Abstract
Cancer dependency maps, which use CRISPR/Cas9 depletion screens to profile the landscape of genetic dependencies in hundreds of cancer cell lines, have identified context-specific dependencies that could be therapeutically exploited. An ideal therapy is both lethal and precise, but these depletion screens cannot readily distinguish between gene effects that are cytostatic or cytotoxic. Here, we employ a diverse panel of functional genomic screening assays to identify NXT1 as a selective and rapidly lethal in vivo-relevant genetic dependency in MYCN-amplified neuroblastoma. NXT1 heterodimerizes with NXF1 and together they form the principle mRNA nuclear export machinery. We describe a previously unrecognized mechanism of synthetic lethality between NXT1 and its paralog NXT2: their common essential binding partner NXF1 is lost only in the absence of both. We propose a potential therapeutic strategy for tumor-selective elimination of a protein that, if targeted directly, is expected to cause widespread toxicity.
Introduction:
The advent of CRISPR/Cas9 genome-editing technology allows for interrogation of gene function at a genome-scale. Dependency maps that profile gene function across hundreds of cancer cell lines have been created, and these data reveal that, as predicted, there are a large number of genes that are not mutated but are nonetheless required for growth and survival of cancer cells (1–3). Some of these previously unstudied dependencies may be tractable drug targets. Indeed, a number of targets identified in these screens have now been validated, demonstrating that the data are robust (4,5). These efforts allow us to expand our search for new therapeutic targets beyond known oncogenes. Coupled with advances in drug development approaches, such as PROTACs (proteolysis targeting chimeras) and molecular glues, which have pushed the limits of what targets are considered “druggable”, dependency maps have the potential to reveal new avenues for selective killing of cancer cells that have not previously been considered or exploited.
Pediatric cancers have a paucity of genetic alterations relative to adult cancers, and therefore have not benefitted from the development of oncogene targeted therapies to the same degree (6–8). As a result, pediatric cancers are still largely treated with conventional chemotherapies, which are broadly cytotoxic drugs that frequently target pan-essential cell proteins and therefore have significant collateral toxicity (9). The application of dependency maps for novel target identification may therefore be particularly helpful for pediatric cancers. Indeed, a first-generation Pediatric Cancer Dependency Map demonstrates that despite harboring fewer oncogenic mutations, pediatric cancers have a similar number of selective genetic dependencies as adult cancers (10).
While these dependency maps enable the identification of selective targets (4,5), prioritizing the most-promising targets remains a challenge. Because these screens assess depletion in vitro at two to three weeks, we cannot readily distinguish between dependencies that slow growth, halt growth, or cause cells to die, nor can we eliminate the subset of the dependencies that could be specific to the cell culture environment and would not be maintained in an in vivo context. The difference between targets in these classes could be very significant when translated clinically. Conventional chemotherapies are largely cytotoxic drugs, but frequently target pan-essential cell proteins and therefore have significant on-target toxicity. Conversely, more recently developed targeted therapies are much more selective in their effects, but these agents are often cytostatic and may not result in as profound a viability effect. We sought to build on dependency map efforts and use additional functional genomic screens to identity in vivo relevant targets that combine the selectivity of a targeted therapy with the cytotoxicity of conventional chemotherapy.
Here, we focused on identifying such targets in neuroblastoma, the most common extra-cranial solid tumor in children. Overall survival for high-risk disease remains unacceptably low, at about 50%, and children who do survive experience significant and sometimes long-term treatment-induced side effects (11). Approximately 20–25% of neuroblastomas harbor genetic amplification of the transcription factor MYCN, which is strongly correlated with high-risk disease, an undifferentiated phenotype, and poor prognosis, and is therefore a disease subset of particular interest (12,13).
We therefore sought to use additional functional genomic screens to prioritize dependencies for therapeutic development in MYCN-amplified neuroblastoma.
Results:
CRISPR screens prioritize rapidly lethal in vivo relevant dependencies
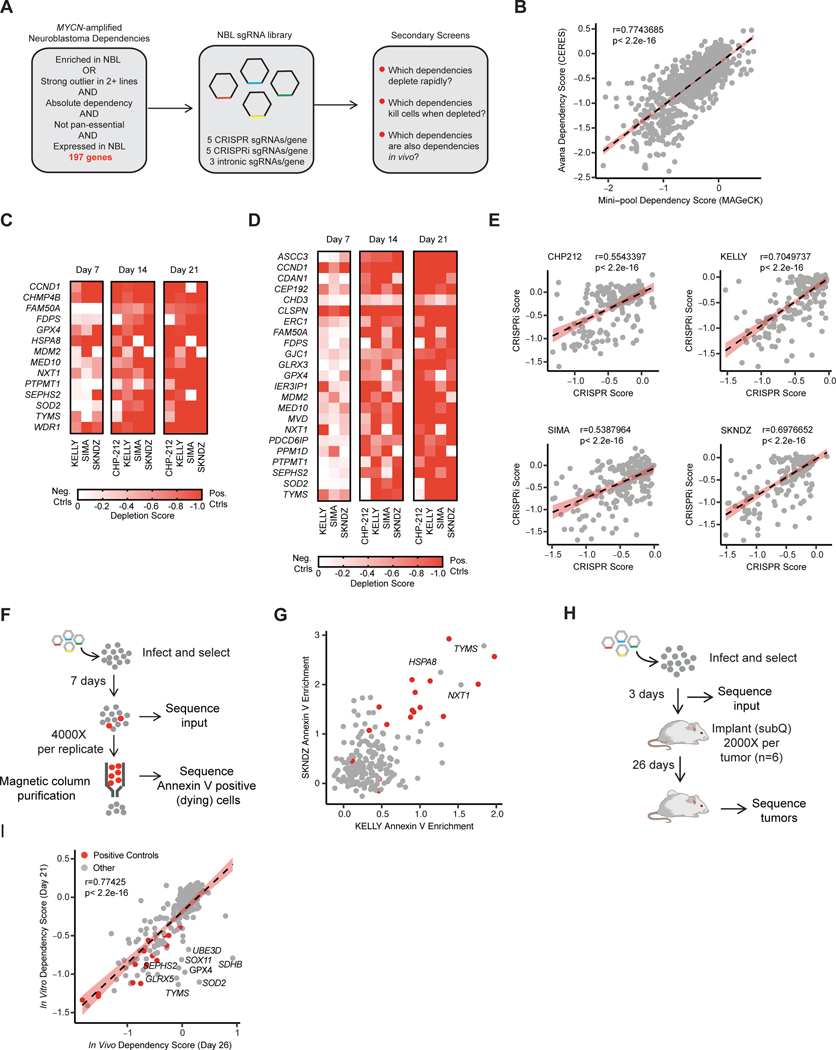
We used the Broad Institute’s Cancer Dependency Map (DepMap) dataset, inclusive of the Pediatric Cancer Dependency Map, to identify 197 putative genetic dependencies that were strong outliers or selective dependencies for MYCN-amplified neuroblastoma and 17 pan-essential genes as positive controls (Figure 1A). We generated a sgRNA library consisting of sgRNAs for both CRISPR-mediated gene knockout (CRISPR) and CRISPR-mediated gene interference (CRISPRi), intronic guides to control for copy number related toxicity effects, and non-targeting and intergenically-targeted negative controls. We performed CRISPR and CRISPRi time-course depletion screens at days 7, 14 and 21 in four neuroblastoma cell lines: SKNDZ, CHP-212, SK-N-BE(2) and KELLY. Notably, our results at 21 days correlated with the depletion observed in the original DepMap screen performed at this timepoint in these cell lines (r=0.774, p < 2.2e −16, Figure 1B, and Supplemental Figure S1A). The additional time point information allowed us to prioritize dependencies that deplete rapidly in CRISPR and CRISPRi (Figure 1C and D) and the correlation between these screening modalities was strong (Figure 1E). Intronic guides accurately identified potential false positives due to the known copy number effect of CRISPR (Supplemental Figure 1B–C) (14). Of note, the intron-targeted MYCN sgRNAs depleted similarly to the gene-targeting sgRNAS for MYCN (Supplemental Figure S1D). An attempt at copy number correction eliminated both effects. MYCN is a well-established dependency in MYCN-amplified neuroblastoma (15,16) and an shRNA screen readily detected this dependency (Supplemental Figure S1E), suggesting that for highly amplified genes such as MYCN, the effect of frequent cutting is so great that additional effect of loss of gene function cannot be accurately assessed in CRISPR depletion screens. While CRISPR suffers from false positives in copy-gained regions, CRISPRi may produce false negatives for these same regions (Supplemental Figure S1F and G). These data provide support for the strategy of therapeutically exploiting highly amplified oncogenes, such as MYCN, with intron targeted sgRNAs. These guide RNAs were as effective as eliminating the oncogene and would be expected to have minimal effect in non-amplified cells as they should not disrupt gene function.
Figure 1. CRISPR screens prioritize rapidly lethal in vivo relevant dependencies.
A, Diagram of gene selection and library design for neuroblastoma specific secondary screens. B, Scaled gene level dependency scores calculated with CERES from the DepMap screen (y-axis) are compared with scaled gene level dependency scores calculated with MAGeCK in our secondary screens at the same time point (21 days). Data for all 214 genes and all four cell lines are plotted. The Pearson correlation coefficient and p-value are shown. Individual cell lines are plotted in Fig. S1A. C, Heat map of scaled MAGeCK depletion scores for all genes that scored as a “strong hit” (median of positive controls in Chronos algorithm, see methods) in at least one cell line in CRISPR. Genes where intronic copy number control guides also scored are excluded. Cell lines and are indicated at the bottom and time points are indicated at the top. Color scale is indicated at right, where a value of −1 is equivalent to the median of the positive controls for that cell line at day 21, and 0 is equivalent to the cutting controls at day 21. Day 7 data for CHP-212 is not included because there was not adequate separation of positive and negative controls at this time-point. D, Heat map of scaled MAGeCK depletion scores for all genes that scored as a “hit” (median of positive controls in Chronos) in any cell line using CRISPRi/dCas9-KRAB. Gene effects are normalized as in B. As in B, Day 7 data for CHP-212 is not included because there was not adequate separation of positive and negative controls to identify hits. E, Correlation of gene effects in CRISPR and CRISPRi screens. The Chronos algorithm was used to calculate a single gene effect score incorporating data from all three time points (see methods). The y-axis indicates this value for each gene in the CRISPRi screens, the x-axis indicates this value for these genes in the CRISPR screens for each cell line. Each dot represents a gene included in the library. The cell line name is indicated at the top, and the Pearson correlation coefficient and p-value are displayed. The best fit line is plotted with standard deviation in red. F, Diagram of Annexin V positive selection screen for lethal dependencies. Briefly, cells were infected, selected, and then 7 days after infection Annexin V positive cells were purified using Annexin V antibody conjugated to magnetic beads and a magnetic column. Both input and the purified population were sequenced. G, The log2 fold change in abundance for guides in the Annexin V positive population relative to input was collapsed to gene level using the STARs algorithm. On the x-axis Annexin V enrichment is plotted for the cell line KELLY. These values are plotted on the y-axis for the SKNDZ cell line. Each dot indicates an individual gene. In gray are the 197 putative neuroblastoma dependencies. In red are the 17 positive control pan-essential genes. Genes that had enrichment scores equal to or greater than the median of the positive controls in both cell lines are labeled with their gene names. H, Diagram of the in vivo subcutaneous xenograft screen I, Correlation between in vitro and in vivo CRISPR screens in KELLY. Gene-level depletion scores were calculated with MAGeCK and the depletion for each gene in vitro at day 21 is indicated on the y-axis. The MAGeCK depletion score for these same genes in the in vivo subcutaneous xenograft model is shown on the x-axis. A subset of genes that depleted in vitro but did not deplete in vivo are labeled with their gene names. Positive control genes are highlighted in red. Pearson correlation coefficient and p-value are shown. The best fit line is plotted with standard error in red.
To further narrow potential dependences to those that are lethal to neuroblastoma when lost, we performed a positive selection screen for Annexin V positive cells, a marker of dying cells (Figure 1F, Supplemental Figure S1H) (17,18). Disruption of only a few of the validated dependency genes induced cell death as measured by this assay, but we observed a striking consistency across cell lines (Pearson r=0.576, p<2.2e-16) with three genes, NXT1, HSPA8, and TYMS scoring as hits in both cell lines (Figure 1G). We next performed an in vivo CRISPR screen in a subcutaneous xenograft model of neuroblastoma to filter out dependencies specific to the cell culture environment (Figure 1H). The overall correlation between in vivo and in vitro in the same cell model was high (Pearson r= 0.774, p<0.0001). However, there was a notable class of outliers that did not deplete in vivo, suggesting that these dependencies may only occur in the context of cell culture (Figure 1I, Supplemental Figure S1I). Notably, the genes that did not validate in vivo were largely related to oxidative stress or metabolism (e.g. SOD2, GPX4, GLRX5, TYMS, SDHB, SEPHS2) suggesting that special care should be taken to validate dependencies in these classes in vivo before conducting further mechanistic experiments.
Nuclear export factor NXT1 is a selective and lethal dependency in neuroblastoma
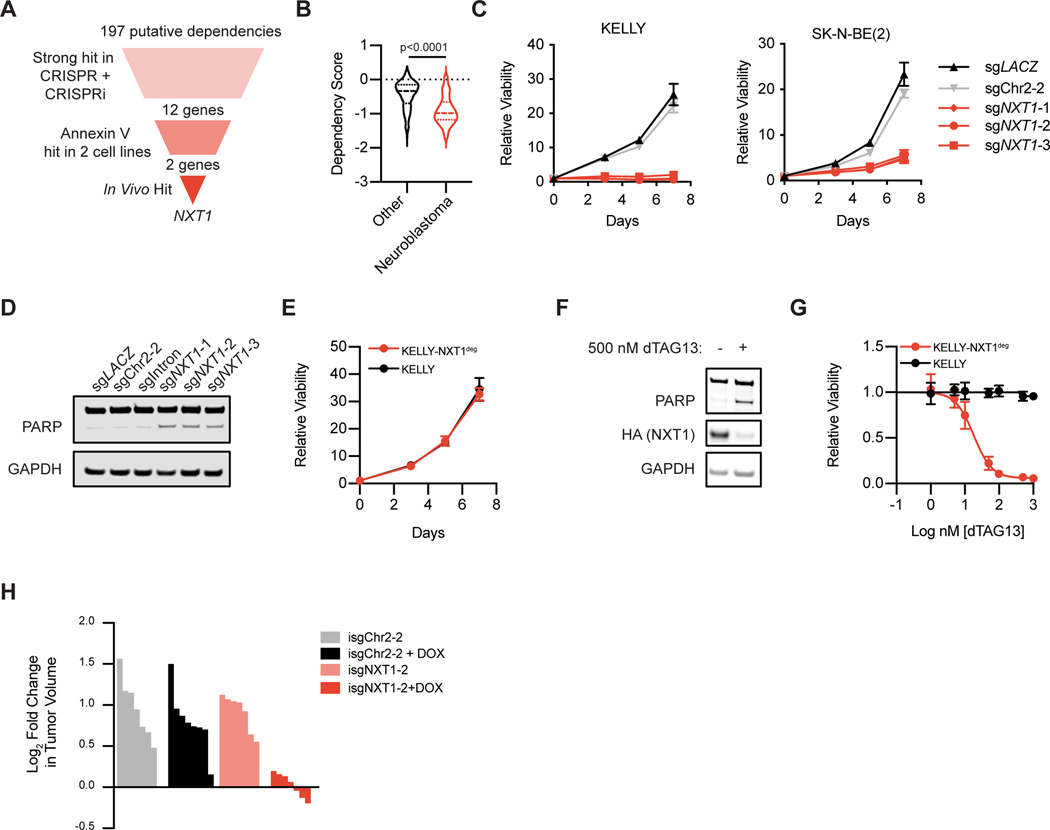
At the intersection of these screens, we identified one dependency gene, NXT1, which scored as a strong hit in both CRISPR and CRISPRi time-course screens, scored in the Annexin V screens in both cell lines, and validated in vivo (Figure 2A). NXT1, the protein encoded by this gene, is a nuclear export factor whose canonical function is as a co-factor for NXF1-mediated export of mRNAs from the nucleus to the cytoplasm where they can be translated into proteins (19). The DepMap dataset has more than doubled since we generated our gene list, but as in the smaller dataset, neuroblastoma cell lines (n=19) are significantly more dependent on NXT1 when compared to all other cancer cell lines (n=720) suggesting this is a selective effect (Figure 2B). We confirmed on-target genomic editing by three CRISPR guides targeting NXT1 (Supplemental Figure S2A) and validated that these sgRNAs induced a profound viability defect in neuroblastoma cell lines compared to control sgRNAs, as well as cell death measured by Annexin V/PI staining and cleavage of PARP (Figure 2C–D, Supplemental Figure S2B).
Figure 2. NXT1 is a selective and lethal dependency in neuroblastoma.
A, Schematic of hit filtering strategy for secondary screens which identifies NXT1 as a top dependency. B, A violin plot of NXT1 dependency scores from the DepMap dataset. In red, are all neuroblastoma cell lines (n=19). In black are all other screened cell lines (n=720). The dashed line indicates the median for each, and the quartiles are shown as dotted lines. A two-tailed Mann-Whitney test was performed to compare the two groups, and the p-value is shown. C, Dependency on NXT1 was validated in two different neuroblastoma cell lines using three constitutive CRISPR guides. Non-targeting (sgLACZ) and cutting control (sgChr2–2) serve as negative controls. Viability was assessed using CellTiter-Glo on the days indicated and normalized to day 0. Data points are mean +/− stdev. D, Western blot showing PARP and GAPDH in SK-N-BE(2) cells 6 days after infection with the indicated constitutive CRISPR guides. sgIntron is an sgRNA that targets an intron in the 5’ UTR of NXT1 and is not expected to disrupt gene function (see methods). E, Relative viability of KELLY parental (black) and KELLY-NXT1deg (red) is shown, as in C. KELLY-NXT1deg is a cell line where endogenous NXT1 has been knocked out with CRISPR and exogenous NXT1 c-terminally tagged with HA and an FKBP12F36V degradation tag is expressed (see methods). F, Western blot of PARP, HA, and GAPDH in KELLY-NXT1deg with and without 24 hours of 500 nM dTAG13 treatment. G, Dose response curve to dTAG13 treatment in KELLY parental (black) and KELLY-NXT1deg (red). Data points indicate mean +/− stdev. H, Loss of NXT1 causes regression in a subset of established tumors. Subcutaneous xenografts with inducible sgRNAs targeting Chr2 or NXT1 as indicated were allowed to form and then randomized to doxycycline (+DOX) or normal chow conditions. Waterfall plot depicts the log2fold change in tumor volume four days after randomization. Each bar represents an individual tumor.
As further validation, we employed the FKBP12-based degron system for inducible protein degradation (20). We created a cDNA of NXT1 in which a silent mutation had been introduced into the PAM site for one of our CRISPR guides (sgNXT1-3) rendering it resistant to CRISPR editing, and it was fused to the FKBP12F36V fragment and an HA tag (NXT1deg) (Supplemental Figure S2C). We generated neuroblastoma cell lines expressing this construct and knocked out endogenous NXT1 using CRISPR-Cas9 (Supplemental Figure S2D and E). Cells with endogenous knockout of NXT1 and exogenous expression of degradable NXT1 grew normally, indicating that tagged NXT1 expression can indeed rescue the effect of NXT1 knockout (Figure 2E). Treatment with the cereblon-recruiting degrader molecule dTAG-13 caused efficient degradation of exogenous NXT1, induction of cleaved PARP, and a profound dose-responsive viability effect (Figure 2F and 2G, Supplemental Figure S2F and G), while a negative control molecule, dTAG-13-NEG, which binds FKBP12F36V but cannot recruit cereblon (21), had no effect on viability (Supplemental Figure S2H and I).
The in vivo screen confirmed that this dependency is not an artifact of cell culture but did not evaluate the role of NXT1 in an established tumor. To address this question, we generated neuroblastoma cells constitutively expressing Cas9 with sgRNAs under the control of a doxycycline inducible promoter (22). In tumors with an inducible sgRNA targeting NXT1, we observed arrest of tumor growth in all tumors and tumor regression in a subset of tumors within four days of doxycycline induction, indicating that there is indeed a profound anti-tumor effect of NXT1 loss (Figure 2H, Supplemental Figure S2J). We do not observe these same effects in doxycycline-treated tumors harboring inducible sgChr2–2, demonstrating that this is an on-target effect of NXT1 loss (Figure 2H and Supplemental Figure S2K). Further supporting that NXT1 loss is incompatible with tumor cell survival, the tumor cells that do grow out after sgNXT1 induction lack expression of Cas9, show a significant selection against edits in NXT1, and among those edits, a significant selection for in-frame edits that are not predicted to be damaging (Supplemental Figure 2L–N).
NXT1 loss leads to loss of the essential protein NXF1
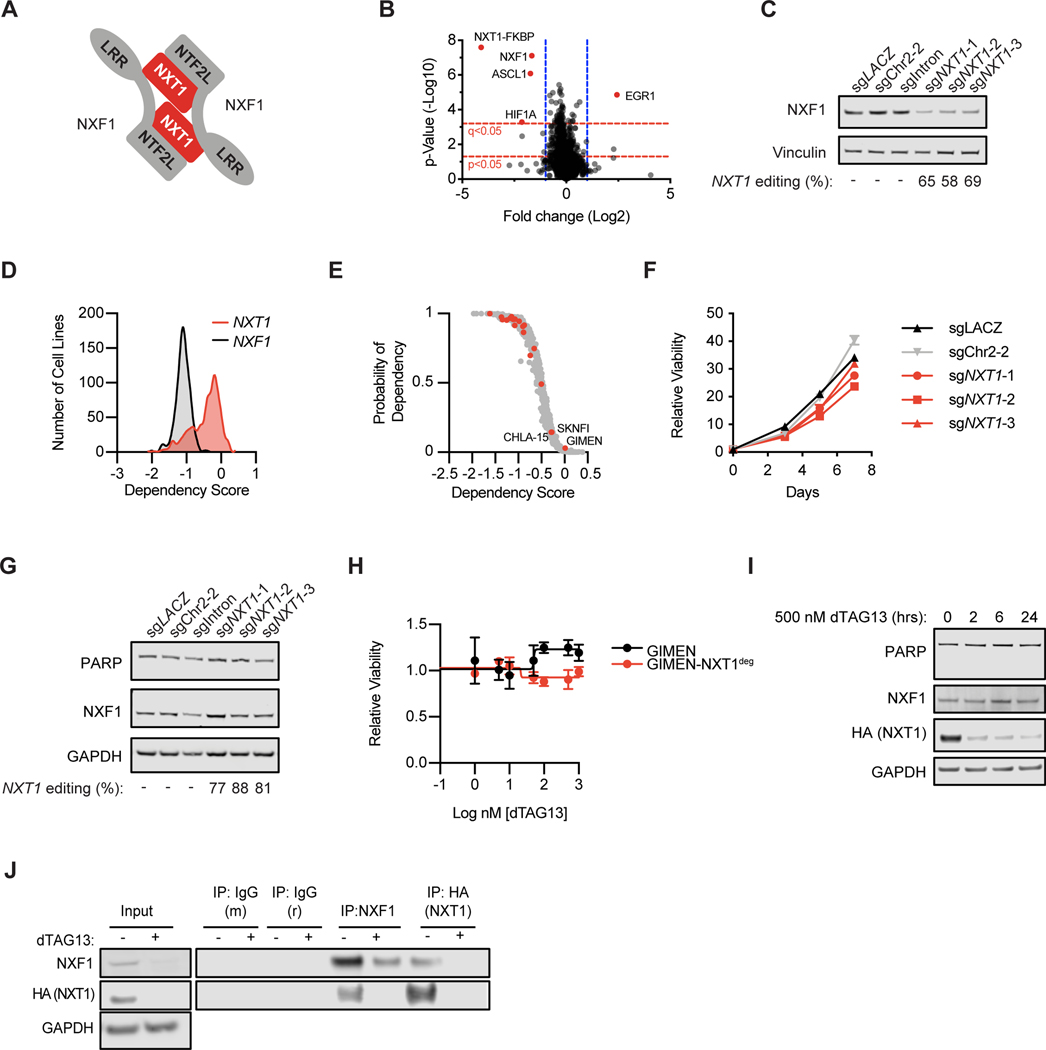
NXT1 binds to the NTF2L domain of NXF1 and together they form a heterodimer that binds mRNA and mediates nuclear export (Figure 3A) (23). Mass spectrometry-based quantitative multiplexed proteomics revealed that in our inducible degron system, when NXT1 is degraded, NXF1 protein levels also decrease rapidly (Figure 3B). NXF1 levels also decrease after CRISPR-mediated knockout of NXT1, demonstrating this observation was not due to off-target degradation (Figure 3C, Supplemental Figure S3A). In the DepMap dataset, NXF1 is a common essential, whereas NXT1 shows a skewed distribution with many lines dependent on NXF1 but not NXT1 (Figure 3D). Therefore, we were interested in whether NXF1 remains stable in cells that are not dependent on NXT1. Indeed, we identified a small number of neuroblastoma lines that are not dependent on NXT1 and determined that NXF1 levels remain stable after CRISPR knockout or inducible degradation of NXT1 (Figure 3E–I, Supplemental Figure S3B–C.) NXT1 loss therefore leads to elimination of the essential protein NXF1 in a context-specific manner, which explains the profound yet selective lethality observed.
Figure 3. NXT1 loss leads to loss of NXF1.
A, Diagram of NXT1 and NXF1 heterodimer (adapted from Aibara et al. (23)). B, Quantitative proteomics after 2 hours of dTAG13 treatment in KELLY-NXT1deg. X-axis indicates log2 of the relative protein abundance of mean dTAG13-treated to mean control (DMSO) samples. Y-axis indicates the -log10 of the p-value. Red dotted lines indicate q- and p-value cut-offs as indicated, and blue dotted lines indicate a cut-off of log2 fold change of 1. Proteins that reach these cut-offs are labeled. C, Western blot of NXF1 and GAPDH levels in SK-N-BE(2) cells five days after infection with constitutive CRISPR guides as indicated. Genetic editing of the NXT1 locus, calculated using TIDE, is shown below. D, Spline curves of dependency score distribution for all 739 cell lines in the DepMap data set. In red is NXT1 and in black is NXF1. Dependency scores are calculated with CERES and scaled such that −1 is equivalent to pan-essentials, and 0 is equivalent to negative control guides. E, NXT1 dependency in the DepMap dataset for neuroblastoma and other cell lines is shown. On the y-axis is the probability of dependency on NXT1 for each cell line, and on the x-axis is the dependency score calculated by CERES. Each point represents an individual cancer cell line. In red, neuroblastoma cell lines (n=19) are shown. In gray, other cell lines (n=720) are shown. Three neuroblastoma cell lines that do not show dependency on NXT1 are labeled with their cell line names. F, GIMEN cells were infected with negative control (sgLACZ or sgChr2–2, black and gray) and three different sgRNAs targeting NXT1 (red). Viability relative to day 0 was determined using CellTiter-Glo on the indicated days and mean +/− stdev of replicates is shown. G, GIMEN cells were stably infected with indicated sgRNAs and a western blot assessing NXF1, PARP, and GAPDH levels was performed. Below, the percent of CRISPR editing at the NXT1 locus calculated with the TIDE algorithm is shown. H, Dose-response curve for GIMEN parental (black) and GIMEN-NXT1deg (red) where endogenous NXT1 has been knocked out, and a degradable exogenous form of NXT1 is expressed (see methods). I, Western blot showing PARP, NXF1, HA-NXT1, and GAPDH levels after treatment with 500 nM dTAG13 for the indicated amount of time in GIMEN-NXT1deg. J, Immunoprecipitations were performed using antibodies targeting mouse and rabbit IgG, NXF1 (mouse) and HA (rabbit) six hours after DMSO or 500 nM dTAG13 treatment, as indicated in the KELLY-NXT1deg cell line. NXF1 and HA levels are shown detecting endogenous NXF1 and HA-tagged NXT1 respectively. Input lysate is shown to the left with NXF1, HA, and GAPDH.
We then sought to understand why NXF1 levels decrease after NXT1 loss in sensitive cells. As NXT1 and NXF1 are known to heterodimerize in other contexts (23), we first confirmed a physical interaction between NXT1 and NXF1 in neuroblastoma by co-immunoprecipitation (Figure 3J and Supplemental Figure 3D). NXF1 mRNA does not decrease after NXT1 loss. Instead, there is a modest early compensatory increase in NXF1 mRNA followed by a return to baseline at 24 hours after NXT1 degradation, consistent with regulation of NXF1 at the protein rather than transcript level (Supplemental Figure 3E–F). We therefore hypothesized that NXF1 might be an obligate heterodimer in this setting and require NXT1 binding for stability. Indeed, when human NXF1 is exogenously expressed in 293 cells, co-expression of exogenous NXT1 significantly increases NXF1 protein levels, supporting a role for NXT1 in NXF1 protein stability (24). To interrogate this possibility, we blocked new protein translation with cycloheximide treatment and assessed the stability of the NXF1 protein. In the presence of NXT1, NXF1 was remarkably stable with sustained protein levels up to 24 hours after cycloheximide treatment, while, in the absence of NXT1, NXF1 protein levels decrease rapidly, consistent with a loss of protein stability (Supplemental Figure S3G). Together these data support a model where NXF1 is an obligate heterodimer, and NXF1 is destabilized at the protein level after NXT1 loss in sensitive cell lines.
Low NXT2 expression is necessary and sufficient for NXT1 dependency in neuroblastoma
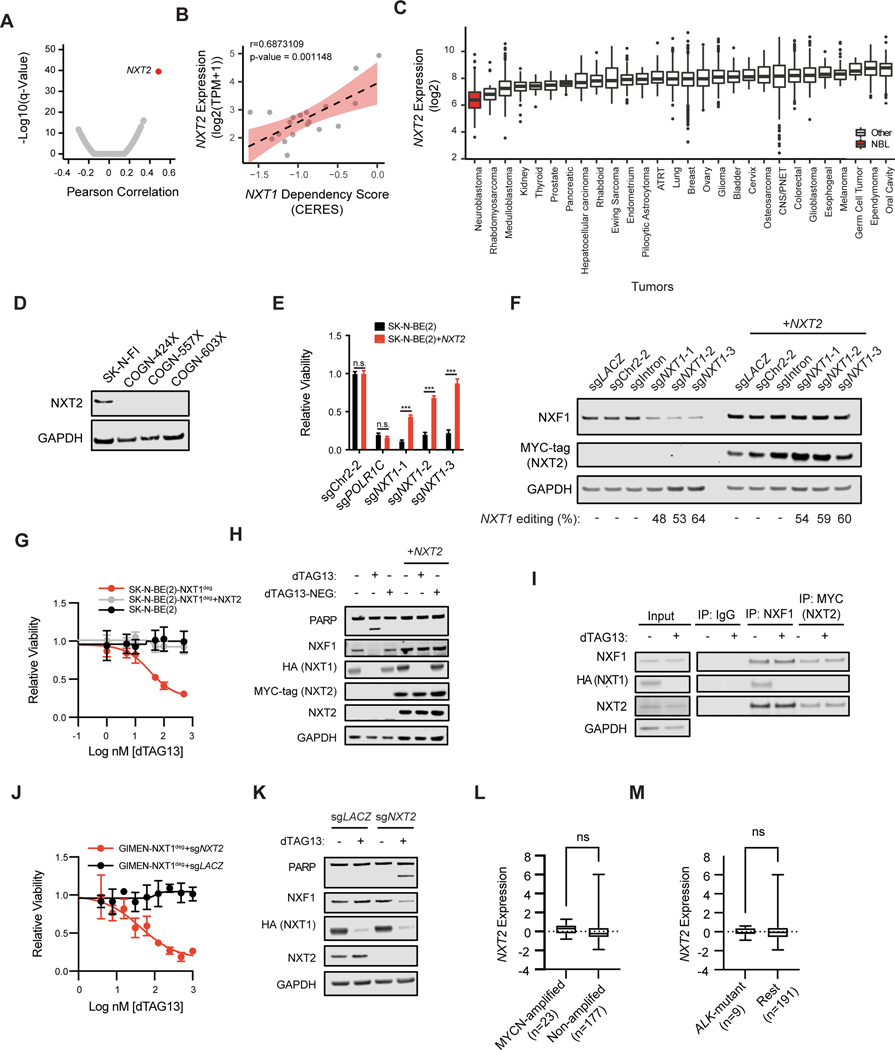
To understand the mechanism underlying this difference in NXF1 stability in sensitive and resistant cell lines, we looked for associations in gene expression with NXT1 dependency across the entire DepMap dataset of over 700 cancer cell lines. Lower expression of NXT2, a paralog of NXT1, is strongly associated with dependency on NXT1, and this relationship is maintained within neuroblastoma cell lines (Figure 4A and B, Supplemental Figure S4A). In an independent large CRISPR dependency dataset generated by the Sanger Institute, this relationship between NXT2 expression and NXT1 dependency is also observed (Supplemental Figure S4B). NXT2 is poorly expressed in neuroblastoma cell lines and primary human tumors relative to other cancer types in multiple primary tumor datasets, consistent with the enrichment of NXT1 dependency in neuroblastoma (Figure 4C and Supplemental Figure S4C and D). Furthermore, we could not detect NXT2 protein in three patient-derived xenograft models of neuroblastoma (Figure 4D). In contrast, NXT2 is expressed in most normal tissues (Figure S4E).
Figure 4. Low NXT2 expression is necessary and sufficient for NXT1 dependency in neuroblastoma.
A, Pearson correlations were calculated for gene expression and NXT1 dependency for all genes and cell lines in the DepMap dataset. Volcano plot indicates the Pearson correlation on the x-axis and the -log10 of the q-value on the y-axis. Each dot indicates a different gene. In red, NXT2, the gene whose expression is most significantly associated with NXT1 dependency is shown. B, The correlation between NXT2 expression and NXT1 dependency in neuroblastoma cell lines in DepMap is shown. The y-axis indicates the expression of NXT2 in log2 transcripts per million (TPM) +1. The x-axis indicates the scaled-CERES score for NXT1. Each data point indicates a different neuroblastoma cell line (n=19). The Pearson correlation coefficient and p-value are shown at the top. The best-fit line is plotted as a dotted line. C, NXT2 expression (log2 transformed, y-axis) was assessed across the R2 database of microarray data (u133p2, MAS5.0) from patient tumor samples. Data sets in the same cancer type were collapsed for clarity. Cancer type is shown on the x-axis. Sample sizes are as follows: Neuroblastoma (n=233), Breast (n=1994), ATRT (n=67), Bladder (n=93), Cervix (n=351), CNS/PNET (n=206), Colorectal (n=2182), Endometrium (n=209), Ependymoma (n=376), Esophogeal (n=40), Ewing Sarcoma (n=154), Germ Cell Tumor (n=13), Glioblastoma (n=200), Glioma (n=607), Hepatocellular Carcinoma (n=91), Kidney (n=261), Lung (n=514), Medulloblastoma (n=350), Melanoma (n=44), Oral Cavity (n=103), Osteosarcoma (n=27), Ovary (n=787), Pancreatic (n=32), Pilocytic Astrocytoma (n=41), Prostate (n=71), Rhabdoid (n=51), Rhabdomyosarcoma (n=58), Thyroid (n=34). D, Western blot showing NXT2 and GAPDH levels in three neuroblastoma patient-derived xenograft models (COGN-424X, COGN-557X, COGN603X). The NXT2-high cell line SK-N-FI serves as a positive control. E, The relative viability effect of the indicated sgRNAs after seven days of growth is shown as the mean +/− stdev. In black is SK-N-BE(2) parental, in red is SK-N-BE(2) over-expressing a MYC-tagged NXT2. sgPOLR1C-1 targets the essential gene POLR1C. n.s. indicates a p-value of >0.05 in a two-way ANOVA followed by Tukey’s multiple comparison test, while *** indicates a p-value of <0.001 (degrees of freedom =50). F, Western blot depicting NXF1, MYC-tagged exogenous NXT2, and GAPDH levels in SK-N-BE(2) cells 5 days after infection with the indicated CRISPR guides. CRISPR editing of the NXT1 locus is shown below as determined using the TIDE algorithm. G, Dose response curve for dTAG13 in parental SK-N-BE(2) (black) SK-N-BE(2)-NXT1deg (red, endogenous NXT1 knocked out, degradable NXT1 exogenously expressed), and SK-N-BE(2)-NXT1deg+NXT2 (endogenous NXT1 knocked out, degradable NXT1 exogenously expressed, and NXT2 exogenously expressed). H, Western blot depicting PARP, NXF1, HA-NXT1, MYC-tagged NXT2, and GAPDH levels after 24 hours of treatment with DMSO, 500 nM dTAG13, or 500 nM of dTAG13-NEG, an inactive form of dTAG13, as indicated at the top. At left is SK-N-BE(2)-NXT1deg, at right SK-N-BE(2)-NXT1deg+NXT2. I, Immunoprecipitations were performed using antibodies targeting mouse IgG, NXF1 and MYC (NXT2) six hours after DMSO or 500 nM dTAG13 treatment, as indicated in the KELLY-NXT1deg cell line overexpressing NXT2. Input lysate is shown to the left. NXF1, NXT2, HA (NXT1), and GAPDH levels are shown. J, Dose response curve for dTAG13 in GIMEN- NXT1deg with a non-targeting sgRNA (sgLACZ, black) and GIMEN-NXT1deg with a sgRNA targeting NXT2 (sgNXT2, red) after 72h of treatment. Y-axis indicates relative viability and x-axis indicates log of nM dose of dTAG13. K, Western blot in GIMEN- NXT1deg after infection with indicated sgRNA 24 hrs after treatment with DMSO or 500 nM dTAG13 as indicated. L, NXT2 expression Z-score for 200 neuroblastoma tumor samples in the Gabriella Miller Kid’s First (GMKF) dataset is shown on the y-axis. Samples are binned according to MYCN copy number as indicated. A t-test with Welch’s correction was not significant (n.s.). M. As in L, NXT2 expression for the GMKF dataset is shown on the y-axis, and samples are binned based on whether they contain a putative driver mutation in ALK. A t-test with Welch’s correction was not significant (n.s.).
NXT2 is a paralog of NXT1, and the two proteins share 73.2% identity and 88.7% similarity (25). NXT2 more closely resembles the gene structure of homologues in other organisms than NXT1, which lacks introns (24). NXT2 is functionally redundant with NXT1 in a constitutive transport element (CTE) containing viral RNA export assay, and NXT2 heterodimerizes with NXF1 with a similar affinity as that of NXT1 (24,26). Complete functional redundancy between these two paralogs has not been established to date. Zebrafish lacking NXT2 have been reported to have cardiac defects as a result of abnormal development; however, mice lacking NXT2 are viable and fertile with no reported cardiac defects (27,28), suggesting that NXT1 may be able to compensate for NXT2 in murine development, indicative of some functional redundancy. When NXT2 is exogenously expressed in three different neuroblastoma cell lines that have low endogenous NXT2 expression, NXF1 levels remain stable and cell viability is restored after NXT1 loss through CRISPR knockout or inducible degradation (Figure 4E–H, Supplemental Figure S4F–I). We confirmed that NXT2 physically interacts with NXF1, and this association is maintained after NXT1 loss in neuroblastoma (Figure 4I and Supplemental Figure S5A), raising the possibility that NXT2 binding could stabilize NXF1 in the absence of NXT1. Indeed, after blocking translation with cycloheximide, NXT2 expression restored NXF1 stability after NXT1 loss, consistent with protein stabilization (Supplemental Figure S5B). Together these data demonstrate that NXT1 and NXT2 have a functionally redundant role in maintaining NXF1 protein stability in neuroblastoma.
We next confirmed that resistant neuroblastoma cell lines express NXT2, and indeed endogenous NXT2 protein was readily detected in cell lines that do not depend on NXT1 but was undetectable in cell lines that do depend on NXT1 (Supplemental Figure S5C). Publicly available epigenetic data revealed H3K4me3 marks at the transcriptional start site for full-length NXT2 and an open chromatin conformation only in neuroblastoma cell lines that highly express NXT2 (Supplemental Figure S5D and E). An internal start site for a shorter isoform of NXT2 is configured similarly across cell lines, but expression of this isoform is uniformly low in neuroblastoma cell lines and not associated with NXT1 dependency, while expression of the full-length isoform is highly variable and was strongly associated with NXT1 dependency in neuroblastoma (Supplemental Figure S5F). Bisulfite sequencing of a previously described string of CpGs in the promoter region of full-length NXT2 uncovered differential methylation patterns between resistant cell lines with high NXT2 expression, which lacked methylation at most CpGs, and sensitive cell lines with low NXT2 expression, which were highly methylated (Supplemental Figure S5G) (29). Notably, one of these differentially methylated CpGs is within an ETS family recognition sequence, and methylation of the CpG within this sequence has been shown to prevent transcription factor binding in other contexts (30–33). Samples from patient-derived xenograft models that lack NXT2 also had methylated CpGs, and the association between methylation of CpGs in this region, as assessed by DNA methylation array, and NXT2 expression is maintained in neuroblastoma tumors (Supplemental Figure S5G–H).
In order to establish whether reduced NXT2 expression is sufficient to render resistant neuroblastoma cells sensitive to NXT1 loss, we knocked out NXT2 in two resistant neuroblastoma lines, GIMEN and SK-N-FI, which have high endogenous NXT2 expression. Loss of NXT2 rendered these cells sensitive to subsequent loss of NXT1 by CRISPR knockout or inducible degradation, and NXF1 protein levels also decreased after NXT1 loss (Figure 4J and K, Supplemental Figure S5I–K). These data support a mechanism of NXT1 selective lethality whereby the cell essential protein NXF1 is lost upon NXT1 loss in cells that do not express NXT2, and that relatively low levels of NXT2 expression are both necessary and sufficient for NXT1 dependency in neuroblastoma.
Neither GIMEN nor SK-N-FI harbor MYCN amplifications but NXT2 loss is sufficient to render these lines dependent on NXT1, which suggests that neuroblastoma with endogenously low NXT2 would depend on NXT1 regardless of MYCN amplification status. We were therefore interested in whether NXT2 expression is associated with any common neuroblastoma genetic alterations, including MYCN amplification. Neither MYCN amplification nor ALK mutations correlate with NXT2 expression in neuroblastoma tumors, suggesting that response to loss of NXT1 may not be restricted by disease subtype (Figure 4L and M, Supplemental Figure S6A) (34,35). Recently described adrenergic and mesenchymal cell states similarly did not correlate with NXT2 expression (Supplemental Figure S6B). These data suggest that non-amplified tumors that lack NXT2 will likely be sensitive to NXT1 loss, although we cannot rule out that MYCN-amplification contributes to the dependence on NXT1 independently of NXT2 expression levels.
Pediatric cancers have low expression of NXT2 and are more dependent on NXT1 than adult cancers
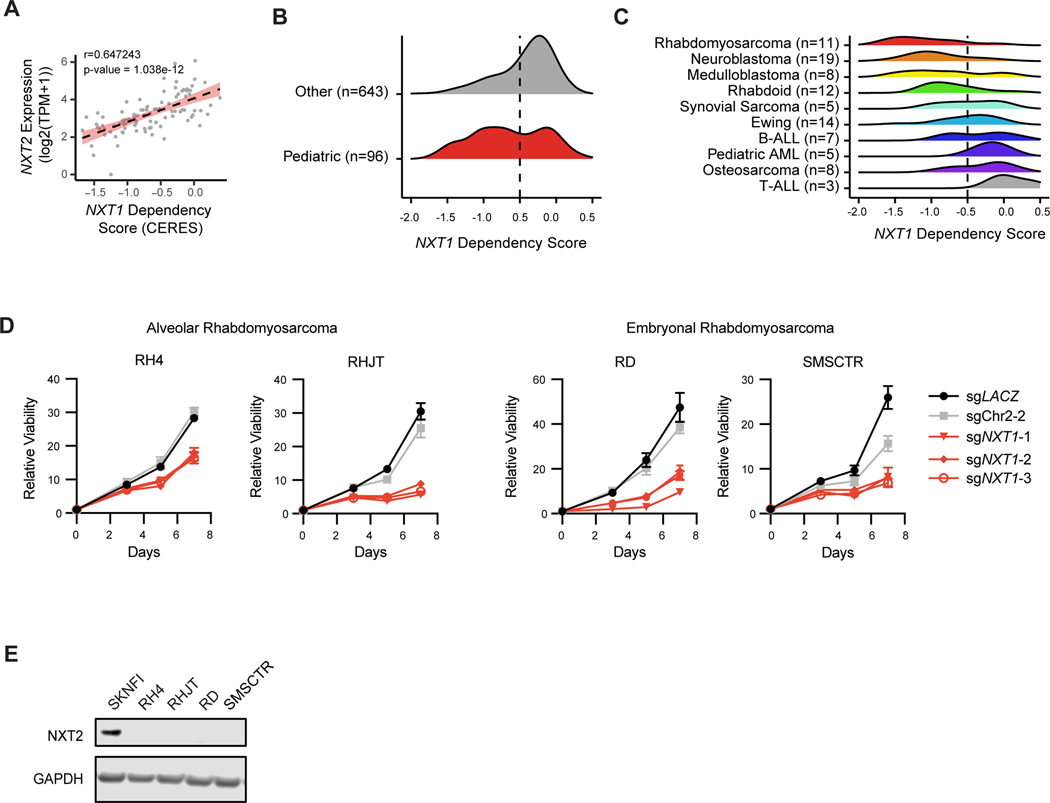
The profound and precise lethality of NXT1 loss in neuroblastoma indicates that it might be a good candidate for therapeutic exploitation, and therefore we sought to determine whether other cancers might also be vulnerable to NXT1 loss. Strikingly, in human tumor expression data, we observed that medulloblastoma and rhabdomyosarcoma, two other pediatric cancers, also had very low expression of NXT2 (Figure 4C and Supplemental Figure S4D). Accordingly, the paralog relationship is conserved in pediatric cancer cell lines (Figure 5A), and pediatric cancer cell lines are more likely to be dependent on NXT1 than adult cancer cell lines (Figure 5B), a difference driven by multiple lineages, including neuroblastoma, rhabdomyosarcoma, medulloblastoma, as well as rhabdoid tumor cell lines (Figure 5C). To further validate this finding, we confirmed that both embryonal and alveolar rhabdomyosarcomas lack NXT2 expression and are dependent on NXT1 (Figure 5D and E).
Figure 5. Other pediatric cancer types have low NXT2 and are dependent on NXT1.
A, Correlation between NXT2 expression (log2 (TPM+1)) and NXT1 dependency in pediatric cancer cell lines in DepMap (n=96). Y-axis indicates expression of NXT2 (log2 (TPM+1)). X-axis indicates scaled-CERES score for NXT1. Pearson correlation coefficient and p-value are shown at the top. The best-fit line is plotted as a dotted line, with standard error in red. B, Gene effect distributions for NXT1 in pediatric cancer cell lines (n=96, black) and adult cancer cell lines (n=643 gray). Dashed red line at −0.5 indicates the gene effect score cut-off to be considered a dependency C, Gene effect distributions for NXT1 are shown for all pediatric cell lines included in B, separated by lineage as indicated. Cell line names are at top, and cell lines are grouped by their disease subtype. D, Viability effects of negative control sgRNAs (sgLACZ and sgChr2–2) and sgRNAs targeting NXT1 in four different rhabdomyosarcoma cell lines. Cell lines were infected with indicated constitutive CRISPR guides and then viability relative to day 0 was assessed using CellTiter-Glo at indicated time points. Mean relative viability +/− stdev of technical replicates is shown. E, Western blot depicting GAPDH and NXT2 levels in rhabdomyosarcoma cell lines. The NXT2 high expressing cell line SK-N-FI serves as a control.
Discussion:
Loss of fitness, as assayed in late time-point genome-scale dropout screens, such as in the DepMap, is a very broad phenotype that can detect minor viability effects caused by disruption of a variety of cellular processes. We provide a framework for mining this data to generate a focused, context-specific library, and for designing follow-up screens that can interrogate precise cell processes. Here we demonstrate that additional CRISPR screening assays can yield critical information about the speed, lethality, and in vivo translatability of many targets simultaneously in a given disease context. Similar approaches could be fruitfully applied to identify genes involved in differentiation or cell cycle arrest with positive selection assays, for example. Critically, our data suggest that the original screens are highly reproducible, and results are mostly representative of an in vivo setting, although attention should be paid to genes involved in oxidative stress or metabolic processes to verify their in vivo validity.
Our functional genomic screens nominate NXT1 as a precision lethal dependency that functions as an essential gene in neuroblastoma but is dispensable in most other cancer cells. Neuroblastoma, like many other pediatric cancers, is currently treated with high-dose cytotoxic chemotherapy, which is curative for a subset of patients but at the cost of both short-and long-term side effects (36). Accordingly, the targets of these chemotherapies include proteins, such as TOP1 and TOP2A, which behave as common essential genes in the DepMap. NXT1, in contrast, is dispensable in most cell lines so therapeutic targeting of NXT1 might be expected to be more tolerable than current curative options without a reduction in efficacy.
An important caveat of these data is that “normal” cells cannot readily be screened in vitro without long-term propagation and modifications rendering them no longer truly representative of normal tissue. The DepMap, therefore, uses all other cancer cell lines as a proxy for assessing pan-essentiality across cell types. However, it is not yet known whether selectivity in this data set is necessarily indicative of selectivity relative to normal tissues in an organism. This is a key question that must be addressed at the organismal level with either conditional knockout animals or a selective molecule targeting NXT1, once it is available. The relatively ubiquitous expression of NXT2 in adult normal tissue, and the selectivity of the target in the DepMap, provide optimism that a therapeutic window can be achieved even if the NXT1-NXT2 synthetic lethal paralog relationship is retained in most normal tissue. Gene expression patterns in normal developing tissues in children have yet to be carefully profiled, and it is possible that NXT2 is not expressed as highly in developing pediatric tissues as it is in fully mature adult tissue. Efforts are underway to create a normal tissue expression database for pediatric tissues, but until such a resource exists, this limitation will continue to impair target discovery efforts in pediatrics (37).
We purposely did not include any filters for “druggability” in the criteria for inclusion in our sgRNA library because the notion of what constitutes a druggable target is rapidly evolving. New approaches, such as targeted protein degradation using a PROTAC or molecular glue approach, as well as protein-protein interaction disruptors, have expanded the realm of druggable targets, and any of these approaches could potentially be employed to selectively target NXT1 (38–40). Developing new therapeutics using these strategies is not trivial, so pursuing the targets most likely to provide maximal anti-cancer activity with a broad therapeutic window is key. We demonstrate the power of genome-scale dependency datasets for identifying such targets in cancer and suggest that creative application of additional functional genomic CRISPR screens to other cancer types could help appropriately harness this resource to direct therapeutic development in cancer.
We demonstrate that NXT1 is in a synthetic lethal paralog relationship with NXT2 due to their regulation of the stability of the essential protein NXF1. NXF1 protein levels only decrease in the absence of both NXT1 and NXT2. NXT1 and NXT2 have a high level of similarity and have been shown to be functionally redundant in mRNA export reporter assays. Exogenously expressed NXF1 in 293 cells can promote mRNA export alone in these assays, but its protein stability and export function are increased with the addition of either exogenous NXT1 or NXT2 (24). We show that NXT1 and NXT2 similarly promote endogenous NXF1 protein stability in neuroblastoma cells. The NXT2 knockout mouse has no overt phenotype, suggesting that NXT1 can functionally replace NXT2 (28). Our data provide evidence that NXT2 can similarly replace NXT1 in maintenance of NXF1 stability. An intron-retaining, truncated isoform of NXF1 has also been proposed to be functionally redundant with NXT1 in mRNA export assays, and whether this or other undiscovered mechanisms of replacing NXT1 function beyond NXT2 occur in other cellular settings will be important to determine (41,42). Our data, however, suggest that within neuroblastoma, expression of either NXT1 or NXT2 is necessary and sufficient for NXF1 stability.
These data support a model for a previously unrecognized mechanism of synthetic lethality whereby a paralog relationship is explained by the loss of stability of a common essential binding partner. The larger DepMap dataset may contain other examples in which modulation of a gene is able to regulate stability of another essential complex member in a context-specific manner and identifying these relationships could yield other promising therapeutic targets. As NXF1 is a common essential gene, direct targeting is anticipated to be broadly toxic, but through exploiting the NXT1-NXT2 paralog relationship we can restrict these effects to cells with low NXT2. We therefore propose a novel therapeutic strategy of indirect but precise targeting of a common essential protein specifically in cancer cells by exploiting this paralog relationship.
Finally, these data suggest that the mechanism of NXT1 dependency is maintained in other low NXT2 tumor types, and if NXT1 can be effectively targeted, this could have broad benefit for children with several different incurable cancers. While adult cancers do not display the same lineage enrichment of low NXT2 expression, there are adult cancer cell lines that have low NXT2, and perhaps a subset of adult cancer patients would similarly benefit from therapeutic exploitation of this target. A key remaining question is why pediatric cancers tend to have low expression of NXT2, while it is more ubiquitously expressed in adult cancers and normal tissues. One possible explanation is that pediatric cancers arise, by definition, through a failure of differentiation, and low NXT2 expression could be more common in undifferentiated cells present during development that give rise to pediatric cancers. Adult cancers, by contrast, generally arise due to an accumulation of mutations in developed tissue where high NXT2 expression may be more common. Future studies will be required to better understand the regulation of NXT2 expression, as well as potential intra-tumoral heterogeneity of expression. Our finding also reinforces the notion that repurposing of adult oncology drugs for pediatric patients will not always be an effective strategy for drug development, as NXT1 dependency is much more frequently observed in pediatric tumors.
Methods
Data Availability
All genome-scale dependency data used in this analysis is publicly available for download at depmap.org and figshare.com [https://figshare.com/articles/dataset/DepMap_20Q1_Public/11791698]. All datasets generated in this study are made publicly available at figshare.com [https://figshare.com/s/85c7d5f316522767ff43].
Cell Lines
All neuroblastoma cell lines were collected by the Cancer Cell Line Encyclopedia (RRID:SCR_013836) and DepMap (Cancer Dependency Map Portal, RRID:SCR_017655) projects as were RD, and SMSCTR. The sources for these lines are listed at DepMap.org, and they can be obtained from their respective sources. Their identities were confirmed by single nucleotide polymorphism (SNP) array. RHJT and RH4 were generously provided by Dr. Thomas Look, at Dana-Farber Cancer Institute, Boston, MA. HT-29s were generously provided by Dr. Karen Cichowski and Brigham and Women’s Hospital, Boston, MA. Cell lines were confirmed negative for mycoplasma infection with a most recent test date of 12-18-20 (Lonza MycoAlert). Cell line identities were re-confirmed by short tandem repeat (STR) profiling (Genetica). For publicly available STR profiles, matching was performed using the DSMZ STR profile database (43). KELLY (DSMZ Cat# ACC-355, RRID:CVCL_2092), GIMEN (DSMZ Cat# ACC-654, RRID:CVCL_1232), RD (ATCC Cat# CRL-7731, RRID:CVCL_1649), SMSCTR (RRID:CVCL_A770), and SIMA (DSMZ Cat# ACC-164, RRID:CVCL_1695) were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS). CHP-212 (ATCC Cat# CRL-2273, RRID:CVCL_1125) and SK-N-BE(2) (ATCC Cat# CRL-2271, RRID:CVCL_0528) were cultured in a 1:1 mix of MEM:F12 supplemented with 10% FBS. SK-N-DZ (ATCC Cat# CRL-2149, RRID:CVCL_1701) and SK-N-FI (ATCC Cat# CRL-2142, RRID:CVCL_1702) were cultured in DMEM supplemented with 10% FBS and non-essential amino acids (10 μM Glycine, 10 μM L-Alanine, 10 μM L-Asparagine, 10 μM L-Aspartic acid, 10 μM L-Glutamic Acid, 10 μM L-Proline, 10 μM L-Serine). RH4 (RRID:CVCL_5916) and RHJT (RRID:CVCL_VU81) were cultured in DMEM supplemented with 10% FBS.
Generation of neuroblastoma specific gene library
To create a gene list of putative dependencies, the CRISPR Avana dependency gene effect scores generated in mid-2017 using the CERES algorithm was used [https://figshare.com/s/85c7d5f316522767ff43]. For the purposes of this analysis only neuroblastoma cell lines with MYCN amplification (n=9) were used. Non-amplified lines were excluded from the analysis (n=2), and all non-neuroblastoma lines (n= 331) were used for comparison. Inclusion criteria was modeled after that in Durbin et al., but modified to be more inclusive (44). Genes had to meet at least one of these criteria: a) >4 sigma dependency in mean centered CERES in at least two MYCN-amplified neuroblastoma cell lines, b) >3 sigma outlier in mean centered CERES in at least three MYCN-amplified neuroblastoma cell lines, c) enriched in MYCN-amplified neuroblastoma with an empiric p-value <0.0005. Genes were then filtered to require that the gene effect score in CERES was <−0.3. Common essentials were defined as genes for which at least 25% of cell lines had a gene effect score <−0.6 in CERES and the 95th percentile was <−0.4 in CERES. Genes meeting this definition were filtered out. Genes were then filtered for expression in neuroblastoma cell lines and excluded if log2(transcripts per million (TPM) +1) in neuroblastoma cell lines in CCLE was <−2. Finally, genes were filtered for expression in human tissue datasets in at least one neuroblastoma sample as follows: Affymetrix microarray expression >6 in GSE12460 (45), log2(reads per million (RPKM)) >2 in GSE49711 (46), or Agilent microarray expression > 10 in GSE73517 (47). For the CRISPR guides, two guide sequences were retained from the Avana library, and three guide sequences were designed de novo. Additionally, three guides targeting intronic regions were generated for each of the 214 genes. Intronic regions were determined using the shared intronic regions across multiple isoforms using NCBI RefSeq UCSC, NCBI REfSeq All (RefSeq, RRID:SCR_003496), GENGODEv24 knownGene. The 30bp closest to known splice sites were avoided. The targeted intron regions were screened to make sure they do not fall in an exon of another gene. For 7 genes, there were no shared introns or they were single exon genes, in which case neighboring regions that did not have other genes were targeted instead. Five CRISPRi guides per gene were designed as well. All CRISPR, CRISPRi, and intronic guide sequences included in the library are available at figshare.com [https://figshare.com/s/85c7d5f316522767ff43]. For the sgRNA library, sgRNAs were cloned into a puromycin selectable sgRNA vector (lentiGuide-Puro, a gift from Feng Zhang (Addgene plasmid # 52963; http://n2t.net/addgene:52963; RRID:Addgene_52963)) as previously described (48). These vectors were infected into stable Cas9 or dCas9-KRAB lines as described above.
Other small guide RNAs
For inducible CRISPR studies, a guide only vector containing a doxycycline inducible sgRNA and constitutive GFP, FgH1tUTG, a gift from Marco Herold (Addgene plasmid # 70183; http://n2t.net/addgene:70183; RRID:Addgene_70183), was used as previously described (22). For NXT2 knockout studies, a hygromycin selectable vector containing Cas9 and the indicated sgRNA (sgLACZ or sgNXT2) was purchased from Vector Builder. For all other experiments the sgRNA was cloned into lentiCRISPR v2, a gift from Feng Zhang (Addgene plasmid # 52961; http://n2t.net/addgene:52961; RRID:Addgene_52961) (49), containing Cas9 and either puromycin or blasticidin resistance markers. For low-throughput experiments, guide sequences can be found in Supplementary Table S1.
CRISPR-Cas9 and CRISPRi-dCas9 essentiality screens
Stable neuroblastoma lines constitutively expressing Streptococcus pyogenes Cas9 have been previously generated and described (2,10). All cell lines were re-assayed for Cas9 activity prior to screening using a GFP Cas9-activity assay. To generate dCas9 lines, parental lines were transduced with a lentivirus expressing a nuclease dead Cas9 (dCas9) fused with a KRAB-transcriptional repressive domain (pLX_311-KRAB-dCas9 was a gift from John Doench & William Hahn & David Root (Addgene plasmid # 96918; http://n2t.net/addgene:96918; RRID:Addgene_96918) {Rosenbluh:2017hg}. The expression of dCas9 was verified by western blot, and activity was assayed using a modified version of the GFP activity assay, in which cells were infected with a lentivirus expressing destabilized EGFP (pLX313 EGFP-mODC) under hygromycin selection, and then infected with lentivirus expressing an sgRNA targeting the transcriptional start site of EGFP (sgCiGFP-2, GACCAGGATGGGCACCACCC). dCas9-KRAB activity was further confirmed using a cell viability readout after infection with an sgRNA directing dCas9-KRAB to the transcriptional start site of the essential gene PSMD. Cas9 or dCas9 expressing cell lines were infected with a single sgRNA library containing both CRISPR and CRISPRi sgRNAs (3985 unique guides) at an MOI of ~30%. Cells were selected with puromycin. For CRISPR screens, cells were maintained under blasticidin and puromycin until puromycin selection was complete (~day 4). For CRISPRi screens cells were grown under puromycin and blasticidin selection for the duration of the screen to prevent loss of guides or dCas9 activity. Representation of 500–1000 cells per guide was achieved at infection and maintained throughout the screen. Cell pellets of 500–1000X representation were frozen down 7, 14, and 21 days post infection. Genomic DNA was extracted from these pellets using the Qiagen DNeasy Blood and Tissue kit (cat # 69506). The sgRNA barcode was PCR amplified and this region was submitted for standard Illumina sequencing as previously described (2). Three replicates were carried out and sequenced for each time point and cell line in each screening assay. No replicates failed or were excluded from the analysis.
Chronos algorithm
To integrate readouts of CRISPR screens at multiple timepoints, we developed the Chronos model. A manuscript describing and benchmarking this method against existing methods in detail is under preparation; here, we describe the model in sufficient detail for reproducibility.
Chronos assumes that cells infected with an sgRNA are divided into two populations: those where the function of the targeted gene remains intact (e.g. due to in-frame INDEL mutations), which continue proliferating at the original rate, and those where the gene was successfully knocked out, which proliferate at some new, potentially negative rate. For an sgRNA i targeting gene g in cell line c, the number of cells Ncj with the sgRNA at time t after infection will be
where pcj is the probability that the sgRNA achieves knockout of its target, Rc is the unperturbed growth rate of the cell line, and rcg is the fractional change in growth rate caused by knockout of the targeted gene. This last term is the quantity which we will call the gene score, and is what we usually want to learn from the experiment. We exclude sgRNAs targeting more than one gene.
A wide range of efficacy for sgRNAs in abrogating protein function has been reported (48). Additionally, we have observed in Project Achilles screens that there is a per-cell line “screen quality” metric (e.g. due to variable Cas9 activity) which determines the overall separation of essential and nonessential genes in the screens (50). We therefore approximate the knockout probability per sgRNA and cell line as the product of a per-line and per-sgRNA factor, both constrained to the interval [0, 1]: pci = pc pi. There is some delay between infection and the emergence of the knockout phenotype, which we will call dg. Finally, we do not observe the number of infected cells Nci directly, but only the proportion of all reads that map to a particular sgRNA, which we assume have expectation equal to the proportion of cells with that sgRNA :〈nci〉= Nci / ∑ i Nci. Let Chronos’ estimation of nci be νci Then,
where Zc is a normalization term:
Chronos infers the parameters on the right to maximize the likelihood of the observed read count fractions nci according to the NB2 parameterization of the negative binomial distribution. The NB2 cost can be written (up to an additive constant)
where νci is the model prediction of the normalized read counts and k enumerates the timepoints measured. The overdispersion parameter ⍺c of the NB2 model is a hyperparameter which may be estimated for each cell line using existing tools such as edgeR (RRID:SCR_012802) (51), or set to a constant. We have found empirically that the value 0.05 produces good results in a variety of metrics.
Confirmation of sgRNA Editing
On-target editing of NXT1 and chromosome 2 (negative control) was confirmed by amplifying the appropriate region of genomic DNA and then performing Sanger sequencing with deconvolution by the TIDE or ICE algorithms. For NXT1, the region targeted by all three guides was amplified using the following primers: Forward 5’ TGGCTGAATCTGTGGATGCAAAAC 3’ Reverse 5’ GCACTGTCTCCGCAACAAC 3’. The gene desert region on chromosome 2 containing the target of sgChr2–2 was amplified with the following primers: forward 5’ TTTGAGGCTTATGGGGGCAG 3’ and reverse 5’ AAGGGCCCCGATTTTCTCAA 3’. PCR products were purified using the QiaQuick PCR purification kit and sent for Sanger sequencing with the following sequencing primers: sgChr2–2 5’ ATGGGTAAGGAATCTGAGCATGG 3’ and sgNXT1–1/2/3 5’ GCCACAGTGGTAATCCCATC 3’. On-target editing was then determined using either the TIDE algorithm https://tide.nki.nl/ (52) or ICE v. 2.0 by Synthego (https://ice.synthego.com/#/) as indicated.
Annexin V positive selection screen
Cas9 expressing cell lines were generated, validated, and infected as described above. Seven days after infection, floating cells were collected, and adherent cells were detached from the plate using Accutase. Cells were combined, and an appropriate number of cells from each replicate (1000X representation) was pelleted and frozen down. The remaining cells (4000X representation) were incubated with magnetic beads (Miltenyi Biotec 130-090-201) and passed over a magnetic column (Miltenyi Biotec 130-042-201). Cells were washed 3X with PBS, and then the bound cells were eluted from the column. The purified cell population was then frozen down by cell pellet. Three biological replicates were collected. A small number of cells were then incubated with fluorescent Annexin V and the purity of the purified population relative to input was assessed by flow cytometry. Genomic DNA was extracted and sequenced as described above. No replicates failed or were excluded from the analysis.
In vivo screening
This study was approved by the Institutional Animal Care and Use Committee (IACUC) of the Broad Institute under animal protocol 0194-01-18. IACUC guidelines on the ethical use and care of animals were followed. KELLY cells constitutively expressing Cas9 were infected with our sgRNA library as described above. Cells were put into puromycin selection 24 hours after infection. Once cells were fully selected, 72 hours after infection, 8.0e6 cells (~2000x library representation) were implanted into the flanks of NRG (NOD-Rag1null IL2rgnull) mice bi-laterally. 26 days after implantation, when tumors were ~100–200 mm3 mice were euthanized and their tumors (n=6) were harvested and flash frozen. Tumors were homogenized with a Precellys tissue homogenizer, and DNA was extracted with a QIAamp DNA Blood Maxi Kit, after tissue lysis in Qiagen buffer ATL. Genomic DNA was sequenced as described above. No tumors failed or were excluded from the analysis.
In vivo inducible sgRNA xenografts
This study was approved by the Institutional Care and Use Committee (IACUC) of Dana-Farber Cancer Institute and performed under protocol 15–029. IACUC guidelines on the ethical use and care of animals were followed. KELLY cells constitutively expressing Cas9 were infected with inducible sgRNAs targeting Chr2–2 or NXT1. 6.0e6 cells were implanted into the flanks of NRG (NOD-Rag1null IL2rgnull) mice. When tumors were ~100–200 mm3 mice were randomized to either normal chow or chow containing doxycycline at 625mg/kg. Tumors were measured by Vernier caliper and volume was determined using the standard formula. Animals were euthanized once they reached a humane endpoint and tumor tissue was flash frozen for later protein and genomic DNA extraction.
sgRNA viability assays
For low-throughput viability assays, cells were transfected with a vector containing Cas9 and the indicated sgRNA as described above, and four days after infection seeded into 384 well plates or 96 well plates. CellTiter-Glo was used according to manufacturer’s instructions to assess the relative viability at days 3, 5, and 7, normalized to day 0. Luminescence was determined using an Envision Plate reader. Experiments were done in triplicate and technical replicates in each experiment were at least triplicate.
dTAG-13 dose response curves
Cells were seeded into 96-well or 384-well plates. 24 hours after plating, compound was added either manually or with a robot (HP D300e digital dispenser) at the concentrations indicated. 72 hours after addition of drug, viability was assessed by CellTiter-Glo according to manufacturer’s instructions. Luminescence was determined using an Envision Plate reader. Viability was normalized to the DMSO condition. dTAG13 and dTAG-13-NEG, the inactive analog, were generously provided by Dr. Nathanael Gray’s laboratory at Dana-Farber Cancer Institute.
Exogenous expression of NXT1 and NXT2
To render it resistant to sgNXT1-3, the PAM sequence in NXT1 was mutated (C→T at position 590). This NXT1 sequence was cloned into pLEX_305-C-dTAG, a gift from James Bradner & Behnam Nabet (Addgene plasmid # 91798; http://n2t.net/addgene:91798; RRID:Addgene_91798) (20). To generate the NXT1deg lines with knockout of endogenous NXT1 and degron-tagged exogenous NXT1, cell lines were co-infected with a puromycin resistant vector containing NXT1 with a c-terminal FKBP12F36V-2XHA tag and a blasticidin resistant vector containing an sgRNA targeting NXT1 (sgNXT1-3). Cell lines were selected with both blasticidin and puromycin. A vector containing the NXT2 orf with a c-terminal MYC tag under neomycin selection was purchased from Origene (RC213204). Cell lines were transfected with this vector and stable integrations were selected with neomycin.
Patient-derived xenograft (PDX) models
Three patient-derived xenograft models (COGN-424X, COGN-557X, and COGN603X) were obtained from the Children’s Oncology Group’s (COG) Childhood Cancer Repository. Written informed consent and IRB approval was obtained according to COG protocols and studies were conducted in accordance with recognized ethical guidelines. Models were propagated in mice and tumors were flash frozen before gDNA or protein was extracted.
Western blotting
Cells were lysed in Cell Signaling Lysis Buffer (9803) supplemented with protease (Roche cat. #11836170001) and phosphatase inhibitors (Roche cat. #04906845001). Lysates were quantified using a BCA assay (Pierce) and normalized. SDS-PAGE gels were used to separate proteins, and proteins were transferred to a PVDF membrane. Primary antibodies used in this study were: NXT2 (Abcam Cat# ab121797, RRID:AB_11127842), GAPDH (Santa Cruz, cat.# sc-47724), PARP (Cell Signaling Technologies, cat# 9542), NXF1 (Abcam, cat.# ab50609), MYC-tag (Cell Signaling Technologies, cat# 9402), HA-tag (Cell Signaling Technologies cat.# 2367 and #3724), CAS9 (Cell Signaling Technologies, cat.# 7A9–3A3), and vinculin (Cell Signaling Technologies, cat.# 4650). Membranes were incubated with secondary antibodies (Licor cat.# 926–68070 and cat.# 926–32211) and imaged on a Licor Odyssey.
Cycloheximide-chase experiments
Neuroblastoma cell lines were plated and 24 hours later treated with 50 μg/mL cycloheximide. Cells were then lysed 2, 6, and 24 hours after cycloheximide treatment and protein levels were assessed by western blot.
Data analysis of CRISPR screens
Time course screen data for the individual time points was collapsed to a gene level score using the MAGeCK algorithm with copy number correction for CRISPR screens (53). No copy number correction was applied to CRISPRi screens. Plasmid pool was used as the reference. To collapse timepoint data to a single gene level score that accounts for rate of depletion we used the Chronos algorithm, without copy number correction (see Chronos algorithm methods section for detailed description). For the in vivo screen, MAGeCK was used to collapse to gene level scores as above, but gDNA from the cells at the time of inoculation was used as the reference. For the Annexin V positive selection screen, the log2 fold change of guides relative to input gDNA was calculated and averaged across replicates. The mean log fold change of the top two guides per gene was considered the Annexin V enrichment score. For all screens, genes were considered a strong hit if they exceeded the effect of the median of the positive controls in that assay. For copy number analysis and corrections gene level copy number calls for each cell line were calculated by taking the log2 of the copy number segment mean of DepMap data (20Q1 Public CN Segments available at ext-link DepMap.org).
Co-immunoprecipitations
Cells were treated as indicated and then the cytoplasmic fraction was lysed in a lysis buffer of 50mM Tris ph7.5, 0.1% NP-40, 1mM EDTA, and 1mM MgCl2 supplemented with protease (Roche cat. #11836170001) and phosphatase inhibitors (Roche cat. #04906845001). The nuclear fraction was pelleted by centrifugation, and then lysed in a nuclear lysis buffer (50mM Tris pH 7.5, 1% NP-40, 1 mM EDTA, 1 mM MgCl2, 300mM NaCl supplemented with protease and phosphatase inhibitors as above). 1 μM DTT was added and 2 μg of antibody. IPs were incubated overnight at 4°C. The next day 30 μL of Protein A or G Dynabeads (Thermo Fisher 10006D and 10007D) were added for 2–4 hours, and then beads were washed 5X in lysis buffer, then eluted in LDS.
Bisulfite sequencing
Genomic DNA was extracted using the Qiagen DNeasy Blood and Tissue kit (cat # 69506). DNA was then bisulfite converted using the Qiagen EpiTect Bisulfite kit (cat # 59104). Bisulfite converted DNA was then amplified with EpiMark HotStart Taq (NEB #M0490) using the following primers: forward 5’ TTGGGAGAATATAAAAGTTTG 3’ and reverse 5’ ATCTCCCTAAAACCAATAAC 3’ from Sung et al., (29). PCR products were purified using the QiaQuick PCR purification kit, and Sanger sequenced with the reverse primer.
Public datasets
To create the list of initial dependencies the CRISPR Avana dependency gene effect scores generated in mid-2017 using the CERES algorithm were used. This data is available for download at figshare.com [https://figshare.com/s/85c7d5f316522767ff43]. We used the CRISPR DepMap Public 20Q1 gene effect, probability, copy number, and CCLE expression and transcript-level expression data datasets; data available for download at depmap.org and figshare.com [https://figshare.com/articles/dataset/DepMap_20Q1_Public/11791698]. We used the Wellcome Sanger Institute Dependency Map data (PROJECT SCORE) processed with the CERES algorithm which can be downloaded from depmap.org and figshare.com [https://figshare.com/authors/Broad_DepMap/5514062]. Human tumor microarray data was downloaded from the R2 database (r2.amc.nl). Human tumor RNA-seq data (Tumor Compendium v11 Public PolyA April 2020) was downloaded from the Treehouse project (https://treehousegenomics.soe.ucsc.edu/). Normal tissue RNA-seq data was downloaded from the Genotype-Tissue Expression project (V8) (GTEXportal.org). ATAC-seq and ChIP-seq data for neuroblastoma cell lines was downloaded from the Gene Expression Omnibus (GEO RRID:SCR_005012) with GEO accession numbers GSE138293 and GSE138314 respectively (54). Gene expression for these lines was downloaded from the GEO database with accession number GSE89413 (55). Tumor mutation, copy number, and expression data for neuroblastoma tumors from the Gabriela Miller Kid’s First provisional dataset (Discovering the Genetic Basis of Human Neuroblastoma (Maris/GMKF dbGaP phs001436.v1.p1, Provisional) was accessed through the pediatric cBioPortal (pedcbioportal.org) (56,57).
Other data analysis and statistics
For statistical tests of significance, the statistical test and p-value are described in the respective figure legends. All t-tests are two-sided unless otherwise indicated. A p-value of 0.05 was used as the cutoff for significance unless otherwise indicated. These values were calculated in GraphPad Prism (RRID:SCR_002798) or R 3.63. Error bars represent standard deviation unless otherwise indicated. All duplicate measures were taken from distinct samples rather than repeated measures of the same sample. For null hypotheses with multiple groups, a two-way ANOVA was used, followed by a Tukey’s multiple comparison test if the ANOVA was significant.
Proteomics
Materials
Isobaric TMT reagents and the BCA protein concentration assay kit were from ThermoFisher Scientific (Rockford, IL). Empore-C18 material for in-house made Stage Tips was from 3 M (Saint Paul, MN). Sep-Pak cartridges (100 mg size) were purchased from Waters (Milford, MA). All solvents used for Liquid chromatography (LC) were purchased from J.T. Baker (Central Valley, PA). Mass spectrometry (MS)-grade trypsin and Lys-C protease were purchased from ThermoFisher Scientific and Wako (Boston, MA), respectively. Complete protease inhibitors were from Millipore Sigma (Saint Louis, MO). Unless otherwise noted, all other chemicals were purchased from ThermoFisher Scientific.
MS sample processing
KELLY cells with endogenous NXT1 knocked-out and expressing degron-tagged exogenous NXT1 (KELLY+ NXT1deg) were treated with 500 nM dTAG-13 for 2 hours. Cells pellets were then collected by cell scraping and frozen at −80°C until future use. Cell pellets were lysed using 8 M urea, 200 mM 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid (EPPS) at pH 8.5 with protease inhibitors (one tablet per 10 mL of lysis buffer). Samples were further homogenized and DNA was sheared via sonication using a probe sonicator (20× 0.5 sec pulses; level 3). Total protein was determined using a BCA assay and proteins were stored at −80°C until future use. A total of 25 μg of protein was aliquoted for each condition and TMT channel for further downstream processing. Protein extracts were reduced using 5 mM tris-(2-carboxyethyl) phosphine (TCEP) for 15 min at room temperature. Next samples were alkylated with 10 mM iodoacetamide for 30 min in the dark at room temperature. To facilitate the removal of incompatible reagents, proteins were precipitated using chloroform and methanol. Briefly, to 100 μL of each sample, 400 μL of methanol was added, followed by 100 μL of chloroform with thorough vortexing. Next, 300 μL of HPLC grade water was added and samples were vortexed thoroughly. Each sample was centrifuged at 14 000 xg for 5 min at room temperature. The upper aqueous layer was removed and the protein pellet was washed twice with methanol and centrifuged at 14 000 xg for 5 min at room temperature. Protein pellets were re-solubilized in 200 mM EPPS buffer and digested overnight with Lys-C (1:100, enzyme:protein ratio) at room temperature. The next day, trypsin (1:100 ratio) was added and incubated at 37°C for an additional 6 hrs in a ThermoMixer set to 1000 RPM.
TMT labeling
To each digested sample, 30% anhydrous acetonitrile was added and 25 μg of peptides were labeled using ~55 μg of TMTPro reagents (TMT1-TMT16) for 1 hour at room temperature with constant agitation. Following labeling, 5% hydroxylamine was added to quench excess TMT reagent. To equalize protein loading a ratio check was performed by pooling ~ 2 μg of each TMT-labeled sample. Samples were pooled and desalted using an in-house packed C18 Stage Tip and analyzed by liquid chromatography (LC) tandem mass spectrometry (MS/MS). Normalization factors derived from the ratio check were used to pool samples 1:1 across all TMT channels and the combined sample was desalted using a 100 mg Sep-Pak solid phase extraction cartridge. Eluted peptides were further fractionated using basic-pH reversed-phase (bRP) on an Agilent 300 extend C18 column and were collected into a 96 deep-well plate. Samples were consolidated into 24 fractions as previously described, and 12 nonadjacent fractions were desalted using Stage Tips prior to analyses using LC-MS/MS (58–60).
Mass spectrometry and data acquisition
All mass spectrometry data were acquired using an Orbitrap Fusion Lumos mass spectrometer in-line with a Proxeon nanoLC-1200 Ultra performance LC (UPLC) system. TMT labeled peptides were separated using an in-house packed 100 μm capillary column with 35 cm of Accucore 150 resin (2.6 μm, 150 Å) (ThermoFisher Scientific) using either a 120 min LC gradient from 4 to 24% acetonitrile in 0.125% formic acid per run. Eluted peptides were acquired using synchronous precursor selection (SPS-MS3) method for TMT quantification. Briefly, MS1 spectra were acquired at 120K resolving power with a maximum of 50 ms ion injection in the Orbitrap. MS2 spectra were acquired by selecting the top 10 most abundant features via collisional induced dissociation (CID) in the ion trap using an automatic gain control (AGC) of 15K, quadrupole isolation width of 0.5 m/z and a maximum ion time of 50 ms. These spectra were passed in real time to the external computer for database searching. Intelligent data acquisition (IDA) using real-time searching (RTS) was performed using Orbiter as previously described (61,62). Peptide spectral matches were analyzed using the Comet search algorithm designed for spectral acquisition speed (63,64). Real-time access to spectral data was enabled by the ThermoFisher Scientific Fusion API. Briefly, peptides were filtered using simple filters that included the following: not a match to a reversed sequence, maximum PPM error 50, minimum XCorr of 0.5, minimum deltaCorr of 0.10 and minimum peptide length of 7. If peptide spectra matched to above criteria, an SPS-MS3 scan was performed using up to 10 b- and y-type fragment ions as precursors with an AGC of 200K for a maximum of 200 ms with a normalized collision energy setting of 45.
Mass spectrometry data analysis
All acquired data were searched using the open-source Comet algorithm using a previously described informatics pipeline (65–67). We acknowledge Dr. Steven Gygi for use of a custom CORE data analysis software as part of the pipeline. Briefly, peptide spectral libraries were first filtered to a peptide false discovery rate (FDR) of less than 1% using linear discriminant analysis employing a target-decoy strategy. Spectral searches were done using a custom fasta formatted database which included common contaminants, reversed sequences (Uniprot Human, 2014) with custom NXT1-FKBP12F36V entries and the following parameters: 50 PPM precursor tolerance, fully tryptic peptides, fragment ion tolerance of 0.9 Da and a static modification by TMT (+304.2071 Da) on lysine and peptide N termini. Carbamidomethylation of cysteine residues (+57.021 Da) was set as a static modification while oxidation of methionine residues (+15.995 Da) was set as a variable modification. Resulting peptides were further filtered to obtain a 1% protein FDR and proteins were collapsed into groups. Reporter ion intensities were adjusted to correct for impurities during synthesis of different TMT reagents according to the manufacturer’s specifications. Lastly, protein quantitative values were column normalized so that the sum of the signal for all protein in each channel was equal to account for sample loading differences and a total sum signal-to-noise of all report ion ions of 100 was required for analysis.
Supplementary Material
Significance:
We provide a framework for identifying new therapeutic targets from functional genomic screens. We nominate NXT1 as a selective lethal target in neuroblastoma and propose a therapeutic approach where the essential protein NXF1 can be selectively eliminated in tumor cells by exploiting the NXT1-NXT2 paralog relationship.
Acknowledgments:
We acknowledge Dr. Steven Gygi for the use of a custom CORE data analysis software as part of the mass spectrometry pipeline. We also thank Andrew Tang for graphic design assistance. This work was funded by NIH R35 CA210030 (KS), the St. Baldrick’s Foundation Robert J. Arceci Innovation Award (KS), Friends for Life (KS) and NIH 1P01 CA217959 (KS), and the Slim Initiative in Genomic Medicine for the Americas (SIGMA) a joint U.S-Mexico project funded by the Carlos Slim Foundation (FV, TRG). CFM was supported by a Helen Gurley Brown Presidential Initiative Fellowship, and by the National Institutes of Health under a Ruth L. Kirschstein National Research Service Award (F32CA243266). NVD was supported by the Julia’s Legacy of Hope St. Baldrick’s Foundation Fellowship.
Declaration of Interests:
KS has funding from Novartis Institute of Biomedical Research, consults for and has stock options in Auron Therapeutics, served as an advisor for Kronos Bio, and consulted for Rigel Pharmaceuticals on topics unrelated to this manuscript. FV and BRP receive research support from Novo Ventures unrelated to this work. TRG was formerly a consultant and equity holder in Foundation Medicine, which was acquired by Roche. TRG is a consultant to GlaxoSmithKline and is a founder and equity holder of Sherlock Biosciences and FORMA Therapeutics. NVD is now an employee of Genentech, Inc., a member of the Roche Group.
References
- 1.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–576.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–6. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Alexe G, Dharia NV, Ross L, Iniguez AB, Conway AS, et al. CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J Clin Invest. American Society for Clinical Investigation; 2018;128:446–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan EM, Shibue T, McFarland JM, Gaeta B, Ghandi M, Dumont N, et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature. 2019;568:551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature. Nature Publishing Group; 2018;555:321–7. [DOI] [PubMed] [Google Scholar]
- 7.Harris MH, DuBois SG, Glade Bender JL, Kim A, Crompton BD, Parker E, et al. Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol. American Medical Association; 2016;2:608–15. [DOI] [PubMed] [Google Scholar]
- 8.Parsons DW, Roy A, Yang Y, Wang T, Scollon S, Bergstrom K, et al. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol. American Medical Association; 2016;2:616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chabner BA, Roberts TG. Timeline: Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5:65–72. [DOI] [PubMed] [Google Scholar]
- 10.Dharia NV, Kugener G, Guenther LM, Malone CF, Durbin AD, Hong AL, Howard TP, Bandopadhayay P, Wechsler CS, Fung I, Warren AC, Dempster JM, Krill-Burger JM, Paolella BR, Moh P, Jha N, Tang A, Montgomery P, Boehm JS, Hahn WC, Roberts CWM, McFarland JM, Tsherniak A, Golub TR, Vazquez F, Stegmaier K. A first-generation pediatric cancer dependency map. Nat Genet. In press. DOI: 10.1038/s41588-021-00819-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maris JM. Recent Advances in Neuroblastoma. N Engl J Med. 2010;362:2202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. Cold Spring Harbor Laboratory Press; 2013;3:a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller S, Matthay KK. Neuroblastoma: biology and staging. Curr Oncol Rep. 2009;11:431–8. [DOI] [PubMed] [Google Scholar]
- 14.Rosenbluh J, Xu H, Harrington W, Gill S, Wang X, Vazquez F, et al. Complementary information derived from CRISPR Cas9 mediated gene deletion and suppression. Nat Commun. Nature Publishing Group; 2017;8:15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burkhart CA, Cheng AJ, Madafiglio J, Kavallaris M, Mili M, Marshall GM, et al. Effects of MYCN antisense oligonucleotide administration on tumorigenesis in a murine model of neuroblastoma. J Natl Cancer Inst. 2003;95:1394–403. [DOI] [PubMed] [Google Scholar]
- 16.Kang J-H, Rychahou PG, Ishola TA, Qiao J, Evers BM, Chung DH. MYCN silencing induces differentiation and apoptosis in human neuroblastoma cells. Biochem Biophys Res Commun. 2006;351:192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arroyo JD, Jourdain AA, Calvo SE, Ballarano CA, Doench JG, Root DE, et al. A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab. 2016;24:875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin SJ, Reutelingsperger CP, McGahon AJ, Rader JA, van Schie RC, LaFace DM, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182:1545–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiegand HL, Coburn GA, Zeng Y, Kang Y, Bogerd HP, Cullen BR. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol Cell Biol. 2002;22:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, et al. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol. Nature Publishing Group; 2018;14:431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nabet B, Ferguson FM, Seong BKA, Kuljanin M, Leggett AL, Mohardt ML, et al. Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. bioRxiv. Cold Spring Harbor Laboratory; 2020;31:2020.03.13.980946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, Milla L, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–32. [DOI] [PubMed] [Google Scholar]
- 23.Aibara S, Katahira J, Valkov E, Stewart M. The principal mRNA nuclear export factor NXF1:NXT1 forms a symmetric binding platform that facilitates export of retroviral CTE-RNA. Nucleic Acids Res. 2015;43:1883–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, et al. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. American Society for Microbiology (ASM); 2000;20:8996–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. Nature Publishing Group; 2017;545:505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Zhang B, Hartenstein PA, Chen J-N, Lin S. NXT2 is required for embryonic heart development in zebrafish. BMC Dev Biol. BioMed Central; 2005;5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan M, Jabeen N, Khan T, Hussain HMJ, Ali A, Khan R, et al. The evolutionarily conserved genes: Tex37, Ccdc73, Prss55 and Nxt2 are dispensable for fertility in mice. Sci Rep. Nature Publishing Group; 2018;8:4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung HY, Choi EN, Ahn Jo S, Oh S, Ahn J-H. Amyloid protein-mediated differential DNA methylation status regulates gene expression in Alzheimer’s disease model cell line. Biochem Biophys Res Commun. 2011;414:700–5. [DOI] [PubMed] [Google Scholar]
- 30.Yokomori N, Kobayashi R, Moore R, Sueyoshi T, Negishi M. A DNA methylation site in the male-specific P450 (Cyp 2d-9) promoter and binding of the heteromeric transcription factor GABP. Mol Cell Biol. American Society for Microbiology Journals; 1995;15:5355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucas ME, Crider KS, Powell DR, Kapoor-Vazirani P, Vertino PM. Methylation-sensitive regulation of TMS1/ASC by the Ets factor, GA-binding protein-alpha. J Biol Chem. 2009;284:14698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens DC, Poon GMK. Differential sensitivity to methylated DNA by ETS-family transcription factors is intrinsically encoded in their DNA-binding domains. Nucleic Acids Res. 2016;44:8671–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lea AJ, Vockley CM, Johnston RA, Del Carpio CA, Barreiro LB, Reddy TE, et al. Genome-wide quantification of the effects of DNA methylation on human gene regulation. Elife. eLife Sciences Publications Limited; 2018;7:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. Nature Publishing Group; 2013;4:e586–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Groningen T, Koster J, Valentijn LJ, Zwijnenburg DA, Akogul N, Hasselt NE, et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat Genet. Nature Publishing Group; 2017;49:1261–6. [DOI] [PubMed] [Google Scholar]
- 36.Laverdière C, Liu Q, Yasui Y, Nathan PC, Gurney JG, Stovall M, et al. Long-term outcomes in survivors of neuroblastoma: a report from the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2009;101:1131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor DM, Aronow BJ, Tan K, Bernt K, Salomonis N, Greene CS, et al. The Pediatric Cell Atlas: Defining the Growth Phase of Human Development at Single-Cell Resolution. Dev Cell. 2019;49:10–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai AC, Crews CM. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. Nature Publishing Group; 2017;16:101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chamberlain PP, Hamann LG. Development of targeted protein degradation therapeutics. Nat Chem Biol. Nature Publishing Group; 2019;15:937–44. [DOI] [PubMed] [Google Scholar]
- 40.Scott DE, Bayly AR, Abell C, Skidmore J. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat Rev Drug Discov. Nature Publishing Group; 2016;15:533–50. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Bor Y-C, Misawa Y, Xue Y, Rekosh D, Hammarskjold M-L. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. Nature Publishing Group; 2006;443:234–7. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Bor Y-C, Fitzgerald MP, Lee KS, Rekosh D, Hammarskjold M-L. An NXF1 mRNA with a retained intron is expressed in hippocampal and neocortical neurons and is translated into a protein that functions as an Nxf1 cofactor. Matera AG, editor. Mol Biol Cell. 2016;27:3903–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dirks WG, MacLeod RAF, Nakamura Y, Kohara A, Reid Y, Milch H, et al. Cell line cross-contamination initiative: an interactive reference database of STR profiles covering common cancer cell lines. Int J Cancer. John Wiley & Sons, Ltd; 2010;126:303–4. [DOI] [PubMed] [Google Scholar]
- 44.Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet. Nature Publishing Group; 2018;50:1240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, de Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. Nature Publishing Group; 2008;455:967–70. [DOI] [PubMed] [Google Scholar]
- 46.Zhang W, Yu Y, Hertwig F, Thierry-Mieg J, Zhang W, Thierry-Mieg D, et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. BioMed Central; 2015;16:133–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henrich K-O, Bender S, Saadati M, Dreidax D, Gartlgruber M, Shao C, et al. Integrative Genome-Scale Analysis Identifies Epigenetic Mechanisms of Transcriptional Deregulation in Unfavorable Neuroblastomas. Cancer Res. American Association for Cancer Research; 2016;76:5523–37. [DOI] [PubMed] [Google Scholar]
- 48.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. Nature Publishing Group; 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. Nature Publishing Group; 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dempster JM, Pacini C, Pantel S, Behan FM, Green T, Krill-Burger J, et al. Agreement between two large pan-cancer CRISPR-Cas9 gene dependency data sets. Nat Commun. 2019;10:5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. BioMed Central; 2014;15:554–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Upton K, Modi A, Patel K, Kendsersky NM, Conkrite KL, Sussman RT, et al. Epigenomic profiling of neuroblastoma cell lines. Sci Data. Nature Publishing Group; 2020;7:116–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harenza JL, Diamond MA, Adams RN, Song MM, Davidson HL, Hart LS, et al. Transcriptomic profiling of 39 commonly-used neuroblastoma cell lines. Sci Data. Nature Publishing Group; 2017;4:170033–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. American Association for Cancer Research; 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. American Association for the Advancement of Science; 2013;6:pl1–pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paulo JA. Nicotine alters the proteome of two human pancreatic duct cell lines. JOP. 2014;15:465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paulo JA, Gygi SP. Nicotine-induced protein expression profiling reveals mutually altered proteins across four human cell lines. Proteomics. 2017;17:1600319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Navarrete-Perea J, Yu Q, Gygi SP, Paulo JA. Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho)proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. J Proteome Res. 2018;17:2226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erickson BK, Mintseris J, Schweppe DK, Navarrete-Perea J, Erickson AR, Nusinow DP, et al. Active Instrument Engagement Combined with a Real-Time Database Search for Improved Performance of Sample Multiplexing Workflows. J Proteome Res. 2019;18:1299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schweppe DK, Eng JK, Yu Q, Bailey D, Rad R, Navarrete-Perea J, et al. Full-Featured, Real-Time Database Searching Platform Enables Fast and Accurate Multiplexed Quantitative Proteomics. J Proteome Res. 2020;:acs.jproteome.9b00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–4. [DOI] [PubMed] [Google Scholar]
- 64.Eng JK, Hoopmann MR, Jahan TA, Egertson JD, Noble WS, MacCoss MJ. A deeper look into Comet--implementation and features. J Am Soc Mass Spectrom. 2015;26:1865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem. 2012;84:7469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–92. [DOI] [PubMed] [Google Scholar]
- 67.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genome-scale dependency data used in this analysis is publicly available for download at depmap.org and figshare.com [https://figshare.com/articles/dataset/DepMap_20Q1_Public/11791698]. All datasets generated in this study are made publicly available at figshare.com [https://figshare.com/s/85c7d5f316522767ff43].