Abstract
Introduction
Evaluating histopathology via machine learning has gained research and clinical interest, and the performance of supervised learning tasks has been described in various areas of medicine. Unsupervised learning of histological images has the advantage of reproducibility for labeling; however, the relationship between unsupervised evaluation and clinical information remains unclear in nephrology.
Methods
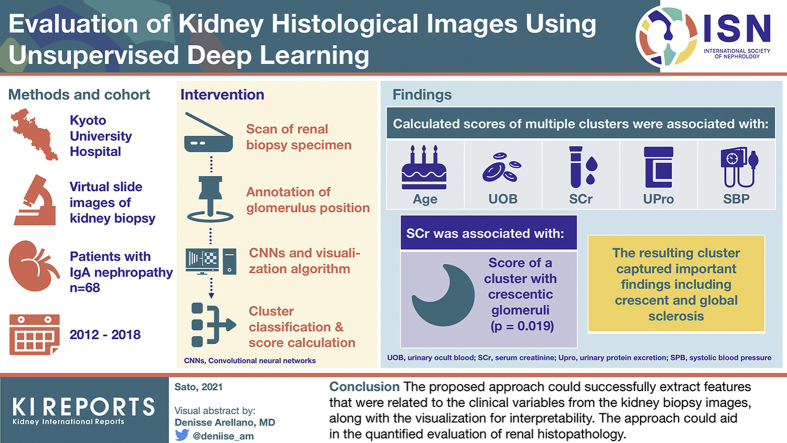
We propose an unsupervised approach combining convolutional neural networks (CNNs) and a visualization algorithm to cluster the histological images and calculate the score for patients. We applied the approach to the entire images or patched images of the glomerulus of kidney biopsy samples stained with hematoxylin and eosin obtained from 68 patients with immunoglobulin A nephropathy. We assessed the relationship between the obtained scores and clinical variables of urinary occult blood, urinary protein, serum creatinine (SCr), systolic blood pressure, and age.
Results
The glomeruli of the patients were classified into 12 distinct classes and 10 patches. The output of the fine-tuned CNN, which we defined as the histological scores, had significant relationships with assessed clinical variables. In addition, the clustering and visualization results suggested that the defined clusters captured important findings when evaluating renal histopathology. For the score of the patch-based cluster containing crescentic glomeruli, SCr (coefficient = 0.09, P = 0.019) had a significant relationship.
Conclusion
The proposed approach could successfully extract features that were related to the clinical variables from the kidney biopsy images along with the visualization for interpretability. The approach could aid in the quantified evaluation of renal histopathology.
Keywords: autoencoder, convolutional neural networks, deep learning, histopathology, machine learning, nephropathology
Graphical abstract
Machine learning algorithms, especially neural network architecture–based convolutional neural networks (CNNs), have achieved breakthrough performance in the classification of images to defined classes1 and are applied in various research areas, including medicine. Furthermore, they have gained considerable attention in the fields of histology and pathology, especially in neoplastic histopathology.2
Generally, in deep learning for histopathological images, supervised learning is performed wherein people decide on labels.3,4 One of the problems with this process is the occasional disagreement between and within pathologists that makes it difficult to obtain the correct labels to be used in the supervised learning tasks. In addition, the labeling of thousands of images is labor intensive. In unsupervised deep learning evaluations, the labeling is automated and reproducible because it is performed by a machine. Therefore, defining the classification labels in an unsupervised manner could be advantageous. However, it remains unclear whether the information obtained in an unsupervised manner is clinically meaningful in nephrology practice. In the present study, we propose an approach to assess the histological findings of biopsy specimens in an unsupervised manner and visualize how deep learning algorithms make these decisions.
In a recent study involving renal pathologies, Ginley et al.5 extracted features from glomerular images, scored them using CNN and recurrent neural networks, and compared them to the findings of pathologists in diabetic nephropathy. Another study showed the preliminary results of classification and visualization of transplant renal biopsy, discriminating the severity of T cell–mediated rejection and antibody-mediated rejection.6 In addition, one study compared the performance discriminating 7 major pathological findings among CNNs and pathologists.7 However, the unsupervised labeling and the association between yielded classification labels and clinical variables have not been examined in nephropathology. Thus, we applied our unsupervised approach to the kidney slide images of patients with immunoglobulin A nephropathy (IgAN) and evaluated if the unsupervised extracted features could relate to clinical information.
Methods
Patient Selection and Covariate Assessment
We retrospectively obtained the available virtual slide images of patients who underwent renal needle biopsy and were diagnosed with IgAN based on findings observed by optical microscopy and immunofluorescence staining between July 2012 and May 2018 at Kyoto University Hospital. We excluded those with a definite concurrent histological diagnosis of other diseases, except for nephrosclerosis. Patients diagnosed with hepatic IgAN were excluded. Patients who underwent multiple biopsy procedures were included. All patients provided written informed consent for the use of specimens in the present study. The study protocol was approved by the Ethics Committee on Human Research of the Graduate School of Medicine, Kyoto University (No. R643-2 and G562), and the study adhered to the Declaration of Helsinki. We collected basic patient demographics, including age, gender, systolic blood pressure (SBP, in mm Hg), laboratory tests comprising serum creatinine value (SCr, in mg/dL), urinary protein excretion level (UPro, in g/day), and the result of a urinary occult blood (UOB) test, which was classified into 5 categories: − (negative), ±, +, 2+, and 3+. These test values were obtained during the stay for renal biopsy in the hospital or at an outpatient visit before the renal biopsy procedure. If a daily urinary protein excretion value was not available during the respective hospital stay, the urinary protein creatinine ratio value was examined. In addition, we obtained the Oxford classification (MEST-C score) based on the definitions in pathological reports of kidney biopsy specimens.8
Extraction of Images and Preprocessing
All renal biopsy specimens were scanned with a NanoZoomer-2.0HT whole-slide imager, digital pathology slide scanner, and the software NDP.scan 3.1.7 (Hamamatsu Photonics, Hamamatsu City, Japan), using a ×40 lens (0.23 μm/pixel). The quality of all the images was checked manually after scanning; if the slides were out of focus, new scans were performed. We stained slides with hematoxylin and eosin (H&E), which is the basic and most commonly used staining protocol. The whole-slide images were stored in NDPI format, and we used OpenSlide9 to extract the PNG images from those files. We obtained the images with the highest resolution. The positions of glomeruli were manually annotated by a nephrologist in the images with lower resolutions and then cropped out from the highest-resolution images. Extracted glomeruli images underwent stain normalization via the method described by Macenko et al.10 The method assumes that every pixel in the image is the linear combination of 2 stain vectors (H&E). The image was first converted to their optical density (OD) values, and OD below the specified threshold was removed. Subsequently, singular value decomposition was calculated, the eigenvectors were obtained, and the plane from the eigenvectors corresponding to the 2 largest singular values was formed. All the OD value was then transformed onto this plane and normalized. Finally, staining intensity was corrected. The detailed method is described in their original article.10 Subsequently, the white areas on the edges in the glomerulus images were removed by the custom function, and in addition, glomerulus images with the proportion of white regions ≥0.2 were removed. For the experiment on the entire glomerulus images, the images were resized to a width of 331 and a height of 331, which is the default input shape for the CNN. These filtered images were used for clustering and training analysis of the CNN, and all the images after the normalization were used for the assessment of the relationship with clinical traits.
Feature Extraction and Dimension Reduction
We used Neural Architecture Search Network (NASNet),11 implemented in keras (NASNet-Large),12 to extract the features. The NASNet searched for the model architecture directly and achieved state-of-the-art performance regarding the classification of ImageNet, which consisted of >14 million images.13 Weights pretrained with ImageNet on keras implementation were used, and the output of the final concatenation layer was averaged by the global average pooling, which yielded 4032 feature vectors obtained per glomerulus image. These vectors were subsequently processed by uniform manifold approximation and projection (UMAP), which is a popular nonlinear dimension reduction algorithm officially implemented in Python library umap-learn.14
Model-based Clustering
We clustered the output of UMAP using mclust, a model-based clustering library using parameterized finite Gaussian mixture models (GMMs) with various covariance structures.15 The models were fitted with components of 1 to 20. The model with the best Bayesian information criterion was selected, and the optimal number of clusters was determined. All other parameters were set to the default, and all the covariances available in mclust were tested. The glomeruli were labeled according to the highest probabilities of belonging to the clusters.
Fine-tuning of the CNN and Calculation of Scores
We subsequently fine-tuned NASNet for the multiclass classification of defined clusters to produce scores robust to the rotation of the glomerulus images and visualize the rationale behind the prediction. We used keras 2.3.1 with tensorflow16 1.15 or 2.2.0 backend to train a model. We constructed a new model using layers of NASNet, from the input layer to the last concatenation layer, and a subsequent global average pooling layer, a dropout layer, and a dense layer with softmax as an activation function. We set the last 10 convolutional layers and the last dense layer to be trainable and set all the other layers to be untrainable. The preprocessed images were split into training and test datasets in a ratio of 8 to 2. Subsequently, the remaining training data were split into training and validation data by the ratio of 8 to 2 for use in the training process, with the stratification of the classes and the fixed seed. We split by stratification of classes, not partitioning by the patients, to preserve class distribution across the dataset. When training, the original images and centered zoomed images were augmented to horizontal, vertical flip, and rotation of 90°, 180°, and 270°, yielding 16 images from 1 raw image. The test and validation images were not augmented. In addition, because there was an imbalance of images between classes, class weights were set when training. The callback function performed early stopping when validation loss did not improve, reducing the learning rate on the plateau, and saving weights with the best validation loss were used during the training. We used categorical cross-entropy as the loss function and Adam as the optimizer.17 The performance was assessed using the area under the receiver operating characteristic curves (AUROCs) with the unaugmented test dataset as input. This was a multiclass classification problem and performance was assessed with a 1-vs-1 pairwise comparison18 and 1-vs-rest comparison with the prevalence weighted average.
Calculation of Histological Scores
The output values of the final layer were calculated using all the images after H&E normalization. The activation function of the last layer was softmax; thus, the scores per glomeruli were summed to 1. The calculated scores served as the histological scores of the respective glomerulus, and the mean scores of all glomerulus images from the slide images of the patient served as the histological scores of the respective patient.
Visualization of the Reason Behind the Prediction
We used score-weighted class activation mapping (Score-CAM) to visualize and highlight the important regions in the images for predicting the respective class.19 Because multiple convolutional layers were batch-normalized and concatenated to 1 layer in the last cell of NASNet, we visualized Score-CAM by obtaining the activation map corresponding to the output of the activation function of the final concatenation layer. In addition, guided backpropagation20 was calculated and multiplied with Score-CAM values to obtain guided Score-CAM to visualize the rationale at higher resolution, and the results of gradient-weighted class activation mapping (Grad-CAM) were visualized.21
Patch-based Analysis
We subsequently conducted the patch-based analysis of each glomerulus image to assess the applicability of the proposed approach for the higher resolution. The patch-based analysis referred to the same workflow applied to the image patches with the width and height of 96 pixels, equally divided into 16 sections from 1 glomerulus image. The patches were filtered beforehand in the same manner as the whole glomerulus images. The convolutional autoencoder with 6 convolutional layers was trained with the extracted patches as input. Subsequently, the output vectors of the encoder were clustered by GMMs, and the encoder was retrained with the defined clusters as same as the whole glomerulus images. In the patch-based analysis, the augmentation was not performed. The scores for each patch were obtained by the output of the retrained encoder, and the scores of all the patches were summed to calculate the glomerulus score. These glomerulus scores were averaged to obtain the histological score for the respective patient. The visualization was obtained by Score-CAM applied for the final layer of the encoder.
Comparison of the Scores in the Patient With Multiple Biopsy Specimens
To assess whether these scores reflect disease progression or regression in the multiple biopsy speciemens obtained from the same patient, we calculated the scores for 2 biopsy specimens from a patient and assessed the relationship between pathological assessment and the changes in the histological scores.
Statistical Analysis
The relationships between continuous variables and histological scores were modeled by linear regression models, and we tested the null hypothesis that the coefficient of the histological scores equals 0. The P values obtained via linear models were corrected using the Bonferroni procedure. The relationship between UOB and histological scores was assessed via one-way analysis of variance, with adjustment using Dunnett’s method22 with the control category as the negative category, performed via R library multcomp. The relationship between the clinical variables of SBP, SCr, and UPro and the MEST-C classification category was assessed by the same methods as the UOB. Adjusted P values < 0.05 were considered statistically significant. Data preprocessing was performed by pandas or tidyverse.23,24 The splitting of training, validation, and test data, the calculation of AUROC scores, and the clustering of patches were performed via the respective functions in scikit-learn.25 The figures were generated using the R libraries ggplot226 and firatheme.27 The visualized significant clusters identified by the algorithm with both the patched and the whole glomerulus images were evaluated first by 3 nephrologists, and the findings were validated by a board-certificated pathologist.
Data Availability
We cannot share the raw slide images because that will potentially breach patient privacy. However, we share the model and weights file used in the study for convolutional autoencoder and NASNet implemented in keras, which can be used to test the glomeruli images from other institutions after the normalization of staining (https://github.com/noriakis/glomerulus-clustering).
Results
Patient Demographics
The demographic information of 68 patients who underwent biopsy procedures at Kyoto University Hospital is summarized in Table 1. The resolution of the slide images was 54,332 ± 36,469 in width and 58,522 ± 14,353 in height (mean ± SD). Overall, 2144 images of glomeruli were obtained from the H&E staining–normalized slide images. The mean number of glomeruli per slide was 31.5 ± 16.8 (mean ± SD; minimum 6, maximum 73). After preprocessing, 1319 images were obtained for the downstream analysis of clustering and training of the CNN. Note that all the images were used in the calculation of histological scores.
Table 1.
Clinical and pathological characteristics of the included patients
| Clinical values | IgAN (n = 68) |
|---|---|
| Age, yr, mean (SD) | 42.28 (18.75) |
| Serum creatinine, mg/dL, mean (SD) | 0.97 (0.53) |
| Urinary protein, g/day or protein/creatinine ratio, mean (SD) | 1.37 (1.92) |
| Systolic blood pressure, mm Hg, mean (SD) | 124.13 (17.23) |
| Male gender, n (%) | 27 (39.7) |
| Urinary occult blood, n (%) | |
| − | 4 (5.9) |
| ± | 5 (7.4) |
| 1+ | 3 (4.4) |
| 2+ | 28 (41.2) |
| 3+ | 28 (41.2) |
| M = 1, n (%) | 28 (41.2) |
| E = 1, n (%) | 9 (13.2) |
| S = 1, n (%) | 52 (76.5) |
| T, n (%) | |
| 0 | 57 (83.8) |
| 1 | 9 (13.2) |
| 2 | 2 (2.9) |
| C, n (%) | |
| 0 | 30 (44.1) |
| 1 | 37 (54.4) |
| 2 | 1 (1.5) |
C, cellular or fibrocellular crescents; E, endocapillary hypercellularity; IgAN, IgA nephropathy; M, mesangial hypercellularity; S, segmental glomerulosclerosis; SD, standard deviation; T, interstitial fibrosis/tubular atrophy.
The Presentation of Workflow and Performance Assessment
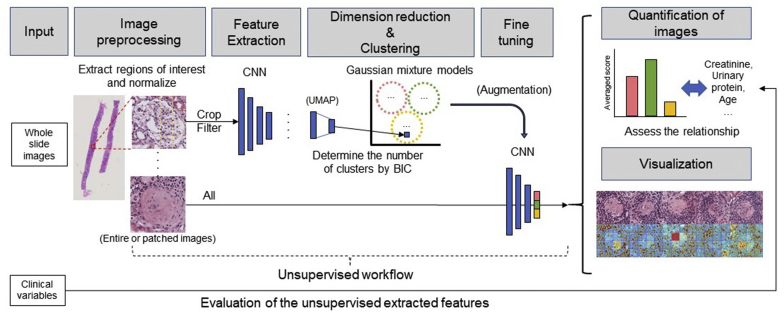
The overall proposed workflow with the selected steps is shown in Figure 1. The complete listing of all the steps is shown in Supplementary Text S1. We first used NASNet to extract the features from the preprocessed and filtered glomeruli images. UMAP was performed on the obtained feature vectors, and using the resulting vectors as inputs the optimal number of clusters was determined by GMMs by Bayesian information criterion. The model with 12 components, with the VVE (ellipsoidal, equal orientation) covariance had the best Bayesian information criterion value; therefore, the number of clusters was determined accordingly. Components 1, 2, and 3 of the UMAP results are shown in Supplementary Figure S1. The number of images in each cluster were 76, 117, 146, 137, 102, 95, 25, 251, 68, 99, 90, and 113, respectively. Using the defined cluster labels as the correct label, we performed fine-tuning of NASNet using the weights trained with ImageNet. The proportion of images of each patient in the training, test, and validation datasets are presented in Supplementary Table S1. The training was performed with 13,504 augmented images. Using the unaugmented test dataset, the 1-vs-1 weighted AUROC average was 0.921, and the 1-vs-rest weighted AUROC average was 0.918. The highest 1-vs-rest AUROC was obtained in cluster 10 (AUROC 0.998) and the lowest was in cluster 5 (AUROC 0.839). Using the weights obtained, we calculated histological scores for all patients using all the glomerulus images. The representative glomeruli for each cluster are presented in Supplementary Figure S2.
Figure 1.
Overall workflow. The overall workflow of the proposed methods is visualized.
Relationship Between Clinical Variables and Histological Scores
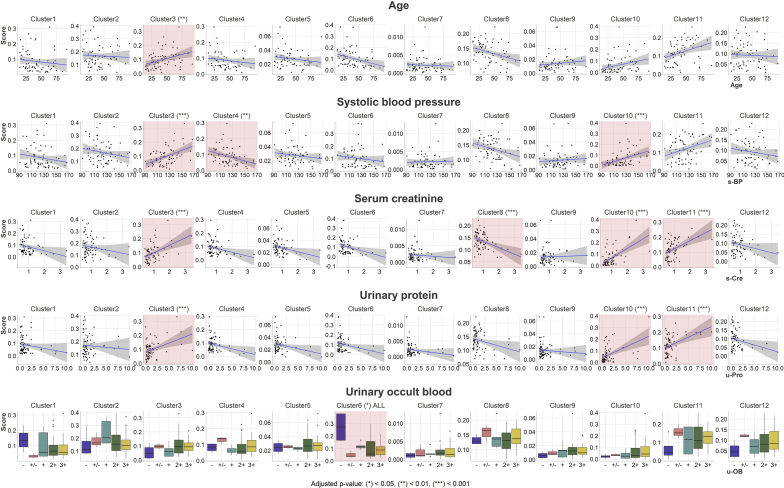
The score of cluster 2 was the highest among all the categories (0.168 ± 0.081). The overall relationships between the histological scores and clinical variables, including age, SBP, SCr, UPro, and the result of UOB test are summarized in Table 2 and Figure 2. The score of cluster 6 was significantly related to UOB, in the way that the negative category had the highest values compared with the other categories. The cluster was presumed to be the clusters containing the glomerulus with normal or minor abnormalities. The score of cluster 10 was significantly associated with SBP, SCr, and UPro, with higher scores indicating higher values. Cluster 11 had a significant relationship between SCr and UPro. The statistical summaries including the coefficients, P values, and R2 values of the linear models are presented in Supplementary Tables S2 and S3. For comparison, we assessed the relationship between the Oxford MEST-C score and clinical variables. For this assessment, the patient with C2 scoring was excluded beforehand. As a result, SCr was significantly associated with M score (coefficient = 0.288, P = 0.027). SBP and UPro had no significant relationship with the MEST-C score in the current cohort.
Table 2.
Statistically significant clusters and their associated variables
| Clinical valuesa | Significant clusterb |
|---|---|
| Age | 3 |
| Systolic blood pressure | 3, 4, and 10 |
| Serum creatinine | 3, 8, 10, and 11 |
| Urinary protein excretion | 3, 10, and 11 |
| Urinary occult blood (significant in ±, +, 2+, and 3+ compared with the negative [−] category) | 6 |
Clinical values tested.
The cluster in which the score is significantly associated with corresponding clinical values after the adjustment of P values.
Figure 2.
Relationship between histological scores and clinical variables. The box plot (urinary occult blood) and line plots (age, systolic blood pressure, serum creatinine, and urinary protein excretion) show the relationship between histological scores and clinical variables. The x axes represent clinical variables, and the y axes represent histological scores. Statistically significant clusters are presented with an asterisk and red background.
Visualization of the Rationale Behind the Prediction
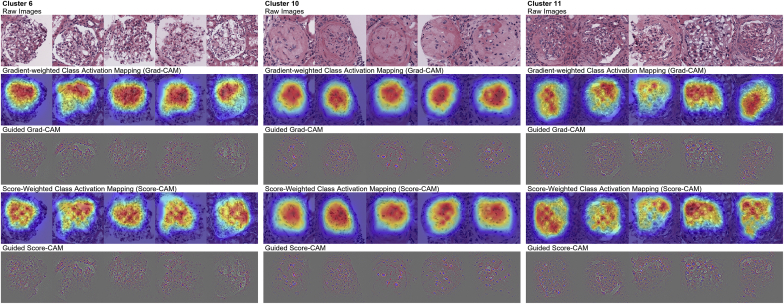
We obtained Score-CAM and guided Score-CAM, along with Grad-CAM and guided Grad-CAM of glomeruli having the highest 5 probabilities of classification of the respective cluster, which served as the rationale for the prediction of each cluster. We present the results of clusters 6, 10, and 11 in Figure 3. Cluster 6 contained glomeruli with mostly minor abnormalities. Cluster 10 contained sclerotic glomeruli. Cluster 11 contained glomeruli with mesangial matrix expansion and mesangial cell proliferation. In addition, the crescentic glomeruli or glomeruli with suspected adhesion and fibrosis were included. Grad-CAM and Score-CAM seemed to correctly point out the structure inside the glomeruli, with attention split to various positions. The guided Grad-CAM and Score-CAM of cluster 6 seemed to indicate that white pixel regions in the images got high attention. In the case of glomerular pathology such as the present study, the white areas are likely to be Bowman’s space or capillary lumen. However, in the other clusters, specific regions such as the regions of the mesangial matrix expansion did not get specific high attention.
Figure 3.
Visualization results of the rationale behind the prediction of each class. The score-weighted class activation mapping, gradient-weighted class activation mapping, and the results obtained by multiplication with guided backpropagation are shown. Clusters 6 (left), 10 (middle), and 11 (right) are shown.
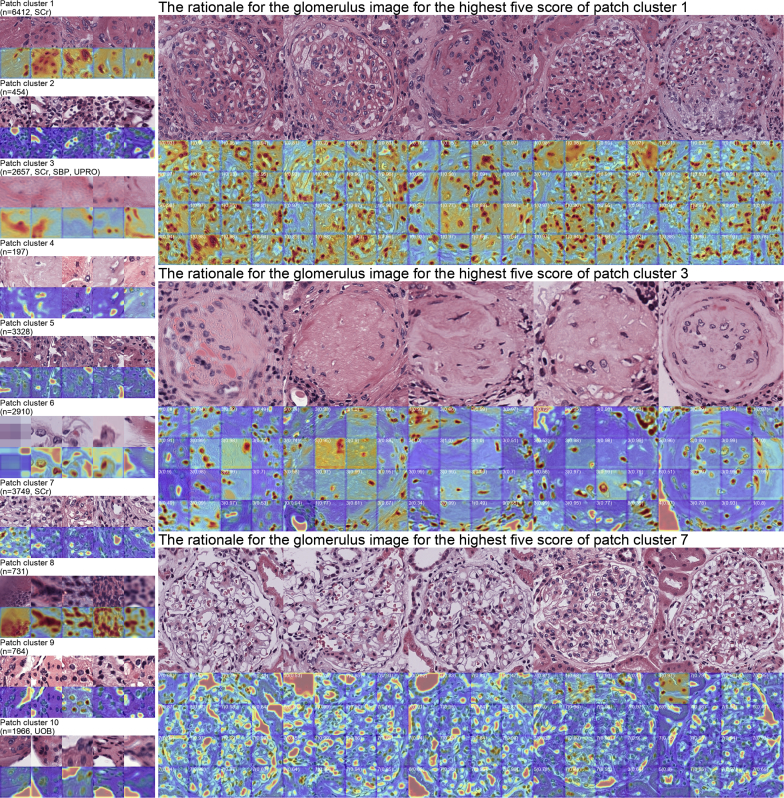
The Result of the Patch-based Analysis
We conducted additional analysis for the equally divided patches using the same workflow to assess the applicability of the approach for the higher resolution. Overall, 23,168 patches extracted from the glomerulus images were analyzed. The results were summarized in Figure 4. The score of patch cluster 1 had a significant relationships with SCr (coefficient = 0.09, P = 0.019), and the score of cluster 3 with SCr (coefficient = 0.249, P < 0.001), SBP (coefficient = 5.71, P = 0.013), and UPro (coefficient = 0.714, P = 0.003). Cluster 7 had a significant relationship with SCr (coefficient = −0.145, P = 0.039). In addition, cluster 10 had a significant relationship with UOB (comparison between negative and ±, 2+, and 3+). All the statistical summaries including the coefficients, P values, and R2 values of the linear models are presented in Supplementary Tables S4 and S5.
Figure 4.
The result summary of the patch-based analysis. The results of the patch-based analysis are shown. The left panel shows the clustered patches and the rationale behind the clustering visualized by score-weighted class activation mapping. The number of patches in the class, along with the clinical variables that had a significant relationship with the score of the patch class, are shown. The right panel shows the rationale for the patches of the glomeruli with the highest scores of the respective cluster of patches. The predicted cluster of each patch is shown in the upper left corner with the prediction probability.
The visualization results suggested that cluster 1 contained crescentic glomeruli. Cluster 3 contained sclerotic glomeruli. Cluster 7 contained glomeruli with mild mesangial matrix expansion and mesangial cell proliferation. The visualization results of Score-CAM suggested that the model gave high attention to cells with mesangial matrix, sclerotic regions, and white regions, such as Bowman’s space or capillary lumen, in clusters 1, 3, and 7, respectively.
Using the patch-based scores as input, we performed the additional analysis comparing the multiple subsequent biopsy specimens of 1 patient. The patient was diagnosed with IgAN after the first biopsy procedure and went through the second biopsy procedure after the Pozzi protocol. The third biopsy specimen was obtained as UPro increased. We performed the analysis on the available virtual slides of the second and third biopsy specimens. The clinical and pathological findings of the second and third biopsy specimens of the respective patients are shown in Supplementary Table S6. As a result, the score of the third biopsy specimen was higher in clusters 1 and 3, which indicated that the proportion of sclerotic and crescentic glomeruli increased. Contrarily, the scores of cluster 10, which gave high attention to mostly the white regions, decreased. This indication matched the pathological reports of the respective biopsy procedures. A summary of the comparison is shown in Supplementary Figure S3.
Discussion
In the present study, we proposed an unsupervised approach to quantitatively assess histological findings and evaluated their relationship with clinical information. In addition, the reason behind the definition of classes was visualized. As a result, the histological scores obtained by unsupervised clustering of the glomerulus image features extracted from the CNN model had significant relationships with the important clinical measurements in patients with IgAN.
Various studies have used machine learning to evaluate renal histopathology. Their main objective is to segment various structures present in the slide images, such as the glomeruli,28 or to detect the glomeruli from the images,29 or to extract novel pathological findings from the glomeruli,30 or to associate defined glomerular features with some pathological findings or clinical variables.5,31 Our study falls into the last category. One study used manually constructed features of the glomerulus images to detect proliferative lesions in the glomerulus, without using deep learning.32 Compared with the study conducted by Ginley et al.5 that used handcrafted features combined with CNN, our approach consisted of no previous knowledge, such as glomerular components, to feed into the network. Rather, we allowed the CNN and clustering algorithm to decide the class of the glomeruli images and patches. This can be advantageous in the sense that we can evaluate how the CNN assesses and classify the glomerular image regardless of the existing knowledge. Conversely, this can also be problematic because the resolution did not reach the expertise of pathologists, thus limiting the use of our model in the clinical setting because of low interpretability.
The CNN could discriminate between the defined clusters of the glomeruli images according to the AUROC. This is an expected result because the correct labels were defined using feature vectors extracted by the same CNN. We used Score-CAM which is a newly developed method to visualize the rationale in CNN architecture that is reported to represent better localization compared with the popular Grad-CAM or Grad-CAM++.33 The clustering results suggested that the proposed model captured some of the important pathological findings and normal findings of IgAN. However, as discussed above, the activation map could not localize the specific pathological changes in the structural components inside the glomeruli, such as adhesion, crescent formation, or mesangial proliferation; rather focusing vaguely in the experiment on the entire glomerulus images. We speculated that one of the reasons for these results is that the H&E stain used in this study was originally difficult to evaluate these findings, compared with periodic acid–Schiff or periodic acid methenamine silver stain. On the patch-based analysis, we found that the proposed approach could correctly give high attention to the structure in the images like cellular components, sclerotic regions, crescentic regions, or capillary lumen, compared with the results that the analysis with the entire glomerulus images as input failed to capture these findings precisely. This suggested that the calculated scores based on the proposed approach could be interpreted by physicians to some extent.
In previous studies investigating the correlation of MEST-C score8 and clinical variables, C-score was associated with SCr and UOB, and S-score with hypertension.34 In addition, global glomerulosclerosis was associated with lower estimated glomerular filtration rate, UPro, SCr, and higher incidence of arterial hypertension,35 and crescentic formation was reported to be associated with estimated glomerular filtration rate and proteinuria.36 We also investigated the relationship between the MEST-C scoring and clinical variables. In our study, the scores reported to be related to clinical variables such as C were not associated with any clinical variables. This is suspected to be the nature of the investigated population from the university hospital, where the severe disease patients tend to admit. Compared with MEST-C scoring, our proposed score could relate to UPro, and the other variables of SBP and UOB as well. In addition, our system is capable of evaluating glomeruli more quantitatively.
The advantage of this study is that we successfully developed a histological assessment workflow, and confirm the calculated scores to relate with clinical indicators in the patients with IgAN, especially UPro and SCr, by examining glomerular image features in an unsupervised manner, independent of nephrologists and pathologists. The weights of this model are publicly available, and thus these could contribute to the standardization of histological assessment. In addition, the CNNs used in the study are replaceable and can easily scale as the new models develop in the future. Moreover, there is a possibility that disease progression could be evaluated quantitatively in the subsequent biopsy specimen in the same patient; however, testing with more patients with the subsequent biopsy specimen is desired. The major limitation of the study was that we could not assess the relationship between histological scores and prognostic information of patients because the observation period was short. In addition, because it involves unsupervised clustering, the results are expected to vary with parameter adjustments. For example, if the number of neighbors in UMAP is reduced in the experiment of the entire glomerulus images as input, the images will split into many more clusters, and it depends on the number of available training images. In addition, the evaluation of the clustering and visualization results remained subjective. Although the association was statistically significant, the R2 values for the linear models were low. This is presumed to be partially because of the variabilities of clinical variables assessed, especially for UOB, which was assessed by dipstick, and UPro, in which the daily urinary protein and urinary creatinine ratio were used.
In summary, we proposed an unsupervised approach to quantitatively evaluate histological findings along with providing the rationale for the evaluation and applied it to the kidney histological images. The obtained scores were related to important clinical variables in patients with IgAN and could be applied to other glomerular diseases or conditions that require evaluation of specific structures inside the slide images.
Disclosure
MY receives research grants from Mitsubishi Tanabe Pharma and Boehringer Ingelheim. All the other authors declared no competing interests.
Acknowledgments
This work was supported by JSPS KAKENHI Grant-in-Aid for Early-Career Scientists Grant No. 19K18321. The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Footnotes
Supplementary Text S1. The complete listing of the steps of the proposed workflow.
Supplementary Table S1. The proportion of images of each patient in the train, test, and validation dataset.
Supplementary Table S2. The statistical summary of the linear models testing the relationship between the scores and the clinical variables.
Supplementary Table S3. The R squared values for the linear models investigating the relationship between the scores and clinical variables.
Supplementary Table S4. The statistical summary of the linear models testing the relationship between the patch-based scores and the clinical variables.
Supplementary Table S5. The R squared values for the linear models investigating the relationship between the patch-based scores and clinical variables.
Supplementary Table S6. The clinical and pathological findings of the second and third biopsy of the patient with multiple biopsies.
Figure S1. The visualization of the UMAP results.
Figure S2. The representative glomeruli for each cluster.
Figure S3. The changes in the patch-based scores for the patient with multiple biopsies.
Supplementary Material
Supplementary Text S1. The complete listing of the steps of the proposed workflow.
Supplementary Table S1. The proportion of images of each patient in the train, test, and validation dataset.
Supplementary Table S2. The statistical summary of the linear models testing the relationship between the scores and the clinical variables.
Supplementary Table S3. The R squared values for the linear models investigating the relationship between the scores and clinical variables.
Supplementary Table S4. The statistical summary of the linear models testing the relationship between the patch-based scores and the clinical variables.
Supplementary Table S5. The R squared values for the linear models investigating the relationship between the patch-based scores and clinical variables.
Supplementary Table S6. The clinical and pathological findings of the second and third biopsy of the patient with multiple biopsies.
Figure S1. The visualization of the UMAP results.
Figure S2. The representative glomeruli for each cluster.
Figure S3. The changes in the patch-based scores for the patient with multiple biopsies.
References
- 1.Krizhevsky A., Sutskever I., Hinton G.E. In: Advances in Neural Information Processing Systems 25. Pereira F., Burges C.J.C., Bottou L., Weinberger K.Q., editors. Curran Associates, Inc.; Red Hook, NY: 2012. ImageNet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 2.Coudray N., Ocampo P.S., Sakellaropoulos T. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iizuka O., Kanavati F., Kato K., Rambeau M., Arihiro K., Tsuneki M. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci Rep. 2020;10:1504. doi: 10.1038/s41598-020-58467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song Z., Zou S., Zhou W. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun. 2020;11:4294. doi: 10.1038/s41467-020-18147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginley B., Lutnick B., Jen K.-Y. Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol. 2019;30:1953–1967. doi: 10.1681/ASN.2018121259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker J.U., Mayerich D., Padmanabhan M. Artificial intelligence and machine learning in nephropathology. Kidney Int. 2020;98:65–75. doi: 10.1016/j.kint.2020.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchino E., Suzuki K., Sato N. Classification of glomerular pathological findings using deep learning and nephrologist-AI collective intelligence approach. Int J Med Inform. 2020;141:104231. doi: 10.1016/j.ijmedinf.2020.104231. [DOI] [PubMed] [Google Scholar]
- 8.Trimarchi H., Barratt J., Cattran D.C. Oxford classification of IgA nephropathy 2016: an update from the IgA Nephropathy Classification Working Group. Kidney Int. 2017;91:1014–1021. doi: 10.1016/j.kint.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Goode A., Gilbert B., Harkes J., Jukic D., Satyanarayanan M. OpenSlide: a vendor-neutral software foundation for digital pathology. J Pathol Inform. 2013;4:27. doi: 10.4103/2153-3539.119005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macenko M., Niethammer M., Marron J.S. In: 2009 IEEE International Symposium on Biomedical Imaging. From Nano to Macro; 2009. A method for normalizing histology slides for quantitative analysis; pp. 1107–1110. [Google Scholar]
- 11.Zoph B., Vasudevan V., Shlens J., Le Q.V. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Learning transferable architectures for scalable image recognition; pp. 8697–8710. [Google Scholar]
- 12.Chollet F. Keras. https://keras.io Available at:
- 13.Russakovsky O., Deng J., Su H. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–252. [Google Scholar]
- 14.McInnes L., Healy J., Saul N., Großberger L. UMAP: uniform manifold approximation and projection. J Open Source Softw. 2018;3:861. [Google Scholar]
- 15.Fraley C., Raftery A. Model-based Methods of Classification: Using the mclust Software in Chemometrics. J Stat Softw. 2007;18(6):1–13. [Google Scholar]
- 16.Abadi M., Agarwal A., Barham P. TensorFlow: large-scale machine learning on heterogeneous systems. http://tensorflow.org/ Available at:
- 17.Kingma D.P., Ba J. Adam: A Method for Stochastic Optimization. arXiv [csLG] http://arxiv.org/abs/1412.6980 Published online December 22, 2014.
- 18.Hand D.J., Till R.J. A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach Learn. 2001;45:171–186. [Google Scholar]
- 19.Wang H., Wang Z., Du M. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops. 2020. Score-CAM: score-weighted visual explanations for convolutional neural networks; pp. 24–25. [Google Scholar]
- 20.Springenberg J.T., Dosovitskiy A., Brox T., Riedmiller M. Striving for simplicity: the all convolutional net. arXiv. http://arxiv.org/abs/1412.6806 Available at:
- 21.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. In: 2017 IEEE International Conference on Computer Vision (ICCV) 2017. Grad-CAM: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 22.Dunnett C.W. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc. 1955;50:1096–1121. [Google Scholar]
- 23.pandas-dev/pandas: Pandas. 10.5281/zenodo.3509134 Available at:
- 24.Wickham H., Averick M., Bryan J. Welcome to the tidyverse. J Open Source Softw. 2019;4:1686. [Google Scholar]
- 25.Pedregosa F., Varoquaux G., Gramfort A. Scikit-learn: machine learning in Python. J Mach Learn Res. https://dl.acm.org/doi/10.5555/1953048.2078195 Available at: [DOI] [PMC free article] [PubMed]
- 26.Wickham H. ggplot2: elegant graphics for data analysis. https://ggplot2.tidyverse.org Available at:
- 27.van Kesteren E.-J. Vankesteren/Firatheme: Firatheme Version 0.2.1. 10.5281/zenodo.3604681 Available at:
- 28.Kannan S., Morgan L.A., Liang B. Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Rep. 2019;4:955–962. doi: 10.1016/j.ekir.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Temerinac-Ott M., Forestier G., Schmitz J. In: Proceedings of the 10th International Symposium on Image and Signal Processing and Analysis. 2017. Detection of glomeruli in renal pathology by mutual comparison of multiple staining modalities; pp. 19–24. [Google Scholar]
- 30.Sheehan S., Mawe S., Cianciolo R.E., Korstanje R., Mahoney J.M. Detection and classification of novel renal histologic phenotypes using deep neural networks. Am J Pathol. 2019;189:1786–1796. doi: 10.1016/j.ajpath.2019.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng C.-H., Nan Y., Xu F. Identification of glomerular lesions and intrinsic glomerular cell types in kidney diseases via deep learning. J Pathol. 2020;252:53–64. doi: 10.1002/path.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barros G.O., Navarro B., Duarte A., Dos-Santos W.L.C. PathoSpotter-K: a computational tool for the automatic identification of glomerular lesions in histological images of kidneys. Sci Rep. 2017;7:46769. doi: 10.1038/srep46769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chattopadhay A., Sarkar A., Howlader P., Balasubramanian V.N. In: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV) 2018. Grad-CAM++: generalized gradient-based visual explanations for deep convolutional networks; pp. 839–847. [Google Scholar]
- 34.Gowrishankar S., Gupta Y., Vankalakunti M. Correlation of Oxford MEST-C scores with clinical variables for IgA nephropathy in South India. Kidney Int Rep. 2019;4:1485–1490. doi: 10.1016/j.ekir.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng W., Tang Y., Tan L., Qin W. Crescents and global glomerulosclerosis in Chinese IgA nephropathy patients: a five-year follow-up. Kidney Blood Press Res. 2019;44:103–112. doi: 10.1159/000498874. [DOI] [PubMed] [Google Scholar]
- 36.Shao X., Li B., Cao L. Evaluation of crescent formation as a predictive marker in immunoglobulin A nephropathy: a systematic review and meta-analysis. Oncotarget. 2017;8:46436–46448. doi: 10.18632/oncotarget.17502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We cannot share the raw slide images because that will potentially breach patient privacy. However, we share the model and weights file used in the study for convolutional autoencoder and NASNet implemented in keras, which can be used to test the glomeruli images from other institutions after the normalization of staining (https://github.com/noriakis/glomerulus-clustering).