Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV -2), a causative agent of COVID-19 disease, poses a significant threat to public health. Since its outbreak in December 2019, Wuhan, China, extensive collection of diverse data from cell culture and animal infections as well as population level data from an ongoing pandemic, has been vital in assessing strategies to battle its spread. Mathematical modelling plays a key role in quantifying determinants that drive virus infection dynamics, especially those relevant for epidemiological investigations and predictions as well as for proposing efficient mitigation strategies. We utilized a simple mathematical model to describe and explain experimental results on viral replication cycle kinetics during SARS-CoV-2 infection of animal and human derived cell lines, green monkey kidney cells, Vero-E6, and human lung epithelium cells, A549-ACE2, respectively. We conducted cell infections using two distinct initial viral concentrations and quantified viral loads over time. We then fitted the model to our experimental data and quantified the viral parameters. We showed that such cellular tropism generates significant differences in the infection rates and incubation times of SARS-CoV-2, that is, the times to the first release of newly synthesised viral progeny by SARS-CoV-2-infected cells. Specifically, the rate at which A549-ACE2 cells were infected by SARS-CoV-2 was 15 times lower than that in the case of Vero-E6 cell infection and the duration of latent phase of A549-ACE2 cells was 1.6 times longer than that of Vero-E6 cells. On the other hand, we found no statistically significant differences in other viral parameters, such as viral production rate or infected cell death rate. Since in vitro infection assays represent the first stage in the development of antiviral treatments against SARS-CoV-2, discrepancies in the viral parameter values across different cell hosts have to be identified and quantified to better target vaccine and antiviral research.
Keywords: COVID-19, Viral dynamics, Parameter estimation, In vitro infection
1. Introduction
Severe acute respiratory syndrome coronavirus 2 or SARS-CoV-2, a causative agent of the coronavirus disease 2019 (COVID-19), is an enveloped, positive-sense, single-stranded RNA virus belonging to the Sarbecovirus subgenus of the Betacoronavirus genus along with several other zoonotic coronaviruses (Hu et al., 2021, Wells et al., 2021). Since December 2019, almost 200 million people have been confirmed to have contracted the virus worldwide and over 4 million fatalities have been reported in relation to COVID-19. Clinically approved antiviral drugs have been tested to be repurposed to treat COIVD-19 and a set of pharmaceutical agents have displayed antiviral activity and inhibited SARS-CoV-2 growth (Hoffmann et al., 2020, Gordon et al., 2020, Pizzorno et al., 2020). One of these therapeutic agents, remdesivir, has already been approved for treatment of patients hospitalized with COVID-19. It showed superiority to placebo in shortening the time to recovery in adults (Beigel et al., 2020). Moreover, monoclonal antibodies have also been assessed as a therapy, but due to their high cost are not widely available (Taylor et al., 2021). Despite the lack of diverse antiviral treatment, recent efforts in vaccine development has resulted in successful mass deployment of several vaccine candidates (Mulligan et al., 2020, Polack et al., 2020, Voysey et al., 2021, Logunov et al., 2021).
Epidemiological investigations of the SARS-CoV-2 spread and possible effects of different mitigation strategies have been conducted by a growing number of scientists around the globe. Public health approaches rely heavily on quantitative analysis of the available data, the results of which can prove essential in identifying and quantifying the most influential determinants of the COVID-19 dynamics (Li et al., 2020, Endo et al., 2020, Ferretti et al., 2020, Dickens et al., 2020). Thus, mathematical modelling is an important tool that can help design efficient strategies to control COVID-19 pandemic.
SARS-CoV-2 infects predominantly epithelium of the human upper and lower respiratory tract (Wölfel et al., 2020), but has been reported to be capable of invading other tissue types and cells including human endocrine and exocrine pancreas (Müller et al., 2021), lung carcinoma, intestinal and liver cell lines (Chu et al., 2020). Thus, there is an urgent need for information on the SARS-CoV-2 replication, incubation period and infectivity in different tissues and cellular hosts. Since cellular tropism as well as genetic variability in the viral RNA can generate differences in the SARS-CoV-2 growth phenotype, obtaining such information is crucial to identify the most promising targets for vaccine development.
In this study, we used a combined approach of mathematical modelling and experimental data to describe replication kinetics of SARS-CoV-2 in animal and human cellular hosts. This approach has been used to explain in vitro and in vivo kinetics of other respiratory viruses, such as influenza A virus (Baccam et al., 2006, Pinilla et al., 2012, Paradis et al., 2015, Heldt et al., 2013, Smith and Ribeiro, 2010, Bai et al., 2019, Schelker et al., 2016, Schmid and Dobrovolny, 2020) and respiratory syncytial virus (Beauchemin et al., 2019, Wethington et al., 2019, González-Parra and Dobrovolny, 2019). We infected the human lung epithelium cell line expressing the ACE2 receptor, A549-ACE2 and the green monkey kidney cell line, Vero-E6, with the SARS-CoV-2 using two distinct multiplicities of infection (MOIs) and tracked the infection progression by measuring the total extracellular viral load over time. We chose the highly permissive Vero-E6 cells because they are commonly used to establish cytopathic assay protocols, characterize the viral kinetics and isolate viruses from clinical samples to generate virus stocks. However, there is a large variation in the SARS-CoV-2 growth phenotype and the Vero-E6-specific findings cannot be extrapolated to other types of host cells (Ogando et al., 2020, Milewska et al., 2020). For this reason, we also infected A549-ACE2 cells with the SARS-CoV-2 as they represent a more realistic model to mathematically study SARS-CoV-2 growth. We fitted the mathematical model to our experimental data and estimated the parameters determining the basic steps of the SARS-CoV-2 replication cycle. We found that the virus infected the A549-ACE2 cells at a significantly reduced rate and had prolonged incubation period compared to Vero-E6 cells. In addition, the associated dynamics of infected cells were significantly different as virus-producing Vero-E6 were removed from the dynamics due to virus-induced death at much earlier times compared to A549-ACE2. These result highly correlate with the findings from in vitro experimental studies on SARS-CoV-2 cellular tropism, replication kinetics and cell damage (Chu et al., 2020, Ogando et al., 2020).
2. Materials and methods
Cells. A549 cells stably expressing ACE2 (A549-ACE2, kindly provided by Dr. Olivier Schwartz) and Vero-E6 cells (Vero 76, clone E6) were maintained and propagated at 37°C, 5% CO2 in DMEM with L-glutamine (Gibco) supplemented with 10% (v/v) FBS (Invitrogen) and 5 units/ml penicillin and 5 g/ml streptomycin (Life Technologies). A549-ACE2 cells were supplemented with 20 g/mL blasticidin S.
Virus. The SARS-CoV-2 strain used (BetaCoV/France/IDF0372/2020 strain) was propagated once in Vero-E6 cells and is a kind gift from the National Reference Centre for Respiratory Viruses at Institut Pasteur, Paris, originally supplied through the European Virus Archive goes Global platform.
RT-qPCR. Viral load was assessed by RT-qPCR. Briefly, the cell culture supernatant was collected, heat inactivated at 95°C for 5 min and used for RT-qPCR analysis. SARS-CoV-2 specific primers targeting the N gene region: 5’-TAATCAGACAAGGAACTGATTA-3’ (Forward) and 5’-CGAAGGTGTGACTT CCATG-3’ (Reverse) were used with the Luna Universal One-Step RT-qPCR Kit (New England Biolabs) in an Applied Biosystems QuantStudio 6 thermocycler, with the following cycling conditions: 55°C for 10 min, 95°C for 1 min, and 40 cycles of 95°C for 10 s, followed by 60°C for 1 min. The number of viral genomes is expressed as PFU equivalents/mL, and was calculated by performing a standard curve with RNA derived from a viral stock with a known viral titer.
Growth curves. 75,000 A549-ACE2 and Vero-E6 cells were seeded per well in a 24-well plate the day before infection and incubated at 37°C, 5% CO2. At the time of infection, the medium was removed and replaced with 250 L virus-containing media (MOI 0.1 or 1 PFU per cell) and incubated at 37°C for 1 h. Following this adsorption period, the virus-containing media was removed, cells washed three times with PBS and replaced with fresh media (0.5 mL). This point is considered time 0 h post infection. At 16, 24, 48, 72 or 96 h p.i., 50 L supernatant from triplicate wells was removed for RT-qPCR analysis. At each point, the medium removed was replaced with fresh media.
Mathematical model. We use a viral dynamics model developed by (Baccam et al., 2006) and extended for a distributed delay in the latent phase of infected cells to capture the time course changes in the accumulation of extracellular virions (Pinilla et al., 2012, Paradis et al., 2015, Kakizoe et al., 2015, Beauchemin et al., 2017, Hurtado and Kirosingh, 2019, Beauchemin et al., 2019). Briefly, susceptible cells S are infected with virus V (expressed in plaque forming unit equivalents per mL, PFUe/mL) at the rate (measured in ), after which they enter the latent phase (Anderson and Watson, 1980, Krylova and Earn, 2013). Since our data suggest a delay in SARS-CoV-2 accumulation (Fig. 1 ), the latent phase, that is the time between the entry of the virus into the cell and the first release of new viral particles out of the cell, is assumed to be separated into stages and the mean duration of the latent phase to follow Erlang distribution (Kakizoe et al., 2015). The latent cells move through each stage at the rate (measured in ). Those in the last stage, , become infectious cells, I, and produce virus at the rate p (measured in ). Here, we consider a simple exponential distribution for the mean duration of the infectious phase, and thus for the infection-induced death rate (measured in ) of infectious cells I. Erlang distributed duration of infectious phase has been suggested, for example, for SHIV in vitro infection (Beauchemin et al., 2017) and supported by kinetic data that revealed temporal changes in the population of infected cells which could be fit to determine the appropriate number of compartments. Since we do not have such cellular data available, we opted for a simple exponentially distributed . Finally, viral particles may degrade at the rate c (measured in ). The full model is as follows
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
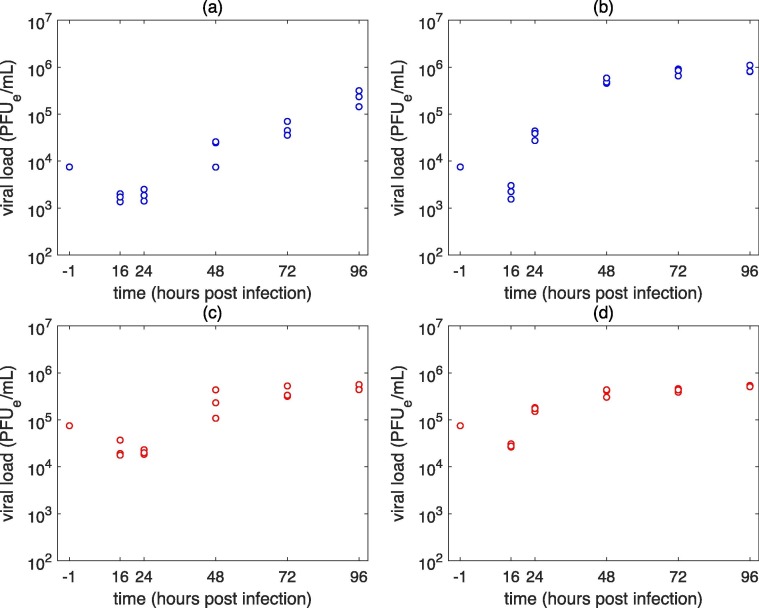
Fig. 1.
Experimental SARS-CoV-2 infection of (a), (c) A549-ACE2 and (b), (d) Vero-E6 cells. Infection was performed at an MOI of (a), (b) 0.1 PFUe/cell (low MOI infection) and (c), (d) 1 PFUe/cell (high MOI infection).
Our experiments revealed (data not presented) that degradation of SARS-CoV-2 particles is negligible over the period of the infection experiment, i.e. 96 h. Thus, we fixed the viral particle degradation rate to zero, h−1.
We take into account the one time procedure during which cells inoculated with virus at the beginning of the experiment are washed off the excess virus that did not enter the cells after one hour after infection. This is modelled by introducing the washing rate into the Eq. (5) as proposed in (Zitzmann et al., 2020) to model the washing procedure during cellular infection with Dengue virus,
| (6) |
The washing procedure is, in the viral dynamics, manifested as a sudden drop in the viral load one hour post infection. The parameter is the ‘strength’ of the washing process which, in general, can exhibit variability for parallel measurements of biologically distinct infection experiments, including those with different initial multiplicity of infection (MOI), despite using the same tools and protocols. Since we infected both A549-ACE2 and Vero-E6 cells with an MOI of 0.1 (low MOI infection) and 1 (high MOI infection) virus per cell, we thus model the washing process separately for low and high MOI infections and consider the respective washing rates to be and , respectively. The strengths of the washing process are, in the case of low and high MOI infection experiments, denoted as and , respectively. The parameter is the timing of the washing process and was set to . The parameter is the standard deviation of the washing process. Since cells were washed for approximately 3 min, was set to .
The initial conditions are set as follows: in the case of low MOI infection and in the case of high MOI infection. The initial number of A549-ACE2 and Vero-E6 cells, , seeded was 75,000. Since 24 h incubation took place, at the time of infection the initial number of cells to be infected doubled and was set to cells.
Virus reproduction ratio, , for the Eqs. (1), (2), (3), (4), (5) is and gives the total number of newly infectious cells that were infected by viral units released from one infectious cell. However, with and , that is assumptions that, as previously described, best reflect the experimental conditions, it follows that . Thus, the infection will always be established.
Experimental data fitting. We used Bayesian inference to estimate the parameters of the model (1), (5) and employed the MATLAB implementation of the Goodman and Weare affine invariant ensemble Markov Chain Monte Carlo (MCMC) sampler, gwmcmc (Goodman and Weare, 2010). Estimates from the minimization of the weighted sum of squared residuals between the logarithms experimental data and the solution of the model (1), (5), given as
| (7) |
were used to initiate the MCMC process. The model was fitted to the low and high MOI infection datasets simultaneously and the individual fits to each dataset were included in the calculation of the weighted sum of squared residuals. The weights and were chosen to be the inverse of the low and high MOI infection data variances calculated from their log-values, respectively, at each measured time. Uniform priors were assumed for all model parameters subject to estimation, i.e. and while the number of compartments, , of the latent phase was kept fixed. The gwmcmc was initiated by an ensemble of walkers within a Gaussian ball centered around the initial estimates. We used a Gaussian likelihood function and maximized its log-likelihood
| (8) |
to evaluate the goodness of the proposed fit in the MCMC process. We initiated (number of fitted parameters) of walkers and posterior distributions were constructed using 30,000 samples with a burn-in of 50% of all drawn samples and thinning by storing every 15th sample. Thus, each posterior parameter distribution comprised of 1000 samples.
Statistical analysis. Statistical significance was quantified using the bootstrap (Efron and Tibshirani, 1993) Mann Whitney U-test. To determine whether there were statistically significant differences between two posterior distributions, we randomly drew 100 samples with replacement out of the total of 1000 accepted posterior samples for each distribution and calculated the Mann Whitney U-test associated P-value using MATLAB’s function ranksum. We repeated the procedure 500 times and calculated the final P-value as the mean of all bootstrap P-values.
3. Results
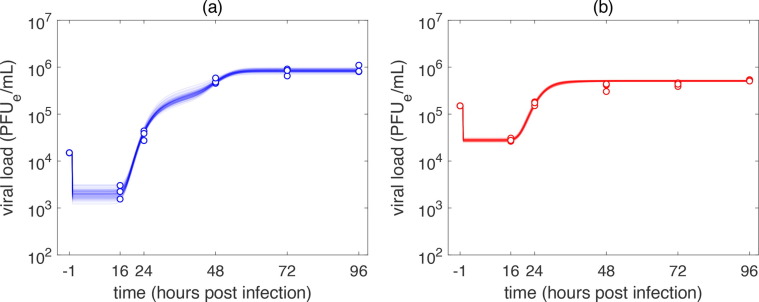
Experimental infection of A549-ACE2 and Vero-E6 cells with SARS-CoV-2. To study the SARS-CoV-2 time course kinetics in vitro and to quantify the basic parameters of its replication cycle, we infected the A549-ACE2 cells and Vero-E6 cells using two distinct initial concentrations of infectious virus or multiplicity of infection (MOI), namely 0.1 and 1 infectious virus per cell (Fig. 1). In Vero-E6 cells, the viral loads increased 6- to 16-fold between 16 and 24 h post infection (h p.i.); whereas the accumulation of viral particles was delayed in A549-ACE2 cells, which had comparable viral loads at these time points for both low and high MOI infections. In Vero-E6 cells infection, the viral load reached its plateau at 48 h for both low and high MOI infections, suggesting that production of new virions would have ceased by that point in time. In A549-ACE2 cells, the plateau was achieved at 72 h p.i. for high MOI infection while in the case of low MOI infection, accumulation of virions was still continuing at 96 h p.i.
Quantification of the SARS-CoV-2 replication cycle parameters. We used the mathematical model (1), (5) to reproduce our experimental data and to quantify the basic parameters of SARS-CoV-2 replication cycle during an infection of A549-ACE2 and Vero-E6 cells, namely the infection rate , the duration of latent phase , the rate of infected cells death and the per-cell virion production rate p. The washing rate parameters characterizing the strength of the washing procedures for the low and high MOI infections, and , were also considered as free parameters. We estimated the parameters of the model (1), (5) using Markov chain Monte Carlo (MCMC) approach (details are in Materials and Methods). This allowed us to extract the parameter posterior distributions and construct the 95% credible regions (CRs).
Since we considered the latent cells to go through stages before entering the infectious phase, we decided by fitting the model (1), (5) to low and high MOI infection data using the MCMC approach. We varied between 1 and 60 with an increment of 10 and determined that the model (1), (5) with yielded statistically equivalent fits for A549-ACE2 cell line and with yielded statistically equivalent fits for Vero-E6 cell line. We thus fixed when fitting the time course infection of A549-ACE2 cells and when fitting the time course infection of Vero-E6 cells. We note that the parameter posterior distributions associated with each two were subject to a bootstrap Mann–Whitney U-test to decide the minimal number of compartments above which the difference between the parameter posterior distributions were not statistically significant (i.e., P-value 0.05, details on the statistical analysis are in Materials and Methods).
Preliminary fitting showed a positive correlation between the infectious cell death rate and virus production rate p and a tendency of the optimization process towards biologically infeasible values of . To avoid ever-increasing values for these two parameters, we put a biologically meaningful upper limit to , so that the mean lifetime of an infectious cell is not shorter than 6 min, i.e. h−1.
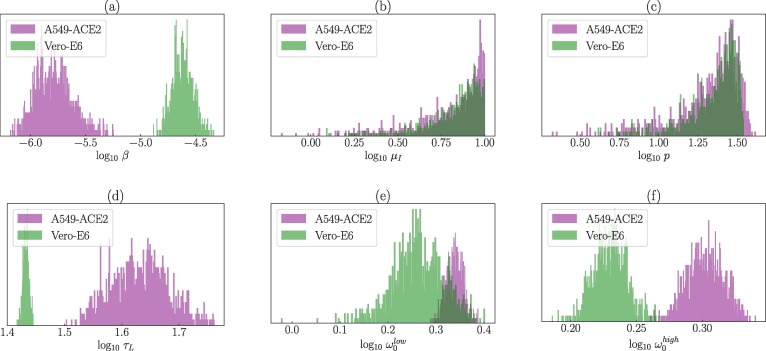
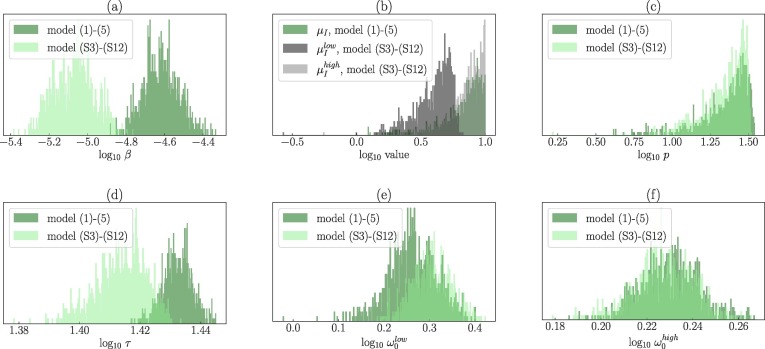
Differences in the SARS-CoV-2 parameters during A549-ACE2 and Vero-E6 cell infections. The fitted parameter values and their associated 95% CRs are in Table 1 . The MCMC inference showed that A549-ACE2 cells are infected by SARS-CoV-2 at a much slower rate (approximately ten times less) than Vero-E6 cells with the median for A549-ACE2 cells and for Vero-E6 cells (Fig. 2 a). We detected statistically significant differences (P-value ) in the inferred posterior distributions of all the model (1), (5) parameters except for the death rate of infectious cells, , and the rate of per-cell virus production p. The medians of the posterior distributions of were for A549-ACE2 cells and for Vero-E6 cells (Fig. 2b). The medians of the rate of per-cell virus production p of infected A549-ACE2 and Vero-E6 cells were determined to be and , respectively, with largely overlapping 95% CRs (Fig. 2c). Because the posterior distributions of were skewed towards the upper limit and due to the strong positive correlation between and p, the posterior distributions of p were also right-skewed. This positive correlation suggests that we cannot separate from p; if one increases the other increases in order to reproduce the data. For this reason, the model does not allow to find the accurate values of and p. Mean durations of the latent phase were significantly different for A549-ACE2 and Vero-E6 cells, with the respective medians equal to and (Fig. 2d). Correlations between all the parameters are depicted in Figures S.1 and S.2 in Supplementary Material.
Table 1.
| Parameter | Description | Units | Median |
95% Credible Region |
||
|---|---|---|---|---|---|---|
| A549-ACE2 | Vero-E6 | A549-ACE2 | Vero-E6 | |||
| infection rate | ()−1 | 1.59 | 2.4 | [8.37, 39.8] | [1.62, 3.66] | |
| infected cell death rate | 7.43 | 7.5 | [1.76, 9.88] | [2.15, 9.87] | ||
| virus production rate | (cell)−1 | 23.91 | 25.13 | [5.62, 35.65] | [7.23, 33.25 | |
| duration of latent phase | 42.74 | 27.07 | [34.82, 53.82] | [26.49, 27.63] | ||
| washing strength (low MOI) | 2.18 | 1.83 | [2.03, 2.35] | [1.40, 2.31] | ||
| washing strength (high MOI) | 2.00 | 1.70 | [1.89, 2.13] | [1.61, 1.81] | ||
Fig. 2.
Posterior parameter distributions associated with the MCMC fits of the model (1), (5) to the time course SARS-CoV-2 infection of A549-ACE2 (purple) and Vero-E6 (green) cells. Parameters: A549-ACE2 (purple): , Vero-E6 (green): .
Statistically significant differences were detected also for the parameters determining the strength of the washing process and . We found statistically significant differences in the posterior distributions of and between A549-ACE2 and Vero-E6 cells with the washing process being more efficient for the Vero-E6 infection experiment (Fig. 2, Fig. 2f). Statistically significant differences were identified also between and within each cell infection experiment. The medians and 95% CRs for the posterior distributions of and are in Table 1. Correlations between and and other parameters are in Figures S.1 and S.2 in Supplementary Material.
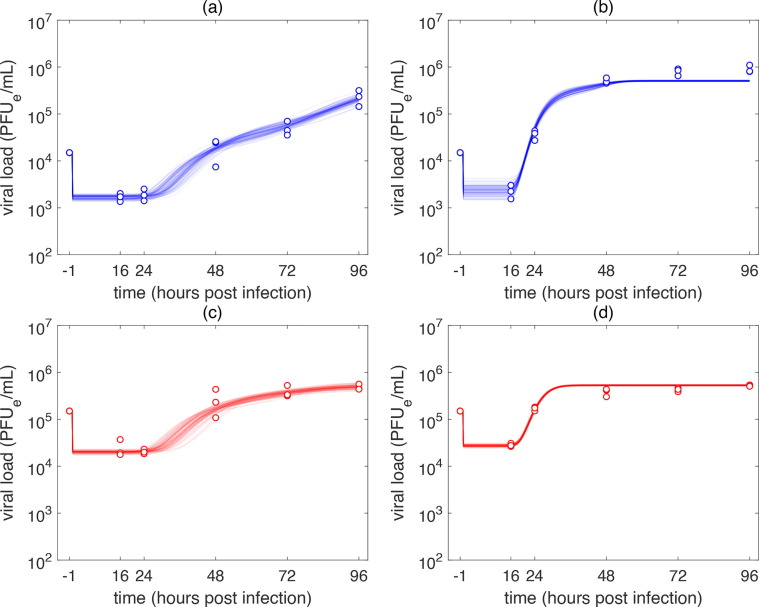
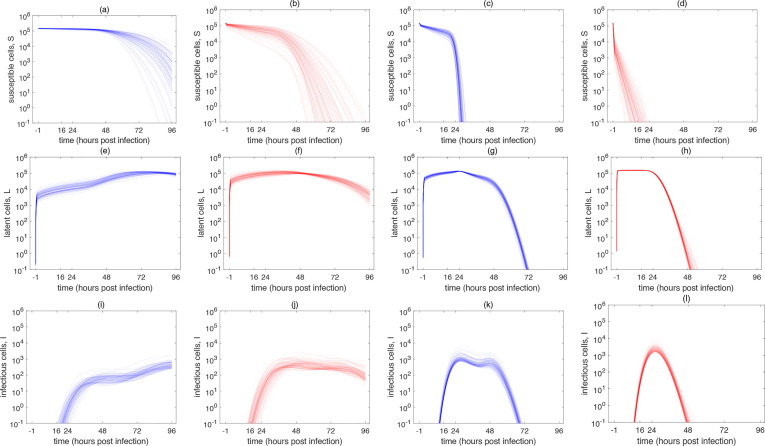
The time course kinetics of the SARS-CoV-2 infection of A549-ACE2 and Vero-E6 cells predicted by the model (1), (5) are depicted in Fig. 3 . The model reproduced our experimental data with high accuracy. The associated changes in the population of cells are captured in Fig. 4 . The population of SARS-CoV-2 infected Vero-E6 cells were predicted to stop viral particle production much earlier than A549-ACE2 cells. In the case of low MOI infection, virion production stopped before 72 h p.i. (Fig. 3b) and in the case of high MOI infection, all infectious Vero-E6 cells ceased virion production at around 48 h p.i. (Fig. 3d). Due to the susceptibility of Vero-E6 cells to early display of cythopatic effects, these dynamics accurately correspond with what is known about biology of SARS-CoV-2 infection of Vero-E6 cells (Ogando et al., 2020).
Fig. 3.
MCMC fits of the model (1), (5) to the time course SARS-CoV-2 infection of (a), (c) A549-ACE2 and (b), (d) Vero-E6 cells. Infection was performed at an MOI of (a), (b) 0.1 PFUe/cell (low MOI infection) and (c), (d) 1 PFUe/cell (high MOI infection). Parameters: (a), (c): , (b), .(d): .
Fig. 4.
Population dynamics of susceptible , latent and infectious cells yielded by the model (1), (5) and associated with the MCMC-accepted parameters for (a), (e), (i) low MOI infection of A549-ACE2 cells, (b), (f), (j) high MOI infection of A549-ACE2 cells, (c), (g), (k) low MOI infection of Vero-E6 cells and (d), (h), (l) high MOI infection of Vero-E6 cells. Parameters: (a), (b), (e), (f), (i), (j): , (c), (d), (g), (h), (k), (l): .
Supernatant sampling of the extracellular SARS-CoV-2 for RT-qPCR quantification does not improve fits of the mathematical model. As described in Materials and Methods, at each measured time, 10% (50 L out of 0.5 mL of the total volume) of the supernatant was extracted for virus quantification by RT-qPCR. This extracted volume was immediately replaced by fresh medium of the same volume. Although such a small sample can be considered negligible compared to the total extracellular viral load, we simulated the punctual extraction of the supernatant at measured times to demonstrate that sampling had only a negligible effect on the resulting viral dynamics. Briefly, we divided the infection time span into sub-intervals delimited by the measured time points t = 0 h, 12 h, 24 h, 48 h, 72 h and 96 h. For each sub-interval, we numerically solved the model (1), (5). Solutions and at each measured time were used as initial conditions for the numerical integration in the next sub-interval with the viral load reduced by the factor 0.1 (i.e. to ), as we assumed the sample to be well mixed.
We again fitted the Eqs. (1), (2), (3), (4), (5) to the experimental data using MCMC, including the extraction of the viral supernatant. We again assumed for A549-ACE2 cell infection and for Vero-E6 cell infection. Statistically significant differences in the posterior likelihood distributions were again determined by the bootstrap Mann–Whitney U-test. In the case of A549-ACE2 cell infection, fits were found to be statistically equivalent (P-value 0.05, Figure S.3a in Supplementary Material). In the case of Vero E6 cell infection, we found that the assumption of sampling worsened the fits of the model (Figure S.3b in Supplementary Material). Thus, we concluded that punctual sampling did not improve the model performance and relaxed this assumption from the model. The time course dynamics of SARS-CoV-2 infection of A549-ACE2 and Vero-E6 cells as predicted by the model (1), (5) assuming sampling is captured in Figure S.4 in Supplementary Material. This corroborates the results we obtained for in vitro Zika virus infection, where sampling of the virus had negligible effect on the overall virus dynamics (Bernhauerová et al., 2020).
Mathematical model with MOI-specific death rate of infectious cellsincreases accuracy of fits to Vero-E6 cell infection data. The model (1), (5) predicted lower SARS-CoV-2 burden in the late phase of the low MOI Vero-E6 infection and slightly higher SARS-CoV-2 burden in the middle phase of the high MOI Vero-E6 infection. This discrepancy in the model fits to the low and high MOI Vero-E6 infection data may be the result of a relatively large difference in the total viral yield at 96 h p.i. for low and high MOI infection (high MOI infection yield was almost 60% lower than that of low MOI infection yield) and an a priori assumption that the model parameters are independent of the initial viral inoculum. However, it has been shown that higher viral concentration can increase cellular cytotoxicity (Saccon et al., 2020, Chu et al., 2020). To asses whether the MOI-specific cellular cytotoxicity could explain and improve the fits to the low MOI Vero-E6 infection data, we introduced the MOI-specific death rates of infectious cells, and , into the mathematical model (Eqs. (A.1), (A.2), (A.3), (A.4), (A.5), (A.6), (A.7), (A.8), (A.9), (A.10) in Appendix A). Fitting the model (A.1), (A.2), (A.3), (A.4), (A.5), (A.6), (A.7), (A.8), (A.9), (A.10) to Vero-E6 infection data and incorporating MOI-specific death rates of infectious cells yielded significantly improved predictions (Fig. 5 ), particularly for the low MOI Vero-E6 infection (Fig. 5a). Statistical comparison of the models (1), (5) and (A.1), (A.10) was performed using the Bayes Information Criterion (Figure S.6, details are in Appendix B).
Fig. 5.
MCMC fits of the model (A.1), (A.2), (A.3), (A.4), (A.5), (A.6), (A.7), (A.8), (A.9), (A.10) to the time course infection of Vero-E6 cells. (a) Low MOI infection, (b) high MOI infection. Paramet.ers: .
Model (A.1), (A.2), (A.3), (A.4), (A.5), (A.6), (A.7), (A.8), (A.9), (A.10) predicted lower values of and higher values of (Fig. 6 , Table 2 ) compared to the model (1), (5). The median for was predicted to be 2.8 times lower, but the median for was only 1.04 times lower than the values predicted by the model (1), (5). Indeed, was statistically significantly lower (median equal to 4.2 h−1) than of (Figs. 6b and S.5 in Supplementary Material). This supports the hypothesis that high viral inoculum may induce increased cytotoxic responses in the host cells. All parameter values are in Table 2 and parameter correlations associated with the model (A.1), (A.2), (A.3), (A.4), (A.5), (A.6), (A.7), (A.8), (A.9), (A.10) are in Figure S.5 in Supplementary Material.
Fig. 6.
Posterior parameter distributions associated with MCMC fits of the models (1), (5) and (A.1), (A.10) to the time course infection of Vero-E6 cells. Paramet.ers: .
Table 2.
Parameter values of the model (A.1), (A.2), (A.3), (A.4), (A.5), (A.6), (A.7), (A.8), (A.9), (A.10) and their 95% credible regions.
| Parameter | Description | Units | Median |
95% Credible Region |
||
|---|---|---|---|---|---|---|
| A549-ACE2 | Vero-E6 | A549-ACE2 | Vero-E6 | |||
| infection rate | ()−1 | 2.0 | 8.65 | [7.07, 36.47] | [5.48, 13.3] | |
| infected cell death rate | 6.6 | 4.3 | [1.83, 9.73]] | [1.82, 5.88] | ||
| infected cell death rate | 6.12 | 7.52 | [1.40, 9.71] | [3.07, 9.94] | ||
| virus production rate | (cell)−1 | 19.55 | 24.35 | [3.96, 35.95] | [9.88, 31.85] | |
| duration of latent phase | 43.87 | 26.0 | [34.71, 52.05] | [24.88, 26.77] | ||
| washing strength (low MOI) | 2.16 | 2.01 | [1.96, 2.34] | [1.67, 2.44] | ||
| washing strength (high MOI) | 2.03 | 1.69 | [1.87, 2.17] | [1.59, 1.78] | ||
4. Discussion
Cellular tropism can generate variation in the time profile of the viral infection, including differences in the slope capturing the exponential phase of the viral growth, time of the infection peak and the slope capturing the post-peak dynamics. The origins of these differences are difficult to identify exclusively from experimental data. Mathematical models have helped to extract such information from time course viral load data and quantify the main determinants of the replication cycle of numerous viruses (Baccam et al., 2006, Zitzmann et al., 2020, Bernhauerová et al., 2020, Wei et al., 1995). In this paper, we performed experimental infections of two host cell lines with the SARS-CoV-2, namely the human lung carcinoma cell line stably expressing the ACE2 receptor, A549-ACE2, and Vero-E6 cell line derived from the green monkey kidney cell line, Vero. We fitted a simple mathematical model to our data in order to numerically evaluate the model parameters. This allowed us to reproduce our experimental observations and compare the viral replication cycles in both types of cells.
We found statistically significant differences between the rates at which the SARS-CoV-2 infects the two cell lines, , and the durations of the latent phase, , in the two cell lines. The SARS-CoV-2 infected Vero-E6 cells at a more than ten times higher rate than A549-ACE2 cells. Moreover, Vero-E6 cells remained in the latent phase for a significantly shorter time than A549-ACE2 cells. This result corresponds with the fact that Vero-E6 is a highly susceptible and permissive cell line and as such is widely used in virus research for plaque assays or rapid multiplication of virus in vaccine development (Ogando et al., 2020, Synowiec et al., 2021). On the other hand, since it is not a credible target for studying human respiratory virus replication in conditions comparable with natural infection or for in vitro drug screening (Milewska et al., 2020, Krammer, 2020, Hoffmann et al., 2020), A549 cells overexpressing ACE2 receptor constitute a more realistic in vitro model (Cagno, 2020, Meyer et al., 2020).
We found no statistically significant differences between the virus production rates, p, and the death rates of infectious cells, , of the two host cell types. Consequently, the total viral burst sizes, given as , produced by the two cell lines were comparable. This was corroborated by viral yields of the high MOI infections to have reached comparable levels at 97 h post infection in both culture types (magnitude of about viral particles per milliliter). This suggests that the observable differences in the viral growth in the two cell lines can be attributed to the differences in the infection rates, , and the duration of the latent phase , specific to each cell line.
Determining the possible relationship between the transmitted infectious virus load and the severity of the disease has been a challenge from the earliest days of the COVID-19 pandemic (Guallar et al., 2020, Karimzadeh et al., 2021). At the in vitro level, the role of the initial MOI as a critical factor influencing viral dynamics has been observed for various viruses, including SARS-CoV-2, and partially explained by mathematical modelling (Li and Handel, 2014, Fain and Dobrovolny, 2020). Our results suggest that SARS-CoV-2 dynamics may depend on the initial viral inoculum and that the particle production may be arrested more rapidly for high initial viral concentrations. On the other hand, we did not observe such a relation in the case of A549-ACE2 infection. In addition, immune responses to high and low viral inoculi may differ extensively. For example, human U937 cells infected at high MOI Sendai virus were shown to induce less subtypes of type I interferon than those infected at low MOI, which affected the viral dynamics (Zaritsky et al., 2015). This is not the case for Vero-E6 cells since they lack type I interferon encoding genes. Thus, MOI dependence of viral dynamics may as well be dependent on the cell type and more experimental work would be desirable to address these issues.
Results from our simulations suggested that by 48 h Vero-E6 cells are no longer producing new SARS-CoV-2 virions. For drug screening assays, the model suggests that Vero-E6 cells are not a suitable system to test drug efficacy if the drug is administered later than a day post-infection. A number of compounds have displayed antiviral activity against SARS-CoV-2 in vitro, such as remdesivir, lopinavir, chloroquine, umifenovir or berberine (Gordon et al., 2020, Pizzorno et al., 2020). We simulated the effects of antiviral administration on the SARS-CoV-2 growth by reducing the viral production rate p by a factor , where is the efficiency of the antiviral treatment and is determined by the administered drug concentration (details are in Appendix C). As expected, when administered at the time of virus inoculation, high drug concentrations caused viral load to decrease in both A549-ACE2 and Vero-E6 cells (Figures S.7 and S.8 in Supplementary Material). However, late introduction of the antiviral drug resulted in a considerably lower reduction of viral load in Vero-E6 cells as opposed to A549-ACE2 cells (Figures S.9 and S.10). Especially for high MOI Vero-E6 infection, administration of a drug later than 24 h p.i. did not have any effect on the SARS-CoV-2 viral load (Figures S.9d and S.10b). This would imply that after infection, the window of opportunity to administer the substance and still detect a sufficient enough effect is rather short. The results would therefore be skewed toward pretreatment regimens where the drug is given before the virus infection, even though in reality, patients are treated considerable time after they are infected. Therefore, it is crucial that for post-infection, repeated or long-term drug screening assays, appropriate cell lines are used that can withstand infections by viruses that induce strong apoptotic effects.
There are several limitations to our study. Fitting a mathematical model to the extracellular viral load data is a common practice when attempting to numerically characterize the virus replication cycle. However, it is important to note that we fitted the model only to the infectious virus equivalents derived from the standard curve capturing the relationship between the CT-values of serial dilutions of the SARS-CoV-2 RNA as determined by qRT-PCR and the number of PFUs of virus present in the sample. Since this technique allows to quantify the concentration of infectious virus only approximately, quantifications by plaque assay would allow to determine specific infectivity of the SARS-CoV-2 in both cell lines and the cell-specific production rates of infectious virus as well as accurately determine . Thus, more diverse data including temporal measurements of the momentary state of the viral and host populations would significantly improve the inference and quantification of the model parameters.
Predictions from this work are limited to the studied SARS-CoV-2 strain (BetaCoV/France/IDF0372/2020) and the cell lines used to measure SARS-CoV-2 replication kinetics, that is, Vero-E6 cells and A549 cells, stably expressing ACE2 receptor. However, viral growth profiles may be different for different SARS-CoV-2 isolates or may be affected by susceptibility and permissivity of various cellular systems (Chu et al., 2020). As a consequence, parameter estimates could vary rather significantly for infections by different variants. Further quantitative investigations are thus needed which would support the development of diverse therapeutics against SARS-CoV-2.
Since December 2019, SARS-CoV-2 has become the focus of extraordinary research activity. Quantitative understanding of the most fundamental stages of the SARS-CoV-2 replication cycle during in vitro infection is thus crucial, particularly in the initial stages of antiviral treatment development when various compounds inhibiting viral growth are tested in vitro in diverse cell types. Such studies can therefore assist in assessing the efficacy of the therapeutics and determining the appropriate doses and times to administer the compound.
Funding
V.B. acknowledges the project number 4EU+/20/F1/9 granted by Charles University. B.L. acknowledges funding by the Priority Research Area DigiWorld under the Strategic Programme Excellence Initiative at the Jagiellonian University. M.V. and V.V.R. acknowledge that the work at Institut Pasteur was funded by Laboratoire d’Excellence grant ANR-10-LABX-62-IBEID and the URGENCE COVID-19 Institut Pasteur fundraising campaign.
CRediT authorship contribution statement
Veronika Bernhauerová: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Bartek Lisowski: Conceptualization, Formal analysis, Investigation, Methodology, Writing - original draft, Writing - review & editing. Veronica V. Rezelj: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing - original draft, Writing - review & editing. Marco Vignuzzi: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank two anonymous reviewers for their helpful comments and suggestions which improved the exposition of the manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jtbi.2021.110895.
Appendix A. Mathematical model with MOI-specific death rates of infectious Vero-E6 cells, and
The mathematical model with MOI-specific death rates of infectious Vero-E6 cells is as follows:
| (A.1) |
| (A.2) |
| (A.3) |
| (A.4) |
| (A.5) |
| (A.6) |
| (A.7) |
| (A.8) |
| (A.9) |
| (A.10) |
The parameters specific to the low and high MOI infections are highlighted in bold.
Appendix B. Bayesian Information Criterion (BIC).
Bayesian Information Criterion (BIC)
| (A.11) |
was used to determine which of the two models, (1), (5) and (A.1), (A.10), is more suitable to describe the experimental data. BIC was calculated for each model and each MCMC-associated log-likelihood and a penalty term , where k is the number of parameters in the candidate model and n is the number of experimental observations. The total number of observations were . The number of free parameters in the models (1), (5), (A.1)–(A.10) were and , respectively.
Appendix C. Antiviral therapy
We assume that the viral production rate p in the Eqs. (5), (A.5), (A.10) is reduced by the inhibition factor , where is modelled by the Hill function
| (A.12) |
where is the maximal inhibition efficacy, C is the administered drug concentration, is the drug concentration at which half of the maximal inhibition efficacy is achieved and N is the Hill coefficient dictating the shape of the curve (A.12) such that for , Eq. (A.12) yields a concave Michaelis–Menten response curve and for , a sigmoid shaped Hill curve is achieved. We assume that for large values of drug concentrations, that is, , inhibition is perfect and set . The half-inhibition constant may vary and depends on the type of antiviral drug. Here, we set as this value was determined by (Pizzorno et al., 2020) from in vitro inhibition assays for remdesivir. Since the value of Hill coefficient was not provided in (Pizzorno et al., 2020), we assume the Michaelis–Menten relationship for (A.12) and set . Changes in the viral dynamics as a result of varying drug concentration C = 0.1, 0.2, 0.5, 1, 2, 5, 10 and 20 are in Figure S.7 for the model (1), (5) and in Figure S.8 for the model (A.12). Changes in the viral dynamics as a result of varying time of drug administration, where the drug was administered at 0 h, 24 h, 36 h and 48 h p.i. are in Figure S.9 for the model (1), (5) and in Figure S.10 for the model (A.1), (A.2), (A.3), (A.4), (A.5), (A.6), (A.7), (A.8), (A.9), (A.10).
Supplementary data
The following are the Supplementary data to this article:
References
- Anderson D., Watson R. On the spread of a disease with gamma distributed latent and infectious periods. Biometrika. 1980;67(1):191–198. doi: 10.2307/2335333. [DOI] [Google Scholar]
- Baccam P., Beauchemin C., Macken C.A., Hayden F.G., Perelson A.S. Kinetics of influenza A virus infection in humans. J. Virol. 2006;80(15):7590–7599. doi: 10.1128/JVI.01623-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F., Huff K.E., Allen L.J. The effect of delay in viral production in within-host models during early infection. J. Biol. Dyn. 2019;13(1):47–73. doi: 10.1080/17513758.2018.1498984. [DOI] [PubMed] [Google Scholar]
- Beauchemin C.A.A., Miura T., Iwami S. Duration of SHIV production by infected cells is not exponentially distributed: Implications for estimates of infection parameters and antiviral efficacy. Sci. Rep. 2017;7(1):42765. doi: 10.1038/srep42765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin C.A.A., Kim Y.-I., Yu Q., Ciaramella G., DeVincenzo J.P. Uncovering critical properties of the human respiratory syncytial virus by combining in vitro assays and in silico analyses. PLOS ONE. 2019;14(4):1–25. doi: 10.1371/journal.pone.0214708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.-D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of COVID-19 – final report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhauerová V., Rezelj V.V., Vignuzzi M. Modelling degradation and replication kinetics of the Zika virus in vitro infection. Viruses. 2020;12(5):1–17. doi: 10.3390/v12050547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagno V. SARS-CoV-2 cellular tropism. The Lancet Microbe. 2020;1(1):e2–e3. doi: 10.1016/s2666-5247(20)30008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F.-W., Yuen T.T.-T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C.-Y., Tsang J.O.-L., Huang X., Chai Y., Yang D., Hou Y., Chik K.K.-H., Zhang X., Fung A.Y.-F., Tsoi H.-W., Cai J.-P., Chan W.-M., Ip J.D., Chu A.W.-H., Zhou J., Lung D.C., Kok K.-H., To K.K.-W., Tsang O.T.-Y., Chan K.-H., Yuen K.-Y. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. The Lancet Microbe. 2020;1(1):e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens B.L., Koo J.R., Lim J.T., Park M., Quaye S., Sun H., Sun Y., Pung R., Wilder-Smith A., Chai L.Y.A., Lee V.J., Cook A.R. Modelling lockdown and exit strategies for COVID-19 in Singapore. The Lancet Regional Health - Western Pacific. 2020;1(1) doi: 10.1016/j.lanwpc.2020.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B. Efron, R.J. Tibshirani, An Introduction to the Bootstrap, 1st Edition, Vol. 57, Chapman and Hall/CRC, Boca Raton Lindon New York Washington, D.C., 1993.
- Endo A., Abbott S., Kucharski A.J., Funk S. Estimating the overdispersion in covid-19 transmission using outbreak sizes outside china. Wellcome Open Research. 2020;5(67):1–18. doi: 10.12688/wellcomeopenres.15842.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain B., Dobrovolny H.M. Initial inoculum and the severity of COVID-19: A mathematical modeling study of the dose-response of SARS-CoV-2 infections. Epidemiologia. 2020;1(1):5–15. doi: 10.3390/epidemiologia1010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L. Ferretti, C. Wymant, M. Kendall, L. Zhao, A. Nurtay, L. Abeler-Dörner, M. Parker, D. Bonsall, C. Fraser, Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing, Science 368 (6491) (2020) 0–8. doi:10.1126/science.abb6936. [DOI] [PMC free article] [PubMed]
- González-Parra G., Dobrovolny H.M. The rate of viral transfer between upper and lower respiratory tracts determines RSV illness duration. J. Math. Biol. 2019;79(2):467–483. doi: 10.1007/s00285-019-01364-1. [DOI] [PubMed] [Google Scholar]
- Goodman J., Weare J. Ensemble samplers with affine invariance. Commun. Appl. Math. Comput. Sci. 2010;5(1):65–80. doi: 10.2140/camcos.2010.5.65. [DOI] [Google Scholar]
- D.E. Gordon, G.M. Jang, M. Bouhaddou, J. Xu, K. Obernier, K.M. White, M.J. O’Meara, V.V. Rezelj, J.Z. Guo, D.L. Swaney, T.A. Tummino, R. Hüttenhain, R.M. Kaake, A.L. Richards, B. Tutuncuoglu, H. Foussard, J. Batra, K. Haas, M. Modak, M. Kim, P. Haas, B.J. Polacco, H. Braberg, J.M. Fabius, M. Eckhardt, M. Soucheray, M.J. Bennett, M. Cakir, M.J. McGregor, Q. Li, B. Meyer, F. Roesch, T. Vallet, A. Mac Kain, L. Miorin, E. Moreno, Z.Z.C. Naing, Y. Zhou, S. Peng, Y. Shi, Z. Zhang, W. Shen, I.T. Kirby, J.E. Melnyk, J.S. Chorba, K. Lou, S.A. Dai, I. Barrio-Hernandez, D. Memon, C. Hernandez-Armenta, J. Lyu, C.J.P. Mathy, T. Perica, K.B. Pilla, S.J. Ganesan, D.J. Saltzberg, R. Rakesh, X. Liu, S.B. Rosenthal, L. Calviello, S. Venkataramanan, J. Liboy-Lugo, Y. Lin, X.-P. Huang, Y. Liu, S.A. Wankowicz, M. Bohn, M. Safari, F.S. Ugur, C. Koh, N.S. Savar, Q.D. Tran, D. Shengjuler, S.J. Fletcher, M.C. O’Neal, Y. Cai, J.C.J. Chang, D.J. Broadhurst, S. Klippsten, P.P. Sharp, N.A. Wenzell, D. Kuzuoglu-Ozturk, H.-Y. Wang, R. Trenker, J.M. Young, D.A. Cavero, J. Hiatt, T.L. Roth, U. Rathore, A. Subramanian, J. Noack, M. Hubert, R.M. Stroud, A.D. Frankel, O.S. Rosenberg, K.A. Verba, D.A. Agard, M. Ott, M. Emerman, N. Jura, M. von Zastrow, E. Verdin, A. Ashworth, O. Schwartz, C. D’Enfert, S. Mukherjee, M. Jacobson, H.S. Malik, D.G. Fujimori, T. Ideker, C.S. Craik, S.N. Floor, J.S. Fraser, J.D. Gross, A. Sali, B.L. Roth, D. Ruggero, J. Taunton, T. Kortemme, P. Beltrao, M. Vignuzzi, A. García-Sastre, K.M. Shokat, B.K. Shoichet, N.J. Krogan, A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature 583 (7816) (2020) 459–468. doi:10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed]
- Guallar M.P., Meiriño R., Donat-Vargas C., Corral O., Jouvé N., Soriano V. Inoculum at the time of SARS-CoV-2 exposure and risk of disease severity. Int. J. Infectious Diseases. 2020;97(1):290–292. doi: 10.1016/j.ijid.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldt F.S., Frensing T., Pflugmacher A., Gröpler R., Peschel B., Reichl U. Multiscale modeling of influenza A virus infection supports the development of direct-acting antivirals. PLoS Comput. Biol. 2013;9(11):1–13. doi: 10.1371/journal.pcbi.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Mösbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Krüger N., Gassen N.C., Müller M.A., Drosten C., Pöhlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585(7826):588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado P.J., Kirosingh A.S. Generalizations of the ‘Linear Chain Trick’: incorporating more flexible dwell time distributions into mean field ODE models. J. Math. Biol. 2019;79(5):1831–1883. doi: 10.1007/s00285-019-01412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizoe Y., Nakaoka S., Beauchemin C.A.A., Morita S., Mori H., Igarashi T., Aihara K., Miura T., Iwami S. A method to determine the duration of the eclipse phase for in vitro infection with a highly pathogenic SHIV strain. Sci. Rep. 2015;5(1):10371. doi: 10.1038/srep10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimzadeh S., Bhopal R., Huy N.T. Review of infective dose, routes of transmission, and outcome of COVID-19 caused by the SARS-COV-2: comparison with other respiratory viruses. Epidemiol. Infect. 2021;149(e96):1–8. doi: 10.1017/S0950268821000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Krylova O., Earn D.J.D. Effects of the infectious period distribution on predicted transitions in childhood disease dynamics. J. R. Soc. Interface. 2013;10(84):20130098. doi: 10.1098/rsif.2013.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Handel A. Modeling inoculum dose dependent patterns of acute virus infections. J. Theor. Biol. 2014;347(1):63–73. doi: 10.1016/j.jtbi.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S., Lau E.H., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/nejmoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., Tukhvatulin A.I., Zubkova O.V., Dzharullaeva A.S., Kovyrshina A.V., Lubenets N.L., Grousova D.M., Erokhova A.S., Botikov A.G., Izhaeva F.M., Popova O., Ozharovskaya T.A., Esmagambetov I.B., Favorskaya I.A., Zrelkin D.I., Voronina D.V., Shcherbinin D.N., Semikhin A.S., Simakova Y.V., Tokarskaya E.A., Egorova D.A., Shmarov M.M., Nikitenko N.A., Gushchin V.A., Smolyarchuk E.A., Zyryanov S.K., Borisevich S.V., Naroditsky B.S., Gintsburg A.L. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. The Lancet. 2021;397(10275):671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B. Meyer, J. Chiaravalli, S. Gellenoncourt, P. Brownridge, D.P. Bryne, L.A. Daly, A. Grauslys, M. Walter, F. Agou, L.A. Chakrabarti, C.S. Craik, C.E. Eyers, P.A. Eyers, Y. Gambin, A.R. Jones, E. Sierecki, E. Verdin, M. Vignuzzi, E. Emmott, Characterisation of protease activity during SARS-CoV-2 infection identifies novel viral cleavage sites and cellular targets for drug repurposing, bioRxiv (2020) 1–39 doi:10.1101/2020.09.16.297945.
- Milewska A., Kula-Pacurar A., Wadas J., Suder A., Szczepanski A., Dabrowska A., Owczarek K., Marcello A., Ochman M., Stacel T., Rajfur Z., Sanak M., Labaj P., Branicki W., Pyrc K., Sandri-Goldin R.M. Replication of severe acute respiratory syndrome coronavirus 2 in human respiratory epithelium. J. Virol. 2020;94(15):e00957–20. doi: 10.1128/JVI.00957-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J.A., Groß R., Conzelmann C., Krüger J., Merle U., Steinhart J., Weil T., Koepke L., Bozzo C.P., Read C., Fois G., Eiseler T., Gehrmann J., van Vuuren J., Wessbecher I.M., Frick M., Costa I.G., Breunig M., Grüner B., Peters L., Schuster M., Liebau S., Seufferlein T., Stenger S., Stenzinger A., MacDonald P.E., Kirchhoff F., Sparrer K.M.J., Walther P., Lickert H., Barth T.F.E., Wagner M., Münch J., Heller S., Kleger A. SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. Nature Metabolism. 2021;3(1):149–165. doi: 10.1038/s42255-021-00347-1. [DOI] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Sahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W., van der Meer Y., Caly L., Druce J., de Vries J.J., Kikkert M., Barcena M., Sidorov I., Snijder E.J. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101(9):925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E.G., Pinilla L.T., Holder B.P., Abed Y., Boivin G., Beauchemin C.A.A. Impact of the H275Y and I223V mutations in the neuraminidase of the 2009 pandemic influenza virus in vitro and evaluating experimental reproducibility. PLOS ONE. 2015;10(5):1–24. doi: 10.1371/journal.pone.0126115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla L.T., Holder B.P., Abed Y., Boivin G., Beauchemin C.A.A. The H275Y neuraminidase mutation of the pandemic A/H1N1 influenza virus lengthens the eclipse phase and reduces viral output of infected cells, potentially compromising fitness in ferrets. J. Virol. 2012;86(19):10651–10660. doi: 10.1128/JVI.07244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Dubois J., Julien T., Traversier A., Dulière V., Brun P., Lina B., Rosa-Calatrava M., Terrier O. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antiviral Res. 2020;181(1) doi: 10.1016/j.antiviral.2020.104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- F.P. Polack, S.J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J.L. Perez, G. Pérez Marc, E.D. Moreira, C. Zerbini, R. Bailey, K.A. Swanson, S. Roychoudhury, K. Koury, P. Li, W.V. Kalina, D. Cooper, R.W. Frenck, L.L. Hammitt, Ö. Türeci, H. Nell, A. Schaefer, S. Ünal, D.B. Tresnan, S. Mather, P.R. Dormitzer, U. Sahin, K.U. Jansen, W.C. Gruber, Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine, New England Journal of Medicine 383 (27) (2020) 2603–2615. doi:10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed]
- E. Saccon, S. Krishnan, B.S. Vinhas, S.N. Byrareddy, A. Mirazimi, U. Neogi, S. Gupta, Replication dynamics and cytotoxicity of SARS-CoV-2 Swedish isolate in commonly used laboratory cell lines, bioRxiv (2020) 1–11 doi:10.1101/2020.08.28.271684.
- Schelker M., Mair C.M., Jolmes F., Welke R.W., Klipp E., Herrmann A., Flöttmann M., Sieben C. Viral RNA degradation and diffusion act as a bottleneck for the influenza A virus infection Efficiency. PLoS Comput. Biol. 2016;12(10):1–23. doi: 10.1371/journal.pcbi.1005075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid H., Dobrovolny H.M. An approximate solution of the interferon-dependent viral kinetics model of influenza. J. Theor. Biol. 2020;498(1) doi: 10.1016/j.jtbi.2020.110266. [DOI] [PubMed] [Google Scholar]
- Smith A.M., Ribeiro R.M. Modeling the viral dynamics of influenza a virus infection. Crit. Rev. Immunol. 2010;30(3):291–298. doi: 10.1615/critrevimmunol.v30.i3.60. [DOI] [PubMed] [Google Scholar]
- Synowiec A., Szczepański A., Barreto-Duran E., Lie L.K., Pyrc K. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a systemic infection. Clin. Microbiol. Rev. 2021;34(2):e00133–20. doi: 10.1128/CMR.00133-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21(6):382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M. Voysey, S.A.C. Clemens, S.A. Madhi, L.Y. Weckx, P.M. Folegatti, P.K. Aley, B. Angus, V.L. Baillie, S.L. Barnabas, Q.E. Bhorat, S. Bibi, C. Briner, P. Cicconi, A.M. Collins, R. Colin-Jones, C.L. Cutland, T.C. Darton, K. Dheda, C.J.A. Duncan, K.R.W. Emary, K.J. Ewer, L. Fairlie, S.N. Faust, S. Feng, D.M. Ferreira, A. Finn, A.L. Goodman, C.M. Green, C.A. Green, P.T. Heath, C. Hill, H. Hill, I. Hirsch, S.H.C. Hodgson, A. Izu, S. Jackson, D. Jenkin, C.C.D. Joe, S. Kerridge, A. Koen, G. Kwatra, R. Lazarus, A.M. Lawrie, A. Lelliott, V. Libri, P.J. Lillie, R. Mallory, A.V.A. Mendes, E.P. Milan, A.M. Minassian, A. McGregor, H. Morrison, Y.F. Mujadidi, A. Nana, P.J. O’Reilly, S.D. Padayachee, A. Pittella, E. Plested, K.M. Pollock, M.N. Ramasamy, S. Rhead, A.V. Schwarzbold, N. Singh, A. Smith, R. Song, M.D. Snape, E. Sprinz, R.K. Sutherland, R. Tarrant, E.C. Thomson, M.E. Török, M. Toshner, D.P.J. Turner, J. Vekemans, T.L. Villafana, M.E.E. Watson, C.J. Williams, A.D. Douglas, A.V.S. Hill, T. Lambe, S.C. Gilbert, A.J. Pollard, M. Aban, F. Abayomi, K. Abeyskera, J. Aboagye, M. Adam, K. Adams, J. Adamson, Y.A. Adelaja, G. Adewetan, S. Adlou, K. Ahmed, Y. Akhalwaya, S. Akhalwaya, A. Alcock, A. Ali, E.R. Allen, L. Allen, T.C.D.S.C. Almeida, M.P.S. Alves, F. Amorim, F. Andritsou, R. Anslow, M. Appleby, E.H. Arbe-Barnes, M.P. Ariaans, B. Arns, L. Arruda, P. Azi, L. Azi, G. Babbage, C. Bailey, K.F. Baker, M. Baker, N. Baker, P. Baker, L. Baldwin, I. Baleanu, D. Bandeira, A. Bara, M.A.S. Barbosa, D. Barker, G.D. Barlow, E. Barnes, A.S. Barr, J.R. Barrett, J. Barrett, L. Bates, A. Batten, K. Beadon, E. Beales, R. Beckley, S. Belij-Rammerstorfer, J. Bell, D. Bellamy, N. Bellei, S. Belton, A. Berg, L. Bermejo, E. Berrie, L. Berry, D. Berzenyi, A. Beveridge, K.R. Bewley, H. Bexhell, S. Bhikha, A.E. Bhorat, Z.E. Bhorat, E. Bijker, G. Birch, S. Birch, A. Bird, O. Bird, K. Bisnauthsing, M. Bittaye, K. Blackstone, L. Blackwell, H. Bletchly, C.L. Blundell, S.R. Blundell, P. Bodalia, B.C. Boettger, E. Bolam, E. Boland, D. Bormans, N. Borthwick, F. Bowring, A. Boyd, P. Bradley, T. Brenner, P. Brown, C. Brown, C. Brown-O’Sullivan, S. Bruce, E. Brunt, R. Buchan, W. Budd, Y.A. Bulbulia, M. Bull, J. Burbage, H. Burhan, A. Burn, K.R. Buttigieg, N. Byard, I. Cabera Puig, G. Calderon, A. Calvert, S. Camara, M. Cao, F. Cappuccini, J.R. Cardoso, M. Carr, M.W. Carroll, A. Carson-Stevens, Y. d. M. Carvalho, J.A.M. Carvalho, H.R. Casey, P. Cashen, T. Castro, L.C. Castro, K. Cathie, A. Cavey, J. Cerbino-Neto, J. Chadwick, D. Chapman, S. Charlton, I. Chelysheva, O. Chester, S. Chita, J.-S. Cho, L. Cifuentes, E. Clark, M. Clark, A. Clarke, E.A. Clutterbuck, S.L.K. Collins, C.P. Conlon, S. Connarty, N. Coombes, C. Cooper, R. Cooper, L. Cornelissen, T. Corrah, C. Cosgrove, T. Cox, W.E.M. Crocker, S. Crosbie, L. Cullen, D. Cullen, D.R.M.F. Cunha, C. Cunningham, F.C. Cuthbertson, S.N.F. Da Guarda, L.P. da Silva, B.E. Damratoski, Z. Danos, M.T.D.C. Dantas, P. Darroch, M.S. Datoo, C. Datta, M. Davids, S.L. Davies, H. Davies, E. Davis, J. Davis, J. Davis, M.M.D. De Nobrega, L.M. De Oliveira Kalid, D. Dearlove, T. Demissie, A. Desai, S. Di Marco, C. Di Maso, M.I.S. Dinelli, T. Dinesh, C. Docksey, C. Dold, T. Dong, F.R. Donnellan, T. Dos Santos, T.G. dos Santos, E.P. Dos Santos, N. Douglas, C. Downing, J. Drake, R. Drake-Brockman, K. Driver, R. Drury, S.J. Dunachie, B.S. Durham, L. Dutra, N.J.W. Easom, S. van Eck, M. Edwards, N.J. Edwards, O.M. El Muhanna, S.C. Elias, M. Elmore, M. English, A. Esmail, Y.M. Essack, E. Farmer, M. Farooq, M. Farrar, L. Farrugia, B. Faulkner, S. Fedosyuk, S. Felle, S. Feng, C. Ferreira Da Silva, S. Field, R. Fisher, A. Flaxman, J. Fletcher, H. Fofie, H. Fok, K.J. Ford, J. Fowler, P.H.A. Fraiman, E. Francis, M.M. Franco, J. Frater, M.S.M. Freire, S.H. Fry, S. Fudge, J. Furze, M. Fuskova, P. Galian-Rubio, E. Galiza, H. Garlant, M. Gavrila, A. Geddes, K.A. Gibbons, C. Gilbride, H. Gill, S. Glynn, K. Godwin, K. Gokani, U.C. Goldoni, M. Goncalves, I.G.S. Gonzalez, J. Goodwin, A. Goondiwala, K. Gordon-Quayle, G. Gorini, J. Grab, L. Gracie, M. Greenland, N. Greenwood, J. Greffrath, M.M. Groenewald, L. Grossi, G. Gupta, M. Hackett, B. Hallis, M. Hamaluba, E. Hamilton, J. Hamlyn, D. Hammersley, A.T. Hanrath, B. Hanumunthadu, S.A. Harris, C. Harris, T. Harris, T.D. Harrison, D. Harrison, T.C. Hart, B. Hartnell, S. Hassan, J. Haughney, S. Hawkins, J. Hay, I. Head, J. Henry, M. Hermosin Herrera, D.B. Hettle, J. Hill, G. Hodges, E. Horne, M.M. Hou, C. Houlihan, E. Howe, N. Howell, J. Humphreys, H.E. Humphries, K. Hurley, C. Huson, A. Hyder-Wright, C. Hyams, S. Ikram, A. Ishwarbhai, M. Ivan, P. Iveson, V. Iyer, F. Jackson, J. De Jager, S. Jaumdally, H. Jeffers, N. Jesudason, B. Jones, K. Jones, E. Jones, C. Jones, M.R. Jorge, A. Jose, A. Joshi, E.A.M.S. Júnior, J. Kadziola, R. Kailath, F. Kana, K. Karampatsas, M. Kasanyinga, J. Keen, E.J. Kelly, D.M. Kelly, D. Kelly, S. Kelly, D. Kerr, R. d. Á. Kfouri, L. Khan, B. Khozoee, S. Kidd, A. Killen, J. Kinch, P. Kinch, L.D.W. King, T.B. King, L. Kingham, P. Klenerman, F. Knapper, J.C. Knight, D. Knott, S. Koleva, M. Lang, G. Lang, C.W. Larkworthy, J.P.J. Larwood, R. Law, E.M. Lazarus, A. Leach, E.A. Lees, N.-M. Lemm, A. Lessa, S. Leung, Y. Li, A.M. Lias, K. Liatsikos, A. Linder, S. Lipworth, S. Liu, X. Liu, A. Lloyd, S. Lloyd, L. Loew, R. Lopez Ramon, L. Lora, V. Lowthorpe, K. Luz, J.C. MacDonald, G. MacGregor, M. Madhavan, D.O. Mainwaring, E. Makambwa, R. Makinson, M. Malahleha, R. Malamatsho, G. Mallett, K. Mansatta, T. Maoko, K. Mapetla, N.G. Marchevsky, S. Marinou, E. Marlow, G.N. Marques, P. Marriott, R.P. Marshall, J.L. Marshall, F.J. Martins, M. Masenya, M. Masilela, S.K. Masters, M. Mathew, H. Matlebjane, K. Matshidiso, O. Mazur, A. Mazzella, H. McCaughan, J. McEwan, J. McGlashan, L. McInroy, Z. McIntyre, D. McLenaghan, N. McRobert, S. McSwiggan, C. Megson, S. Mehdipour, W. Meijs, R.N. Á. Mendonça, A.J. Mentzer, N. Mirtorabi, C. Mitton, S. Mnyakeni, F. Moghaddas, K. Molapo, M. Moloi, M. Moore, M.I. Moraes-Pinto, M. Moran, E. Morey, R. Morgans, S. Morris, S. Morris, H.C. Morris, F. Morselli, G. Morshead, R. Morter, L. Mottal, A. Moultrie, N. Moya, M. Mpelembue, S. Msomi, Y. Mugodi, E. Mukhopadhyay, J. Muller, A. Munro, C. Munro, S. Murphy, P. Mweu, C.H. Myasaki, G. Naik, K. Naker, E. Nastouli, A. Nazir, B. Ndlovu, F. Neffa, C. Njenga, H. Noal, A. Noé, G. Novaes, F.L. Nugent, G. Nunes, K. O’Brien, D. O’Connor, M. Odam, S. Oelofse, B. Oguti, V. Olchawski, N.J. Oldfield, M.G. Oliveira, C. Oliveira, A. Oosthuizen, P. O’Reilly, P. Osborne, D.R.J. Owen, L. Owen, D. Owens, N. Owino, M. Pacurar, B.V.B. Paiva, E.M.F. Palhares, S. Palmer, S. Parkinson, H.M.R.T. Parracho, K. Parsons, D. Patel, B. Patel, F. Patel, K. Patel, M. Patrick-Smith, R.O. Payne, Y. Peng, E.J. Penn, A. Pennington, M.P. Peralta Alvarez, J. Perring, N. Perry, R. Perumal, S. Petkar, T. Philip, D.J. Phillips, J. Phillips, M.K. Phohu, L. Pickup, S. Pieterse, J. Piper, D. Pipini, M. Plank, J. Du Plessis, S. Pollard, J. Pooley, A. Pooran, I. Poulton, C. Powers, F.B. Presa, D.A. Price, V. Price, M. Primeira, P.C. Proud, S. Provstgaard-Morys, S. Pueschel, D. Pulido, S. Quaid, R. Rabara, A. Radford, K. Radia, D. Rajapaska, T. Rajeswaran, A.S.F. Ramos, F. Ramos Lopez, T. Rampling, J. Rand, H. Ratcliffe, T. Rawlinson, D. Rea, B. Rees, J. Reiné, M. Resuello-Dauti, E. Reyes Pabon, C.M. Ribiero, M. Ricamara, A. Richter, N. Ritchie, A.J. Ritchie, A.J. Robbins, H. Roberts, R.E. Robinson, H. Robinson, T.T. Rocchetti, B.P. Rocha, S. Roche, C. Rollier, L. Rose, A.L. Ross Russell, L. Rossouw, S. Royal, I. Rudiansyah, S. Ruiz, S. Saich, C. Sala, J. Sale, A.M. Salman, N. Salvador, S. Salvador, M. Sampaio, A.D. Samson, A. Sanchez-Gonzalez, H. Sanders, K. Sanders, E. Santos, M.F.S. Santos Guerra, I. Satti, J.E. Saunders, C. Saunders, A. Sayed, I. Schim van der Loeff, A.B. Schmid, E. Schofield, G. Screaton, S. Seddiqi, R.R. Segireddy, R. Senger, S. Serrano, R. Shah, I. Shaik, H.E. Sharpe, K. Sharrocks, R. Shaw, A. Shea, A. Shepherd, J.G. Shepherd, F. Shiham, E. Sidhom, S.E. Silk, A.C. da Silva Moraes, G. Silva-Junior, L. Silva-Reyes, A.D. Silveira, M.B.V. Silveira, J. Sinha, D.T. Skelly, D.C. Smith, N. Smith, H.E. Smith, D.J. Smith, C.C. Smith, A. Soares, T. Soares, C. Solórzano, G.L. Sorio, K. Sorley, T. Sosa-Rodriguez, C.M. Souza, B.S.D.F. Souza, A.R. Souza, A.J. Spencer, F. Spina, L. Spoors, L. Stafford, I. Stamford, I. Starinskij, R. Stein, J. Steven, L. Stockdale, L.V. Stockwell, L.H. Strickland, A.C. Stuart, A. Sturdy, N. Sutton, A. Szigeti, A. Tahiri-Alaoui, R. Tanner, C. Taoushanis, A.W. Tarr, K. Taylor, U. Taylor, I.J. Taylor, J. Taylor, R. te Water Naude, Y. Themistocleous, A. Themistocleous, M. Thomas, K. Thomas, T.M. Thomas, A. Thombrayil, F. Thompson, A. Thompson, K. Thompson, A. Thompson, J. Thomson, V. Thornton-Jones, P.J. Tighe, L.A. Tinoco, G. Tiongson, B. Tladinyane, M. Tomasicchio, A. Tomic, S. Tonks, J. Towner, N. Tran, J. Tree, G. Trillana, C. Trinham, R. Trivett, A. Truby, B.L. Tsheko, A. Turabi, R. Turner, C. Turner, M. Ulaszewska, B.R. Underwood, R. Varughese, D. Verbart, M. Verheul, I. Vichos, T. Vieira, C.S. Waddington, L. Walker, E. Wallis, M. Wand, D. Warbick, T. Wardell, G. Warimwe, S.C. Warren, B. Watkins, E. Watson, S. Webb, A. Webb-Bridges, A. Webster, J. Welch, J. Wells, A. West, C. White, R. White, P. Williams, R.L. Williams, R. Winslow, M. Woodyer, A.T. Worth, D. Wright, M. Wroblewska, A. Yao, R. Zimmer, D. Zizi, P. Zuidewind, Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK, The Lancet 397 (10269) (2021) 99–111. doi:10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed]
- Wei X., Ghosh S.K., Taylor M.E., Johnson V.A., Emini E.A., Deutsch P., Lifson J.D., Bonhoeffer S., Nowak M.A., Hahn B.H., Saag M.S., Shaw G.M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373(6510):117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- Wells H.L., Letko M., Lasso G., Ssebide B., Nziza J., Byarugaba D.K., Navarrete-Macias I., Liang E., Cranfield M., Han B.A., Tingley M.W., Diuk-Wasser M., Goldstein T., Johnson C.K., Mazet J.A.K., Chandran K., Munster V.J., Gilardi K., Anthony S.J. The evolutionary history of ACE2 usage within the coronavirus subgenus Sarbecovirus. Virus Evolution. 2021;7(1):1–22. doi: 10.1093/ve/veab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wethington D., Harder O., Uppulury K., Stewart W.C., Chen P., King T., Reynolds S.D., Perelson A.S., Peeples M.E., Niewiesk S., Das J. Mathematical modelling identifies the role of adaptive immunity as a key controller of respiratory syncytial virus in cotton rats. J. R. Soc. Interface. 2019;16(160):1–14. doi: 10.1098/rsif.2019.0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Zaritsky L.A., Bedsaul J.R., Zoon K.C. Virus multiplicity of infection affects type I interferon subtype induction profiles and interferon-stimulated genes. J. Virol. 2015;89(22):11534–11548. doi: 10.1128/jvi.01727-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitzmann C., Schmid B., Ruggieri A., Perelson A.S., Binder M., Bartenschlager R., Kaderali L. A coupled mathematical model of the intracellular replication of Dengue virus and the host cell immune rsponse to infection. Front. Microbiol. 2020;11(April):725. doi: 10.3389/fmicb.2020.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.