was quantified following a single centrifugation spin at 1,000 g for 15 min. Therefore, compared to studies that used a second faster centrifugation step, results may include more cf-mtDNA derived from cellular debris and large platelets, capturing a greater range of cf-mtDNA species and reducing its biological specificity.
In summary, the psychopathology cf-mtDNA literature provides mixed results. This may in part be driven by technical differences including different centrifugation protocols that yield different forms of transport of cf-mtDNA and levels of platelet contamination. In small pediatric studies, there is limited evidence of increased serum cf-mtDNA in pediatric cases of ASD (Zhang et al., 2010b; Tsilioni and Theoharides, 2018). In contrast, larger studies from the adult psychopathology literature suggests moderate-to-large elevations of cf-mtDNA in MDD and suicide attempters (Lindqvist et al., 2018, 2016c). But the highly heterogeneous blood processing methodology precludes a direct comparison among most studies. Further, many studies had small sample sizes, which may overestimate effect sizes. Larger epidemiological studies with comparable cf-DNA isolation procedures (discussed below in Section 6) are needed to confirm increased cf-mtDNA levels in different clinical populations. We also note that all but one study (Lindqvist et al., 2018) collected blood and measured serum or plasma cf-mtDNA at one timepoint, precluding any conclusions regarding the stability of potential group differences. In addition, it is not possible to rule out potential publication bias (i.e., studies with negative results not being published) that may limit the number of negative findings.
3.1.2. Acute psychological stress
Two studies identified through the search query directly tested the hypothesis that acute psychological stress triggers cf-mtDNA release (see Fig. 2 and Table 1) (Hummel et al., 2018; Trumpff et al., 2019a). These studies were experimental laboratory studies that used a repeated-measures design to quantify cf-mtDNA levels before and after socio-evaluative stress. Hummel et al. found that a 10–15 min psychosocial stress task (the Trier Social Stress Test (TSST) (Kirschbaum et al., 1993) in healthy young men (n = 20) caused a 1.6-fold increase in plasma cf-mtDNA two minutes after the task was performed (d = 0.5) (Hummel et al., 2018). In line with these findings, Trumpff et al. found in a sample of middle aged women and men (n = 53) that a brief psychosocial stress task (5 min) induced a 2–3 fold increase in serum cf-mtDNA within 30 min (d = 0.85–1.23) (Trumpff et al., 2019a). In this study, stress-induced cf-mtDNA release was measured on two occasions separated by one month, yielding similar results on both occasions. A follow up analysis showed that while there is some degree of stability in cf-mtDNA responses at the group level, some individuals exhibited large differences in cf-mtDNA responses between sessions, which were largely predicted by baseline cardiovascular and mood measures (Trumpff et al., 2019b). These two studies, one in plasma and one in serum, showed rapid increases in cf-mtDNA following acute psychosocial stress. However, they did not include a non-stressed control group, which is a limitation for future studies to address.
Although they differed in their pre-analytical protocol and blood fraction used, a shared strength of these studies is that both circulating nuclear DNA (cf-nDNA) and cf-mtDNA were measured in parallel (using duplex qPCR). Synchronous release of both nuclear and mitochondrial genomes could indicate cell death or damage, whereas cf-mtDNA release without cf-nDNA indicates a selective, and possibly regulated, process. The study in plasma observed a parallel 2-fold cf-nDNA increase immediately after stress (Hummel et al., 2018), whereas the study in serum found no concomitant cf-nDNA release (Trumpff et al., 2019a). An common strength of these studies is the use of a second centrifugation spin to limit cf-mtDNA derived from cellular debris and platelets, therefore increasing the biological specificity of their results.
To summarize, while these findings need to be replicated, they suggest that acute psychological stress alone can lead to a robust increase in cf-mtDNA in both plasma and serum. The exact kinetics for stress-induced cf-mtDNA increase and its association with stress hormones remain to be determined. In Hummel et al. (Hummel et al., 2018), plasma cf-mtDNA levels peaked 12–17 min after the onset of the psychological stress. In Trumpff et al., serum cf-mtDNA concentration was only measured 35 min after the onset of the psychological stress (Trumpff et al., 2019a). Repeated quantification of cf-mtDNA throughout the stress response is necessary to more closely establish the true kinetics of blood cf-mtDNA.
3.2. Exercise
A total of six studies, summarized below, met the inclusion criteria of measuring cf-mtDNA in response to exercise training or acute physical activity. Table 2 summarizes the study design, method and findings and Fig. 3 presents a summary of effect sizes of the included studies. All studies examined were conducted on plasma samples, and only three measured both cf-mtDNA and cf-nDNA.
3.2.1. Exercise training
Nasi et al. measured changes in plasma cf-mtDNA concentration every two months over two athletic seasons (8 timepoints) in professional volleyball players (n = 12) and compared them to baseline cf-mtDNA concentrations of controls (n = 20) (Nasi et al., 2016). Except for baseline measures during the first season, control subjects had significantly higher cf-mtDNA concentrations compared to the athlete group at all time points. The volleyball players’ cf-mtDNA concentrations decreased significantly during the first season while remaining stable during the second season. This suggests regular physical activity may be associated with lower levels of circulating mtDNA. The authors found no correlation between cf-mtDNA levels and complete blood count parameters, electrolyte balance, iron metabolism and cortisol and testosterone levels but because of their small sample size, they might not have been powered (n = 12) to look at these questions.
3.2.2. Acute physical activity
Shockett et al. found that in moderately-trained men (n = 7), aerobic exercise decreased plasma cf-mtDNA concentrations at 54 and 90 min post-exercise (cf-nDNA was not measured) (Shockett et al., 2016). Four studies assessed the effects of exhaustive exercise on cf-mtDNA levels. Beiter et al. studied male athletes (n = 9) and found that a 30-minute treadmill test did not change plasma cf-mtDNA concentrations, but did increase cf-nDNA by 14-fold immediately and 4.2-fold 30 min after exercise (Beiter et al., 2011). In line with these findings, Helmig et al. studied a small group of men (n = 5) and found that an incremental treadmill test did not significantly change cf-mtDNA levels measured just after, as well as 10, 30 and 90 min later, while cf-nDNA was increased by 7.9 fold post-exercise (Helmig et al., 2015). In contrast, Stawski et al. studied a group of healthy men (n = 11) and found that three repeated bouts of exhaustive exercise increased both cf-mtDNA (1.3–2.6-fold, d = 0.39–2.11) and cf-nDNA levels (11.3–17.3-fold, d 2.43–3.01) just after the task, with the increase in cf-nDNA levels being on average 7 times larger than the increase in cf-mtDNA (Stawski et al., 2017). Similarly, Hummel et al. found that 15 min of an incremental treadmill test in 20 healthy men increased cf-mtDNA by 1.6-fold after 2 min post-exercise (d = 0.66), while cf-nDNA was increased by 4-fold (Hummel et al., 2018).
In summary, one study found that regular exercise training is associated with lower cf-mtDNA levels and another study found reduced cf-mtDNA levels one hour after moderate exercise (Shockett et al., 2016). Some – the ones with larger sample size – but not all studies, suggest that cf-mtDNA increases directly after exhaustive exercise. Three studies showed that exercise decreased cf-mtDNA levels acutely (Nasi et al., 2016; Shockett et al., 2016; Stawski et al., 2017) and one suggests that regular training may be associated with lower cf-mtDNA over time (Nasi et al., 2016), suggesting that exercise training may reduce baseline circulating cf-mtDNA levels. Importantly, most of the existing studies have small sample size and present major differences in the blood processing for DNA isolation and the timing of the blood sampling (see Table 2). While the effect of acute exercise on cf-mtDNA levels is mixed, all studies that measured cf-nDNA found that exhaustive exercise provokes a large (up to 10-fold) increase in cf-nDNA, with larger effects when measured directly after the task (Beiter et al., 2011; Helmig et al., 2015; Stawski et al., 2017). These results are in line with previous findings showing cf-nDNA increases following incremental exercise testing (i.e., increasing intensity over time) (Breitbach et al., 2014), high-intensity interval training (Ferrandi et al., 2018), marathon running (Atamaniuk et al., 2004), ultramarathon running (Atamaniuk et al., 2008), and rowing (Velders et al., 2014). The observed difference between cf-mtDNA and cf-nDNA may be due to different release and clearing mechanisms for nDNA and mtDNA (see Section 5.6).
3.3. Conclusions of the systematic review
Overall, this systematic review reveals three major knowledge gaps: First, cf-mtDNA can be rapidly released into the blood in response to stimuli, but its kinetics and mechanisms of regulation remain poorly defined. Second, some studies suggest that cf-mtDNA may be chronically elevated in populations with psychiatric morbidity, but this should be confirmed in adequately powered future studies incorporating standardized blood processing procedures. Third, cf-mtDNA detection methods currently used are highly heterogeneous and many are inconsistent with the known biological properties of cf-mtDNA, highlighting the need to harmonize protocols across cf-mtDNA psychopathology research. Steps needed towards protocol unification include the systematic assessment of both cf-mtDNA and cf-nDNA and, more importantly, standardized method of blood processing. Studies with larger sample sizes are needed to increase the robustness of the findings, and to evaluate the influence of potential moderators and confounding variables. Below we address physiological and technical considerations that provide an empirical foundation to address these important knowledge gaps.
4. Physiological considerations
4.1. Sex differences in cf-mtDNA concentration
A significant sex difference was reported in cf-mtDNA release in response to psychological stress, with middle aged men exhibiting 2.1-fold larger stress-induced responses than women (Trumpff et al., 2019a). Importantly, existing reports on the effect of acute exercise on cf-mtDNA did not include women (see Table 2). Future studies should assess the effect of acute exercise in both women and men. Indeed, there are sex differences in energy metabolism (Mittelstrass et al., 2011); and studies of various aspects of mitochondrial biology in model organisms suggest that mitochondria in males exhibit functional differences that increase vulnerability to apoptotic signaling and reactive oxygen species, as well as decreased mitochondrial content and ATP production (Ventura-Clapier et al., 2017). No study that we know of has so far systematically examined human sex differences in cf-mtDNA levels nor the potential effect of sex hormones (e.g., estrogen, testosterone) on cf-mtDNA levels. Therefore, whether baseline or stress-induced cf-mtDNA levels differ by sex or gender remains undetermined.
4.2. Inter and intra-individual variation
A crucial aspect to establishing the utility of cf-mtDNA as a biomarker of health and disease is to understand its stability within individuals. In two studies that assessed baseline cf-mtDNA levels in the same participants on two different occasions, we calculated inter-individual and intra-individual variation (Hummel et al., 2018; Trumpff et al., 2019a). In plasma, on two different occasions over 2 days apart, the coefficient of variation (C.V.) of baseline cf-mtDNA inter-individual variation was 71.5%, while the range of intra-individual variation was 62.0%, indicating that the magnitude of change over time in the same person is comparable to the difference between different people (Fig. 4A). In serum cf-mtDNA measured on two different occasions over 1 month apart, the C.V. of inter-individual variation was larger (110.5%) than the range of intra-individual variation (34.0%), suggesting moderate stability of cf-mtDNA levels. However, the within-person variation over time was still substantial; cf-mtDNA in some individuals increased or decreased several folds, up to an order of magnitude, over this period (Fig. 4B). While further studies are needed to define cf-mtDNA inter- and intra-individual variation, these initial results suggest large intra-individual cf-mtDNA variation within days to months. Moreover, how the same individual’s cf-mtDNA levels react to psychological stress (i.e., one’s reactivity) also changes from one occasion of testing to another (Trumpff et al., 2019b), highlighting the potentially changeable nature of cf-mtDNA regulation. Together, these findings emphasize the need to characterize the magnitude of intra-individual variation over time – over both short and long time scales – and to define physiological factors responsible for these variations.
Fig. 4. Analysis of inter- and intra- individual variation in cf-mtDNA levels.
(A) Results from inter- and intra-individual variability in baseline cf-mtDNA levels quantified by the coefficient of variation (C.V.) of fold change in plasma (Hummel et al., 2019), and (B) in serum (Trumpff et al., 2019a). Each line represents a healthy participant sampled twice (visits 1 and 2). Note that cf-mtDNA can vary widely (i.e., more than double, in some case ± 80%) across both visits, suggesting that cf-mtDNA exhibits state properties. (C) Diagram representing known kinetics of trait and state variables typically assessed in clinical and psychobiological studies. Current evidence suggests that cfmtDNA levels vary within minutes to hours in healthy individuals. (D) Hypothetical intra-individual variation in cf-mtDNA across a normal day, highlighting the absence of information about potential diurnal variation and minimal information about the influence of acute psychosocial stress on cf-mtDNA dynamics. (E) Illustrative example of three occasions of sampling between two subjects exhibiting normal variation, resulting in three qualitatively different conclusions. As for other neuroendocrine factors (e.g., cortisol), mapping intra-individual cf-mtDNA dynamics and repeated-measures study designs may be required to achieve robust estimates of inter-individual differences.
4.3. Sampling time and frequency
As noted in the systematic review, cf-mtDNA levels can significantly change within 15–30 min in response to acute psychological or exercise stress (Hummel et al., 2018; Trumpff et al., 2019a). Combined with the substantial within-person variation described above, evidence of rapid reactivity in cf-mtDNA levels suggests that cf-mtDNA is more likely to represent a state marker than a stable individual trait (Fig. 4C). Such cf-mtDNA temporal dynamics position cf-mtDNA in the same category as cortisol, which exhibits stress-inducible characteristics (Kirschbaum et al., 1993). We note that cortisol and other neuroendocrine factors also exhibit diurnal variation (Krieger, 1972; Fukushima et al., 1970; Rose et al., 1972), but that no study has yet examined potential diurnal variation in cf-mtDNA (Fig. 4D). Knowledge on the diurnal variation of cf-mtDNA appears critical to properly design studies (e.g., avoid potential awakening response as is recommended for cortisol (Stalder et al., 2016; Kirschbaum and Hellhammer, 1989)) to investigate how acute and chronic factors influence cf-mtDNA in people.
Previous reports suggest that the upper and lower boundaries within which cf-mtDNA varies within-person might be influenced by chronic biobehavioral factors such as regular physical activity, age (Pinti et al., 2014; Zhang et al., 2020a), chronic disease, or psychopathology (Boyapati et al., 2017; Lindqvist et al., 2016, 2018; Nasi et al., 2016) (Fig. 4E). As intra-individual variation in cf-mtDNA appears large, studies designed to detect inter-individual and group differences might require particularly large sample sizes to achieve a sufficient signal-to-noise ratio. The use of repeated-measures designs (within the same individual) can also be leveraged to provide more stable individual-level estimates, as for other neuroendocrine measurements (Segerstrom and Miller, 2004). Overall, the major questions around cf-mtDNA dynamics that remain to be addressed include the temporal stability/variability of cf-mtDNA, whether it exhibits diurnal variation, and to what extent it is influenced by behavioral (diet, physical activity, sleep) or psychosocial factors.
5. Outstanding questions about cf-mtDNA biological regulation and signaling
While a detailed exploration of molecular mechanisms underlying cf-mtDNA regulation is beyond the primary focus of this review, it is important for the translational scientist to have some fundamental knowledge of the various potential forms of cf-mtDNA, and of the available evidence of its functional implications. Potential functions of cf-mtDNA involve the prominent topic of inflammation and some emerging physiological behavior of extracellular, cell-free mitochondria, such as transcellular (i.e., cell-to-cell) mitochondrial transfer. While scientists can generate high-quality reproducible data without these additional considerations, the sections that follow provide useful context for the interpretation of cf-mtDNA results.
5.1. Biological forms of cf-mtDNA
Understanding the form of mtDNA that is detected in the plasma is important to establish both the source of cf-mtDNA and the potential downstream physiological effects (see Fig. 1A,B) (Miliotis et al., 2019). Regarding the transport of cf-mtDNA in blood, the compartmentalization of cf-DNA associated with health, disease, or acute stress is a major knowledge gap. It is not clear if the cf-mtDNA that is detected in specific blood preparations consist in naked DNA, DNA encapsulated in lipid-based vesicular structures, in whole mitochondria, or in larger containers. Most research aiming to understand the physiological effects of cf-mtDNA involves using purified mtDNA, which include DNA isolated from enriched mitochondria, PCR products, or DNA isolated from plasma (e.g., (Hu et al., 2019). However, as stated above, the vast majority of naturally occurring physiological forms of cf-mtDNA may exist inside lipid-based vesicles (Al Amir Dache et al., 2020; Stephens et al., 2020; Helmig et al., 2015; Chiu et al., 2003).
In plasma, mtDNA can be packaged in vesicles with vastly different in size, including apoptotic bodies, platelets, microvesicles, or exosomes (Boudreau et al., 2014; Jiang et al., 2017; Kahlert et al., 2014; Cai et al., 2013). The DNA structure of the mtDNA in vesicular structures could be intact or fragmented, and could be oxidized or not (Al Amir Dache et al., 2020; Song et al., 2020; Chiu et al., 2003; Zhang et al., 2020a; Ma et al., 2019). The downstream actions of cf-mtDNA could also be modified by associated proteins, but this has not been thoroughly investigated. Compared to naked DNA that is directly accessible to degradation, mtDNA encapsulated in particles are physically protected from circulating DNAses, which may prolong their presence in the circulation. Further studies are required to determine which forms of cf-mtDNA are most prevalent, pro-inflammatory, inducible, and most relevant to specific psychopathological and physiological stressors.
5.2. Effects on target cells: Is cf-mtDNA pro-inflammatory?
Cf-mtDNA detected in patients has been assumed to have a pro-inflammatory function in humans based on four main lines of evidence: i) the correlation between cf-mtDNA, septic shock, and mortality in intensive care unit (ICU) patients; ii) experimental administration of naked (i.e, purified) mtDNA inducing inflammation in vivo; iii) immune cell co-stimulation experiments of bona-fide pro-inflammatory molecules with added synthetic or purified naked mtDNA in vitro, and iv) innate immune gene-expression responses to intracellular mtDNA signaling within the cell cytoplasm (see Fig. 5). However, each of these lines of evidence assumes that cf-mtDNA is accessible to pro-inflammatory receptors, such as TLR9, to trigger inflammatory responses. If cf-mtDNA is packaged in lipid-bound vesicles or whole mitochondria, cf-mtDNA would be accessible by intracellular TLR9 receptors only after endocytosis.
Fig. 5. Intracellular and extracellular mtDNA release in relation to pro-inflammatory signaling.
(A) Existing data from experiments on the pro-inflammatory effects of cf-mtDNA have used laboratory DNA extraction methods to remove the membrane and proteins and obtain purified mtDNA, or synthetic (PCR-amplified) oligomers. Therefore, isolated mtDNA using conventional techniques is not biologically equivalent to cf-mtDNA in human circulation. Left: depicted are the most prominent forms of cf-mtDNA in human plasma, which are either whole mitochondria or membrane-encapsulated forms of cf-mtDNA. Additional work is necessary to establish their immunogenic potential. Right: purified or synthetic forms of mtDNA used for in vivo and in vitro adjuvant and co-stimulation experiments, which can act as a DAMPs under certain conditions. Beyond their potential pro-inflammatory effects, physiological forms of cf-mtDNA and cell-free mitochondria could play other signaling roles that remains to be elucidated Section 5. (B) Intracellular release of mtDNA in the cytoplasm cause increased expression of interferon-related pro-inflammatory gene expression through known pathways (see (West and Shadel, 2017) and (Riley and Tait, 2020) for in depth reviews). (C) In humans, the association between physiological forms of cf-mtDNA and inflammation is correlational, and causality has not been established. Studies by Zhang et al. (Zhang et al., 2010a); Pinti et al. (Pinti et al., 2014), Nasi et al. (Nasi et al., 2020) and Kim et al. (Kim et al., 2020) show that purified/naked mtDNA can potentiate the pro-inflammatory effects of bona fide immune agonists like lipopolysaccharide (LPS) and N-Formylmethionyl-leucyl-phenylalanine (fMLF), but found that on its own, extracellular cf-mtDNA is not sufficient to induce pro-inflammatory gene expression or cytokine release.
We also note that in healthy individuals, total blood cf-mtDNA is abundant even in the absence of systemic inflammation (Al Amir Dache et al., 2020; Song et al., 2020; unpublished data). By recent estimates (Al Amir Dache et al., 2020; Stephens et al., 2020), there are between 200,000 and 3.7 million intact, cell-free mitochondria circulating per ml of plasma in healthy women and men, which could represent >90% of total cf-mtDNA in healthy individuals. While a subset of cf-mtDNA may be inflammatory, the fact that cf-mtDNA is a normal blood constituent suggests other potential physiological roles for extracellular DNA.
Below we evaluate these four lines of evidence previously advanced to support the popular notion that cf-mtDNA is pro-inflammatory. An analysis of the underlying data suggests that not all forms of cf-mtDNA are, by themselves, pro-inflammatory, and that more research is needed to support this hypothesis.
5.2.1. Correlational evidence in the ICU
In hospitalized critically ill patients with acute respiratory distress syndrome (ARDS) or sepsis (a systemic inflammatory condition), as noted above, elevated plasma cf-mtDNA levels are prospectively associated with a 4–8-fold increased risk of mortality (Nakahira et al., 2013). Another study in patients with ARDS found that cf-mtDNA was a significant predictor of mortality, and that addition of cf-mtDNA to traditional prognostic indicators significantly improved the prediction accuracy for mortality (Scozzi et al., 2020). A study in ICU patients also found that elevated cf-mtDNA levels was a stronger predictor of mortality in patients with elevated toll-like receptor-9 (TLR-9) expression (Krychtiuk et al., 2015), consistent with the idea that the mortality risk linked to cf-mtDNA levels might be associated with TLR-9 signaling. Together these studies suggest a potential link but not causality between cf-mtDNA and inflammatory pathologies.
Some studies have also reported associations between cf-mtDNA levels and canonical inflammatory markers. In a study of patients with rheumatoid arthritis, the presence of cf-mtDNA in synovial fluid was significantly correlated with the presence of rheumatoid factor (Hajizadeh et al., 2003). A study of patients on hemodialysis also showed that individuals in the higher tertile of cf-mtDNA levels had elevated concentrations of the inflammatory markers CRP and TNF-α compared to those in the lower tertile (Kim et al., 2020). However, these analyses were not adjusted for covariates nor for other factors (e.g., multiorgan failure and/or tissue damage) that could explain why sick individuals have elevated levels of both cf-mtDNA and cytokines, without a direct link between them. Similarly, a study of patients with COVID-19 found cf-mtDNA was positively correlated with the following cytokines: IL-6 (r = 0.39), MIG (r = 0.31), MCP-1 (r = 0.25), IP-10 (r = 0.25), IL-1RA (r = 0.43), IL-2R (r = 0.28), and HGF (r = 0.47) (Scozzi et al., 2020). In healthy adults, we have found that correlations between serum cf-mtDNA and IL-6 at three time points ranged from r = −0.19 to r = 0.13 (mean r = 0.01, all ns) (unpublished data from (Trumpff et al., 2019a)). In relation to psychopathology, one study reported small positive correlations between cf-mtDNA and the cytokines GM-CSF (r 0.23), IL-2 (r = 0.22) and IL-4 (r = 0.38), but not with IL-6 (r = 0.03, ns) (Kageyama et al., 2017). As mentioned above, the plasma in this latter study was likely heavily driven by platelets (due to centrifugation at low g-forces) and should be considered with caution. Overall, these correlations are weak to moderate and indicate that cf-mtDNA may account for only a small fraction of the variance in cytokine levels. We note that these estimates also do not account for potential publication bias (Easterbrook et al., 1991), which likely favors the reporting of positive cf-mtDNA-cytokine associations.
Among studies of life-threatening inflammatory conditions often cited as indication of the pro-inflammatory effect of cf-mtDNA, multiple co-morbid and pharmacological confounds naturally exist, complicating any conclusions around the physiologic role of cf-mtDNA. ICU patients are studied in an unique setting where patients are frequently given large doses of medications that disrupt physiological functions and mitochondrial respiratory chain activity (Mitsui et al., 2002), and their circadian regulation is often perturbed by the lack of light cues and meals (Mundigler et al., 2002; Gazendam et al., 2013). Hospitalized patients can naturally experience high levels of psychological distress from the uncertainty and the life-threatening nature of their medical condition (Griffiths et al., 2007). As discussed above, psychological stress alone can induce cf-mtDNA release, and can also trigger pro-inflammatory states (Marsland et al., 2017). Therefore, it is not possible to separate the relative contribution of biological injury from the effects of psychological distress in the elevation in cf-mtDNA levels in the ICU. Moreover, the possibility that both cf-mtDNA and inflammatory markers are independently induced by a third stress-related factor cannot be ruled out, and ICU-based clinical studies only provide correlational evidence linking inflammation with cf-mtDNA. Moreover, the effect size estimates indicate that if the naturally-occurring forms of cf-mtDNA were to be pro-inflammatory, it would account for only a small fraction of systemic inflammation levels in humans. Thus, the evidence of the pro-inflammatory nature of cf-mtDNA in clinical studies is currently not well-supported.
5.2.2. In vivo experiments in animals
One of the earliest studies in vivo showed that intra-articular injection of PCR-amplified mtDNA induced arthritis and TNF-α secretion in mice (Collins et al., 2004). Towards understanding the downstream role of mtDNA release after LPS-triggered release, intraperitoneal injection of mtDNA similarly caused acute lung injury and provoked systemic inflammation, which was attenuated by TLR9 inhibitors (Zhang et al., 2016). Another experiment in rats found that mitochondrial lysates triggered inflammation when injected into the circulation (Zhang et al., 2010b). By contrast, injection of supra-physiological levels of mtDNA and mitochondrial DAMPs did not induce proteinuria or kidney injury in rodents (He et al., 2015). These experiments used either mitochondrial homogenates, where mtDNA and other immunogenic components such as cardiolipin and N-formylated peptides are present, or naked synthetic DNA (Grazioli and Pugin, 2018).
When not associated with proteins nor encapsulated in vesicles, mtDNA, particularly when oxidized (Zhong et al., 2018), unambiguously acts as damage-associated molecular pattern (DAMPs) and cause sterile inflammation (Gong et al., 2020; Krysko et al., 2011). But as described above, the accessibility of DNA to receptors may be critical for determining whether the DNA can activate a signaling pathway. Even in experimental settings where inflammation is apparently triggered in a TLR9-dependent manner (implicating DNA as the ligand and primary stimulus) in the mouse heart, the inflammatory response was not affected by perfusion with DNA-degrading enzyme (DNase) (Kitazume-Taneike et al., 2019). While that data has been interpreted to indicate that extracellular cf-mtDNA does not play a role in TLR9 activation, an equally likely explanation is that the cf-DNA is encapsulated and protected from the action of DNAse. More work is greatly needed to better define the differentiating factors among these results, so caution is warranted in the interpretation of these experiments in relation to circulating cf-mtDNA in humans.
Overall, much remains to be understood about how the nature of cf-mtDNA influences its downstream functions. Among in vivo studies, there are differences between purified PCR fragments and native cf-mtDNA particles (Fig. 5A). Whether mtDNA is purified to be devoid of lipid and protein or not, the route of administration (i.e., intraperitoneal, intravenous, intratracheal), fragmentation, oxidation, and co-stimulation may be significant modifiers of inflammatory potential. Likewise, the metrics for inflammation (systemic vs tissue cytokine production, immune cell counts, which tissues are tested for damage, etc.) have not proven to be consistent between studies. For these reasons, experimental in vivo evidence using synthetic DNA or mtDNA from mitochondrial lysates are unlikely to reflect the nature and physiological signaling effects of naturally occurring cf-mtDNA in humans.
5.2.3. In vitro co-stimulation experiments
In vitro studies have shown that extracellular mtDNA potentiates, but does not independently induce, cellular inflammatory responses in cultured cells. In a study using human serum, EVs from ASD pediatric patients stimulated human microglia to secrete higher IL-1β levels than EVs from healthy controls (Tsilioni and Theoharides, 2018). However, the heterogeneity of cargo within EVs (they include mtDNA but also other cargos) does not mechanistically isolate the influence of cf-mtDNA relative to other potential pro-inflammatory cargos. Several studies have found that purified mtDNA and the potent pro-inflammatory agonists lipopolysaccharide (LPS) or N-Formylmethionyl-leucyl-phenylalanine (fMLF) trigger pro-inflammatory responses measured by either TNF-α gene expression or cytokine (e.g., IL-6, IL-8) release (Zhang et al., 2010b; Pinti et al., 2014). In a LPS co-stimulation study in monocytes, purified cf-mtDNA isolated from HepG2 cells potentiated the production of pro-inflammatory cytokines TNF-α by 1.4-fold (Pinti et al., 2014). However, mtDNA by itself did not by itself trigger TNF-α gene expression or cytokine production (Pinti et al., 2014). In another study on polymorphonuclear leukocytes, mtDNA combined with the pro-inflammatory agonist fMLF significantly induced IL-8 by 4-fold relative to fMLF alone (Zhang et al., 2010b). Synthetic mtDNA alone was not sufficient to induce robust IL-8 expression (Zhang et al., 2010b). In bone marrow-derived macrophages, purified or synthetic mtDNA combined with LPS also potentiated TNF-α by 1.1-fold and IL-6 by 2.7-fold compared to LPS alone (Kim et al., 2020). And in human microglia cells (HMC3), mtDNA alone also did not upregulate pro-inflammatory gene expression or cytokine production (Nasi et al., 2020).
Thus, although some forms of isolated DNA can induce cytokines in some circumstances (Hu et al., 2019), available extracellular co-stimulation studies indicate that naked extracellular mtDNA (not membrane encapsulated) potentiates inflammation, but it may not be sufficient by itself to provoke cytokine production.
5.2.4. Intracellular mtDNA signaling of innate immune responses
A number of in vitro studies have demonstrated that mtDNA released into the cytosol triggers the innate immune system through the intracellular DNA-sensing systems including cGAS (West et al., 2015), TLR9, AIM2, and NLRP3 (reviewed in (West and Shadel, 2017; Riley and Tait, 2020; Liu et al., 2018)). For example, cytosolic mtDNA activates the inflammasome and pro-inflammatory cytokine release in monocytes and macrophages (Nakahira et al., 2011; Shimada et al., 2012). In fibroblasts and cancer cells, genotoxin-induced cytosolic mtDNA release also triggers interferon-stimulated gene expression (Wu et al., 2019; Rai et al., 2021). And in skeletal muscle, mitochondrial instability triggers NF-kB and proinflammatory gene expression in a TLR-9 and mtDNA-dependent manner (Rodríguez-Nuevo et al., 2018). Activation of the classical NLRP3 inflammasome pathway even appears to require mtDNA synthesis (Zhong et al., 2018), illustrating the potential evolutionary overlap between mtDNA signaling and innate immune activation within the cell. While these data provide unambiguous evidence of a pro-inflammatory signaling function of mtDNA in the cytosol of various cell types (Fig. 5B), extruded cytosolic mtDNA is a distinct biological entity than extracellular, blood-based forms of cf-mtDNA that also acts in a different biological context (Fig. 5C). Therefore, it provides little direct evidence about the potential pro-inflammatory effects of blood cf-mtDNA.
5.2.5. Summary of evidence for pro-inflammatory function of cf-mtDNA
In conclusion, the current dogma that cf-mtDNA is ubiquitously pro-inflammatory appears mostly derived from i) the misinterpretation of correlational clinical studies, ii) how exogenously applied extracellular mtDNA potentiates, but does not appear sufficient to trigger, inflammation, and iii) experimental studies demonstrating how cytosolic (i.e., from inside the cell) mtDNA triggers antiviral signaling. Thus, the hypothesis that physiologically-occurring forms of blood cf-mtDNA in humans is obligatorily pro-inflammatory should be re-evaluated, and other possible functions of cf-mtDNA should be considered.
5.2.6. Possible functions of cf-mtDNA beyond inflammation
Extracellular cf-mtDNA could have different functions within the organism. For example, one study suggested that extruded mtDNA has antibacterial effects: in response to bacterial exposure with IL-5 and LPS adjuvants, eosinophils from the gastrointestinal tract actively release mtDNA that form extracellular structures able to bind and kill bacteria, therefore acting as an antibacterial defense system (Yousefi et al., 2008). Alternatively, cell-free whole mitochondria released from traumatized mouse brains can also stimulate platelet activation (Zhao et al., 2020), reflecting non-canonical effects of mtDNA and whole mitochondria on target circulating cells.
Growing evidence also suggests that whole mitochondria are transferred between cells. This phenomenon may serve two main purposes. The first purpose is the clearance of dysfunctional mitochondria. For example, functional neurons can release and transfer damaged whole mitochondria to neighboring astrocytes for either recycling or clearance (Chung-ha et al., 2014), effectively outsourcing the degradation of dysfunctional mitochondria. Cardiomyocytes also extrude dysfunctional mitochondria out of the cell in small membrane-bound structures called exophers, which are absorbed and degraded by resident macrophages, contributing to maintaining cardiac homeostasis (Nicolás-Ávila et al., 2020).
The second purpose of cell-to-cell mitochondrial transfer appears related to communication and/or bioenergetic rescue. Whole mitochondria are transferred between cells in vivo, including between bone marrow-derived stromal cells, pulmonary alveoli, and breast cancer cells where they appear to contribute to energetics (Sansone et al., 2017; Islam et al., 2012). In vitro, several cell types can extrude or uptake extracellular mitochondria, including endothelial progenitor cells (Hayakawa et al., 2018), astrocytes (Hayakawa et al., 2016), lymphocytes (Anker et al., 1975), neutrophils (Yousefi et al., 2009), eosinophils (Yousefi et al., 2008), platelets (Aucamp et al., 2018), and cardiomyocytes (Shin et al., 2017; Cowan et al., 2017) suggesting that mitochondrial transfer may serve a physiological function. Indeed, mammalian cells have the ability to internalize exogenous (i.e., purified or free-floating) extracellular mitochondria (Clark and Shay, 1982; Kitani et al., 2014; Caicedo et al., 2015), making it plausible that extracellular mitochondria could be exchanged between cells. Recently, isolated mitochondria have even been applied for therapeutic application in humans, being injected in the myocardium where this appeared to improve clinical outcomes (Emani et al., 2017). Collectively, this body of work has led to the emergence of the field of mitochondrial transplantation (Bertero et al., 2018).
Therefore, together with recent observations that cf-mtDNA may be largely contained within whole mitochondria circulating in human blood (Al Amir Dache et al., 2020; Song et al., 2020; Stephens et al.,2020); it is conceivable that cell-free mitochondria could be absorbed by cells via micropinocytosis, become resident within those cells and affect their phenotype (Miliotis et al., 2019). Until it becomes possible to directly manipulate different forms of cf-mtDNA in humans to study their cellular and physiological effects, mapping specific forms of transport of cf-mtDNA levels at high temporal resolution, in parallel with other biomarkers and health outcomes, will help to understand the functional significance of cf-mtDNA in humans.
5.3. Triggers of mtDNA release into the circulation
Several in vivo and in vitro studies have assessed potential triggers of cytosolic and extracellular mitochondrial release. In immune cells, mtDNA extrusion may be triggered by an increase in reactive oxygen species (ROS) production (Nakahira et al., 2011). mtDNA extrusion could occur through mitochondrial outer membrane permeabilization caused by the assembly of pores containing the protein voltage-dependent anion channel (VDAC), leading to the release of short mtDNA fragments (Kim et al., 2019b). During apoptotic cell death, cytoplasmic mtDNA release also occurs through mitochondrial herniation that requires the pore-forming proteins Bax and Bak (McArthur et al., 2018), indicating the existence of multiple mechanisms whereby whole or fragmented mtDNA molecules can translocate from the matrix to the cytoplasm. However, these mechanisms do not explain how whole mitochondria are released extracellularly, which represents an active area of research. Which forms of cf-mtDNA are released, by what mechanism, and in what cells, remains an important area of research.
To understand what triggers cf-mtDNA release in response to real world psychosocial and physical stress, it is also relevant to identify potential upstream systemic signals, neuroendocrine or metabolic, that could trigger the release of different forms of transport of cf-mtDNA. Two studies have investigated this question. In a study of 20 healthy men, there were no significant associations between stress- or exercise-induced changes cf-mtDNA levels and cortisol, while cf-mtDNA was associated with epinephrine 15 min after exercise (Hummel et al., 2018). Another study in human-cultured fibroblast tested the effect of the glucocorticoid receptor agonist dexamethasone, which triggered mtDNA extrusion into the cytosol within 30 min through an unknown process (Trumpff et al., 2019ba). Therefore, additional work is required to identify the triggers and mechanisms that connect, within minutes, subjective psychological states to the cf-mtDNA release into circulation.
5.4. Mechanisms of cf-mtDNA release
Generally, mtDNA can be released either through cellular death/breakdown processes (passive release) or from regulated processes in living cells (active release) (for a review see (Aucamp et al., 2018) (see Fig. 1C). “Passive” release mechanisms of mtDNA include apoptosis, an active programmed cell-death process of autonomous cellular dismantling generally not associated with inflammation, but which does produce apoptotic bodies that may enter the circulation. In contrast, necrosis is passive, accidental cell death accompanied by the uncontrolled release of inflammatory cellular components. Apoptosis is thought to be the main passive release source of plasma cf-mtDNA and cf-nDNA (Jahr et al., 2001; Rostami et al., 2020; Choi et al., 2005; Suzuki et al., 2008), although there is no direct evidence to rule out other contributing processes. While necrosis is a possible source of plasma cf-mtDNA and cf-nDNA in cancer (Jahr et al., 2001; Rostami et al., 2020), whether it contributes to circulating DNA in plasma from healthy subjects is unclear (Aucamp et al., 2018).
Under physiological conditions where cell death is not a major factor, cf-mtDNA may be derived from an active regulated process. As noted in Section 5.5, mitochondria are released extracellularly or transcellularly by a variety of cell types. Active release mechanisms secrete cf-mtDNA via microvesicles, mitochondria-derived vesicles, and neutrophil extracellular traps (NETs) (Hu et al., 2019; Boudreau et al., 2014; Yousefi et al., 2009). However, as noted above, the most abundant form of cf-mtDNA in plasma from healthy humans is possibly contained within mitochondria (Al Amir Dache et al., 2020; Song et al., 2020), for which the mechanisms of release remains undefined.
In relation to psychopathology and stress, given the: i) lack of mechanical stress or cellular injury associated with psychosocial challenges, ii) selective mtDNA release in response to psychosocial stress (Trumpff et al., 2019a), and iii) larger mtDNA release relative to nDNA in response to psychosocial stress compared to exercise stress (Hummel et al., 2018), it appears likely that psychosocial stress-induced cf-mtDNA is actively released. This point remains to be confirmed by future studies.
5.5. Cellular origin of blood cf-mtDNA
The cellular source(s) of physiological cf-mtDNA largely remain to be identified. A variety of cell types have the ability to release mitochondria or mtDNA under experimental conditions. This includes platelets (Boudreau et al., 2014), leukocytes (Thurairajah et al., 2018), endothelial cells (Hayakawa et al., 2018), astrocytes (Hayakawa et al., 2016), primary human fibroblasts (unpublished data), and immortalized cell lines (Al Amir Dache et al., 2020; Song et al., 2020; Hayakawa et al., 2018; Munakata et al., 2019; Stigliani et al., 2019; Lin et al., 2015; Kansaku et al., 2019). A recent study in healthy individuals found that sorted circulating cf-mtDNA show platelet (11%), endothelial (50%), and leukocyte (9%) cell-surface markers, suggesting that human cf-mtDNA may also be derived from bone marrow-derived cellular sources in vivo (Stephens et al., 2020). Unlike specific cargoes that are secreted by single cell types, such as insulin (from beta cells) and cortisol (adrenal cortex, zona fasciculata), cell-free mitochondria and cf-mtDNA are potentially released by multiple cell types.
Identifying the sources of cell-free mitochondria in humans is an important research goal as it will enable to determine what triggers cf-mtDNA release and to understand its function(s). It is also likely that cf-mtDNA exists as different forms of transport (naked, exosome, microvesicles, whole mitochondria, etc.) that have various origins and physiological functions. Furthermore, it appears crucial to understand whether stress-induced elevation in cf-mtDNA is due to increased cf-mtDNA secretion into the extracellular space, or due to changes in their uptake by target tissues, passive elimination, or active degradation that contribute to its removal.
5.6. Removal of cf-mtDNA
For the various potential forms of cf-mtDNA to be regulated, every trigger event needs to have a removal mechanism to restore homeostatic levels. For example, after challenges such as exercise or psychological stress, cf-mtDNA levels peak within 15 to 30 min and subsequently decrease (e.g., (Hummel et al., 2018; Beiter et al., 2011)) showing not only that cf-mtDNA is inducible, but also implying the existence of mechanisms to reduce cf-mtDNA levels. Based on the limited evidence discussed above, the time frame for cf-mtDNA removal is similar to its induction, and is similar to other inducible hormones, including cortisol and cytokines. Two major mechanisms could actively contribute to cf-mtDNA removal: i) circulating DNases could degrade cf-mtDNA in the blood; ii) cf-mtDNA could be uptaken or sequestered by target cells for use or removal.
DNases are produced by various organs that are secreted into blood, and other body fluids, to hydrolyze circulating DNA molecules that are later cleared by the liver and kidney (Velders et al., 2014; Saukkonen et al., 2008; Napirei et al., 2000; Keyel, 2017). DNases are either categorized as endonucleases that cleave nucleotides within the DNA or as exonucleases that cleave nucleotides from free 5′ or 3′ ends. Physiologically, DNases are triggered by i) apoptotic signals in response to infection (either bacterial or viral), ii) clearance of excess circulating RBCs during regular developmental processes (Hotz et al., 2018; Jacobson et al., 1997; Strasser et al., 2009), and by iii) high-intensity exercise (Velders et al., 2014; Kawane et al., 2014). Interestingly, almost two decades ago it was shown that fetal cf-DNA, which is foreign to the maternal plasma, is rapidly cleared from the circulation with a half-life of about 16 min by undefined plasma nucleases (Lo et al., 1999). While DNases within the mitochondria are mostly involved in mtDNA replication and repair (Kazak et al., 2013; Uhler and Falkenberg, 2015), it is unclear if they are stimulated once mtDNA escapes into the cytoplasm.
Differences in the magnitude and dynamics of circulating DNAs between different physical and psychological stressors could in part be related to differences in clearance dynamics. For example, some populations of cf-mtDNA – those in whole mitochondria – could be protected from the action of DNases by the mitochondrial membranes. In other cases, mtDNA, if released in complex with proteins like TFAM or as oxidized-mtDNA adduct (Lood et al., 2016; Caielli et al., 2016), may evade DNases due to inaccessibility of the DNA (Sisirak et al., 2016), exhibiting a longer half-life than non-membrane encapsulated DNA fragments.
Regarding non DNAse-related cf-mtDNA uptake or removal, membrane-associated or non-membrane-associated cf-mtDNA may also be eliminated via the liver, spleen, or kidney, as has been shown for other forms of circulating cf-DNAs (Yu et al., 2013; Gauthier et al., 1996; Botezatu et al., 2000). Elevated cf-mtDNA levels in urine, but not plasma, is an indicator of mitochondrial defects and kidney injury in septic patients (Hu et al., 2018), suggesting urine as a viable path of cf-mtDNA elimination from the circulation and contributor to overall plasma cf-mtDNA dynamics (see also Zhang et al., 2020b). While many studies have documented the kinetics of cf-DNA clearance in general, understanding the specific mechanisms of their removal will provide a more robust quantitative mapping of cf-mtDNA dynamics. For example, activation of removal systems could mask or diminish the apparent release of cf-mtDNA in circulation.
Moreover, the potential physiological uptake of cell-free mitochondria (and therefore cf-mtDNA, discussed above in Section 5.2.5) remains a possible source of removal from the circulation. Understanding the relative contribution of the rates of i) release and ii) removal on observed variation in specific forms of cf-mtDNA levels will help resolve its role in stress adaptation and psychobiological processes.
6. Technical considerations
The methodologies employed by previous studies differ in several significant ways, including the choice of biological sample type (serum vs. plasma), the timing and procedures for blood collection, as well as pre-extraction centrifugation conditions, DNA extraction methods, and qPCR protocols (Fig. 6A). However, as noted previously, pre-analytical methods, in particular the centrifugation conditions, have a large influence on cf-mtDNA levels and are crucial to ensure the biological specificity of the measurements (Meddeb et al., 2019; El Messaoudi et al., 2013; Chiu et al., 2003; Zhong et al., 2000). To promote more robust and reproducible results, we: i) have integrated methodological guidelines and experiences from the hematological and mitochondrial biology literatures, ii) recommend methodological guidelines to selectively isolate cf-mtDNA in its different forms, iii) propose methods to quantify both cell-free mitochondrial and nuclear genomes (cf-mtDNA and cf-nDNA) in the same samples, and iv) provide easy-to-follow minimum reporting criteria with a simple reporting checklist.
Fig. 6. Recommended protocol for cf-mtDNA quantification in human blood.
(A) Experimental steps to quantify cf-mtDNA and cf-nDNA. (B) Left: electron micrograph showing platelets isolated from a healthy individual’s plasma. Middle: magnified image of a single human platelet containing several visible mitochondria (mtDNA) but no nucleus (no nDNA). Right: Single platelet mitochondrion showing the characteristic electron-dense outer and inner membranes, and cristae. (C) Diagram illustrating how platelet abundance influences mtDNA and nDNA quantification and increases cf-mtDNA levels detected in human plasma. (D) Quantitative analysis of studies in the systematic review, showing reported copies of cf-mtDNA (copies per ml) relative to the maximal centrifugation conditions used in each study. Relationship between centrifugation time and speed with the amount of mtDNA detected in healthy subjects at baseline (log transformed plot), linear regression results not statistically significant. (E) Suggested minimum reporting guidelines and (F) standardized technical procedures to increase the robustness and reproducibility of cf-mtDNA research in humans. In addition to these steps, which yield total cf-mtDNA levels (multiple forms of transport), additional centrifugation steps can be applied to to the final clean plasma to isolate additional vehicular and potential non-vesicular forms of transport of cf-mtDNA.
6.1. Plasma versus serum
Serum is the liquid fraction of blood obtained after clotting, whereas plasma is obtained from anti-coagulated blood, and thus represents the liquid fraction of whole blood. If the researchers’ goal is to measure blood cf-mtDNA levels of the individuals being studied, the most appropriate approach is to rapidly process plasma, as it most directly reflects the circulating un-coagulated blood (Al Amir Dache et al., 2020). In contrast, serum collection requires a coagulation period (typically 30 to 45 min at room temperature) during which time platelets become activated and actively release respiratory competent mitochondria and mtDNA in the serum (Boudreau et al., 2014). In a direct plasma-serum comparison study of 20 individuals, serum cf-mtDNA and cf-nDNA were respectively 12-fold and 45-fold higher than plasma (Xia et al., 2009). Another study found serum cf-mtDNA values to be 2.6-fold higher than plasma (Rosa et al., 2020). Thus, rapidly processed plasma most directly reflects what is in a person’s bloodstream at the time of collection, whereas serum composition is altered by cellular activities during coagulation.
6.2. Platelet contamination and other vesicular bodies
Whether plasma or serum, the liquid fraction of blood is a complex biofluid that contains various membrane-bound structures, including platelets, microvesicles, and exosomes. This complex liquid fraction is separated from most blood cells by low-speed centrifugation at 300–1000 g for 5–15 min (1,500–4,500 g min) on freshly collected blood. During centrifugation, whole blood separates into three layers: red blood cells (bottom), the buffy coat (middle, containing most leukocytes), and the liquid fraction on top. This initial separation removes leukocytes, which harbor 200–600 mtDNA copies per cell (Rausser et al., 2020). However, excessive centrifugation speeds while cells are still in suspension in fresh blood can cause the shearing and release of genomic material from leukocytes (Enjeti et al., 2008), which would artificially inflate cf-mtDNA and cf-nDNA during preparation (Meddeb et al., 2019). Thus, the initial centrifugation of blood should be performed at <1,000 g.
This resulting plasma contains platelets and platelet-derived extracellular vesicles, which are a heterogeneous population of membranes with diverse morphologies, sizes, and contents (Gasecka et al., 2019). Because platelets and their vesicles can contain mitochondria and therefore large numbers of mtDNA copies (Fig. 6B and C), it will be important to determine what proportion of cf-mtDNA variability is attributable to variation in the size and number of these platelet microparticles.
6.3. cf-mtDNA isolation and protocol standardization
Among studies in our systematic review, the more centrifugation force (time and speed or g-force) was applied, the less cf-mtDNA was detected in the final samples (Fig. 6D). This inverse relationship is expected when cf-mtDNA is membrane encapsulated rather than non-membrane encapsulated. As larger structures, such as platelets, pellet under the action of centrifugal force, DNA concentrations in the final sample supernatant decrease with centrifugation force. The goal of differential centrifugation is to eliminate as much of the mtDNA contained within cells and platelets, while preserving the entire cell-free fraction.
Three bodies of literature define the sedimentation characteristics of blood components: platelet biology and hematology (Mustard et al., 1972), mitochondrial isolation protocols (Frezza et al., 2007), and extracellular vesicles (EV) isolation guidelines (Momen-Heravi, 2017; Mateescu et al., 2017). Platelets, which are smaller than leukocytes and easily activated (which can impair their sedimentation), pellet at ~2,200 g × 10–16 min (22,000–35,200 g × min) (Cazenave et al., 2004; Hechler et al., 2019), although platelets greatly vary in size and these parameters may not eliminate platelets of all sizes. Whole mitochondria pellet at ~10,000 g × 10 min (100,000 g × min) (Graham, 2001; Djafarzadeh and Jakob, 2017). Endothelial-derived microparticles as well as large intermediate-sized vesicular bodies are removed by centrifugation at ~10,000–20,000 g × 30 min (300,000 – 600,000 g × min) (Szatanek et al., 2015). And smaller vesicular bodies (exosomes) are removed at >100,000 g × 70 min (7,000,000 g × min) (Livshits et al., 2015).
Standardized conditions sensitive to sedimentation characteristics of clinically-meaningful blood components will help clarify these biologically and clinically-meaningful questions, as well as increase reproducibility. Because recent evidence suggests that >75–90% of cf-mtDNA is bound within membranes that include whole mitochondria (Song et al., 2020; Stephens et al., 2020; Zhang et al., 2020a), preserving the whole-mitochondria fraction with intermediate isolation conditions is desirable to capture the major physiological form of cf-mtDNA in healthy individuals.
For practical reasons, collected samples are generally frozen for storage prior to batch measurements of cf-mtDNA (and other analytes). But freezing plasma samples prior to centrifugation may damage the membranes of platelets and other potential vesicular bodies (El Messaoudi et al., 2013). The released materials that would have otherwise been removed during centrifugation remain in suspension with cell-free content. Freezing prior to processing may inflate cf-mtDNA levels by up to 8-fold when the thawed samples are centrifuged after (rather than before) freezing (Cox et al., 2020). Therefore, all depletion centrifugation should be performed on freshly collected plasma prior to freezing.
6.4. Recommended cf-mtDNA isolation procedures
Based on evidence reviewed above, we recommend the following procedure to quantify cf-mtDNA levels in humans. The recommended reporting guidelines and procedures are illustrated in Fig. 6E and F. They build but differ from previous recommendations (Meddeb et al., 2019) and consider the vast diversity of forms of transport for cf-mtDNA in human plasma. A detailed blood processing protocol, validated qPCR primers and probes, and a sample methods section with methodologies to standardize cf-mtDNA isolation and quantification across laboratories is available in the Supplemental Document of this article.
We recommend that anticoagulated blood should be immediately centrifuged after draw (within 1–2 min) at 1,000 g × 5 min (10,000 g × min) [first centrifugation]. This step eliminates whole cells and a good proportion of platelets, producing cell-free but platelet-rich plasma. Subsequently, about 80% of the plasma volume from the top fraction is transferred to a conical tube and centrifuged a second time at 5,000 g × 10 min (50,000 g × min) [second centrifugation]. This second step specifically reduces platelets and large vesicular bodies. The top 80% of the volume is then transferred to a new tube [clean plasma] and can be used for total cf-mtDNA quantification by qPCR or sequencing.
To gain additional information about which forms contain cf-mtDNA, additional differential centrifugation steps can further be applied to the supernatant to more finely discriminate among EV populations. For example, after an aliquot of the 50,000 g min plasma is obtained for total cf-mtDNA quantification, an additional centrifugation step at 18,000 g for 30 min will deplete whole mitochondria and some large EVs, leaving only smaller forms of transport including small microparticles and small EVs, which includes exosomes, and non-membrane bound cf-mtDNA. Subsequent (ultra)centrifugations (>100,000g) can also be applied to eliminate all membrane-bound forms of transport and focus exclusively on naked DNA, for example.
Once plasma is obtained using the desired isolation conditions, methods applied to isolate cf-DNA from plasma should be optimized for maximal quantitative recovery of cf-mtDNA and cf-nDNA. Because mtDNA and nDNA vary in their size and nucleoprotein compositions, they may not both be optimally extracted by certain DNA isolation methods. For an experimental analysis of optimal mtDNA isolation methods, see (Jorgez et al., 2006). To avoid bias in the extraction of either mtDNA or nDNA using standard DNA isolation kits (Guo et al., 2009; Fazzini et al., 2018), lysis based methods which can be combined with DNA retrieval methods (Kolesar et al., 2012) appear to be optimal. Optimized high-throughput methods have recently been reported that increase the efficiency of mtDNA recovery by 95-fold and nDNA recovery by 20-fold (Ware et al., 2020).
6.5. Quantification of cf-mtDNA and cf-nDNA
Two technical factors related to cf-mtDNA quantification can further increase the interpretability and robustness of the findings. To determine whether total cellular genomic material is indiscriminately released or if the mitochondrial genome is preferentially released, both cf-mtDNA and cf-nDNA may be measured in parallel. In most cases, both mtDNA and nDNA amplicons can be used in a duplex Taqman qPCR reaction for parallel quantification (Trumpff et al., 2019a). In sequencing-based methods, reads can also be mapped to both genomes. To increase measurement robustness, two different mtDNA amplicons (regions of the mitochondrial genome, such as the ND1 and COX1 genes) can also be quantified (e.g., (Trumpff et al., 2019a)) to increase the robustness of cf-mtDNA estimates. If the relative cf-mtDNA values for a specific sample, or the change over time or in response to challenges in a given individual based on both mtDNA amplicons are similar, it decreases the probability that sequence variations (e.g., single nucleotide polymorphisms in the mtDNA sequence) would impair amplification efficiency, or that the preferential release of sub-genomic regions would bias the results. When amplicons yield substantially different results (>10–20% divergence), it could theoretically reveal sequence variation or the release of specific mtDNA fragments.
Absolute cf-mtDNA concentration in plasma can be obtained using a reference standard curve validated by digital PCR (e.g., (Trumpff et al., 2019a)) or from an engineered reference plasmid DNA (Fazzini et al., 2018). For this analysis, it is important to use reference standards that can be applied across plates to account for batch effects, and that are sequence insensitive. We have used pooled DNA samples from multiple sources to dilute sequence divergence and extend the number of applications to which the standard can be used. Estimated cf-mtDNA levels should be computed in copies of mtDNA per unit of volume (e.g., copies per ml) of original plasma.
7. Summary
The presence of the mitochondrial genome passively and actively released in the bloodstream adds to the repertoire of mitochondrial signaling behavior. Because heterotypic stressors appear to change blood cf-mtDNA levels, including abnormal psychological states in psychopathology and socio-evaluative stress, cf-mtDNA must now be considered an emerging stress biomarker, which is certainly in need of validation in properly-controlled studies. Our current ignorance of the specific form(s) of transport of cf-mtDNA – in health, disease, and with acute stress – is an important knowledge gap. Likewise, we still know little about the potential triggers and mechanisms that connect, within minutes, subjective psychological states to the cf-mtDNA release into circulation.
In the absence of high temporal resolution data or of a mechanistic understanding of cf-mtDNA regulation, it has been assumed that cf-mtDNA is mostly a stable trait-like biomarker. However, our analysis (see Fig. 4A, B) shows large day-to-day and month-to-month changes in resting cf-mtDNA within individuals. Together with the inducibility of cf-mtDNA within minutes to hours, this positions cf-mtDNA as a state measure similar to other neuroendocrine stress mediators.
In relation to physiological significance, we conclude that the current notion that cf-mtDNA is pro-inflammatory is mostly derived from the misinterpretation of correlational clinical studies, as well as from experimental studies demonstrating how cytoplasmic (i.e., from inside the cell) mtDNA triggers antiviral signaling, and how exogenously applied purified forms of mtDNA potentiates but, in most cases, does not appear sufficient to systematically trigger inflammation. Thus, the hypothesis that physiological forms of cf-mtDNA in humans have pro-inflammatory effects must be re-evaluated and tested in relevant systems, using naturally occurring forms of cf-mtDNA. Other possible functions of cf-mtDNA should also be considered, including but not limited to the potential cell-to-cell transfer of cf-mtDNA and whole mitochondria (Miliotis et al., 2019).
In conclusion, based on the interdisciplinary body of work reviewed here, we propose that the most valuable insights into the role of cf-mtDNA in human psychobiology are likely to emerge from future studies that: i) quantify cf-mtDNA dynamics over multiple time points, thereby capturing true intra-individual variation from inter-individual and between-group differences, ii) adopt standard methods to isolate specific biological forms of plasma cf-mtDNA (whole mitochondria, EV-associated mtDNA, or naked DNA), which will increase the biological specificity and interpretability of findings, and iii) establish changes in cf-mtDNA in parallel with other well-known markers of normal physiological function and disease biomarkers, including hormones, cytokines, and metabolites. Elucidating the mechanisms and forms of transport of cf-mtDNA under different health states and (psycho)pathological conditions is an important, immediate challenge for the field. The implementation of standardized, robust, and scalable experimental procedures grounded in the principles of mitochondrial biology should produce new insights into the significance of cf-mtDNA for human health.
Supplementary Material
Acknowledgments
This work was supported by NIH grant Nos. GM119793 to MP, MH119336 to MP and BAK, the Wharton Fund and a NARSAD young investigator grant to CT. Daniel Lindqvist was supported by the Swedish Research Council (registration number 2020-01428) and the province of Scania (Sweden) state grants (ALF).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mito.2021.04.002.
References
- Al Amir Dache Z, Otandault A, Tanos R, et al. , 2020. Blood contains circulating cell-free respiratory competent mitochondria. FASEB J. 34, 3616–3630. [DOI] [PubMed] [Google Scholar]
- Anker P, Stroun M, Maurice PA, 1975. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 35 (9), 2375–2382. [PubMed] [Google Scholar]
- Atamaniuk J, Vidotto C, Tschan H, Bachl N, Stuhlmeier KM, Müller MM, 2004. Increased concentrations of cell-free plasma DNA after exhaustive exercise. Clin. Chem 50 (9), 1668–1670. [DOI] [PubMed] [Google Scholar]
- Atamaniuk J, Stuhlmeier KM, Vidotto C, Tschan H, Dossenbach-Glaninger A, Mueller MM, 2008. Effects of ultra-marathon on circulating DNA and mRNA expression of pro-and anti-apoptotic genes in mononuclear cells. Eur. J. Appl. Physiol 104 (4), 711–717. [DOI] [PubMed] [Google Scholar]
- Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ, 2018. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol. Rev. Camb. Philos. Soc 93 (3), 1649–1683. [DOI] [PubMed] [Google Scholar]
- Beiter T, Fragasso A, Hudemann J, Nieß AM, Simon P, 2011. Short-term treadmill running as a model for studying cell-free DNA kinetics in vivo. Clin. Chem 57 (4), 633–636. [DOI] [PubMed] [Google Scholar]
- Bertero E, Maack C, O’Rourke B, 2018. Mitochondrial transplantation in humans:“magical” cure or cause for concern? J. Clin. Investig 128 (12), 5191–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagirath VC, Dwivedi DJ, Liaw PC, 2015. Comparison of the Proinflammatory and Procoagulant Properties of Nuclear, Mitochondrial, and Bacterial DNA. Shock 44 (3), 265–271. [DOI] [PubMed] [Google Scholar]
- Boeck C, Koenig AM, Schury K, et al. , 2016. Inflammation in adult women with a history of child maltreatment: the involvement of mitochondrial alterations and oxidative stress. Mitochondrion 30, 197–207. [DOI] [PubMed] [Google Scholar]
- Boeck C, Gumpp AM, Calzia E, et al. , 2018. The association between cortisol, oxytocin, and immune cell mitochondrial oxygen consumption in postpartum women with childhood maltreatment. Psychoneuroendocrinology 96, 69–77. [DOI] [PubMed] [Google Scholar]
- Botezatu I, Og S, Potapova G, et al. , 2000. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin. Chem 46 (8), 1078–1084. [PubMed] [Google Scholar]
- Boudreau LH, Duchez AC, Cloutier N, et al. , 2014. Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A2 to promote inflammation. Blood 124 (14), 2173–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyapati RK, Tamborska A, Dorward DA, Ho G-T, 2017. Advances in the understanding of mitochondrial DNA as a pathogenic factor in inflammatory diseases. F1000. Research 6 (169). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbach S, Sterzing B, Magallanes C, Tug S, Simon P, 2014. Direct measurement of cell-free DNA from serially collected capillary plasma during incremental exercise. J. Appl. Physiol 117 (2), 119–130. [DOI] [PubMed] [Google Scholar]
- Cai N, Chang S, Li Y, et al. , 2015. Molecular signatures of major depression. Curr. Biol 25 (9), 1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Han Y, Ren H, et al. , 2013. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J. Mol. Cell. Biol 5 (4), 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo A, Fritz V, Brondello J-M, et al. , 2015. MitoCeption as a new tool to assess the effects of mesenchymal stem/stromal cell mitochondria on cancer cell metabolism and function. Sci. Rep 5, 9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caielli S, Athale S, Domic B, et al. , 2016. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J. Exp. Med jem. 20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardamone MD, Tanasa B, Cederquist CT, et al. Mitochondrial retrograde signaling in mammals is mediated by the transcriptional cofactor GPS2 via direct mitochondria-to-nucleus translocation. Molecular cell 2018; 69(5): 757–72. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazenave J-P, Ohlmann P, Cassel D, Eckly A, Hechler B, Gachet C. Preparation of washed platelet suspensions from human and rodent blood. Platelets and megakaryocytes: Springer; 2004: 13–28. [DOI] [PubMed] [Google Scholar]
- Cherix A, Larrieu T, Grosse J, et al. , 2020. Metabolic signature in nucleus accumbens for anti-depressant-like effects of acetyl-L-carnitine. Elife 9, e50631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RW, Chan LY, Lam NY, et al. , 2003. Quantitative analysis of circulating mitochondrial DNA in plasma. Clin. Chem 49 (5), 719–726. [DOI] [PubMed] [Google Scholar]
- Choi JJ, Reich CF III., Pisetsky DS, 2005. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology 115 (1), 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Pell VR, Gaude E, et al. , 2014. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515 (7527), 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung-ha OD, Kim K-Y, Bushong EA, et al. , 2014. Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci 111 (26), 9633–9638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA, Shay JW, 1982. Mitochondrial transformation of mammalian cells. Nature 295 (5850), 605–607. [DOI] [PubMed] [Google Scholar]
- Collins LV, Hajizadeh S, Holme E, Jonsson IM, Tarkowski A, 2004. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J. Leukoc. Biol 75 (6), 995–1000. [DOI] [PubMed] [Google Scholar]
- Cowan DB, Yao R, Thedsanamoorthy JK, Zurakowski D, Pedro J, McCully JD, 2017. Transit and integration of extracellular mitochondria in human heart cells. Sci. Rep 7 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR, Wong BK, Yang L, et al. High Speed Centrifugation Before Frozen Storage of Plasma Is Critical for Quantitative Analysis of Mitochondrial-Derived Cell-Free DNA. Clinical chemistry 2020: hvaa127. [DOI] [PubMed] [Google Scholar]
- Daniels TE, Olsen EM, Tyrka AR. Stress and Psychiatric Disorders: The Role of Mitochondria. Annual Review of Clinical Psychology 2020; 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Borowski LS, et al. , 2018. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560 (7717), 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djafarzadeh S, Jakob SM, 2017. Isolation of intact mitochondria from skeletal muscle by differential centrifugation for high-resolution respirometry measurements. JoVE (J. Visualized Experiments) 121, e55251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi DJ, Toltl LJ, Swystun LL, et al. , 2012. Prognostic utility and characterization of cell-free DNA in patients with severe sepsis. Crit Care 16 (4), R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterbrook PJ, Gopalan R, Berlin J, Matthews DR, 1991. Publication bias in clinical research. The Lancet 337 (8746), 867–872. [DOI] [PubMed] [Google Scholar]
- Eisner V, Lenaers G, Hajnoczky G, 2014. Mitochondrial fusion is frequent in skeletal muscle and supports excitation-contraction coupling. J. Cell. Biol 205 (2), 179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Messaoudi S, Rolet F, Mouliere F, Thierry AR, 2013. Circulating cell free DNA: preanalytical considerations. Clin. Chim. Acta 424, 222–230. [DOI] [PubMed] [Google Scholar]
- Emani SM, Piekarski BL, Harrild D, Pedro J, McCully JD, 2017. Autologous mitochondrial transplantation for dysfunction after ischemia-reperfusion injury. The Journal of Thoracic and Cardiovascular Surgery 154 (1), 286–289. [DOI] [PubMed] [Google Scholar]
- Enjeti AK, Lincz LF, Seldon M, 2008. In: Microparticles in health and disease. © Thieme Medical Publishers, 2008, pp. 683–691. [DOI] [PubMed] [Google Scholar]
- Farge G, Falkenberg M, 2019. Organization of DNA in mammalian mitochondria. Int. J. Mol. Sci 20 (11), 2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattal O, Link J, Quinn K, Cohen BH, Franco K, 2007. Psychiatric comorbidity in 36 adults with mitochondrial cytopathies. CNS Spectr. 12 (6), 429–438. [DOI] [PubMed] [Google Scholar]
- Fazzini F, Schöpf B, Blatzer M, et al. , 2018. Plasmid-normalized quantification of relative mitochondrial DNA copy number. Sci. Rep 8 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandi PJ, Fico BG, Whitehurst M, et al. , 2018. Acute high-intensity interval exercise induces comparable levels of circulating cell-free DNA and Interleukin-6 in obese and normal-weight individuals. Life Sci. 202, 161–166. [DOI] [PubMed] [Google Scholar]
- Filograna R, Mennuni M, Alsina D, Larsson NG, 2020. Mitochondrial DNA copy number in human disease: the more the better? FEBS Lett. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin Y-F, Rosin N, Pellegrino MW, Haynes CM, 2016. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol 26 (15), 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L, 2007. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured filroblasts. Nat. Protoc 2 (2), 287. [DOI] [PubMed] [Google Scholar]
- Fukushima DK, Bradlow HL, Hellman L, Gallagher TF, 1970. Cortisol metabolism in the morning and evening; relation to cortisol secretion rate measurements. Steroids 16 (5), 603–610. [DOI] [PubMed] [Google Scholar]
- Gasecka A, Nieuwland R, Siljander P-R-M, 2019. Platelet-derived extracellular vesicles. Platelets: Elsevier; 401–416. [Google Scholar]
- Gauthier VJ, Tyler LN, Mannik M, 1996. Blood clearance kinetics and liver uptake of mononucleosomes in mice. J. Immunol 156 (3), 1151–1156. [PubMed] [Google Scholar]
- Gazendam JA, Van Dongen HP, Grant DA, Freedman NS, Zwaveling JH, Schwab RJ, 2013. Altered circadian rhythmicity in patients in the ICU. Chest 144 (2), 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong T, Liu L, Jiang W, Zhou R, 2020. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol 20 (2), 95–112. [DOI] [PubMed] [Google Scholar]
- Graham JM, 2001. Isolation of Golgi membranes from tissues and cells by differential and density gradient centrifugation. Curr. Protocol. Cell Biol 10(1), 3.9. 1–3.9. 24. [DOI] [PubMed] [Google Scholar]
- Grazioli S, Pugin J, 2018. Mitochondrial damage-associated molecular patterns: from inflammatory signaling to human diseases. Front. Immunol 9, 832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Fortune G, Barber V, Young JD, 2007. The prevalence of post traumatic stress disorder in survivors of ICU treatment: a systematic review. Intensive Care Med. 33 (9), 1506–1518. [DOI] [PubMed] [Google Scholar]
- Gumpp AM, Boeck C, Behnke A, et al. , 2020. Childhood maltreatment is associated with changes in mitochondrial bioenergetics in maternal, but not in neonatal immune cells. Proc. Natl. Acad. Sci 117 (40), 24778–24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH, 2009. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion 9 (4), 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajizadeh S, DeGroot J, TeKoppele JM, Tarkowski A, Collins LV, 2003. Extracellular mitochondrial DNA and oxidatively damaged DNA in synovial fluid of patients with rheumatoid arthritis. Arthritis Res. Ther 5 (5), R234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K, Esposito E, Wang X, et al. , 2016. Transfer of mitochondria from astrocytes to neurons after stroke. Nature 535 (7613), 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K, Chan SJ, Mandeville ET, et al. , 2018. Protective Effects of Endothelial Progenitor Cell-Derived Extracellular Mitochondria in Brain Endothelium. Stem Cells 36 (9), 1404–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Lu Y, Xia H, et al. , 2015. Circulating mitochondrial DAMPs are not effective inducers of proteinuria and kidney injury in rodents. PLoS ONE 10 (4), e0124469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechler B, Dupuis A, Mangin PH, Gachet C, 2019. Platelet preparation for function testing in the laboratory and clinic: Historical and practical aspects. Res. Pract. Thrombos. Haemost 3 (4), 615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmig S, Frühbeis C, Krämer-Albers E-M, Simon P, Tug S, 2015. Release of bulk cell free DNA during physical exercise occurs independent of extracellular vesicles. Eur. J. Appl. Physiol 115 (11), 2271–2280. [DOI] [PubMed] [Google Scholar]
- Hollis F, van der Kooij MA, Zanoletti O, Lozano L, Cantó, C., Sandi, C., 2015. Mitochondrial function in the brain links anxiety with social subordination. Proc. Natl. Acad. Sci 112 (50), 15486–15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holper L, Ben-Shachar D, Mann J, 2019. Multivariate meta-analyses of mitochondrial complex I and IV in major depressive disorder, bipolar disorder, schizophrenia, Alzheimer disease, and Parkinson disease. Neuropsychopharmacology 44 (5), 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz MJ, Qing D, Shashaty MG, et al. , 2018. Red blood cells homeostatically bind mitochondrial DNA through TLR9 to maintain quiescence and to prevent lung injury. Am. J. Respir. Crit. Care Med 197 (4), 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Ren J, Ren H, et al. , 2018. Urinary mitochondrial DNA identifies renal dysfunction and mitochondrial damage in sepsis-induced acute kidney injury. Oxid. Med. Cell. Longev 2018, 8074936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Shi H, Zeng T, et al. , 2019. Increased neutrophil extracellular traps activate NLRP3 and inflammatory macrophages in adult-onset Still’s disease. Arthritis research & therapy 21 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel E, Hessas E, Müller S, et al. , 2018. Cell-free DNA release under psychosocial and physical stress conditions. Transl. Psychiatry 8 (1), 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Das SR, Emin MT, et al. , 2012. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat Med 18 (5), 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC, 1997. Programmed cell death in animal development. Cell 88 (3), 347–354. [DOI] [PubMed] [Google Scholar]
- Jahr S, Hentze H, Englisch S, et al. , 2001. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61 (4), 1659–1665. [PubMed] [Google Scholar]
- Jiang L, Paone S, Caruso S, et al. , 2017. Determining the contents and cell origins of apoptotic bodies by flow cytometry. Sci. Rep 7 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgez CJ, Dang DD, Simpson JL, Lewis DE, Bischoff FZ, 2006. Quantity versus quality: optimal methods for cell-free DNA isolation from plasma of pregnant women. Genet. Med 8 (10), 615–619. [DOI] [PubMed] [Google Scholar]
- Kageyama Y, Kasahara T, Kato M, et al. , 2017. The relationship between circulating mitochondrial DNA and inflammatory cytokines in patients with major depression. J. Affect. Disord [DOI] [PubMed] [Google Scholar]
- Kahlert C, Melo SA, Protopopov A, et al. , 2014. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem 289 (7), 3869–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansaku K, Munakata Y, Shirasuna K, Kuwayama T, Iwata H, 2019. Mitochondrial cell-free DNA secreted from porcine granulosa cells. Zygote 27 (5), 272–278. [DOI] [PubMed] [Google Scholar]
- Kasahara T, Kubota M, Miyauchi T, et al. , 2006. Mice with neuron-specific accumulation of mitochondrial DNA mutations show mood disorder-like phenotypes. Mol. Psychiatry 11 (6), 577–593. [DOI] [PubMed] [Google Scholar]
- Kawane K, Motani K, Nagata S, 2014. DNA degradation and its defects. Cold Spring Harbor Perspect. Biol 6 (6), a016394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Reyes A, Duncan AL, et al. , 2013. Alternative translation initiation augments the human mitochondrial proteome. Nucleic Acids Res. 41 (4), 2354–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyel PA, 2017. Dnases in health and disease. Dev. Biol 429 (1), 1–11. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Xiao J, Wan J, Cohen P, Yen K, 2017. Mitochondrially derived peptides as novel regulators of metabolism. J. Physiol 595 (21), 6613–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Son JM, Benayoun BA, Lee C. The mitochondrial-encoded peptide MOTS-c translocates to the nucleus to regulate nuclear gene expression in response to metabolic stress. Cell metabolism 2018; 28(3): 516–24. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Moon H, Lee YH, et al. , 2019a. Clinical relevance of cell-free mitochondrial DNA during the early postoperative period in kidney transplant recipients. Sci. Rep 9 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Gupta R, Blanco LP, et al. , 2019b. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 366 (6472), 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Jung SW, Cho W-H, et al. , 2020. Associations between Cell-Free Mitochondrial DNA and Inflammation, and Their Clinical Implications for Patients on Hemodialysis: A Prospective Multicenter Cohort Study. Blood Purif. 1–8. [DOI] [PubMed] [Google Scholar]
- Kirschbaum C, Hellhammer DH, 1989. Salivary cortisol in psychobiological research: an overview. Neuropsychobiology 22 (3), 150–169. [DOI] [PubMed] [Google Scholar]
- Kirschbaum C, Pirke K-M, Hellhammer DH, 1993. The ‘Trier Social Stress Test’–a tool for investigating psychobiological stress responses in a laboratory setting. Neuropsychobiology 28 (1–2), 76–81. [DOI] [PubMed] [Google Scholar]
- Kitani T, Kami D, Matoba S, Gojo S, 2014. Internalization of isolated functional mitochondria: involvement of macropinocytosis. J. Cell Mol. Med 18 (8), 1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazume-Taneike R, Taneike M, Omiya S, et al. , 2019. Ablation of Toll-like receptor 9 attenuates myocardial ischemia/reperfusion injury in mice. Biochem. Biophys. Res. Commun 515 (3), 442–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesar JE, Wang CY, Taguchi YV, Chou S-H, Kaufman BA, 2012. Two-dimensional intact mitochondrial DNA agarose electrophoresis reveals the structural complexity of the mammalian mitochondrial genome. Nucleic Acids Res. 41 (4), e58-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger DT, 1972. Circadian corticosteroid periodicity: critical period for abolition by neonatal injection of corticosteroid. Science 178 (4066), 1205–1207. [DOI] [PubMed] [Google Scholar]
- Krychtiuk KA, Ruhittel S, Hohensinner PJ, et al. , 2015. Mitochondrial DNA and toll-like receptor-9 are associated with mortality in critically ill patients. Crit. Care Med 43 (12), 2633–2641. [DOI] [PubMed] [Google Scholar]
- Krysko DV, Agostinis P, Krysko O, et al. , 2011. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 32 (4), 157–164. [DOI] [PubMed] [Google Scholar]
- Kung C-T, Hsiao S-Y, Tsai T-C, et al. , 2012. Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room. Journal of translational medicine 10 (1), 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam NY, Rainer TH, Chiu RW, Joynt GM, Lo YD, 2004. Plasma mitochondrial DNA concentrations after trauma. Clin. Chem 50 (1), 213–216. [DOI] [PubMed] [Google Scholar]
- Lazo S, Noren Hooten N, Green J, et al. , 2020. Mitochondrial DNA in extracellular vesicles declines with age. Aging Cell, e13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Han J, 2017. Mitochondrial nucleoid: shield and switch of the mitochondrial genome. Oxid. Med. Cell. Longevity 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Zeng J, Drew BG, et al. , 2015. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 21 (3), 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hann HW, Wan S, et al. , 2016. Cell-free circulating mitochondrial DNA content and risk of hepatocellular carcinoma in patients with chronic HBV infection. Sci. Rep 6, 23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-Y, Liou C-W, Chen S-D, et al. , 2015. Mitochondrial transfer from Wharton’s jelly-derived mesenchymal stem cells to mitochondria-defective cells recaptures impaired mitochondrial function. Mitochondrion 22, 31–44. [DOI] [PubMed] [Google Scholar]
- Lindqvist D, Fernström J, Grudet C, et al. , 2016. Increased plasma levels of circulating cell-free mitochondrial DNA in suicide attempters: associations with HPA-axis hyperactivity. Transl. Psychiatry 6 (12), e971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist D, Wolkowitz OM, Picard M, et al. , 2018. Circulating cell-free mitochondrial DNA, but not leukocyte mitochondrial DNA copy number, is elevated in major depressive disorder. Neuropsychopharmacology 43 (7), 1557–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang D, Hu D, Zhou X, Zhou Y, 2018. The role of mitochondria in NLRP3 inflammasome activation. Mol. Immunol 103, 115–124. [DOI] [PubMed] [Google Scholar]
- Livshits MA, Khomyakova E, Evtushenko EG, et al. , 2015. Isolation of exosomes by differential centrifugation: theoretical analysis of a commonly used protocol. Sci. Rep 5, 17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YD, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM, 1999. Rapid clearance of fetal DNA from maternal plasma. The American Journal of Human Genetics 64 (1), 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lood C, Blanco LP, Purmalek MM, et al. , 2016. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med 22 (2), 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozoya OA, Martinez-Reyes I, Wang T, et al. , 2018. Mitochondrial nicotinamide adenine dinucleotide reduced (NADH) oxidation links the tricarboxylic acid (TCA) cycle with methionine metabolism and nuclear DNA methylation. PLoS Biol. 16 (4), e2005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M-J-L, Zhang H, Jiang P, et al. , 2019. Topologic analysis of plasma mitochondrial DNA reveals the coexistence of both linear and circular molecules. Clin. Chem 65 (9), 1161–1170. [DOI] [PubMed] [Google Scholar]
- Marsland AL, Walsh C, Lockwood K, John-Henderson NA, 2017. The effects of acute psychological stress on circulating and stimulated inflammatory markers: a systematic review and meta-analysis. Brain Behav. Immun 64, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B, Kowal EJ, van Balkom BW, et al. , 2017. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA–an ISEV position paper. J. Extracel. Vesic 6 (1), 1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur K, Whitehead LW, Heddleston JM, et al. , 2018. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 359 (6378), eaao6047. [DOI] [PubMed] [Google Scholar]
- Meddeb R, Dache ZAA, Thezenas S, et al. , 2019. Quantifying circulating cell-free DNA in humans. Sci. Rep 9 (1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miliotis S, Nicolalde B, Ortega M, Yepez J, Caicedo A, 2019. Forms of extracellular mitochondria and their impact in health. Mitochondrion 48, 16–30. [DOI] [PubMed] [Google Scholar]
- Minocherhomji S, Tollefsbol TO, Singh KK, 2012. Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics 7 (4), 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui T, Umaki Y, Nagasawa M, et al. , 2002. Mitochondrial damage in patients with long-term corticosteroid therapy: development of oculoskeletal symptoms similar to mitochondrial disease. Acta Neuropathol. 104 (3), 260–266. [DOI] [PubMed] [Google Scholar]
- Mittelstrass K, Ried JS, Yu Z, et al. , 2011. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 7 (8), e1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman D. Group TP, Oxman A, Cook D, Guyatt G, Swingler G, Volmink J, Ioannidis J, Young C, Horton R, et al. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med Public Library of Science 2009; 6: e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F, 2017. In: Isolation of extracellular vesicles by ultracentrifugation. Extracellular Vesicles: Springer, pp. 25–32. [DOI] [PubMed] [Google Scholar]
- Morava E, Gardeitchik T, Kozicz T, et al. , 2010. Depressive behaviour in children diagnosed with a mitochondrial disorder. Mitochondrion 10 (5), 528–533. [DOI] [PubMed] [Google Scholar]
- Munakata Y, Shirasuna K, Kuwayama T, Iwata H, 2019. Cell-free DNA in medium is associated with the maturation ability of in vitro cultured oocytes. Journal of Reproduction and Development 2018–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundigler G, Delle-Karth G, Koreny M, et al. , 2002. Impaired circadian rhythm of melatonin secretion in sedated critically ill patients with severe sepsis. Crit. Care Med 30 (3), 536–540. [DOI] [PubMed] [Google Scholar]
- Mustard J, Perry D, Ardlie N, Packham MA, 1972. Preparation of suspensions of washed platelets from humans. Br. J. Haematol 22 (2), 193–204. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, et al. , 2011. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol 12 (3), 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Kyung S-Y, Rogers AJ, et al. , 2013. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 10 (12), e1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Möröy T, 2000. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat. Genet 25 (2), 177–181. [DOI] [PubMed] [Google Scholar]
- Nasi M, Cristani A, Pinti M, et al. , 2016. Decreased circulating mtDNA levels in professional male volleyball players. International journal of sports physiology and performance 11 (1), 116–121. [DOI] [PubMed] [Google Scholar]
- Nasi M, De Gaetano A, Bianchini E, et al. , 2020. Mitochondrial damage-associated molecular patterns stimulate reactive oxygen species production in human microglia. Mol. Cell. Neurosci 108, 103538. [DOI] [PubMed] [Google Scholar]
- Nicolás-Ávila JA, Lechuga-Vieco AV, Esteban-Martínez L, et al. A network of macrophages supports mitochondrial homeostasis in the heart. Cell 2020; 183(1): 94–109. e23. [DOI] [PubMed] [Google Scholar]
- Ono T, Isobe K, Nakada K, Hayashi JI, 2001. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat. Genet 28 (3), 272–275. [DOI] [PubMed] [Google Scholar]
- Pei L, Wallace DC, 2018. Mitochondrial etiology of neuropsychiatric disorders. Biol.Psychiatry 83 (9), 722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, McEwen BS, 2018. Psychological Stress and Mitochondria: A Systematic Review. Psychosom Med 80 (2), 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, Zhang J, Hancock S, et al. , 2014. Progressive increase in mtDNA 3243A >G heteroplasmy causes abrupt transcriptional reprogramming. Proc. Natl. Acad. Sci. U S A 111 (38), E4033–E4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, McManus MJ, Csordas G, et al. , 2015a. Trans-mitochondrial coordination of cristae at regulated membrane junctions. Nat. Commun 6, 6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, McManus MJ, Gray JD, et al. , 2015b. Mitochondrial functions modulate neuroendocrine, metabolic, inflammatory, and transcriptional responses to acute psychological stress. Proc. Natl. Acad. Sci 112 (48), E6614–E6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, Prather AA, Puterman E, et al. , 2018. A mitochondrial health index sensitive to mood and caregiving stress. Biol. Psychiatry 84 (1), 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, Trumpff C, Burelle Y, 2019. Mitochondrial psychobiology: foundations and applications. Current Opinion Behav. Sci 28, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinti M, Cevenini E, Nasi M, et al. , 2014. Circulating mitochondrial DNA increases with age and is a familiar trait: Implications for “inflamm-aging”. Eur. J. Immunol 44 (5), 1552–1562. [DOI] [PubMed] [Google Scholar]
- Rai P, Janardhan KS, Meacham J, et al. , 2021. IRGM1 links mitochondrial quality control to autoimmunity. Nat. Immunol 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausser S, Trumpff C, McGill MA, et al. , 2020. Mitochondrial profiling reveals dynamic, sex-and age-specific mitochondrial phenotypes in human immune cell subtypes. bioRxiv. [Google Scholar]
- Reynolds JC, Bwiza CP, Lee C, 2020a. Mitonuclear genomics and aging. Hum. Genet 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JC, Lai RW, Woodhead JS, et al. , 2020b. MOTS-c is an exercise-induced mitochondrial-encoded regulator of age-dependent physical decline and muscle homeostasis. bioRxiv 2020: 2019.12. 22.886432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley JS, Tait SW, 2020. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 21 (4), e49799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Nuevo A, Díaz-Ramos A, Noguera E, et al. , 2018. Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. The EMBO journal 37 (10), e96553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa HS, Ajaz S, Gnudi L, Malik AN, 2020. A case for measuring both cellular and cell-free mitochondrial DNA as a disease biomarker in human blood. FASEB J. 34 (9), 12278–12288. [DOI] [PubMed] [Google Scholar]
- Rose RM, Kreuz LE, Holaday JW, Sulak KJ, Johnson CE, 1972. Diurnal variation of plasma testosterone and cortisol. J Endocrinol 54 (1), 177–178. [DOI] [PubMed] [Google Scholar]
- Rossignol D, Frye R, 2012. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol. Psychiatry 17 (3), 290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami A, Lambie M, Caberry WY, Stambolic V, Waldron JN, Bratman SV, 2020. Senescence, Necrosis, and Apoptosis Govern Circulating Cell-free DNA Release Kinetics. Cell Reports 31 (13), 107830. [DOI] [PubMed] [Google Scholar]
- Sagan L, 1967. On the origin of mitosing cells. J. Theor. Biol 14 (3), 225–IN6. [DOI] [PubMed] [Google Scholar]
- Sansone P, Savini C, Kurelac I, et al. , 2017. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci 114 (43), E9066–E9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JH, 2020. Mitochondria signaling to the epigenome: A novel role for an old organelle. Free Radical Biol. Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saukkonen K, Lakkisto P, Pettila V, et al. , 2008. Cell-free plasma DNA as a predictor of outcome in severe sepsis and septic shock. Clin. Chem 54 (6), 1000–1007. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DS, Pantel K, 2011. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11 (6), 426–437. [DOI] [PubMed] [Google Scholar]
- Scozzi D, Cano M, Lina M, et al. Circulating Mitochondrial DNA is an Early Indicator of Severe Illness and Mortality from COVID-19. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstrom SC, Miller GE, 2004. Psychological stress and the human immune system: a meta-analytic study of 30 years of inquiry. Psychol. Bull 130 (4), 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj V, Stocco DM, Clark BJ, 2018. Current knowledge on the acute regulation of steroidogenesis. Biol. Reprod 99 (1), 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, et al. , 2012. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36 (3), 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin B, Cowan DB, Emani SM, Pedro J, McCully JD, 2017. Mitochondrial transplantation in myocardial ischemia and reperfusion injury. Mitochondrial Dynamics in Cardiovascular Medicine: Springer; 595–619. [DOI] [PubMed] [Google Scholar]
- Shockett PE, Khanal J, Sitaula A, et al. , 2016. Plasma cell-free mitochondrial DNA declines in response to prolonged moderate aerobic exercise. Physiological reports 4 (1), e12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisirak V, Sally B, D’Agati V, et al. , 2016. Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell 166 (1), 88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Hu W, Yu H, et al. , 2020. Existence of circulating mitochondria in human and animal peripheral blood. Int. J. Mol. Sci 21 (6), 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder T, Kirschbaum C, Kudielka BM, et al. , 2016. Assessment of the cortisol awakening response: Expert consensus guidelines. Psychoneuroendocrinology 63, 414–432. [DOI] [PubMed] [Google Scholar]
- Stawski R, Walczak K, Kosielski P, et al. , 2017. Repeated bouts of exhaustive exercise increase circulating cell free nuclear and mitochondrial DNA without development of tolerance in healthy men. PLoS ONE 12 (5), e0178216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens OR, Grant D, Frimel M, et al. , 2020. Characterization and origins of cell-free mitochondria in healthy murine and human blood. Mitochondrion 54, 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz L, Fries G, Rosa A, et al. , 2015. Damage-associated molecular patterns and immune activation in bipolar disorder. Acta Psychiatr. Scand 132 (3), 211–217. [DOI] [PubMed] [Google Scholar]
- Stigliani S, Orlando G, Massarotti C, et al. , 2019. Non-invasive mitochondrial DNA quantification on day 3 predicts blastocyst development: a prospective, blinded, multi-centric study. Mol. Hum. Reprod 25 (9), 527–537. [DOI] [PubMed] [Google Scholar]
- Strasser A, Jost PJ, Nagata S, 2009. The many roles of FAS receptor signaling in the immune system. Immunity 30 (2), 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Kamataki A, Yamaki J, Homma Y, 2008. Characterization of circulating DNA in healthy human plasma. Clin. Chim. Acta 387 (1–2), 55–58. [DOI] [PubMed] [Google Scholar]
- Szatanek R, Baran J, Siedlar M, Baj-Krzyworzeka M, 2015. Isolation of extracellular vesicles: Determining the correct approach. Int. J. Mol. Med 36 (1), 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurairajah K, Briggs GD, Balogh ZJ, 2018. The source of cell-free mitochondrial DNA in trauma and potential therapeutic strategies. Eur. J. Trauma. Emerg. Surg 44 (3), 325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, et al. , 2016. Mitochondrial stress induces chromatin reorganization to promote longevity and UPRmt. Cell 165 (5), 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpff C, Marsland AL, Basualto-Alarcon C, et al. , 2019a. Acute psychological stress increases serum circulating cell-free mitochondrial DNA. Psychoneuroendocrinology 106, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpff C, Marsland AL, Sloan RP, Kaufman BA, Picard M, 2019b. Predictors of ccf-mtDNA reactivity to acute psychological stress identified using machine learning classifiers: A proof-of-concept. Psychoneuroendocrinology 107, 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilioni I, Theoharides TC, 2018. Extracellular vesicles are increased in the serum of children with autism spectrum disorder, contain mitochondrial DNA, and stimulate human microglia to secrete IL-1beta. J Neuroinflam. 15 (1), 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler JP, Falkenberg M, 2015. Primer removal during mammalian mitochondrial DNA replication. DNA Repair 34, 28–38. [DOI] [PubMed] [Google Scholar]
- van der Kooij MA, Hollis F, Lozano L, et al. , 2018. Diazepam actions in the VTA enhance social dominance and mitochondrial function in the nucleus accumbens by activation of dopamine D1 receptors. Mol. Psychiatry 23 (3), 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varhaug KN, Vedeler CA, Myhr K-M, Aarseth JH, Tzoulis C, Bindoff LA, 2017. Increased levels of cell-free mitochondrial DNA in the cerebrospinal fluid of patients with multiple sclerosis. Mitochondrion 34, 32–35. [DOI] [PubMed] [Google Scholar]
- Velders M, Treff G, Machus K, Bosnyak E, Steinacker J, Schumann U, 2014. Exercise is a potent stimulus for enhancing circulating DNase activity. Clin. Biochem 47 (6), 471–474. [DOI] [PubMed] [Google Scholar]
- Ventura-Clapier R, Moulin M, Piquereau J, et al. , 2017. Mitochondria: a central target for sex differences in pathologies. Clin. Sci 131 (9), 803–822. [DOI] [PubMed] [Google Scholar]
- Wang Y, Picard M, Gu Z, 2016. Genetic evidence for elevated pathogenicity of mitochondrial DNA heteroplasmy in autism spectrum disorder. PLoS Genet. 12 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware SA, Desai N, Lopez M, et al. , 2020. An automated, high throughput methodology optimized for quantitative cell-free mitochondrial and nuclear DNA isolation from plasma. J. Biol. Chem jbc. RA120, 015237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Shadel GS, 2017. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol 17 (6), 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, et al. , 2015. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520 (7548), 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Oeck S, West AP, et al. , 2019. Mitochondrial DNA stress signalling protects the nuclear genome. Nat. Metabol 1 (12), 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Radpour R, Zachariah R, et al. , 2009. Simultaneous quantitative assessment of circulating cell-free mitochondrial and nuclear DNA by multiplex real-time PCR. Genetics and molecular biology 32 (1), 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi S, Kudo D, Yamada M, Miyagawa N, Furukawa H, Kushimoto S, 2013. Plasma mitochondrial DNA levels in patients with trauma and severe sepsis: time course and the association with clinical status. J. Crit. Care 28 (6), 1027–1031. [DOI] [PubMed] [Google Scholar]
- Yen K, Mehta HH, Kim S-J, et al. , 2020. The mitochondrial derived peptide humanin is a regulator of lifespan and healthspan. Aging (Albany NY) 12 (12), 11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Gold JA, Andina N, et al. , 2008. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med 14 (9), 949–953. [DOI] [PubMed] [Google Scholar]
- Yousefi S, Mihalache C, Kozlowski E, Schmid I, Simon H-U, 2009. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 16 (11), 1438. [DOI] [PubMed] [Google Scholar]
- Yu SC, Lee SW, Jiang P, et al. , 2013. High-resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin. Chem 59 (8), 1228–1237. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Raoof M, Chen Y, et al. , 2010a. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464 (7285), 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Angelidou A, Alysandratos K-D, et al. , 2010b. Mitochondrial DNA and anti-mitochondrial antibodies in serum of autistic children. J. Neuroinflam 7 (1), 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Deng S, Zhao S, et al. , 2016. Intra-peritoneal administration of mitochondrial DNA provokes acute lung injury and systemic inflammation via toll-like receptor 9. Int. J. Mol. Sci 17 (9), 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hubal MJ, Kraus VB, 2020a. Immune cell extracellular vesicles and their mitochondrial content decline with ageing. Immunity & Ageing 17 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WZ, Rice MC, Hoffman KL, et al. , 2020b. Association of urine mitochondrial DNA with clinical measures of COPD in the SPIROMICS cohort. JCI insight 5 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zhou Y, Hilton T, et al. , 2020. Extracellular mitochondria released from traumatized brains induced platelet procoagulant activity. Haematologica 105 (1), 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Liang S, Sanchez-Lopez E, et al. , 2018. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560 (7717), 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong F, Liang S, Zhong Z, 2019. Emerging Role of Mitochondrial DNA as a Major Driver of Inflammation and Disease Progression. Trends Immunol. [DOI] [PubMed] [Google Scholar]
- Zhong S, Ng MC, Lo YD, Chan JC, Johnson PJ, 2000. Presence of mitochondrial tRNA Leu (UUR) A to G 3243 mutation in DNA extracted from serum and plasma of patients with type 2 diabetes mellitus. J. Clin. Pathol 53 (6), 466–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.