Abstract
Objective:
The classical allergic march model posits that atopy begins in infancy with atopic dermatitis and progresses to asthma and allergic rhinitis in a subset of individuals. The growing prevalence and severity of allergic diseases has prompted renewed interest in refining this model. This review outlines epidemiologic evidence for the existence of allergic march trajectories (distinct paths of atopy development in individuals), reviews the roles that genetics, environment, and disease endotypes play in determining trajectory outcomes, and discusses the clinical utility of the trajectory model.
Data Sources:
PubMed search of English-language articles and reviews without date limits pertaining to the epidemiology, genetics, and immunologic mechanisms of allergic march trajectories and disease endotypes.
Study Selections:
Studies and reviews were selected based on their high quality and direct relevance to the review topic.
Results:
Recent work in the field has shown that IgE-mediated food allergy and eosinophilic esophagitis are components of the allergic march. Further, the field is acknowledging that variability exists in the number and sequence of allergic manifestations that individuals develop. These allergic march pathways, or trajectories, are influenced by genetic, environmental, and psychosocial factors that are incompletely understood.
Conclusion:
Continued elucidation of the landscape and origins of allergic march trajectories will inform efforts to personalize allergic disease prevention, diagnosis, and treatment.
Keywords: Allergic march, allergic march trajectories, atopic dermatitis, food allergy, asthma, allergic rhinitis, eosinophilic esophagitis
Introduction
Allergic diseases are among the most prevalent and increasing chronic medical conditions in both children and adults.1 The allergic (or atopic) march serves as an important paradigm for understanding the natural progression of T helper type 2 (TH2) cell-mediated allergic diseases. Classically, the march begins in infancy with atopic dermatitis (AD) and progresses to IgE-mediated food allergy (IgE-FA), asthma, and allergic rhinitis (AR). Recent work indicates that eosinophilic esophagitis (EoE) is also a manifestation of the allergic march that is typically diagnosed towards the latter portion of the progression.2 This paradigm has provided important insights into the relationship between these highly prevalent and immunologically-linked conditions.
Though it has some utility in gauging a child’s risk of developing allergic multimorbidity,3 the classical march sequence also has limitations.4 Most notably, it was historically informed by population-level studies which suggested a stereotyped progression that began with AD.5 To the practicing allergist, however, it has long been apparent that there is considerable variability in the number and sequence of allergic conditions that individuals develop.3,5 The basis for this heterogeneity in allergic march pathways, or trajectories, is not well understood and is likely the result of a complex interplay of environmental, genetic, and psychosocial factors (Figure 1).
Figure 1: Allergic march trajectories.
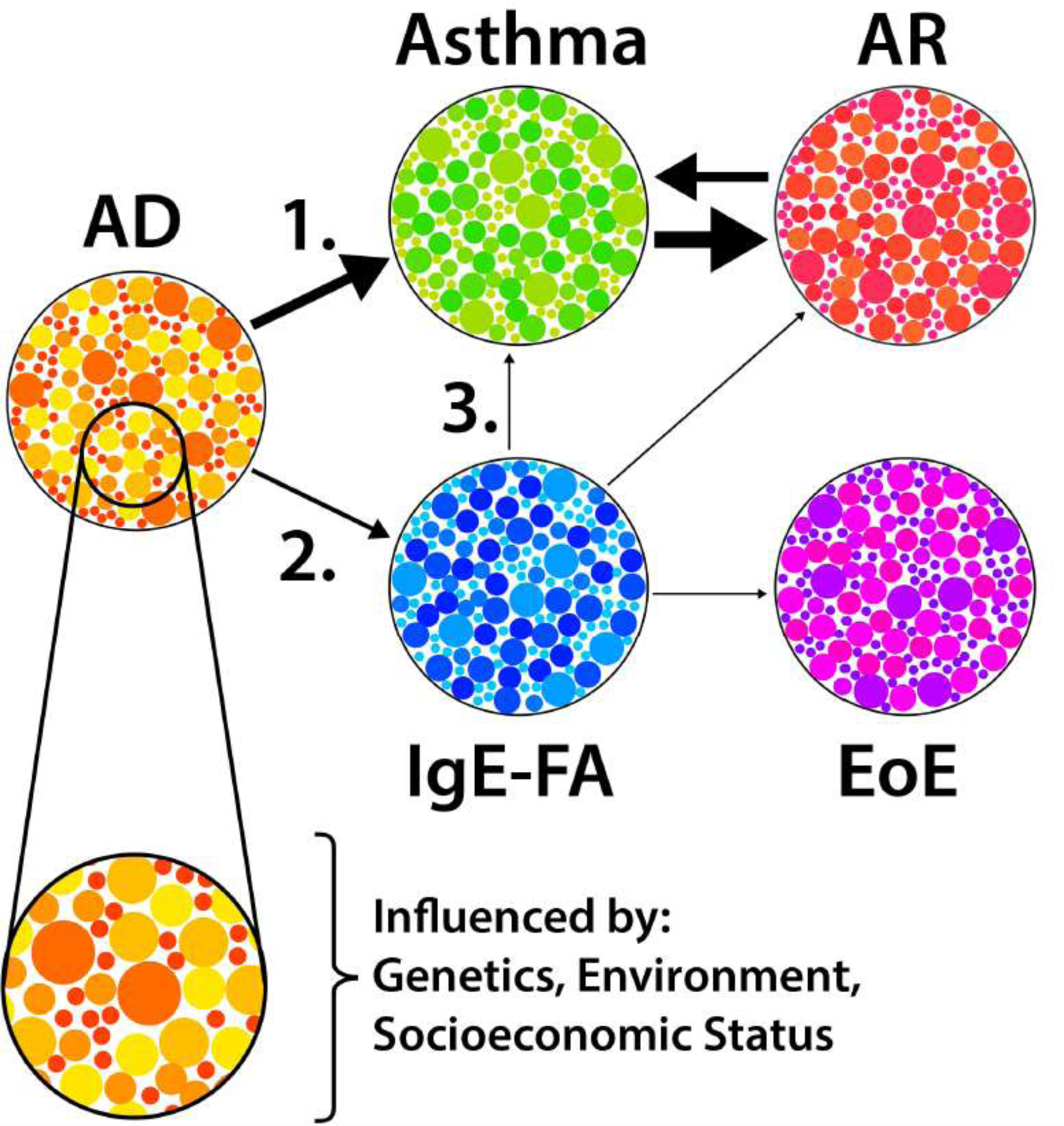
Allergic diseases have distinct endotypes (small circles), and developmental trajectories (arrows) that are influenced by genetic, environmental, and psychosocial factors. The most common trajectories are shown. Arrow weight represents relative prevalence at our institution. AD, atopic dermatitis; IgE-FA, IgE-mediated food allergy; AR, allergic rhinitis; EoE, eosinophilic esophagitis.
Here we review epidemiologic evidence supporting the existence of distinct allergic march trajectories, as well as insights into the major genetic and environmental factors that influence allergic outcomes. We discuss clinical applications for this paradigm and highlight important knowledge gaps including those relating to mechanisms of allergic sensitization and allergic march progression, as well as the need for a more nuanced understanding of the role that allergic disease endotypes play in determining trajectory outcomes. This review is relevant to both clinicians and experimental scientists, as it provides a framework for understanding march trajectories and pursuing investigations of their cause and consequence.
Epidemiologic evidence for allergic trajectories
A seminal study that highlighted heterogeneity in allergic developmental profiles used Bayesian machine learning and latent class analysis to analyze data from two pediatric birth cohorts.6 The authors reported eight distinct developmental profiles of AD, wheeze, and AR. While 48.3% of children exhibited some form of allergic disease, only 7% followed trajectory profiles resembling the classical allergic march. Limitations of this study included cohort size, survey-based nature of diagnosis, and number of allergic conditions considered. Nevertheless, this study was among the first to highlight the existence of distinct allergic trajectories among individuals.
A number of epidemiologic studies that utilize heterogenous methods and disease definitions have since been published that further support the concept of allergic march trajectories. As AD onset often occurs early in life, most longitudinal prospective cohort studies of children have considered AD as the origin of the allergic march and considered trajectories that emerge after AD diagnosis. Supporting this approach, AD was found to be the first allergic manifestation in 60.7% of UK children who were followed through age 18.7 In the same study, AD-asthma-AR was the most common allergic trajectory, though IgE-FA and EoE were not considered. Of note, this study identified 14 additional trajectories, with asthma and AR occurring as the first allergic manifestation in 20.4% and 10.5% of allergic children, respectively.7 A second pediatric cross-sectional analysis also supported a strong relationship between AD and subsequent development of respiratory allergy, as the authors observed that AD, asthma, and AR coexist to a greater degree than would be expected due to chance alone.8 Moreover, this study found that allergic comorbidity at age four was associated with comorbidity four years later in a dose-dependent fashion.8 Together, these studies support the sequence of AD to respiratory allergy (asthma and/or AR) as a predominant allergic trajectory (Figure 1), while simultaneously challenging the notion that it is the only pattern of allergy development.
Subsequent studies have provided more nuanced information regarding potential modifiers of the AD-to-respiratory allergy trajectory. In a prospective pediatric birth cohort study, very early-onset AD and persistent AD significantly increased risk of developing asthma and AR.9 Interestingly, late-onset AD in this study also increased risk of asthma, but not AR. In another prospective study examining children with AD, 43% developed asthma and 45% developed AR by age seven.10 In this case, severe and early-onset AD were both associated with increased risk of sensitization to food and aeroallergens.10 These findings echo those of a multi-ethnic population-based prospective cohort study that found that early and persistent AD increased risk of physician-diagnosed food allergy.11 There are also alterations to the skin that occur in the absence of clinically apparent AD that may facilitate allergic sensitization.12,13 For example, compared to non-allergic individuals, subjects with peanut allergy and no AD history have reduced skin content of cis-urocanic acid and pyrrolidone carboxylic acid, features typically seen in non-lesional skin of AD patients.13 Together, these observations suggest that certain AD endotypes may alter progression on the march (Figure 1).
Allergic landscapes in adulthood are less clear than in childhood. Conceptually, childhood allergic disease that is outgrown may be distinct from disease that either develops or persists later in life. It is likely that non-allergic co-morbidities influence allergic trajectories and outcomes in adulthood. These complexities are highlighted in a latent class analysis of subjects aged seven to 53 with asthma and comorbid allergies (AD, IgE-FA, and/or AR) in the Tasmanian Longitudinal Health Study.14 Data on lung function and presence of chronic obstructive pulmonary disease (COPD) was also considered. In this study, five trajectories with varying severities and durations of asthma and allergies were reported. Two trajectories (“early-onset persistent asthma and allergies” and “late-onset asthma and allergies”) were specifically associated with COPD. The latter was also associated with the presence of other co-morbidities such as psychiatric illness, hypertension, and gastroesophageal reflux disease.14 This study highlights the need to account for comorbid disease, and the possibility of allergic disease misdiagnosis, when studying the course of allergic trajectories in adulthood.
Although allergic asthma and AR are often clinically associated, the existence of comorbid AD, asthma, and AR is rare in adults. A recent study of Swedish adults showed that about 2% of the general population have concomitant AD, asthma, and AR, a rate that increased to about 6% in sensitized adults.15 The finding that sensitization modifies disease risk was also found in a multiplex component analysis of European adults with AR, in which total IgE levels and IgE polysensitization to aeroallergens were most predictive of allergic multimorbidity (AR plus AD and/or asthma).16 Polysensitization was associated with allergic multimorbidity (AD, asthma, and AR) to a larger extent in children than in adults in a Polish cohort.17 In a separate study, latent class analysis identified five patterns of sensitization and pediatric allergic morbidity (AD, asthma, or AR).18 Early onset and persistent sensitization were associated with AD, while early onset and transient sensitization were associated with increased asthma risk. Finally, sensitization to house dust mite allergen was linked to increased risk for both asthma and AR.18 Together, these studies further demonstrate the importance of age of onset and the modulatory role of allergic sensitization in predicting allergic multimorbidity.
Characterizing respiratory allergy endotypes has recently been facilitated through the use of cluster analyses. Examining children with differential profiles of wheeze and/or asthma, Tang et al identified a high-risk group with persistent wheeze, very early allergic sensitization, and frequent respiratory infections.19 Two lower-risk clusters were collectively associated with lower IgE levels, minimal allergic sensitization, and infrequent wheeze.19 Recapitulating the challenge of characterizing allergic disease outcomes in adults, a recent cluster analysis identified three distinct endotypes (C1–C3) of adult asthmatics.20 C1 asthmatics exhibited active asthma, poor lung function, elevated body mass index, and high blood neutrophil count and had poor outcomes. In contrast, C2 asthmatics exhibited mild asthma and rhinitis symptoms, while C3 asthmatics had inactive or mild untreated childhood-onset asthma with high IgE levels.20 The distinctive characteristics reported for pediatric versus adult respiratory allergy endotypes further support the concept of heterogeneity in allergic trajectories and highlight the need to identify clinical and molecular features that correlate with specific disease outcomes.
While initial definitions of the allergic march did not consider IgE-FA, it is now clear that it is an important constituent. In a systematic review, a strong and dose-dependent association was observed between AD and IgE-FA.21 In some population studies, clinically proven IgE-FA occurred in up to 15% of subjects with AD.21 AD has been linked to sensitization to food allergens, which occurs earlier in life as compared with sensitization to aeroallergens.22 IgE-FA may also be an allergic trajectory modifier. In a Canadian birth cohort study, there was a strong positive additive interaction between AD and IgE-FA.23 In addition, AD increased risk for asthma seven-fold but only in individuals sensitized to food or aeroallergens.23 A subsequent study found that food sensitization in the first two years of life was associated with increased risk of asthma and AR, regardless of aerosensitization status.24
The fact that some children with IgE-FA, without a history of AD, progress on the allergic march begs the question of whether IgE-FA is an alternative mode of initiation on the march (Figure 1). Indeed, IgE-FA is an independent risk factor for AR and asthma, in particular IgE-FA to peanut, milk, and egg.25 Tolerance at the gut is mediated by a distinct milieu of immune cells (e.g. tolerogenic dendritic cells) and cytokines that collectively favor differentiation of T regulatory (TReg) cells and suppression of allergic responses (Figure 2A).26,27 When this system of tolerance fails, immune mechanisms similar to those observed in cutaneous sensitization become relevant, including roles for the cytokines thymic stromal lymphopoietin (TSLP), interleukin (IL)-25, and IL-33 (Figure 2B,C).27–29 The prospect of allergic sensitization at the gut as a distinct mechanism for allergy would expand the popular dual allergen exposure hypothesis that posits that food allergen-skin interactions promote IgE-FA, whereas food allergen-gut interactions yield allergic tolerance.25,28 However, the genetic and/or environmental factors (e.g. microbiome, dietary exposures) that skew immune responses to favor gut sensitization necessitate further investigation.
Figure 2. Mechanisms of tolerance and sensitization.
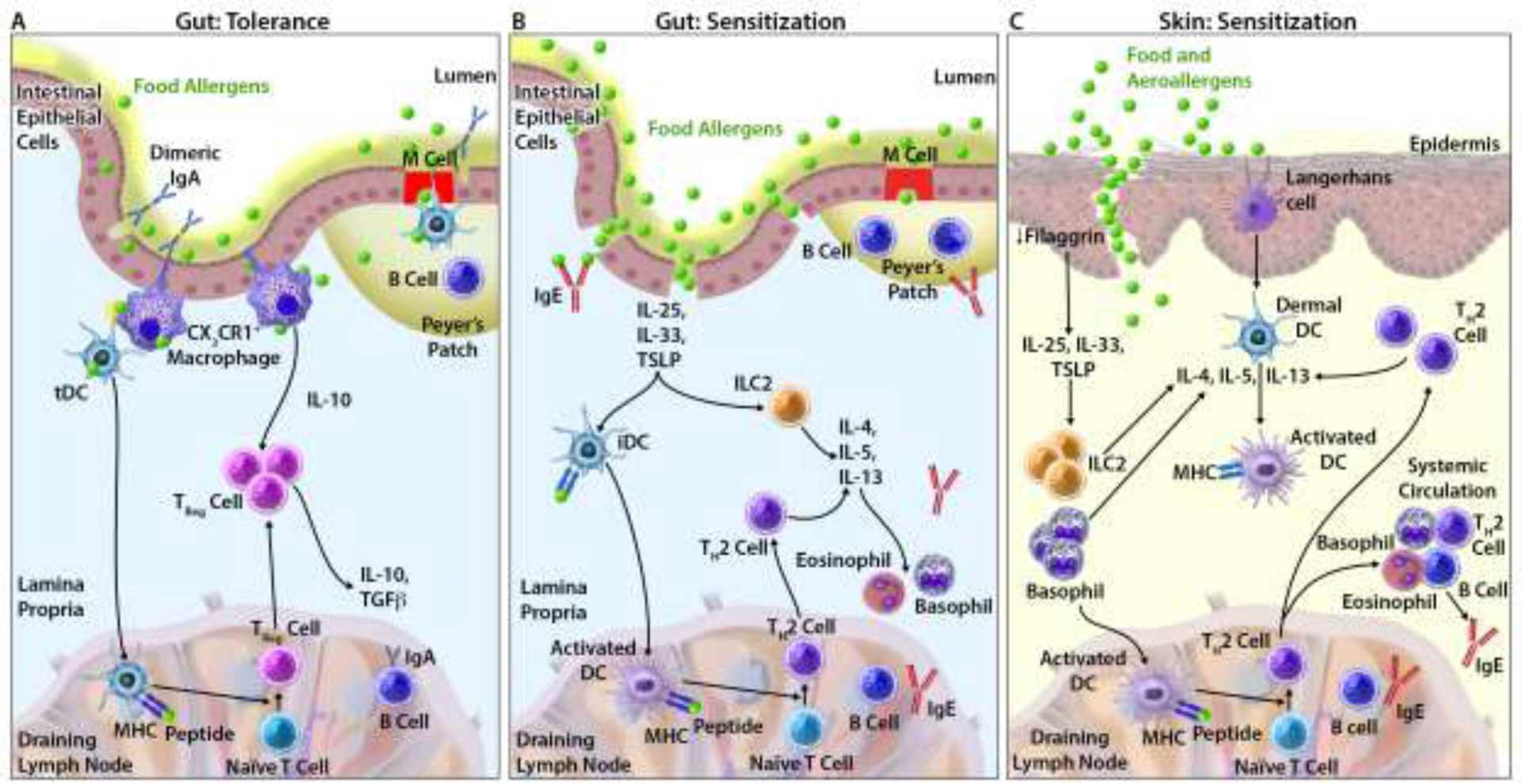
(A) Oral tolerance in the gut. (B) Allergic sensitization in the gut. (C) Allergic sensitization via the skin.
Finally, recent studies have uncovered a particularly strong relationship between IgE-FA and the chronic esophageal disorder EoE. In a pediatric virtual birth cohort study, children with IgE-FA exhibited a nine-fold increased risk of ultimately developing EoE.2 The study also found that personal history of AD, IgE-FA, and asthma independently and cumulatively associated with increased rate of developing EoE, findings that collectively support a role for EoE as a late-diagnosed member of the allergic march.2 The increased prevalence of the major allergic conditions in patients with EoE is corroborated by recent studies, including a meta-analysis and a multisite registry analysis examining children and adults with EoE.30,31 Continued investigations of the immunopathology of EoE will likely uncover additional mechanistic links between it and the other allergic manifestations.
Influence of genetic factors on allergic trajectories
The importance of inherited genetic factors in predisposing to atopy is well established, although it is not fully clear how they contribute to specific allergic trajectories. In one study, mother-child dyads demonstrated a dose-dependent association between maternal and childhood allergic diseases; the strongest associations were seen for identical allergic disease (asthma and AR), whereas non-identical diseases exhibited weaker associations.32 In another prospective cohort study performed in Sweden, history of allergy in both biological parents predicted allergy development at 10 years of age, though heredity did not associate with allergic disease burden.33 These studies highlight the role of genetics as a critical, though not exclusive, determinant of allergic outcomes.
The availability of large-scale genome wide association studies (GWAS) has revolutionized the ability to identify allergic risk loci. To date, these have uncovered an array of genes and intergenic variants with predicted functions that encompass innate and adaptive immunity, epithelial barrier integrity, cellular signaling, and gene regulation (Table 1).34–37 Of note, many genetic variants identified in these studies are associated with multiple allergic manifestations. Despite this, it is challenging to determine the impact of specific genetic variants on distinct allergic trajectories, as individuals with various combinations of allergic manifestations are often grouped into a single disease outcome category. Additionally, temporal disease relationships are unclear, disease phenotypes are often self-reported, and all known allergic march manifestations are rarely studied simultaneously.
Table 1.
Susceptibility genes associated with two or more allergic march manifestations.a
| Allergic diseases | Susceptibility genes |
|---|---|
| AD, IgE-FA | CCDC80, DEXI, IL21, KIF3A, NLRP10, OR10A3 |
| AD, IgE-FA, Asthma, AR | CLEC16A, FLG, GLB1, IL2, LRRC32, OVOL1, SLC25A46, TNFRSF6B, ZNF365, ZNF652 |
| AD, IgE-FA, Asthma, AR, EoE | EMSY (C11orf30), STAT6, TSLP, WDR36 |
| AD, IgE-FA, AR | TMEM232 |
| AD, Asthma, AR | AAGAB, ABCB9, ABO, AC004893.11, ACTR1A, ADAMTS4, AHI1, AK056081, ALOX15, APC1, APOBR, AQP5, ARHGAP15, ARHGAP27, ARL3, ARL6IP4, ASCL2, ATXN2L, B4GALT3, BATF3, BCL2L11, BCL6, BOLL, C1orf54, C22orf46, C5orf56, CAMK4, CCDC134, CCL20, CCR7, CD200R1L, CD247, CEP57, COL15A1, CRTC3, CSDC2, CSF2RB, CTC-551A13.2, CXCR5, D2HGDH, DC86, DLEU1, DYNAP, EAF2, EFEMP2, ERBB3, ERMP1, ETS1, F11R, FAM105A, FAM177A1, FAM76B, FBXW2, FCER1G, FOSL2, FOXO1, GFRA3, GNGT2, GPANK1, GSAP, GSDMB, HDAC3, HDAC7, HHEX, HIS1H2BD, HLA-C, HLA-DQA1, IKZF2, IL13, IL18R1, IL18RAP, IL1B, IL1RL1, IL27, IL2RA, IL4R, IL6R, IL7R, INPP5D, IPCEF1, IQCB1, IQGAP1, ITGB8, ITPKA, JAZF1, KIAA0391, KIAA1109, KLF2, LAT, LINC00284, LINC00299, LINC00393, LPP, MANBA, MAP3K14-AS1, MARS2, MCCD1P1, MEI1, MFSD13A, MFSD9, MIR146A, MYC, MYL6B, NAB2, NCF4, NDFIP1, NDFIP1, NEK6, NFATC2, NFKBIA, NHP2L1, NOD2, NRROS, NSMCE1, NUDT12, OGFOD2, OR10J5, ORMDL3, PAG4, PAPOLG, PHF5A, PIGN, PITPNM2, PLCL1, PMM1, POLI, PPCDC, PPOX, PPP2R1B, PPP2R3C, PRR5L, PRRC2A, PSMD5, PTGER4, PTPRC, PTPRK, PVALB, PVT1, RAB24, RAD51B, RASA2, RASGEF1A, RBM15B, REL, RERE, RFTN2, RGS14, RIN3, RORA, RORC, RP11-132N15.1, RP11-132N15.2, RP11-24N18.1, RP11-264B17.4, RP11-534L20.5, RP11-554D20.1, RP11-770G2.5, RP11-94L15.2, RP4-1115A15.1, RPRD2, RPS26, RTEL1, RTF1, RUNX3, RYBP, SBNO1, SDK1, SENP7, SERPINB7, SH2B3, SIK2, SLC15A2, SLC22A4, SLC22A5, SMAD4, SMAD7, SNX32, SPATA32, SPNS1, SPPL3, STAT5B, STMN3, SULT1A1, SUOX, TARS2, TBL1XR1, THEM4, TIAM2, TLR1, TMEM180, TNC, TNFAIP3, TNFAIP8, TNFRSF14, TNFRSF8, TNFSF4, TRAF3, USF1, VDAC1, VPRBP, ZBTB38, ZDHHC12, ZNF217, ZPBP2 |
| AD, AR | ANKRD46, ATXN2, BNC2, ETV7, HSPE1, IL31, LRRC43, MOB4, NOS3, TLR10 |
Gene-disease associations were identified in previous genome-wide association studies.34–39 Individuals with gene variants did not necessarily exhibit all listed allergic manifestations for a given gene. AD, atopic dermatitis; IgE-FA, IgE-mediated food allergy; AR, allergic rhinitis; EoE, eosinophilic esophagitis.
Despite these shortcomings, important insights into how genetic factors may shape allergic trajectories can be gleaned from existing genomic studies. For example, focusing on susceptibility variants previously associated with AD and EoE, Hirota et al investigated their associations with childhood IgE-FA. 14 genetic loci exhibited significant associations (Table 1).38 The strongest association for subjects with AD and IgE-FA occurred for a KIF3A/IL13 variant, consistent with the critical role of IL-13 in IgE-mediated allergic responses.38 Notably, the IL13-dependent locus CAPN13 has been more recently associated exclusively with EoE.39 It is plausible that such differences in gene pleiotropy contribute to differences in allergic outcomes and, thus, heterogenous allergic trajectories.
Focusing on susceptibility to AD, asthma, and AR, a series of seminal GWASs have uncovered a shared set of risk genes with functions that include immune cell signaling and epidermal differentiation (Table 1). In two separate GWASs, Ferreira et al previously reported over 150 associated genetic variants.35,36 Remarkably, very few variants (affecting FLG, IL2RA, GSDMB, and several intergenic regions) exhibited differential disease-specific effects.35 The observed association between AD and variants of FLG, which encodes the keratin-binding protein filaggrin, corroborate the knowledge that FLG variants are permissive for progression of atopy specifically in the setting of AD.40,41 Another GWAS performed by Johansson et al identified 41 additional novel loci.37 While most risk loci were associated with the combined phenotype of AD, asthma, and AR, 20 loci were more significantly associated with AD and AR, and 16 loci had stronger effects on asthma.37 Among the variants with differential disease effects were two SNPs in the intron of IL2RA; rs12722547 was more strongly associated with asthma than rs61839660.37 These studies indicate that a set of immunologically important genes is shared in predisposition to multiple allergic manifestations, with some genetic variants exhibiting differential effects on allergic outcomes.
Using a reductionist approach, a recent analysis examined genotype associations with pediatric allergic trajectories that begin with AD.42 Four single nucleotide polymorphisms (SNPs) were associated with distinct allergic trajectories and were also linked to specific demographic features, including self-identified race and sex.42 Interestingly, all identified SNPs affect non-coding parts of the genome, suggesting regulatory functions. One of the SNPs, rs60242841, was associated with the AD-to-asthma trajectory and was more common in individuals of African ancestry than European ancestry.42 This study sheds light on the complex role of genetics in driving heterogenous allergic outcomes. As race is a social construct, the study also highlights the need to consider concomitant modifying factors, including psychosocial factors. In sum, while important progress has been made in identifying allergic risk loci, studies that examine genotype associations with specific allergic trajectories of interest are warranted.
Influence of environmental factors on allergic trajectories
The growing prevalence of allergic disease has sparked interest in understanding how exogenous and potentially modifiable factors facilitate allergy. As infants are uniquely vulnerable to the effects of dysbiosis,43 there has been particular interest in identifying early-life modifiers of microbial colonization that predispose to allergy. While there are varying degrees of evidence for vaginal delivery, breastmilk-based diet, and early-life avoidance of antibiotics and/or antacids in preventing atopy,43–46 there is paucity of data on how these factors impact specific allergic disease trajectories. In a recent study examining effects of these factors on allergic multimorbidity (AD, IgE-FA, asthma, and/or AR), vaginal delivery, breastmilk feeding, and avoidance of antibiotics and antacids were associated with reduced rate of developing at least three allergic manifestations.47 Whether early-life factors favor specific allergic trajectories, and what their impacts are on EoE, remain to be determined.
The effects of dysbiosis extend beyond the gastrointestinal tract. For example, analysis of Staphylococcus aureus colonization in the nares revealed a direct correlation between colonization and sensitization to either egg or peanut, regardless of AD severity.48 Other examples of dysbiosis in allergy include the esophageal microbiome in EoE, which exhibits high prevalence of Haemophilus species.49 In the respiratory tract, pharyngeal colonization with Streptococcus pneumoniae, Haemophilus influenzae, and/or Moraxhella catarrhalis has been associated with early-life asthma.50 Longitudinal analyses with sampling at various mucosal sites will be needed to further dissect microbial effects on specific allergic trajectories.
Another important environmental factor that can influence allergic trajectories is infant diet and timing of allergic food introduction. The hallmark Learning Early About Peanut (LEAP) Allergy trial motivated recommendations for early peanut introduction in patients with high risk of peanut allergy.51 The success of this approach is likely due to anatomical differences in immunologic responses that mediate tolerance versus sensitization (Figure 2). While early study of the effects of peanut consumption by at-risk infants does not appear to support a role in reducing allergic disease burden beyond peanut allergy,51 further studies of early food introduction cohorts is warranted.
Various ambient exposures (e.g. pollutants, endotoxin, dust, and viruses) have also been found to modulate allergic susceptibility.52 For example, air pollution associated with urban environments is thought to cause epigenetic changes and immune dysregulation that lead to increased risk of AD and asthma.52,53 Conversely, elevated endotoxin levels observed in rural environments and in the setting of early-life animal exposure are associated with lower risk of respiratory allergy and allergic sensitization.52 This effect was validated in a landmark study that examined allergic outcomes in Hutterites and the Amish, who share a similar genetic background but have distinct farming practices.54 As further evidence of the importance of microbial exposures to allergic outcomes, asthma risk is decreased in children living in urban areas where microbial composition of dust resembles that of children living in rural areas.55
It is well established that allergic diseases are associated with significant economic costs and impairments in quality of life, factors that disproportionately affect socioeconomically disadvantaged groups.56 Highlighting an important role for racial disparities in driving differential health outcomes, black children suffer from higher allergic burden than white children, even when accounting for socioeconomic status.57 Further, structural determinants (e.g. housing policies) are known to contribute to allergic health disparities.58 Relevant future directions for the field include standardizing methods used to assess disease burden, considering impacts of structural health determinants on differential allergic trajectories, and ensuring that future investigations of allergic march trajectories maximize socioeconomic and racial diversity.
Relevance of allergic march trajectories to patient risk stratification and treatment
How genetic and environmental factors dictate allergic progression is a question of considerable scientific and clinical interest, as it is relevant to determining disease risk, prevention, and treatment. The ability to accurately model gene-environment interactions to predict an individual’s allergic profile represents a holy grail of personalized medicine that may be coming within reach. This is primarily due to the recent revolution in “big data” approaches that have allowed the identification of novel allergic disease risk factors, endotypes, and biomarkers.59 These data are especially valuable when partnered with unbiased machine learning methods that reveal otherwise unappreciated and testable patterns.42 Through these efforts, it is conceivable that the next decade will see the introduction of a new generation of clinical risk assessment and disease stratification systems. In the following section, we provide some examples of clinical practices supported by current evidence and identify areas where more research is needed to inform clinical decision making.
Given that AD is the most common initiation point on the march, there is particular focus on identifying interventions that alter allergic progression of children with AD.60,61 Compelling evidence demonstrates that risk of allergic sensitization to foods and environmental allergens correlates with AD presence and severity.10–11,62 Further, multiple studies have now shown that early introduction of allergenic foods in children with AD prevents IgE-FA.51,62 Despite this, evidence showing a benefit of AD treatment has been more elusive. A recent Cochrane review found that skin care interventions during infancy did not change an individual’s risk of developing AD or the amount of time to onset of AD.63 Recent studies specifically examining the role of AD treatment in preventing allergic progression demonstrate mixed results.64,65 It is possible that such studies have not focused on the appropriate disease endotype(s) as outcomes. It is even less clear how such efforts will influence march trajectory outcomes later in life.
The realization that IgE-FA is a manifestation of the allergic march has increased the complexity and utility of the paradigm. For example, IgE-FA increases risk for being diagnosed with asthma and AR later in life,23 which may help with patient risk stratification. Further, the risk relationship between IgE-FA and EoE is particularly strong, an observation that is useful both clinically and with regard to understanding the immunopathology of this condition.66,67 As EoE is sometimes a complication of sublingual immunotherapy (SLIT) and oral immunotherapy (OIT), it should be considered in children who develop consistent symptoms during these treatments.68 Given that some patents exhibit IgE-FA as their first allergic manifestation, it is conceivable that IgE-FA unpreceded by AD represents a distinct IgE-FA endotype. It is not known if early introduction of allergenic foods modifies risk of development of allergic manifestations other than IgE-FA. Specifically designed prospective studies are needed to better understand how various IgE-FA endotypes associate with distinct allergic trajectories.69
Immunomodulation has long been an approach used in clinical allergy, and there is evidence that its benefits may extend beyond the primary condition(s) being treated. Indeed, a systematic review and meta-analysis of allergen immunotherapy observed a short-term benefit in preventing asthma in children with AR.70 The introduction of several new biologics that block inflammatory pathways shared among multiple allergic manifestations holds additional promise for modifying allergic march trajectories. For example, the anti-IL4-alpha receptor antibody dupilumab has demonstrated efficacy in the treatment of AD, asthma, and EoE, and is being investigated for use in treating peanut allergy.71,72 Similarly, the anti-TSLP monoclonal antibody tezepelumab is in clinical trials for the treatment of asthma.73 As TSLP is an early regulator of TH2 responses, it is conceivable that it may confer additional clinical benefits. Finally, there is an ongoing trial assessing the efficacy of the anti-IgE biologic omalizumab in preventing asthma in high-risk 2–3-year-olds,74 the results of which are eagerly awaited. In sum, the potential for immunomodulatory therapies to affect, and possibly halt, the allergic march is high, though more detailed studies will be needed to ascertain effects on specific allergic trajectories.
Conclusion
The concept of the allergic march is critical for understanding the natural patterns of allergic disease development as well as the pathophysiologic relationships shared among major allergic manifestations. The heterogeneity of allergic march pathways is attributable to the existence of multiple allergic disease endotypes, which reflect a critical and incompletely understood interplay between hereditary and exogenous factors. Despite this, evidence for two predominant trajectories is emerging: 1) AD to respiratory allergy (asthma and/or AR) and 2) AD to food allergy (IgE-FA and/or EoE). There may also be reason to consider trajectories starting with IgE-FA as a distinct entity. Several questions remain about the mechanistic underpinnings and clinical implications of allergic trajectories:
To what extent is allergic sensitization in the gut responsible for distinct disease endotypes and march trajectories?
How do genetic variants interact with each other and with environmental factors to yield specific allergic outcomes?
Which patients warrant close surveillance for disease development, and how can the march be prevented?
Such questions will benefit from systems medicine studies that integrate epidemiologic, clinical, and “omics” data and further inform mechanistic experimental studies. It is also essential that future investigations include prospective, longitudinal studies that consider geographically and demographically diverse patient populations. Collectively, these efforts have the potential to revolutionize approaches in allergic disease prevention, diagnosis, and treatment.
Supplementary Material
Learning Objectives.
Recognize epidemiologic evidence supporting the existence of allergic march trajectories.
Be aware of the major genetic and environmental determinants of allergic march trajectories.
Funding source:
Work in the Hill Lab is supported by the National Institutes of Health (grant no. K08DK116668), the American College of Allergy Asthma and Immunology, the American Academy of Allergy Asthma and Immunology, the American Partnership for Eosinophilic Disorders, and a Children’s Hospital of Philadelphia Research Institute Development Award. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- AD
atopic dermatitis
- AR
allergic rhinitis
- COPD
chronic obstructive pulmonary disease
- EoE
eosinophilic esophagitis
- GWAS
genome wide association study
- IgE-FA
IgE-mediated food allergy
- IL
interleukin
- LEAP
Learning Early About Peanut
- OIT
oral immunotherapy
- SLIT
sublingual immunotherapy
- SNP
single nucleotide polymorphism
- TH2
T helper type 2
- TSLP
thymic stromal lymphopoietin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors have no conflicts of interest relevant to this work.
References
- 1.Pawankar R, et al. “Allergic Diseases as a Global Public Health Issue.” White Book on Allergy, World Allergy Organization; (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill DA, et al. “Eosinophilic esophagitis is a late manifestation of the allergic march.” The Journal of Allergy and Clinical Immunology: In Practice 6.5 (2018): 1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill DA, et al. “A march by any other name.” Annals of Allergy, Asthma and Immunology 121.1 (2018): 137–138. [DOI] [PubMed] [Google Scholar]
- 4.Busse WW. “The atopic march: fact or folklore?.” Annals of Allergy, Asthma and Immunology 120.2 (2018): 116–118. [DOI] [PubMed] [Google Scholar]
- 5.Paller AS, et al. “The atopic march and atopic multimorbidity: Many trajectories, many pathways.” Journal of Allergy and Clinical Immunology 143.1 (2019): 46–55. [DOI] [PubMed] [Google Scholar]
- 6.Belgrave DCM, et al. “Developmental profiles of eczema, wheeze, and rhinitis: two population-based birth cohort studies.” PLoS Med 11.10 (2014): e1001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Punekar YS and Sheikh A. “Establishing the sequential progression of multiple allergic diagnoses in a UK birth cohort using the General Practice Research Database.” Clinical and Experimental Allergy 39.12 (2009): 1889–1895. [DOI] [PubMed] [Google Scholar]
- 8.Pinart M, et al. “Comorbidity of eczema, rhinitis, and asthma in IgE-sensitised and non-IgE-sensitised children in MeDALL: a population-based cohort study.” The Lancet Respiratory Medicine 2.2 (2014): 131–140. [DOI] [PubMed] [Google Scholar]
- 9.Lowe AJ, et al. “Age at onset and persistence of eczema are related to subsequent risk of asthma and hay fever from birth to 18 years of age.” Pediatric Allergy and Immunology 28.4 (2017): 384–390. [DOI] [PubMed] [Google Scholar]
- 10.Gustafsson DO, et al. “Development of allergies and asthma in infants and young children with atopic dermatitis–a prospective follow-up to 7 years of age.” Allergy 55.3 (2000): 240–245. [DOI] [PubMed] [Google Scholar]
- 11.Hu C, et al. “Eczema phenotypes and risk of allergic and respiratory conditions in school age children.” Clinical and Translational Allergy 10.1 (2020): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Splunter M, et al. “Mechanisms Underlying the Skin-Gut Cross Talk in the Development of IgE-Mediated Food Allergy.” Nutrients 12.12 (2020): 3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berdyshev E, et al. “Unique skin abnormality in patients with peanut allergy but no atopic dermatitis.” Journal of Allergy and Clinical Immunology 147.1 (2021): 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bui DS, et al. “Trajectories of asthma and allergies from 7 years to 53 years and associations with lung function and extrapulmonary comorbidity profiles: a prospective cohort study.” The Lancet Respiratory Medicine (2020). [DOI] [PubMed] [Google Scholar]
- 15.Pullerits T, et al. “The triad of current asthma, rhinitis and eczema is uncommon among adults: Prevalence, sensitization profiles, and risk factors.” Respiratory Medicine 176 (2021): 106250. [DOI] [PubMed] [Google Scholar]
- 16.Blöndal V, et al. “Study of atopic multimorbidity in subjects with rhinitis using multiplex allergen component analysis.” Clinical and Translational Allergy 10.1 (2020): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raciborski F, et al. “Dissociating polysensitization and multimorbidity in children and adults from a Polish general population cohort.” Clinical and Translational Allergy 9.1 (2019): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabet S, et al. “Asthma and allergic rhinitis risk depends on house dust mite specific IgE levels in PARIS birth cohort children.” World Allergy Organization Journal 12.9 (2019): 100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang HF, et al. “Trajectories of childhood immune development and respiratory health relevant to asthma and allergy.” Elife 7 (2018): e35856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadif R, et al. “Endotypes identified by cluster analysis in asthmatics and non-asthmatics and their clinical characteristics at follow-up: the case-control EGEA study.” BMJ Open Respiratory Research 7.1 (2020): e000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsakok T, et al. “Does atopic dermatitis cause food allergy? A systematic review.” Journal of Allergy and Clinical Immunology 137.4 (2016): 1071–1078. [DOI] [PubMed] [Google Scholar]
- 22.Nissen SP, et al. “The natural course of sensitization and allergic diseases from childhood to adulthood.” Pediatric Allergy and Immunology 24.6 (2013): 549–555. [DOI] [PubMed] [Google Scholar]
- 23.Tran MM, et al. “Predicting the atopic march: results from the Canadian healthy infant longitudinal development study.” Journal of Allergy and Clinical Immunology 141.2 (2018): 601–607. [DOI] [PubMed] [Google Scholar]
- 24.Alduraywish SA, et al. “Is there a march from early food sensitization to later childhood allergic airway disease? Results from two prospective birth cohort studies.” Pediatric Allergy and Immunology 28.1 (2017): 30–37. [DOI] [PubMed] [Google Scholar]
- 25.Hill DA, et al. “The epidemiologic characteristics of healthcare provider-diagnosed eczema, asthma, allergic rhinitis, and food allergy in children: a retrospective cohort study.” BMC Pediatrics 16.1 (2016): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worbs T, et al. “Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells.” Journal of Experimental Medicine 203.3 (2006): 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W, et al. “Food allergy: immune mechanisms, diagnosis and immunotherapy.” Nature Reviews Immunology 16.12 (2016): 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khodoun MV, et al. “Prevention of food allergy development and suppression of established food allergy by neutralization of thymic stromal lymphopoietin, IL-25, and IL-33.” Journal of Allergy and Clinical Immunology 141.1 (2018): 171–179. [DOI] [PubMed] [Google Scholar]
- 29.Brough HA, et al. “Epicutaneous sensitization in the development of food allergy: What is the evidence and how can this be prevented?.” Allergy 75.9 (2020): 2185–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González-Cervera J, et al. “Association between atopic manifestations and eosinophilic esophagitis: a systematic review and meta-analysis.” Annals of Allergy, Asthma and Immunology 118.5 (2017): 582–590. [DOI] [PubMed] [Google Scholar]
- 31.Chehade M, et al. “Phenotypic characterization of eosinophilic esophagitis in a large multicenter patient population from the consortium for food allergy research.” The Journal of Allergy and Clinical Immunology: In Practice 6.5 (2018): 1534–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C-H, et al. “Shared prenatal impacts among childhood asthma, allergic rhinitis and atopic dermatitis: a population-based study.” Allergy, Asthma & Clinical Immunology 15.1 (2019): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Björkander S, et al. “The allergic phenotype during the first 10 years of life in a prospective cohort.” Immunity, Iinflammation and Disease 7.3 (2019): 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portelli MA, et al. “Genetic risk factors for the development of allergic disease identified by genome-wide association.” Clinical and Experimental Allergy 45.1 (2015): 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira MA, et al. “Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology.” Nature Genetics 49.12 (2017): 1752–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferreira MAR, et al. “Eleven loci with new reproducible genetic associations with allergic disease risk.” Journal of Allergy and Clinical Immunology 143.2 (2019): 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson A, et al. “Genome-wide association analysis of 350 000 Caucasians from the UK Biobank identifies novel loci for asthma, hay fever and eczema.” Human Molecular Genetics 28.23 (2019): 4022–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirota T, et al. “Association study of childhood food allergy with genome-wide association studies–discovered loci of atopic dermatitis and eosinophilic esophagitis.” Journal of Allergy and Clinical Immunology 140.6 (2017): 1713–1716. [DOI] [PubMed] [Google Scholar]
- 39.Kottyan LC, et al. “Replication and Meta-Analyses Nominate Numerous Eosinophilic Esophagitis Risk Genes.” Journal of Allergy and Clinical Immunology (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thyssen JP, et al. “Filaggrin gene mutations are not associated with food and aeroallergen sensitization without concomitant atopic dermatitis in adults.” Journal of Allergy and Clinical Immunology 135.5 (2015): 1375–1378. [DOI] [PubMed] [Google Scholar]
- 41.Clark H, et al. “Differential associations of allergic disease genetic variants with developmental profiles of eczema, wheeze and rhinitis.” Clinical and Experimental Allergy 49.11 (2019): 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabryszewski SJ, et al. “Unsupervised modeling and genome-wide association identify novel features of allergic march trajectories.” Journal of Allergy and Clinical Immunology (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sbihi H, et al. “Thinking bigger: How early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease.” Allergy (2019). [DOI] [PubMed] [Google Scholar]
- 44.Kim H, et al. “Birth mode, breastfeeding, pet exposure, and antibiotic use: associations with the gut microbiome and sensitization in children.” Current Allergy and Asthma reports19.4 (2019): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuppari C, et al. “Mode of delivery and risk for development of atopic diseases in children.” Allergy Asthma Proc. Vol. 36. No. 5. 2015. [DOI] [PubMed] [Google Scholar]
- 46.Mitre E, et al. “Association between use of acid-suppressive medications and antibiotics during infancy and allergic diseases in early childhood.” JAMA Pediatrics 172.6 (2018): e180315–e180315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabryszewski SJ, et al. “Early-life environmental exposures associate with individual and cumulative allergic morbidity.” Pediatric Allergy and Immunology (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsilochristou O, et al. “Association of Staphylococcus aureus colonization with food allergy occurs independently of eczema severity.” Journal of Allergy and Clinical Immunology 144.2 (2019): 494–503. [DOI] [PubMed] [Google Scholar]
- 49.Dellon ES. “The esophageal microbiome in eosinophilic esophagitis.” Gastroenterology 151.2 (2016): 364–365. [DOI] [PubMed] [Google Scholar]
- 50.Bisgaard H, et al. “Childhood asthma after bacterial colonization of the airway in neonates.” New England Journal of Medicine 357.15 (2007): 1487–1495. [DOI] [PubMed] [Google Scholar]
- 51.Du Toit G, et al. “Allergen specificity of early peanut consumption and effect on development of allergic disease in the Learning Early About Peanut Allergy study cohort.” Journal of Allergy and Clinical Immunology 141.4 (2018): 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peden DB. “The “envirome” and what the practitioner needs to know about it.” Annals of Allergy, Asthma and Immunology 123.6 (2019): 542–549. [DOI] [PubMed] [Google Scholar]
- 53.Wang B, et al. “Why do intrauterine exposure to air pollution and cigarette smoke increase the risk of asthma?.” Frontiers in Cell and Developmental Biology 8 (2020): 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein MM, et al. “Innate immunity and asthma risk in Amish and Hutterite farm children.” New England Journal of Medicine 375.5 (2016): 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirjavainen PV, et al. “Farm-like indoor microbiota in non-farm homes protects children from asthma development.” Nature Medicine 25.7 (2019): 1089–1095. [DOI] [PubMed] [Google Scholar]
- 56.Dierick BJH, et al. “Burden and socioeconomics of asthma, allergic rhinitis, atopic dermatitis and food allergy.” Expert Review of Pharmacoeconomics & Outcomes Research 20.5 (2020): 437–453. [DOI] [PubMed] [Google Scholar]
- 57.Sitarik A, et al. “Racial disparities in allergic outcomes persist to age 10 years in black and white children.” Annals of Allergy, Asthma and Immunology 124.4 (2020): 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan K and Thakur N. “Structural and social determinants of health in asthma in developed economies: a scoping review of literature published between 2014 and 2019.” Current Allergy and Asthma Reports 20.2 (2020): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Breiteneder H, et al. “Future research trends in understanding the mechanisms underlying allergic diseases for improved patient care.” Allergy 74.12 (2019): 2293–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irvine AD and Mina-Osorio P. “Disease trajectories in childhood atopic dermatitis: an update and practitioner’s guide.” British Journal of Dermatology 181.5 (2019): 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lowe AJ, et al. “The skin as a target for prevention of the atopic march.” Annals of Allergy, Asthma & Immunology 120.2 (2018): 145–151. [DOI] [PubMed] [Google Scholar]
- 62.Matsumoto K, et al. “Are both early egg introduction and eczema treatment necessary for primary prevention of egg allergy?.” Journal of Allergy and Clinical Immunology 141.6 (2018): 1997–2001. [DOI] [PubMed] [Google Scholar]
- 63.Kelleher MM, et al. “Skin care interventions in infants for preventing eczema and food allergy.” Cochrane Database of Systematic Reviews 2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bawany F, et al. “Halting the march: primary prevention of atopic dermatitis and food allergies.” J Allergy Clin Immunol: In Practice 8.3 (2020): 860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perkin MR, et al. “Association of frequent moisturizer use in early infancy with the development of food allergy.” J Allergy Clin Immunol 147.3 (2021): 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Capucilli P and Hill DA. “Allergic comorbidity in eosinophilic esophagitis: mechanistic relevance and clinical implications.” Clinical reviews in Allergy and Immunology 57.1 (2019): 111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruffner MA, et al. “Screening children for eosinophilic esophagitis: allergic and other risk factors.” Expert Review of Clinical Immunology 15.4 (2019): 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cafone J, et al. “Eosinophilic esophagitis during sublingual and oral allergen immunotherapy.” Current Opinion in Allergy and Clinical Immunology 19.4 (2019): 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baker MG and Sampson HA. “Phenotypes and endotypes of food allergy: A path to better understanding the pathogenesis and prognosis of food allergy.” Annals of Allergy, Asthma and Immunology 120.3 (2018): 245–253. [DOI] [PubMed] [Google Scholar]
- 70.Kristiansen M, et al. “Allergen immunotherapy for the prevention of allergy: a systematic review and meta-analysis.” Pediatric Allergy and Immunology 28.1 (2017): 18–29. [DOI] [PubMed] [Google Scholar]
- 71.Brar KK, et al. “Biologics for the Treatment of Food Allergies.” Immunology and Allergy Clinics 40.4 (2020): 575–591. [DOI] [PubMed] [Google Scholar]
- 72.Hirano I, et al. “Efficacy of dupilumab in a phase 2 randomized trial of adults with active eosinophilic esophagitis.” Gastroenterology 158.1 (2020): 111–122. [DOI] [PubMed] [Google Scholar]
- 73.Dorey-Stein ZL and Shenoy KV. “Tezepelumab as an Emerging Therapeutic Option for the Treatment of Severe Asthma: Evidence to Date.” Drug Design, Development and Therapy 15 (2021): 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phipatanakul W, et al. “Preventing asthma in high risk kids (PARK) with omalizumab: Design, rationale, methods, lessons learned and adaptation.” Contemporary Clinical Trials 100 (2020): 106228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


