SUMMARY
Alphaviruses cause severe arthritogenic or encephalitic disease. The E1 structural glycoprotein is highly conserved in these viruses and mediates viral fusion with host cells. However, the role of antibody responses to the E1 protein in immunity is poorly understood. We isolated E1-specific human monoclonal antibodies (mAbs) with diverse patterns of recognition for alphaviruses (ranging from Eastern equine encephalitis virus (EEEV)-specific to alphavirus cross-reactive) from survivors of natural EEEV infection. Antibody binding patterns and epitope mapping experiments identified differences in E1 reactivity based on exposure of epitopes on the glycoprotein through pH-dependent mechanisms or presentation on the cell surface prior to virus egress. Therapeutic efficacy in vivo of these mAbs corresponded with potency of virus egress inhibition in vitro and did not require Fc-mediated effector functions for treatment against subcutaneous EEEV challenge. These studies reveal the molecular basis for broad and protective antibody responses to alphavirus E1 proteins.
Keywords: Alphavirus; Antibodies, Human Monoclonal; Antibodies, Non-Neutralizing; Antibodies, Cross-Reactive; Post-Exposure Prophylaxis
Graphical Abstract
eTOC blurb
Broadly reactive alphavirus E1 antibodies to cryptic epitopes obtained from survivors of natural Eastern equine encephalitis virus infection inhibit virus egress in vitro and protect against infection by encephalitic (Eastern equine encephalitis) and arthritogenic (chikungunya) alphaviruses in mice.
INTRODUCTION
Alphaviruses are positive-sense single-stranded RNA viruses that are members of the Togaviridae family. These viruses are classified based on antigenic complex, historical geographic distribution (New or Old World), genetic relatedness, or disease state (encephalitic or arthritogenic) (Ronca et al., 2016). The New World encephalitic alphaviruses include Eastern equine encephalitis virus (EEEV), Venezuelan equine encephalitis virus (VEEV), and Western equine encephalitis virus (WEEV), which cause a febrile illness that can lead to encephalomyelitis (Lindsey et al., 2018; Weaver et al., 2012; Honnold et al., 2015; Griffin, 2010). EEEV is the most virulent encephalitic alphavirus, with a human case fatality rate of 30–75% (Lindsey et al., 2018; Ayres and Feemster, 1949; Armstrong and Andreadis, 2013). On average, 11 human cases are reported each year, however, in 2019, an outbreak occurred with 38 reported human EEEV cases and 19 deaths, raising concern for EEEV as an emergent arbovirus (Morens et al., 2019; Lindsey et al., 2020; CDC). The arthritogenic (historically Old World) alphaviruses associated with human disease include Barmah Forest virus, chikungunya virus (CHIKV), Mayaro virus (MAYV), O’nyong-nyong virus, Ross River virus, Semliki Forest virus (SFV), and Sindbis virus (SINV) (Mostafavi et al., 2019). These viruses lead to an acute or chronic arthritogenic and musculoskeletal disease characterized by rash, arthralgia, myalgia, and joint swelling (Diagne et al., 2020; Mostafavi et al., 2019; Ganesan et al., 2017). Globally, CHIKV is the most morbid alphavirus causing millions of human cases (Mavalankar et al., 2008; Weaver et al., 2012; Jin and Simmons, 2019). In addition to natural transmission by mosquito vectors, EEEV, VEEV, WEEV, and CHIKV are classified as NIAID Category B priority pathogens and USDA/CDC Select Agents due to potential as bioterrorism agents through aerosol spread (Sidwell and Smee, 2003). No human vaccines or antiviral drugs for alphaviruses are approved for public use (Morens et al., 2019; Trobaugh et al., 2019).
Alphavirus E2 and E1 glycoproteins heterodimerize to form 80 trimeric spikes on the viral surface and interact with the capsid protein underneath the viral membrane (Li et al., 2010; Voss et al., 2010; Zhang et al., 2011; Chen et al., 2020; Brown et al., 2018; Byrd et al., 2019; Sun et al., 2013). The E2 glycoprotein contains three main Ig-like domains: A, B, and C (Hasan et al., 2018; Voss et al., 2010; Li et al., 2010; Kielian et al., 2010). E1 is a class II fusion protein and consists of three β-sheet structural domains: domain I (DI; central domain), domain II (DII; contains a highly conserved fusion loop at its distal tip), and domain III (DIII; Ig-like fold that connects to the stem region and transmembrane domain) (Hasan et al., 2018; Gibbons et al., 2004; Lescar et al., 2001; Mukhopadhyay et al., 2006; Roussel et al., 2006; Kielian, 2014). On the virion surface, the fusion loop is occluded by domains A and B of E2 under neutral pH conditions (Kielian, 2014; Voss et al., 2010). Two main E1 interdomains are also present, the flexible DI/DII hinge region and the DI/DIII linker region (Lescar et al., 2001; Roussel et al., 2006; Sahoo et al., 2020; Kielian, 2014).
Alphaviruses enter cells by receptor-mediated endocytosis upon recognition of attachment factors (Gardner et al., 2011; Klimstra et al., 2003) and entry receptors (Zhang et al., 2018; Ma et al., 2020; Rose et al., 2011; Holmes et al., 2020). A drop in pH within the endosome leads to dissociation of the E2 glycoprotein from E1, insertion of the fusion loop with host cell membranes, and formation of E1 homotrimers (Wahlberg and Garoff, 1992; Justman et al., 1993; Wahlberg et al., 1992; Gibbons et al., 2000; Gibbons et al., 2004; Roussel et al., 2006). Substantial movement of the DI/DII hinge region and DI/DIII linker helps stabilize the trimer to enable a “fold-back” mechanism of DIII and the stem region against the core trimer. This movement brings the cell and viral membranes together, resulting in fusion and release of the nucleocapsid into the cytosol (Gibbons et al., 2004; Mukhopadhyay et al., 2006; Kielian, 2014; Zheng et al., 2011). E2 and E1 are translated from the subgenomic mRNA, processed in the ER and Golgi apparatus, and trafficked to the plasma membrane for encapsidation and egress from the cell (Brown et al., 2018). During these processes, large rearrangements and different conformational states of the surface proteins occur (Fuller et al., 1995; Li et al., 2010; Mukhopadhyay et al., 2006; Sahoo et al., 2020; Voss et al., 2010), likely exposing cryptic epitopes (Fong et al., 2014; Gibbons et al., 2004; Schmaljohn et al., 1983). Murine WEEV, SINV, and human CHIKV E1-specific mAbs previously identified some of these transitional epitopes at various stages of virus maturation (i.e., native virions or the surface of infected cells). Treatment strategies with detergent, elevated temperature, acidic pH, reducing agents, or fixation strategies induced epitope exposure (Fong et al., 2014; Gibbons et al., 2004; Hunt and Roehrig, 1985; Meyer et al., 1992; Schmaljohn et al., 1983; Ahn et al., 1999). Furthermore, epitopes have been identified for murine anti-E1 mAbs that protect against CHIKV, MAYV, VEEV, WEEV, SINV, or SFV infection (Boere et al., 1984; Hunt and Roehrig, 1985; Mendoza et al., 1988; Pal et al., 2013; Schmaljohn et al., 1983; Earnest et al., 2019; Mathews and Roehrig, 1982). However, moderately neutralizing and non-neutralizing cross-reactive human anti-E1 mAbs did not protect against lethal CHIKV infection (Fong et al., 2014; Quiroz et al., 2019).
Here, we outline the molecular basis for human antibody interaction with the alphavirus E1 glycoprotein through isolation and characterization of naturally occurring human E1-specific mAbs from survivors of EEEV infection. From this collection, we identified mAbs specific to EEEV and cross-reactive mAbs to EEEV, VEEV, WEEV, CHIKV, and/or MAYV. We determined the antigenic sites recognized by these mAbs on the EEEV E1 glycoprotein, some of which become more exposed under acidic-pH conditions or treatment with detergent. In addition, we found that mAb binding to EEEV-infected cells inhibited virus egress, which corresponded with therapeutic efficacy in vivo.
RESULTS
Binding characterization of human E1-specific mAbs isolated from naturally infected EEEV survivors.
We isolated human mAbs from B cells of individuals who had been naturally infected with EEEV. Twenty mAbs were selected based on reactivity in ELISA to EEEV E1 glycoprotein, EEEV virus-like particles (VLPs), or SINV/EEEV virions (Kim et al., 2019). We assessed binding to EEEV, VEEV, or WEEV VLPs, recombinant E1 glycoproteins (EEEV, CHIKV, or MAYV), and recombinant EEEV E2 glycoprotein in ELISA in the presence of the nonionic detergent Tween®20 (Figure 1). As the E1 glycoprotein on the virion surface is relatively obscured by the overlying E2 glycoprotein, exposure of epitopes for E1-specific mAbs can require pre-treatment conditions. Tween®20 was included in buffers during the incubation and wash steps to enhance exposure of epitopes that might otherwise be obstructed in the native virus structure. We also assessed binding breadth to the structural proteins of a diverse panel of alphavirus subtypes (EEEV, Madariaga virus [MADV], VEEV, WEEV, and CHIKV) using a cell surface antigen display method, in which we transfected Expi293F cells with a plasmid encoding the structural proteins (capsid-E3-E2–6K-E1) (Figures 2 and S1).
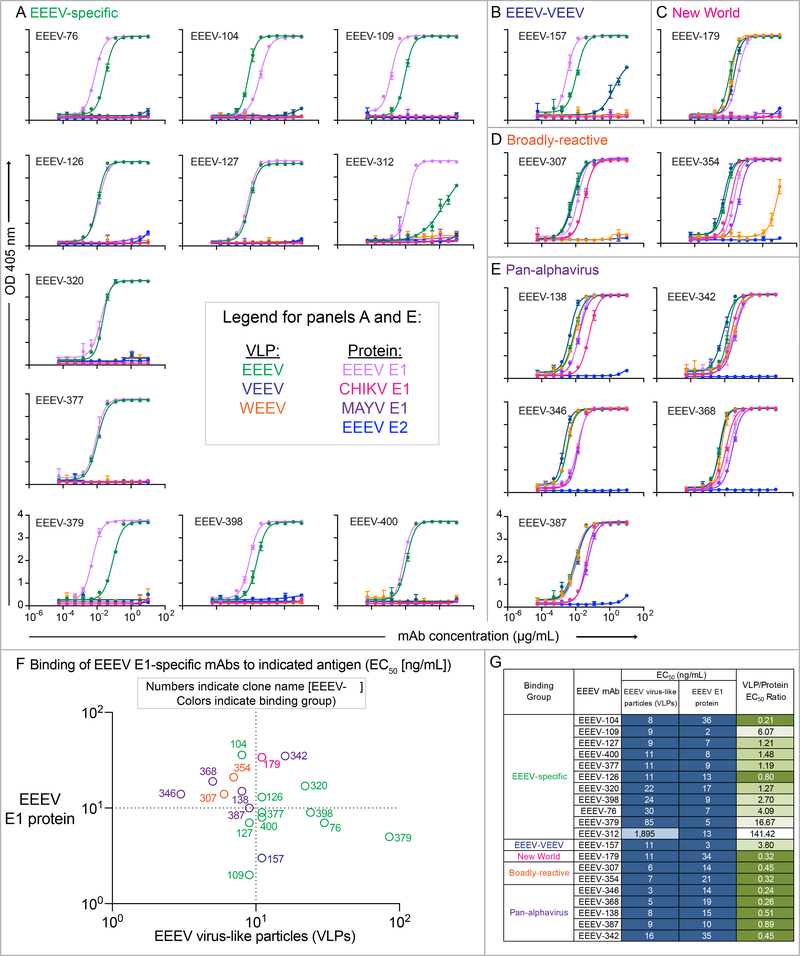
Figure 1. Binding reactivity of human E1-specific mAbs to virus-like particles (VLPs) or recombinant E1 glycoproteins in the presence of Tween®20 for exposure of potentially cryptic E1 epitopes.
A to E. Representative binding curves for human E1-specific mAbs binding to VLPs for EEEV (green), VEEV (dark blue), or WEEV (orange) VLPs, or recombinant E1 glycoprotein for EEEV (light purple), CHIKV (magenta), or MAYV (purple), or recombinant EEEV E2 glycoprotein (blue) in the presence of Tween®20. MAb concentration (μg/mL) is on the x-axis and optical density at 405 nm on the y-axis. Binding curves are ordered numerically and grouped according to binding reactivity (EEEV-specific [A; green], ‘EEEV-VEEV’ [B; blue], ‘New World’ [C; magenta], ‘broadly-reactive’ [D; orange], or ‘pan-alphavirus’ [E; purple]). F. EC50 (ng/mL) binding ratio of EEEV-specific (green) and cross-reactive (‘pan-alphavirus [purple], ‘broadly-reactive’ [orange], ‘New World’ [magenta], ‘EEEV-VEEV’ [blue]) mAbs to display binding patterns to EEEV VLPs (x-axis) versus recombinant EEEV E1 glycoprotein (y-axis). The dotted line indicates 10 ng/mL EC50 values for binding, and open circles are labeled with antibody clone name [EEEV- #]. G. EC50 values (ng/mL) for mAb binding to EEEV VLPs or recombinant EEEV E1 glycoprotein in the presence of the nonionic detergent Tween®20. The mAbs are grouped according to binding reactivity and are listed in order of increasing EC50 value for binding to EEEV VLPs. EC50 value in ng/mL is indicated by the blue fill color (≤100.00 [dark blue], 101–500 [light blue], and 501 to 5,000 [lightest blue]). Ratio of binding to EEEV VLPs compared to EEEV E1 glycoprotein is indicated as the ratio of EC50 values. Increasing depth of green color (≤1.00 [dark green], 1.01–2.00 [green], 2.01 to 5.00 [light green], 5.01 to 100 [lightest green], and >100 [white]) indicates lower binding ratios and suggests greater dependence on virus particle-specific epitopes. Data in A to G represent mean ± SD of technical triplicates and are representative of 2 independent experiments.
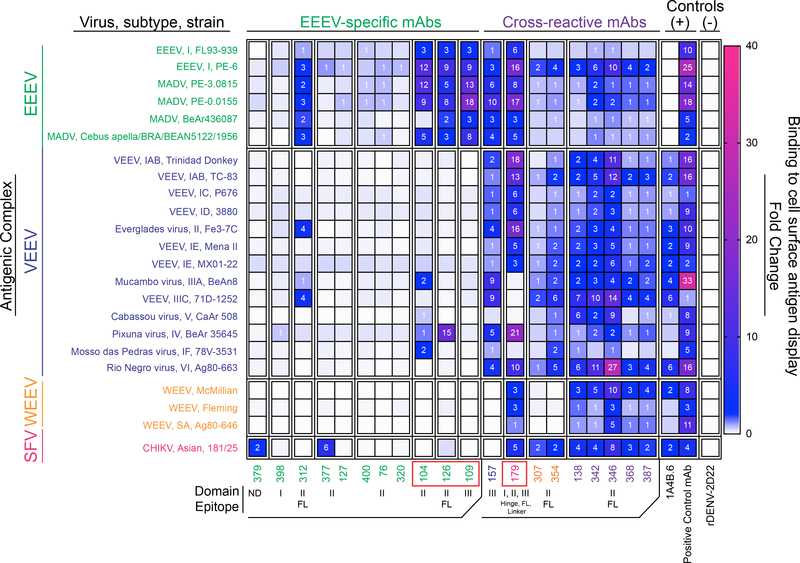
Figure 2. Binding breadth of human E1-specific mAbs to diverse alphavirus subtype structural proteins displayed on the surface of cells.
Heatmap of fold-change for mAb binding to alphavirus subtypes (labeled with virus, subtype, and strain; grouped by antigenic complex for either EEEV (green), VEEV (blue), WEEV (orange), or Semliki Forest virus (magenta)). The relative fold-change for mAb binding to each subtype was calculated by background subtraction of median fluorescence to Expi293F cells and normalized relative to the negative control mAb rDENV-2D22. The alphavirus group E1-reactive mouse mAb,1A4B-6 (Roehrig et al., 1990) served as a positive control for E1 mAb binding. The following additional positive control mAbs were used: rEEEV-97 IgG (human mAb; EEEV E2-specific; Williamson et al., 2020), 1A3B-7 (mouse mAb; VEEV E2-specific; Roehrig and Mathews, 1985), 2A3D-5 (mouse mAb; WEEV E1-specific; Hunt and Roehrig, 1985), and mouse anti-CHIKV ascites fluid (CHIKV; ATCC). The mAbs are shown in order based on antigen-specificity (EEEV-specific or cross-reactive) and epitope (as defined in Figure 3). SINV/EEEV egress-inhibiting mAbs are indicated by the red boxes (as defined in Figure 5). Data represents mean ± SD of technical triplicates and are representative values of 2 independent biological replicates. See Figure S1 for multiple sequence alignment and percent E1 amino acid identity of the alphavirus subtypes tested.
Using ELISA and cell surface antigen display methods, we defined antigen binding groups within the panel of mAbs. Two classes emerged, defined by mAb antigen specificity of either EEEV-specific or cross-reactive mAbs (Figures 1 and 2). In the first class of mAbs, EEEV-specific mAbs bound specifically to EEEV VLPs or recombinant EEEV E1 glycoprotein (Figure 1A) in an ELISA under the conditions tested. Binding to the structural proteins of EEEV and MADV subtypes displayed at the cell surface was observed for the EEEV-specific mAbs, EEEV-104, −109, −126, and −312, as defined by a >2-fold change in mAb binding compared to the dengue virus (DENV)-specific negative control mAb, rDENV-2D22 (Figure 2) (Fibriansah et al., 2015). Several mAbs displayed little or no binding. Low binding reactivity for the positive control alphavirus-group E1-specific mouse mAb 1A4B.6 also was observed. In contrast, the positive control neutralizing EEEV E2-specific human mAb, rEEEV-97 IgG, strongly bound to the EEEV and MADV subtypes tested (Williamson et al., 2020). The difference in binding between an E2-specific and E1-specific mAb suggests that some E1 epitopes are not fully exposed on the surface of transfected cells, compared to more surface exposed E2 epitopes. Thus, the human EEEV-specific mAbs may display differences in recognition between exposed and cryptic E1 epitopes.
The second class of mAbs exhibited cross-reactivity profiles, with similar binding profiles identified by ELISA and cell surface antigen display. The broadest subgroup of cross-reactive mAbs (designated here as ‘pan-alphavirus’), comprised of mAbs EEEV-138, −342, −346, −368, and −387, recognized EEEV, VEEV, WEEV, and CHIKV subtypes (Figures 1E and 2). Recombinant MAYV E1 also was recognized by the ‘pan-alphavirus’ subgroup in ELISA (Figure 1E). Another subgroup of cross-reactive mAbs (designated here as ‘broadly-reactive’), comprised by mAbs EEEV-307 and −354, recognized EEEV, VEEV, and CHIKV subtypes but not WEEV in either format (Figures 1D and 2). Recombinant MAYV E1 also was recognized by the ‘broadly-reactive’ subgroup by ELISA (Figure 1D). A third subgroup (designated here as ‘New World’) formed by EEEV-179, recognized EEEV, VEEV, and WEEV subtypes (Figures 1C and 2). EEEV-179 also recognized the CHIKV subtype tested via cell surface antigen display, which contrasted with the ELISA results (Figures 1C and 2). The increase in binding breadth of EEEV-179 to CHIKV suggests either a difference in the conformational state of the epitope between intact virus particles and the surface of infected cells or a dependence on a quaternary epitope. Lastly, EEEV-157 exhibited a ‘EEEV-VEEV’ pattern, recognizing EEEV and VEEV subtypes (Figures 1B and 2). None of the E1-reactive mAbs isolated bound to EEEV E2 (Figure 1).
Both classes avidly bound (EC50 values <100 ng/mL) to their respective antigens by ELISA (Figure 1G and Table S1). EEEV-157, however, bound to VEEV VLPs only weakly (EC50 value 1.7 μg/mL) (Table S1). We also noted differences in binding strength to VLPs or recombinant E1, which suggests preferential binding to either virus particles or protein exposed epitopes (Figures 1F and 1G).
Identification of antigenic sites on the EEEV E1 glycoprotein.
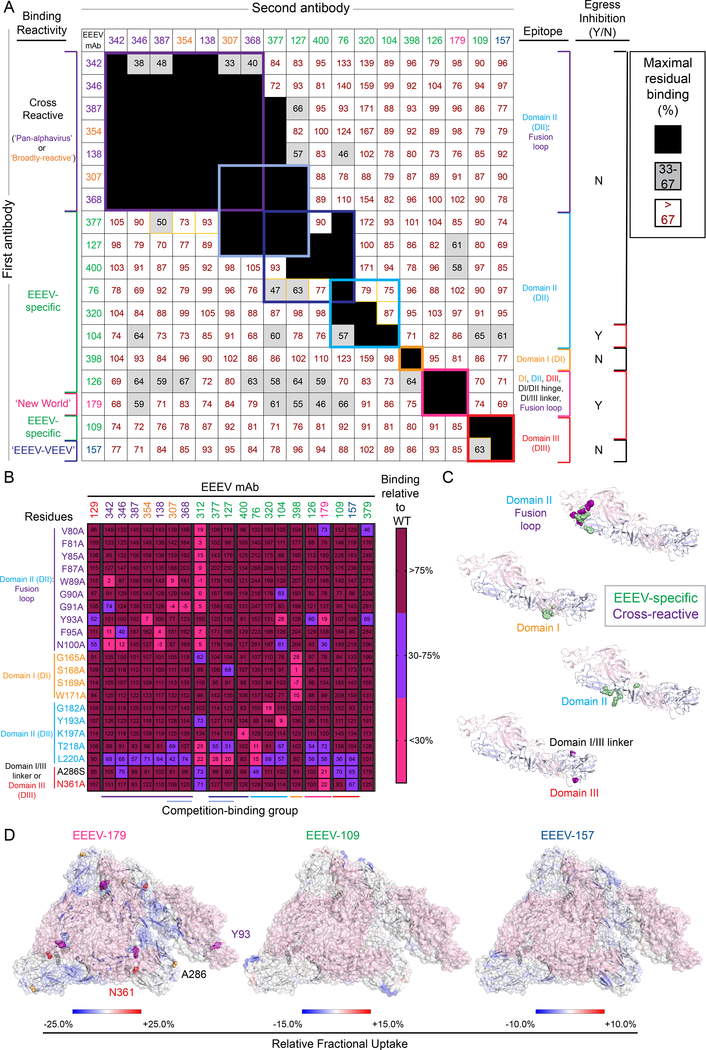
We performed competition-binding studies with EEEV VLPs by ELISA, in which we observed at least seven binding groups (Figure 3A). The ‘pan-alphavirus’ and ‘broadly-reactive’ mAbs grouped together, suggesting a common antigenic site for these mAbs. The ‘New World’ or ‘EEEV-VEEV’ mAbs, EEEV-179 or EEEV-157, respectively, did not compete with the ‘pan-alphavirus’ or ‘broadly-reactive’ mAbs or with each other, indicating that these mAbs recognize conserved sites on E1 with distinct cross-reactivity profiles. Thus, up to 3 antigenic sites may be present on E1 for recognition by cross-reactive mAbs (‘pan-alphavirus’ and ‘broadly-reactive, ‘New World’, or ‘EEEV-VEEV’). The EEEV-specific mAbs primarily bound to different competition-binding groups, some (EEEV-126, −127, and −377) of which competed with certain cross-reactive mAbs.
Figure 3. Human E1-specific mAbs recognize cryptic and exposed epitopes on the EEEV E1 glycoprotein.
A. Competition-binding groups as determined by ELISA using EEEV VLPs. Results for a total of 18 mAbs are shown. EEEV-312 and −379 are not shown due to minimal binding to EEEV VLP under the conditions tested. The first mAb (10 μg/mL) incubated with EEEV VLPs is shown in the left-hand column and the second mAb (biotinylated; 0.5 μg/mL) is shown in the top row. Black boxes indicate competition (reduction in maximal binding to <33%), grey boxes indicate intermediate competition (33 to 67% maximal residual binding), and white boxes indicate no competition (>67% maximal residual binding). Competition-binding groups are indicated by the respective colored boxes. Yellow boxes indicate unidirectional competition. Each mAb is colored according to binding reactivity (EEEV-specific [green], cross-reactive: ‘pan-alphavirus’ [purple], ‘broadly-reactive’ [orange], ‘New World’ [magenta], and ‘EEEV-VEEV’ [blue]). Data are representative values of 3 independent experiments. Epitopes identified for each mAb (as described in B through D) are indicated in the right-hand column. Y = inhibition of SINV/EEEV egress and N = no observed inhibition of SINV/EEEV egress (as described in Figure 5). B. Heatmap of mAb binding to critical interaction residues identified through alanine-scanning mutagenesis library analysis of the EEEV E1 glycoprotein. The average percent binding for each mAb is indicated for the residues identified (<30% binding of mAb relative to wild-type (WT), in which the positive control mAb, rEEEV-129 IgG (Williamson et al., 2020), or at least five mAbs exhibited >75% binding to control for expression. The heatmap indicates >75% (maroon), 30 to 75% (light blue), or <30% (magenta) mAb binding relative to WT. Residues are colored based on E1 domain or region (purple [DII, fusion loop]), cyan [DII], orange [DI], red [DIII], and black [DI/DIII linker]). Each mAb is ordered to correspond with the competition-binding groups (A) as defined by the colored boxes and respective lines at the bottom of the heat map. Data represent mean of at least 2 independent experiments. C. Epitope mapping of critical interaction residues identified by alanine-scanning (B) for mAb binding to the EEEV E1 glycoprotein. Critical interaction residues were mapped onto the cryo-EM reconstruction of SINV/EEEV (PDB: 6MX4). A side view of the EEEV E2-E1 heterodimer (E2 – light pink, E1 – light blue) is shown with critical interaction residues indicated by spheres for the EEEV-specific (green) or cross-reactive (purple) E1-specific mAbs. D. Hydrogen-deuterium exchange mass spectrometry for EEEV-179, −109, and −157 Fab molecules binding to the EEEV E1 glycoprotein. Relative fractional deuterium uptake difference at the 10,000 s time point is mapped onto the cryo-EM reconstruction of EEEV VLP (PDB: 6XO4; E2 – light pink). E1 regions are colored from blue to red based on the relative fractional deuterium uptake difference (%). The color scale corresponds to the highest observed difference and is adjusted to −25% to 25%, −15% to 15%, or −10% to 10% for EEEV-179, −109, or −157 Fab molecules, respectively. Regions with no deuterium uptake difference or with no peptide measurements are shown in white. See Figure S2 for analysis of W89A, F95A, and N100A CHIKV E2/E1 and full panel of EEEV E1 alanine mutants tested. See Figure S3 for HDX-MS analysis summary for binding of EEEV-109, −126, −157, and −179 Fab molecules to the EEEV E1 glycoprotein. See Figure S4 and Table S2 for summary of epitope mapping results.
Cross-reactive (‘pan-alphavirus’ and ‘broadly-reactive’) mAbs recognize the highly conserved fusion loop.
The fusion loop of E1 is highly conserved among alphaviruses (Figure S1). We hypothesized that the ‘pan-alphavirus’ and ‘broadly-reactive’ mAbs might recognize the fusion loop. To test this hypothesis, we examined loss-of-binding phenotype(s) to CHIKV proteins containing alanine substitution mutants in the E1 fusion loop that were identified previously (W89A, F95A, and N100A; Figure S2) in the context of mAb mapping experiments (Fong et al., 2014). For the ‘pan-alphavirus’ and ‘broadly-reactive’ mAbs, these mutations caused a loss-of-binding phenotype, supporting the hypothesis that these mAbs recognize a shared antigenic site within the highly conserved fusion loop. To test for additional critical interaction residues, we generated a complete alanine-scanning mutagenesis library of residues within EEEV E1 and transfected cells with cDNAs encoding the structural proteins (capsid-E3-E2–6K-E1), each containing a separate individual mutant (Figures 3B, 3C, and S2; Table S2). The ‘pan-alphavirus’ and ‘broadly-reactive’ mAbs showed markedly reduced binding when mutations were introduced at residues within the fusion loop (residues W89, G91, Y93, F95, and N100 [cd loop]). The residues identified were relatively consistent between the two libraries. One key difference was the lack of a loss-of-binding phenotype observed for F95A in the EEEV library, whereas this phenotype was observed for the CHIKV F95A mutant (Figure S2).
To determine the binding footprint of EEEV-157 on EEEV E1, we performed hydrogen-deuterium exchange mass spectrometry (HDX-MS). A potential interacting peptide (residues 353–366) within DIII was identified by comparing deuterium uptake of EEEV-157 Fab bound versus unbound EEEV E1 glycoprotein (Figures 3D and S3).
EEEV-179 (‘New World’) recognizes a quaternary epitope on the E1 glycoprotein.
The ‘New World’ reactive mAb EEEV-179 binds to the encephalitic alphaviruses by ELISA and the arthritogenic alphavirus CHKV in a cell-surface antigen display assay (Figures 1 and 2). The critical interaction residues in alanine scanning mutagenesis for EEEV-179 were within the DI/DIII linker (A286), DIII (N361), and proximal to the fusion loop (Y93 [cd loop]). Next, we performed HDX-MS studies to map potential interacting residues. Multiple peptides were identified with a ≥ 0.4 Da of relative deuterium uptake of EEEV-179 Fab bound versus unbound EEEV E1 glycoprotein, separated by ± 2.4 standard deviations for at least one labeling time point (Figure S3). These peptides span several domains of EEEV E1 (i.e., residues 37–45 [DI and DI/DII hinge region], 80–92 [fusion loop: cd loop], 107–113 [DII: cd loop], 141–146 [DI], 147–162 [DI], 191–201 [DII: central β sheet – gh loop], 202–214 [DII: central β sheet – gh loop], 221–228 [DII: ij loop], 229–254 [DII: ij loop], 262–271 [DII: kl β hairpin and DI/DII hinge region], and 288–300 [DIII and DI/DIII linker]). The large binding footprint of EEEV-179 to EEEV E1 suggests it recognizes a quaternary epitope between two adjacent E1 protomers of neighboring trimeric spikes (Figure 3D).
Human EEEV-specific mAbs recognize multiple epitopes on E1.
We next assessed for a loss-of-binding phenotype with the EEEV alanine-scanning mutagenesis library (Figures 3B, 3C, and S2; Table S2). The critical interaction residues identified were consistent with the competition-binding groups. EEEV-312 recognized residues within and proximal to the fusion loop (V80, F81, Y85, F87, W89, G90, G91, F95, N100 [cd loop]; T218 [DII: central β sheet] and L220 [DII: ij loop]). While several residues were like those identified for the cross-reactive mAbs, additional residues were identified in this region that may account for the EEEV specificity of EEEV-312. For several mAbs, residues were identified at the interface of E2 and E1 or on the membrane proximal side of the heterodimer. EEEV-377 and −127 lost binding with a L220A [DII: ij loop] substitution in E1. These EEEV-specific mAbs compete with some of the fusion loop cross-reactive mAbs, which is expected given their proximity of binding to this epitope. EEEV-76, −104, −320, and −400 partially compete with EEEV-377 and −127 but do not compete with the fusion loop cross-reactive mAbs. These mAbs recognize additional critical residues (G182 [DII: central β sheet; EEEV-320], Y193 [DII: central β sheet – gh loop; EEEV-104], K197 [DII: central β sheet – gh loop; EEEV-400], and T218 [DII: central β sheet; EEEV-76]), which suggests a distinct antigenic site that does not hinder the fusion loop cross-reactive mAbs from binding. EEEV-398 recognizes a distinct antigenic site (DI: G165, S168, S169, and W171) within DI.
Binding residues were not identified for EEEV-109 or −126 using the alanine mutagenesis approach. To determine the binding footprint of EEEV-109 and EEEV-126, we again performed HDX-MS. Two potential interacting peptides (residues 289–300 and 348–353) within DIII were identified for EEEV-109 (Figures 3D and S3). This finding supports the results of competition-binding observed with EEEV-157, which recognizes an adjacent epitope within DIII of EEEV E1. No potential interacting peptide was identified for EEEV-126, due to the lack of sequence peptide coverage. We considered Q79 as a potential interaction residue based on the pattern of binding to alphavirus subtypes in cell-surface antigen display, but a peptide containing this residue could not be deuterated detectably (Figures 2, S1, and S3).
Acidic pH exposes cryptic epitopes on virus particles for binding of human E1-specific mAbs.
To define how human E1-specific mAbs recognize potentially occluded E1 epitopes, we assessed conditions that might expose epitopes for mAb binding like the nonionic detergent Tween®20, as described above. One condition involves elevated temperature, which increases binding of E1 mAbs (Fong et al., 2014). We compared binding to VLPs or recombinant protein by ELISA at room temperature (~22°C), 37°C, or 42°C (Figure S5) in the absence of detergent. Temperature did not affect binding of E1-specific mAbs, as shown by the similar EC50 values for binding at diverse temperatures (Figures S5H and I; Table S3).
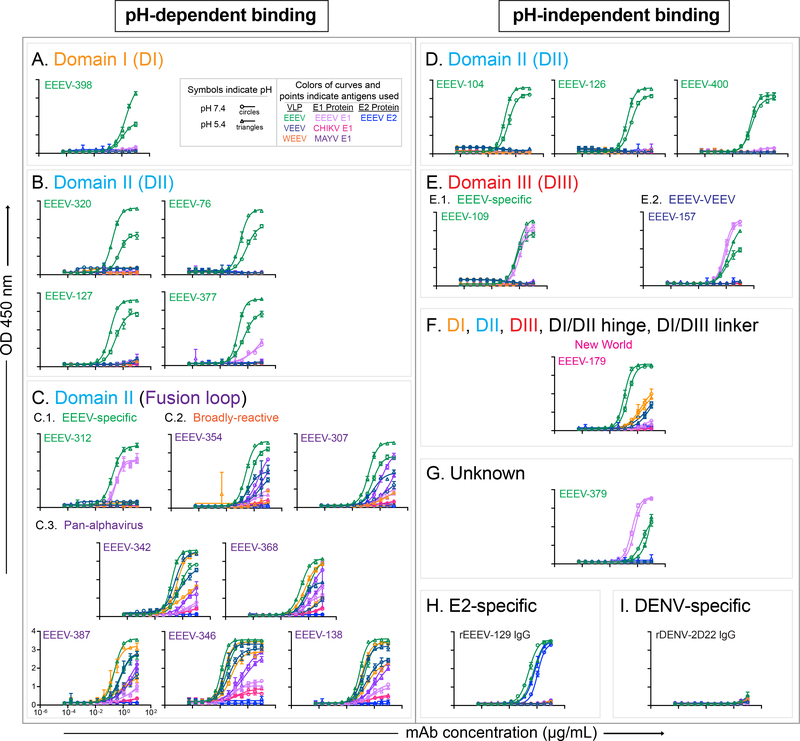
Another condition that might expose E1 epitopes is acidic pH, which mimics the endosomal environment during viral fusion. This procedure can increase recognition of virus particles by some E1 mAbs (Gibbons et al., 2004; Meyer et al., 1992; Schmaljohn et al., 1983). We tested binding to VLPs or recombinant proteins under neutral or acidic pH conditions in the absence of detergent by ELISA (Figure 4 and Table S4). Several binding phenotypes emerged, which corresponded with epitope specificity. For most mAbs that recognize DI or DII of E1 (i.e., fusion loop, E2/E1 interface, or on the membrane proximal side), an acidic pH of 5.4 increases the binding strength compared to neutral pH 7.4 conditions.
Figure 4. Acidic pH-dependent or -independent binding reactivity of human E1-specific mAbs to virus-like particles (VLPs) or recombinant E1 glycoproteins.
A to C. Representative binding curves of acidic pH-dependent EEEV-specific (green labels) and cross-reactive (‘pan-alphavirus’ [purple labels] and ‘broadly-reactive’ [orange labels]) mAbs to EEEV (green), VEEV (dark blue), or WEEV (orange) VLPs, recombinant EEEV (light purple), CHIKV (magenta), or MAYV (purple) E1 glycoproteins, and recombinant EEEV E2 (blue) glycoprotein at 1× DPBS pH 5.4 (open circles) or 1× DPBS pH 7.4 (open triangles) with mAb concentration (μg/mL) on the x-axis and optical density at 450 nm on the y-axis. Binding curves are ordered by decreasing dependence on pH 5.4 for binding and grouped according to epitope and binding reactivity (DI (A) [EEEV-specific], DII (B) [EEEV-specific], and fusion loop (C) [EEEV-specific (C.1.), ‘broadly-reactive’ (C.2.), and ‘pan-alphavirus’ (C.3.)]. D to G. Representative binding curves of pH-independent EEEV-specific (green labels) and cross-reactive (‘New World’ [magenta labels], ‘EEEV-VEEV’ [blue labels]) mAbs to respective antigens as described in A. Binding curves are ordered numerically and grouped according to epitope and binding reactivity (DII (D) [EEEV-specific], DIII (E) [EEEV-specific (E.1.) and ‘EEEV-VEEV’ (E.2.)], a quaternary epitope including DI, DII, DIII, DI/DII hinge region, and the DI/DIII linker (F) [‘New World’], or an unknown epitope (G). H and I. Representative binding curves of a pH-independent positive control neutralizing, EEEV E2-specific mAb (rEEEV-129 IgG [H]; Williamson et al., 2020) and the negative control DENV-specific mAb (rDENV-2D22 IgG [I]; Fibriansah et al., 2015). Data in A to I represent mean ± SD of technical triplicates and are representative of 2 independent experiments. See Figure 1 for mAb binding by ELISA in the presence of the nonionic detergent Tween®20. See Figures 3 and S4 for epitope summary. See Table S4 for acidic pH-dependent or pH-independent binding summary.
For the EEEV-specific mAbs, several binding groups emerged with differences in acidic pH dependence and antigen recognition (Figure 4). At neutral pH, EEEV-312 did not bind to EEEV VLPs and preferentially bound recombinant EEEV E1 (Figure 1). However, at acidic pH, EEEV-312 also bound to EEEV VLPs, which suggests that acidic pH is necessary for mAb binding to the fusion loop of EEEV VLPs (Figure 4C.1). Furthermore, one group of mAbs (EEEV-76, −127, −320, −377, and −398) bound to EEEV VLPs with greater binding strength at acidic pH conditions (Figures 4A and 4B). In contrast, another group of mAbs, EEEV-104 and −400, specifically bound to EEEV VLPs in a pH-independent manner (Figure 4D). Lastly, EEEV-109 and −379, recognized EEEV VLPs and EEEV E1 with similar binding strength irrespective of pH (Figures 4E.1 and G).
The ‘pan-alphavirus’ mAbs (EEEV-138, −342, −346, −368, and −387), bound to EEEV, VEEV, WEEV, CHIKV, and MAYV antigens (VLPs and/or recombinant E1) with greater strength at acidic pH conditions (Figure 4C.3). The ‘broadly-reactive’ mAbs, EEEV-307 and −354, bound to EEEV, VEEV, and MAYV (VLPs and/or recombinant E1) more avidly under acidic pH conditions. Some binding reactivity was observed to WEEV VLPs at acidic but not neutral pH. These results suggest that the epitope for EEEV-307 and −354 is less exposed in WEEV virions than on EEEV or VEEV virions (Figure 4C.2). In contrast, EEEV-179 (‘New World’: EEEV, VEEV, and WEEV) and EEEV-157 (‘EEEV-VEEV) bound in a pH-independent manner to their respective antigens (Figures 4E.2 and 4F).
Human E1-specific mAbs do not potently neutralize virus entry.
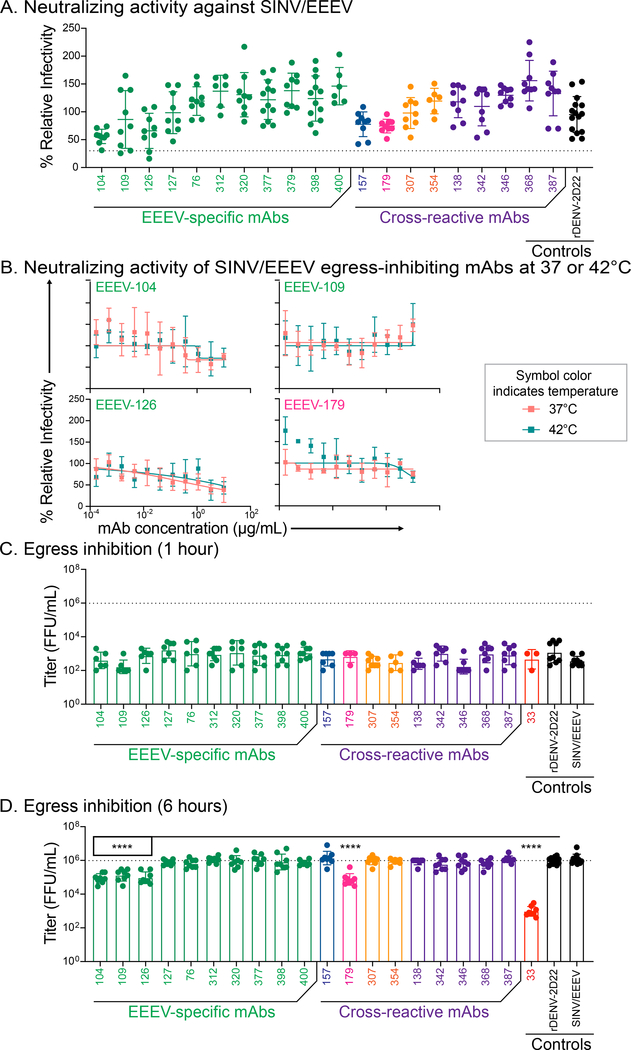
Initially, we tested neutralization activity against SINV/EEEV at an antibody concentration of 10 μg/mL. Most E1-specific mAbs did not show evidence of SINV/EEEV neutralization (as defined by <70% reduction in relative infection) at the concentration tested (Figure 5A and Table S5). Several mAbs (EEEV-104, −109, and −126) that exhibited some modest reduction in SINV/EEEV infectivity were tested at higher assay temperatures (i.e., 42°C) but still showed little inhibitory activity (Figure 5B).
Figure 5. Human E1-specific mAbs that bind exposed, pH-independent E1 epitopes inhibit viral egress.
A. Neutralization activity (10 μg/mL) against SINV/EEEV with mAb name on the x-axis and % relative infectivity on the y-axis. The dotted line indicates 30% relative infectivity as an arbitrary cutoff for neutralization activity. Each mAb is colored based on binding reactivity (EEEV-specific [green], ‘EEEV-VEEV’ [blue], ‘New World’ [magenta], ‘broadly-reactive’ [orange], ‘pan-alphavirus’ [purple], or negative control [black]. Data represent mean ± SD of technical triplicates and of at least 2 independent experiments. B. Neutralization curves for SINV/EEEV egress-inhibiting mAbs, EEEV-104, −109, −126, and −179, (C) against SINV/EEEV at 37°C (light orange squares) or 42°C (teal squares) with mAb concentration (μg/mL) on the x-axis and % relative infectivity on the y-axis. Data represent mean ± SD of technical triplicates of a focus reduction neutralization test experiment. C and D. Focus-forming units (FFUs) of supernatant harvested at either 1 hour (C) or 6 hours (D) following mAb addition in an egress inhibition assay. Reduction in SINV/EEEV FFUs were compared to the negative control mAb, rDENV-2D22, using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test (**** p<0.0001). The positive control neutralizing EEEV E2-specific mAb, EEEV-33 (Williamson et al., 2020), was included. Data represent mean ± SD of technical duplicates and of 2 independent experiments. See Figure S6 for neutralization activity of cross-reactive mAbs against several alphaviruses. See Table S5 for neutralization activity summary.
We also assessed neutralization activity of the cross-reactive mAbs for SINV/EEEV, SINV/VEEV, SINV/WEEV, CHIKV, or MAYV at 37 or 42°C (Figure S6 and Table S5). These mAbs did not neutralize SINV/EEEV, SINV/WEEV, CHIKV, or MAYV at either temperature. However, EEEV-342, −346, −354, and −387 moderately neutralized SINV/VEEV (IC50: 2 to 5 μg/mL). This finding suggests that there may be differential exposure of the fusion loop on native virion particles during morphogenesis or in solution allowing mAb accessibility.
Human EEEV-specific and EEEV-179 (‘New World’) mAbs inhibit SINV/EEEV egress.
The presentation of E2 and E1 on the cell surface prior to virus egress is not well understood. Some E1 mAbs were found to bind epitopes exposed at the plasma membrane (Schmaljohn et al., 1983), which may inhibit virus egress from the cell. To assess this possibility, we performed an egress inhibition assay (Fox et al., 2015). In this method, virus is added to cells prior to addition of mAb, and then supernatant is collected at 1 or 6 hours after mAb addition to capture virus that has egressed from the cell. Given that our human E1-specific mAbs do not appreciably neutralize SINV/EEEV in the standard focus reduction neutralization test (FRNT) assay (Figure 5 and S6), we measured egress by determining the titer of virus present in the supernatant using a focus forming assay (FFA). We observed inhibition of SINV/EEEV egress by several human EEEV-specific mAbs (EEEV-104, −109, and −126) and the ‘New World’ mAb EEEV-179 (Figure 5). This finding suggests that epitopes are exposed prior to virus budding, enabling engagement by mAbs and blockade of virus egress. Also, these data suggest that the standard FRNT assay is likely ‘entry-biased’ and does not capture the activity of egress inhibition even in the presence of mAb.
EEEV-109 (‘EEEV-specific’) exhibits Fc polyfunctionality.
Some alphavirus antibodies require Fc functionality for optimal in vivo efficacy (Earnest et al., 2019; Fox et al., 2019). We evaluated whether the human E1-specific mAbs can mediate Fc effector functions, including antibody-dependent cellular phagocytosis (ADCP), neutrophil phagocytosis (ADNP), complement deposition (ADCD), and natural killer cell activation (ADNKA), in vitro using bead-based assays with recombinant EEEV E1 (Figure S7).
Overall, the mAbs exhibited a higher level of ADCP activity compared to ADNP, suggesting a greater dependence on monocytes than neutrophils for phagocytosis. Additionally, NK activation was observed for many mAbs (EEEV-109, −138, −157, −307, −312, −368, −377, −379, and −387) as reflected by higher percentage of MIP-1β and CD107a positive cells. Few IFNγ positive cells were detected for all mAbs compared to the irrelevant Ebola-virus-specific negative control mAb 13C6. EEEV-138 (‘pan-alphavirus’) and EEEV-354 (‘broadly-reactive’) exhibited high levels of ADCD activity. EEEV-109 (‘EEEV-specific’) exhibited several Fc-mediated effector functions (ADCP, ADNP, ADCD, and ADNKA) with a >2-fold change compared to 13C6. The relatively high Fc activities observed for EEEV-109 suggests these functions might contribute to in vivo efficacy. The remainder of the mAbs did not appear to facilitate Fc activities, as they caused a <2-fold change in the assay readout compared to 13C6 (Figure S7).
EEEV-109 (‘EEEV-specific’) treats EEEV-induced disease after subcutaneous challenge in a Fc-independent manner.
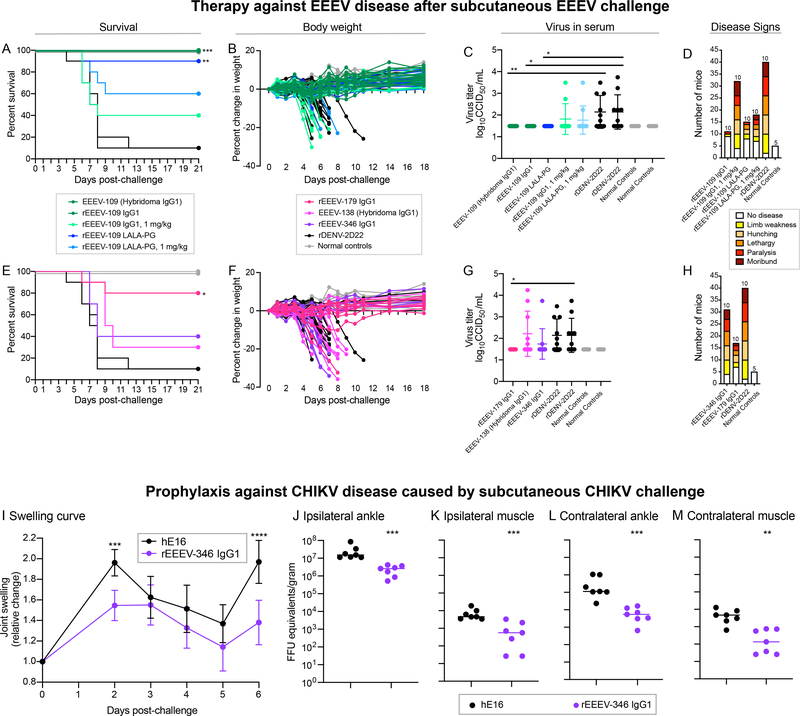
We assessed the therapeutic efficacy of EEEV-109 against EEEV infection following subcutaneous (s.c.) challenge. Administering mAb 24 hours after virus inoculation provides a stringent model for evaluation of mAb in vivo efficacy and more realistic application of potential immunotherapy administration against EEEV. EEEV-109 was selected for study based on its high level of binding to different EEEV and MADV subtypes in the cell-surface antigen display method (Figure 2), pH-independent binding (Figure 4), SINV/EEEV egress inhibition activity (Figure 5), and Fc polyfunctionality (Figure S7). EEEV was inoculated s.c. into C57BL/6 mice at a dose of 103.3 50% cell culture infectious dose (CCID50), which normally results in mice succumbing to neurological disease by about nine days post-inoculation. Twenty-four hours later, EEEV-109 was administered by intraperitoneal (i.p.) injection at a dose of 200 μg mAb/mouse (10 mg/kg), and mice were monitored for 21 days. 100% of the mice treated with EEEV-109 (hybridoma-derived or recombinantly expressed IgG1) survived infection, whereas only 10% of the animals given rDENV-2D22 (negative isotype control) mAb survived (Figure 6A). A dose-response effect was observed, as reduced survival (40%) was observed with a 20 μg mAb/mouse (1 mg/kg) dose compared to rDENV-2D22 (10% survival) (Figure 6A). Given the Fc polyfunctionality observed for EEEV-109, we also assessed the therapeutic efficacy of a Fc knockout variant, LALA-PG (Lo et al., 2017). When administered at 200 μg (10 mg/kg) or 20 μg (1 mg/kg) mAb/mouse, rEEEV-109 LALA-PG exhibited 90% or 60% survival, respectively, compared to rDENV-2D22 (Figure 6A). The minimal reduction in in vivo efficacy observed for the LALA-PG variant suggests that in this model, Fc-mediated effector functions of EEEV-109 are not required. Body weight measurements corresponded with the survival data (Figure 6B). EEEV-109 (WT IgG1 or LALA-PG; 10 mg/kg) also reduced viremia to the limit of detection, whereas most rDENV-2D22-treated animals had virus in the serum 3 days post inoculation (dpi) (Figure 6C). In addition, a reduction in disease signs (limb weakness, hunching, lethargy, paralysis, or moribund) was observed for the rEEEV-109 IgG1 (10 mg/kg) and LALA-PG (10 or 1 mg/kg) mAb treatment groups compared to rDENV-2D22 (Figure 6D).
Figure 6. SINV/EEEV egress inhibiting mAbs, EEEV-109 (‘EEEV-specific’) and EEEV-179 (‘New World’), treat EEEV-induced disease.
A to H. C57BL/6 mice (5 to 7-weeks old) were inoculated subcutaneously (s.c.) with 103.3 CCID50 of EEEV (strain FL93–939) 24 hours prior to mAb administration intraperitoneally (i.p.) at 20 or 200 μg/mouse (1 mg/kg or 10 mg/kg, respectively; n=10). A mock control was included (n=5; grey). A. EEEV-109 (hybridoma-derived) or rEEEV-109 IgG1 (10 mg/kg; green), rEEEV-109 IgG1 (1 mg/kg; light green), rEEEV-109 LALA-PG (10 mg/kg; blue), or rEEEV-109 LALA-PG (1 mg/kg; light blue) mediated 100, 40, 90, or 60%, respectively, therapeutic survival compared to the negative control mAb rDENV-2D22 (10 mg/kg; black). Survival curves were compared using the log-rank test with Bonferroni multiple comparison correction (**padj<0.01, ***padj<0.001). B and F. Percent body weight change of mAb or mock-treated C57BL/6 mice over the course of 18 days after EEEV inoculation. C and G. Virus titer (log10CCID50/mL; y-axis) in serum collected 3 days post-inoculation was determined by an infectious cell culture assay. mAb or mock-treated controls are indicated on the x-axis. Virus titer in the serum for the treatment groups were compared to rDENV-2D22 using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test (*p<0.05, **p<0.01). D and H. Disease signs (limb weakness [yellow], hunching [light orange], lethargy [orange], paralysis [red], and moribund [dark red]) of mAb or mock-treated C57BL/6 mice over the course of 21 days after EEEV inoculation. E. rEEEV-179 IgG1 (10 mg/kg; magenta), EEEV-138 (10 mg/kg; pink), or rEEEV-346 IgG1 (10 mg/kg; purple) mediated 80, 40, or 30%, respectively, therapeutic survival compared to rDENV-2D22 (10 mg/kg; black). Survival curves were compared using the log-rank test with Bonferroni multiple comparison correction (*padj<0.05). I to M. C57BL/6 mice (4-weeks-old) were administered 200 μg/mouse (10 mg/kg; n=7) of rEEEV-346 IgG1 (purple) or the West Nile virus-specific negative control human mAb hE16 (black), via the i.p. route 24 hours prior to s.c. footpad inoculation with 103 FFU of CHIKV strain LR 2006 OPY1. I. rEEEV-346 IgG1 reduced joint swelling at 2 and 6 dpi. Joint swelling in the rEEEV-346 IgG1 treatment group was compared to hE16 using a two-way ANOVA with Dunnett’s multiple comparisons test (***p<0.001, ****p<0.0001). J to M. Viral RNA levels were assessed in the ipsilateral or contralateral ankles (J and L) and muscles (K and M) 6 dpi. Viral RNA levels present within the ipsilateral or contralateral ankles or muscles of the rEEEV-346 IgG1 treatment group were compared to hE16 using a one-way ANOVA with Dunnett’s multiple comparisons test (**p<0.01, ***p<0.001).
EEEV-179 (‘New World’) treats EEEV-induced disease after subcutaneous challenge.
We assessed the therapeutic efficacy of the human cross-reactive mAbs EEEV-138, −179, and −346 against EEEV infection. These three mAbs were selected for study based on their functional activities. EEEV-179 (‘New World’) was selected due to its broad and strong binding reactivity by ELISA-based and cell surface antigen display methods (Figures 1 and 2), potential recognition of a quaternary epitope (Figure 3), pH-independent binding (Figure 4), and ability to inhibit SINV/EEEV egress (Figure 5). EEEV-346 was selected because of its strong ‘pan-alphavirus’ cross-reactivity to the encephalitic (EEEV, VEEV, and WEEV) and arthritogenic (CHIKV) alphaviruses (Figures 1, 2, 4, and S5). EEEV-138 was selected due to its ‘pan-alphavirus’ cross-reactivity (Figure 1) and high level of ADCD activity (Figure S7). In this model, EEEV was inoculated s.c. in C57BL/6 mice at a dose of 103.3 CCID50. Twenty-four hours later, 200 μg mAb/mouse (10 mg/kg) of EEEV-138, −179, or −346 were administered by i.p. injection, and survival was monitored for 21 days. EEEV-138, −179, and −346 resulted in 30%, 80%, or 40% survival, respectively, compared to rDENV-2D22 (10% survival) (Figure 6E). The survival benefit mediated by EEEV-179 was significant (padj<0.05). Again, survival results were consistent with body weights (Figure 6F). Viremia was observed in the serum of mice treated with EEEV-138 or EEEV-346. In contrast, viremia was reduced to the limit of detection in the serum 3 dpi and disease signs (limb weakness, hunching, lethargy, paralysis, or moribund) were reduced for the mice treated with EEEV-179 compared to rDENV-2D22 (Figures 6G and 6H).
EEEV-346 reduces CHIKV-induced joint swelling and viral RNA presence.
To determine if EEEV-346 can confer cross-protection, we assessed the efficacy of this mAb against CHIKV-induced joint swelling and infection. We chose this prophylactic model due to its standard evaluation in the field to assess the potency of anti-CHIKV mAbs against musculoskeletal disease (Fox et al., 2015; Zhou et al., 2020). EEEV-346 was selected because of its strong ‘pan-alphavirus’ cross-reactivity to CHIKV in ELISA and cell surface antigen display methods (Figures 1 and 2). Four-week-old C57BL/6 mice were administered a single 200 μg (10 mg/kg) dose by the i.p. route 24 hours prior to inoculation with 103 FFU CHIKV LR 2006 OPY1. Mice were monitored for 6 days. EEEV-346 reduced joint swelling (Figure 6I) and viral RNA levels in the ipsilateral and contralateral muscles and ankles (Figures 6J–M) compared to the isotype control human mAb WNV E16. Thus, EEEV-346 cross-protects against CHIKV-induced disease in mice.
DISCUSSION
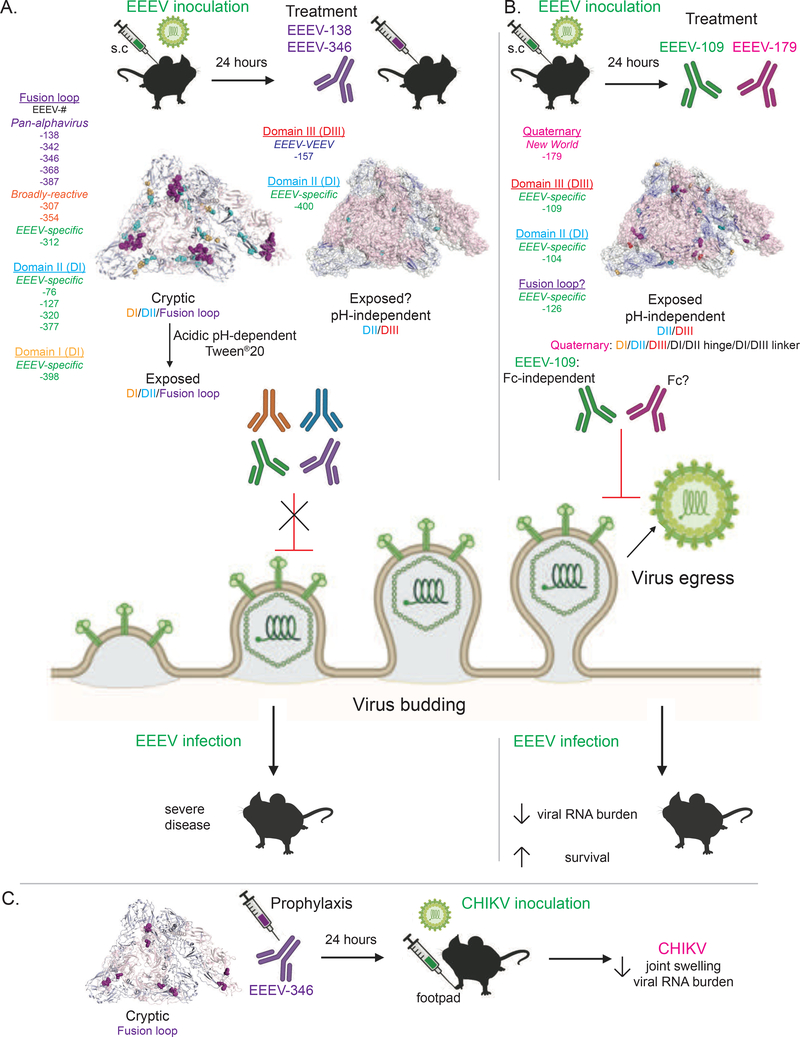
In this study, we identified human E1-specific mAbs isolated from the B cells of EEEV-immune individuals with specificity to EEEV and broad cross-reactivity for the encephalitic and arthritogenic alphaviruses. Human E1-specific mAbs that recognize exposed epitopes avidly bound alphavirus proteins present on the cell surface and inhibited virus egress. This activity in vivo likely contributes to the therapeutic efficacy observed for EEEV-109 and EEEV-179 against EEEV infection in a Fc-independent manner. In contrast, human E1-specific mAbs that recognize cryptic epitopes on intact virions require exposure for stronger mAb recognition (i.e., acidic pH and Tween®20). At the cell surface, the ability for mAb binding to occur suggests the structural proteins are not in the same conformation as on intact virus particles. However, inhibition of SINV/EEEV egress was not observed. In vivo, conditions needed for greater epitope exposure may not naturally occur, which may account for the low therapeutic efficacy observed for EEEV-138 and EEEV-346 against EEEV infection. However, protection was observed by EEEV-346 against CHIKV-induced joint swelling and infection (Figure 7).
Figure 7. Model for mechanism of action of human E1-specific mAbs.
A. Human E1-specific mAbs that target cryptic or partially exposed epitopes do not inhibit SINV/EEEV egress. Exposure for cryptic epitopes depends on pre-treatment conditions, such as acidic pH or addition of the nonionic detergent Tween®20. Treatment efficacy (EEEV-138 and EEEV-346) of EEEV infection following s.c. challenge is minimally significant due to low survival efficacy and presence of viral RNA levels in the serum of treated mice. B. Human E1-specific mAbs that target exposed, pH-independent epitopes can inhibit SINV/EEEV egress. Quaternary epitopes between two adjacent E1 proteins of neighboring trimeric spikes may aid in binding to infected cells for inhibition of virus egress and enable broad alphavirus cross-reactivity (EEEV-179 [‘New World’]). Treatment efficacy (EEEV-109 and −179) of EEEV infection following s.c. challenge corresponds with SINV/EEEV egress inhibition potency due to 100 to 80% survival, respectively, reduction in viral RNA levels in the serum, and disease signs of treated mice. The Fc-mediated effector functions of EEEV-109 did not fully contribute to the treatment efficacy observed. C. A ‘pan-alphavirus’ mAb, EEEV-346, provided cross-protection against CHIKV footpad inoculation through a significant reduction in joint swelling and viral RNA presence. Created with BioRender.com.
The E2 glycoprotein occludes a majority of the E1 glycoprotein in the heterodimer complex, such that exposure of some E1 epitopes may require conformational changes for exposure (Voss et al., 2010; Gibbons et al., 2004; Gibbons et al. 2000). To understand how E1-specific mAbs recognize potentially cryptic E1 epitopes, we initially assessed binding in the presence of the nonionic detergent Tween®20 to disrupt lipid membrane interactions and increase the solubility of hydrophobic proteins. We observed varying degrees of binding reactivity of the mAbs to virus-like particles (VLPs) and recombinant E1 in the presence of Tween®20 by ELISA, with some mAbs showing specificity within the encephalitic alphaviruses (EEEV-specific, ‘EEEV-VEEV’, and ‘New World’), and some displaying cross-reactivity to the arthritogenic alphaviruses (‘broadly-reactive’ and ‘pan-alphavirus’). To mimic more native-like conditions, we tested different treatment conditions to expose potential E1 epitopes in the absence of Tween®20 by ELISA. While we did not detect a difference in binding at elevated temperatures, acidic pH exposed epitopes for several mAbs, indicating occlusion of their epitopes at neutral pH. Acidic pH mimics the endosomal environment during the fusion process, which can result in movement of the flexible B domain of the E2 glycoprotein and dissociation of E2 from E1 (Gibbons et al., 2000; Gibbons et al., 2004; Mukhopadhyay et al., 2006). We mapped the pH-dependent epitopes through complementary techniques (i.e., competition-binding, EEEV and CHIKV alanine scanning mutagenesis, and HDX-MS analyses) to cryptic sites in the E1 glycoprotein, such as the highly conserved fusion loop, residues within the E2/E1 interface, and membrane proximal regions. Human E1-specific mAbs that recognize more exposed E1 epitopes (i.e., DIII and the DII interface or quaternary epitopes involving two E1 proteins of neighboring trimeric spikes) similarly bound irrespective of pH.
In addition to the fusion process, cryptic E1 epitopes may be exposed during assembly of new progeny virions on the surface of infected cells. The observed preference of some E1 mAbs for binding to infected cells compared to native virions, suggests a difference in presentation of the E1 glycoprotein prior to virus budding (Schmaljohn et al., 1983; Meyer et al., 1992). To mimic the antigen display on the surface of cells, we transfected cells with the structural proteins (capsid, E3, E2, 6K, and E1) of multiple alphavirus subtypes. Several EEEV-specific mAbs (EEEV-104, −109, −126, and −312), EEEV-157 (‘EEEV-VEEV’), EEEV-179 (‘New World’), and EEEV-346 (‘pan-alphavirus’) avidly bound respective alphavirus structural proteins. These mAbs (except for EEEV-312 and −346) did not depend on acidic pH or elevated temperatures for epitope exposure on EEEV VLPs by ELISA. Additionally, these mAbs (except for EEEV-157, −312, and −346) inhibited SINV/EEEV egress, which suggests that at the cell surface the epitope is exposed to allow for binding and inhibition of virus budding. Through mapping techniques, the epitopes recognized by these egress-inhibiting mAbs localized to sites within DIII (EEEV-109) and DII proximal to fusion loop (EEEV-104, −126). Thus, transfection of Expi293F cells with different structural proteins may recapitulate the orientation of these proteins observed on the cell surface during virus infection.
EEEV-179 bound to EEEV, VEEV, WEEV, and CHIKV subtypes in cell surface antigen display methods but did not bind to CHIKV E1 in ELISA-based methods. Epitope mapping studies using alanine-scanning mutagenesis described critical interaction residues for EEEV-179 binding to residues proximal to the fusion loop (Y93), the DI/III linker (A286), and DIII (N361) of the E1 glycoprotein. A286 is conserved amongst the encephalitic (EEEV, VEEV, and WEEV) and arthritogenic (CHIKV) alphaviruses, which supports the cross-reactivity binding profile of EEEV-179 to these viruses. A286 was previously identified for a moderately neutralizing CHIKV E1-reactive human mAb (DC2.315; Quiroz et al., 2019, and Kim et al, 2021- Cell companion paper in same issue), which suggests a common epitope or an allosteric effect induced upon mAb binding. DC2.315, a ‘pan-alphavirus’ mAb, also was mapped to a proximal region of the fusion loop on E1. Competition-binding analysis suggests a similar epitope is recognized by EEEV-126. Binding reactivity to EEEV, MADV, and Pixuna virus subtypes in cell surface antigen display methods identified Q79 as a possible critical residue for recognition by EEEV-126. Q79 lies proximal to the fusion loop. Furthermore, HDX-MS studies identified potential interacting peptides within DI, II, and III, DI/DII hinge region, and the DI/III linker with EEEV-179. The large antibody footprint and complementary epitope mapping results identified for EEEV-179 binding to EEEV E1 supports recognition of a quaternary epitope that may be important for broadly cross-reactive E1-specific mAbs that inhibit virus egress. Furthermore, analysis of the ability of EEEV-179 Fab molecules to inhibit virus egress could indicate whether cross-linking of the E1 glycoproteins via binding to the quaternary epitope occurs as has been observed for many neutralizing E2-specific mAbs (Williamson et al., 2020; Fox et al., 2015; Porta et al., 2014).
Fc-mediated effector functions of antibodies also may contribute to clearance of alphavirus infection in vivo. In mouse models the optimal clearance of infection and reduction of joint swelling for CHIKV and decreased MAYV-induced musculoskeletal disease required Fc-FcyR interactions on monocytes (Fox et al., 2019, Earnest et al., 2019). The phagocytic activity exhibited by THP-1 monocytes described here suggests that the human E1-specific mAbs might interact with FcγRs on monocytes to aid in clearance of alphavirus infection in vivo. Removal of effector functions in EEEV-109 via the Fc mutation LALA-PG, however, did not affect therapeutic efficacy against EEEV s.c. challenge.
EEEV-346, a ‘pan-alphavirus’ fusion loop mAb, moderately neutralized SINV/VEEV, with IC50 values of 2 or 3 μg/mL at 37 or 42°C, respectively. Furthermore, EEEV-346 displayed strong binding to ‘pan-alphavirus’ subtypes through ELISA-based and cell surface antigen display methods. Given these criteria, we selected EEEV-346 for in vivo protection studies, in which we observed 40% survival of mice compared to the negative control rDENV-2D22 (10% survival) in an EEEV s.c. challenge model. Viremia was reduced but not significantly. We also selected EEEV-138, due to its high level of ADCD activity, to compare potential Fc-mediated effector functions for the ‘pan-alphavirus’ mAbs. The low efficacy of treatment observed for EEEV-346 and EEEV-138 compared to EEEV-179 and EEEV-109 in this model highlights the importance of egress inhibition as a correlate of protection. In contrast, EEEV-346 reduced joint swelling and CHIKV RNA levels in a s.c. challenge model of CHIKV, thus establishing cross-protection. Although the two murine models cannot be directly compared, EEEV-346 may reduce viral load when given as prophylaxis but not once viral infection is established.
The antibodies described here could be used as tools to elucidate the structural conformation states between native intact virions, cell surface, and subtype differences through structural analyses. The therapeutic efficacy and moderate neutralizing activity through inhibition of virus egress observed here suggests E1-specific mAbs could serve as correlates of protection and could be used as antiviral therapeutic candidates. Antibody cocktail therapeutics or bispecific mAbs combining either two E1-specific mAbs or an E1 mAb and a neutralizing E2-specific mAb may further increase the efficacy of mAbs in vivo against alphavirus-induced disease through cooperative effects and decreased ability for viral escape mutant viruses to emerge. Additionally, isolation of the cross-reactive mAbs described here suggests that immunization with E1 protein could elicit such antibodies in vivo for protection against alphaviruses. Overall, these studies define the basis for molecular recognition of EEEV-specific and ‘pan-alphavirus’ cross-reactive mAbs, which may aid in identifying conserved targets of antibody recognition for alphaviruses that are useful for future development of diagnostic tests, vaccines, and therapeutics.
LIMITATIONS OF THE STUDY
The involvement of Fc-mediated effector functions for in vivo efficacy is not clear. Here we measured the in vitro Fc activities for the panel of human E1-specific mAbs, which may not completely relate to the contribution of Fc in vivo. The therapeutic efficacy of EEEV-109 expressed as human IgG1 (100%) or LALA-PG (90%) against EEEV infection supports this. Further analysis of Fc-mediated effector functions is warranted. Additionally, we observed differences in the efficacy of EEEV-346 in the animal models tested, which may be due to factors such as timing of mAb administration. More study is needed to define the importance of egress inhibition as a correlate of protection for cross-reactive mAbs to other alphaviruses.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information or requests for reagents may be directed to and fulfilled by the Lead Contact: Dr. James E. Crowe, Jr. (james.crowe@vumc.org).
Materials Availability
Materials described in this paper are available from the Lead Contact for distribution under the Uniform Biological Material Transfer Agreement.
Data and Code Availability
Data. All relevant data are included within the manuscript and are available upon request from the Lead Contact.
Code. This study did not generate any code.
Additional information. This study did not generate any additional information.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subject information.
EEEV-76, EEEV-104, EEEV-109, EEEV-126, EEEV-127, EEEV-138, EEEV-157, EEEV-179 were isolated from one research subject as previously described (Williamson et al., 2020). EEEV-307, EEEV-312, EEEV-320, EEEV-342, EEEV-346, EEEV-354, EEEV-368, EEEV-377, EEEV-379, EEEV-387, EEEV-398, and EEEV-400 were isolated from a second research subject. The second research subject was a 60-year-old female subject who succumbed to infection ~4–5 days following mosquito bite(s) at the end of July 2014. The subject was hospitalized for ~1 month. Peripheral blood was collected 17 months after infection. Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood samples by density gradient purification and cryopreserved until use. Written informed consent was given from the subjects and protocols were approved by the Institutional Review Board (IRB) at Vanderbilt University Medical Center for the recruitment and collection of blood samples used in this study.
Mouse model.
Male and female C57BL/6 mice were purchased from Jackson Laboratories. For the EEEV studies, mice were 5 to 7-weeks old; for the CHIKV studies, the mice were 4-weeks old. All animal procedures were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance no. A3381–01) and the Utah State University IACUC protocol #10025.
Cell lines.
Cell lines were maintained as previously described (Williamson et al., 2020). Briefly, BHK-21 (hamster, male origin; ATCC) and Vero (monkey, female origin; ATCC) cells were maintained in DMEM (Gibco) and 10% heat-inactivated FBS (Thermo Fisher Scientific) at 37°C in a humidified atmosphere of 5% CO2. The HMAA 2.5 heteromyeloma cell line (female mouse-female human; kindly provided by Lisa Cavacini) is a non-secreting myeloma cell line and was maintained in ClonalCell™-HY Medium A (Stem Cell Technologies) at 37°C in a humidified atmosphere of 7% CO2, as previously described (Yu et al., 2008a; Yu et al., 2008b). To collect supernatants containing Epstein-Barr virus (EBV), B95.8 (monkey, female; ATCC) cells were cultured in Medium A. Expi293F (human, female origin; Thermo Fisher Scientific) cells were maintained in Expi293 expression medium (Gibco) or Freestyle F17 medium (Gibco) supplemented with 4 mM L-glutamine (Gibco) and 0.1% pluronic F-68 (Gibco) at 125 rpm, 37°C in a humidified atmosphere of 8% CO2. Expi293F cells were authenticated by the ATCC cell line authentication service using Short Tandem Repeat (STR) analysis. ExpiCHO-S cells (hamster, female origin; Thermo Fisher Scientific) were maintained at 37°C in a humidified atmosphere of 8% CO2 in ExpiCHO Expression Medium (Thermo Fisher Scientific). Routine mycoplasma detection was performed using a universal mycoplasma detection kit (ATCC). THP-1 (human, male origin; ATCC) human monocytic cells were maintained in RPMI-1640 (Sigma) containing 10% heat-inactivated FBS (Thermo Fisher Scientific) at 37°C in a humidified atmosphere of 5% CO2. HEK293T (human, female origin; ATCC) cells were grown at 37°C in DMEM supplemented with 10% FBS, 10 mM HEPES pH 7.4, 1 mM Na pyruvate, and 2 mM L-alanyl-L glutamine.
Viruses.
The chimeric viruses Sindbis virus (SINV; TR339)/Eastern equine encephalitis virus (EEEV; strain FL93–939) were described previously (Kim et al., 2019), SINV/Venezuelan equine encephalitis virus (VEEV; strain Trinidad Donkey), and SINV/Western equine encephalitis virus (WEEV; strain McMillian) were kindly provided by William B. Klimstra. Briefly, the structural proteins genes of SINV TR339 were replaced with the structural protein genes of EEEV (FL93–939), VEEV (Trinidad Donkey), and WEEV (McMillian) under control of the SINV 26S subgenomic promoter in the cDNA clone. MAYV (strain TR VL-4675; UTMB) was obtained from the World Reference Center for Emerging Viruses and Arboviruses at UTMB as previously described (Powell et al., 2020). CHIKV (strain 18½5) and the CHIKV LR2006 OPY1 strain for animal studies was produced from an cDNA clone as described previously (Tsetsarkin et al., 2006). The EEEV FL93–939 strain for animal studies was obtained from Dr. Robert Tesh, World Reference Center for Emerging Viruses and Arboviruses (University of Texas Medical Branch), Galveston, TX. The virus was passaged twice in Vero cells prior to use in mice.
Plasmids.
The plasmid for recombinant EEEV E1 ectodomain (strain FL93–939; amino acids Y1-S409) was previously described (Williamson et al., 2020). EEEV subtype I (strains FL93–939, PE-6), MADV (formally EEEV subtypes II-IV [strains PE-3.0815, PE-0.0155, BeAr436087, Cebus/apella/BRA/BEAN5122/1956]), VEEV subtype IAB (strains Trinidad Donkey, TC-83), VEEV subtype IC (strain P676), VEEV subtype ID (strain 3880), VEEV subtype IE (strains Mena II, MX01–22), Mosso das Pedras virus (VEEV subtype IF [strain 78V-3531]), Everglades virus (VEEV subtype II [strain Fe3–7C]), Mucambo virus (VEEV subtype IIIA [strain BeAn 8]), VEEV subtype IIIC (strain 71D-1252), Pixuna virus (VEEV subtype IV [strain BeAr 35645]), Cabassou virus (VEEV subtype V [strain CaAr 508]), Rio Negro virus (VEEV subtype VI [strain Ag80–663]), WEEV (strain McMillian), WEEV (strain Fleming), WEEV (strain Ag80–646), and CHIKV (strain 18½5) structural protein genes (capsid-E3-E2–6K-E1) were codon-optimized, synthesized and cloned into the mammalian expression vectors pcDNA3.1(+) or pTwist CMV BetaGlobin for expression of the WT EEEV or the different alphavirus structural proteins, respectively. Residues Y1-H441 of the EEEV E1 structural protein were mutated to alanine or alanine residues to serine for expression of the EEEV E1 mutants for alanine-scanning mutagenesis library analyses. Recombinant human mAb variable genes were synthesized and cloned into either a pTwist CMV Betaglobin WPRE Neo mammalian expression vector containing the IgG1-specific constant region or a monocistronic vector, pTwist_mCis_hG1 (Zost et al., 2020) (Twist Bioscience Inc).
Recombinant proteins.
Recombinant EEEV E2 glycoprotein (strain v105) was purchased from IBT Bioservices and contains a mixture of the p62 (E3E2) and E2 glycoproteins as previously described (Williamson et al., 2020). Recombinant CHIKV E1 and MAYV E1 proteins were purchased from Meridian Life Science. EEEV, VEEV, and WEEV virus-like particles (VLPs) were kindly provided by Dr. John Mascola at the NIH/NIAID Vaccine Research Center (Ko et al., 2019).
METHOD DETAILS
Virus production.
BHK-21 cells were plated the day before using 3×107 cells per T-225 cm2 flask (Corning). The following day, cells were inoculated with SINV/EEEV, SINV/VEEV, SINV/WEEV, or MAYV at a MOI of 0.2 in DMEM/2% FBS/10 mM HEPES. CHIKV was kindly provided from Dr. Michael S. Diamond’s laboratory. After incubation at 37°C 5% CO2 for 48 hours, virus was harvested by clarification of infected BHK-21 cell supernatants through a 0.2-μm pore size filter (Nalgene). Virus then was used fresh or stored at −80 °C until use.
Recombinant EEEV E1 ectodomain expression.
EEEV E1 ectodomain was produced as previously described (Williamson et al., 2020) using the ExpiFectamine 293 transfection kit (Gibco) and cell supernatant purified through a HisTrap excel column (Cytiva Life Sciences) according to the manufacturer’s protocol on an ÄKTA pure 25M chromatography system.
Human hybridoma and monoclonal antibody (mAb) generation.
Human mAbs were generated as previously described (Williamson et al., 2020). Briefly, cryopreserved PBMCs were thawed, transformed with Epstein-Barr virus (EBV), and fused with the HMMA 2.5 myeloma cell line to generate hybridomas. Hybridomas were cloned by single-cell fluorescence-activated cell sorting and mAbs were purified from hybridoma supernatant filtrate using HiTrap Protein G (Cytiva Life Sciences) or HiTrap MabSelect SuRe (Cytiva Life Sciences) columns on an ÄKTA Pure 25M chromatography system. Antibodies were concentrated using 50K MWCO Amicon® Ultra centrifugal filter units (Millipore) followed by desalting and buffer exchange with 7K MWCO Zeba desalting columns (Thermo Fisher Scientific). The humanized WNV E16 mAb has been described previously (Oliphant et al., 2005) and was purified by protein A affinity chromatography.
Hybridoma supernatant ELISA.
EEEV-76, EEEV-104, EEEV-109, EEEV-126, EEEV-127, EEEV-138, EEEV-157, EEEV-179 were selected as previously described (Williamson et al., 2020). EEEV-307, EEEV-312, EEEV-320, EEEV-342, EEEV-346, EEEV-354, EEEV-368, EEEV-377, EEEV-379, EEEV-387, EEEV-398, and EEEV-400 were screened by ELISA. Recombinant EEEV E1 ectodomain (2 μg/mL; strain FL93–939), CHIKV E1 protein (2 μg/mL; Meridian Life Science), or EEEV, VEEV, WEEV virus-like particles (VLPs; 2 μg/mL) were diluted in 1× D-PBS to coat 384-well ELISA plates (Thermo Fisher Scientific) and incubated at 4°C overnight. For SINV/EEEV, a capture mouse mAb (EEEV-66; (Kim et al., 2019)) was diluted to 0.5 μg/mL in 1× D-PBS to coat 384-well ELISA plates (Thermo Fisher Scientific) and incubated at 4°C overnight. The plates were aspirated and blocked for 1 hour at room temperature with blocking solution (2% non-fat dry milk (Bio-Rad), 2% goat serum (Gibco) in 1× D-PBS-T (Cell Signaling Technology)). Plates then were washed 3× with D-PBS-T and EBV-transformed B cell or hybridoma cell line supernatant was added. For SINV/EEEV, clarified SINV/EEEV supernatant (approximately 1 × 106 to 1 × 107 ffu/mL as determined through focus reduction test (FRT) with BHK-21 cells) diluted in 1× D-PBS was added prior to addition of cell supernatant and incubated at room temperature for 1 to 2 hours. Cell supernatant was then added following 6× washes with D-PBS-T (the first 2 to 3 washes were conducted under BSL2 conditions in a laminar flow biosafety cabinet). Following incubation with cell supernatant for 2 hours at room temperature or overnight at 4°C, plates were washed 3× with D-PBS-T and incubated for 1 hour at room temperature with a suspension of secondary antibodies (goat anti-human IgG-AP (Meridian Life Science) and goat anti-human IgA-AP (Southern Biotech)) at a 1:4,000 dilution in blocking solution (1% non-fat dry milk, 1% goat serum). Plates were washed 4× with 1× D-PBS-T and incubated with alkaline phosphatase substrate solution in the dark for 2 hours at room temperature. Optical density at 405 nm was read with a Biotek™ plate reader.
5′ RACE nucleotide sequence analysis.
Antibody heavy and light-chain variable region genes were sequenced from hybridoma clones generated as described above and as previously described (Williamson et al., 2020). Briefly, total RNA was extracted using the RNeasy Mini kit (Qiagen), amplified with a modified 5′RACE (Rapid Amplification of cDNA Ends) approach, prepared according to the Pacific Biosciences Multiplex SMRT Sequencing protocol, and sequenced on a Pacific Biosciences Sequel platform. The Pacific Biosciences SMRT Analysis tool suite was used to demultiplex and determine the circular consensus sequences (CCS) from the raw sequencing data. Gene segments and germline mutations were determined using the IMGT database or PyIR software (Soto et al., 2020).
Recombinant human IgG1 and Fab production.
Recombinant human EEEV-specific and cross-reactive mAbs and Fabs were produced as previously described (Williamson et al., 2020) using the ExpiFectamine 293 transfection kit (Gibco). rDENV-2D22 was produced as previously described (Zost et al., 2020) using the ExpiCHO Expression System (Gibco). Cell supernatant was purified through HiTrap MabSelect SuRe (Cytiva Life Sciences) according to the manufacturer’s protocol on an ÄKTA pure 25M chromatography system. Antibodies were processed as described for hybridoma mAb generation above.
Digestion of recombinant human IgG1 for Fab generation.
For EEEV-109, EEEV-126, and EEEV-157 Fab molecules, hybridoma mAb or recombinant IgG1 were cleaved using the enzyme FabALACTICA® (Genovis) according to the manufacturer’s protocol. Briefly, up to 0.5 mg of IgG was cleaved with FabALACTICA® for 16 to 18 hours overnight at room temperature. Collection of Fab from uncleaved IgG and cleaved Fc was performed by incubation with CaptureSelect™ Fc resin (Genovis) or CaptureSelect™ IgG-Fc affinity matrix (Thermo Fisher Scientific) for 30 minutes at room temperature.
Protein EC50 ELISA.
Recombinant EEEV E3E2/E2 glycoprotein (strain V105; IBT Bioservices), EEEV E1 ectodomain (strain FL93–939), CHIKV E1 protein (Meridian Life Science), MAYV E1 protein (Meridian Life Science) or EEEV, VEEV, WEEV virus-like particles (VLPs) were diluted to 2 μg/mL in 1× D-PBS to coat 384-well ELISA plates (Thermo Fisher Scientific) and incubated at 4°C overnight. A protein screening ELISA was performed as described above. However, instead of hybridoma supernatant, purified mAb was diluted to 10 μg/mL in blocking solution (1% non-fat dry milk, 1% goat serum in 1× D-PBS-T) and incubated for 2 hours at room temperature. For temperature-dependent binding ELISA, purified mAb was incubated with respective antigen for 1 hour at room temperature (~22°C), 37°C, or 42°C in blocking solution (1% non-fat dry milk, 1% goat serum in 1× D-PBS). For pH-dependent binding ELISA, purified mAb was incubated with respective antigen for 1 hour at room temperature in 1× D-PBS pH 5.4 or 7.4. Additionally, for temperature- and pH-dependent binding ELISAs, all wash and incubation steps were performed with 1× D-PBS and a suspension of secondary antibodies (goat anti-human kappa-HRP (Southern Biotech) and goat anti-human lambda-HRP (Southern Biotech)) at a 1:4,000 dilution in blocking solution (1% non-fat dry milk, 1% goat serum in 1× D-PBS) was added for 1 hour at room temperature. One-step Ultra-TMB ELISA substrate solution (Thermo Fisher Scientific) was added and incubated at room temperature for 10–30 minutes. The reaction was stopped with 1N HCl (Thermo Fisher Scientific) and then read at an optical density of 450 nm with a Biotek™ plate reader.
Cell surface display alphavirus subtype binding.
Transient transfection of Expi293F cells with plasmids containing the structural proteins (capsid-E3-E2–6K-E1) of EEEV subtype I (strains FL93–939, PE-6), MADV (formally EEEV subtypes II-IV [strains PE-3.0815, PE-0.0155, BeAr436087, Cebus/apella/BRA/BEAN5122/1956]), VEEV subtype IAB (strains Trinidad Donkey, TC-83), VEEV subtype IC (strain P676), VEEV subtype ID (strain 3880), VEEV subtype IE (strains Mena II, MX01–22), Mosso das Pedras virus (VEEV subtype IF [strain 78V-3531]), Everglades virus (VEEV subtype II [strain Fe3–7C]), Mucambo virus (VEEV subtype IIIA [strain BeAn 8]), VEEV subtype IIIC (strain 71D-1252), Pixuna virus (VEEV subtype IV [strain BeAr 35645]), Cabassou virus (VEEV subtype V [strain CaAr 508]), Rio Negro virus (VEEV subtype VI [strain Ag80–663]), WEEV (strain McMilian), WEEV (strain Fleming), WEEV (strain Ag80–646), or CHIKV (strain 18½5) was performed using the ExpiFectamine 293 transfection kit (Gibco) according to the manufacturer’s protocol as previously described (Williamson et al., 2020). Cells were incubated at 37°C in a humidified atmosphere of 8% CO2 and harvested 24 hours after transfection via centrifugation at 400 × g for 5 minutes. Cells were resuspended in Expi293F expression medium (Gibco) and 10% DMSO for storage at −80°C and/or in the vapor phase of liquid nitrogen. Untransfected Expi293F cells served as a negative control for non-specific mAb binding and were used fresh following incubation at 37°C in a humidified atmosphere of 8% CO2 at 125 rpm. For analysis of mAb binding, cryopreserved cells were thawed and washed twice with FACS buffer (1× D-PBS, 2% FBS, 2 mM EDTA). Cells were plated at 25,000–50,000 cells/well in 96-well V-bottom plates (Corning). Human EEEV-specific and cross-reactive E1-specific mAbs, positive control mAbs: 1A4B-6 (alphavirus group-reactive (Millipore)), rEEEV-97 IgG (Williamson et al., 2020), 1A3B-7 (VEEV E2-specific (Millipore)), 2A3D-5 (WEEV E1-specific (Millipore)), or the irrelevant mAb negative control, rDENV-2D22, were diluted to 10 μg/mL in FACS buffer and incubated with the cells for 30 minutes at 4°C. Cells were washed with FACS buffer and then incubated with secondary antibodies (anti-human IgG-PE (Southern Biotech) or anti-mouse IgG-PE (Southern Biotech)) diluted 1:1,000 in FACS buffer for 30 minutes at 4°C. Cells were washed with FACS buffer and resuspended in 1:1,000 dilution of DAPI stain in FACS buffer at 25 to 30 μL/well. Number of events were collected on an IntelliCyt® iQue Screener PLUS flow cytometer (Sartorius). For analysis, binding of the mAb to untransfected Expi293F was subtracted to account for any non-specific mAb binding. Fold change for mAb binding to the different alphavirus subtypes was calculated by normalization of the median fluorescence relative to the irrelevant negative control mAb, rDENV-2D22.
Competition-binding ELISA.
EEEV virus-like particles (VLPs) were diluted to 2 μg/mL in 1× D-PBS to coat 384-well plates (Thermo Fisher Scientific) and incubated at 4°C overnight. The plates were aspirated and blocked for 1 hour at room temperature with blocking solution (2% non-fat dry milk, 2% goat serum in 1× D-PBS-T). After blocking, the plates then were washed 3× with 1× D-PBS-T and the first antibody (10 μg/mL) in blocking solution (1% non-fat dry milk, 1% goat serum in 1× D-PBS-T) was added at 20 μL/well for 1 hour at room temperature. Biotinylated second antibody (2.5 μg/mL; final concentration of 0.5 μg/mL) was added in blocking solution (1% non-fat dry milk, 1% goat serum in 1× D-PBS-T) at 5 μL/well for 1 hour at room temperature. The plates then were washed 3× with 1× D-PBS-T and incubated with a solution of secondary antibodies (mouse anti-biotin-HRP (Southern Biotech)) diluted 1:4,000 in blocking solution 1% non-fat dry milk, 1% goat serum in 1× D-PBS-T) for 1 hour at room temperature. The plates then were washed 4× with 1× D-PBS-T followed by addition of One-step Ultra-TMB ELISA substrate solution (Thermo Fisher Scientific). The reaction was stopped with 1N HCl (Thermo Fisher Scientific) and then read at an optical density of 450 nm with a Biotek™ plate reader. Percent binding of each antibody was normalized to optical density value for binding in the presence of the irrelevant negative control mAb, rDENV-2D22. A reduction in signal to <33%, 33 to 67%, or >67% in the presence of rDENV-2D22 was considered full competition, intermediate competition, or no competition, respectively.
Alanine-scanning mutagenesis analysis.
Analysis of EEEV-specific and cross-reactive mAb binding to WT EEEV (strain FL93–939) and EEEV E1 mutant structural proteins (capsid-E3-E2–6K-E1) was performed as previously described (Williamson et al., 2020). Residues were selected for analysis based on previously described epitopes within the Immune Epitope Database (IEDB). Briefly, the WT and E1 mutant structural proteins were expressed through transient transfection of Expi293F cells according to the manufacturer’s protocol. Cells were harvested 24 hours after transfection, stained with LIVE/DEAD™ Fixable Violet Dead Cell Stain Kit (Invitrogen™), and fixed with 1% PFA/PBS. Cells were washed twice with 1× D-PBS and stored at 4°C in FACS buffer until use. Cells were plated at 50,000 cells/well in 96-well V-bottom plates (Corning). Human EEEV-specific and cross-reactive E1-specific mAbs, or the irrelevant mAb negative control, rDENV-2D22, were diluted to either 1, 0.5, 0.1 μg/mL in FACS buffer and incubated with the cells for 1 hour at 4°C. Cells were washed with FACS buffer and then incubated with secondary antibodies (goat anti-human IgG-PE (Southern Biotech) or a mixture of goat anti-human kappa-PE or goat anti-human lambda-PE (Southern Biotech)) diluted 1:1,000 in FACS buffer for 1 hour at 4°C. Cells were washed with FACS buffer and resuspended in 30 μL/well FACS buffer. Number of events were collected on an IntelliCyt® iQue Screener PLUS flow cytometer (Sartorius). Mock transfected Expi293F cells were included as a negative control to subtract background mAb binding to Epxi293F cells. Percent binding of each mAb to the E1 mutants was normalized to the WT EEEV structural protein control. Anti-alphavirus mAb or Fabs were screened at 1 μg/mL for binding to WT CHIKV and CHIKV E2/E1 mutant constructs expressed in HEK 293T cells with binding detected by flow cytometry as previously described (Fong et al., 2014). Antibody reactivity against each mutant protein clone was calculated relative to WT protein reactivity by subtracting the signal from mock-transfected controls and normalizing to the signal from WT-transfected controls.
Hydrogen-deuterium exchange mass spectrometry (HDX-MS).
EEEV E1 glycoprotein and Fabs were prepared at 20 pmol/μL. Labeling occurred in PBS pH 7.4 in D2O at 20°C for 10 seconds, 100 seconds, 1,000 seconds, or 10,000 seconds. The reaction was quenched in 6 M guanidinium/HCl, 500 mM tris (2-carboxyethyl) phosphine in 0.1% formic acid to a pH of 2.3, 0°C. Automated HDX incubations, quenches, and injections were performed using an HDX-specialized nano-ACQUITY UPLC ultraperformance liquid chromatography (UPLC) system coupled to a Xevo G2-XS mass spectrometer. Online digestion was performed at 15°C using an immobilized-pepsin column with generated peptides immediately trapped at 0°C on a VanGuard BEH C18 1.7 μm guard column. Peptides were eluted using 5%–35% acetonitrile, 0.1% formic acid in H2O, and separated on a ACQUITY UPLC BEH C18 1.7 μm, 1 mm × 100 mm column with data being acquired using an MSe data independent analysis (DIA) strategy. Peptide identification was performed using Waters ProteinLynx Global Server 3.0.3 software and deuterium uptake calculated using DynamX 3.0 software.
Focus forming assay (FFA).
Virus titration was performed as previously described (Williamson et al., 2020). Briefly, BHK-21 or Vero cells were inoculated with serial ten-fold dilutions of virus in DMEM/2% FBS/10 mM HEPES (Corning). The virus and cells were incubated at 37°C in 5% CO2 for 1.5 hours followed by addition of a 2% methylcellulose (Sigma-Aldrich):2x DMEM (Millipore):4% FBS:20 mM HEPES overlay. After incubation at 37°C in 5% CO2 for 18 hours, plates were fixed with 1% PFA (Alfa Aesar) for at least 1 hour at room temperature. Plates then were washed 3× with 1× D-PBS followed by 1× Perm Wash (1× D-PBS, 0.1% saponin (Sigma-Aldrich), 0.1% BSA (Sigma-Aldrich)). Immune EEEV, VEEV, WEEV, CHIKV, or MAYV ascites fluid (ATCC) at 1:6,000 dilution in 1× Perm Wash then was added and plates were incubated either at room temperature for 2 hours with rocking or overnight at 4°C. Plates were washed 3 times with 1× D-PBS-T prior to addition of a suspension of secondary antibodies (anti-mouse IgG-Fc-specific-HRP (Jackson ImmunoResearch)) at 1:2,000 dilution in 1× Perm Wash. Plates were incubated for 1 hour at room temperature with rocking then washed 3 times with 1× D-PBS-T. TrueBlue™ peroxidase substrate solution (SeraCare) then was added and plates were incubated for ~15 minutes at room temperature. Plates were gently rinsed with MilliQ water and air dried. Plates were imaged on an ImmunoSpot S6 Universal machine (CTL).
Temperature-dependent focus reduction neutralization test (FRNT).
Neutralization activity of EEEV-specific and cross-reactive E1-specific mAbs was performed as previously described (Williamson et al., 2020). Briefly, a FFA was performed as described above. However, prior to addition of virus to cells, purified mAb was diluted to 20 μg/mL (final concentration 10 μg/mL) and serially diluted three-fold in DMEM/2% FBS/10 mM HEPES. MAb-only dilutions were separated to serve as a negative control. Virus then was diluted to 2,000 ffu/mL (~50 to100 ffu/well) in DMEM/2% FBS/10 mM HEPES and added to the mAb serial dilutions. The mAb:virus mixture was incubated at 37°C or 42°C in 5% CO2 for 1 hour. After cells were washed 2× with 1× D-PBS, the mAb:virus mixture was then added to the cells and incubated at 37°C in 5% CO2 for 1.5 hours. A 2% methylcellulose:2× DMEM or 2× EMEM:4% FBS:20 mM HEPES overlay was then added to the cells and incubated at 37°C in 5% CO2 for 18 hours. Plates were fixed and immunostained as described for FFA.
Egress inhibition assay.
BHK-21 cells were plated in 12-well plates (Corning) at 3 × 105 cells/well in DMEM/5% FBS/10 mM HEPES at 800 μL/well. Cells were incubated at 37°C in 5% CO2 overnight. SINV/EEEV was diluted to MOI 1 in DMEM/2% FBS/10 mM HEPES and added to cells for incubation at 37°C in 5% CO2 for 2 hours. Cells then were washed 5× with DMEM/2% FBS/10 mM HEPES followed by a wash with DMEM/2% FBS/10 mM HEPES/25 mM NH4Cl. Purified mAb was diluted to 10 μg/mL in DMEM/2% FBS/10 mM HEPES/25 mM NH4Cl and added to cells at 37°C in 5% CO2 for up to 6 hours. Supernatant was harvested following 1 hour and 6 hours incubation period. Virus presence was determined through FFA at 37°C for 1 hour as described above.
Antibody-dependent cellular phagocytosis (ADCP) assay.
The ADCP assay was adapted from Ackerman et al. 2011. Briefly, antigen was biotinylated using sulfo-NHS LC-LC biotin, coupled to yellow-green fluorescent Neutravidin 1 μm beads (Invitrogen™) for 2 hours at 37°C and washed two times in 0.1% BSA in PBS. 10 μL/well of coupled beads were added to 96-well plates with 100 μL/well of antibodies at a concentration of 5, 1, 0.2, 0.04, 0.008 or 0.0016 μg/mL for 2 hours at 37°C to form immune complexes. After incubation, the immune complexes were spun down and the supernatant was removed. THP-1 cells were added at a concentration of 2.5 × 104 cells/well and incubated for 18 hours at 37°C. After incubation, the plates were spun down, the supernatant was removed, and cells were fixed with 4% PFA for 10 minutes. Fluorescence was acquired with an Intellicyt iQue Screener. Phagocytic score was calculated using the following formula: (percentage of FITC+ cells) * (the geometric mean fluorescent intensity (gMFI) of the FITC+ cells)/10,000.
Antibody-dependent neutrophil phagocytosis (ADNP) assay.
The ADNP assay was adapted from Karsten et al., 2019. Antigens were coupled to beads and immune complexes were formed as described for ADCP. Neutrophils were isolated from fresh whole blood. Erythrocytes were lysed with ammonium-chloride potassium (ACK) lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA, pH 7.4) and leukocytes were separated out by centrifugation. Leukocytes were washed with cold PBS, resuspended in R10, and added to plates at a concentration of 5 × 104 cells/well. The plates were incubated for 1 hour at 37°C. The neutrophil marker CD66b (Pacific Blue conjugated anti-CD66b; BioLegend) was used to stain cells. Cells were fixed for 10 minutes in 4% PFA. Fluorescence was acquired with an Intellicyt iQue Screener and phagocytic score was calculated as described for ADCP.
Antibody-dependent complement deposition (ADCD) assay.
The ADCD assay was adapted from Fischinger et al. 2019. Antigen was coupled to red fluorescent Neutravidin 1 μm beads (Invitrogen™) as described for ADCP. Immune complexes were formed by incubating 10 μL of coupled beads with 50 μL of antibody at concentrations of 100, 20, 4, 0.8, 0.16 or 0.032 μg/mL for 2 hours at 37°C. Plates were spun down, and immune complexes were washed with PBS. Lyophilized guinea pig complement (Cedarlane Labs) was resuspended in 1 mL of cold water, diluted 1:50 in GVB++ (gelatin veronal buffer and additional Ca2+ and Mg2+, Boston BioProducts) and added to the immune complexes. The plates were incubated for 20 minutes at 37°C and the reaction was stopped by washing the plates twice with 15 mM EDTA in PBS. To detect complement deposition, plates were incubated with fluorescein-conjugated goat anti-guinea pig complement C3 (MP Biomedicals) for 15 minutes in the dark. Fluorescence was acquired with an Intellicyt iQue Screener.
Antibody-dependent NK cell activation (NK activation).
Human NK cells were isolated from buffy coats using RosetteSep NK cell enrichment kit (STEMCELL Technologies) and Ficoll separation. The isolated NK cells were rested overnight at 1.5 × 106 cells/mL in IL-15 at 37°C. ELISA plates were coated with antigen at 300 ng/well and incubated for 2 hours at 37°C. Plates were blocked with 5% BSA in PBS overnight at 4°C. The next day, 100 μL of antibodies at a concentration of 5, 1 or 0.2 μg/mL, were added to the plates. Plates were incubated for 2 hours at 37°C to form immune complexes. After the incubation, NK cells were added to the plates at 5 × 104 cells/well in R10 supplemented with anti-CD107a PE-Cy5, BFA and GolgiStop (BD Biosciences). Plates were incubated for 5 hours at 37°C. Following the incubation, NK cells were stained for surface markers with anti-CD56 PE-Cy7, anti-CD16 APC-Cy7 and anti-CD3 Pacific Blue (BD Biosciences). NK cells were fixed and permeabilized with Fix & Perm cell permeabilization kit (Invitrogen™). Cells were incubated with anti-MIP1β PE and anti-IFNγ FITC (BD Biosciences) to stain for intracellular markers. Cells were acquired on an Intellicyt iQue Screener.
Mouse subcutaneous challenge with EEEV.
Male and female mice 5–7 weeks of age were challenged with EEEV via bilateral SC injections. Animals were treated with antibodies at doses of 10 mg/kg or 1 mg/kg via a single IP injection 24 hours post-virus challenge. Animals were monitored until 21 days post-virus inoculation (dpi) for disease signs (limb weakness, hunching, lethargy, paralysis, and moribund) and survival. Early euthanasia criteria were used where possible, but some animals succumbed to virus infection between well checks. Criteria for early euthanasia included inability for the animal to right itself, no mobility or ability to access food and water, or non-responsiveness to stimuli. Individual weights were recorded daily 0–10 dpi and on 14 and 18 dpi. Serum was collected from all mice 3 dpi for assessment of serum viremia. Viral titers on 3 dpi were the most consistent of any timepoint tested, even though the highest average titers often occur on 2 dpi.
EEEV Infectious cell culture assay.
Virus titer was quantified using an infectious cell culture assay where a specific volume of either tissue homogenate or serum was added to the first tube of a series of dilution tubes. Serial dilutions were made and added to Vero cells. Three days later cytopathic effect (CPE) was used to identify the end-point of infection. Four replicates were used to calculate the 50% cell culture infectious doses (CCID50) per mL of plasma or gram of tissues.
Mouse subcutaneous challenge with CHIKV.
Male and female C57BL/6 mice in equal numbers at 4 weeks of age were housed in groups of up to 5 mice per cage. Age- and sex-matched mice were inoculated subcutaneously in the footpad with 103 FFU of CHIKV. Joint swelling was measured using a digital caliper as described previously (Fox et al., 2019).
Measurement of CHIKV burden.
CHIKV-infected mice were euthanized at 6 dpi and perfused extensively with 20 mL of PBS. Tissues were harvested, weighed, and homogenized with zirconia beads in a MagNA Lyser instrument (Roche Life Science) in 500 μL of DMEM media supplemented with 2% heat-inactivated FBS. All tissue homogenates were clarified by centrifugation at 10,000 rpm for 5 min and stored at −80°C. RNA was extracted using an Applied Biosystems 5x MagMax RNA v96 viral isolation kit (Thermo Fisher Scientific) and a Kingfisher duo prime extraction machine (Thermo Fisher Scientific). CHIKV RNA levels were determined by one-step quantitative reverse transcriptase PCR (qRT-PCR) using an Applied Biosystems Taqman RNA-to-Ct 1-step kit (Thermo Fisher Scientific) on an ABI 7500 Fast Instrument using standard cycling conditions and previously designed primer/probe sets for CHIKV (Fox et al., 2015). Viral burden was expressed on a log10 scale as CHIKV RNA equivalents per gram or mL after comparison with a standard curve produced using RNA isolated from viral stocks as a standard curve to determine FFU equivalents, as previously described (Fox et al., 2019).
QUANTIFICATION AND STATISTICAL ANALYSIS
Details about statistical analyses can be found in the figure legends. Half-maximal effective concentration (EC50) and half-maximal inhibitory concentration (IC50) values were determined after log transformation of concentration values and non-linear regression analysis using sigmoidal dose-response (variable slope). One-way ANOVA with Dunnett’s multiple comparisons test was used to compare virus titer in the serum of EEEV inoculated mice (*p<0.05, **p<0.01). Joint swelling in the CHIKV mouse model was compared using a two-way ANOVA with Dunnett’s multiple comparisons test (***p<0.001, ****p<0.0001). CHIKV RNA levels were analyzed with a one-way ANOVA with Dunnett’s multiple comparisons test (**p<0.01, ***p<0.001). Survival curves were compared using the log-rank test with Bonferroni multiple comparison correction (*padj<0.05, **padj<0.01, ***padj<0.001; padj = 1-(1-porig)n; n = number of comparisons [in Figure 6 n = 8]). Statistical analyses were performed with GraphPad Prism v8.
Supplementary Material
Figure S1, related toFigure 2. Sequence alignment of the E1 protein for diverse alphavirus subtypes. A. Multiple amino acid sequence alignment of the E1 protein for alphavirus subtypes (virus, subtype, and strain) labeled in the left-hand column (made with Geneious software). Alphavirus subtypes are grouped by antigenic complex for EEEV (green), VEEV (blue), WEEV (orange), or Semliki Forest virus (magenta). Dots represent conserved amino acids. Amino acids that are not conserved are colored based on polarity (cyan [polar, uncharged], orange [non-polar], red [polar, acidic], or dark blue [polar, basic]). The consensus sequence is shown at the top of the sequence alignment. Respective domains and regions of interest are labeled accordingly (domain I [light orange], domain II [blue], domain III [red], fusion loop [purple], domain I/domain II hinge and domain I/III linker [black]). B. Amino acid percent identity of the E1 protein for alphavirus subtypes (virus, subtype, and strain). Alphavirus subtypes are grouped by antigenic complex for EEEV (green), VEEV (blue), WEEV (orange), or Semliki Forest virus (magenta). The heat map corresponds to amino acid percent identity between alphavirus subtypes, ranging from 40–99.9% (black to light orange). White boxes with a cross include the amino acid percent identity for the same alphavirus subtype (100%).
Figure S2, related toFigure 3. Human cross-reactive E1-specific mAbs recognize the fusion loop. A. Binding of human cross-reactive E1-specific mAbs to CHIKV E2/E1 clones. MAbs or Fabs were screened at a 1 μg/mL concentration for binding to CHIKV E2/E1 constructs expressed in HEK 293T cells. Binding was detected by flow cytometry (as described previously; Fong et al., 2014). Binding to each mutant protein clone was calculated relative to that for wild-type (WT) protein by subtracting the signal from mock-transfected controls and normalizing to the signal from WT-transfected controls. Data represent the mean (standard deviation) of four replicate data points. B. Epitope mapping of critical residues identified by alanine-scanning (A) for human cross-reactive E1-specific mAbs binding to the CHIKV E1 glycoprotein. Critical residues were mapped onto the CHIKV E2/E1 heterodimer (PDB ID: 3N42). A side view of the CHIKV E2/E1 heterodimer (E2 – light pink, E1 – light blue) is shown with W89, F95, and N100 critical interaction residues (purple spheres) indicated for the cross-reactive E1-specific mAbs. C. Heatmap of the EEEV E1 glycoprotein alanine mutant residues tested for human E1-specific mAbs. Residues were selected for analysis based on previously described epitopes within the Immune Epitope Database (IEDB). The average percent binding is displayed as a heatmap with >75% (maroon), 30 to 75% (light blue), or <30% (magenta) mAb binding relative to WT. Each mAb is colored according to binding reactivity (neutralizing EEEV E2-specific mAb [red], ‘pan-alphavirus’ [purple], ‘broadly-reactive’ [orange], ‘New World’ [magenta], ‘EEEV-VEEV’ [blue], EEEV-specific [green]) and is ordered to correspond with the competition-binding groups as defined in Figure 3A.
Figure S3, related toFigure 3. Hydrogen-deuterium exchange mass spectrometry (HDX-MS) analysis of EEEV-109, −126, −157, and −179 Fab molecules binding footprint to the EEEV E1 glycoprotein. A. E1 peptide sequence coverage map for EEEV-126 Fab bound EEEV E1 glycoprotein. Peptides covered during HDX-MS analysis are indicated by the blue lines. B. Potential interacting peptides for EEEV-179 (‘New World’ [magenta]), −109 (EEEV-specific [green]), and −157 (‘EEEV-VEEV’ [blue]), as defined by at least one time point with ≥ 0.4 Da ± 2.4 standard deviations relative deuterium uptake (y-axis) of Fab bound versus unbound EEEV E1 glycoprotein over the course of the 10,000 s exposure time (x-axis). The labels at each time point indicate the change in relative deuterium uptake of Fab bound versus unbound EEEV E1 glycoprotein.
Figure S4, related toFigure 3. Epitope mapping of human E1-specific mAbs. A to C. Epitope mapping from cell surface antigen display, EEEV and CHIKV E1 alanine-scanning mutagenesis, and hydrogen-deuterium exchange mass spectrometry (HDX-MS) analyses of human E1-specific mAbs binding to the E1 glycoprotein. Critical interaction residues were mapped onto the cryo-EM reconstruction of SINV/EEEV (PDB: 6MX4). Potential antibody footprint peptides identified through HDX-MS were mapped onto the cryo-EM reconstruction of EEEV VLP (PDB: 6XO4). A top trimeric view with an asymmetric unit of a neighboring spike (E2 – light pink, E1 – light blue) is shown with critical interaction residues (spheres) indicated for the E1-specific mAbs. Residues are labeled and colored based on E1 domain or region of interest (domain I (DI) [light orange], domain II (DII) [cyan], fusion loop [purple], domain III (DIII) [red], DI/II hinge and DI/DIII linker [black]) Each mAb is grouped according to antigen binding reactivity and epitope (A – DII: ‘pan-alphavirus’ [purple], ‘broadly-reactive’ [orange], ‘New World’ [magenta], EEEV-specific [green]; B – DIII: ‘EEEV-VEEV’ [blue], EEEV-specific [green]; C – DI: EEEV-specific [green]) as described in Figure 3.
Figure S5, related toFigures 1, 4, and S6. Temperature-independent binding of human E1-specific mAbs to virus-like particles (VLPs) or recombinant E1 glycoproteins. A through G. Representative binding curves for human E1-specific mAbs binding to EEEV (green), VEEV (dark blue), or WEEV (orange) VLPs, recombinant EEEV (light purple), CHIKV (magenta), or MAYV (purple) E1 glycoproteins, or recombinant EEEV E2 (blue) glycoprotein at room temperature (open circles), 37°C (open triangles), or 42°C (open squares) with mAb concentration (μg/mL) on the x-axis and optical density at 450 nm on the y-axis. Binding curves are ordered numerically and grouped according to binding reactivity and strength (EEEV VLP and E1 protein-reactive [A; VLP/protein EC50 ratio <1], EEEV VLP > EEEV E1 protein [B; VLP/protein EC50 ratio = not applicable (NA) to EEEV E1 protein], EEEV E1 protein > EEEV VLP [C; VLP/protein EC50 ratio >1 or NA to EEEV VLP], ‘EEEV-VEEV’ [D], ‘New World’ [E], ‘broadly-reactive’ [F], or ‘pan-alphavirus’ [G]). H. EC50 values (ng/mL) for binding of human EEEV-specific E1-specific mAbs to EEEV VLPs or recombinant EEEV E1 glycoprotein at room temperature, 37 °C, or 42°C. The human EEEV-specific mAbs are grouped according to binding strength (EEEV VLP and E1 protein-reactive, EEEV VLP > EEEV E1 protein, and EEEV E1 protein > EEEV VLP) and listed in order of increasing EC50 value for binding to EEEV VLPs for each respective group. EC50 value in ng/mL is indicated by the blue fill color (≤100.00 [dark blue], 101 to 500 [light blue], 501 to 5,000 [lightest blue], and >5,001). Ratio of binding to EEEV VLPs compared to EEEV E1 glycoprotein at room temperature is indicated as the ratio of EC50 values. Increasing depth of green color indicates lower ratios and thus more dependence on virus particle-specific epitopes. I. EC50 values (ng/mL) for binding of human cross-reactive E1-specific mAbs to EEEV VLPs or recombinant EEEV E1 glycoprotein at room temperature, 37°C, or 42°C. The human cross-reactive mAbs are grouped according to binding cross-reactivity (‘EEEV-VEEV’, ‘New World’, ‘broadly-reactive, and ‘pan-alphavirus’) and listed in order of increasing EC50 value for binding to EEEV VLPs for each respective group. EC50 value in ng/mL is indicated by blue fill color (≤100.00 [dark blue], 101 to 501 [light blue], 501 to 5,000 [lightest blue], and >5,001). Ratio of binding to EEEV VLPs compared to EEEV E1 glycoprotein at room temperature is indicated as the ratio of EC50 values. A VLP/protein EC50 ratio of <1 suggests a greater dependence on virus particle-specific epitopes. Data in A to I represent mean ± SD of technical triplicates and are representative of two independent experiments. See Figure 1 for ELISA binding of human E1-specific mAbs in the presence of the nonionic detergent Tween®20. See Table S1 for summary of human E1-specific mAbs binding to VLPs and recombinant proteins in the presence of the nonionic detergent Tween®20. See Table S3 for temperature-independent binding summary for human E1-specific mAbs binding to VLPs and recombinant proteins.
Figure S6, related toFigure 5. Human cross-reactive E1-specific mAbs moderately neutralize SINV/VEEV. Neutralization curves of human cross-reactive E1-specific mAbs against SINV/EEEV, SINV/VEEV, SINV/WEEV, CHIKV, or MAYV at 37°C (light pink squares) or 42°C (teal squares) with mAb concentration (μg/mL) on the x-axis and % relative infectivity on the y-axis. Each mAb is grouped according to cross-reactivity (‘EEEV-VEEV’ [blue], ‘New World’ [magenta], ‘broadly-reactive’ [orange], ‘pan-alphavirus’ [purple]) and ordered numerically within each group. Half-maximal inhibitory concentration (IC50) values (ng/mL) for moderately neutralizing mAbs (EEEV-342, −346, −354, and −387) against SINV/VEEV are indicated at each temperature (light pink - 37°C, teal - 42°C). Data represent mean ± SD of technical triplicates of a focus reduction neutralization test (FRNT) experiment.
Figure S7, related toFigure 6. Fc-mediated effector function activities for human E1-specific mAbs. A and B. Monocyte and neutrophil phagocytosis (antibody-dependent cellular phagocytosis [ADCP; maroon diamonds] and antibody-dependent neutrophil phagocytosis [ADNP; light blue squares], respectively), antibody-dependent complement deposition (ADCD; teal triangles), and positive NK cells (antibody-dependent natural killer cell activation [ADNKA; MIP-1β (orange circles), CD107a (open purple circles), and IFNγ (open magenta squares)] activity was assessed for human EEEV-specific (A) or cross-reactive (B) mAbs using recombinant EEEV E1-glycoprotein-coated beads. Each mAb is colored based on binding reactivity (EEEV-specific [green], ‘EEEV-VEEV’ [blue], ‘New World’ [magenta], ‘broadly-reactive’ [orange], ‘pan-alphavirus’ [purple]). The corresponding graphs represent mAb concentration (μg/mL) on the x-axis and phagocytic score (in thousands [ADCP and ADNP]), median fluorescence intensity of anti-C3-FITC binding (in thousands [ADCD]), or % positive cells (ADNKA) on the y-axis. The Ebola virus-specific negative control mAb 13C6 is shown in red squares. The red dotted line represents the mean y-axis value. The black dotted line indicates the mean y-axis value for a no-mAb negative control. Data represent mean ± SD of two independent experiments.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| EEEV-76 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-104 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-109 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-126 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-127 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-138 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-157 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-179 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-307 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-312 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-320 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-342 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-346 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-354 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-368 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-377 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-379 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-387 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-398 (hybridoma-produced IgG1) | This paper | N/A |
| EEEV-400 (hybridoma-produced IgG1) | This paper | N/A |
| rEEEV-97 IgG (recombinant Expi293F-produced IgG1) | Williamson et al., 2020 | N/A |
| rEEEV-109 IgG (recombinant Expi293F-produced IgG1) | This paper | N/A |
| rEEEV-109 LALA-PG (recombinant Expi293F-produced IgG1) | This paper | N/A |
| rEEEV-126 IgG (recombinant Expi293F-produced IgG1) | This paper | N/A |
| rEEEV-157 IgG (recombinant Expi293F-produced IgG1) | This paper | N/A |
| rEEEV-346 IgG (recombinant Expi293F-produced IgG1) | This paper | N/A |
| rDENV-2D22 IgG (recombinant ExpiCHO-produced IgG1) | Fibriansah et al., 2015 | N/A |
| Murine mAb: EEEV-66 | Michael S. Diamond | Kim et al., 2019 |
| Eastern equine encephalomyelitis immune ascites fluid | ATCC | Cat# VR-1242AF |
| Venezuelan equine encephalitis virus hyperimmune ascitic fluid | ATCC | Cat# VR-1249AF |
| Western equine encephalomyelitis virus immune ascitic fluid | ATCC | Cat# VR-1251AF |
| Chikungunya virus immune ascitic fluid | ATCC | Cat# VR-1241AF |
| Mayaro virus immune ascitic fluid | ATCC | Cat# VR-1277AF |
| 1A4B-6 | Millipore | Cat# MAB8754; RRID:AB_11211668 |
| 1A3B-7 | Millipore | Cat# MAB8755; RRID:AB_95409 |
| 2A3D-5 | Millipore | Cat#MAB8746; RRID:AB_95399 |
| Goat anti-human IgG-AP | Meridian Life Science | Cat# W90088A; RRID:AB_205090 |
| Goat anti-human IgA-AP | Southern Biotech | Cat# 2050-04; RRID:AB_2795704 |
| Goat anti-human Kappa-HRP | Southern Biotech | Cat# 2060-05; RRID:AB_2795720 |
| Goat anti-human Lambda-HRP | Southern Biotech | Cat# 2070-05; RRID:AB_2795753 |
| Goat anti-mouse IgG-HRP | Jackson ImmunoResearch | Cat# 115035008; RRID:AB_2313585 |
| Goat anti-human IgG-PE | Southern Biotech | Cat# 2040-09; RRID:AB_2795648 |
| Goat anti-mouse IgG-PE, human adsorbed | Southern Biotech | Cat# 1030-09S; RRID:AB_2794298 |
| Mouse anti-biotin-HRP | Southern Biotech | Cat# 6404-05; RRID:AB_2796299 |
| Pacific Blue™ anti-human CD66b antibody | BioLegend | Cat# 305112; RRID:AB_2563294 |
| Anti-guinea pig complement C3 goat IgG fraction, fluorescein-conjugated | MP Biomedicals | Cat# 0855385; RRID:AB_2334913 |
| PE-Cy™5 Mouse Anti-Human CD107a | BD Biosciences | Cat# 555802; RRID:AB_396136 |
| PE-Cy™7 Mouse Anti-Human CD56 | BD Biosciences | Cat# 557747; RRID:AB_396853 |
| APC-Cy™7 Mouse Anti-Human CD16 | BD Biosciences | Cat# 557758; RRID:AB_396864 |
| Pacific Blue™ Mouse Anti-Human CD3 | BD Biosciences | Cat# 558124; RRID:AB_397044 |
| PE Mouse Anti-Human MIP-1β | BD Biosciences | Cat# 550078; RRID:AB_393549 |
| FITC Mouse Anti-Human IFN-y | BD Biosciences | Cat# 340449; RRID:AB_400425 |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| Epstein-Barr virus (EBV) | This paper | N/A |
| Sindbis (TR339)/Eastern equine encephalitis virus (FL93-939) (SINV/EEEV) | Michael S. Diamond | Kim et al., 2019 |
| Sindbis (TR339)/Venezuelan equine encephalitis virus (Trinidad Donkey) (SINV/VEEV) | William B. Klimstra | N/A |
| Sindbis (TR339)/Western equine encephalitis virus (McMillian) (SINV/WEEV) | William B. Klimstra | N/A |
| CHIKV (181/25) | Michael S. Diamond | N/A |
| MAYV (TR VL-4675) | World Reference Center for Emerging Viruses and Arboviruses (UTMB) | Powell et al., 2020 |
| EEEV FL93-939 | World Reference Center for Emerging Viruses and Arboviruses (UTMB) | N/A |
| CHIKV LR2006 OPY1 | Tsetsarkin et al., 2006 | N/A |
|
| ||
| Biological Samples | ||
|
| ||
| Peripheral blood mononuclear cells (PBMCs) from EEEV survivors | This paper; Williamson et al., 2020 | N/A |
| Human PBMCs, from whole blood leukofiltration filters | Nashville Red Cross | N/A |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| Chk2 Inhibitor II Hydrate | Sigma-Aldrich | Cat# C3742 |
| Cyclosporin A | Sigma-Aldrich | Cat# C1832 |
| Ouabain Octahydrate | Sigma-Aldrich | Cat# O3125 |
| 50 × HAT Medium Supplement | Sigma-Aldrich | Cat# H0262 |
| IL-15 | StemCell Technologies, Inc. | Cat# 78031.3 |
| DAPI (4’, 6-diamidino-2-phenylindole, dihydrochloride) | Thermo Fisher Scientific | Cat# D1306 |
| LIVE/DEAD™ Fixable Violet Dead Cell Stain Kit | Thermo Fisher Scientific | Cat# L34964 |
| Fix/Perm cell permeabilization kit | Invitrogen | Cat# GAS004 |
| Paraformaldehyde | Alfa Aesar | Cat# J61899-AP |
| Saponin | Sigma-Aldrich | Cat# 47036 |
| Methylcellulose | Sigma-Aldrich | Cat# M7027 |
| Ammonium chloride | Sigma-Aldrich | Cat# A9434 |
| 2 × Dulbecco’s Modified Eagle’s Medium (DMEM) | Millipore | Cat# SLM202B |
| 2 × Eagle’s Minimal Essential Medium (EMEM) | Quality Biological | Cat# 115-073-101 |
| TrueBlue™ Peroxidase Substrate Solution | SeraCare | Cat# 5510-0030 |
| L-Glutamine (200 mM) | GIBCO | Cat# 25030081 |
| Pluronic™ F-68 Non-ionic Surfactant (100X) | GIBCO | Cat# 24040032 |
| Fetal Bovine Serum, ultra-low IgG | Thermo Fisher Scientific | Cat# 16250078 |
| Low-Tox® Guinea Pig Complement | CEDARLANE | Cat# CL4051 |
| Gelatin Veronal Buffer | Boston BioProducts | Cat# IBB-300X |
| Expi293 Expression Medium | GIBCO | Cat# A1435101 |
| Freestyle F17 Expression Medium | GIBCO | Cat# A1383502 |
| Opti-MEM™ I Reduced Serum Medium | GIBCO | Cat# 31985088 |
| ExpiFectamine 293 Transfection Kit | GIBCO | Cat# A14525 |
| ExpiCHO Expression System Kit | GIBCO | Cat# A29133 |
| RosetteSep™ Human NK Cell Enrichment Cocktail | Stem Cell Technologies | Cat# 15025 |
| ClonaCell™-HY Medium A | Stem Cell Technologies | Cat# 03081 |
| ClonaCell™-HY Medium E | Stem Cell Technologies | Cat# 03085 |
| Hybridoma SFM | GIBCO | Cat# 12045084 |
| 1-Step Ultra TMB-ELISA | Thermo Fisher Scientific | Cat# 34029 |
| Recombinant EEEV E2 GP (E3E2) | IBT Bioservices | Cat# 0560-001 |
| CHIKV E1 protein | Meridian Life Science | Cat# R01653 |
| MAYV E1 protein | Meridian Life Science | Cat# R01779 |
| EEEV E1 glycoprotein | Williamson et al., 2020 | N/A |
| EEEV virus-like particles (VLPs) | NIH Vaccine Research Center | Ko et al., 2019 |
| VEEV virus-like particles (VLPs) | NIH Vaccine Research Center | Ko et al., 2019 |
| WEEV virus-like particles (VLPs) | NIH Vaccine Research Center | Ko et al., 2019 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| Universal Mycoplasma Detection Kit | ATCC | Cat# 30-1012K |
| Cell Authentication Service Short Tandem Repeat (STR) analysis | ATCC | N/A |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| Human: Expi293F | Thermo Fisher Scientific | Cat# A14527; RRID:CVCL_D615 |
| Human: HEK293T | ATCC | Cat# CRL-3216; RRID:CVCL_0063 |
| Monkey: Vero | ATCC | Cat# CCL-81; RRID:CVCL_0059 |
| Monkey: B95.8 | ATCC | Cat# CRL-1612 (discontinued); RRID:CVCL_1953 |
| Hamster: BHK-21 | ATCC | Cat# CCL-10; RRID:CVCL_1915 |
| Hamster: ExpiCHO | Thermo Fisher Scientific | Cat# A29127;RRID:CVCL_5J31 |
| Mouse-human HMMA 2.5 myeloma cell line | Dr. L. Cavacini | N/A |
| THP-1 cells | ATCC | Cat# TIB-202;RRID:CVCL_0006 |
| EEEV-76 hybridoma clone | This paper | N/A |
| EEEV-104 hybridoma clone | This paper | N/A |
| EEEV-109 hybridoma clone | This paper | N/A |
| EEEV-126 hybridoma clone | This paper | N/A |
| EEEV-127 hybridoma clone | This paper | N/A |
| EEEV-138 hybridoma clone | This paper | N/A |
| EEEV-157 hybridoma clone | This paper | N/A |
| EEEV-179 hybridoma clone | This paper | N/A |
| EEEV-307 hybridoma clone | This paper | N/A |
| EEEV-312 hybridoma clone | This paper | N/A |
| EEEV-320 hybridoma clone | This paper | N/A |
| EEEV-342 hybridoma clone | This paper | N/A |
| EEEV-346 hybridoma clone | This paper | N/A |
| EEEV-354 hybridoma clone | This paper | N/A |
| EEEV-368 hybridoma clone | This paper | N/A |
| EEEV-377 hybridoma clone | This paper | N/A |
| EEEV-379 hybridoma clone | This paper | N/A |
| EEEV-387 hybridoma clone | This paper | N/A |
| EEEV-398 hybridoma clone | This paper | N/A |
| EEEV-400 hybridoma clone | This paper | N/A |
|
| ||
| Experimental Models: Organisms/Strains | ||
|
| ||
| Mouse: C57BL/6 | Jackson Laboratories | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| CpG (ZOEZOEZZZZZOEEZOEZZZT oligonucleotide) | Invitrogen | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: pCDNA3.1(+)-EEEV (FL93-939) E1 ectodomain (Y1-S409) | Williamson et al., 2020 | GenScript Biotech |
| Plasmid: pCDNA3.1(+)-EEEV (FL93-939) structural protein (capsid-E3-E2-6K-E1) | Williamson et al., 2020 | GenScript Biotech |
| Plasmid: pCDNA3.1(+)-EEEV (FL93-939) structural protein (capsid-E3-E2-6K-E1) E1 mutants (Y1-H441) | This paper; GenBank: AF159554 | Twist Bioscience Inc. |
| Plasmid: pChikV-E3E2E1-V5 [S27] WT-CHIKV | Fong et al., 2014 | N/A |
| Plasmid: CHIKV E1 mutants W89A, F95A, N100A | Fong et al., 2014 | N/A |
| Plasmid: pTwist-EEEV (PE-6) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAU95735 | Twist Bioscience Inc. |
| Plasmid: pTwist-MADV (PE-3.0815) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: ABB45866 | Twist Bioscience Inc. |
| Plasmid: pTwist-MADV (PE-0.0155) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: ABB45868 | Twist Bioscience Inc. |
| Plasmid: pTwist-MADV (BeAr436087) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: ABL84689 | Twist Bioscience Inc. |
| Plasmid: pTwist-MADV (Cebus/apella/BRA/BEAN5122/1956) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: YP_009020571 | Twist Bioscience Inc. |
| Plasmid: pTwist-VEEV IAB (Trinidad Donkey) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAC19322 | Twist Bioscience Inc. |
| Plasmid: pTwist-VEEV IAB (TC-83) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAB02517 | Twist Bioscience Inc. |
| Plasmid: pTwist-VEEV IC (P676) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: NP_040824 | Twist Bioscience Inc. |
| Plasmid: pTwist-VEEV ID (3880) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAC19325 | Twist Bioscience Inc. |
| Plasmid: pTwist-VEEV IE (Mena II) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAD14553 | Twist Bioscience Inc. |
| Plasmid: pTwist-VEEV IE (MX01-22) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAW30006 | Twist Bioscience Inc. |
| Plasmid: pTwist-Mosso das Pedras virus (78V-3531) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAD14563 | Twist Bioscience Inc. |
| Plasmid: pTwist-Everglades virus (Fe3-7C) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAD14551 | Twist Bioscience Inc. |
| Plasmid: pTwist-Mucambo virus (BeAn 8) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAD14555 | Twist Bioscience Inc. |
| Plasmid: pTwist-VEEV IIIC (71D-1252) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAD14559 | Twist Bioscience Inc. |
| Plasmid: pTwist-Pixuna virus (BeAr 35645) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAD14561 | Twist Bioscience Inc. |
| Plasmid: pTwist-Cabassou virus (CaAr 508) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAD14567 | Twist Bioscience Inc. |
| Plasmid: pTwist-Rio Negro virus (Ag80-663) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAD14565 | Twist Bioscience Inc. |
| Plasmid: pTwist-WEEV (McMillian) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: ACT75276 | Twist Bioscience Inc. |
| Plasmid: pTwist-WEEV (Fleming) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: ABD57956 | Twist Bioscience Inc. |
| Plasmid: pTwist-WEEV (Ag80-646) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: ACT75288 | Twist Bioscience Inc. |
| Plasmid: pTwist-CHIKV (181/25) structural protein (capsid-E3-E2-6K-E1) | This paper; GenBank: AAA53256 | Twist Bioscience Inc. |
| Plasmid (pTwist): EEEV-97 rIgG1 heavy chain | Williamson et al., 2020 | Twist Bioscience Inc. |
| Plasmid (pTwist): EEEV-97 light chain | Williamson et al., 2020 | Twist Bioscience Inc. |
| Plasmid (pTwist): EEEV-126 rIgG1 heavy chain | This paper | Twist Bioscience Inc. |
| Plasmid (pTwist): EEEV-126 light chain | This paper | Twist Bioscience Inc. |
| Plasmid (pTwist): EEEV-346 monocistronic IgG1 | This paper | Twist Bioscience Inc. |
| Plasmid (pTwist): EEEV-109 monocistronic IgG1 | This paper | Twist Bioscience Inc. |
| Plasmid (pTwist): EEEV-109 monocistronic IgG1 LALA-PG | This paper | Twist Bioscience Inc. |
| Plasmid (pTwist): EEEV-157 rIgG1 heavy chain | This paper | Twist Bioscience Inc. |
| Plasmid (pTwist): EEEV-157 light chain | This paper | Twist Bioscience Inc. |
|
| ||
| Software and Algorithms | ||
|
| ||
| Prism | GraphPad | v8 |
| ImMunoGeneTics (IMGT) database | Brochet et al., 2008, Giudicelli et al., 2011 | N/A |
| PyMOL | Schrödinger | v2.3.0 |
| PyIR | Soto et al., 2020 | N/A |
| Geneious Prime | Geneious | 2020.1 |
|
| ||
| Other | ||
|
| ||
| PacBio Sequel System | Pacific Biosciences | N/A |
| BioStack™ 3 Microplate Stacker | BioTek™ | Cat# BIOSTACK3WR |
| EL406 Washer Dispenser | BioTek™ | Cat# 406SUB3 |
| BioTek™ PowerWave™ Microplate Spectrophotometer | BioTek™ | Cat# BT-RPRWI |
| ImmunoSpot® S6 Universal | Cellular Technology Limited | Cat# S6UNV12 |
| IntelliCyt® iQue Screener PLUS | Sartorius | N/A |
| 6-well G-rex® plates | Wilson Wolf | Cat# 80240M |
| AKTA Pure 25M Chromatography System | Cytiva Life Sciences | Cat# 29018226 |
| HisTrap Excel | Cytiva Life Sciences | Cat# 17371205 |
| HiTrap Protein G HP Columns | Cytiva Life Sciences | Cat# 17040503 |
| HiTrap MabSelect SuRe | Cytiva Life Sciences | Cat# 11003493 |
| Amicon® Ultra-15 Centrifugal Filter Unit 50K MWCO | Millipore | Cat# UFC905008 |
| Zeba™ Spin Desalting Columns, 7K MWCO | Thermo Fisher Scientific | Cat# 89894 or 89890 |
| EZ-Link™ NHS-PEG4-Biotin, No-Weigh™ Format | Thermo Fisher Scientific | Cat# A39259 |
| Sulfo-NHS-LC-LC Biotin | Thermo Fisher Scientific | Cat# 21338 |
| FluoSpheres™ NeutrAvidin™-Labeled Microspheres, 1.0 μm, yellow-green fluorescent (505/515), 1% solids | Invitrogen™ | Cat# F8776 |
| FluoSpheres™ NeutrAvidin™-Labeled Microspheres, 1.0 μm, red fluorescent (580/605), 1% solids | Invitrogen™ | Cat# F8775 |
| FabALACTICA Fab kit Microspin | Genovis | Cat# A2-AFK-005 |
| CaptureSelect™ IgG-Fc (ms) affinity matrix | Thermo Fisher Scientific | Cat# 191285505 |
| CaptureSelect™ CH1-XL pre-packed column | Thermo Fisher Scientific | Cat# 494346201 |
Highlights.
Human E1 mAbs recognize the conserved fusion loop for ‘pan-alphavirus’ reactivity
Cryptic E1 epitopes depend on acidic pH for exposure
Human E1 mAbs can inhibit virus egress
Egress inhibition serves as a correlate of protection for EEEV-induced disease
ACKNOWLEDGEMENTS
We thank P. Gilchuk, C. Slaughter, and S. Zost at Vanderbilt and A. Bandyopadhyay at Purdue University for helpful discussions and training. We thank D. Yousif, M. Jensen, N. Murphy, M. Goff, I. Kovach, W. Reichard, M. Majedi, and R. Troseth for technical support. We thank M. Leksell, M. Mayo, K. Moton, T. Martin, V. Morris, A. Bunnell, and A. Jordan for administrative assistance and R. Irving for lab management. L.E.W. was supported by NIH grants T32 HL069765 and F31 AI145189. E.S.W. is supported by T32 AI007163. This project received support from the U.S. Defense Threat Reduction Agency-Joint Science and Technology Office for Chemical and Biological Defense (Grant no. HDTRA1–13-1–0034), by CTSA award UL1 TR000445 from the U.S. National Center for Advancing Translational Sciences, and by NIAID grant U19 AI142790. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
DECLARATION OF INTERESTS
J.E.C. has served as a consultant for Luna Biologics, is a member of the Scientific Advisory Board of Meissa Vaccines and is Founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received unrelated sponsored research agreements from IDBiologics and AstraZeneca. M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory at Washington University School of Medicine has received sponsored research agreements from Emergent BioSolutions, Moderna and Vir Biotechnology. All other authors report no conflicts. M.E.F., E.D. and B.J.D. are employees of Integral Molecular and B.J.D. is a shareholder. G.A. is a founder and the Chief Scientific Officer (Consulting) of Seromyx.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ackerman ME, Moldt B, Wyatt RT, Dugast A-S, McAndrew E, Tsoukas S, Jost S, Berger CT, Sciaranghella G, Liu Q, Irvine DJ, Burton DR, and Alter G (2011). A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J Immunol Methods 366, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar PV, Estrada-Franco JG, Navarro-Lopez R, Ferro C, Haddow AD, and Weaver SC (2011). Endemic Venezuelan equine encephalitis in the Americas: hidden under the dengue umbrella. Future Virol 6, 721–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Luis MA, Del Valle-Mendoza J, Silva-Caso W, Gil-Ramirez T, Levy-Blitchtein S,Bazan-Mayra J, Zavaleta-Gavidia V, Cornejo-Pacherres D, Palomares-Reyes C, and Del Valle LJ (2020). An emerging public health threat: Mayaro virus increases its distribution in Peru. Int J Infect Dis 92, 253–258. [DOI] [PubMed] [Google Scholar]
- Ahn A, Klimjack MR, Chatterjee PK, Kielian M (1999). An epitope of the Semliki Forest virus fusion protein exposed during virus-membrane fusion. J Virol 73(12): 10029–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong PM, and Andreadis TG (2013). Eastern equine encephalitis virus--old enemy, new threat. N Engl J Med 368, 1670–1673. [DOI] [PubMed] [Google Scholar]
- Ayres JC, and Feemster RF (1949). The sequelae of eastern equine encephalomyelitis. N Engl J Med 240, 960–962. [DOI] [PubMed] [Google Scholar]
- Bergren NA, Auguste AJ, Forrester NL, Negi SS, Braun WA, and Weaver SC (2014). Western equine encephalitis virus: evolutionary analysis of a declining alphavirus based on complete genome sequences. J Virol 88, 9260–9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergren NA, Haller S, Rossi SL, Seymour RL, Huang J, Miller AL, Bowen RA, Hartman DA, Brault AC, and Weaver SC (2020). “Submergence” of Western equine encephalitis virus: Evidence of positive selection argues against genetic drift and fitness reductions. PLoS Pathog 16, e1008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boere WA, Harmsen T, Vinje J, Benaissa-Trouw BJ, Kraaijeveld CA, and Snippe H (1984). Identification of distinct antigenic determinants on Semliki Forest virus by using monoclonal antibodies with different antiviral activities. J Virol 52, 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RS, Wan JJ, and Kielian M (2018). The Alphavirus Exit Pathway: What We Know and What We Wish We Knew. Viruses 10: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd EA, Kielian M (2019). The Alphavirus E2 membrane-proximal domain impacts capsid interaction and glycoprotein lattice formation. J Virol 93(4):e01881–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) website, www.cdc.gov/easternequineencephalitis/tech/epi.html, accessed on December 15th, 2020.
- Chen CL, Hasan SS, Klose T, Sun Y, Buda G, Sun C, Klimstra WB, and Rossmann MG (2020). Cryo-EM structure of eastern equine encephalitis virus in complex with heparan sulfate analogues. Proc Natl Acad Sci USA 117: 8890–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagne CT, Bengue M, Choumet V, Hamel R, Pompon J, Missé D (2020). Mayaro virus pathogenesis and transmission mechanisms. Pathogens 9(9):738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest JT, Basore K, Roy V, Bailey AL, Wang D, Alter G, Fremont DH, and Diamond MS (2019). Neutralizing antibodies against Mayaro virus require Fc effector functions for protective activity. J Exp Med 216, 2282–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Ibarra KD, Ng TS, Smith SA, Tan JL, Lim XN, Ooi JS, Kostyuchenko VA, Wang J, de Silva AM, et al. (2015). DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischinger S, Fallon JK, Michell AR, Broge T, Suscovich TJ, Streeck H, and Alter G 2019. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J Immunol Methods 473: 112630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong RH, Banik SS, Mattia K, Barnes T, Tucker D, Liss N, Lu K, Selvarajah S, Srinivasan S, Mabila M, et al. (2014). Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. J Virol 88, 14364–14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JM, Long F, Edeling MA, Lin H, van Duijl-Richter MKS, Fong RH, Kahle KM, Smit JM, Jin J, Simmons G, et al. (2015). Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell 163, 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JM, Roy V, Gunn BM, Huang L, Edeling MA, Mack M, Fremont DH, Doranz BJ, Johnson S, Alter G, et al. (2019). Optimal therapeutic activity of monoclonal antibodies against chikungunya virus requires Fc-FcγR interaction on monocytes. Science Immunology 4, eaav5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller SD, Berriman JA, Butcher SJ, and Gowen BE (1995). Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell 81, 715–725. [DOI] [PubMed] [Google Scholar]
- Ganesan VK, Duan B, Reid SP (2017). Chikungunya virus: pathophysiology, mechanism, and modeling. Viruses. 9(12):368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner CL, Ebel GD, Ryman KD, and Klimstra WB (2011). Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc Natl Acad Sci U S A 108, 16026–16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Ahn A, Chatterjee PK, and Kielian M (2000). Formation and characterization of the trimeric form of the fusion protein of Semliki Forest Virus. J Virol 74, 7772–7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Ahn A, Liao M, Hammar L, Cheng RH, and Kielian M (2004). Multistep regulation of membrane insertion of the fusion peptide of Semliki Forest virus. J Virol 78, 3312–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild SA, O’Brien LM, Steven J, Muller MR, Lanning OJ, Logue CH, D’Elia RV, Phillpotts RJ, and Perkins SD (2011). A humanised murine monoclonal antibody with broad serogroup specificity protects mice from challenge with Venezuelan equine encephalitis virus. Antiviral Res 90, 1–8. [DOI] [PubMed] [Google Scholar]
- Griffin DE (2010). Recovery from viral encephalomyelitis: immune-mediated noncytolytic virus clearance from neurons. Immunol Res 47(1–3):123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Sun C, Kim AS, Watanabe Y, Chen CL, Klose T, Buda G, Crispin M, Diamond MS, Klimstra WB, et al. (2018). Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization. Cell Rep 25, 3136–3147 e3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AC, Basore K, Fremont DH, and Diamond MS (2020). A molecular understanding of alphavirus entry. PLoS Pathog 16, e1008876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnold SP, Mossel EC, Bakken RR, Lind CM, Cohen JW, Eccleston LT, Spurgers KB, Erwin-Cohen R, Glass PJ, Maheshwari RK (2015). Eastern equine encephalitis virus in mice II: pathogenesis is dependent on route of exposure. J Virol 12:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AR, Frederickson S, Maruyama T, Roehrig JT, and Blair CD (2010). The first human epitope map of the alphaviral E1 and E2 proteins reveals a new E2 epitope with significant virus neutralizing activity. PLoS Negl Trop Dis 4, e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AR, and Roehrig JT (1985). Biochemical and biological characteristics of epitopes on the E1 glycoprotein of western equine encephalitis virus. Virology 142, 334–346. [DOI] [PubMed] [Google Scholar]
- Jin J and Simmons G (2019). Antiviral functions of monoclonal antibodies against Chikungunya virus. Viruses 11, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justman J, Klimjack MR, and Kielian M (1993). Role of spike protein conformational changes in fusion of Semliki Forest virus. J Virol 67: 7597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten CB, Mehta N, Shin SA, Diefenbach TJ, Slein MD, Karpinski W, Irvine EB, Broge T, Suscovich TJ, and Alter G (2019). A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J Immunol Methods 471: 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M, Chanel-Vos C, Liao M (2010). Alphavirus entry and membrane fusion. Viruses 2(4):796–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M (2014). Mechanisms of virus membrane fusion proteins. Annu Rev Virol 1(1):171–89. [DOI] [PubMed] [Google Scholar]
- Kim AS, Austin SK, Gardner CL, Zuiani A, Reed DS, Trobaugh DW, Sun C, Basore K, Williamson LE, Crowe JE Jr., et al. (2019). Protective antibodies against Eastern equine encephalitis virus bind to epitopes in domains A and B of the E2 glycoprotein. Nat Microbiol 4, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra WB, Nangle EM, Smith MS, Yurochko AD, and Ryman KD (2003). DC-SIGN and L-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J Virol 77, 12022–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SY, Akahata W, Yang ES, Kong WP, Burke CW, Honnold SP, Nichols DK, Huang YS, Schieber GL, Carlton K, et al. (2019). A virus-like particle vaccine prevents equine encephalitis virus infection in nonhuman primates. Sci Transl Med 11. [DOI] [PubMed] [Google Scholar]
- Langsjoen RM, Haller SL, Roy CJ, Vinet-Oliphant H, Bergren NA, Erasmus JH, Livengood JA, Powell TD, Weaver SC, and Rossi SL (2018). Chikungunya Virus Strains Show Lineage-Specific Variations in Virulence and Cross-Protective Ability in Murine and Nonhuman Primate Models. mBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky JA, White SK, Mavian CN, El Badry MA, Telisma T, Salemi M, BA OK, Beau De Rochars VM, and Morris JG Jr. (2019). Emergence of Madariaga virus as a cause of acute febrile illness in children, Haiti, 2015–2016. PLoS Negl Trop Dis 13, e0006972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Wengler G, and Rey FA (2001). The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105, 137–148. [DOI] [PubMed] [Google Scholar]
- Li L, Jose J, Xiang Y, Kuhn RJ, and Rossmann MG (2010). Structural changes of envelope proteins during alphavirus fusion. Nature 468, 705–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey NP, Staples JE, and Fischer M (2018). Eastern equine encephalitis virus in the United States, 2003–2016. Am J Trop Med Hyg 98:1472–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey NP, Martin SW, Staples JE, and Fischer M (2020). Notes from the Field: Multistate Outbreak of Eastern Equine Encephalitis Virus - United States, 2019. MMWR Morb Mortal Wkly Rep 69, 50–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M, Kim HS, Tong RK, Bainbridge TW, Vernes JM, Zhang Y, et al. (2017). Effector-attenuating substitutions that maintain antibody stability and reduce toxicity in mice. J Biol Chem 292, 3900–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Kim AS, Kafai NM, Earnest JT, Shah AP, Case JB, Basore K, Gilliland TC, Sun C, Nelson CA, et al. (2020). LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 588, 308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews JH, Roehrig JT (1982). Determination of the protective epitopes on the glycoproteins of Venezuelan equine encephalomyelitis virus by passive transfer of monoclonal antibodies. J Immunol 129(6):2763–7. [PubMed] [Google Scholar]
- Mavalankar D, Shastri P, Bandyopadhyay T, Parmar J, Ramani KV (2008). Increased mortality rate associated with chikungunya epidemic, Ahmedabad, India. Emerg Infect Dis 14(3):412–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza QP, Stanley J, and Griffin DE (1988). Monoclonal antibodies to the E1 and E2 glycoproteins of Sindbis virus: definition of epitopes and efficiency of protection from fatal encephalitis. J Gen Virol 69 ( Pt 12), 3015–3022. [DOI] [PubMed] [Google Scholar]
- Meyer WJ, Gidwitz S, Ayers VK, Schoepp RJ, and Johnston RE (1992). Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J Virol 66, 3504–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, Folkers GK, and Fauci AS (2019). Eastern Equine Encephalitis Virus - Another Emergent Arbovirus in the United States. N Engl J Med 381, 1989–1992. [DOI] [PubMed] [Google Scholar]
- Mostafavi H, Abeyratne E, Zaid A, Taylor A (2019). Arthritogenic alphavirus-induced immunopathology and targeting host inflammation as a therapeutic strategy for alphaviral disease. Viruses 11(3):290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, Strauss JH, Baker TS, Kuhn RJ, and Rossmann MG (2006). Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noval MG, Rodriguez-Rodriguez BA, Rangel MV, and Stapleford KA (2019). Evolution-Driven Attenuation of Alphaviruses Highlights Key Glycoprotein Determinants Regulating Viral Infectivity and Dissemination. Cell Rep 28: 460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, and Diamond MS (2005) Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med 11: 522–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I, Akahata W, Nabel GJ, Richter MK, et al. (2013). Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog 9, e1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta J, Jose J, Roehrig JT, Blair CD, Kuhn RJ, and Rossmann MG (2014). Locking and blocking the viral landscape of an alphavirus with neutralizing antibodies. J Virol 88, 9616–9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LA, Fox JM, Kose N, Kim AS, Majedi M, Bombardi R, Carnahan RH, Slaughter JC, Morrison TE, Diamond MS, et al. (2020). Human monoclonal antibodies against Ross River virus target epitopes within the E2 protein and protect against disease. PLoS Pathog 16, e1008517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz JA, Malonis RJ, Thackray LB, Cohen CA, Pallesen J, Jangra RK, Brown RS, Hofmann D, Holtsberg FW, Shulenin S, et al. (2019). Human monoclonal antibodies against chikungunya virus target multiple distinct epitopes in the E1 and E2 glycoproteins. PLoS Pathog 15, e1008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehrig JT, and Mathews JH (1985). The neutralization site on the E2 glycoprotein of Venezuelan equine encephalomyelitis (TC-83) virus is composed of multiple conformationally stable epitopes. Virology 142, 347–356. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, Hunt AR, Chang GJ, Sheik B, Bolin RA, Tsai TF, and Trent DW (1990) Identification of monoclonal antibodies capable of differentiating antigenic varieties of eastern equine encephalitis viruses. Am J Trop Med Hyg 42: 394–8. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, and Bolin RA (1997). Monoclonal antibodies capable of distinguishing epizootic from enzootic varieties of subtype 1 Venezuelan equine encephalitis viruses in a rapid indirect immunofluorescence assay. J Clin Microbiol 35, 1887–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronca SE, Dineley KT, and Paessler S (2016). Neurological Sequelae Resulting from Encephalitic Alphavirus Infection. Front Microbiol 7, 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Hanna SL, Spiridigliozzi A, Wannissorn N, Beiting DP, Ross SR, Hardy RW, Bambina SA, Heise MT, and Cherry S (2011). Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host Microbe 10, 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel A, Lescar J, Vaney MC, Wengler G, Wengler G, and Rey FA (2006). Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14, 75–86. [DOI] [PubMed] [Google Scholar]
- Sahoo B, Gudigamolla NK, and Chowdary TK (2020). Acidic pH-Induced Conformational Changes in Chikungunya Virus Fusion Protein E1: a Spring-Twisted Region in the Domain I-III Linker Acts as a Hinge Point for Swiveling Motion of Domains. J Virol 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn AL, Kokubun KM, and Cole GA (1983). Protective monoclonal antibodies define maturational and pH-dependent antigenic changes in Sindbis virus E1 glycoprotein. Virology 130, 144–154. [DOI] [PubMed] [Google Scholar]
- Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney M-C, Lavenir R, Pardigon N, Reynes J-M, Pettinelli F, et al. (2006). Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 3: e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidwell RW, and Smee DF (2003). Viruses of the Bunya- and Togaviridae families: potential as bioterrorism agents and means of control. Antiviral Res 57, 101–111. [DOI] [PubMed] [Google Scholar]
- Soto C, Finn JA, Willis JR, Day SB, Sinkovits RS, Jones T, Schmitz S, Meiler J, Branchizio A, and Crowe JE (2020). PyIR: a scalable wrapper for processing billions of immunoglobulin and T cell receptor sequences using IgBLAST. BMC Bioinformatics 21: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleford KA, Coffey LL, Lay S, Bordería AV, Duong V, Isakov O, Rozen-Gagnon K, Arias-Goeta C, Blanc H, Beaucourt S, et al. (2014). Emergence and transmission of arbovirus evolutionary intermediates with epidemic potential. Cell Host Microbe 15: 706–16. [DOI] [PubMed] [Google Scholar]
- Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, Diamond MS, Nabel GJ, and Rossmann MG (2013). Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. Elife 2:e00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobaugh DW, Sun C, Dunn MD, Reed DS, and Klimstra WB (2019). Rational design of a live-attenuated eastern equine encephalitis virus vaccine through informed mutation of virulence determinants. PLoS Pathog 15, e1007584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K, Higgs S, McGee CE, De Lamballerie X, Charrel RN, and Vanlandingham DL (2006). Infectious clones of Chikungunya virus (La Réunion isolate) for vector competence studies. Vector Borne Zoonotic Dis 6, 325–337. [DOI] [PubMed] [Google Scholar]
- Vazeille M, Moutailler S, Coudrier D, Rousseaux C, Khun H, Huerre M, Thiria J, Dehecq J-S, Fontenille D, Schuffenecker I, et al. (2007). Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One 2: e1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, and Rey FA (2010). Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712. [DOI] [PubMed] [Google Scholar]
- Wahlberg JM and Garoff H (1992). Membrane fusion process of Semliki Forest virus. I: Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol 116: 339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg JM, Bron R, Wilschut J, and Garoff H (1992). Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J Virol 66: 7309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Winegar R, Manger ID, Forrester NL (2012). Alphaviruses: population genetics and determinants of emergence. Antiviral Res 94(3):242–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G, Wengler G, and Rey FA (1999). The isolation of the ectodomain of the alphavirus E1 protein as a soluble hemagglutinin and its crystallization. Virology 257, 472–482. [DOI] [PubMed] [Google Scholar]
- Williamson LE, Gilliland T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RE, Durie CL, Armstrong E, et al. (2020). Human antibodies protect against aerosolized Eastern equine encephalitis virus infection. Cell S0092–8674: 31528–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap ML, Klose T, Urakami A, Hasan SS, Akahata W, and Rossmann MG (2017). Structural studies of Chikungunya virus maturation. Proc Natl Acad Sci U S A 114, 13703–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, McGraw PA, House FS, and Crowe JE Jr. (2008a). An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Methods 336, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, et al. (2008b). Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455, 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, and Chiu W (2011). 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J 30, 3854–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Kim AS, Fox JM, Nair S, Basore K, Klimstra WB, Rimkunas R, Fong RH, Lin H, Poddar S, et al. (2018). Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Sánchez-San Martín C,Qin ZL, Kielian M (2011). The domain I-domain III linker plays an important role in the fusogenic conformational change of the alphavirus membrane fusion protein. J Virol 85(13): 6334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QF, Fox JM, Earnest JT, Ng TS, Kim AS, Fibriansah G, Kostyuchenko VA, Shi J, Shu B, Diamond MS, Lok SM (2020). Structural basis of Chikungunya virus inhibition by monoclonal antibodies. Proc Natl Acad Sci USA 117(44): 27637–27645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Shafer A, Reidy JX, Trivette A, Nargi RS, et al. (2020). Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584: 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related toFigure 2. Sequence alignment of the E1 protein for diverse alphavirus subtypes. A. Multiple amino acid sequence alignment of the E1 protein for alphavirus subtypes (virus, subtype, and strain) labeled in the left-hand column (made with Geneious software). Alphavirus subtypes are grouped by antigenic complex for EEEV (green), VEEV (blue), WEEV (orange), or Semliki Forest virus (magenta). Dots represent conserved amino acids. Amino acids that are not conserved are colored based on polarity (cyan [polar, uncharged], orange [non-polar], red [polar, acidic], or dark blue [polar, basic]). The consensus sequence is shown at the top of the sequence alignment. Respective domains and regions of interest are labeled accordingly (domain I [light orange], domain II [blue], domain III [red], fusion loop [purple], domain I/domain II hinge and domain I/III linker [black]). B. Amino acid percent identity of the E1 protein for alphavirus subtypes (virus, subtype, and strain). Alphavirus subtypes are grouped by antigenic complex for EEEV (green), VEEV (blue), WEEV (orange), or Semliki Forest virus (magenta). The heat map corresponds to amino acid percent identity between alphavirus subtypes, ranging from 40–99.9% (black to light orange). White boxes with a cross include the amino acid percent identity for the same alphavirus subtype (100%).
Figure S2, related toFigure 3. Human cross-reactive E1-specific mAbs recognize the fusion loop. A. Binding of human cross-reactive E1-specific mAbs to CHIKV E2/E1 clones. MAbs or Fabs were screened at a 1 μg/mL concentration for binding to CHIKV E2/E1 constructs expressed in HEK 293T cells. Binding was detected by flow cytometry (as described previously; Fong et al., 2014). Binding to each mutant protein clone was calculated relative to that for wild-type (WT) protein by subtracting the signal from mock-transfected controls and normalizing to the signal from WT-transfected controls. Data represent the mean (standard deviation) of four replicate data points. B. Epitope mapping of critical residues identified by alanine-scanning (A) for human cross-reactive E1-specific mAbs binding to the CHIKV E1 glycoprotein. Critical residues were mapped onto the CHIKV E2/E1 heterodimer (PDB ID: 3N42). A side view of the CHIKV E2/E1 heterodimer (E2 – light pink, E1 – light blue) is shown with W89, F95, and N100 critical interaction residues (purple spheres) indicated for the cross-reactive E1-specific mAbs. C. Heatmap of the EEEV E1 glycoprotein alanine mutant residues tested for human E1-specific mAbs. Residues were selected for analysis based on previously described epitopes within the Immune Epitope Database (IEDB). The average percent binding is displayed as a heatmap with >75% (maroon), 30 to 75% (light blue), or <30% (magenta) mAb binding relative to WT. Each mAb is colored according to binding reactivity (neutralizing EEEV E2-specific mAb [red], ‘pan-alphavirus’ [purple], ‘broadly-reactive’ [orange], ‘New World’ [magenta], ‘EEEV-VEEV’ [blue], EEEV-specific [green]) and is ordered to correspond with the competition-binding groups as defined in Figure 3A.
Figure S3, related toFigure 3. Hydrogen-deuterium exchange mass spectrometry (HDX-MS) analysis of EEEV-109, −126, −157, and −179 Fab molecules binding footprint to the EEEV E1 glycoprotein. A. E1 peptide sequence coverage map for EEEV-126 Fab bound EEEV E1 glycoprotein. Peptides covered during HDX-MS analysis are indicated by the blue lines. B. Potential interacting peptides for EEEV-179 (‘New World’ [magenta]), −109 (EEEV-specific [green]), and −157 (‘EEEV-VEEV’ [blue]), as defined by at least one time point with ≥ 0.4 Da ± 2.4 standard deviations relative deuterium uptake (y-axis) of Fab bound versus unbound EEEV E1 glycoprotein over the course of the 10,000 s exposure time (x-axis). The labels at each time point indicate the change in relative deuterium uptake of Fab bound versus unbound EEEV E1 glycoprotein.
Figure S4, related toFigure 3. Epitope mapping of human E1-specific mAbs. A to C. Epitope mapping from cell surface antigen display, EEEV and CHIKV E1 alanine-scanning mutagenesis, and hydrogen-deuterium exchange mass spectrometry (HDX-MS) analyses of human E1-specific mAbs binding to the E1 glycoprotein. Critical interaction residues were mapped onto the cryo-EM reconstruction of SINV/EEEV (PDB: 6MX4). Potential antibody footprint peptides identified through HDX-MS were mapped onto the cryo-EM reconstruction of EEEV VLP (PDB: 6XO4). A top trimeric view with an asymmetric unit of a neighboring spike (E2 – light pink, E1 – light blue) is shown with critical interaction residues (spheres) indicated for the E1-specific mAbs. Residues are labeled and colored based on E1 domain or region of interest (domain I (DI) [light orange], domain II (DII) [cyan], fusion loop [purple], domain III (DIII) [red], DI/II hinge and DI/DIII linker [black]) Each mAb is grouped according to antigen binding reactivity and epitope (A – DII: ‘pan-alphavirus’ [purple], ‘broadly-reactive’ [orange], ‘New World’ [magenta], EEEV-specific [green]; B – DIII: ‘EEEV-VEEV’ [blue], EEEV-specific [green]; C – DI: EEEV-specific [green]) as described in Figure 3.
Figure S5, related toFigures 1, 4, and S6. Temperature-independent binding of human E1-specific mAbs to virus-like particles (VLPs) or recombinant E1 glycoproteins. A through G. Representative binding curves for human E1-specific mAbs binding to EEEV (green), VEEV (dark blue), or WEEV (orange) VLPs, recombinant EEEV (light purple), CHIKV (magenta), or MAYV (purple) E1 glycoproteins, or recombinant EEEV E2 (blue) glycoprotein at room temperature (open circles), 37°C (open triangles), or 42°C (open squares) with mAb concentration (μg/mL) on the x-axis and optical density at 450 nm on the y-axis. Binding curves are ordered numerically and grouped according to binding reactivity and strength (EEEV VLP and E1 protein-reactive [A; VLP/protein EC50 ratio <1], EEEV VLP > EEEV E1 protein [B; VLP/protein EC50 ratio = not applicable (NA) to EEEV E1 protein], EEEV E1 protein > EEEV VLP [C; VLP/protein EC50 ratio >1 or NA to EEEV VLP], ‘EEEV-VEEV’ [D], ‘New World’ [E], ‘broadly-reactive’ [F], or ‘pan-alphavirus’ [G]). H. EC50 values (ng/mL) for binding of human EEEV-specific E1-specific mAbs to EEEV VLPs or recombinant EEEV E1 glycoprotein at room temperature, 37 °C, or 42°C. The human EEEV-specific mAbs are grouped according to binding strength (EEEV VLP and E1 protein-reactive, EEEV VLP > EEEV E1 protein, and EEEV E1 protein > EEEV VLP) and listed in order of increasing EC50 value for binding to EEEV VLPs for each respective group. EC50 value in ng/mL is indicated by the blue fill color (≤100.00 [dark blue], 101 to 500 [light blue], 501 to 5,000 [lightest blue], and >5,001). Ratio of binding to EEEV VLPs compared to EEEV E1 glycoprotein at room temperature is indicated as the ratio of EC50 values. Increasing depth of green color indicates lower ratios and thus more dependence on virus particle-specific epitopes. I. EC50 values (ng/mL) for binding of human cross-reactive E1-specific mAbs to EEEV VLPs or recombinant EEEV E1 glycoprotein at room temperature, 37°C, or 42°C. The human cross-reactive mAbs are grouped according to binding cross-reactivity (‘EEEV-VEEV’, ‘New World’, ‘broadly-reactive, and ‘pan-alphavirus’) and listed in order of increasing EC50 value for binding to EEEV VLPs for each respective group. EC50 value in ng/mL is indicated by blue fill color (≤100.00 [dark blue], 101 to 501 [light blue], 501 to 5,000 [lightest blue], and >5,001). Ratio of binding to EEEV VLPs compared to EEEV E1 glycoprotein at room temperature is indicated as the ratio of EC50 values. A VLP/protein EC50 ratio of <1 suggests a greater dependence on virus particle-specific epitopes. Data in A to I represent mean ± SD of technical triplicates and are representative of two independent experiments. See Figure 1 for ELISA binding of human E1-specific mAbs in the presence of the nonionic detergent Tween®20. See Table S1 for summary of human E1-specific mAbs binding to VLPs and recombinant proteins in the presence of the nonionic detergent Tween®20. See Table S3 for temperature-independent binding summary for human E1-specific mAbs binding to VLPs and recombinant proteins.
Figure S6, related toFigure 5. Human cross-reactive E1-specific mAbs moderately neutralize SINV/VEEV. Neutralization curves of human cross-reactive E1-specific mAbs against SINV/EEEV, SINV/VEEV, SINV/WEEV, CHIKV, or MAYV at 37°C (light pink squares) or 42°C (teal squares) with mAb concentration (μg/mL) on the x-axis and % relative infectivity on the y-axis. Each mAb is grouped according to cross-reactivity (‘EEEV-VEEV’ [blue], ‘New World’ [magenta], ‘broadly-reactive’ [orange], ‘pan-alphavirus’ [purple]) and ordered numerically within each group. Half-maximal inhibitory concentration (IC50) values (ng/mL) for moderately neutralizing mAbs (EEEV-342, −346, −354, and −387) against SINV/VEEV are indicated at each temperature (light pink - 37°C, teal - 42°C). Data represent mean ± SD of technical triplicates of a focus reduction neutralization test (FRNT) experiment.
Figure S7, related toFigure 6. Fc-mediated effector function activities for human E1-specific mAbs. A and B. Monocyte and neutrophil phagocytosis (antibody-dependent cellular phagocytosis [ADCP; maroon diamonds] and antibody-dependent neutrophil phagocytosis [ADNP; light blue squares], respectively), antibody-dependent complement deposition (ADCD; teal triangles), and positive NK cells (antibody-dependent natural killer cell activation [ADNKA; MIP-1β (orange circles), CD107a (open purple circles), and IFNγ (open magenta squares)] activity was assessed for human EEEV-specific (A) or cross-reactive (B) mAbs using recombinant EEEV E1-glycoprotein-coated beads. Each mAb is colored based on binding reactivity (EEEV-specific [green], ‘EEEV-VEEV’ [blue], ‘New World’ [magenta], ‘broadly-reactive’ [orange], ‘pan-alphavirus’ [purple]). The corresponding graphs represent mAb concentration (μg/mL) on the x-axis and phagocytic score (in thousands [ADCP and ADNP]), median fluorescence intensity of anti-C3-FITC binding (in thousands [ADCD]), or % positive cells (ADNKA) on the y-axis. The Ebola virus-specific negative control mAb 13C6 is shown in red squares. The red dotted line represents the mean y-axis value. The black dotted line indicates the mean y-axis value for a no-mAb negative control. Data represent mean ± SD of two independent experiments.
Data Availability Statement
Data. All relevant data are included within the manuscript and are available upon request from the Lead Contact.
Code. This study did not generate any code.
Additional information. This study did not generate any additional information.